Abstract
In machine learning for image-based medical diagnostics, supervised convolutional neural networks are typically trained with large and expertly annotated datasets obtained using high-resolution imaging systems. Moreover, the network’s performance can degrade substantially when applied to a dataset with a different distribution. Here, we show that adversarial learning can be used to develop high-performing networks trained on unannotated medical images of varying image quality. Specifically, we used low-quality images acquired using inexpensive portable optical systems to train networks for the evaluation of human embryos, the quantification of human sperm morphology and the diagnosis of malarial infections in the blood, and show that the networks performed well across different data distributions. We also show that adversarial learning can be used with unlabelled data from unseen domain-shifted datasets to adapt pretrained supervised networks to new distributions, even when data from the original distribution are not available. Adaptive adversarial networks may expand the use of validated neural-network models for the evaluation of data collected from multiple imaging systems of varying quality without compromising the knowledge stored in the network.
Image analysis, which is a fundamental component of medical diagnostics, has undoubtedly benefited from human- or super-human levels of feature recognition, anomaly detection and localization due to advances in supervised deep learning over the past decade1–3. However, supervised learning models—the most widely used deep learning approach in medical image analysis—are often dependent on large expertly annotated datasets and are usually limited to the training data distribution4 (Fig. 1a). In medicine, such limitations can have dire consequences where, for example, networks that were developed using one brand of an instrument can observe substantial decreases in performance when tested on data that were collected using a different brand/instrument of imaging system to the one used during training5–8. Furthermore, high-quality medical images are critical for human interpreters to annotate, limiting most of the current supervised machine learning approaches to cost-prohibitively expensive state-of-the-art imaging hardware, making the use of these technologies considerably more challenging particularly in low- and middle-income countries9.
Fig. 1 |. Schematics of the use of adversarial domain adaptive neural networks for medical image analysis.
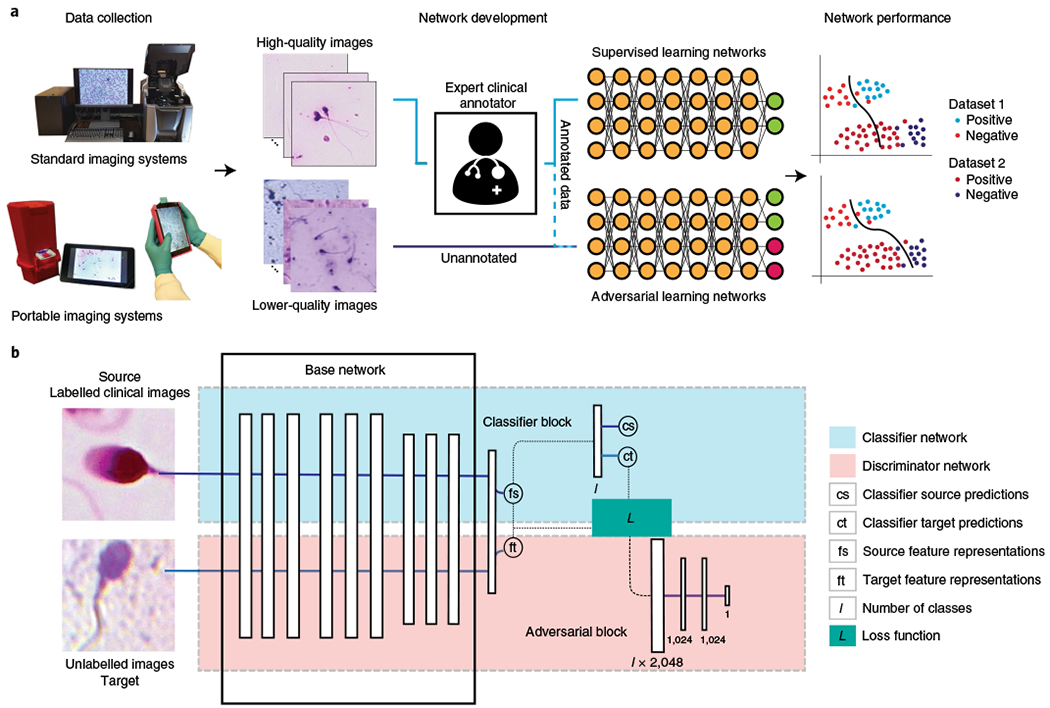
a, Supervised learning networks for medical image analysis are limited to fully expert-annotated datasets for training and are generally unable to adapt to unseen distributions of data collected using different imaging systems used in different clinical settings. Clinical expert staff may not be able to reliably annotate medical images obtained through portable point-of-care optical systems that are usually of lower quality compared with bulky and expensive benchtop microscopes. However, adversarial learning networks can be used to utilize standardized annotated image datasets obtained from one distribution (source) to adapt themselves with unannotated data obtained from a different distribution (target) towards a substantially more generalized neural network. b, Schematic of the general framework of the adversarial domain adaptive medical neural networks (MD-nets). The base network layers can be replaced using any regular custom neural network architecture. Additional elements for pseudolabelling can be added to enable the network to achieve adaptation in the absence of source data.
In this Article, we present a deep learning framework for achieving unsupervised domain adaptation between various microscopy imaging systems in medical image analysis tasks without the need for any additional domain-specific information, including explicit annotations of the domain-shifted images, the magnifications and fields-of-view of the imaging system, optical and image resolutions, lighting and exposures, and optical image corrections (Fig. 1a). To achieve this, we made use of adversarial learning, a powerful learning technique that is most popular for its generative-variant capable of realistic image synthesis10. For domain adaptation, these adversarial learning schemes can be repurposed to refine the neural network’s learning process such that common features specific to each target class, across the different domains, are prioritized in its decision making11,12. Here, we used the gamified learning technique to achieve adaptation across unseen shifted distributions of potentially impossible-to-annotate microscopy cellular images with diagnostic applications in medicine (Fig. 1b). To demonstrate the success of the network’s decisions for medical image analysis tasks, we compared the network’s inferences using shifted data against a human interpreter’s overall decisions made using conventional imaging systems (for example, benchtop high-resolution microscopy).
We evaluated the performance of the developed medical domain adaptive neural network (MD-net) framework in cellular image analysis of biological samples with applications in infertility and infectious diseases as clinical models. First, we used datasets of non-shifted and shifted embryo images for semisupervised learning and unsupervised adaptation and compared the performance of MD-nets with models learned through supervised training, and alternative domain adaptation strategies. Second, we evaluated MD-net’s ability to quantify morphological defects among sperm cells in smeared semen samples across shifted domains, where individual sperm annotations on the shifted datasets cannot be established by human readers, to evaluate its performance against conventional clinical analyses. Finally, we used models that were originally trained under supervision to achieve unsupervised adaptation—even in the absence of data from the original domain that was used during supervised training—through MD-nets, on shifted datasets of whole-blood slides in qualitatively identifying the presence of malaria-infected cells in thin whole-blood smears, evaluating the network’s suitability in point-of-care clinical applications. We demonstrated the versatility of MD-nets by achieving adaptation to new domains with and without the need to access the original medical image data used for training the network in the original domain (that is, source data) and with relatively higher image processing performance compared with conventional supervised convolutional neural networks (CNNs) and other domain adaptation strategies in both shifted and non-shifted datasets.
Results
Comparison of supervised and domain adaptation methods in medical image analysis.
To effectively evaluate the performance of MD-nets as a suitable alternative to supervised methods for medical image analysis, a clinically relevant medical image analysis task for which supervised networks have shown promising results was selected. Supervised neural networks have been studied in evaluating human embryos on the basis of their morphology for applications in in vitro fertilization (IVF)5,13–16. These supervised neural networks have shown very high efficiencies and, in some cases, have outperformed highly trained embryologists in embryo assessment14. Deep supervised neural networks are currently being evaluated in IVF for their applicability in clinical settings and have the potential to improve and standardize clinical assisted reproductive practices. Most previously developed neural networks were evaluated using datasets collected using a single instrument17. However, one study indicated a drop in performance when evaluated using data collected through the same model of the imaging systems from different fertility centres, although, in that case, the effect of domain shift was not very well studied5. Thus, to evaluate the performance of MD-net compared to supervised networks, we classified embryo images on the basis of their developmental status (two classes; blastocyst/non-blastocyst) recorded at 113 h after insemination (fifth day of embryo development) using different microscopy instruments operated by different users at multiple fertility centres15. Blastocysts have fluid-filled cavities and two distinguishable cell lineages—the trophectoderm and the inner cell mass. Advanced-staged blastocysts, based on the expansion of the blastocoel cavity and the quality of the trophectoderm and inner cell mass, are preferred for transfer during a clinical IVF cycle18.
Embryos were imaged using a commercial time-lapse imaging system (ED4), various clinical microscopic systems (ED3), an inexpensive and portable 3D-printed microscope (ED2) and a smartphone-based microscope (ED1) (Fig. 2a). The collected data were of varying quality and, in an effort to highlight the shift in image quality with regard to computational image analysis, we measured the average number of interest points available in each dataset and class through scale-invariant feature transform (SIFT) measurements (Fig. 2a,b). The mean SIFT values for blastocyst images recorded using ED4, ED3, ED2 and ED1 were 193.8 (95% confidence interval (CI) = 189.9–197.8; n = 491), 551.8 (95% CI = 498.7–604.8; n = 117), 165.1 (95% CI = 134.7–195.6; n = 56) and 4.9 (95% CI = 4.4–5.4; n = 197), respectively (Fig. 2b). The mean SIFT values for non-blastocyst images recorded using ED4, ED3, ED2 and ED1 were 169.5 (95% CI = 164.2–174.9; n = 251), 607.8 (95% CI = 476.2–748.3; n = 141), 57.1 (95% CI = 28.3–85.8; n = 13) and 0.4 (95% CI = 0.2–0.5; n = 99), respectively (Fig. 2b).
Fig. 2 |. Comparison of supervised CNNs and domain adaptation methods for the morphological analysis of human embryo images.
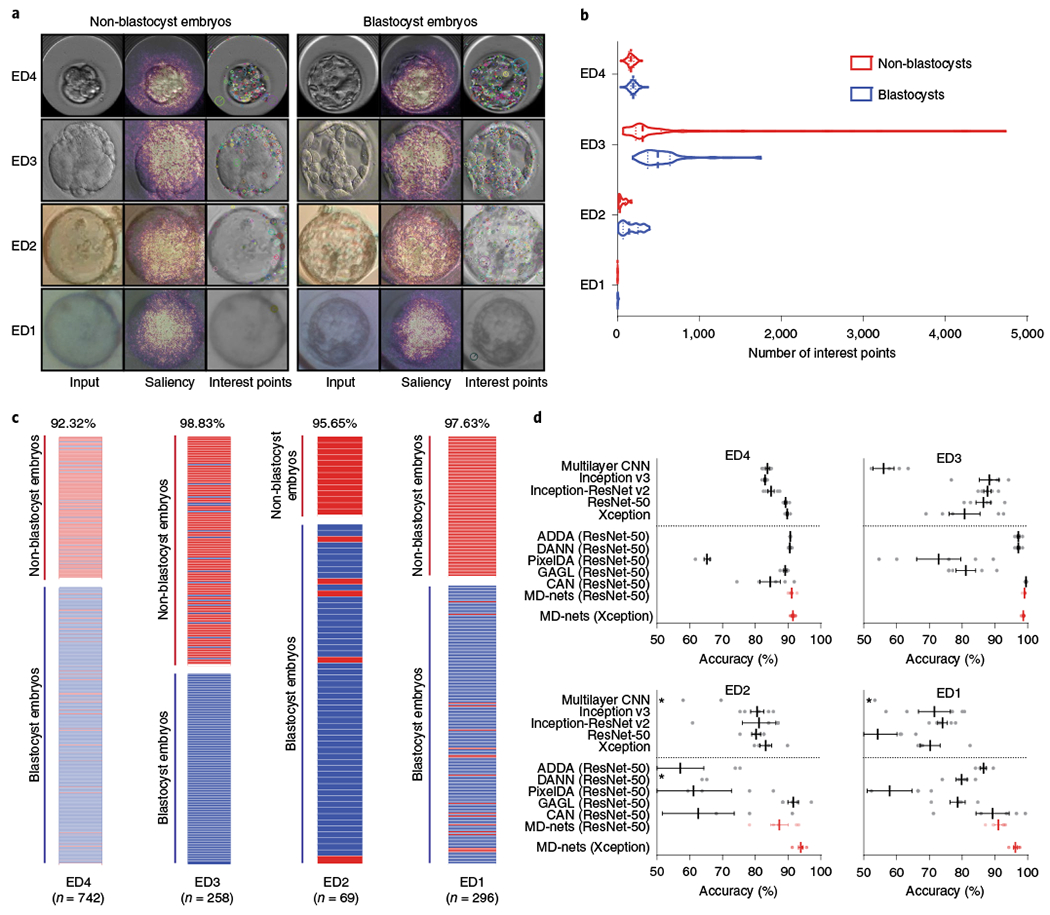
a, The embryo image datasets were collected from a clinical time-lapse system (ED4), various clinical brightfield inverted microscopes (ED3), a portable 3D-printed imaging system (ED2) and a 3D-printed smartphone-based imaging system (ED1). The overlaid saliency maps help to visualize pixels that are most utilized by the MD-nets in their decision-making. The interest points are examples of strong features of different embryo images identified by the SIFT algorithm. b, The distribution of feature points in non-blastocyst and blastocyst-stage embryo images collected from ED4 (n = 251 and n = 491), ED3 (n = 141 and n = 117), ED2 (n = 13 and n = 56) and ED1 (n = 99 and n = 197). The dashed lines represent the median and dotted lines represent quartiles. c, The performance of MD-nets in evaluating embryo images collected using different imaging systems on the basis of their developmental stage. The red bars represent non-blastocysts and the blue bars represent blastocysts. d, The performance of MD-nets in embryo image classification compared to different supervised learning models trained with only the ED4 dataset (source) and unsupervised domain adaptation strategies implemented with ResNet-50, when tested on target test datasets of ED4 (n = 742), ED3 (n = 258), ED2 (n = 69) and ED1 (n = 296). The dotted line separates the domain adaptation methods from the supervised models. Each result represents the average of five random initialization seeds and the error bars represent the s.e.m. The asterisks indicate performance averages of below 50%. The dots represent the individual performance values of each model.
Using 1,698 ED4 embryo images, we trained and validated five CNN architectures (de novo multilayer CNN, Inception v3, ResNet-50, Xception and Inception-ResNet v2) through supervised learning and the Xception-based MD-net through semisupervised learning. Domain adaptation to ED3, ED2 and ED1 using MD-nets was achieved in an unsupervised manner. The networks were initially tested with a hold-out test set of 742 ED4 embryo images, followed by tests with 258, 69 and 296 embryo images from the ED3, ED2 and ED1 datasets, respectively (Supplementary Fig. 1). As with all test sets, the annotations were performed blinded to networks during evaluation.
The best MD-net model was able to classify ED4 images into blastocysts and non-blastocysts with a high accuracy of 92.32% (95% CI = 90.16–94.13%; n = 742) (Fig. 2c). The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were 92.67% (95% CI = 89.99–94.81%), 91.63% (95% CI = 87.50–94.75%), 95.59% (95% CI = 93.50–97.03%) and 86.47% (95% CI = 82.32–89.76%), respectively (Supplementary Fig. 2a and Supplementary Table 1). When evaluating domain-shifted embryo images (ED3, ED2 and ED1), MD-net was able to adapt itself without supervision to new and unseen data recorded using different imaging systems. Once adapted, the performance of MD-nets was excellent with classification accuracies of 98.84% (95% CI = 96.64–99.76%; n = 258) for the ED3 dataset, 95.65% (95% CI = 87.82–99.09%; n = 69) for the ED2 dataset and 97.63% (95% CI = 95.19–99.04%; n = 296) for the ED1 dataset (Fig. 2c). The sensitivity, specificity, PPV and NPV were 100% (95% CI = 96.90–100%), 97.87% (95% CI = 93.91–99.56%), 97.50% (95% CI = 92.72–99.17%) and 100% for the ED3 dataset; 94.64% (95% CI = 85.13–98.88%), 100% (95% CI = 75.29–100%), 100% and 81.25% (95% CI = 59.04–92.87%) for the ED2 dataset; 98.48% (95% CI = 95.61–99.68%), 95.96% (95% CI = 89.98–98.89%), 97.98% (95% CI = 94.89–99.22%) and 96.94% (95% CI = 91.15–98.98%) for the ED1 dataset, respectively (Supplementary Fig. 2b–d and Supplementary Table 1).
The performance of the evaluated supervised CNNs and MD-net in assessing/classifying the source (ED4) embryo image dataset was similar (Fig. 2d and Supplementary Table 2). MD-net performed with an average accuracy of 91.51% with a coefficient of variation (%CV) of 0.77% and the supervised CNNs, de novo multilayer CNN, Inception v3, ResNet-50, Xception and Inception-ResNet v2, performed with average accuracies of 83.72% (%CV = 1.59%), 82.99% (%CV = 0.78%), 89.27% (%CV = 0.85%), 89.78% (%CV = 0.65%) and 84.80% (%CV = 2.60%), respectively (n = 742 ED4 embryo images, n = 5 seeds) (Fig. 2d and Supplementary Table 2). However, in contrast to MD-nets, the supervised networks performed worse in analysing domain-shifted target data (ED3, ED2 and ED1). MD-net performed with an average accuracy of 98.68% (%CV = 0.35%) whereas de novo multilayer CNN, Inception v3, ResNet-50, Xception and Inception-ResNet v2 performed with average accuracies of 56.05% (%CV = 12.70%), 88.29% (%CV = 7.64%), 86.51% (%CV = 5.75%), 80.78% (%CV = 13.05%) and 87.75% (%CV = 2.80%), respectively, when the ED3 embryo dataset was used (n = 258 images and n = 5 seeds; Fig. 2d and Supplementary Table 2). When the ED2 dataset was analysed, MD-net performed with an average accuracy of 93.91% (%CV = 1.69%) and the supervised CNNs de novo multilayer CNN, Inception v3, ResNet-50, Xception and Inception-ResNet v2 performed with average accuracies of 48.12 (%Cv = 35.85%), 80.58% (%CV = 5.49%), 80.29% (%CV = 3.74%), 83.19 (%CV = 4.87%) and 81.16% (%CV = 14.06%), respectively (n = 69 images and n = 5 seeds; Fig. 2d and Supplementary Table 2). Finally, with the ED1 dataset, MD-net performed with an average accuracy of 96.28% (%CV = 1.38%) and the supervised CNNs de novo multilayer CNN, Inception v3, ResNet-50, Xception and Inception-ResNet v2 performed with average accuracies of 42.91% (%Cv = 16.59%), 71.55% (%CV = 15.25%), 54.26% (%CV = 24.25%), 70.27 (%CV = 9.70%), and 74.05% (%CV = 4.45%), respectively (n = 296 images and n = 5 seeds; Fig. 2d and Supplementary Table 2). In addition to the decrease in accuracy of the tested supervised networks, we also observed large variances in the performance of such networks when tested with domain-shifted data for different initialization seeds used during training with the source data (ED4), although no substantial deviance in the loss and accuracies was observed during the evaluations with ED4 for these models (Fig. 2d and Supplementary Table 2).
We also evaluated the effect of adversarial training on the performance of MD-net in analysing the source test data and compared it to the effect of traditional transfer learning in supervised models when using the Xception architecture. The supervised networks showed a substantial reduction in performance on source data (ED4) after transfer learning, whereas MD-net showed a minimal decrease in classification performance using the source data. Xception performed with an accuracy of 89.78% with a s.d. of 0.58% (n = 5 seeds) on the ED4 test set (n = 742 images) when trained with the ED4 data, but its performance significantly dropped to 69.40% (t = 78.21, P < 0.001), 85.18% (t = 17.67, P < 0.001) and 77.22% (t = 48.21, P < 0.001) after transfer learning with ED3, ED2 and ED1, respectively (two-sided one-sample t-tests with d.f. = 4; Supplementary Fig. 3a). By contrast, MD-net (Xception) performed with an average accuracy of 91.51% (s.d. = 0.71%; n = 5 seeds) on ED4, while adaptive training with ED3, ED2 and ED1 led to average accuracies of 91.64% (t = 0.41, P = 0.70), 88.41 (t = 9.81, P < 0.001) and 89.08 (t = 7.67, P = 0.002) on the ED4 test set (n = 742), retaining most of the relevant weights (two-sided one-sample t-tests with d.f. = 4; Supplementary Fig. 3a). These adversarial networks perform well across different domains and are more robust than supervised learning networks owing to their ability to continuously adapt to different distributions, even with unlabelled datasets. We evaluated whether MD-nets can perform equally well on a target data distribution when they are not continuously adapting to the unlabelled test data by freezing the network weights after an initial set of examples. For this purpose, we truncated our test sets and used a portion of the data as an initial set of examples (approximately 50% training and 50% test) to train and fix the final weights. We compared the performance of a network trained as such on the target datasets with and without fixing weights. When evaluating MD-nets trained on ED4, ED3, ED2 and ED1 with fixed weights, the networks performed with accuracies of 90.35% (n = 373), 90.77% (n = 130), 91.67% (n = 36) and 90% (n = 150), respectively (Supplementary Fig. 3b). The networks performed with the same accuracies when allowed to update themselves for all datasets except ED1, where the network benefitted from the additional training, with an accuracy gain of 5.33% (Supplementary Fig. 3b). Furthermore, we probed MD-nets by visualizing the distribution of the network-utilized features using t-distributed stochastic neighbour embedding (t-SNE) and using saliency maps in identifying activations. The t-SNE plots indicated that the adapted MD-net made use of features that were more class specific and similar across domains (Supplementary Fig. 4). Saliency maps of the feature activations helped to confirm that the features used by the neural networks in their decision-making are associated with key embryonic features and were relevant to the classification task (Fig. 2a).
Thus far, our experiments evaluated the advantages of domain adaptation methods, such as MD-nets, over traditional supervised methods that are commonly used in medical image analysis. Although MD-net was developed specifically to assist in medical image analysis tasks, it was also important to evaluate its domain adaptation performance using well-known datasets to benchmark and compare the network against other high-performance unsupervised domain adaptation strategies that exist in the literature. We made use of the Office-31 dataset for a comparative analysis of different domain adaptation strategies19. All of the implemented networks were trained with the ResNet-50 architecture to make comparisons with previously reported results. All six Office-31 domain adaptation tasks were evaluated, and the average performance values were compared. The mean values of three experiments with random seeds were used for the comparison of all of the domain adaptation methods implemented in this study. We compared the strategies of Adversarial Discriminative Domain Adaptation (ADDA), Domain-Adversarial Neural Networks (DANN), Deep Adaptation Networks (DAN), Pixel-level Domain Adaptation (PixelDA), Conditional Domain Adversarial Networks (CDAN), Generative Adversarial Guided Learning (GAGL), and the current state-of-the-art Contrastive Adaptation Network (CAN) and were able to confirm the relatively high performance of MD-nets in adaptation to new domains11,12,20–24.
MD-nets (ResNet-50) achieved an overall average accuracy of 90.7% for all six adaptation tasks available using the Office-31 dataset (Supplementary Table 3). For individual tasks, A → W, D → W, W → D, A → D, D → A and W → A, MD-nets achieved adaptation performance accuracies with s.e.m. values of 95.2 ± 0.5%, 99.2 ± 0.04%, 100 ± 0%, 94.2 ± 0.3%, 77.2 ± 0.3% and 78.2 ± 0.2%, respectively. By contrast, ADDA, DANN, DAN, PixelDA, CAN, CDAN and GAGL achieved average accuracies of 82.9%, 82.2%, 80.4%, 10.6%, 90.6%, 87.7% and 77.7%, respectively (Supplementary Table 3). We observed that MD-nets showed a marginal improvement over the evaluated methods in four out of the six tasks (A → W, D → W, W → D, W → A), placing the evaluated approach among the highest performing domain adaptation strategies reported to date.
Although Office-31 served as a valuable benchmarking dataset, it does not capture the complexity of the real-world medical image analysis tasks addressed in our study. Medical image analysis tasks usually involve images that share a high degree of feature overlap across classes (for example, high-quality blastocyst versus blastocyst), in contrast to the Office-31 natural image dataset, in which the classes are well-defined (for example, laptop versus table). Thus, we used some of the evaluated high-performance domain adaptation alternatives in another comparative analysis using our embryo datasets, which we originally used to emphasize the benefits of MD-nets over supervised networks. All of the implemented domain adaptation methods were trained with the ResNet-50 architecture. All four domain adaptation tasks were evaluated, and the average performance values were compared. The mean values of five experiments with random seeds were used for the comparison of all domain adaptation methods implemented in this study. We compared MD-nets, which makes use of feature space adaptation, against other approaches that utilize strategies for adaptation at the image space (PixelDA), the feature space (DANN, ADDA, CAN) and both image and feature spaces (GAGL)25. The relative performances of ADDA, DANN, PixelDA, GAGL, CAN and MD-nets on the embryo datasets highlighted through example images from the datasets of ED4, ED3, ED2 and ED1 are provided in the Supplementary Information (Supplementary Figs. 5–8). In our evaluations, we observed that MD-nets outperformed all of the other networks, with MD-net (ResNet-50) models achieving an average accuracy of 92.2% for embryo developmental quality-based classification tasks15 (Fig. 2d and Supplementary Table 4). By contrast, DANN, PixelDA, CAN, ADDA and GAGL performed with average accuracies of 78.4%, 64.3%, 84.0%, 82.1% and 85.1%, respectively (Fig. 2d and Supplementary Table 4). The Xception variant of MD-nets was relatively more stable and was able to achieve further gains in the developmental quality-based classification task with an average accuracy of 95.1%. Overall, these results highlight the suitability of the MD-net-based approach for unsupervised domain adaptation, especially in medical image analyses.
Evaluation of MD-net in quantitative morphological assessments of human biological samples.
Quantification of the target cellular morphologies, which is usually performed with some form of a contrast enhancer, is a crucial task in applications involving medical image analysis, such as in pathology. Relative-frequency estimates are often used for the diagnosis of diseases and in the determination of a treatment regimen. Many such estimations are performed through manual microscopy analyses that are subjective and time-consuming. For example, the morphology of individual sperm cells is routinely assessed clinically as part of semen analysis, the primary test in evaluating male factor of couples who experience difficulties in conceiving. Grading criteria used in assessing sperm morphology are extremely tedious and take into consideration the size and shape of head, neck and tail, along with the presence or absence of different cellular features within the head such as the acrosome and vacuoles in defining the morphological quality of each individual cell26. Manual image-based sperm morphology assessment continues to be the gold-standard modality in clinical analysis, and all of the proposed alternative technologies have been either too expensive or too inaccurate for clinical cost-effectiveness27,28. Unsurprisingly, sperm morphology evaluations using such grading criteria are time-consuming, subjective and labour intensive. Although supervised deep learning-based approaches have shown promise in automated morphological analyses of sperm, they have been limited to only high-quality images, as reliable manual interpretation in noisy data is not possible29,30.
To perform automated sperm morphology assessment, we used MD-net (Xception-based) trained with sperm images collected by the American Association of Bioanalysts (AAB) Proficiency Testing Service (PTS) and annotated by trained andrologists (SD4) (Supplementary Fig. 9). Sperm cells were extracted from microscopy images during sample preprocessing (Methods) and were filtered using a CNN trained to identify sperm with 90.07% accuracy (n = 1,340) (Supplementary Fig. 10a). The network, in separating sperm and non-sperm images, performed with a PPV of 84.42% (95% CI = 81.56–87.00%) and NPV of 96.62% (95% CI = 94.88% to 97.89%) (n = 1,340). The sensitivity and specificity of the network were 96.66% (95% CI = 94.99–97.78%) and 84.27% (95% CI = 81.87–86.40%), respectively (n = 1,340). Extracted sperm images were classified into normal and abnormal morphological quality sperm using the MD-net trained with 2,899 annotated sperm images. In our evaluations, the network performed with an accuracy of 86.99% (95% CI = 83.37–90.07%) in classifying sperm on the basis of their morphology (n = 415; Supplementary Fig. 10b).
We randomly selected 200 sperm images from each slide to estimate the morphological score of the tested sample. The measured score was compared against the national averages reported by AAB PTS for each sample. The morphological scores measured for samples 1–10 by MD-net were 2.895%, 9.782%, 5.1%, 8.3%, 4.7%, 10.0%, 7.0%, 9.3%, 8.2% and 5.1%, respectively, while the national average scores reported through the AAB PTS were 3.4%, 10.1%, 4%, 8.6%, 6%, 7.6%, 6.8%, 6%, 8.3% and 3.4%, respectively. A high correlation coefficient of 0.82 (95% CI = 0.38–0.95) was observed between the two methods of measurements (P = 0.004) (Fig. 3a). Overall, the morphological quality measures obtained through MD-net were not largely different from the average score measured by the different technicians who have participated in the AAB PTS across the United States with an average absolute difference of 0.6% (s.d. = 1.5%; Fig. 3b).
Fig. 3 |. Assessment of the performance of MD-nets in quantitatively evaluating cell morphology using human sperm cells as a clinical model.
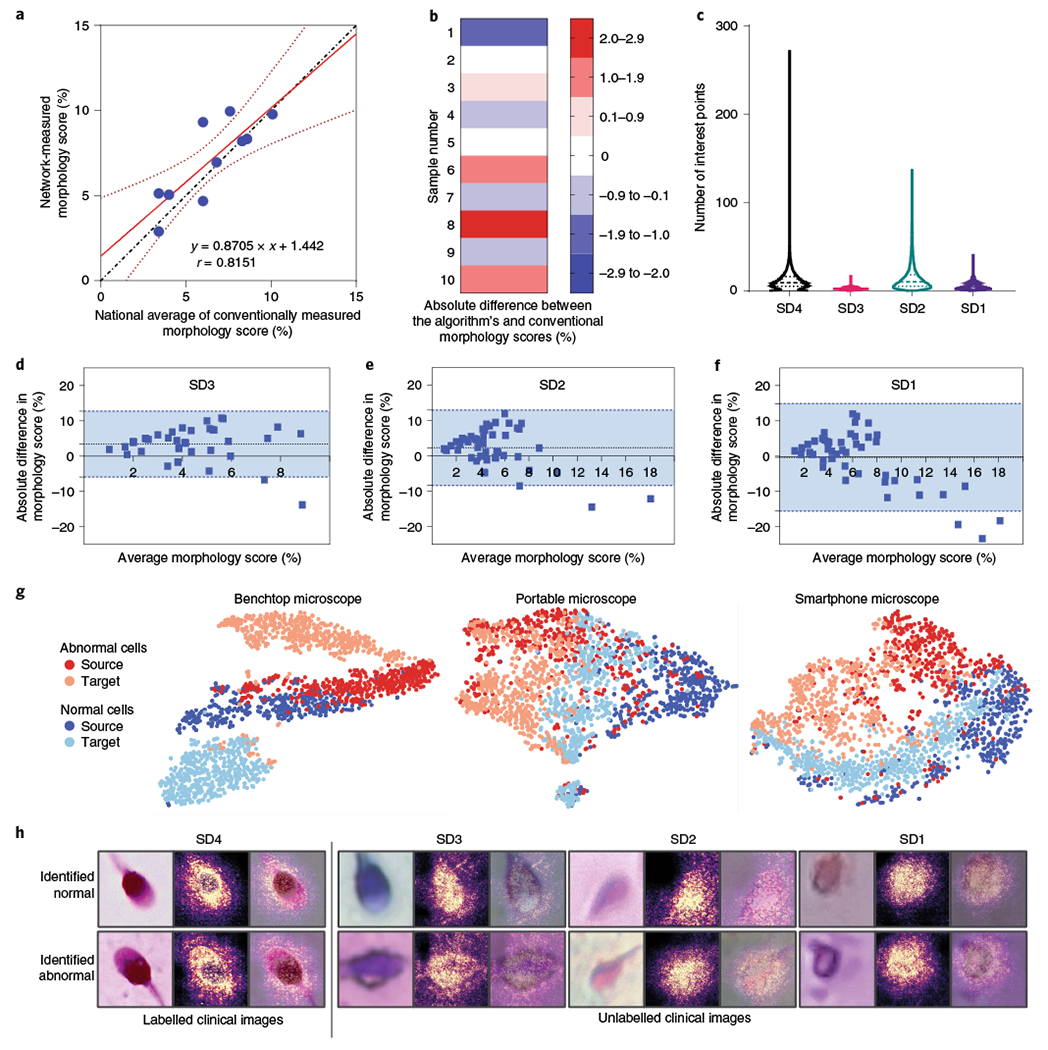
a, Linear regression plot (n = 10). The dotted line represents the line of identity. The solid red line represents the best-fitting straight line on the available data points identified using a least-squares analysis. The dotted red line represents the 95% confidence interval of the fitted line. The equation represents the line equation of the fitted line and r represents the Pearson’s correlation coefficient (P = 0.004). b, The absolute difference between morphological scores of human semen samples (SD4) when measured using automated MD-net and manual-based assessment (national average) performed using conventional manual microscopy. n = 10. c, Comparison of distributions of SIFT feature points per image identified across the various domain-shifted datasets of microscopic sperm images. SD4, n = 86,440; SD3, n = 18,972; SD2, n = 19,668; and SD1, n = 20,554. Feature estimates are not separated by classes as manual annotations are unavailable. The dashed lines represent the median and the dotted lines represent quartiles. d–f, Bland-Altman tests comparing morphology scores estimated on the basis of manual microscopy analysis and MD-net using sperm samples imaged using a benchtop microscope (d) (SD3, n = 40); a portable 3D-printed microscope (e) (SD2, n = 48); and a portable smartphone-based microscope (f) (SD1, n = 47). The dotted lines indicate the mean bias and the blue region within the blue dashed lines indicates the 95% limits of agreement. g, t-SNE plots illustrating source and target clustering achieved by MD-net for the four different sperm datasets. h, Examples of sperm images that were collected using the different optical instruments along with the associated saliency maps obtained from the MD-net feature extractor.
Once a high-performance model that was trained and tested with SD4 was available, we adapted the network to classify images of sperm collected using a benchtop microscope (SD3), a 3D-printed inexpensive and portable microscope (SD2) and a portable smartphone-based imaging system (SD1) (Supplementary Fig. 9). We collected data from over 40 clinical semen samples by imaging them with different imaging systems. Conventionally, sperm morphology assessments involve manual evaluation of individual sperm cells after staining and fixing using a ×100 objective (SD4)26. The low-cost imaging systems produced images at much lower magnifications, poorer optical resolutions and with higher distortions and aberrations compared with the laboratory-grade microscopes (Supplementary Fig. 11). The clinical experts who were involved in this study were unable to annotate such low-resolution images and, therefore, the annotations for individual sperm samples across all systems were not available. SIFT scores were also measured in an effort to describe the images collected using the different imaging systems. The mean SIFT features recorded in sperm images amounted to 11.68 (95% CI = 11.61–11.75), 2.33 (95% CI = 2.31–2.35), 14.56 (95% CI = 14.35–14.77) and 4.69 (95% CI = 4.64–4.74) for SD4 (n = 86,440), SD3 (n = 18,972), SD2 (n = 19,668) and SD1 (n = 20,554), respectively (Fig. 3c).
In our evaluations with sperm samples, the network’s predictions for each isolated sperm cell (at least 200 sperm cells for most samples) were taken into account to measure an overall morphology score for the semen sample. As individual sperm annotations through manual assessment were not possible, a clinical expert manually evaluated replicate slides of each semen sample under a benchtop microscope to estimate the morphology score for each semen sample, which was used in a comparative analysis to establish the network’s performance. Bland–Altman analyses were used to compare the overall agreement between the two approaches (Fig. 3d–f). For SD3, the analysis showed a mean bias of 3.43% (s.d. = 4.77%) with limits ranging from −5.91% to 12.77% (n = 40) (Fig. 3d). Bland–Altman tests for SD2 showed a mean bias of 2.47% (s.d. = 5.52%) with limits ranging from −8.36% to 13.29% (n = 46) (Fig. 3e). For SD1, the Bland–Altman test showed a mean bias of −0.30% (s.d. = 7.84%) with limits ranging from −15.66% to 15.06% (n = 47) (Fig. 3f). The tests revealed that, for SD3, SD2 and SD1, MD-nets did not possess any systematic biases. SD3 did not suffer from proportional biases either; however, SD2 and SD1 possessed proportional biases. Sufficient adaptation was verified through t-SNE plots and saliency maps (Fig. 3g,h). Additional example images of sperm cells from the different domain-shifted datasets, which the developed networks consider to be cells of normal and abnormal morphologies, are provided in the Supplementary Information (Supplementary Fig. 12). Sperm morphology testing at the point-of-care using such systems has remained an unsolved challenge for years even after the advent of mobile-based sperm testing methods31,32. The results suggest that MD-nets, which emphasize retaining source information through unsupervised domain adaptation, can enable the development of inexpensive and portable image analysis-based screening tools for such point-of-care clinical applications.
Using pretrained supervised models for unsupervised adaptation in the absence of source data.
Although supervised networks can be adapted to different distributions through transfer learning using additional annotated data from the target domain, the adapted supervised networks may not necessarily utilize clinically relevant features that were identified by the original supervised network (Supplementary Fig. 3a). Furthermore, the additional annotation required for network adaptation can be both time-consuming and expensive. Adversarial networks can be used to develop frameworks that are capable of working sufficiently well across shifted domains that share relevant features. In the above sections of this Article, we demonstrated the performance of MD-nets trained from scratch across different domains. Here, we intended to investigate the possibility of using saved weights obtained from a pretrained supervised model for unsupervised domain adaptation (Figs. 1b and 4a). To evaluate the suitability of MD-nets for such medical use cases, we used the diagnosis of the blood samples of patients infected with malaria (Plasmodium falciparum) as a clinical model. The original supervised network was developed using a large expertly annotated clinical dataset (27,558 images) and was designed to differentiate between parasitized and non-parasitized blood cells33.
Fig. 4 |. Performance of MD-nets (NoS) in the evaluation of malaria-infected samples.
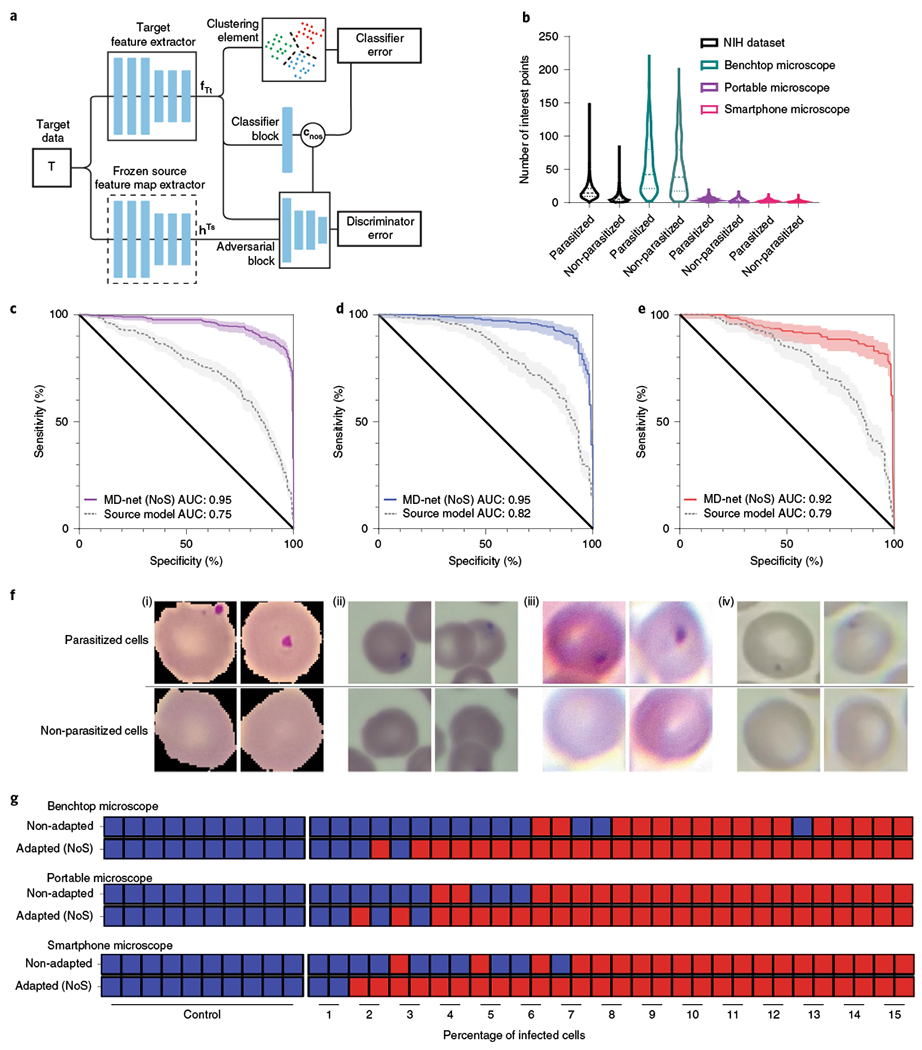
a, Schematic of the adversarial neural network scheme showing the preloaded frozen layers and trainable layers. This version of MD-nets makes use of a clustering element and pretrained source weights and is designed to not use any source data during adaptation while using only unlabelled target data. fTt and hTs represent the feature representations of the target feature extractor and multilinear maps of the frozen source feature extractor, respectively. cnos represents the classifier predictions. b, Comparison of the distributions of SIFT feature points per image identified across the various domain-shifted datasets of microscopy images of parasitized and non-parasitized cells. Samples from non-parasitized MD3 (n = 601), MD2 (n = 688), MD1_t (n = 1,896) and MD1_s (n = 12,401), and parasitized MD3 (n = 601), MD2 (n = 688), MD1_t (n = 1,896) and MD1_s (n = 12,401) were used in the SIFT feature measurements. The dashed lines represent the median and the dotted lines represent quartiles. NIH, National Institutes of Health. c–e, Receiver operator characteristics analyses were conducted to compare the performance of MD-net (NoS) before and after adaptation with preloaded weights on MD3 (c), MD2 (d) and MD1_t (e). The shaded regions represent the 95% confidence intervals. f, Example images of parasitized and non-parasitized cells from the different malaria datasets MD1_s (i), MD2 (ii), MD3 (iii) and MD1_t (iv). g, Qualitative diagnostic performance of MD-net (NoS) before and after no-source adaptation to domain-shifted data collected using a benchtop microscope, a portable microscope and a smartphone microscope (n = 40). The blue squares represent image sets that were qualitatively predicted to be negative for malaria and the red squares represent those that were predicted to be positive for malaria.
Malaria is a major public health problem in countries of the tropical and subtropical areas of the world, affecting an estimated population of 219 million people worldwide and causing nearly half a million deaths annually34. There is a substantial social and economic cost due to malaria infection management in the affected countries, with an estimated direct cost reaching US$12 billion per year in Africa alone, due to disease morbidity, mortality and treatment35. P. falciparum, which is one of the deadliest parasites to humans, is usually the cause of severe malarial parasitaemia (>5% parasitized red blood cells)36,37. Early detection of malaria can help to rapidly identify individuals with or at risk of malaria infection, which can lead to a dramatic reduction in morbidity and mortality rates worldwide. Furthermore, the United States Centers for Disease Control and Prevention (CDC) recommends aggressive intravenous treatment interventions for severe parasitaemia36. The availability of quality-assured microscopy at the point-of-care within the first 2 h of a patient presenting for treatment can contribute to a reduction in the time-to-initiation of antimalarial treatment based solely on clinical grounds, meeting current World Health Organisation (WHO) recommendations34,38. However, owing to limitations on the availability of skilled technicians at the point-of-care, it has recently been proposed that a reliable, low-cost and simple solution for malaria diagnosis in resource-poor settings could be a portable microscopy system associated to powerful machine-learning-based analysis to classify and quantify parasites and blood cells39. In brief, the previous research made use of CNNs in classifying images of parasitized and non-parasitized blood cells, computationally segmented from thin blood smear data collected through a smartphone-attached light microscope (MD1_s)33. We have used a model equivalent to the previously best-reported model to evaluate isolates of blood cell images with and without malarial infections and that were recorded using different imaging hardware.
Thin blood smears were imaged using a desktop microscope (MD3), an inexpensive 3D-printed microscope (MD2) and a smartphone-attached microscope similar to the original reported work (MD1_t) (Supplementary Fig. 13). The SIFT features recorded in non-parasitized cell images, on average, amounted to 50.77 (s.d. = 41.74), 3.23 (s.d. = 2.80), 1.15 (s.d. = 1.42) and 7.83 (s.d. = 8.22) for MD3 (n = 601), MD2 (n = 688), MD1_t (n = 1,896) and MD1_s (n = 12,401), respectively (Fig. 4b). SIFT features recorded in infected cell images, on average, amounted to 54.53 (s.d. = 40.86), 4.65 (s.d. = 2.91), 1.81 (s.d. = 1.88) and 16.84 (s.d. = 11.55) for MD3 (n = 601), MD2 (n = 688), MD1_t (n = 1896) and MD1_s (n = 12,401), respectively (Fig. 4b). To showcase the transferability of learning through unsupervised adaptive scheme, we made use of the previously published dataset and developed a supervised CNN (ResNet-50) model (Methods) for the analysis of malaria infections in red blood cells33. The ResNet-50 network (source only) developed with the supervised learning scheme using MD1_s performed with an area under the curve (AUC) of 0.994 (95% CI = 0.991–0.997) (P < 0.001) in differentiating between parasitized and non-parasitized cells when evaluating MD1_s test images (n = 2,756), similar to the originally reported AUC measure (0.990 ± 0.004) (Supplementary Fig. 14a). Without adaptation, the network experienced a performance drop, as expected, in all shifted datasets with AUCs of 0.751 (95% CI = 0.719–0.782), 0.815 (95% CI = 0.768–0.855), 0.785 (95% CI = 0.736–0.829) for datasets MD3 (n = 766), MD2 (n = 330) and MD1_t (n = 321), respectively (Fig. 4c–e).
Initially, in an effort to minimize the dependence on source dataset requirements, we tried evaluating the original MD-net approach using a limited source dataset (source validation and source test data) (Fig. 1b). The weights of the source-only model were used to initialize an MD-net (ResNet-50) model. The initial layers of the network were frozen, and the network was retrained with the limited source data and target data to adapt the neural network to the target dataset without supervision (Supplementary Fig. 14b). We compared the performance of the network, initialized with the source-only model weights (MD-nets SW), on the target’s test dataset before and after adaptation to evaluate the gain in performance. On the domain-shifted datasets MD3 (n = 766), MD2 (n = 330) and MD1_t (n = 321), the adapted MD-nets (SW) performed with higher AUCs of 0.945 (95% CI = 0.927–0.960), 0.974 (95% CI = 0.951–0.988) and 0.870 (95% CI = 0.828–0.905) in differentiating blood cells on the basis of the infection status (Supplementary Fig. 14c–e) compared with the network that was not adapted (source-only) (Fig. 4c–e). Adaptation to all three domains was also confirmed through t-SNEs, which revealed the ability of the network to separate the common classes from the different domains sufficiently well even with the limited dataset (Supplementary Fig. 14f). We also confirmed that there is no target performance drop due to the source weight initialization, limited dataset and frozen layers during adaptation. The modified MD-net approach (MD-nets SW) achieved classification performances on the target datasets that were not inferior to the unmodified MD-net approach, which was trained from scratch. The unmodified MD-nets approach achieved AUCs of 0.952 (95% CI = 0.935–0.966), 0.932 (95% CI = 0.899–0.956) and 0.848 (95% CI = 0.804–0.886) for the MD3 (n = 766), M2 (n = 330) and MD1_t (n = 321) datasets, respectively (Supplementary Fig. 14c–e).
Although we have shown that MD-nets can also be used with minimal source data when the source weights are available, one limitation, especially for medical image analysis tasks, is still the need for getting access to an annotated source dataset during adaptation. A suitable solution for the medical domain would involve the complete removal of any dependence on the regulated clinical source data while using unlabelled target data collected from different centres/instruments for adaptation. We therefore expanded the framework MD-nets to include a clustering element to generate pseudolabels during adaptation (Fig. 4a). The updated framework utilized pretrained and frozen weights loaded onto one feature extractor and adapted an MD-net model, which was also preloaded with the source weights, to the target distribution. The updated MD-nets framework, MD-nets no-source (MD-nets (NoS)), used only unlabelled target distribution data for network adaptation and did not use any data from the source distribution. To verify and benchmark the adaptation ability of MD-nets (NoS) to shifted distributions, we evaluated the network for its domain adaptation performance using Office-31 (Supplementary Table 3). MD-net (NoS), with ResNet-50 as the feature extractor, achieved a relatively high average adaptation accuracy of 88.4%.
In the experiments involving malaria datasets, when preloaded with the network weights from the source-only ResNet-50 model and trained with the shifted datasets, the resultant models were able to classify parasitized and non-parasitized red blood cells with AUCs of 0.952 (95% CI = 0.935–0.966), 0.954 (95% CI = 0.926–0.974) and 0.920 (95% CI = 0.885–0.947) using the MD3 (n = 766), M2 (n = 330) and MD1_t (n = 321) datasets, respectively (Fig. 4c–e). Example images of parasitized and non-parasitized cells from the different domain-shifted datasets that were categorized by the developed network have been provided (Fig. 4f and Supplementary Fig. 15). While these results showcase the advantage of adversarial training for network adaptation across domains and distributions, without the need for source data, the cell-wise classification performance does not provide a clear perspective on the clinical benefit of adapting the network to the target domain distribution.
To evaluate the efficacy of MD-nets (NoS) for clinical use-cases of such a system, we used the supervised source-only model and MD-nets (NoS) models, to identify simulated samples on the basis of the presence of parasitized cells. A total of 40 sample sets of annotated images was prepared (Methods) such that the ratio of parasitized to non-parasitized cells in each set reflected clinically relevant ranges (0–15%)36,37. For image sets of MD3 (benchtop microscope), the adapted MD-net (NoS) model performed with a diagnostic accuracy of 90%, whereas the non-adapted network performed with 65% accuracy (Fig. 4g). For MD2 image sets (Portable microscope), the adapted MD-net (NoS) model performed with an overall diagnostic accuracy of 90%, and the non-adapted network performed with a 77.5% accuracy (Fig. 4g). For MD1_t image sets (Smartphone microscope), the adapted MD-net (NoS) model performed with an overall diagnostic accuracy of 95%, and the non-adapted network performed with 75% accuracy (Fig. 4g). In this evaluation using the 120 prepared sample sets, the adapted networks were able to identify all cases of severe parasitaemia (>5% infected cells) correctly across all datasets, while the performance of non-adapted networks was variable.
Discussion
Data available at different medical clinics can be skewed or may be divergent from the overall distribution due to localization of disease prevalence, practice-dependent technical procedures, variations in the quality and model of data acquisition systems, and variations in patient populations. As most deep learning models are limited by their confinement to the training data domain, the data collected from a single clinical centre may not be generalizable across different facilities or instruments40. Most studies using artificial intelligence (AI) for image-based medical diagnoses do not evaluate performance across centres, let alone provide solutions for adaptation at different centres41. Furthermore, clinical data are highly regulated and are therefore not easily available for research or AI-based product development. The development of highly robust machine learning models that are suitable for multiple centres is therefore more difficult due to logistical constraints. Although networks can be adapted to different distributions under supervision through additional training using transfer learning with site-specific data, the lack of control on features used by the new network may not be well suited for medical image analysis tasks6,7,42. Such networks would need additional stringent validations that require resources and experts in machine learning and clinical staff, making it difficult for most and impossible for some centres. Even when training using the same dataset, different supervised models, trained identically, tend to perform unpredictably when tested on a shifted distribution (Fig. 2d and Supplementary Table 2). A recent study has also identified this problem and has indicated that this unpredictability is due to underspecification—the availability of numerous equivalent solutions for a given task and dataset43. Thus, although such networks might perform very well during development and initial validation, they may not hold up well when handling shifted or real-world distributions. This problem is likely to worsen with both larger networks and smaller datasets, as is the case with most medical image analysis tasks. The reported MD-nets approach presents a promising solution for such problems with domain dependence in medical image analysis tasks, where reliability is paramount.
The adversarial network scheme utilized by MD-nets uses feature alignment between the source and target datasets during its training phase. This aspect of the network allows for the validation of models, developed using site-specific target data, with known standardized datasets that can be used as a part of the source dataset. It also helps the network to minimize overfitting on either source or target-specific features and, in theory, mostly limits feature utilization by the network to shared features between the two domains. As medical tasks usually involve evaluating classes that have strong similarities between them and possess a limited number of distinctive features that networks can utilize, conditioning on class information is vital. Furthermore, the prioritization of network update protocols on the basis of uncertainty helps to minimize learning failures that may be induced by the class-conditioning step. We expect that such a controlled learning and adaptation methodology is highly suitable for clinical tasks and our evaluations that make use of clinical embryo data highlight its benefit over supervised learning (Fig. 2 and Supplementary Fig. 3). Such an approach is particularly useful in the development of automated point-of-care systems that make use of inexpensive hardware with relatively poorer imaging capabilities. As the collection and annotation of large datasets with new systems would be extremely difficult, the reported approach will aid the development and adaptation of machine learning models that were developed using standardized datasets to work with unannotated data collected using the newer inexpensive systems. Even when the data are collected using the same instrument, annotation of medical datasets, in particular, is a highly resource-intensive and expert-dependent task that considerably hinders the development of robust medical AI models. Owing to its suitability for semi-supervised applications, the reported MD-nets approach also offers an additional benefit over traditional supervised networks by enabling effective network training using largely unannotated datasets, with only a limited number of annotated examples.
To date, studies that made use of adversarial learning schemes for medical image analysis have primarily focused on utilizing their generative capabilities for tasks such as image reconstruction and synthesis among others44–48. In fact, generative approaches have been examined towards converting subpar image data collected from different inexpensive systems to resemble high-quality human-readable microscopy images45,49. Although these methods show substantial promise in certain areas of medicine, they are also limited to the training data domains and utilize a complex matched-image training process during model development, which, depending on the task, may not be possible45,48,49. Such models, depending on the training methodologies used, may be highly susceptible to hallucinations as the generator actively tries to outperform the discriminator, which limits its value for use with lossy image datasets such as those observed with point-of-care systems50. By contrast, the adversarial approach used here, MD-net, lacks a generative element and focuses on minimizing the performance of the discriminator between the images from the different data distributions, effectively forcing the network to utilize features that are available in both source and target domains. The relatively easy adaptation to different distributions and lack of need for matched image training greatly improves the developmental process of such generalizable networks and with simpler and inexpensive systems. Although, generative variants of domain adaptation strategies, such as PixelDA and GAGL, have been reported to be suitable using the Modified National Institute of Standards and Technology database (MNIST)22,23, it has been suggested that such an approach may not be suitable for tasks/datasets when label pollution is a potential concern, such as in Office-31, and is therefore probably unsuitable for most real-world medical datasets22. Furthermore, generative models could also struggle from the limited dataset availability for image generation during the adaptation process51.
Current deep learning approaches in medicine primarily focus on developing models for a particular task that work with one type of imaging system, usually the most commonly available system due to data dependency issues, and rarely focus on adaptability to different systems. The use of such deep learning approaches is therefore limited for newer and more efficient systems of the future or those that are still in the developmental pipeline. The reported MD-net approach can aid in adaptability from earlier systems to newer or other similar systems with potentially lower target data requirements. Such an approach can be very useful in improving access to care by facilitating the adaptation of high-performance models to smaller clinics and medical centres, where large, structured clinical datasets suitable for the development of reliable deep learning models may not be available. Furthermore, the reported methods can be used in repurposing models for clinical image analysis tasks performed at different centres and practices and similar tasks that can be performed using different biological samples (for example, sperm assessment in semen for fertility screening and sperm identification in tissue samples during microscopic testicular sperm extraction procedures).
Most clinical datasets used in the development of high-performance models are unavailable for external and research use due to regulatory limitations on getting access to patient information/data. MD-nets (NoS) utilizes previously developed models to adapt to datasets from different clinics without the need to access the original clinical data used for system training and validation and without the need for additional data annotation. Advancements provided through MD-nets open the possibility of augmenting most supervised models to be efficiently and reliably repurposed and reused using an adversarial learning scheme. We envision that such an approach has federated learning applications and could potentially save a considerable amount of time and resources while leading to the wider utility of well-trained and highly validated deep learning models. Potentially, knowledge from multicentre datasets can be synchronized under a single unified model/knowledge library, while protecting confidential information.
While the MD-nets (NoS) approach, which makes use of unlabelled target dataset and without direct access to original source dataset, can be of enormous benefit to medical and biomedical communities, the source model weights of the originally trained network, which are needed for domain adaptation, have a strong influence on the adaptation process. Supervised models carrying data biases and a high degree of task-irrelevant feature information can affect the network’s performance on the target dataset adversely. Furthermore, such a no-source approach, in its current form, makes use of a small number of labelled target data in validating the trained model and may not be suitable for tasks, such as the sperm morphology assessment described in this work, where target data labelling and manual verification are not feasible. When used properly, MD-nets can help to extend the effective life of many supervised models given that the original network’s learning can be utilized on newer and less common data domains, such as in the development of point-of-care imaging systems.
Point-of-care systems such as those presented in our study using smartphones can greatly improve access-to-care and present pathways for surveillance diagnostics and disease monitoring systems52. A crucial aspect for the feasibility of such point-of-care devices is their use costs, especially for low- and middle-income countries. In such point-of-care optical diagnostic systems, the image quality may not be suitable for reliable human inference due to factors such as limited imaging capabilities, noisy signal and looser control of imaging parameters. As shown in this study with sperm morphology assessments, images that cannot be reliably evaluated by human experts can be analysed using MD-nets given that features relevant for the system’s classification in the source are also available in the target domain. MD-nets analyse images of a target domain for classification without the need for any additional expert intervention, making it more suitable for point-of-care systems.
Furthermore, MD-nets can detect unseen distributions through the activity of its discriminative element, and automated adaptation to a target domain can be achieved if the network weights are not fixed (Supplementary Fig. 3b). In the case that the network does not recognize a shift, there will be a negligible change in the network’s discriminator loss and the model weights will not be updated. This can be useful for a non-expert end user who may not recognize shifts in the image data used by the network during the post-development phase. Thus, MD-nets can greatly contribute to the development and utility of automated point-of-care imaging systems for diagnostic applications.
MD-nets were developed taking into consideration the limits and preferences of the medical and healthcare communities. The approach makes use of a source-constrained, class-specific feature space-based adversarial adaptation strategy in developing robust and high-performing deep learning models for biomedical applications. The versatility of the approach enables the use of MD-net in both semisupervised and unsupervised learning scenarios to effectively capitalize on the largely unlabelled medical datasets. Finally, its ability to adapt pretrained clinical models developed at one medical centre to the dataset composition of another centre without the need for any data sharing and labelling can be of high value within the healthcare and industrial circles.
Methods
Dataset preparation.
In this study, we collected image data for three clinical image analysis tasks, namely: differentiating human embryos on the basis of their developmental status, human sperm on the basis of their morphological quality and red blood cells on the basis of their status of malarial infections. For each task, samples were imaged using both commercially available and non-commercially available imaging systems. Datasets were designed and named on the basis of the quality of images produced by the imaging system (Supplementary Information). In brief, datasets ranged from a quality level of 4 to 1, with 4 being the highest and, usually, the clinical imaging set-up used by the expert annotators. Malaria datasets carry 1_s and 1_t categories, where 1_s, used as the source, was externally collected and annotated, although both 1_s and 1_t were imaged using a smartphone attached to a benchtop microscope.
Human embryo image datasets.
The highest quality embryo dataset (ED4) comprises 2,440 images of embryos captured at 113 h after insemination of embryo culture from 374 patients at the Massachusetts General Hospital (MGH) fertility centre in Boston, Massachusetts during routine clinical care and has been used in this study under an institutional review board approval (IRB#2017P001339) (Supplementary Fig. 1). The embryos were imaged using a commercial time-lapse imaging system at MGH (Vitrolife Embryoscope) and only embryo images that were collected at 113 h after insemination were used for this study. There is no universal grading system for embryos, and the annotators used a five-quality grade system that is specific to our dataset as defined by the Massachusetts General Hospital fertility centre, which uses a modified Gardener blastocyst grading system13,15. A two-category embryo classification (blastocyst; non-blastocyst) based on the blastocyst status is more commonly recognized worldwide. The two-category system is a condensed version of the five-category system, where classes 1 and 2 of the five-category systems belong to one class (non-blastocyst), and classes 3, 4 and 5 belong to the other class (blastocyst). Images were therefore annotated by MGH embryologists on the basis of their developmental grade, and the annotated data were used for training on the basis of the previously described five-class system focused on embryo morphological features with inferences made at a two-class level (blastocyst; non-blastocyst). ED4 was used with a split of 1,188, 510 and 742 for training, validation and testing, respectively (Supplementary Fig.1). ED3 comprises 258 images of embryos recorded using various clinical benchtop microscopes and was originally collected by the Society for Reproductive Biologists and Technologists (SRBT) for the Embryo ATLAS project (Supplementary Fig. 1). The images were categorized into two different classes, namely blastocysts and non-blastocysts by eight director-level embryologists from eight fertility practices across the United States. The 69 images of ED2 were recorded by imaging embryos designated for discard or research using a portable stand-alone optical system (Supplementary Information and Supplementary Figs. 11 and 16), while the 296 images of ED1 were recorded by imaging embryos designated for discard or research using a portable smartphone-based optical system (Supplementary Information and Supplementary Figs. 11 and 17). All imaging tasks, including data collection and annotation, were performed by staff at the MGH fertility centre under institutional review board approval (IRB#2017P001339, IRB#2019P001000 and IRB#2019P002392). Images were categorized into five different classes by MGH technical staff and consolidated into two inference classes similar to the ED4 data preparation. All of the experiments were performed in compliance with the relevant laws and institutional guidelines of the Massachusetts General Hospital, Brigham and Women’s Hospital and Mass General Brigham. Data split for each individual domain adaptation task evaluated in this study are available in the Supplementary Information (Supplementary Table 5).
Human sperm image datasets.
The highest-quality human sperm image dataset (SD4), which was used in this study as the source dataset, was obtained from images of ten slides of smeared and stained human sperm samples. The images of these slides were obtained from the American Association of Bioanalysts (AAB) and were imaged using ×100 microscopes. The resolution of these images in their stitched form (full slide image) was as high as 266,000 × 180,000 px. A total of 322,081 individual cells/objects was extracted from these images using a template matching algorithm (Supplementary Information and Supplementary Fig. 18). A CNN classifier was used to differentiate between sperm cells and non-sperm cells to refine the dataset before use29. The refinement process yielded a total of 197,283 individual sperm images, which were later used mostly for testing the developed network (Supplementary Figs. 9 and 18). A custom-built mobile application was used to facilitate individual sperm image annotations, which comprised the following classes: normal sperm, head defect, neck defect and tail defect (Supplementary Information and Supplementary Fig. 18). A total of 4,142 sperm images was annotated by five clinicians at MGH, which made up the annotated source data. The data were split into three sets, namely training, validation and test of sizes 2,899, 828 and 415, respectively, during the development of the initial network (Supplementary Fig. 9). The network used for the evaluation of the AAB sperm slides utilized 4,142 annotated sperm image data as the source and 193,141 as the target (testing) in estimating the overall morphological score of each sperm slide. We measured the average sperm morphology score of each patient sperm slide (n = 10) by analysing 193,141 sperm images using MD-net and compared the results with the national average sperm morphology score measured by at least 90 technicians across the country as reported by AAB. The sperm image data used for SD3, SD2 and SD1 domains were obtained from 47, 48 and 48 patient semen slides that were collected as part of the participant’s routine clinical practice at the MGH fertility centre (Supplementary Fig. 9) (IRB#2019P001015). Clinically, the datasets were prepared by imaging smeared semen samples on glass slides and stained using the Romanowsky staining method. The sperm image data used for SD3, SD2 and SD1 domains were recorded using a benchtop Keyence microscope at ×60 magnification, a 3D-printed portable imaging system (Supplementary Information and Supplementary Figs. 11 and 16) and a 3D-printed smartphone-based imaging system (Supplementary Information and Supplementary Figs. 11 and 17), respectively. The overall morphology score for each slide was measured using conventional manual microscopy by MGH fertility centre technical staff. Individual cells were extracted from these images using a template matching algorithm. As a result, 100,892 sperm images were collected for SD3; 19,668 sperm images were collected for SD2 and 20,554 sperm images were collected for SD1, which were eventually used in the development and evaluation of the MD-nets (Supplementary Fig. 9). A total of 7, 2 and 1 slides was used in calibrating the network for performance with SD3, SD2 and SD1, respectively, during network development and these slides were therefore not used during the system evaluation. Data split for each individual domain adaptation task evaluated in this study are available in the Supplementary Information (Supplementary Table 5).
Malaria image datasets.
The externally annotated source dataset (MD1_s) comprises 27,558 single blood cell images of Giemsa-stained thin-blood smear slides, which were collected from 150 patients with P. falciparum infection and 50 healthy controls (Supplementary Fig. 13). The thin-smear slides were imaged using a smartphone camera attached to a benchtop bright-field microscope, and segmentation was performed to isolate individual red blood cell images. All images were manually annotated between infected (parasitized) and non-infected (non-parasitized) cells by a single expert slide reader from Mahidol-Oxford Tropical Medicine Research Unit in Bangkok, Thailand. The dataset is publicly available and is hosted by the National Library of Medicine (NLM) of the NIH33. The MD1_s dataset was split into 19,290, 5,512 and 2,756 for training, validation and testing, respectively, with equal class distributions for each set. The malaria blood cell data used for MD3, MD2 and MD1 domains were obtained from eight patients who were positive for malarial infection, confirmed using an immunochromatographic antigen test (Abbott BinaxNOW), during routine clinical care at MGH. Thin-smear slides were prepared and stained with Giemsa. These deidentified slides were approved for secondary research use by the institutional review board at BWH (IRB#2020P000644). Slides were imaged using a benchtop microscope, a portable stand-alone 3D-printed microscope and a smartphone-based microscope (Supplementary Information), and individual cells were extracted from these images using a template-matching algorithm (Supplementary Information and Supplementary Figs. 11, 16 and 19). As parasitized cells of these slides that were originally confirmed by clinical staff presented very distinct morphological patterns, such as chromatin dots under microscopic investigations, the annotators were asked to differentiate images collected from these slides on the basis of the presence of such dots in the individual red blood cell images53. These samples were manually annotated by three members of our research group into two different classes, namely P. Falciparum infected (parasitized) and uninfected (non-parasitized) cells. Only cells with a perfect agreement between all three annotators were used in this study. MD3 comprised 3,792 blood cell images recorded using a benchtop microscope. The data were split into a validation set of 3,026 and a test set of 766 (S13). MD2 comprised 1,645 blood cell images recorded using a portable imaging system. The data were split into 1,315 and 330 for validation and testing, respectively (Supplementary Fig. 13). MD1_t comprised 1,603 blood cell images recorded using a smartphone attached to a benchtop microscope, similar to data imaging performed for MD1_s. The data were split into 1,282 and 321 for validation and testing, respectively (Supplementary Fig. 13). Data split for each individual domain adaptation task evaluated in this study are available in the Supplementary Information (Supplementary Table 5).
Malaria image set preparations for simulated patient cases.
To quantitatively evaluate the performance of MD-nets (NoS) in detecting malaria-infected blood cells in samples, we prepared a new set of data using malaria slides received from MGH. A total of 12,866, 9,752 and 8,843 malaria images recorded from MGH malaria slides were included in the MD1_t, MD2 and MD3 datasets, respectively. The new images were also annotated manually by three staff on the basis of the status of malarial infection. Using the updated datasets, 40 image sets per domain, composed of different clinically relevant proportions (0–15%) of parasitized blood cell images, were prepared for the simulated diagnostic evaluation of MD-nets (NoS). Each image set contained a total of 1,500 unique cell images. Ten image sets (for all three datasets) contained only non-infected blood cells (0%). The other 30 cases contained varying levels of P. falciparum-infected blood cells with a ratio of infected cells ranging from 1% to 15% (2 cases for every 1% increase) (for all three datasets). The models were then tested using these prepared simulated cases and, in each dataset, all of the samples that reported a higher number of positives than the control samples were considered to be positive.
Network development and training.
The general MD-net design includes a base network architecture with a final flattened layer connected to a classifier block and an adversarial block (Fig. 1b). MD-nets utilize two different training strategies based on the availability of source data. With the availability of source data, MD-nets capitalize on a more traditional approach taking into consideration best practices suited for the medical domain with a strong emphasis on source performance. When source datasets are unavailable, the pretrained source model is utilized by MD-nets, in combination with a clustering element to generate pseudolabels, with an emphasis on retaining source information (Fig. 4a).
MD-nets.
During the training phase, the images from both the source and the target datasets are transformed into the respective feature representations by the feature extractor of the base network. The feature representations are utilized by the classifier and adversarial blocks during training to effectively classify between the different classes and differentiate between the different domain distributions, respectively. Borrowing ideas from previous work, MD-nets are trained by minimizing the classification loss generated using the source data by the classification block, while maximizing the discriminator loss (transfer loss), increasing the domain confusion11,12,24,54 (Fig. 1b). More specifically, the traditional approach builds on the concepts and ideas utilized by DANN and CDAN11,12. We conditioned the discriminative block using the class labels to improve the transfer of class-specific information between the domains. The domain discriminator, which is trained to discriminate source and target features conditioned by class information, makes use of the class predictions from the SoftMax function of the classifier network to compute the conditional distribution. Moreover, to improve the adaptation performance, imbalance in the training data (class imbalance) was addressed by balancing data through random oversampling and under sampling distributions such that the resultant distribution of labels in each epoch is balanced. We performed hyperparameter tuning by computing reverse validation risk. The stoppage of network training in MD-nets was defined by monitoring performance on source data to minimize overfitting on the target. Furthermore, MD-nets included weight normalizations at the SoftMax layer to improve class separation among distributions, and batch normalization was added to the feature representation to help in reducing domain discrepancy (by reducing internal covariate shift).
To adapt a network trained using a source data distribution Ds for a particular task to a shifted target data distribution Dt for the same task, both Ds and Dt were passed through the MD-net’s base network (specific to the task) to iteratively obtain the feature representations fs and ft for every data point of Ds and Dt. Here, Ds and Dt are represented by and , where x is the datapoint (image) and y is the associated classification label for n number of images. ResNet-50 was used for all sections of the study involving comparisons. For the cell classification tasks based on embryo and sperm morphologies, we used the Xception as the base network as Xception, especially for embryo image analysis, resulted in a more robust performance15,55. Xception models did not include additional weight and batch normalizations, although it is probable that the performance will improve with their inclusion. The 2,048 features from the flattened layer of these networks were used to obtain fs and ft from xs and xs for every training step. These representations are passed to the classifier block where the conditional probability vectors cs and ct are generated using a SoftMax function. For the classification tasks involving sperm and red blood cells, the probability vector length was limited to 2, while embryo tasks used a vector length of 5 and the length was condensed to 2 through summation15. The source classifier error is minimized to guarantee lower source risk and is defined as
where L() represents cross-entropy loss and C() is the classifier network.
In parallel, during the adaptation process, the discriminator error is maximized. The discriminator network, D, utilizes the common base network along with the adversarial block which consists of three layers with rectified linear units, activations and dropouts. In the discriminator error calculation, weighted entropy conditioning is utilized along with a multilinear feature map h. The computation of h(f,c) is a multilinear map, formed by the tensor product of feature representation f and classifier prediction c. Where c for k classes is given by c = [c1, c2, c3, … , ck] and f for l dimensions is given by f = [f1, f2, f3, … , fl]. The resultant multilinear map h is expressed as
The combination of f and c, performed as a conditioning step, helps to preserve class-specific information across domains. Furthermore, entropy was used as a metric of uncertainty in the classifier predictions to improve the classification performance on target distribution by encouraging the high confidence predictions in the unlabelled target domain. The uncertainty of the predictions, H(c), was defined as,
Where n is the total number of training classes and ci is the probability vector with each class. Each training example at the discriminator is weighted with
Therefore, the discriminator error ε(D) is given by,
The overall MD-net training is achieved by minimizing source risk and maximizing the discriminator error for distance reduction between the source and target distributions, which is achieved by minimizing the overall cost function given by,
where λ is a trade-off between discriminator error and source-risk.
MD-nets (NoS).
MD-nets (NoS) were developed for specific scenarios in which high-quality clinical source data are unavailable and source model weights with only the unlabelled target dataset were available. This version of MD-nets carries all of the elements of the original MD-nets, along with an additional frozen feature map extractor initialized with source weights and a DeepCluster-based clustering element56 (Fig. 4a). As there are no source data available during network training, MD-net (NoS) makes use of feature maps, fTs, generated by the frozen source feature map extractor along with pseudolabels generated by the clustering element, when using the unlabelled target data for adaptation. The target feature extractor and classifier block, also initialized with the source weights, along with the adversarial block are updated throughout training. However, the clustering element is updated periodically at regular intervals, which is treated as a hyperparameter for the different tasks. In the NoS version, MD-nets are trained by minimizing the discrepancy between the pseudolabels generated by the clustering element and the target classifier, which is treated as the classifier error, ε(Cnos). Furthermore, while minimizing the classifier error, we maximize the discriminator error similar to the MD-nets design. In this approach, during adaptation with the unlabelled target examples, the discriminator helps to stabilize the adaptation process by acting as a regularizer, restricting the target feature maps, fTt, in substantially deviating from the frozen source feature maps, fTs.
The classifier error is minimized to match the generated pseudolabels obtained from the clustering element56. For a given set of target images , once the initial labels, assigned based on the classifier predictions , the initial centroids are calculated using
Once all of the centroids for each class are obtained, we compute the initial pseudolabels, , by finding the nearest centroid cluster by obtaining the minimum cosine distance between the feature map and the centroids.
Using the generated pseudolabels, we calculate the centroids and generate pseudolabels once more,
The newly generated pseudolabels are utilized in the calculation of the classifier error during training. The NoS classifier error ε(Cnos) is defined as
where Lnos() represents cross-entropy loss and Cnos() is the NoS target classifier network.
As there are no source images, the discriminator error ε(D) is given by
The overall MD-net (NoS) training is achieved similar to the original approach, by minimizing classifier error and maximizing the discriminator error,
where λ is a trade-off between discriminator error and classifier error.
Embryo classification network parameters.
MD-net primary embryo classification models were trained with ED4 as the source, and ED3, ED2 and ED1 as the target. ED4 was divided into source training and source validation when used for adaptation to other distributions (Supplementary Table 5). These embryo classification models used Xception as the base network. The models were trained to classify embryo images recorded at 113 h after insemination into blastocysts and non-blastocysts as two major clinically relevant classes of embryos. Hyperparameters were evaluated manually (Supplementary Table 6). Learning rates between 0.1 and 0.0001 and batch sizes of 32 and 64 were evaluated when developing the best performing neural network (Supplementary Table 6). Data balancing was achieved through augmentation and stratified batch distributions. Data augmentation was performed by random horizontal or vertical flipping and random 0–359° rotations of the images. Early stoppage was set at 2,000 iterations after the lowest source validation loss (Supplementary Table 6). Reverse validation risk for each model trained was also evaluated when picking the final models used in the study. For the variability experiments (Fig. 2d), the supervised networks were trained using the Xception, Inception v3, ResNet-50, Inception-ResNet v2 and de novo 40-layer CNN55,57–59. These networks were trained using ED4 data and their hyperparameters were evaluated manually to obtain the best performing model. The dimensions of all images used in network training were resized to 210 × 210 px. All training was performed within the Keras environment. We picked the best model on the basis of the best validation loss and tested the performance on unseen target datasets (ED4, ED3, ED2 and ED1) by randomly changing the seed (5 seeds). All networks were trained with hyperparameters of the best performing model and using different seeds (5 seeds) (Supplementary Table 2).
We envision the use of such approaches for medical image analysis tasks to have different benefits. While continuous adaptation offers the potential to adapt regardless of domain shifts and in the absence of any knowledge of the shifts, freezing the weights helps in utilizing an adapted network for rapid testing. In our study, we wanted to investigate the feasibility of both approaches for medical image analysis tasks if their test performances would be satisfactory. We therefore utilized our Embryo datasets (ED4, ED3, ED2 and ED1) in evaluating MD-net performance when the network weights were frozen, and when they were continuously adapting. The original target test sets of the four datasets were split into 50%–50% (t1–t2) (ED4 = 373, ED3 = 130, ED2 = 36 and ED1 = 150) for this experiment. In the continuous adaptation arm of the experiment, both test sets were used during adaptation, that is, the network was trained with the unlabelled target using both sets (t1 and t2), and the test result was obtained by calculating the network performance on t2. In the fixed weights arm, the MD-net was trained with t1 as the unlabelled target and its weights were frozen once the best performing model on source was obtained. The network with its frozen weights, which now behaves as a classifier, was used to evaluate t2 and the performance was calculated for comparison with the other arm. For continuously adapting MD-nets, training was terminated with the early stoppage set at 2,000 iterations, and the model with the best validation loss was saved.
In medical tasks, assuming the original network training source data are well validated, it is preferable that networks developed for newer domains utilize features that are shared by both domains while performing sufficiently well in both domains. High network performance when using such common features between the domains helps confirm that these networks developed for the target domain make use of medically task-relevant features during classification. Thus, in our study with embryo images, we evaluated the effect of transfer learning compared to our adversarial approach. For the experiment comparing adversarial performance and supervised adaptation performance, MD-nets (Xception) were trained with ED4 as the source data and unlabelled target data as described earlier. A baseline supervised network (Xception) was trained using ED4 data and tested on a held-out ED4 test set. The network was then, through transfer learning, adapted to the different domains (ED3, ED2 and ED1) with the use of the target labels. The networks were then tested using the ED4 test set. Similarly, the baseline network weights were used in the MD-net (Xception) framework and the network was trained using the target sets ED3, ED2 and ED1 without their target labels. The MD-net models were also tested using the common ED4 held-out test set.
For comparison with other methods, we reimplemented MD-nets with ResNet-50. Hyperparameters were optimized through manual evaluations and selection. Learning rates between 0.1 and 0.0001 and batch sizes of 32 and 64 were evaluated when developing the best performing neural network model (Supplementary Table 6). Data balancing was achieved through augmentation and stratified batch distributions. Data augmentation was performed by random horizontal or vertical flipping and random 0–359° rotations of the images. The early stoppage was set at 5,000 iterations after the lowest source validation loss. We also implemented DANN, ADDA, PixelDA, GAGL and CAN with ED4 as source, and ED3, ED2 and ED1 as the target. These networks were trained and optimized based on the practices and guidelines suggested by the original authors. A list of the evaluated hyperparameters and best performing models is provided in the Supplementary Information (Supplementary Table 6).
Sperm classification network parameters.
MD-nets trained for the classification of sperm cells on the basis of their morphology was a binary classification model with SD4 as the source dataset and SD3, SD2 and SD1 as target datasets (Supplementary Table 5). Hyperparameters were evaluated manually. Learning rates between 0.01 and 0.00001 and batch sizes of 8, 16, 32 and 64 were evaluated when developing the best performing neural network (Supplementary Table 6). Data balancing was achieved through augmentation and stratified batch distributions. Early stoppage was set at 2,000 iterations after the lowest source validation loss (Supplementary Table 6). Reverse validation risk for each model trained was also evaluated when picking the final models used in the study.
Malaria classification network parameters.
Initially, a supervised neural network classifier was developed using the NIH dataset to replicate a previously reported neural network. Using the same architecture mentioned in their work (ResNet-50), we trained a binary classifier on MD1_s33. The images were resized to 100 × 100 px and normalized during training. Data augmentation was performed by random horizontal or vertical flipping and random 0–359° rotations of the images. Hyperparameter tuning was performed by a manual search of learning rate (0.001–0.0000001) and the best model was selected on the basis of the lowest validation loss.
For the development of MD-nets (SW), the base network was initialized with the trained weights of the supervised model (ResNet-50). The initial layers of the base network were then frozen and the whole network was retrained with MD1_s as the source, and MD3, MD2 and MD1_t as the target (Supplementary Table 5). Only the last 7 layers, which include the fully connected layer and six sets of convolution layers with batch normalization from the classifier and discriminator, were used during training by MD-nets. Hyperparameters were evaluated manually (Supplementary Table 6). Learning rates between 0.01 and 0.00001 and batch sizes of 16 and 32 were evaluated when developing the best performing neural network. Data balancing was achieved through augmentation and stratified batch distributions. Patience of 5,000 iterations was used to allow adaptation after the lowest source validation loss is achieved (Supplementary Tables 5 and 6). Similarly, MD-nets were also trained using the source and target datasets in the traditional manner for comparison (Supplementary Tables 5 and 6).
For the development of MD-nets (NoS), the target images were passed through the target feature extractor and the frozen source feature map extractor, which were both initialized with the pretrained weights of the supervised model (ResNet-50). The images were resized to 100 × 100 px and normalized during training. Hyperparameters were evaluated manually. Learning rates between 0.01 and 0.0001 and batch sizes of 8, 16 and 32 were evaluated when developing the best performing neural network, we also fine-tuned classifier trade-off λ (0–1).
MD-nets Office-31.
Office-31 is a widely used publicly available dataset for visual domain adaptation, with 4,652 images and 31 categories collected from three distinct domains: Amazon (A), Webcam (W) and DSLR (D)19. We evaluate all methods on six transfer tasks A → W, D → W, W → D, A → D, D → A and W → A. In this study, we implemented PixelDA, GAGL, MD-nets and MD-nets (NoS) models. We use all source domain data and all unlabelled target data used for training and picking the model. For all implemented models, we divide the data into 90% source training and a 10% source validation dataset (Supplementary Table 5). PixelDA, MD-net (NoS) and GAGL used a target data validation (Supplementary Table 5). MD-net models were finalized based on their performance on the source validation loss. The images were resized to 256 × 256 px and were cropped to 224 × 224 px. The images were normalized during training. Hyperparameters were evaluated manually (Supplementary Table 6). Data balancing was performed for all methods except MD-nets (NoS) on the source data through augmentation and stratified batch distributions. We used ResNet-50 as our base architecture with batch normalization, weight normalization and an additional bottleneck layer.
Specifically, for MD-nets (NoS), a supervised classifier neural network was trained on all of the datasets, Amazon (A), Webcam (W), and DSLR (D), using ResNet-50. The images were also resized and cropped to 244 × 224 px and normalized during training. Hyperparameters were evaluated manually (Supplementary Table 6).
Statistical information.
Statistical analyses and measures such as the two-tailed one-sample t-tests, diagnostic sensitivity, specificity and accuracy were performed using MedCalc (v.19.1) and GraphPad Prism (v.8.4.0). AUC values were calculated through receiver operator characteristic analyses using the Python library Scikit-learn library (v.0.23.1).
Supplementary Material
Acknowledgements
We thank the staff members of the Massachusetts General Hospital (MGH) IVF laboratory and the MGH clinical pathology laboratory for their support and assistance in data collection and annotation; the American Association of Bioanalysts Proficiency Testing Services for providing sperm image data and clinical performance information; and staff at the Massachusetts General Hospital and Brigham and Women’s Hospital Centre for Clinical Data Science (CCDS) for providing access to additional compute power. The work reported here was partially supported by the National Institutes of Health under award numbers R01AI118502, R01AI138800 and R61AI140489; the Brigham and Women’s Hospital through the Precision Medicine Development Grant; and the Mass General Brigham through Partners Innovation Discovery grant.
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
The codes and algorithms developed for this study, in particular MD-nets and its variants, are available at GitHub (https://github.com/shafieelab/Medical-Domain-Adaptive-Neural-Networks). Some custom software and scripts that are supplementary in nature and specific to some of the subsections of the study (in particular, the smartphone application for sperm annotation) are available from the corresponding author on reasonable request.
Competing interests
M.K.K., P.T., C.L.B. and H.S. have submitted patent applications (WO2019068073) and invention disclosures related to this work through Brigham and Women’s Hospital and Mass General Brigham. All other authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-021-00733-w.
Data availability
Deidentified data collected and annotated for this study are available for research use online (https://osf.io/3kc2d/). The public datasets used in this study can be accessed via information in the relevant cited publications.
References
- 1.Esteva A et al. A guide to deep learning in healthcare. Nat. Med 25, 24–29 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Topol EJ High-performance medicine: the convergence of human and artificial intelligence. Nat. Med 25, 44–56 (2019). [DOI] [PubMed] [Google Scholar]
- 3.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Morvant E Advances in Domain Adaptation Theory: Available Theoretical Results (Elsevier, 2019). [Google Scholar]
- 5.Khosravi P et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit. Med 2, 21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zech JR et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 15, e1002683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badgeley MA et al. Deep learning predicts hip fracture using confounding patient and healthcare variables. NPJ Digit. Med 2, 31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beede E et al. A human-centered evaluation of a deep learning system deployed in clinics for the detection of diabetic retinopathy. In Proc. 2020 CHI Conference on Human Factors in Computing Systems 1–12 (Association for Computing Machinery, 2020). [Google Scholar]
- 9.Hosny A & Aerts HJWL Artificial intelligence for global health. Science 366, 955–956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodfellow IJ et al. Generative adversarial networks. In Adv. Neural Inf. Process. Syst (eds Ghahramani Z et al. ) (Curran Associates, Inc., 2014). [Google Scholar]
- 11.Long M, Cao Z, Wang J & Jordan MI Conditional adversarial domain adaptation. In Adv. Neural Inf. Process. Syst (eds Bengio S et al. ) (Curran Associates, Inc., 2018). [Google Scholar]
- 12.Ganin Y et al. Domain-adversarial training of neural networks. J. Mach. Learn. Res 17, 1–35 (2016). [Google Scholar]
- 13.Kanakasabapathy MK et al. Development and evaluation of inexpensive automated deep learning-based imaging systems for embryology. Lab Chip 19, 4139–4145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bormann CL et al. Consistency and objectivity of automated embryo assessments using deep neural networks. Fertil. Steril 113, 781–787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thirumalaraju P et al. Evaluation of deep convolutional neural networks in classifying human embryo images based on their morphological quality. Heliyon 7, e06298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bormann CL et al. Performance of a deep learning based neural network in the selection of human blastocysts for implantation. eLife 9, e55301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curchoe CL & Bormann CL Artificial intelligence and machine learning for human reproduction and embryology presented at ASRM and ESHRE 2018. J. Assist. Reprod. Genet 36, 591–600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardarson T, Van Landuyt L & Jones G The blastocyst. Hum. Reprod 27, i72–i91 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Saenko K, Kulis B, Fritz M & Darrell T Adapting visual category models to new domains. In 11th European Conference on Computer Vision (eds Daniilidis K et al. ) 213–226 (Springer Berlin Heidelberg, 2010). [Google Scholar]
- 20.Tzeng E, Hoffman J, Saenko K & Darrell T Adversarial discriminative domain adaptation. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2962–2971 (IEEE, 2017). [Google Scholar]
- 21.Long M, Cao Y, Wang J & Jordan MI Learning transferable features with deep adaptation networks. In Proc. 32nd International Conference on Machine Learning (eds Francis B & David B) 97–105 (PMLR, 2015). [Google Scholar]
- 22.Bousmalis K, Silberman N, Dohan D, Erhan D & Krishnan D Unsupervised pixel-level domain adaptation with generative adversarial networks. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 95–104 (IEEE, 2017). [Google Scholar]
- 23.Wei K-Y & Hsu C-T Generative adversarial guided learning for domain adaptation. In British Machine Vision Conference 2018 100 (BMVA Press, 2018). [Google Scholar]
- 24.Kang G, Jiang L, Yang Y & Hauptmann AG Contrastive adaptation network for unsupervised domain adaptation. In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 4888–4897 (IEEE, 2019). [Google Scholar]
- 25.Wilson G & Cook DJ A survey of unsupervised deep domain adaptation. ACM Trans. Intell. Syst. Technol 11, 51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO Laboratory Manual for the Examination and Processing of Human Semen (WHO, 2010). [PubMed] [Google Scholar]
- 27.Kose M, Sokmensuer LK, Demir A, Bozdag G & Gunalp S Manual versus computer-automated semen analysis. Clin. Exp. Obstet. Gynecol 41, 662–664 (2014). [PubMed] [Google Scholar]
- 28.Mortimer ST, van der Horst G & Mortimer D The future of computer-aided sperm analysis. Asian J. Androl 17, 545–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thirumalaraju P et al. Automated sperm morpshology testing using artificial intelligence. Fertil. Steril 110, e432 (2018). [Google Scholar]
- 30.Thirumalaraju P et al. Human sperm morphology analysis using smartphone microscopy and deep learning. Fertil. Steril 112, e41 (2019). [Google Scholar]
- 31.Kanakasabapathy MK et al. An automated smartphone-based diagnostic assay for point-of-care semen analysis. Sci. Transl. Med 9, eaai7863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal A et al. Home sperm testing device versus laboratory sperm quality analyzer: comparison of motile sperm concentration. Fertil. Steril 110, 1277–1284 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Rajaraman S et al. Pre-trained convolutional neural networks as feature extractors toward improved malaria parasite detection in thin blood smear images. PeerJ 6, e4568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Malaria Report 2018 (WHO, 2018). [Google Scholar]
- 35.Parasites—Malaria (CDC, 2019); https://www.cdc.gov/parasites/malaria/index.html [Google Scholar]
- 36.Treatment of Malaria: Guidelines For Clinicians (United States) (CDC, 2020); https://www.cdc.gov/malaria/diagnosis_treatment/clinicians1.html [Google Scholar]
- 37.Guidelines for the Treatment of Malaria (WHO, 2015). [Google Scholar]
- 38.Global Technical Strategy for Malaria 2016–2030. Library Cataloguing-in-Publication Data (WHO, 2015). [Google Scholar]
- 39.Poostchi M, Silamut K, Maude RJ, Jaeger S & Thoma G Image analysis and machine learning for detecting malaria. Transl. Res 194, 36–55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G & King D Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim DW, Jang HY, Kim KW, Shin Y & Park SH Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J. Radiol 20, 405–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winkler JK et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol. 155, 1135–1141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Amour A et al. Underspecification presents challenges for credibility in modern machine learning. Preprint at https://arxiv.org/abs/2011.03395 (2020).
- 44.Kazeminia S et al. GANs for medical image analysis. Artif. Intell. Med 109, 101938 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Rivenson Y et al. Deep learning enhanced mobile-phone microscopy. ACS Photonics 5, 2354–2364 (2018). [Google Scholar]
- 46.Shin H-C et al. in Simulation and Synthesis in Medical Imaging Vol. 11037 (eds Gooya A et al. ) 1–11 (Springer, 2018). [Google Scholar]
- 47.Ghorbani A, Natarajan V, Coz D & Liu Y DermGAN: synthetic generation of clinical skin images with pathology. In Proc. Machine Learning for Health NeurIPS Workshop (eds Dalca Adrian, V. et al.) 155–170 (PMLR, 2020). [Google Scholar]
- 48.Rivenson Y et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng 3, 466–477 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Rivenson Y, Wu Y & Ozcan A Deep learning in holography and coherent imaging. Light Sci. Appl. 8, 85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belthangady C & Royer LA Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215–1225 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Sankaranarayanan S, Balaji Y, Castillo CD & Chellappa R Generate to adapt: aligning domains using generative adversarial networks. In 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 8503–8512 (IEEE, 2018). [Google Scholar]
- 52.Wood CS et al. Taking connected mobile-health diagnostics of infectious diseases to the field. Nature 566, 467–474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DPDx—Laboratory Identification of Parasites of Public Health Concern (CDC, 2020); https://www.cdc.gov/dpdx/malaria/index.html [Google Scholar]
- 54.Mirza M & Osindero S Conditional generative adversarial nets. Preprint at https://arxiv.org/abs/1411.1784 (2014).
- 55.Chollet F Xception: deep learning with depthwise separable convolutions. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 1800–1807 (IEEE, 2017). [Google Scholar]
- 56.Caron M, Bojanowski P, Joulin A & Douze M Deep clustering for unsupervised learning of visual features. In 15th European Conference on Computer Vision (eds Ferrari V et al. ) 139–156 (Springer, 2018). [Google Scholar]
- 57.Szegedy C, Vanhoucke V, Ioffe S, Shlens J & Wojna Z Rethinking the inception architecture for computer vision. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2818–2826 (IEEE, 2016). [Google Scholar]
- 58.He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (IEEE, 2016). [Google Scholar]
- 59.Szegedy C, Ioffe S, Vanhoucke V & Alemi A Inception-v4, Inception-ResNet and the impact of residual connections on learning. In Proc. Thirty-First AAAI Conference on Artificial Intelligence 4278–4284 (AAAI Press, 2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified data collected and annotated for this study are available for research use online (https://osf.io/3kc2d/). The public datasets used in this study can be accessed via information in the relevant cited publications.


