Abstract
Hepatocellular carcinoma (HCC) is the most frequent primary liver cancer, being the sixth most commonly diagnosed cancer and the fourth leading cause of cancer-related death. As other heterogeneous solid tumours, HCC results from a unique synergistic combination of genetic alterations mixed with epigenetic modifications.
In HCC the patterns and frequencies of somatic variations change depending on the nearby chromatin. On the other hand, epigenetic alterations often induce genomic instability prone to mutations. Epigenetics refers to heritable states of gene expression without alteration to the DNA sequence itself and, unlike genetic changes, the epigenetic modifications are reversible and affect gene expression more extensively than genetic changes. Thus, studies of epigenetic regulation and the involved molecular machinery are greatly contributing to the understanding of the mechanisms that underline HCC onset and heterogeneity. Moreover, this knowledge may help to identify biomarkers for HCC diagnosis and prognosis, as well as future new targets for more efficacious therapeutic approaches.
In this comprehensive review we will discuss the state-of-the-art knowledge about the epigenetic landscape in hepatocarcinogenesis, including evidence on the diagnostic and prognostic role of non-coding RNAs, modifications occurring at the chromatin level, and their role in the era of precision medicine.
Apart from other better-known risk factors that predispose to the development of HCC, characterization of the epigenetic remodelling that occurs during hepatocarcinogenesis could open the way to the identification of personalized biomarkers. It may also enable a more accurate diagnosis and stratification of patients, and the discovery of new targets for more efficient therapeutic approaches.
Keywords: HCC, Epigenetics, Methylation, Non-coding RNAs, Histone modifications, Epidrugs
Background
Hepatocellular carcinoma (HCC) is currently one of the most leading causes of death worldwide, being the sixth most commonly diagnosed cancer and the fourth leading cause of cancer-related death, with approximately 854.000 new cases in 2015 [1, 2].
HCC corresponds to 75–85% of primary liver cancers [1]. The bad incidence of HCC is attributed to its highly complex heterogeneity and to the plurality of the aetiologies. Among the well-defined risk factors for HCC development there are cirrhosis, hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, alcohol abuse and non-alcoholic steatohepatitis (NASH). Moreover, obesity, diabetes, insulin resistance, smoking, and dietary exposure to aflatoxins are relevant cofactors that contribute to the onset of HCC [3]. HCC commonly arises in patients with underlying liver disease, mostly as a result of HBV or HCV infection or alcohol abuse. However, the increase in non-alcoholic fatty liver disease (NAFLD) will soon become the major leading cause of liver cancer in Western countries [4]. Currently, more and more HCC cases, including the paediatric ones, occur in the absence of cirrhosis mainly in the context of NAFLD/NASH or chronic HBV infection [5–7].
Apart imaging examinations and biopsies, the biomarker commonly used for the screening of HCC is the circulating α-fetoprotein (AFP) that, however, has an unsatisfied diagnostic efficiency in the screening of early-stage HCC for its elevations in patients with liver benign diseases, such as cirrhosis and chronic hepatitis [8]. Therefore, the research of new and more accurate biomarkers for HCC becomes urgent, especially for an early diagnosis and to monitor the outcome of the disease after treatment [9, 10].
The HCC is highly refractory to treatments and continues to have a high recurrence rate, but the scenario of available therapies for HCC has improved during the last five years [11]. Indeed, the recent technological progresses for conventional therapeutic options, as well as the development of new promising targeted systemic therapy and immune checkpoint inhibitors have increased the life-expectancy of patients with advanced HCC [11, 12]. Despite this step forward, it has been reported an high incidence of recurrent de novo HCC tumours, which progress into incurable, advanced-stage condition for which the available medical therapies could have only marginal survival benefit and are not cost-effective [13].
The understanding why some HCCs are refractory and recur after the current treatments remains as open question. However, HCC heterogeneity, which depends on genetics, epigenetics, transcriptomics, proteomics, and metabolomics, could be the answer to this question [14–16]. Moreover, HCC consists of malignant cells embedded in a tumour microenvironment that is critical for either inhibiting or enhancing the efficiency of all the steps of carcinogenesis. Indeed, the close interaction between the malignant cells and the tumour microenvironment paves the way to epigenetic processes able to modify chromatin allowing DNA accessibility, thus modifying the gene expression. Therefore, the HCC heterogeneity could be the result of a unique synergistic combination of genetics mixed with epigenetics [17]. Epigenetics includes three main heritable regulatory systems that determine chromatin remodelling and gene transcription regulation: non-coding RNAs, DNA methylation and chromatin remodelling, and the functional crosstalk among these epigenetic processes determines cell phenotype [18].
All these lines of evidences highlight that studies of the epigenetics may give a great contribute to the comprehension of the mechanisms that underline HCC onset and progression, thus revealing new targets for future more efficacious therapeutic approaches or new biomarkers for HCC diagnosis.
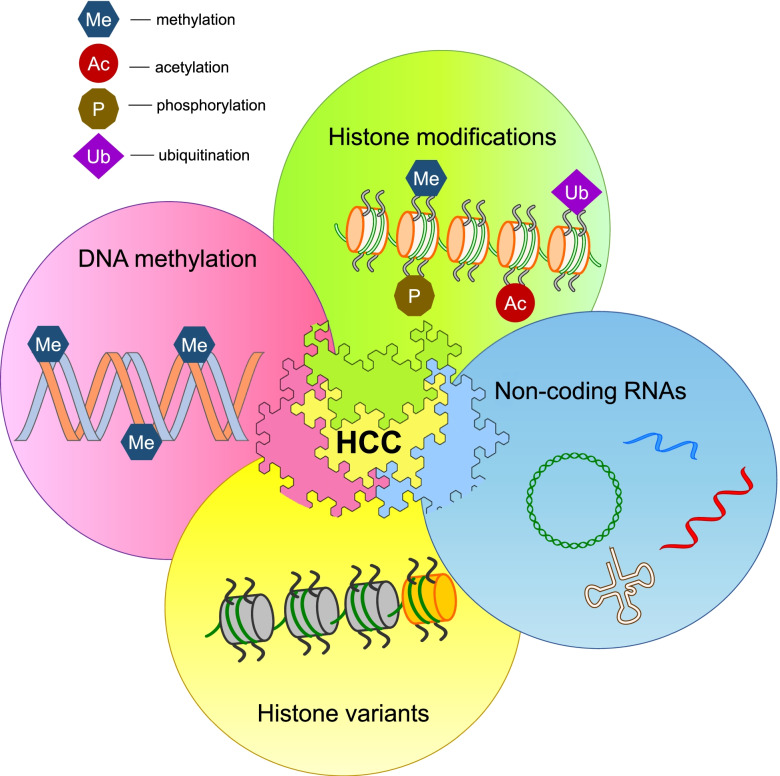
In this comprehensive review we discuss the epigenetic landscape investigated in the last decade in human HCC (Fig. 1).
Fig. 1.
The regulatory systems involved in the epigenetic landscape of human HCC. The epigenetic marks in HCC include histone tails modifications (acetylation, methylation, phosphorylation, etc.), non-coding RNAs, the presence of abnormal histone variants, and DNA promoter hypo- or hyper-methylation. The post-translational modifications of histone tails can modulate chromatin organization and alter the control of gene expression. Non-coding RNAs have emerged as regulators of chromatin structure. Non-canonical histone variants can affect chromatin remodelling and histone post-translational modifications. Alterations in the methylation of gene promoter regions cause altered gene expression
Non-coding RNAs
Non-coding RNAs (ncRNAs) comprise multiple classes of RNAs that are not translated into proteins. They are a heterogeneous group of molecules that have emerged as regulators of chromatin structure and function with crucial roles in both physiological and pathological conditions [19]. Altered expression of ncRNAs has been reported in many disease settings, as well as in many types of cancer, including HCC [20]. They are classified into two major group types based on length. The first group is represented by small/short ncRNAs (sncRNAs) with a length < 200 nucleotides, mainly represented by microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), endogenous short interfering RNAs (endo-siRNAs) and small nucleolar RNAs (snoRNAs). The second group includes ncRNAs with a length > 200 nucleotides known as long ncRNA (lncRNAs) [21]. Relatively recent is the discovery of a further class of ncRNAs called circular RNAs (circRNAs) [22].
Hepatic and circulating miRNAs and HCC
The most widely studied ncRNAs are miRNAs, approximately 22 nucleotides-long single-stranded RNAs, which mostly function as negative gene regulators at post-transcriptional level. Defects in the complex process of miRNA biogenesis and exportation may lead to a global deregulation of miRNA expression, as well as that observed in human HCC [23, 24]. miRNAs can regulate gene expression by inducing cleavage/translational repression of their complementary to the 3′-untranslated region (3ʹUTR) of mRNA targets [25]. Multiple profiling studies have highlighted that miRNA expression is substantially different between human tumour and non-tumour livers. miRNAs that normally exert tumour-suppressive effects by repressing oncogenes are recurrently down-regulated in HCC, such as miR-15a/16–1, miR-122, miR-139 miR-192, miR-199a/b-3p and miR-275 [26–34]. On the other hand, the miRNAs known as ‘oncomiRs’ promote oncogenesis by negatively regulating important tumour-suppressive protein-coding genes, and they are aberrantly increased in HCC. Among the most representative oncomiRs there are miR-21, miR-103a, miR-221, miR-222 [28–30, 35]. However, in HCC, miRNAs may target also several cellular signalling hubs involved in innate and adaptive immune cell responses and tumour microenvironment [36–39]. Interestingly, in patient-based studies several hepatic miRNAs have been associated to HCC outcome, suggesting their potential use as biomarkers. In particular, the up-regulation of DLK1-DIO3 miRNA cluster and miR-10b, and the down-regulation of miR-199a/b and miR-122 have been correlated with poor prognosis; while the up-regulation of miR-135a, and down-regulation of miR-99a have been correlated with poor survival (see the review by Gupta and Akhtar) [36].
During the last decade also several circulating miRNAs, released from cancerous tissues as free miRNAs or contained within the exosomes, have been identified and tested for their efficiency as biomarkers in HCC diagnosis and prognosis (Table 1) [40–50].
Table 1.
List of the main sets of circulating miRNAs described as modulated in HCC
| miRNAs sets | Pattern of regulation in HCC patients compared to controls | Reference |
|---|---|---|
| miR-122 | Up | [40] |
| miR-21 | Up | |
| miR-223 | Down | |
| miR-885-5p | Up | [41] |
| miR-574-3p | Up | |
| miR-224 | Up | |
| miR-215 | Up | |
| miR-146a | Up | |
| miR-15b | Up | [42] |
| miR-130b | Up | |
| miR-206 | Up | [43] |
| miR-141-3p | Up | |
| miR-433-3p | Up | |
| miR-1228-5p | Up | |
| miR-199a-5p | Down | |
| miR-122-5p | Down | |
| miR-192-5p | Down | |
| miR-26a-5p | Down | |
| miR-29a | Up | [44] |
| miR-29c | Up | |
| miR-133a | Up | |
| miR-143 | Up | |
| miR-145 | Up | |
| miR-192 | Up | |
| miR-505 | Up | |
| miR-939 | Up | [45] |
| miR-595 | Up | |
| miR-519d | Up | |
| miR-494 | Up | |
| miR-224 | Up | [46, 47] |
| miR-92-3p | Up | [48] |
| miR-107 | Up | |
| miR-3126-5p | Down | |
| miR-1972 | Up | [49] |
| miR-193a-5p | Up | |
| miR-214-3p | Up | |
| miR-365a-3p | Up | |
| miR-1246 | Up | [50] |
| miR-122 | Up | [51] |
| miR-155 | Down | |
| miR-885-5p | Up | |
| miR-221-3p | Down | [52] |
| miR-223-3p | Down | |
| miR-26a-5p | Down | |
| miR-30c-5p | Down | |
| miR-365a-3p | Up | |
| miR-423-3p | Up |
Some studies highlighted that altered circulating miRNAs may be biomarkers for liver injury but not specific for HCC [40, 41, 45, 49], and revealed that the serum levels of miR-21, miR-122 and miR-223 were found elevated in HCC but also in chronic hepatitis B patients. However, the large part of the literature identified specific circulating miRNA signatures in HCC compared with healthy or cirrhotic livers in monocentric or multicentric studies [42–44, 46–48, 50, 51, 53]. Interestingly, a systematic review highlighted that plasma levels of miR-224 in HCC patients were higher than in healthy volunteers, and were significantly correlated with larger tumour size and recurrence. The authors also highlighted that the plasma miR-224 levels could accurately identify small tumours less than 18 mm in diameter [46]. Lin et al. confirmed the high serum level of miR-224 in early-stage HCC respect to cirrhosis, chronic hepatitis B and C [47]. In addition, Yamamoto et al. [51], performed the analysis of circulating miRNAs by microarrays in serum samples from 345 patients with HCC, 46 patients with chronic hepatitis, 93 patients with liver cirrhosis, and 1.033 healthy individuals. The authors developed a detection model of HCC, including early stage, based on a set of 52 selected miRNAs that had altered expression according to disease progression status. More recently, Wong et al. [53] showed a six miRNA signature, including miR-221-3p, miR-223-3p, miR-26a-5p, miR-30c-5p, miR-365a-3p, and miR-423-3p, which were significantly differentiated in HCC compared to healthy individuals.
Finally, several authors have demonstrated the value of circulating miRNAs also as biomarkers of the follow-up after different types of treatments [52, 54, 55].
What is evident from the survey by Shen et al. [56] is that miRNA signatures in HCC are dependent on etiological factors and ethnic groups, thus explaining why miRNAs are different across the cohorts studied, and their application as biomarkers requires further investigation.
piRNAs and HCC
piRNAs include a large family of small (26–31 nucleotides), single stranded ncRNAs that interact with the PIWI subfamily of argonaute proteins to form functional RNA-protein complexes that may play a role in cancer [57, 58].
However, only a few studies have clarified the role of piRNAs in HCC (Table 2). Law et al. [59] found the up-regulation of piR-Hep1 in 46.6% of hepatic tumours when compared with the corresponding adjacent tissues. Rizzo et al. [60] investigated piRNAs expression by RNA-Seq in cirrhotic and HCC samples, and stringent analyses revealed a set of 125 piRNAs that characterize HCC from matched cirrhotic nodules, and correlating also to microvascular invasion. Moreover, the same authors showed a specific expression of 24 piRNAs in dysplastic nodules respect to cirrhotic liver and/or progressed HCC. The latest study by Koduru et al. [32] recognized 128 piRNAs in HCC (83 up-regulated and 45 down-regulated). The top five up-regulated piRNAs include piR-32,299, piR-23,670, piR-24,684, piR-28,488, and piR-7239; while, the top five down-regulated were piR-952, piR-820, piR-28,525, piR-5938, and piR-5937. The most relevant piRNAs emerged by the above mentioned studies are listed in Table 2.
Table 2.
List of the main piRNAs described as modulated in HCC
| piRNAs | Pattern of regulation in HCC patients compared to controls | Reference |
|---|---|---|
| piR-Hep1 | Up | [59] |
| hsa-piR-001078 | Up | [60] |
| hsa-piR-001207 | Up | |
| hsa-piR-001346 | Up | |
| hsa-piR-017061 | Up | |
| hsa-piR-017295 | Up | |
| hsa-piR-019420 | Up | |
| hsa-piR-020450 | Up | |
| hsa-piR-001170 | Down | |
| hsa-piR-016975 | Down | |
| hsa-piR-017724 | Down | |
| hsa-piR-019951 | Down | |
| hsa-piR-020828 | Down | |
| hsa-piR-020829 | Down | |
| piR-LLi-7 | Down | |
| piR-LLi-54 | Down | |
| piR-LLi-1977 | Down | |
| piR-LLi-13,491 | Down | |
| piR-LLi-24,259 | Down | |
| piR-LLi-28,197 | Down | |
| piR-LLi-30,517 | Down | |
| piR-LLi-30,581 | Down | |
| hsa-piR-020498 | Down | |
| piR-LLi-30,552 | Down | |
| piR-LLi-24,894 | Down | |
| hsa-piR-013306 | Down | |
| piR-32,299 | Up | [32] |
| piR-23,670 | Up | |
| piR-24,684 | Up | |
| piR-28,488 | Up | |
| piR-7239 | Up | |
| piR-952 | Down | |
| piR-820 | Down | |
| piR-28,525 | Down | |
| piR-5938 | Down | |
| piR-5937 | Down |
However, most of the studies investigate the role of genes involved in piRNAs machinery, such as Piwi and Hiwi [61–63].
snoRNAs and HCC
snoRNAs are a highly conserved subclass of sncRNAs of 60–300 bp localized into the nucleolus, where they contribute to maturation of RNA molecules through chemical modifications targeting mainly rRNAs, tRNAs, and sncRNAs [64]. In the recent years, have demonstrated that dysregulated expression of snoRNAs may play a crucial role in HCC development (Table 3).
Table 3.
List of the main snoRNAs described as modulated in HCC
Xu et al. found that over-expression of SNORD113–1 suppressed tumorigenicity through inactivation of MAPK/ERK and TGF-β pathways in vitro and in vivo HCC models [65]. A study by Ma et al. [66] found that SNORD78 was up-regulated in HCC tissues respect to adjacent non-neoplastic tissues. Furthermore, their results showed that high expression of SNORD78 was associated with aggressive tumour phenotype and poor outcome of HCC patients. In addition, loss-of-functional analyses indicated that SNORD78 silencing inhibited cell growth, invasion, and migration in vitro, suggesting that SNORD78 was associated with development and progression of HCC.
Wu et al. [67] demonstrated that the snoRNA ACA11 was higher expressed in HCC tissues than in adjacent non-tumour tissues, correlating also with shorter overall survival and higher recurrence rates. This study provides a novel evidence of a potential diagnostic role of ACA11 in HCC.
To date, new studies are discovering new snoRNAs differential expression signature in HCC tissues. Indeed, in tumours compared with non-tumour tissues, it has been found a down-regulation of SNORD113–1 associated with worse survival, and an up-regulation of SNORA47, SNORD126, SNORD78 and SNORD76 linked to the aggressive phenotype and poor prognosis of HCC [68].
In addition, more recently it has been reported that up-regulated snoU2_19 was associated with HCC progression and aggressive phenotypes [69].
Furthermore, the biological role of SNORD76 has been clarified in HCC development, demonstrating for the first time that SNORD76 can be used as a prognostic biomarker. Indeed, the authors found a significant up-regulation of SNORD76 in 66 HCC specimens compared to non-tumour tissues that associated with poor survival [70].
Recently, McMahon et al. [71] provided evidence that one snoRNA plays a direct role in specific steps of cancer development. The study revealed that SNORA24 levels, which mediates two distinct pseudouridine modifications in the small 40S subunit of the ribosome, were decreased in HCC and is implicated in a tumour suppressor program that blocks cellular transformation. Moreover, human and mouse data support a role for SNORA24 dysfunction in tumour initiation and in maintenance of RAS-driven cancers. Interestingly, in the context of HCC, SNORA24 appears to be unnecessary for protein synthesis, but it is involved in more selective functions, for example towards controlling translation of specific mRNAs.
In a recent study Mao et al. [72] revealed that the knockdown of lncRNA-LALR1 up-regulated the snoRNA SNORD72 via binding with SNORD72, and stabilized ID2 mRNA. SNORD72 was over-expressed in HCC tissues and enhanced cells proliferation, colony formation and invasion. Interestingly, a recent comprehensive analysis provided a classification of the snoRNAs correlated to this tumour [73].
lncRNAs and HCC
lncRNAs, with a length > 200 nucleotides, exert different biological functions depending on their localization into specific cellular compartments [74]. Nuclear lncRNAs can act as a guide for chromatin-modifying-complexes or transcription factors; cytoplasmatic lncRNAs often function as regulators of protein levels, either by directly controlling mRNA stability or by acting as the so-called competing endogenous RNAs, that sequester miRNAs.
A large amount of studies demonstrated that lncRNAs may play an important role in the development and progression of HCC, thus they could be promising targets for therapy, but also promising biomarkers with high accuracy and efficiency for diagnosis and prognosis [75–77]. Since the role of hepatic lncRNAs has been extensively investigated and deserves a separate discussion, here we focused only on the circulating lncRNAs that differed in patients with HCC respect to healthy subjects (Table 4) [55, 76–86].
Table 4.
Circulating lncRNAs altered in HCC
Among the circulating lncRNAs, RP11–160H22.5, LOC149086, and XLOC_014172 were found up-regulated in HCC [78]. Li et al. [79] reported that the plasma expression levels of HULC and LINC00152 may discriminate between HCC and controls. Also the lncRNA CTBP was significantly up-regulated in serum, thus it can serve as a potential biomarker for HCC diagnosis [80]. Again, LINC00152, RP11-160H22.5, and XLOC014172 were found up-regulated in the plasma of patients with HCC by Yuan et al. [81]. Sun et al. [82] demonstrated that LINC00161, up-regulated in the serum of patients with HCC, may be a novel biomarker for this type of tumour. Moreover, Habieb et al. [83] found in HCC patients increased serum levels of TSIX, a lncRNA antisense for X-inactive-specific transcript. The plasma levels of GAS5 were significantly lower in HCC patients compared to healthy subjects [84]. Furthermore, this lncRNA, as well as the exosomal H19, could be counted as a biomarker for response to treatment with sorafenib [55, 85]. Finally, recently the circulating levels of some lncRNAs have been correlated with metastasis and poor prognosis in HCC [86].
CircRNAs and HCC
circRNAs are particular non-coding RNAs formed by back-splicing events, a “head-to-tail” attachment, leading to covalently closed loop structures, thus without the presence of 5′ and 3′ ends [87, 88]. They are highly stable and can be detected in cell-free compartments such as exosomes and plasma, suggesting circRNAs as biomarkers for cancer diagnosis. High-throughput RNA sequencing technology has highlighted a large number of circRNAs, which are endogenous, abundant and stable in mammalian cells, and that may be involved in many diseases including cancer [89, 90]. Recently, many reports suggested that circRNAs had special effects on the occurrence and development of liver disease [91]. Many circRNAs are differentially expressed in HCC tissues, indicating a potential function in their process [92].
Among the most up-regulated circRNAs in HCC tissues there are CDR1as/ciRS-7 (a circular RNA sponge for antisense of microRNAs-7 or CDR1) and hsa_circ_0005075, while among the most down-regulated are included circZKSCAN1, hsa_circ_0001649, hsa_circ_0005986 and hsa_circ_0004018 [93]. CDR1 inhibits the expression of microRNAs-7, which can stop the growth of cancer cells and promote apoptosis, thereby increasing the expression of its target genes. As a result, it can promote the initiation and development of cancer [94]. The up-regulation of CDR1 in HCC tissues was observed in patients whose AFP level ≥ 400 ng/ml, suggesting CDR1 as a valuable biomarker for HCC treatment [95].
The up-regulation of hsa_circ_0005075 and the down-regulation of hsa- circ_0001649 expression were correlated with HCC tumour size [96]. Circ-0001649 served as a competing endogenous RNA (ceRNA) to sponge miR-127-5p, miR-612 and miR-4688, thus activating SHPRH, a gene ubiquitously expressed, containing motifs of some DNA repair proteins, transcriptional factors, and helicases [97]. The absence of circZKSCAN1 endowed several malignant properties, including cancer stemness, and tightly correlated with worse overall and recurrence-free survival rate in HCC [98].
More recently, it has been described a cluster of 70 circRNAs specifically expressed in HCC specimens, which included 26 circRNAs up-regulated and 44 circRNAs down-regulated. The most five up-regulated circRNAs were: circR-0021905, circR-0016456, circR-0015637, circR-0087119, and circR-0014848. On the contrary, the most five circRNAs down-regulated were: circR-0035409, circR-0035407, circR 0076872, circR-0092360, and circR-0069970 [32]. Noteworthy, two very recent studies highlighted the role of circ-LRIG3 and circSOD2 as interactor/regulators of other epigenetic circuits involved in the development and progression of HCC [99, 100].
Chromatin remodelling
The chromatin organization as a dynamic structure consisting of DNA and histone proteins was described for the first time by Kornberg and Thomas [101]. The basic units of chromatin are represented by nucleosomes, histone octamer cores around which DNA is wrapped, and consisting of two copies each of histones H2A, H2B, H3, and H4 [102]. The formation of specific nucleosome arrays along the genome confers to local regions different structural and functional chromatin states, such as euchromatin (transcriptionally active open form) or heterochromatin (transcriptionally silent closed form) [103]. These chromatin conformations are essentially controlled by chemical modifications on DNA (i.e. DNA methylation) and histones that partly define the epigenome. N-terminal tails of histone proteins, protruding out of the nucleosome, are subjected to a variety of post-translational modifications (PTMs), for example methylation and acetylation of arginine and lysine residues, phosphorylation of serine and threonine residues, ubiquitination and sumoylation [104]. All PTMs are catalysed by different families of enzymes (e.g. acetyltransferases, deacetylases, methyl-transferases and demethylases, etc.) that, according to their effect on gene transcription, are classified as activating or repressing enzymes [105].
DNA methylation and HCC
Methylation is a heritable enzyme-mediated chemical modification that occurs on the 5 position of the Cytosine ring and is mediated by DNA methyltransferases (DNMTs) [106]. DNA methylation mainly affects the Cytosine base (C) when is followed by a Guanine (G), the so-called CpG sites. In human DNA, 70–80% of CpG sites are methylated; however, methylation is mostly found in areas where CpG density is low, or at repetitive DNA sites. On the contrary, CpG islands (CpGIs) are completely unmethylated in healthy individuals, and represent approximately 1 to 2% of human genome [107]. In general, methylation in the gene promoter region causes transcriptional repression, while methylation in the gene body promotes gene expression, as reported in most cancers including HCC [108]. Among the DNMTs, DNMT1 is essential for the maintenance of methylation pattern in the genome, by regulating gene expression but also other epigenetic regulators (e.g. miRNAs) in HCC [109–111].
Defects in DNA methylation and its machinery are closely associated with cancer, including HCC [112]. Three types of alterations in DNA methylation occur in cancer: hypermethylation of the CpGIs in the promoter regions of tumour suppressor genes, aberrant expression of DNMTs, and global hypomethylation of genes and repetitive sequences leading to genomic instability and oncogene activation [113].
In the last decade, numerous studies highlighted different methylome patterns in HCC, compared to normal tissues through genome-wide DNA methylation profiling. The majority of these studies in HCC focused on finding significantly hypermethylated genes, and among these CDKN2A, RASSF1, APC and SMAD6 emerged [16, 107, 108, 113–125]; while fewer studies described hypomethylated genes, such as IGF2, CCL20, NQO1 [107, 118, 120, 121]. The main hyper- or hypomethylated genes in HCC are listed in Table 5.
Table 5.
The most relevant hyper- and hypomethylated genes in HCC
| Genes | DNA Methylation pattern in HCC |
Reference |
|---|---|---|
| HHIP, PTGR1, TMEM106A, MT1M, MT1E, CPS1, CDKN2A | Hyper | [16] |
| CDKN2A, MLH1, FZD7, SOCS1, APC, RASSF1 | Hyper | [107] |
| HIST1H2AJ, SPDYA | Hyper | [108] |
| HRNBP3 | Hypo | |
| CDKN2A | Hyper | [113] |
| FZD7, CDKN2A, APC, RASSF1 | Hyper | [114] |
| SOCS1, RASSF1 | Hyper | [115] |
| CHFR, MGMT, RASSF1, GSTP1 | Hyper | [116] |
| CDKN2A, BMP4, NFATC1, GSTP1 | Hyper | [117] |
| APC, RASSF1 | Hyper | [118] |
| IGF2 | Hypo | |
| SOCS1, APC, GSTP1 | Hyper | [119] |
| SMAD6, IFITM1, LRRC4, CHST4, TBX15 | Hyper | [120] |
| CCL20, NQO1 | Hypo | |
| p16, CDH1, p15, RUNX3, SFRP1, PRDM2, p14, RARβ, DAPK1, SOCS1, APC, RASSF1, MGMT, GSTP1 | Hyper | [121] |
| OCIAD2 | Hyper | [122] |
| RASSF1A | Hyper | [123] |
| CDCA1, CDCA3, CDCA4, CDCA5, CDCA6, CDCA8 | Hypo | [124] |
| CDCA2, CDCA7 | Hyper | |
| MTFR2 | Hyper | [125] |
Nowadays, the new generation of sequencing platforms allows the preparation of genomic maps of DNA methylation, permitting the identification of DNA methylation patterns highly rearranged in HCC. Indeed, Meunier et al. identified various processes remodelling HCC methylomes and revealed the epigenetic and transcriptional impact of different alterations [126].
Histone modifications and their specific impact on HCC
As said, DNA is organised into chromatin, composed by nucleosomes wrapped around a histone octamer (Fig. 2). The tails of the histones can be modified and these PTMs can modulate chromatin compaction and control the recruitment of remodelling complexes and transcription factors. This is achieved through the recognition of histone PTMs by proteins harbouring specialized domains that recognize specific covalent modifications, the so-called epigenetic readers. Alterations in histone modifying enzymes cause the deregulation of histone marks, and consequently the control of chromatin-associated processes, which further contribute to malignant cancer hallmarks [18].
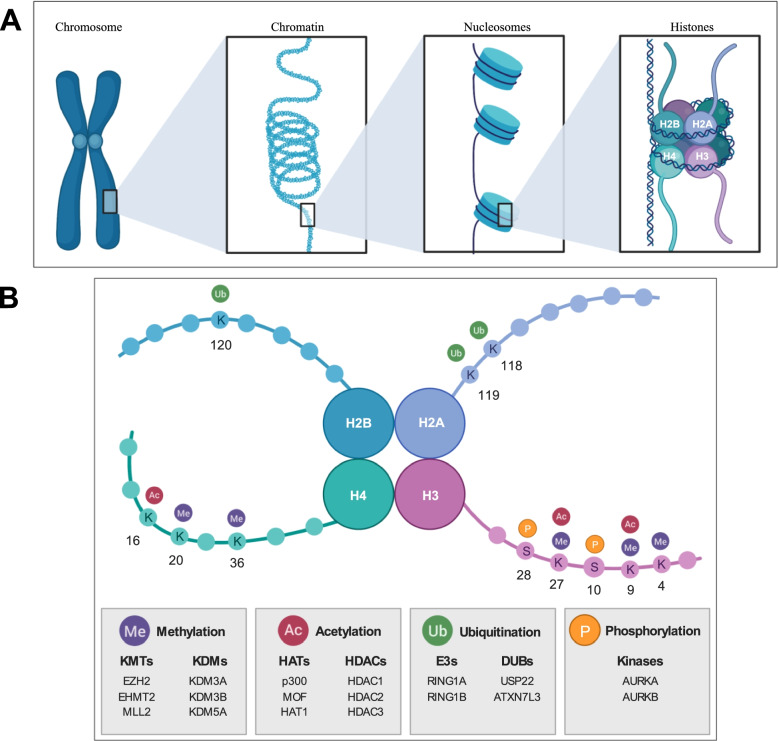
Fig. 2.
Histone modifications in HCC. A Schematic representation of DNA-histone protein complexes. Human chromosomal DNA is made by looped chromatin fibers, tightly coiled around nucleosomes. Each nucleosome is composed of DNA and eight histone proteins (H2A, H2B, H3 and H4 dimers). B Schematic representation of the nucleosome. The main PTMs (methylation, acetylation, ubiquitination and phosphorylation) and their localization on the tails of the 4 main histones are reported. Enzymes involved in the mentioned modifications are listed in gray boxes. KMTs: Lysine Methyltransferases; KDMs: Lysine Demethylases; HATs: Histone Acetyltransferases; HDACs: Histone Deacetylases; E3s: E3 Ubiquitin Ligases; DUBs: Deubiquitinating enzymes. Created with BioRender.com (https://biorender.com7), accessed 10 Feb. 2022
Histone methylation
Methylation of lysines on histones is controlled by various methyltransferases and demethylases and is an important component of epigenetic histone modifications. Abnormal regulation of such enzymes is frequently found in HCC, thus comporting an altered regulation of gene expression. In fact, a recent study identified 11 methyltransferases and demethylases, such as EZH2, EHMT2, SETDB1 and SETD2 associated with the clinical characteristics of tissues from HCC patients respect to normal liver tissues, confirming the fundamental role of histone methylation regulation [127].
The major sites of histones methylation are lysine 4, 9 and 27 of histone H3. Generally, histone 3 lysine 4 methylation (H3K4me) is associated with gene activation, while histone 3 lysine 9 methylation (H3K9me) and histone 3 lysine 27 methylation (H3K27me) result in gene repression [128].
H3K4 can be mono-, di- or tri-methylated by different enzymes. SET7 is the only methyltransferase which can specifically monomethylate H3K4 and plays critical roles in various diseases, including breast cancer, HCV, atherosclerotic vascular disease, diabetes, prostate cancer and HCC [129].
H3K9 methylation is another epigenetic marker commonly associated with the pathogenesis of HCC, where the global levels of H3K9me2 and H3K9me3 are commonly found higher in cancer tissues than in normal liver tissues, and both H3K9me2/3 are positively correlated with the degree of tumour differentiation and with poor prognosis [130].
Regarding H3K27 methylation, this modification plays a crucial role in repressing gene transcription during developmental stage and cellular differentiation, and is mediated by polycomb group proteins, that consequently have a key role in oncogenesis of many types of cancer, including HCC [131]. The polycomb proteins are assembled into polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2) to generate two different histone modifications: monoubiquitination of lysines 118 and 119 in histone H2A (H2AK118ub and H2AK119ub) and H3K27me1/2/3, respectively. The PRC2 complex, responsible for H3K27 methylation, is composed by enhancer of zeste homolog 1 and 2 (EZH1/2), suppressor of zeste 12 (SUZ12), the embryonic ectoderm development (EED) and retinoblastoma-binding protein 4/7 (RBBP4/7) [132]. Among these proteins, EZH2 is the most studied. In line with this, Hung et al. [133] showed an up-regulation of different histone modifications markers in 50 paired tumour tissues respect to adjacent non-cancerous tissues and, among others, EZH2 was associated with tumour stage and survival. EZH2-mediated H3K27me3 represents a major oncogenic chromatin modification, but is not well known how it modulates the signalling pathways. A recent study showed that sorafenib-induced AKT inhibition in HCC resulted in an increase of H3K27me3 levels through a decrease of EZH2 phosphorylation, generating a mechanism of sorafenib-resistance in HCC. This study suggests that a combined treatment of sorafenib plus EZH2 inhibitors may represent a novel therapeutic approach in HCC [134]. We observed that in human HCC the up-regulation of EZH2 was correlated with increased levels of focal adhesion kinase (FAK) and H3K27me3. This study evidenced a new molecular nexus between EZH2 and FAK that may control HCC growth [135, 136], and that has been correlated with poor prognosis in paediatric HCC [137, 138].
Other examples of altered histone H4 lysine methylation in HCC involve lysines 20 and 36. A recent study demonstrated that the development of NASH-related HCC is characterized by a global loss of histone H4 lysine 20 trimethylation (H4K20me3), and global and gene-specific deacetylation of histone H4 lysine 16 (H4K16). As a consequence, altered gene expression resulted in inhibited cell death [139].
Histone acetylation
Among the epigenetic changes that regulate gene expression, histone acetylation is one of the most significant and has taken clinical diagnostic significance in various types of cancer, including HCC [140]. Histone acetylation is controlled by two groups of enzymes, histone acetyltransferases (HATs) and histone deacetylases (HDACs), working in an accurate and regulated manner to open the chromatin and transcriptionally activate the expression of determined genes [141].
p300, the HAT most studied, was found to mediate H3K9 acetylation (H3K9ac) after phosphorylation elicited by EGF in HCC cells. This signalling pathway was correlated with high expression of High-mobility group protein A2 (HMGA2), frequently up-regulated in HCC and associated with metastasis and poor survival, and the knockdown of p300 reversed the EGF-induced HMGA2 expression and histone H3K9ac [142]. Another HAT, called MOF, plays critical roles in the transcription activation by acetylating histone H4K16. MOF was found down-regulated in HCC, and this down-regulation correlated with poor overall survival. Moreover, MOF knockdown promoted HCC growth, whilst MOF over-expression reduced it, suggesting that MOF participates to HCC progression [143]. The role of MOF has also been studied in association with the process of vascular invasion in HCC. In fact, H4K16ac was identified as a biomarker of microvascular invasion in HCC, and a study showed that MOF down-regulation caused a decrease of expression in genes and pathways involved in vascular invasion in HCC cells. Furthermore, MOF down-regulation significantly reduced tumour cell intravasation and metastasis [144]. Among the HATs, also the expression of HAT1 was up-regulated in HCC specimens respect to normal liver tissues, and its protein level was increased with tumour stage [145].
Interestingly, the H3K27ac serves in part to counteract PRC2 action, since acetylation precludes methylation by PRC2 at this site. Accordingly, we recently showed in HCC that a combined therapy with a FAK inhibitor plus sorafenib caused a decrease of the global levels of H3K27me3, of the protein levels of HDAC1 and 2, and consequently H3K27ac increased [146].
Because of the fundamental role of histone acetylation, inhibitors targeting HDACs and HATs have been developed and are used in different preclinical and clinical studies for cancer therapy, including HCC, where several evidences suggest that epigenetic therapies could be used for its treatment, and numerous studies employed inhibitors with promising results against tumour growth [147].
Histone ubiquitination
As previously mentioned, H2AK118ub and H2AK119ub are modifications caused by PRC1. As for PRC2, the catalytic activity of PRC1 is associated with transcription repression, especially the formation of H2AK119ub [148].
A recent study suggested that Hepatitis C virus infection, which is cause of liver pathologies including HCC, impairs H2AK119ub in the homeobox (HOX) gene promoter by destabilizing RNF2, a PRC1 component, and thus inducing HOX genes expression. These findings reveal a novel mechanism of Hepatitis C virus-related histone modification that may contribute to HCC induction [149].
Another epigenetic marker involved in HCC progression is the Ubiquitin-specific protease 22 (USP22), responsible of the deubiquitination of both histones H2A and H2B. Histone deubiquitination is often linked to transcriptional activation, epigenetic regulation and cancer progression. Tang et al. showed that the expression level of USP22 was significantly higher in HCC than in normal liver tissues, and it was significantly correlated with clinical stage, tumour grade and shortened survival time [150]. Moreover, different studies demonstrated that USP22 levels were responsible for the altered HCC drug-resistant phenotype [151–153]. Furthermore, it has been shown that the H2B deubiquitination by ATXN7L3 protein enhanced SMAD7 expression, which acts to inhibit tumour growth in HCC [154].
Histone phosphorylation
Like histone acetylation, histone phosphorylation has been associated with active gene transcription, but it has not been well characterized so far. Aurora kinases are responsible for the phosphorylation of most of serine/threonine residues of the histone H3. Aurora A (centrosome-associated) is frequently over-expressed in HCC patients, correlating with high grade tumours. It has been shown that it is able to reduce radiotherapy-induced apoptosis by activating NF-κB signalling, thereby contributing to HCC radioresistance [155]. Aurora B (chromosomal passenger protein) directly phosphorylates histone H3 at Ser10 (pH3S10) and Ser28 (pH3S28), thus contributing to chromosome instability and mitotic chromosome condensation. Aurora B is considered an independent molecular marker, predicting tumour growth and invasion of HCCs [156]. Liu et al. observed that Aurora A and B tended to be parallelly over-expressed, and their concurrent over-expression associated with the worst prognosis, being an independent predictor for the survival [157]. It has been demonstrated that the inhibition of Aurora B through the use of butein, that has showed anti-tumour activities in different cancers, induced arrest of the cell cycle in G2/M phase and apoptosis in a HCC preclinical study [158].
Notably, pH3S10 was found in 70.6% of HCCs and, together with pH3S28, was up-regulated in aflatoxin B1 (AFB1)-transformed hepatocytes, highlighting a key role of persistent pH3S10 or pH3S28 in chemical carcinogenesis. In fact, the down-regulation of pH3S10 was able to confer resistance to AFB1-induced cell transformation through regulating the transcription of DNA damage response genes [159].
Finally, phosphorylation plays a key role also in regulating other histones. As an example, Li et al. [160] described a novel axis in HCC composed by the Metastasis-associated 1 (MTA1) protein, functioning as an oncogene and promoting HCC progression by regulating the DNA protein kinase proteasomal degradation, and consequent inhibition of phosphorylation of the histone cluster 1 H1 family member c (H1.2) at T146 [160].
Others histone post-transcriptional modifications
Protein glycosylation with O-linked N-acetylglucosamine (O-GlcNAc) is a PTM highly sensitive to changes in the cellular environment that interacts with other PTMs, playing an essential role. O-GlcNAc is a reversible PTM of serine and threonine residues through a process controlled by O-GlcNActransferases (OGTs) and O-GlcNAcases (OGAs) enzymes, which add or remove the O-GlcNAc to the free hydroxyl group of serine or threonine residues [161].
OGT interacts with various other proteins involved in histone modifications and DNA methylation. Studies have indicated that the O-GlcNAc and OGT are highly expressed in liver cancer, and are associated with HCC progression [162], but literature is lacking about histone glycosylation in HCC.
A study in HCC cells identified the histones H2 and H3 among 110 proteins that showed significant glycosylation, detected through mass spectrometry technology. Histone glycosylation sites were H3K115 and H2BK108, which can also undergo other modifications. However, the interrelationship among glycosylation and other PTMs, and the biological effects of this mark are currently unknown [163].
Histone lysine crotonylation is a relatively new PTM identified by Tan et al. a decade ago [164]. Histone crotonylation, like acetylation, can affect chromatin structure and is enriched on active gene promoters. Its function in cancer is not well known but its levels have been found down-regulated in liver, stomach and kidney carcinomas; and up-regulated in thyroid, oesophagus, colon, pancreas and lung carcinomas. Moreover, in HCC histone lysine crotonylation levels are correlated with metastasis and are increased following HDACs depletion [165]. These results encourage investigations to better understand the biological role of this recently discovered histone modification.
Citrullination is a PTM catalysed by the peptidyl arginine deiminase (PAD) enzyme family that is stimulated by an increased calcium concentration and that can occur in many proteins, such as vimentin, keratin, and some histones like H1, H2A, H3 and H4. PAD prevents histone arginine methylation by directly citrullinating arginine or converting a monomethyl arginine to citrulline, suggesting that histone citrullination antagonizes arginine methylation, thus promoting chromatin decondensation [166].
In hepatitis B virus-associated HCC tissues, a study detected a correlation between the citrullination of histone H3 (H3cit) and Beclin1, an autophagy regulator. The average levels of H3cit and Beclin1 mRNA in HCC were higher than those in non-tumour tissues, and were significantly correlated with vascular invasion and serum AFP levels, suggesting that H3cit could be related to the increase of Beclin1 expression [167]. Another very recent study showed that in HCC histone citrullination by PADI4 may also stabilize the binding of hypoxia-inducible factors to their response elements by increasing access to DNA or by altering interactions with histones [168].
Finally, SUMOylation is a PTM that involves the covalent, but reversible addition of a small ubiquitin-like modifier (SUMO) group to proteins. Due to its capacity to regulate protein activity, this modification has been shown to have profound effects on carcinogenesis and multidrug resistance in HCC [169]. SUMO-specific proteases (SENPs, from 1 to 7) process SUMO precursors to mature forms and also remove SUMO from its targets [170]. In the human genome three functional SUMO isoforms (SUMO-1, SUMO-2, and SUMO-3) are encoded [171]. SUMO-1 was found to be highly expressed in HCC cell lines and HCC specimens respect to non-neoplastic liver tissues [172], and SUMO-2 expression is strongly correlated with patient survival rate [169].
The cellular targets of SUMO proteins favouring HCC progression are multifaceted. SUMO-1 can modify the activity of methyltransferase-like 3, promoting HCC progression via regulating Snail (an epithelial to mesenchymal transition effector) mRNA homeostasis [173]. SUMO-2 regulates liver kinase B1 (LKB1) and exportin-5 (XPO-5) oncogenic activity in HCC [174, 175]. Moreover, SUMOylation appears to play a key role in HCC multidrug resistance. Quin et al. [176] observed that SUMO-1 was significantly increased in 20 HCC compared with matched adjacent non-neoplastic controls. Moreover, the authors demonstrated that multidrug-resistant HCC cell lines showed a markedly high level of SUMO-1-conjugated proteins, associated with an up regulation of SENP1. High level of SENP1 involved HGF/c-Met signals [177]. Silencing of SENP1 in HCC cells reduces the HGF-induced proliferation, invasion and migration, sustaining cell apoptosis and growth arrest. Besides, protein depletion altered the epithelial-to-mesenchymal transition process, with increase of E-cadherin expression, decrease of fibronectin and N-cadherin expression. Moreover, SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation [178, 179]. Recently, SENP5 has been recognized as crucial factor in promoting tumorigenesis process in HCC via regulating DNA damage response [180]. The mRNA and protein levels of SENP5 were over-expressed in 10 pairs of HCC samples and demonstrated that SENP5 was required for the proliferation of HCC cells and tumour growth in vivo.
Histone variants
Histone variants are non-canonical (non-allelic) variants of histones, presenting one or a few amino acid differences – with specific expression, localization and species-distribution patterns, that affect chromatin remodelling and histone PTMs [181]. Histone H2A is the canonical histone displaying the highest number of variants: H2A.X, H2A.Z.1, H2A.J, H2A.Z.2.1, H2A.Z.2.2, H2A.Bbd, macroH2A1.1, macroH2A1.2 and macroH2A2 [182]. Hereby, we will summarize selected literature on H2A.Z, macroH2A1 and H2A.X, and HCC.
H2A.Z is one of the histones H2A variants that has 60% similarity with canonical histone H2A in mammalian cells, existing in vertebrates as two distinct genes: H2A.Z.1, encoded by H2AFZ, and H2A.Z.2 subtypes, encoded by H2AFV [183]. Interestingly, in vitro studies demonstrated a potential oncogenic role for H2A.Z isoforms in hepatocarcinogenesis [184–186].
In a large cohort of HCC patients and in HCC cell lines (Hep3B, HepG2, Huh7, PLC/PRF/5, SK-Hep1, SNU182, SNU-368, SNU-449 and SNU-475), H2A.Z.1 was significantly up-regulated, and this correlated with poor prognosis. H2A.Z.1 knockdown suppressed HCC cell growth by transcriptional deregulation of cell cycle proteins, and reduced the metastatic potential of HCC cells by selectively regulating cell cycle components. H2A.Z.1 also promoted proliferation by selectively modulating tumour microenvironment regulatory proteins such as E-cadherin and fibronectin that were elevated [184].
Histone variant macroH2A1 contains a domain that shows 66% homology with histone H2A, which is conserved across various functionally unrelated proteins throughout the animal kingdom, vertebrates and some invertebrates [182]. This variant stands out because of its unique structure, whereby a C-terminal linker connects the histone fold domain to a macrodomain. This macro domain protrudes from the compact nucleosome structure and likely affects the function and organization of the surrounding chromatin. Once in the nucleosome, macroH2A1 may contribute to different cellular processes, including cell cycle regulation, stem cell differentiation, and DNA repair and transcription in somatic and cancer cells [187–191]. MacroH2A1 exists as two alternatively exon-spliced isoforms, macroH2A1.1 and macroH2A1.2. There is also an unrelated variant, called macroH2A2, which is encoded by an independent gene. The protein levels of macroH2A1 splice variants were significantly up regulated in the livers of rodent and human HCC [192]. In particular, in response to the treatment with chemotherapeutic and DNA-demethylating agent 5-aza-deoxycytidine, macroH2A1 expression prevented the acquisition of senescent-like phenotype in HCC cell lines, through induction of global DNA hypomethylation. On the other hand, it has been shown that macroH2A1 knocked down cell lines acquire cancer stem cells features: enhancement of the tumorigenic potential, slow proliferation, resistance to chemotherapy treatments, increasing in mRNA expression of reprogramming genes, and increased glycolysis, reflecting the same phenotype of human undifferentiated and aggressive HCCs, which express a low level of macroH2A1 [193]. Moreover, macroH2A1 was able to rewires carbohydrate and lipid metabolism of HCC cells towards cancer stem cells with increased lipid accumulation through activation of the LXR pathway [194]. Finally, macroH2A1 depleted HCC cells display specific lipidomic and paracrine signatures, which may allow them to escape recognition by the adaptive immune system [192, 194]. Collectively these findings highlighted the critical role of macroH2A1 in HCC diagnosis and prognosis [188, 192, 195].
Phosphorylation of the Ser139 residue of the histone variant H2A.X, forming γ-H2A.X, is an early cellular response to the induction of DNA double-strand breaks. Detection of this phosphorylation event has emerged as a highly specific and sensitive molecular marker for monitoring DNA damage initiation and resolution. Several authors detected an increase of phosphorylated histone variant γ-H2A.X in HCC tissue, highlighting the possibility of use γ-H2A.X also as potential indicator for HCC surveillance [196, 197]. By monitoring the expression levels of γ-H2A.X in HCC tissue samples, obtained from 58 patients (7 cases with hepatitis B virus-positive, 35 with hepatitis C virus-positive, and 16 with unknown aetiology) that underwent hepatic resection, the authors discovered a major enrichment of γ-H2A.X in dysplastic nodules and in non-tumours tissues of HCC as compared with liver cirrhosis without HCC. This accumulation in the pre-neoplastic lesions of HCC supported the idea of an inverse correlation between histological grades of HCC and the level of γ-H2A.X that might play a critical role especially during the early stages of carcinogenesis. It seems that γ-H2A.X could act in promoting HCC angiogenesis via modulation of γ-H2A.X/EGFR/HIF1α/VEGF signalling [198], suggesting a possible combination of γ-H2A.X/EGFR/HIF-1α as a novel marker in the prognosis of HCC and as potential therapeutic target.
Perspective on application in therapy
At variance with genetic mutations, epigenetic mechanisms such as DNA and histones PTMs, are highly flexible and dynamic. As discussed above, these processes involve reversible enzymatic reactions and specific protein-protein interactions, which make them susceptible to pharmacological manipulation. Indeed, an increasing number of small molecules are being developed to inhibit the deposition of chromatin marks, the elimination of these marks and the interactions of chromatin modifiers and transcriptional regulators with these marks. These new families of compounds are collectively named “epidrugs”, and are showing a great potential for cancer treatment [199, 200]. Interestingly, numerous compounds originally designed for other indications, and that failed to prosper towards their original clinical applications, are being repurposed as promising epidrugs for cancer therapy [201]. Inhibitors of DNMTs and HDACs were among the first epidrugs approved by the Food and Drug Administration for the treatment of haematological malignancies [202, 203]. The anti-tumour activity of these compounds may take place at different levels, including the reprogramming of cancer stemness, the inhibition of tumour initiation and progression, the restoration of cancer cells sensitivity to targeted and conventional chemotherapeutics, and the interaction with the tumour microenvironment [199, 204–207].
DNA methylation inhibitors
The effects of DNMT inhibitors in HCC in vitro and in vivo models have been studied (Fig. 3). Treatment with first-generation DNMT inhibitors such as 5-azacytidine (5-aza) and 5-aza-2′-deoxycytidine (decitabine) induced the expression of tumour suppressor genes (TSGs), partially recovered HCC cells differentiation, reduced tumorigenicity and enhanced the sensitivity to sorafenib [208, 209]. More advanced DNMT inhibitors with improved pharmacokinetic properties such as guadecitabine (SGI-110) have been developed. Guadecitabine was also shown to induce TSG expression, and to down-regulate the expression of pro-tumorigenic genes through the demethylation of gene body regions [210]. Guadecitabine was clinically tested as a single agent on patients with advanced HCC who failed prior treatment with sorafenib. No disease control was observed, and dose had to be reduced due to toxicity (Table 6). Besides increasing the efficacy of chemotherapy, DNMT inhibitors (DNMTi) may also leverage the efficacy of immunotherapy [211]. This has been experimentally shown for guadecitabine, and ongoing clinical trials are evaluating its effects in combination with immune checkpoints inhibitors (ICI) such as durvalumab in digestive tumours including HCC (Table 6) [212].
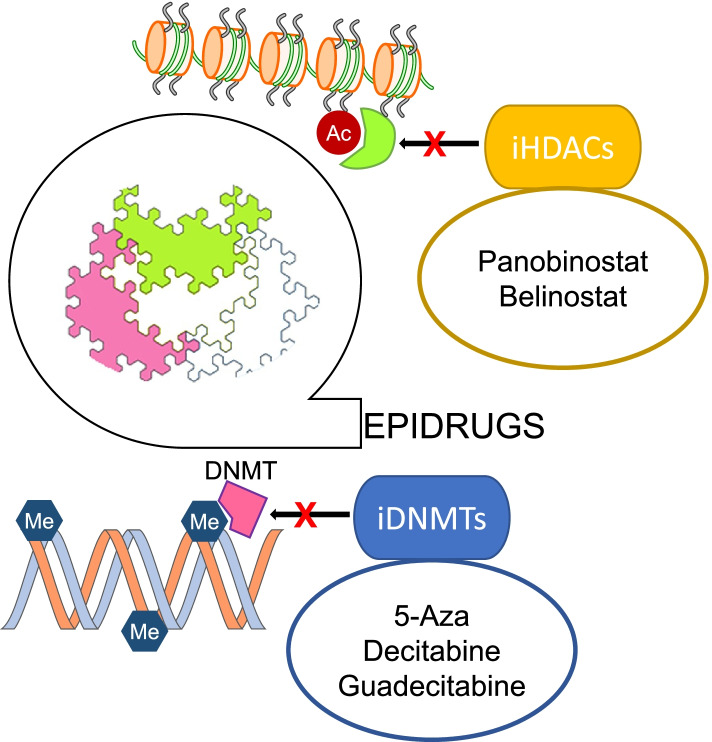
Fig. 3.
Epigenetic drugs in HCC. Schematic representation of the mechanisms of action of epigenetic drugs tested for HCC therapy
Table 6.
Epigenetics drugs in trial for HCC
| DRUGS | Phase | Reference |
|---|---|---|
| DNMTi | ||
| Guadecitabine (SGI-110) after sorafenib | Phase II | NCT01752933 |
| Guadecitabine (SGI-110) + durvalumab | Phase Ib | NCT03257761 |
| HDACi | ||
| Belinostat (PDX-101) | Phase I/II | NCT00321594 |
| Tefinostat | Phase I/II | NCT02759601 |
| Vorinostat + FOLFIRI | Phase I | NCT00537121 |
| Panobinostat + sorafenib | Phase I | NCT00823290 |
| Panobinostat + sorafenib | Phase I | NCT00873002 |
| Resminostat + sorafenib | Phase II | NCT00943449 |
| Resminostat + sorafenib | Phase I/II | NCT02400788 |
| Vorinostat + sorafenib | Phase I | NCT01075113 |
HDAC inhibitors
The efficacy of HDAC inhibitors (HDACi) has also been tested in experimental HCC (Fig. 3). Non-selective HDACi like panobinostat reduced HCC cells proliferation, reprogrammed cancer cells metabolism and reduced tumour angiogenesis [213]. Another pan-HDACi, belinostat, also demonstrated preclinical efficacy, and some activity when tested as second line treatment in HCC patients with unresectable tumours (Table 6) [214]. Interestingly, combination of belinostat with ICI resulted in enhanced efficacy of immunotherapy in an experimental model of HCC [215]. Tefinostat, another HDACi, has been also tested as single agent in HCC patients in a dose escalation trial but no results are still available (Table 6). Vorinostat, in combination with irinotecan, fluorouracil and leucovorin (FOLFIRI), was also evaluated in patients with digestive cancers and HCC (Table 6). The HDACi panobinostat has been clinically evaluated in combination with sorafenib in a limited number of patients with advanced HCC, but no results are available (Table 6). The combination of the HDACi resminostat with sorafenib was evaluated as second-line therapy in patients with advanced HCC. This combination was safe and showed early signs of efficacy [216]. A subsequent Asian study compared first-line therapy with resminostat plus sorafenib versus sorafenib monotherapy [217]. This approach did not achieve any improvement over sorafenib monotherapy, however some benefit was appreciated in a subgroup of patients including those with hepatitis B virus-related HCC (Table 6). The combination of vorinostat with sorafenib has also been evaluated. Although some patients had durable disease control, the inclusion of vorinostat led to toxicity in a majority of patients [218] (Table 6).
Other epigenetic drugs
The development of other epidrugs, such as inhibitors of histones methyltransferases, as cancer therapies is also actively pursued [219]. For instance, GSK126, an EZH2 inhibitor already used in trial against different types of lymphomas, has been shown to enhance natural killer cell-mediated HCC cell death [220]. Targeting of other histone methyltransferases like G9a (EHMT2) has also shown preclinical efficacy in HCC models [221]. As can be inferred from all the information summarized along this review, epigenetic mechanisms are quite complex and very often work in concert in the normal and also pathological regulation of gene expression. These notions have led to the development of new types of molecules combining dual inhibitory activities acting on different epigenetic targets. In the context of hepatocarcinogenesis, one recent example of this new family of compounds is CM272, a dual inhibitor of DNMT1 and G9a, enzymes that co-ordinately mediates the silencing of TSG and promote cancer growth [222]. Combined inhibition of DNMT1 and G9a by CM272 showed anti-tumour activity in experimental HCC, and also has a strong effect on the fibrogenic reaction, which is closely linked to HCC development and progression [223, 224]. Pharmacological targeting of epigenetic readers is also attracting much attention [225]. The expression of the acetylated H3 and H4 reader BRD4 is enhanced in HCC [225, 226], and treatment with the BRD4 inhibitor JQ-1 reduced HCC cell growth and survival [227, 228]. Importantly, targeting BRD4 also enhanced ICI efficacy in experimental HCC [229]. Taken together, these findings support the potential of epigenetic strategies for the treatment of HCC. Nevertheless, the efficacy and safety of these approaches cannot be properly established until our knowledge of the fundamental intricacies of epigenetic regulation is not further advanced. The evaluation of safer and more efficacious combination regimens with conventional therapies, and ICI-based therapies, need also to be carefully addressed.
Conclusion
As described in the present review, epigenetic marks can be either indicators of the presence of disease and/or specific biomarkers for disease stage and prognosis. Also, technical approaches and results from epigenetic studies performed in adult HCC may be used to explore the most common aberrant molecular pathways, to stratify tumour subtypes, and to guide research efforts in paediatric liver cancer as well, as recently reported [230–232].
Most preclinical studies demonstrated that epigenetic remodelling is potentially reversible by drugs. The list of available epigenetic modifiers and inhibitors is progressively growing. Therefore, in the era of precision medicine the unravelling of the epigenetic modifications occurring in HCC may provide new tips for the screening of potential therapeutic targets and the study of personalized interventions strategies for the management of this tumour. When considering combination therapies with conventional chemotherapeutics, other targeted agents, or ICIs, the optimization of scheduling strategies is a very relevant issue. In order to maximize efficacy, potentiate synergisms and overcome resistances, the simultaneous, sequential or alternate administration of epigenetic drugs needs to be taken into account [233]. As mentioned above, toxicity has been observed in the few clinical trials performed on HCC patients. Therefore, adverse events and dosage must be carefully monitored, particularly in patients with an underlying liver disease like most HCC patients. Finally, the identification of predictive biomarkers of response is also a challenge, particularly when mutations in epigenetic targets are not frequent in HCC. Further molecular analyses of samples from patients enrolled in clinical trials may facilitate biomarker discovery.
Acknowledgments
Not applicable.
Abbreviations
- HCC
Hepatocellular carcinoma
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- NASH
non-alcoholic steatohepatitis
- NAFLD
non-alcoholic fatty liver disease
- AFP
α-fetoprotein
- ncRNAs
Non-coding RNAs
- sncRNAs
small/short noncoding RNAs
- miRNAs
microRNAs
- piRNAs
piwi-interacting RNAs
- endo-siRNAs
endogenous short interfering RNAs
- snoRNAs
small nucleolar RNA
- lncRNAs
long noncoding RNA
- circRNA
circular RNA
- 3’UTR
3′-untranslated region
- ceRNA
competing endogenous RNA
- PTMs
post-translational modifications
- DNMTs
DNA methyltransferases
- C
Cytosine base
- G
Guanine base
- CpGIs
CpG islands
- H3K4me
histone 3 lysine 4 methylation
- H3K9me
histone 3 lysine 9 methylation
- H3K27me
histone 3 lysine 27 methylation
- PRC1
polycomb repressive complex 1
- PRC2
polycomb repressive complex 2
- H2AK118ub
histone H2A lysine 118 monoubiquitination
- H2AK119ub
histone H2A lysine 119 monoubiquitination
- EZH1/2
enhancer of zeste homolog 1 and 2
- SUZ12
suppressor of zeste 12
- EED
embryonic ectoderm development
- RBBP4/7
retinoblastoma-binding protein 4/7
- FAK
focal adhesion kinase
- H4K20me3
histone H4 lysine 20 trimethylation
- H4K16
histone H4 lysine 16
- HATs
histone acetyltransferases
- HDACs
histone deacetylases
- H3K9ac
H3K9 acetylation
- HMGA2
High-mobility group protein A2
- HOX
homeobox
- USP22
Ubiquitin-specific protease 22
- pH3S10
histone H3 serine 10 phosphorylation
- pH3S28
histone H3 serine 28 phosphorylation
- AFB1
aflatoxin B1
- MTA1
Metastasis-associated 1
- H1.2
histone cluster 1 H1 family member c
- O-GlcNAc
O-linked N-acetylglucosamine
- OGTs
O-GlcNActransferases
- OGAs
O-GlcNAcases
- PAD
peptidyl arginine deiminase
- H3cit
citrullination of histone H3
- SUMO
small ubiquitin-like modifier
- SENPs
SUMO-specific proteases
- LKB1
liver kinase B1
- XPO-5
exportin-5
- 5-aza
5-azacytidine
- TSGs
Tumour suppressor genes
- ICI
Immune checkpoints inhibitors
- HDACi
HDAC inhibitor
Authors’ contributions
AA was responsible of conception and guidance of this manuscript. MRB, OLR, IR, MGFB, BB, SP wrote the manuscript. RR, MV, MAA, AA edited and critical revised the manuscript. All authors read and approved the final manuscript.
Funding
The study was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC, MFAG12936), Fondazione Bambino Gesù-ONLUS, Italian Ministry of Health (Ricerca Corrente 2019) to AA, and by the European Commission Horizon 2020 Framework Program (Project 856871—TRANSTEM) and by the European Social Fund and European Regional Development Fund – Project MAGNET (No. CZ.02.1.01/0.0/0.0/15_003/0000492) to MV. AIRC grant 15312 to RR, and SP is a Fellow of “Fondazione Veronesi”. FEDER/Ministerio de Ciencia, Innovación y Universidades-Agencia Estatal de Investigación (PID2019-104878RB-100 and PI02020-120387RB-100 to MAA and MGFB, respectively), “Fundación Científica de la Asociación Española Contra el Cáncer” (AECC Scientific Foundation: “AECC Lab call 2020” to MGFB), and MGFB is a Fellow of the Ramón y Cajal Program RYC-2018-024475-1.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol. 2020;72:250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Prim. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 4.Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 5.Bellentani S. The epidemiology of non-alcoholic fatty liver disease. Liver Int. 2017;37:81–84. doi: 10.1111/liv.13299. [DOI] [PubMed] [Google Scholar]
- 6.Mittal S, El-Serag HB, Sada YH, Kanwal F, Duan Z, Temple S, et al. Hepatocellular carcinoma in the absence of cirrhosis in United States veterans is associated with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2016;14:124–131. doi: 10.1016/j.cgh.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauzay C, Petit A, Bourgeois A-M, Barbare J-C, Chauffert B, Galmiche A, et al. Alpha-foetoprotein (AFP): a multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta. 2016;463:39–44. doi: 10.1016/j.cca.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Khanna R, Verma SK. Pediatric hepatocellular carcinoma. World J Gastroenterol. 2018;24:3980–3999. doi: 10.3748/wjg.v24.i35.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng LX, Mehta N. Does hepatocellular carcinoma surveillance increase survival in at-risk populations? Patient selection, biomarkers, and barriers. Dig Dis Sci. 2020;65:3456–3462. doi: 10.1007/s10620-020-06550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adeniji N, Dhanasekaran R. Current and emerging tools for hepatocellular carcinoma surveillance. Hepatol Commun. 2021;5:1972–1986. doi: 10.1002/hep4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo XY, Wu KM, He XX. Advances in drug development for hepatocellular carcinoma: clinical trials and potential therapeutic targets. J Exp Clin Cancer Res. 2021;40:172. doi: 10.1186/s13046-021-01968-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su T, Hsu S, Kao J. Paradigm shift in the treatment options of hepatocellular carcinoma. Liver Int. 2021. 10.1111/liv.15052. [DOI] [PubMed]
- 13.Fujiwara N, Friedman SL, Goossens N, Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68:526–549. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hlady R, Robertson K. Genetic and epigenetic heterogeneity in Normal liver homeostasis and its implications for liver disease and hepatocellular Cancer. Semin Liver Dis. 2018;38:041–050. doi: 10.1055/s-0037-1621712. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Liu Z, Xu X. Molecular subtyping of hepatocellular carcinoma: a step toward precision medicine. Cancer Commun. 2020;40:681–693. doi: 10.1002/cac2.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ally A, Balasundaram M, Carlsen R, Chuah E, Clarke A, Dhalla N, et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S-M, Kim-Ha J, Choi W-Y, Lee J, Kim D, Lee J, et al. Interplay of genetic and epigenetic alterations in hepatocellular carcinoma. Epigenomics. 2016;8:993–1005. doi: 10.2217/epi-2016-0027. [DOI] [PubMed] [Google Scholar]
- 18.Nebbioso A, Tambaro FP, Dell’Aversana C, Altucci L. Cancer epigenetics: moving forward. PLoS Genet. 2018;14:e1007362. doi: 10.1371/journal.pgen.1007362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Jiménez E, Andrés-León E. The implications of ncRNAs in the development of human diseases. Non-Coding RNA. 2021;7:17. doi: 10.3390/ncrna7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T, et al. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109:401–407. doi: 10.1016/j.ygeno.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, Urushidate T, et al. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013;104:543–551. doi: 10.1111/cas.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caruso S, Calderaro J, Letouzé E, Nault J-C, Couchy G, Boulai A, et al. Germline and somatic DICER1 mutations in familial and sporadic liver tumors. J Hepatol. 2017;66:734–742. doi: 10.1016/j.jhep.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 26.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Negrini M, Gramantieri L, Sabbioni S, M. Croce C. microRNA Involvement in Hepatocellular Carcinoma. Anti Cancer Agents Med Chem. 2011;11:500–521. doi: 10.2174/187152011796011037. [DOI] [PubMed] [Google Scholar]
- 28.Wong C-M, Tsang FH-C, Ng IO-L. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 29.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang J, Li L, Huang W, Sui C, Yang Y, Lin X, et al. MiR-429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial–mesenchymal transition. Cancer Lett. 2015;364:33–43. doi: 10.1016/j.canlet.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Thurnherr T, Mah W-C, Lei Z, Jin Y, Rozen SG, Lee CG. Differentially expressed miRNAs in hepatocellular carcinoma target genes in the genetic information processing and metabolism pathways. Sci Rep. 2016;6:20065. doi: 10.1038/srep20065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koduru SV, Leberfinger AN, Kawasawa YI, Mahajan M, Gusani NJ, Sanyal AJ, et al. Non-coding RNAs in various stages of liver disease leading to hepatocellular carcinoma: differential expression of miRNAs, piRNAs, lncRNAs, circRNAs, and sno/mt-RNAs. Sci Rep. 2018;8:7967. doi: 10.1038/s41598-018-26360-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai H-T, Wang S-T, Chen B, Tang K-Y, Li N, Wen C-Y, et al. microRNA-375 inhibits the malignant behaviors of hepatic carcinoma cells by targeting NCAPG2. Neoplasma. 2022;69:16–12. doi: 10.4149/neo_2021_210318N358. [DOI] [PubMed] [Google Scholar]
- 34.Liu N, Chang CW, Steer CJ, Wang XW, Song G. MicroRNA-15a/16-1 prevents hepatocellular carcinoma by disrupting the communication between Kupffer cells and regulatory T cells. Gastroenterology. 2022;162:575–589. doi: 10.1053/j.gastro.2021.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Zhang Y, Xiao B, Tang N, Hu J, Liang S, et al. MiR-103a promotes tumour growth and influences glucose metabolism in hepatocellular carcinoma. Cell Death Dis. 2021;12:618. doi: 10.1038/s41419-021-03905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta M, Akhtar J, Sarwat M. MicroRNAs: regulators of immunological reactions in hepatocellular carcinoma. Semin Cell Dev Biol. 2021. 10.1016/j.semcdb.2021.05.025. [DOI] [PubMed]
- 37.Liu N, Steer CJ, Song G. MicroRNA-206 enhances antitumor immunity by disrupting the communication between malignant hepatocytes and regulatory T cells in c-Myc mice. Hepatology. 2021. 10.1002/hep.32182. [DOI] [PubMed]
- 38.Liu N, Wang X, Steer CJ, Song G. MicroRNA-206 promotes the recruitment of CD8 + T cells by driving M1 polarisation of Kupffer cells. Gut. 2021. 10.1136/gutjnl-2021-324170. [DOI] [PMC free article] [PubMed]
- 39.Shi T, Morishita A, Kobara H, Masaki T. The role of long non-coding RNA and microRNA networks in hepatocellular carcinoma and its tumor microenvironment. Int J Mol Sci. 2021;22:10630. doi: 10.3390/ijms221910630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 41.Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, et al. Serum microRNA characterization identifies miR - 885 - 5p as a potential marker for detecting liver pathologies. Clin Sci. 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu AM, Yao T-J, Wang W, Wong K-F, Lee NP, Fan ST, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, et al. A serum MicroRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. doi: 10.1371/journal.pone.0107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X-J, Chong Y, Guo Z-W, Xie C, Yang X-J, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16:804–815. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 45.Fornari F, Ferracin M, Trerè D, Milazzo M, Marinelli S, Galassi M, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 2015;10:e0141448. doi: 10.1371/journal.pone.0141448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okajima W, Komatsu S, Ichikawa D, Miyamae M, Kawaguchi T, Hirajima S, et al. Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget. 2016;7:53820–53836. doi: 10.18632/oncotarget.10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin L, Lu B, Yu J, Liu W, Zhou A. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2016;40:397–404. doi: 10.1016/j.clinre.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma X, et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e5642. doi: 10.1097/MD.0000000000005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Y, Wong YS, Goh BKP, Chan CY, Cheow PC, Chow PKH, et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci Rep. 2019;9:10464. doi: 10.1038/s41598-019-46872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuma M, Toyoda H, Matsuzaki J, Saito Y, Kumada T, Tada T, et al. Circulating microRNA-1246 as a possible biomarker for early tumor recurrence of hepatocellular carcinoma. Hepatol Res. 2019;49:810–822. doi: 10.1111/hepr.13338. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto Y, Kondo S, Matsuzaki J, Esaki M, Okusaka T, Shimada K, et al. Highly sensitive circulating MicroRNA panel for accurate detection of hepatocellular carcinoma in patients with liver disease. Hepatol Commun. 2020;4:284–297. doi: 10.1002/hep4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratama MY, Visintin A, Crocè LS, Tiribelli C, Pascut D. Circulatory miRNA as a biomarker for therapy response and disease-free survival in hepatocellular carcinoma. Cancers (Basel) 2020;12:2810. doi: 10.3390/cancers12102810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong VC-L, Wong M-I, Lam C-T, Lung ML, Lam K-O, Lee VH-F. Hallmark microRNA signature in liquid biopsy identifies hepatocellular carcinoma and differentiates it from liver metastasis. J Cancer. 2021;12:4585–4594. doi: 10.7150/jca.59933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manganelli M, Grossi I, Ferracin M, Guerriero P, Negrini M, Ghidini M, et al. Longitudinal circulating levels of miR-23b-3p, miR-126-3p and lncRNA GAS5 in HCC patients treated with Sorafenib. Biomedicines. 2021;9:813. doi: 10.3390/biomedicines9070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelizzaro F, Cardin R, Sartori A, Imondi A, Penzo B, Aliberti C, et al. Circulating MicroRNA-21 and MicroRNA-122 as prognostic biomarkers in hepatocellular carcinoma patients treated with Transarterial chemoembolization. Biomedicines. 2021;9:890. doi: 10.3390/biomedicines9080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen S, Lin Y, Yuan X, Shen L, Chen J, Chen L, et al. Biomarker MicroRNAs for diagnosis, prognosis and treatment of hepatocellular carcinoma: a functional survey and comparison. Sci Rep. 2016;6:38311. doi: 10.1038/srep38311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 58.Han Y-N, Li Y, Xia S-Q, Zhang Y-Y, Zheng J-H, Li W. PIWI proteins and PIWI-interacting RNA: emerging roles in Cancer. Cell Physiol Biochem. 2017;44:1–20. doi: 10.1159/000484541. [DOI] [PubMed] [Google Scholar]
- 59.Law PT-Y, Qin H, Ching AK-K, Lai KP, Co NN, He M, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 60.Rizzo F, Rinaldi A, Marchese G, Coviello E, Sellitto A, Cordella A, et al. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget. 2016;7:54650–54661. doi: 10.18632/oncotarget.10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang J, Zhang H, Tang Q, Hao B, Shi R. Expression of HIWI in human hepatocellular carcinoma. Cell Biochem Biophys. 2011;61:53–58. doi: 10.1007/s12013-011-9160-1. [DOI] [PubMed] [Google Scholar]
- 62.Zhao Y-M, Zhou J-M, Wang L-R, He H-W, Wang X-L, Tao Z-H, et al. HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer. 2012;118:2708–2717. doi: 10.1002/cncr.26524. [DOI] [PubMed] [Google Scholar]
- 63.Wang N, Tan H-Y, Lu Y, Chan Y-T, Wang D, Guo W, et al. PIWIL1 governs the crosstalk of cancer cell metabolism and immunosuppressive microenvironment in hepatocellular carcinoma. Signal Transduct Target Ther. 2021;6:86. doi: 10.1038/s41392-021-00485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, Zhang J, Diao L, Han L. Small non-coding RNAs in human cancer: function, clinical utility, and characterization. Oncogene. 2021;40:1570–1577. doi: 10.1038/s41388-020-01630-3. [DOI] [PubMed] [Google Scholar]
- 65.Xu G, Yang F, Ding C-L, Zhao L-J, Ren H, Zhao P, et al. Small nucleolar RNA 113–1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. 2014;13:216. doi: 10.1186/1476-4598-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma P, Wang H, Han L, Jing W, Zhou X, Liu Z. Up-regulation of small nucleolar RNA 78 is correlated with aggressive phenotype and poor prognosis of hepatocellular carcinoma. Tumor Biol. 2016;37:15753–15761. doi: 10.1007/s13277-016-5366-6. [DOI] [PubMed] [Google Scholar]
- 67.Wu L, Zheng J, Chen P, Liu Q, Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;90:705–712. doi: 10.1016/j.biopha.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 68.Baral D, Wu L, Katwal G, Yan X, Wang Y, Ye Q. Clinical significance and biological roles of small nucleolar RNAs in hepatocellular carcinoma. Biomed Reports. 2018;8:319–324. doi: 10.3892/br.2018.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Ma P, Liu P, Chen B, Liu Z. Small nucleolar RNA U2_19 promotes hepatocellular carcinoma progression by regulating Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;500:351–356. doi: 10.1016/j.bbrc.2018.04.074. [DOI] [PubMed] [Google Scholar]
- 70.Wu L, Chang L, Wang H, Ma W, Peng Q, Yuan Y. Clinical significance of C/D box small nucleolar RNA U76 as an oncogene and a prognostic biomarker in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2018;42:82–91. doi: 10.1016/j.clinre.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 71.McMahon M, Contreras A, Holm M, Uechi T, Forester CM, Pang X, et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife. 2019. 10.7554/eLife.48847. [DOI] [PMC free article] [PubMed]
- 72.Mao L-H, Chen S-Y, Li X-Q, Xu F, Lei J, Wang Q-L, et al. LncRNA-LALR1 upregulates small nucleolar RNA SNORD72 to promote growth and invasion of hepatocellular carcinoma. Aging (Albany NY) 2020;12:4527–4546. doi: 10.18632/aging.102907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shuwen H, Xi Y, Quan Q, Yin J, Miao D. Can small nucleolar RNA be a novel molecular target for hepatocellular carcinoma? Gene. 2020;733:144384. doi: 10.1016/j.gene.2020.144384. [DOI] [PubMed] [Google Scholar]
- 74.Chen L-L. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 75.Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, et al. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:198. doi: 10.1186/s13046-019-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D, Chen F, Zeng T, Tang Q, Chen B, Chen L, et al. Comprehensive biological function analysis of lncRNAs in hepatocellular carcinoma. Genes Dis. 2021;8:157–167. doi: 10.1016/j.gendis.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan D, Chen Y, Li X, Li J, Zhao Y, Shen J, et al. Long non-coding RNAs: potential biomarkers and targets for hepatocellular carcinoma therapy and diagnosis. Int J Biol Sci. 2021;17:220–235. doi: 10.7150/ijbs.50730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505–4515. doi: 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, et al. HULC and Linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem. 2015;37:687–696. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 80.El-Tawdi AHF, Matboli M, Shehata HH, Tash F, El-Khazragy N, Azazy AE-SM, et al. Evaluation of circulatory RNA-based biomarker panel in hepatocellular carcinoma. Mol Diagn Ther. 2016;20:265–277. doi: 10.1007/s40291-016-0200-9. [DOI] [PubMed] [Google Scholar]
- 81.Yuan W, Sun Y, Liu L, Zhou B, Wang S, Gu D. Circulating LncRNAs serve as diagnostic markers for hepatocellular carcinoma. Cell Physiol Biochem. 2017;44:125–132. doi: 10.1159/000484589. [DOI] [PubMed] [Google Scholar]
- 82.Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9:2631–2639. doi: 10.7150/jca.24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Habieb A, Matboli M, El-Tayeb H, El-Asmar F. Potential role of lncRNA-TSIX, miR-548-a-3p, and SOGA1 mRNA in the diagnosis of hepatocellular carcinoma. Mol Biol Rep. 2019;46:4581–4590. doi: 10.1007/s11033-019-04810-x. [DOI] [PubMed] [Google Scholar]
- 84.Faranda T, Grossi I, Manganelli M, Marchina E, Baiocchi G, Portolani N, et al. Differential expression profiling of long non-coding RNA GAS5 and miR-126-3p in human cancer cells in response to sorafenib. Sci Rep. 2019;9:9118. doi: 10.1038/s41598-019-45604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang D, Xing N, Yang T, Liu J, Zhao H, He J, et al. Exosomal lncRNA H19 promotes the progression of hepatocellular carcinoma treated with Propofol via miR-520a-3p/LIMK1 axis. Cancer Med. 2020;9:7218–7230. doi: 10.1002/cam4.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S, Li C, Chang W, Cheng W, Yen C, Tu W, et al. Exosome-derived differentiation antagonizing non-protein coding RNA with risk of hepatitis C virus-related hepatocellular carcinoma recurrence. Liver Int. 2021;41:956–968. doi: 10.1111/liv.14772. [DOI] [PubMed] [Google Scholar]
- 87.Cortés-López M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89:527–537. [PMC free article] [PubMed] [Google Scholar]
- 88.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta - Gene Regul Mech. 2016;1859:163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 89.Zhao Z-J, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song M, Xia L, Sun M, Yang C, Wang F. Circular RNA in liver: health and diseases. Adv Exp Med Biol. 2018;1087:245–257. doi: 10.1007/978-981-13-1426-1_20. [DOI] [PubMed] [Google Scholar]
- 92.Qiu L, Wang T, Ge Q, Xu H, Wu Y, Tang Q, et al. Circular RNA Signature in Hepatocellular Carcinoma. J Cancer. 2019;10:3361–3372. doi: 10.7150/jca.31243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu J, Li P, Song Y, Ge Y, Meng X, Huang C, et al. Progress and prospects of circular RNAs in hepatocellular carcinoma: novel insights into their function. J Cell Physiol. 2018;233:4408–4422. doi: 10.1002/jcp.26154. [DOI] [PubMed] [Google Scholar]
- 94.Liu Q, Cai Y, Xiong H, Deng Y, Dai X. CCRDB: a cancer circRNAs-related database and its application in hepatocellular carcinoma-related circRNAs. Database. 2019;2019:baz063. doi: 10.1093/database/baz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143:17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 97.Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (Albany NY) 2019;11:3362–3375. doi: 10.18632/aging.101988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhu Y-J, Zheng B, Luo G-J, Ma X-K, Lu X-Y, Lin X-M, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9:3526–3540. doi: 10.7150/thno.32796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Z, Song J, Tang B, Fang S, Zhang D, Zheng L, et al. CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J Exp Clin Cancer Res. 2020;39:259. doi: 10.1186/s13046-020-01769-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun S, Gao J, Zhou S, Li Y, Wang Y, Jin L, et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2020;39:252. doi: 10.1186/s13046-020-01779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kornberg RD, Thomas JO. Chromatin structure: oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 102.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 103.Hauer MH, Gasser SM. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017;31:2204–2221. doi: 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cutter AR, Hayes JJ. A brief review of nucleosome structure. FEBS Lett. 2015;589:2914–2922. doi: 10.1016/j.febslet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ngo-Yin Fan D, Ho-Ching Tsang F, Hoi-Kam Tam A, Leung-Kuen AS, Chak-Lui Wong C, Wei L, et al. Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57:637–647. doi: 10.1002/hep.26083. [DOI] [PubMed] [Google Scholar]
- 106.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wahid B, Ali A, Rafique S, Idrees M. New insights into the epigenetics of hepatocellular carcinoma. Biomed Res Int. 2017;2017:1–16. doi: 10.1155/2017/1609575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bhat V, Srinathan S, Pasini E, Angeli M, Chen E, Baciu C, et al. Epigenetic basis of hepatocellular carcinoma: a network-based integrative meta-analysis. World J Hepatol. 2018;10:155–165. doi: 10.4254/wjh.v10.i1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 110.Gao X, Sheng Y, Yang J, Wang C, Zhang R, Zhu Y, et al. Osteopontin alters DNA methylation through up-regulating DNMT1 and sensitizes CD133+/CD44+ cancer stem cells to 5 azacytidine in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:179. doi: 10.1186/s13046-018-0832-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu B, Chen JJ, Feng Y, Yang JL, Huang H, Chung WY, et al. DNMT1-induced miR-378a-3p silencing promotes angiogenesis via the NF-κB signaling pathway by targeting TRAF1 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2021;40:352. doi: 10.1186/s13046-021-02110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nakamura M, Chiba T, Kanayama K, Kanzaki H, Saito T, Kusakabe Y, et al. Epigenetic dysregulation in hepatocellular carcinoma: an up-to-date review. Hepatol Res. 2019;49:3–13. doi: 10.1111/hepr.13250. [DOI] [PubMed] [Google Scholar]
- 113.Zheng Y, Lu X, Zhang X, Guan B. The landscape of DNA methylation in hepatocellular carcinoma. J Cell Physiol. 2019;234:2631–2638. doi: 10.1002/jcp.27077. [DOI] [PubMed] [Google Scholar]
- 114.Hernandez-Vargas H, Lambert M-P, Le Calvez-Kelm F, Gouysse G, McKay-Chopin S, Tavtigian SV, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS One. 2010;5:e9749. doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Um T-H, Kim H, Oh B-K, Kim MS, Kim KS, Jung G, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011;54:939–947. doi: 10.1016/j.jhep.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 116.Li Z, Zhang H, Yang J, Hao T, Li S. Promoter hypermethylation of DNA damage response genes in hepatocellular carcinoma. Cell Biol Int. 2012;36:427–432. doi: 10.1042/CBI20100851. [DOI] [PubMed] [Google Scholar]
- 117.Song M-A, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761. doi: 10.1371/journal.pone.0055761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945–1956. doi: 10.1002/hep.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu M, Cui L-H, Li C-C, Zhang L. Association of APC, GSTP1 and SOCS1 promoter methylation with the risk of hepatocellular carcinoma. Eur J Cancer Prev. 2015;24:470–483. doi: 10.1097/CEJ.0000000000000121. [DOI] [PubMed] [Google Scholar]
- 120.Gao F, Liang H, Lu H, Wang J, Xia M, Yuan Z, et al. Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing approach. Clin Epigenetics. 2015;7:86. doi: 10.1186/s13148-015-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang C, Li J, Huang T, Duan S, Dai D, Jiang D, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget. 2016;7:81255–81267. doi: 10.18632/oncotarget.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu D, Yang X, Peng H, Guo D, Zhao W, Zhao C, et al. OCIAD2 suppressed tumor growth and invasion via AKT pathway in Hepatocelluar carcinoma. Carcinogenesis. 2017;38:910–919. doi: 10.1093/carcin/bgx073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu G, Zhou X, Xing J, Xiao Y, Jin B, Sun L, et al. Identification of RASSF1A promoter hypermethylation as a biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20:547. doi: 10.1186/s12935-020-01638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y, Yang Y, Gao H, Ouyang T, Zhang L, Hu J, et al. Comprehensive analysis of CDCAs methylation and immune infiltrates in hepatocellular carcinoma. Front. Oncol. 2021. 10.3389/fonc.2020.566183. [DOI] [PMC free article] [PubMed]
- 125.Li D, Ji Y, Guo J, Guo Q. Upregulated expression of MTFR2 as a novel biomarker predicts poor prognosis in hepatocellular carcinoma by bioinformatics analysis. Future Oncol. 2021;17:3187–3201. doi: 10.2217/fon-2020-1160. [DOI] [PubMed] [Google Scholar]
- 126.Meunier L, Hirsch TZ, Caruso S, Imbeaud S, Bayard Q, Roehrig A, et al. DNA methylation signatures reveal the diversity of processes remodeling hepatocellular carcinoma Methylomes. Hepatology. 2021;74:816–834. doi: 10.1002/hep.31796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zheng Y, Tang L, Chen G, Liu Z. Comprehensive bioinformatics analysis of key Methyltransferases and Demethylases for histone Lysines in hepatocellular carcinoma. Technol Cancer Res Treat. 2020;19:153303382098328. doi: 10.1177/1533033820983284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Y, Chen X, Lu C. The interplay between DNA and histone methylation: molecular mechanisms and disease implications. EMBO Rep. 2021. 10.15252/embr.202051803. [DOI] [PMC free article] [PubMed]
- 129.Hou Z, Min W, Zhang R, Niu A, Li Y, Cao L, et al. Lead discovery, chemical optimization, and biological evaluation studies of novel histone methyltransferase SET7 small-molecule inhibitors. Bioorg Med Chem Lett. 2020;30:127061. doi: 10.1016/j.bmcl.2020.127061. [DOI] [PubMed] [Google Scholar]
- 130.Qian Y, Li Y, Zheng C, Lu T, Sun R, Mao Y, et al. High methylation levels of histone H3 lysine 9 associated with activation of hypoxia-inducible factor 1α (HIF-1α) predict patients’ worse prognosis in human hepatocellular carcinomas. Cancer Genet. 2020;245:17–26. doi: 10.1016/j.cancergen.2020.04.077. [DOI] [PubMed] [Google Scholar]
- 131.Au SL-K, Ng IO-L, Wong C-M. Epigenetic dysregulation in hepatocellular carcinoma: focus on polycomb group proteins. Front Med. 2013;7:231–241. doi: 10.1007/s11684-013-0253-7. [DOI] [PubMed] [Google Scholar]
- 132.Gall Trošelj K, Novak Kujundzic R, Ugarkovic D. Polycomb repressive complex’s evolutionary conserved function: the role of EZH2 status and cellular background. Clin Epigenetics. 2016;8:55. doi: 10.1186/s13148-016-0226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hung S-Y, Lin H-H, Yeh K-T, Chang J-G. Histone-modifying genes as biomarkers in hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 134.Kusakabe Y, Chiba T, Oshima M, Koide S, Rizq O, Aoyama K, et al. EZH1/2 inhibition augments the anti-tumor effects of sorafenib in hepatocellular carcinoma. Sci Rep. 2021;11:21396. doi: 10.1038/s41598-021-00889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gnani D, Romito I, Artuso S, Chierici M, De Stefanis C, Panera N, et al. Focal adhesion kinase depletion reduces human hepatocellular carcinoma growth by repressing enhancer of zeste homolog 2. Cell Death Differ. 2017. 10.1038/cdd.2017.34. [DOI] [PMC free article] [PubMed]
- 136.Panera N, Crudele A, Romito I, Gnani D, Alisi A. Focal adhesion kinase: insight into molecular roles and functions in hepatocellular carcinoma. Int J Mol Sci. 2017;18:99. doi: 10.3390/ijms18010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Francalanci P, Giovannoni I, De Stefanis C, Romito I, Grimaldi C, Castellano A, et al. Focal adhesion kinase (FAK) over-expression and prognostic implication in pediatric hepatocellular carcinoma. Int J Mol Sci. 2020;21:5795. doi: 10.3390/ijms21165795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Romito I, Panera N, D’Ermo G, Alisi A. Letter to the editor: focal adhesion kinase/β-catenin network may act as a regulator of hepatocellular carcinoma epigenetics. Hepatology. 2019;70:1494–1495. doi: 10.1002/hep.30849. [DOI] [PubMed] [Google Scholar]
- 139.de Conti A, Dreval K, Tryndyak V, Orisakwe OE, Ross SA, Beland FA, et al. Inhibition of the cell death pathway in nonalcoholic Steatohepatitis (NASH)-related Hepatocarcinogenesis is associated with histone H4 lysine 16 Deacetylation. Mol Cancer Res. 2017;15:1163–1172. doi: 10.1158/1541-7786.MCR-17-0109. [DOI] [PubMed] [Google Scholar]
- 140.Shanmugam MK, Arfuso F, Arumugam S, Chinnathambi A, Jinsong B, Warrier S, et al. Role of novel histone modifications in cancer. Oncotarget. 2018;9:11414–11426. doi: 10.18632/oncotarget.23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Z, Shilatifard A. Epigenetic modifications of histones in cancer. Genome Biol. 2019;20:245. doi: 10.1186/s13059-019-1870-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liang C, Niu J, Wang X, Zhang Z, Yang R, Yao X, et al. P300-dependent acetylation of histone H3 is required for epidermal growth factor receptor-mediated high-mobility group protein A2 transcription in hepatocellular carcinoma. Cancer Sci. 2021;112:679–690. doi: 10.1111/cas.14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang J, Liu H, Pan H, Yang Y, Huang G, Yang Y, et al. The histone acetyltransferase hMOF suppresses hepatocellular carcinoma growth. Biochem Biophys Res Commun. 2014;452:575–580. doi: 10.1016/j.bbrc.2014.08.122. [DOI] [PubMed] [Google Scholar]
- 144.Poté N, Cros J, Laouirem S, Raffenne J, Negrão M, Albuquerque M, et al. The histone acetyltransferase hMOF promotes vascular invasion in hepatocellular carcinoma. Liver Int. 2020;40:956–967. doi: 10.1111/liv.14381. [DOI] [PubMed] [Google Scholar]
- 145.Jin X, Tian S, Li P. Histone Acetyltransferase 1 promotes cell proliferation and induces Cisplatin resistance in hepatocellular carcinoma. Oncol Res Featur Preclin Clin Cancer Ther. 2017;25:939–946. doi: 10.3727/096504016X14809827856524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Romito I, Porru M, Braghini MR, Pompili L, Panera N, Crudele A, et al. Focal adhesion kinase inhibitor TAE226 combined with Sorafenib slows down hepatocellular carcinoma by multiple epigenetic effects. J Exp Clin Cancer Res. 2021;40:364. doi: 10.1186/s13046-021-02154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fernández-Barrena MG, Arechederra M, Colyn L, Berasain C, Avila MA. Epigenetics in hepatocellular carcinoma development and therapy: the tip of the iceberg. JHEP Reports. 2020;2:100167. doi: 10.1016/j.jhepr.2020.100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Piunti A, Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol. 2021;22:326–345. doi: 10.1038/s41580-021-00341-1. [DOI] [PubMed] [Google Scholar]
- 149.Kasai H, Mochizuki K, Tanaka T, Yamashita A, Matsuura Y, Moriishi K. Induction of HOX genes by hepatitis C virus infection via impairment of histone H2A Monoubiquitination. J Virol. 2021. 10.1128/JVI.01784-20. [DOI] [PMC free article] [PubMed]
- 150.Tang B, Tang F, Li B, Yuan S, Xu Q, Tomlinson S, et al. High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget. 2015;6:12654–12667. doi: 10.18632/oncotarget.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ling S, Li J, Shan Q, Dai H, Lu D, Wen X, et al. USP22 mediates the multidrug resistance of hepatocellular carcinoma via the SIRT1/AKT/MRP1 signaling pathway. Mol Oncol. 2017;11:682–695. doi: 10.1002/1878-0261.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang J, Luo N, Tian Y, Li J, Yang X, Yin H, et al. USP22 knockdown enhanced chemosensitivity of hepatocellular carcinoma cells to 5-Fu by up-regulation of Smad4 and suppression of Akt. Oncotarget. 2017;8:24728–24740. doi: 10.18632/oncotarget.15798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–1334. doi: 10.1136/gutjnl-2019-319616. [DOI] [PubMed] [Google Scholar]
- 154.Sun N, Zhong X, Wang S, Zeng K, Sun H, Sun G, et al. ATXN7L3 positively regulates SMAD7 transcription in hepatocellular carcinoma with growth inhibitory function. EBioMedicine. 2020;62:103108. doi: 10.1016/j.ebiom.2020.103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shen Z-T, Chen Y, Huang G-C, Zhu X-X, Wang R, Chen L-B. Aurora-a confers radioresistance in human hepatocellular carcinoma by activating NF-κB signaling pathway. BMC Cancer. 2019;19:1075. doi: 10.1186/s12885-019-6312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lin Z-Z, Jeng Y-M, Hu F-C, Pan H-W, Tsao H-W, Lai P-L, et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B overexpression in HCC. BMC Cancer 2010. 10.1186/1471-2407-10-461. [DOI] [PMC free article] [PubMed]
- 157.Liu F, Wang G, Wang X, Che Z, Dong W, Guo X, et al. Targeting high Aurora kinases expression as an innovative therapy for hepatocellular carcinoma. Oncotarget. 2017;8:27953–27965. doi: 10.18632/oncotarget.15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou Y, Li M, Yu X, Liu T, Li T, Zhou L, et al. Butein suppresses hepatocellular carcinoma growth via modulating Aurora B kinase activity. Int J Biol Sci. 2018;14:1521–1534. doi: 10.7150/ijbs.25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu X, Li D, Zhang Z, Zhu W, Li W, Zhao J, et al. Persistent phosphorylation at specific H3 serine residues involved in chemical carcinogen-induced cell transformation. Mol Carcinog. 2017;56:1449–1460. doi: 10.1002/mc.22605. [DOI] [PubMed] [Google Scholar]
- 160.Li Y-H, Zhong M, Zang H-L, Tian X-F. MTA1 promotes hepatocellular carcinoma progression by Downregulation of DNA-PK-mediated H1.2T146 phosphorylation. Front Oncol. 2020. 10.3389/fonc.2020.00567. [DOI] [PMC free article] [PubMed]
- 161.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18:452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhu Q, Zhou L, Yang Z, Lai M, Xie H, Wu L, et al. O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med Oncol. 2012;29:985–993. doi: 10.1007/s12032-011-9912-1. [DOI] [PubMed] [Google Scholar]
- 163.Sanghvi VR, Leibold J, Mina M, Mohan P, Berishaj M, Li Z, et al. The oncogenic action of NRF2 depends on De-glycation by Fructosamine-3-kinase. Cell. 2019;178:807–819. doi: 10.1016/j.cell.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone Marks and histone lysine Crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wan J, Liu H, Ming L. Lysine crotonylation is involved in hepatocellular carcinoma progression. Biomed Pharmacother. 2019;111:976–982. doi: 10.1016/j.biopha.2018.12.148. [DOI] [PubMed] [Google Scholar]
- 166.Zhu D, Zhang Y, Wang S. Histone citrullination: a new target for tumors. Mol Cancer. 2021;20:90. doi: 10.1186/s12943-021-01373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lu M, Zhang X, Xu Y, He G, Liu Q, Zhu J, et al. Elevated histone H3 citrullination is associated with increased Beclin1 expression in HBV-related hepatocellular carcinoma. J Med Virol. 2020;92:1221–1230. doi: 10.1002/jmv.25663. [DOI] [PubMed] [Google Scholar]
- 168.Wang Y, Lyu Y, Tu K, Xu Q, Yang Y, Salman S, et al. Histone citrullination by PADI4 is required for HIF-dependent transcriptional responses to hypoxia and tumor vascularization. Sci Adv. 2021. 10.1126/sciadv.abe3771. [DOI] [PMC free article] [PubMed]
- 169.Tomasi ML, Ramani K. SUMOylation and phosphorylation cross-talk in hepatocellular carcinoma. Transl Gastroenterol Hepatol. 2018;3:20. doi: 10.21037/tgh.2018.04.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nayak A, Müller S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014;15:422. doi: 10.1186/s13059-014-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 172.Guo W, Yuan L, Xiao Z, Liu D, Zhang J. Overexpression of SUMO-1 in hepatocellular carcinoma: a latent target for diagnosis and therapy of hepatoma. J Cancer Res Clin Oncol. 2011;137:533–541. doi: 10.1007/s00432-010-0920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu H, Wang H, Zhao W, Fu S, Li Y, Ni W, et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020;10:5671–5686. doi: 10.7150/thno.42539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zubiete-Franco I, García-Rodríguez JL, Lopitz-Otsoa F, Serrano-Macia M, Simon J, Fernández-Tussy P, et al. SUMOylation regulates LKB1 localization and its oncogenic activity in liver cancer. EBioMedicine. 2019;40:406–421. doi: 10.1016/j.ebiom.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin D, Fu Z, Yang G, Gao D, Wang T, Liu Z, et al. Exportin-5 SUMOylation promotes hepatocellular carcinoma progression. Exp Cell Res. 2020;395:112219. doi: 10.1016/j.yexcr.2020.112219. [DOI] [PubMed] [Google Scholar]
- 176.Qin Y, Bao H, Pan Y, Yin M, Liu Y, Wu S, et al. SUMOylation alterations are associated with multidrug resistance in hepatocellular carcinoma. Mol Med Rep. 2014;9:877–881. doi: 10.3892/mmr.2014.1882. [DOI] [PubMed] [Google Scholar]
- 177.Zhang W, Sun H, Shi X, Wang H, Cui C, Xiao F, et al. SENP1 regulates hepatocyte growth factor-induced migration and epithelial-mesenchymal transition of hepatocellular carcinoma. Tumour Biol. 2016;37:7741–7748. doi: 10.1007/s13277-015-4406-y. [DOI] [PubMed] [Google Scholar]
- 178.Cui C-P, Wong CC-L, Kai AK-L, Ho DW-H, Lau EY-T, Tsui Y-M, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut. 2017;66:2149–2159. doi: 10.1136/gutjnl-2016-313264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Conigliaro A, Tripodi M, Parola M. SENP1 activity sustains cancer stem cell in hypoxic HCC. Gut. 2017;66:2051–2052. doi: 10.1136/gutjnl-2017-313946. [DOI] [PubMed] [Google Scholar]
- 180.Jin Z-L, Pei H, Xu Y-H, Yu J, Deng T. The SUMO-specific protease SENP5 controls DNA damage response and promotes tumorigenesis in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20:3566–3573. [PubMed] [Google Scholar]
- 181.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 182.Giallongo S, Lo Re O, Vinciguerra M. Macro histone variants: emerging rheostats of gastrointestinal cancers. Cancers (Basel). 2019;11:676. doi: 10.3390/cancers11050676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Subramanian V, Fields PA, Boyer LA. H2A.Z: a molecular rheostat for transcriptional control. F1000Prime Rep. 2015. 10.12703/P7-01. [DOI] [PMC free article] [PubMed]
- 184.Yang HD, Kim P-J, Eun JW, Shen Q, Kim HS, Shin WC, et al. Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016;7:11412–11423. doi: 10.18632/oncotarget.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yuan Y, Cao W, Zhou H, Qian H, Wang H. H2A.Z acetylation by lincZNF337-AS1 via KAT5 implicated in the transcriptional misregulation in cancer signaling pathway in hepatocellular carcinoma. Cell Death Dis. 2021;12:609. doi: 10.1038/s41419-021-03895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tang S, Huang X, Wang X, Zhou X, Huang H, Qin L, et al. Vital and distinct roles of H2A.Z isoforms in hepatocellular carcinoma. Onco Targets Ther. 2020;13:4319–4337. doi: 10.2147/OTT.S243823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lo Re O, Vinciguerra M. Histone MacroH2A1: a chromatin point of intersection between fasting, senescence and cellular regeneration. Genes (Basel) 2017;8:367. doi: 10.3390/genes8120367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Giallongo S, Rehakova D, Raffaele M, Lo Re O, Koutna I, Vinciguerra M. Redox and epigenetics in human pluripotent stem cells differentiation. Antioxid Redox Signal. 2021;34:335–349. doi: 10.1089/ars.2019.7983. [DOI] [PubMed] [Google Scholar]
- 189.Chiodi V, Domenici MR, Biagini T, De Simone R, Tartaglione AM, Di Rosa M, et al. Systemic depletion of histone macroH2A1.1 boosts hippocampal synaptic plasticity and social behavior in mice. FASEB J. 2021. 10.1096/fj.202100569R. [DOI] [PubMed]
- 190.Bereshchenko O, Lo Re O, Nikulenkov F, Flamini S, Kotaskova J, Mazza T, et al. Deficiency and haploinsufficiency of histone macroH2A1.1 in mice recapitulate hematopoietic defects of human myelodysplastic syndrome. Clin. Epigenetics. 2019;11:121. doi: 10.1186/s13148-019-0724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lo Re O, Mazza T, Giallongo S, Sanna P, Rappa F, Vinh Luong T, et al. Loss of histone macroH2A1 in hepatocellular carcinoma cells promotes paracrine-mediated chemoresistance and CD4 + CD25 + FoxP3 + regulatory T cells activation. Theranostics. 2020;10:910–924. doi: 10.7150/thno.35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Borghesan M, Fusilli C, Rappa F, Panebianco C, Rizzo G, Oben JA, et al. DNA Hypomethylation and histone variant macroH2A1 synergistically attenuate chemotherapy-induced senescence to promote hepatocellular carcinoma progression. Cancer Res. 2016;76:594–606. doi: 10.1158/0008-5472.CAN-15-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lo Re O, Fusilli C, Rappa F, Van Haele M, Douet J, Pindjakova J, et al. Induction of cancer cell stemness by depletion of macrohistone H2A1 in hepatocellular carcinoma. Hepatology. 2018;67:636–650. doi: 10.1002/hep.29519. [DOI] [PubMed] [Google Scholar]
- 194.Rivas Serna IM, Romito I, Maugeri A, Lo Re O, Giallongo S, Mazzoccoli G, et al. A Lipidomic signature complements Stemness features acquisition in liver Cancer cells. Int J Mol Sci. 2020;21:8452. doi: 10.3390/ijms21228452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jueliger S, Lyons J, Cannito S, Pata I, Pata P, Shkolnaya M, et al. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics. 2016;11:709–720. doi: 10.1080/15592294.2016.1214781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Matsuda Y, Wakai T, Kubota M, Osawa M, Takamura M, Yamagiwa S, et al. DNA damage sensor γ -H2AX is increased in Preneoplastic lesions of hepatocellular carcinoma. Sci World J. 2013;2013:1–7. doi: 10.1155/2013/597095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kim H, Oh B-K, Roncalli M, Park C, Yoon S-M, Yoo JE, et al. Large liver cell change in hepatitis B virus-related liver cirrhosis. Hepatology. 2009;50:752–762. doi: 10.1002/hep.23072. [DOI] [PubMed] [Google Scholar]
- 198.Xiao H, Tong R, Ding C, Lv Z, Du C, Peng C, et al. γ-H2AX promotes hepatocellular carcinoma angiogenesis via EGFR/HIF-1α/VEGF pathways under hypoxic condition. Oncotarget. 2015;6:2180–2192. doi: 10.18632/oncotarget.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lu Y, Chan Y-T, Tan H-Y, Li S, Wang N, Feng Y. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19:79. doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Aydin C, Kalkan R. Cancer treatment: an epigenetic view. Glob Med Genet. 2020;07:003–007. doi: 10.1055/s-0040-1713610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Montalvo-Casimiro M, González-Barrios R, Meraz-Rodriguez MA, Juárez-González VT, Arriaga-Canon C, Herrera LA. Epidrug repurposing: discovering new faces of old acquaintances in Cancer therapy. Front Oncol. 2020. 10.3389/fonc.2020.605386. [DOI] [PMC free article] [PubMed]
- 202.Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 203.Bennett RL, Licht JD. Targeting epigenetics in Cancer. Annu Rev Pharmacol Toxicol. 2018;58:187–207. doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tsui Y-M, Chan L-K, Ng IO-L. Cancer stemness in hepatocellular carcinoma: mechanisms and translational potential. Br J Cancer. 2020;122:1428–1440. doi: 10.1038/s41416-020-0823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yu X, Li M, Guo C, Wu Y, Zhao L, Shi Q, et al. Therapeutic targeting of Cancer: epigenetic homeostasis. Front Oncol. 2021. 10.3389/fonc.2021.747022. [DOI] [PMC free article] [PubMed]
- 206.Oronsky BT, Oronsky AL, Lybeck M, Oronsky NC, Scicinski JJ, Carter C, et al. Episensitization: defying Time's arrow. Front Oncol. 2015. 10.3389/fonc.2015.00134. [DOI] [PMC free article] [PubMed]
- 207.Ravindran Menon D, Hammerlindl H, Torrano J, Schaider H, Fujita M. Epigenetics and metabolism at the crossroads of stress-induced plasticity, stemness and therapeutic resistance in cancer. Theranostics. 2020;10:6261–6277. doi: 10.7150/thno.42523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu J, Liu Y, Meng L, Liu K, Ji B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol Rep. 2017;38:899–907. doi: 10.3892/or.2017.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gailhouste L, Liew LC, Yasukawa K, Hatada I, Tanaka Y, Nakagama H, et al. Differentiation therapy by epigenetic reconditioning exerts antitumor effects on liver Cancer cells. Mol Ther. 2018;26:1840–1854. doi: 10.1016/j.ymthe.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu M, Zhang L, Li H, Hinoue T, Zhou W, Ohtani H, et al. Integrative epigenetic analysis reveals therapeutic targets to the DNA methyltransferase inhibitor Guadecitabine (SGI-110) in hepatocellular carcinoma. Hepatology. 2018;68:1412–1428. doi: 10.1002/hep.30091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Villanueva L, Álvarez-Errico D, Esteller M. The contribution of epigenetics to Cancer immunotherapy. Trends Immunol. 2020;41:676–691. doi: 10.1016/j.it.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 212.Sanceau J, Gougelet A. Epigenetic mechanisms of liver tumor resistance to immunotherapy. World J Hepatol. 2021;13:979–1002. doi: 10.4254/wjh.v13.i9.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Toh TB, Lim JJ, Chow EK. Epigenetics of hepatocellular carcinoma. Clin Transl Med. 2019. 10.1186/s40169-019-0230-0. [DOI] [PMC free article] [PubMed]
- 214.Zhao J, Gray SG, Greene CM, Lawless MW. Unmasking the pathological and therapeutic potential of histone deacetylases for liver cancer. Expert Rev Gastroenterol Hepatol. 2019;13:247–256. doi: 10.1080/17474124.2019.1568870. [DOI] [PubMed] [Google Scholar]
- 215.Llopiz D, Ruiz M, Villanueva L, Iglesias T, Silva L, Egea J, et al. Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol Immunother. 2019;68:379–393. doi: 10.1007/s00262-018-2283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bitzer M, Horger M, Giannini EG, Ganten TM, Wörns MA, Siveke JT, et al. Resminostat plus sorafenib as second-line therapy of advanced hepatocellular carcinoma - the SHELTER study. J Hepatol. 2016;65:280–288. doi: 10.1016/j.jhep.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 217.Tak WY, Ryoo BY, Lim HY, Kim DY, Okusaka T, Ikeda M, et al. Phase I/II study of first-line combination therapy with sorafenib plus resminostat, an oral HDAC inhibitor, versus sorafenib monotherapy for advanced hepatocellular carcinoma in east Asian patients. Investig New Drugs. 2018;36:1072–1084. doi: 10.1007/s10637-018-0658-x. [DOI] [PubMed] [Google Scholar]
- 218.Gordon SW, McGuire WP, 3rd, Shafer DA, Sterling RK, Lee HM, Matherly SC, et al. Phase I study of Sorafenib and Vorinostat in advanced hepatocellular carcinoma. Am J Clin Oncol. 2019;42:649–654. doi: 10.1097/COC.0000000000000567. [DOI] [PubMed] [Google Scholar]
- 219.Bhat KP, Ümit Kaniskan H, Jin J, Gozani O. Epigenetics and beyond: targeting writers of protein lysine methylation to treat disease. Nat Rev Drug Discov. 2021;20:265–286. doi: 10.1038/s41573-020-00108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bugide S, Green MR, Wajapeyee N. Inhibition of enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc Natl Acad Sci. 2018;115:E3509–E3518. doi: 10.1073/pnas.1802691115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wei L, Chiu DK-C, Tsang FH-C, Law C-T, Cheng CL-H, Au SL-K, et al. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol. 2017;67:758–769. doi: 10.1016/j.jhep.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 222.Tomaselli D, Lucidi A, Rotili D, Mai A. Epigenetic polypharmacology: a new frontier for epi-drug discovery. Med Res Rev. 2020;40:190–244. doi: 10.1002/med.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bárcena-Varela M, Caruso S, Llerena S, Álvarez-Sola G, Uriarte I, Latasa MU, et al. Dual targeting of histone methyltransferase G9a and DNA-methyltransferase 1 for the treatment of experimental hepatocellular carcinoma. Hepatology. 2019;69:587–603. doi: 10.1002/hep.30168. [DOI] [PubMed] [Google Scholar]
- 224.Barcena-Varela M, Paish H, Alvarez L, Uriarte I, Latasa MU, Santamaria E, et al. Epigenetic mechanisms and metabolic reprogramming in fibrogenesis: dual targeting of G9a and DNMT1 for the inhibition of liver fibrosis. Gut. 2020. 10.1136/gutjnl-2019-320205. [DOI] [PubMed]
- 225.Tsang FH, Law C, Tang TC, Cheng CL, Chin DW, Tam WV, et al. Aberrant super-enhancer landscape in human hepatocellular carcinoma. Hepatology. 2019. 10.1002/hep.30544. [DOI] [PubMed]
- 226.Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8:24–36. doi: 10.1158/2159-8290.CD-17-0605. [DOI] [PubMed] [Google Scholar]
- 227.Bayo J, Fiore EJ, Dominguez LM, Real A, Malvicini M, Rizzo M, et al. A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J Hepatol. 2019;71:78–90. doi: 10.1016/j.jhep.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 228.Jühling F, Hamdane N, Crouchet E, Li S, El Saghire H, Mukherji A, et al. Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut. 2021;70:157–169. doi: 10.1136/gutjnl-2019-318918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Liu M, Zhou J, Liu X, Feng Y, Yang W, Wu F, et al. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69:365–379. doi: 10.1136/gutjnl-2018-317257. [DOI] [PubMed] [Google Scholar]
- 230.Cui X, Liu B, Zheng S, Dong K, Dong R. Genome-wide analysis of DNA methylation in hepatoblastoma tissues. Oncol Lett. 2016;12:1529–1534. doi: 10.3892/ol.2016.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Carrillo-Reixach J, Torrens L, Simon-Coma M, Royo L, Domingo-Sàbat M, Abril-Fornaguera J, et al. Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol. 2020;73:328–341. doi: 10.1016/j.jhep.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nagae G, Yamamoto S, Fujita M, Fujita T, Nonaka A, Umeda T, et al. Genetic and epigenetic basis of hepatoblastoma diversity. Nat Commun. 2021;12:5423. doi: 10.1038/s41467-021-25430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Morel D, Jeffery D, Aspeslagh S, Almouzni G, Postel-Vinay S. Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat Rev Clin Oncol. 2020;17:91–107. doi: 10.1038/s41571-019-0267-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.