Abstract
Small non-coding microRNAs (miRNAs) have the ability to target and bind to many mRNAs within the cytosol resulting in reduced protein expression and modulation of a number of cellular pathways and networks. In addition to the cytosol, miRNAs have been identified in other cellular compartments and organelles, including the mitochondria. While a few mitochondria-associated miRNAs (mitomiRs) are predicted to be derived from the mitochondrial genome, the majority appear to be transcribed from nuclear DNA and somehow transported into the mitochondria. These findings raise interesting questions about why miRNAs are located in the mitochondria and if they play a role in regulating processes within these organelles. Previously published work from our laboratory showed that miR-181a/b can regulate osteogenesis, in part, by enhancing mitochondrial metabolism. In other published studies, miR-181 paralogs and many other miRNAs have been identified in mitochondrial extracts derived from common cell lines and specific primary cells and tissues. Taken together, we were motivated to identify mitomiR expression profiles during in vitro osteogenesis. Specifically, we obtained RNA from purified mitochondrial extracts of human bone marrow-derived mesenchymal stem/stromal cells (MSCs) and from whole cell extracts of MSCs at day 0 or following osteogenic induction for 3, 7 and 14 days. Utilizing Affymetrix GeneChip™ miRNA 4.0 arrays, mitomiR expression signatures were determined at each time point. Based on the Affymetrix detection above background algorithm, the total number of miRNAs detected in MSC mitochondria extracts was 527 (non-induced MSCs), 627 (day 3 induced), 372 (day 7 induced) and 498 (day 14 induced). In addition, we identified significantly differentially-expressed mitomiRs at day 7 and day 14 of osteogenic induction when compared to day 0 (fold change ≥1.5; adjusted p value <0.05). In general, the most pronounced and highly significant changes in mitomiR expression during osteogenesis were observed at the day 7 time point. Interestingly, most miRNAs found to be differentially-expressed in mitochondria extracts did not show significantly altered expression in whole cell extracts at the same time points during osteoblast differentiation. This array study provides novel information on miRNAs associated with the mitochondria in MSCs during differentiation toward the osteoblast phenotype. These findings will guide future research to identify new miRNA candidates that may function in regulating mitochondrial function and/or bone formation, homeostasis or repair.
Keywords: MicroRNA, Mitochondria, MitomiR, Osteogenesis, Bone, Metabolism
1. Introduction
MicroRNAs (miRNAs) are small non-coding RNAs (ncRNAs) that function as epigenetic regulators by binding to specific regions of target mRNAs resulting in reduced protein expression. They are transcribed from genomic DNA as large (~1000 nt) primary RNAs (pri-miRs) which are processed in the nucleus by a Drosha-containing microprocessor complex into smaller (~70–100 nt) precursor miRNAs (pre-miRNAs) and subsequently exported into the cytosol. In the cytosol, pre-miRNAs are processed further by Dicer to form a mature miRNA duplex. The functional 5p or 3p strand of this duplex can associate with the RNA-induced silencing complex (RISC) where it binds, via its short seed sequence, to a complementary site in the 3’UTR of a target mRNA resulting in either mRNA degradation or inhibition of translation [1]. It is also known that miRNAs may target tens to hundreds of mRNAs within a given cell [2] thereby resulting in modulation of many cellular pathways and networks, a feature that renders these ncRNAs as attractive therapeutic targets.
Interestingly, miRNAs, as well as proteins involved in their biogenesis, are not confined to the cytosol. They are also located in other sub-cellular compartments including the nucleus, nucleolus, P-bodies, the ER, exosomes and late endosomes [3]. In addition, a number of reports have identified miRNAs enriched in or associated with the mitochondria of various mouse, rat and human cell types and tissues [4–10]. Mitochondria are double membrane-bound organelles that play a crucial role in regulating energy metabolism as well as other processes including cell death, autophagy, fatty acid oxidation and calcium homeostasis. The biogenesis and function of mitochondria depend on products derived from both the nuclear and mitochondrial genome. Mitochondrial DNA contains only 37 genes encoding 22 tRNAs, 2 rRNAs and 13 protein subunits that make up respiratory complexes I, III, IV and V of the electron transport chain. The majority of proteins in the mitochondria (> 1000) are derived from genomic DNA. Thus, cross talk with the nucleus provides mitochondria with imported nuclear-encoded proteins as well as ncRNAs that are required for mitochondrial homeostasis and function [11,12].
Why miRNAs exist inside or associated with mitochondria, and how they may potentially function in this context, is not well understood. One hypothesis involves the ability of some of these miRNAs to regulate mitochondrial function by targeting specific mRNAs derived from the mitochondrial genome. The fact that Ago 2, a critical protein component of the RISC, has been identified in mitochondria [4,13,14] suggests that miRNAs may interact with Ago 2 in this location to regulate mitochondrial mRNA stability or translation. From previous studies identifying mitochondrial miRNAs in rat liver or human primary muscle cells, computational approaches predicted that miR-130a may target mitochondria-derived cytochrome C oxidase subunit 3 (mt-COX3) mRNA or that miR-133a may target mRNA encoding mitochondria-derived NADH-ubiquinone oxidoreductase chain 1 (mt-ND1), a component of the NADH dehydrogenase complex [5,7]. Other studies have since confirmed, via biochemical approaches, that specific miRNAs can target genes derived from the mitochondrial genome, suggesting that miRNAs may function within the mitochondria [15–19]. For example, studies have demonstrated that miR-181c can target and suppress mt-COX1 expression [13,20], miR-378 can target ATP6 [10], miR-21 targets mt-Cytb [21], while miR-1 can target mt-COX1 and mt-ND1 [22]. These findings demonstrate another layer of complexity with respect to miRNA function and mechanism that is dependent on both the cell type as well as sub-cellular location. These reports also suggest additional mechanisms by which cellular metabolism, specifically the electron transport chain controlling mitochondrial respiration, may be regulated.
Recent studies in our laboratory showed that the miR-181a/b-1 cluster can enhance in vitro osteogenesis, in part, by increasing mitochondrial metabolism [23]. In this work, we reported that the cytosolic role of this miRNA cluster, in modulating PI3K/AKT signaling, was one mechanism by which mitochondrial metabolism could be regulated during osteogenesis. However, the discovery that miR-181 paralogs (i.e. miR-181a, b, c, d) were found to be associated with the mitochondria of other cell types [5,9,13,20] was interesting within the context of our studies. We thus postulated that if miR-181 family members are also be present in the mitochondria of osteo-progenitor cells and differentiating osteoblasts, we may discover additional miRNA-mediated mechanisms that regulate mitochondrial metabolism and hence, osteogenesis. Given that there is nothing known about mitochondria-associated miRNA (mitomiR) expression or function in bone biology, we were thus motivated to design a study to determine mitomiR expression signatures in an established in vitro osteogenesis assay system. Specifically, a miRNA array was carried out on RNA extracted from purified mitochondria or from whole cell extracts of human bone marrow-derived mesenchymal stem/stromal cells (MSCs) at day 0 or following induction toward the osteoblast lineage at different time points (days 3, 7, 14). Note that the term “mitomiR” defines mitochondria-associated miRNAs that are derived from either genomic DNA or mitochondrial DNA, the latter of which are poorly described. The mitomiRs reported in this study are primarily known miRNAs derived from nuclear genomic DNA.
Here, we present novel microarray profiling data describing the most abundantly-expressed mitomiRs in non-induced MSCs and in MSCs at specific time points of osteogenic induction. We also list mitomiRs that were found to be significantly differentially-expressed between MSCs at day 0 and either day 7 or day 14 of osteogenic induction. Notably, the day 7 time point revealed the highest number of differentially-expressed mitomiRs. Expression re-analysis of specific mitomiRs by qPCR confirmed the patterns detected by microarray, particularly those changes detected at day 7. Interestingly, the profiles of differentially-expressed miRNAs from mitochondria extracts were mostly distinct from those found to be altered in whole cell extracts at a specific time point of osteoblast differentiation. Findings from this array study may aid in identifying new functional miRNA candidates to pursue in the future as a means to determine novel strategies to regulate mitochondrial metabolism/function as well as bone formation, homeostasis or repair.
2. Materials and methods
2.1. Osteogenic differentiation of human MSCs for miRNA array analysis
Three human bone marrow-derived mesenchymal stem/stromal (MSC) lines from different donors were purchased from RoosterBio Inc. [i. MSC-049 from male 23 yr old (MSC-003 vial; Lot: 00049); ii. MSC-139 from male 25 yr old (MSC-003 vial, Lot: 00139; iii) MSC-177 from male 22 yr old (MSC-001 vial, Lot: 00177]. Cells were cultured in T150 flasks (5 × 106 cells / flask) in growth medium (DMEM low glucose, 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 1 ng/mL bFGF) until approximately 80% confluency. Cells were then cultured in a standard osteogenic induction medium (αMEM containing 10% FBS, 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 mM β-glycerol phosphate, 50 μM ascorbic acid, 10 nM dexamethasone) for 3, 7 or 14 days. Non-induced cells (day 0) or osteogenic-induced cells (day 3, 7, or 14) were collected for isolation of whole cell extracts or purified mitochondria as described below. To confirm the osteogenic potential of each cell line, MSCs were also cultured in T75 flasks for 3 weeks (3 × 106 cells / flask) in osteogenic induction medium. To examine matrix mineralization, MSCs were fixed in 4% paraformaldehyde and stained for 30 min with 1% Alizarin Red ethanol solution.
2.2. Purification of mitochondria from MSCs
Two T150 flasks of non-induced MSCs (day 0) or osteogenic-induced MSCs (day 3, 7 or 14) were trypsinized, washed in cold PBS and lysed. Cell lysates were also passed through a sterile 26G needle twenty times to ensure complete disruption of the cells. Mitochondria were isolated from cell lysates using the human Mitochondria Isolation Kit (Milteny Biotec) following the manufacturer’s instructions. This kit utilizes magnetic antibody cell sorting (MACS®) technology. Specifically, magnetic microbeads conjugated to an anti-TOM22 monoclonal antibody were used to magnetically label mitochondria in cell lysates. (Note-the TOM22 antibody binds to the translocase of the outer mitochondrial membrane 22 protein of human mitochondria). The labeled cell lysate was loaded onto a MACS column and placed in the magnetic field of a MACS separator. Magnetically-labeled mitochondria retained within the column were eluted and collected by centrifugation at 13,000g for 2 min at 4 °C. Prior to RNA extraction, mitochondrial extract pellets were treated with 300 μL RNAse A (16 μg/mL) on ice for 20 min to remove cytosolic RNA contamination that may be present on the surface of mitochondria.
2.3. Imaging of purified mitochondria by transmission electron microscopy
For transmission electron microscopy (TEM), pellets of isolated mitochondria were resuspended in fixative solution containing 2% paraformaldehyde and 2.5% glutaraldehyde in 0.15 M cacodylate buffer with 2 mM CaCl2, pH 7.4 and kept at 4 °C overnight with gentle agitation. Samples were then rinsed in cacodylate buffer 3 times and subjected to a secondary fixation for 1 h in 1% osmium tetroxide/1.5% potassium ferrocyanide in cacodylate buffer. Following this, samples were rinsed in ultrapure water 3 times and pellets were enrobed in 2% ultra-low gelling temperature agarose (Sigma-Aldrich). Enrobed pellets were cut into smaller pieces and stained overnight in an aqueous solution of 1% uranyl acetate at 4 °C. Then, samples were washed in ultrapure water 3 times for 10 min in each step, dehydrated in a graded ethanol series (50%, 70%, 90%, 100% x3) for 10 min in each step, and infiltrated with microwave assistance (Pelco BioWave Pro, Redding, CA) into Epon resin. Samples were then cured in an oven at 60 °C for 72 h and post-curing, 70 nm thin sections were cut from the resin blocks, post-stained with uranyl acetate and Reynold’s lead and imaged on a Transmission Electron Microscope (JEOL JEM-1400 Plus, Tokyo, Japan) operating at 120 KeV.
2.4. Assessment of functional mitochondria by TMRE staining and FACS analysis
To assess the functional integrity of isolated mitochondria, TMRE (tetramethylrhodamine, ethyl ester) was used which is a red-orange dye that readily accumulates in active mitochondria. Mitochondria from three independent MSC cultures were isolated using the Mitochondria Isolation Kit (Milteny Biotec) as described in Section 2.2. Purified mitochondria from each time point were pooled to ensure sufficient numbers for analysis. Following the manufacturer’s instructions, mitochondria extracts were resuspended in 1 mL storage buffer and then incubated in 100 nM TMRE (ThermoFisher; T669) for 20 min on ice while protected from light. Stained mitochondria were centrifuged at 12,000g for 2 min and washed once in ice cold storage buffer. Unstained mitochondria were included as a control. Stained or unstained mitochondria pellets were resuspended in 1 mL of storage buffer and incubated on ice prior to analysis on a BD LSRFortessa™ X-20 Flow Cytometer (BD Biosciences). The flow cytometer sample flow rate was set to record at 1000 events per second. The population of unstained mitochondria and TMRE-stained mitochondria were analyzed using FlowJo (v10.7) software.
2.5. RNA isolation and RT-qPCR
RNA was extracted from whole cells or purified mitochondria using the Total RNA Purification Kit (Norgen) followed by DNase I treatment (1 U/μg RNA) at 37 °C for 15 min. RNA concentration was determined by a spectrophotometer (Nanodrop®; Thermo Scientific) and removal of DNA contamination was confirmed in the mitochondrial RNA samples by Bioanalyzer analysis. We analyzed expression of mt-COX1 and mt-ND1 in whole cells or mitochondria extracts as well as RUNX2, alkaline phosphatase (ALP) and osteocalcin (OCN) expression in whole cell extracts from osteogenic cultures to confirm differentiation. Isolated mRNAs were reverse-transcribed using Superscript RT II (Life Technologies), and qPCR was performed using PowerUp SYBR master mix (Life Technologies). PCR primer sequences are: COX1-Forward: 5’-CTCAGACGCTCAGGAAATAGAA-3′ and COX1-Reverse: 5’-TCGTTGACCTCGTCTGTTATG-3′; ND1-Forward: 5’-GAAGTCACCCTAGCCATCATTC-3′; ND1-Reverse: 5′-GCAGGAGTAATCAGAGGTGTTC; RUNX2-Forward: 5’-CATCACTGTCCTTTGGGAGTAG-3′; RUNX2-Reverse: 5’-ATGTCAAAGGCTGTCTGTAGG-3′; ALP-Forward: 5’-GGAGTATGAGAGTGACGAGAAAG-3′; ALP-Reverse: 5’-GAAGTGGGAGTGCTTGTATCT-3′; OCN-Forward: 5’-AGGAGGGCAGCGAGGTAG-3’ OCN-Reverse: 5’-GAAAGCCGATGTGGTCAGC-3′; PPIA-Forward: 5′-TCCTGGCATCTTGTCCATG-3′; PPIA-Reverse: 5’-CCATCCAACCACTCAGTCTTG-3′.
To attempt to validate expression profiles of specific miRNAs in the same mitochondria and whole cell extract samples used for array analysis, TaqMan miRNA assays, containing an RT primer and a TaqMan probe and primer set specific for amplification of a miRNA of interest, were used with the TaqMan miRNA reverse transcription kit and TaqMan master mix with no UNG following the manufacturer’s instructions (ThermoFisher). The following TaqMan assays were utilized to amplify: miR-320a (assay ID #002277), RNU44 (assay ID # 001094), miR-4485-3p (assay ID #462832), miR-26a-5p (assay ID #000405), miR-20a-5p (assay ID #000580), miR-181a-5p (assay ID #000480), miR-138-5p (assay ID #002284). RNU44 was used to normalize miRNA expression in whole cell extracts. Given RNU44 is not present to any significant degree in mitochondria, we utilized miR-320a to normalize mitomiR expression in mitochondrial extracts. This miRNA was found to be abundantly expressed in mitochondrial extracts and its expression did not significantly change at each time point during osteogenesis. A Student’s t-test was used to determine significant changes in expression of selected miRNAs at day 7 or day 14 when compared only to day 0 samples.
2.6. Western blot
Lysates were prepared from whole cells or purified mitochondria using RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton-X-100, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with 1× cOmplete protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Pierce). Total protein concentration was measured using the Bio-Rad Protein Assay kit. Proteins were resolved by SDS-PAGE and transferred onto a PVDF membrane. Protein-containing membranes were blocked with Odyssey Blocking Buffer (Licor) containing 0.1% Tween 20. Western blots were performed with either rabbit polyclonal anti-RUNX2 (Cell Signaling Technology #8486), monoclonal anti-COX1 (mitochondria-specific protein marker; Abcam, ab14705) or the monoclonal antibody KDEL (Lys-Asp-Glu-Leu: an ER-specific peptide marker; Abcam, ab12223). To normalize RUNX2 expression from whole cell lysates, mouse anti-β-actin antibodies were used (Abcam, ab6267). All primary antibodies were used at a 1:1000 dilution. Secondary antibodies (IRDye® 800CW-labeled anti-rabbit; IRDye® 680RD-labeled anti-mouse) (Licor) were used at a 1:10000 dilution following the manufacturer’s recommendation. Resulting protein bands were imaged and quantified using the Licor Odyssey software. To confirm successful purification of mitochondria, the ratio of COX1: KDEL was analyzed in lysates from whole cells and purified mitochondria samples.
2.7. MicroRNA array and data analysis
Total RNA extracted from purified mitochondria (400 ng) or whole cell lysates (500 ng) of MSCs or osteogenic-induced MSCs, was labeled with the Flashtag™ Biotin HSR RNA labeling kit (ThermoFisher Scientific) following the manufacturer’s instructions. Labeled RNA was hybridized at 48 °C for 42 h at 60 rpm on an Affymetrix GeneChip™ human miRNA 4.0 array (ThermoFisher Scientific). This array contains 2578 human mature miRNA probe sets and 2025 human pre-miRNA (stem-loop) probe sets. Probe sets (a total of 1996) to determine the presence of human snoRNA and scaRNAs are also included in this array. GeneChips were scanned using the Affymetrix GeneChip scanner G3000 7G with standard setting to capture signal intensities for the miRNAs. The raw intensity data was imported into Affymetrix Expression Console software (v1.4.1.46) for signal pre-processing including background correction utilizing the robust multi-array average (RMA) algorithm, median polish summarization from probe to probe set level of signal values, and the quantile method to normalize across multiple arrays. A detection call on the strength of miRNA signal was made using the Affymetrix “Detection Above Background” (DABG) algorithm, which generates a p-value for signal above background probability. Only those miRNA probe sets with a “present” call at p < 0.05 in at least 2 of 3 replicates at any of the 4 time points (day 0, 3, 7, and 14) were kept for further analysis. The normalized log2 intensity values were analyzed for differential expression between the different timepoints using the R software “limma” package which uses a moderated t-test with linear modeling and empirical Bayes statistics [24]. The significance for differential expression was set at fold change absolute ≥ 1.5 and adjusted p < 0.05. The nominal p-values were subjected to the multi-test correction false discovery rate procedure using the Benjamini-Hochberg method [25]. Note that all miRNAs listed in each Table within the main body of this manuscript were cross-checked on the miRBase website to confirm that they are currently defined as miRNAs and have not been removed from the database as “dead entries”. Raw data files were submitted to the GEO data depository (online at https://www.ncbi.nlm.nih.gov/geo/) and assigned the accession codes: GSE134946 (mitochondria-associated miRNAs) and GSE148049 (whole cell extract miRNAs).
3. Results
3.1. Confirmation of osteogenic differentiation and purification of functional mitochondria from osteogenic-induced MSC cultures
We routinely carry out in vitro osteogenesis of primary human MSC lines and always achieve successful differentiation as determined by the generation of a mineralized extracellular matrix in addition to expression of key osteoblast markers such as transcription factor RUNX2. Fig. 1A shows Alizarin Red stained MSC cultures following 3 weeks of osteogenic induction and Fig. 1B shows an increase in RUNX2 protein expression at days 3, 7 and 14 of osteogenesis when compared to control non-induced cultures. This data corresponds to the human MSC “Sample 1” cell line (MSC-S1) and similar results were found for Sample 2 and Sample 3 MSC lines that were also used for the miRNA array study (results not shown). Fig. 1C–D shows increased gene expression of RUNX2 and alkaline phosphatase (ALP) over time during osteogenesis and Fig. 1E shows increased expression of osteocalcin (OCN) at day 14 only. Note that we selected days 3, 7, and 14 for mitomiR analysis to attempt to determine potential changes in expression levels during early commitment of MSCs (day 3) as well as at stages where cells are committed toward the osteoblast lineage (days 7 and 14). At these two later time points, we always observe a distinctive change in cell phenotype indicating successful differentiation (results not shown).
Fig. 1.
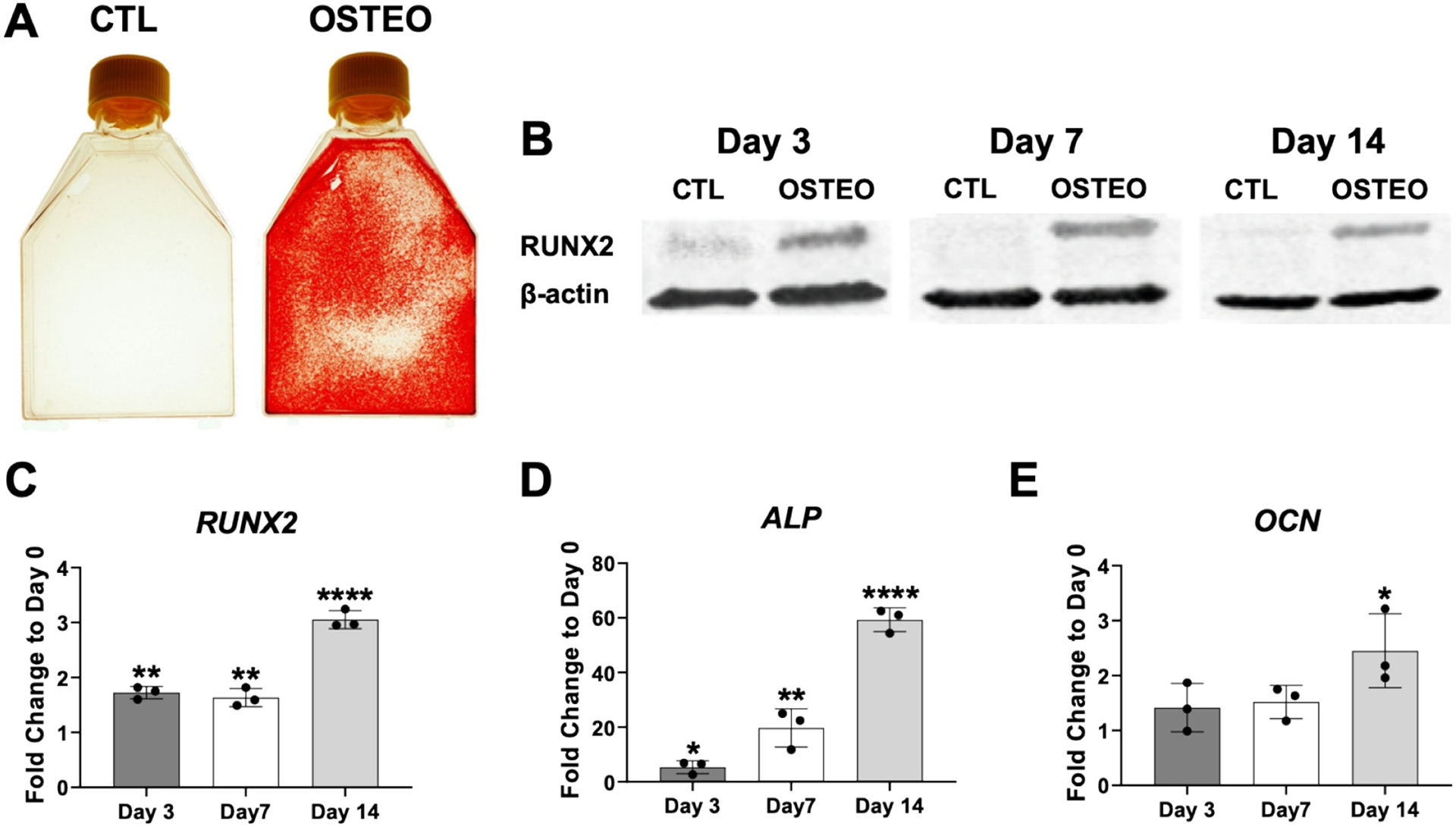
Confirmation of osteogenic differentiation. Panel (A) shows positive Alizarin Red staining following osteogenic differentiation (OSTEO) of human bone marrow-derived MSCs at 3 weeks. No staining was detected in control (CTL) MSC cultures grown at the same time in regular growth medium. Panel (B) shows increased levels of RUNX2 in protein lysates harvested from MSCs at days 3, 7 and 14 of osteogenic induction when compared to control, non-induced cultures. Panels C-E show expression patterns for RUNX2, alkaline phosphatase (ALP) and osteocalcin (OCN) at different time points during osteogenesis (fold change expression relative to day 0). Data in A, B is from MSC-Sample 1 (MSC-S1) cell line that was used in the microarray study. Similar results were shown for MSC-S2 and MSC-S3 samples (results not shown). Data in C-E is from the three MSC samples used for the microarray (data expressed +/− SD, n = 3; *p < 0.05; ** p < 0.01; **** p < 0.0001).
To confirm efficient purification of mitochondria from MSCs, qPCR was carried out to determine the expression of two genes derived from the human mitochondrial genome, COX1 and ND1. In addition, Western blotting was carried out to determine the expression of COX1 protein as well as the ER-specific peptide, KDEL, in whole cell lysates and mitochondria extracts. Fig. 2A, B shows enrichment of COX1 and ND1 expression in RNA samples derived from purified mitochondria. We used PPIA as a normalizing gene here since it has been found to be associated with mitochondria [26] and Ct values were fairly similar in RNA samples from both mitochondria (Ct ~23) and whole cell lysates Ct ~ 21). Fig. 2C shows an enrichment in COX1 protein expression in mitochondria-derived protein lysates compared to whole cell lysates. Fig. 2D shows expression of KDEL in whole cell lysates whereas levels of this ER-specific peptide were almost undetectable in the mitochondrial extracts. Note that we could not use common normalizing proteins (e.g. β-actin, GAPDH, tubulin) since their expression levels are substantially different between lysates of whole cells and purified mitochondria. Instead, we ensured loading of equal amounts of total protein per lane. Taken together, our data demonstrates that we were able to obtain highly purified mitochondrial extracts from MSC cultures for subsequent downstream expression analysis of RNA and protein.
Fig. 2.
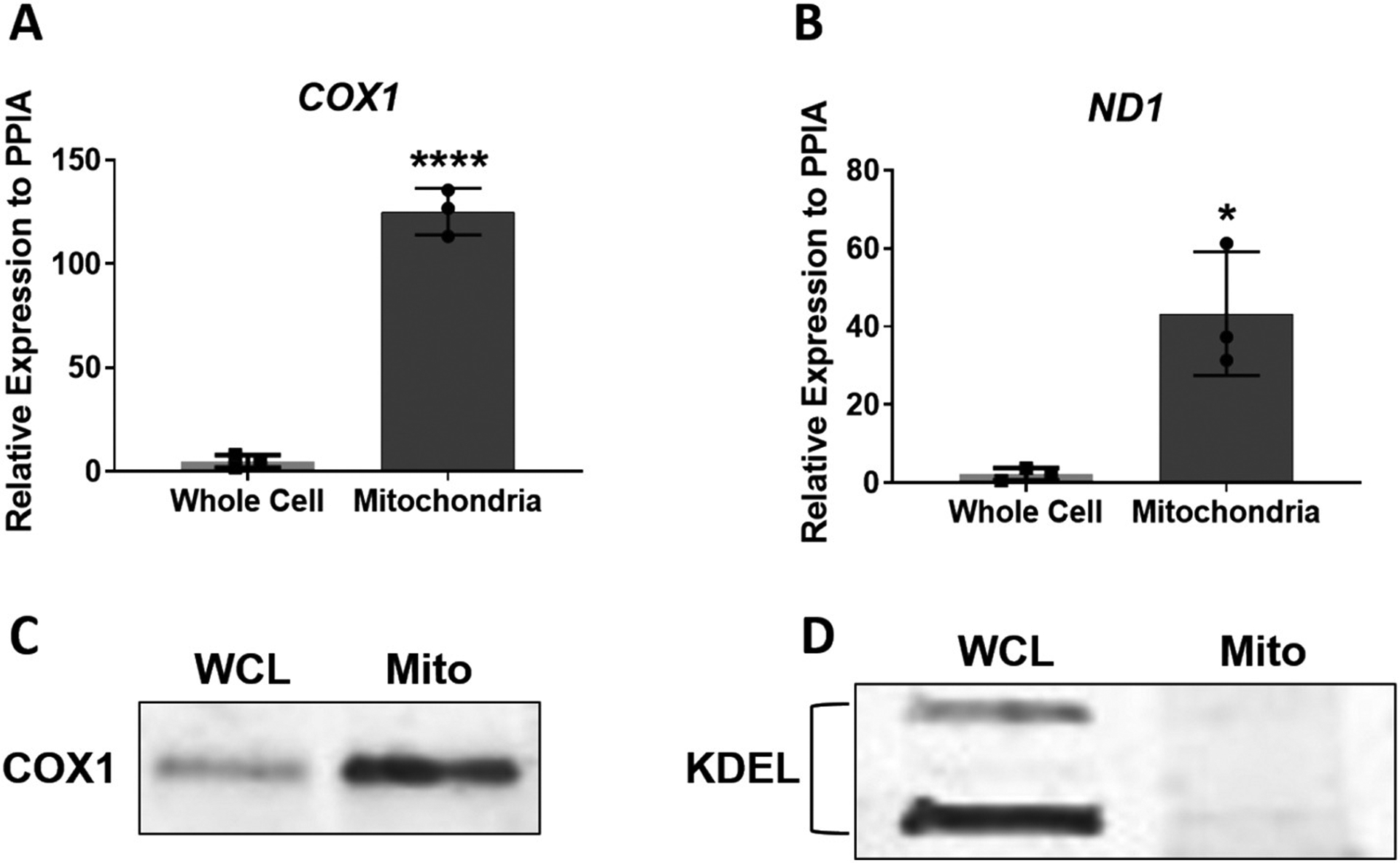
Analysis of purified mitochondria extracts from MSCs. Panels (A) and (B) show enrichment in expression of two mitochondria-derived genes, COX1 and ND1, in purified MSC mitochondria extracts when compared to whole cell extracts. Panel (C) shows enriched expression of COX1 protein in mitochondrial extracts (Mito) compared to whole cell protein lysates (WCL). Panel (D) shows expression of the ER-specific peptide, KDEL, in whole cell lysates, but not in protein lysates isolated from purified MSC mitochondria extracts. Data in (A) and (B) are expressed ± SD; n = 3. *p < 0.05; ****p < 0.0001.
In addition, transmission electron microscopy (TEM) was carried out to analyze the ultrastructure of mitochondria in our purified preparations. Representative images in Supplemental Fig. 1 shows an abundance of normal, healthy-looking mitochondria in MSCs at day 0, day 7, and day 14 of osteogenic induction, as indicated by the presence of highly folded inner membrane cristae structures. These images also show some mitochondria that appear “swollen” which can occur during the purification process as well as additional processing required for EM analysis. However, addition of TMRE to our purified mitochondria extracts followed by FACS analysis showed that, at all time points analyzed, the majority of mitochondria stained positive for TMRE, indicating functionally active mitochondria (Fig. 3).
Fig. 3.
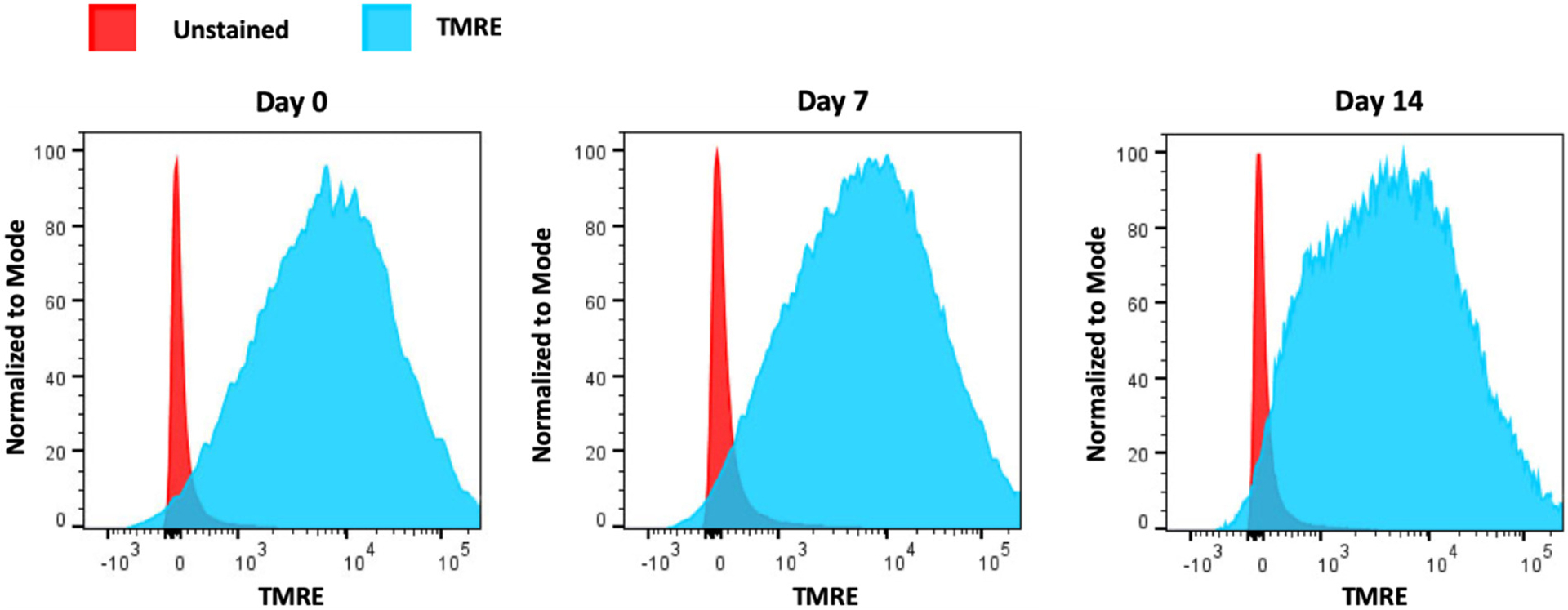
Functional integrity assessment of purified mitochondria harvested from MSCs. Three independent cultures of MSCs were utilized and mitochondria were purified from cells at day 0 or at day 7 and 14 following osteogenic induction. Mitochondria from each time point were pooled and stained (or not) with TMRE. Histograms from FACS analysis revealed the distribution of unstained (red) and TMRE-stained (blue) mitochondria. Utilizing corresponding FACS analysis software, we calculated that approximately 92%, 90% and 86% of mitochondria stained positive for TMRE in day 0, day 7 or day 14 samples, respectively.
3.2. Quality control analysis of RNA and corresponding microarray data
RNA obtained from purified mitochondria (mtRNA) had an OD A260/A280 reading within the range 1.93–2.03 and RNA obtained from whole cells extracts (wcRNA) had an OD A260/A280 within the range 1.89–1.95. Bioanalyzer data showed that DNA contamination was successfully removed from the mtRNA samples (Supplemental Fig. 2). It should be noted that RNA Integrity Number (RIN) values obtained from the Bioanalyzer are not informative for mitochondrial RNA analysis. RIN values are calculated primarily by the detection of cytoplasmic 28S and 18S rRNA, which are not present in the mitochondria. Only two rRNA species (12S and 16S) are encoded by the mitochondrial genome [27]. The fact that no peaks corresponding to 28S or 18S RNA were observed (Supplemental Fig. 2) further demonstrates the purity of our mitochondria extracts. However, peaks corresponding to 28S and 18S were detected in wcRNA samples (Supplemental Fig. 3) and RIN values ranged from ~5–7. Lower RIN values were typically observed for day 14 wcRNA samples most likely due to the presence of a mineralized extracellular matrix in osteogenic cultures at this time point, which likely negatively affects the RNA isolation process.
Quality control analysis showed that, overall, the miRNA arrays were performed well and generated reliable data. The five vendor spike-in control probes all had a signal above 1000 units, which passed the vendor-defined QC cut-off (results not shown). Supplemental Figs. 4–5 shows that, on average, strong signals were obtained for each sample (blue bars) when compared to background chip readings (grey bars). These signal values were used to calculate the average signal-to-noise ratio (SNR) for each sample (red dots). SNRs ranged from 1.89–2.51 (mtRNA) and 2.08–2.75 (wcRNA), which we consider translates to a strong chip signal. Taking into account these signal intensity readings, approximately 13% - 22% of human miRNA probe sets were detectable on the chips (orange bars) for both mtRNA and wcRNA. Note that RNA analyses and quality control assessment identified MSC-S1 day 14 wcRNA sample as an outlier.
3.3. Expression signatures of miRNAs and pre-miRNAs in purified mitochondria extracts and whole cell extracts from MSCs during osteogenesis
Based on the Affymetrix “detection above background” algorithm (see Section 2.5), Supplemental Table 1 shows the entire list of detectable mature mitomiRs in MSCs prior to differentiation (day 0; 574 miRNAs) and in MSCs at day 3, 7, or 14 of osteogenic induction (day 3: 627 miRNAs; day 7: 372 miRNAs; day 14: 489 miRNAs). From these lists, Table 1 shows the top 30 most abundant mature miRNAs associated with the mitochondria in non-induced MSCs and osteogenic-induced MSCs. Among these are members of the let-7 family and, specifically, let-7b-5p was found to be the most abundantly expressed mitomiR in MSCs at all time points analyzed. Other highly-expressed mitomiRs expressed in MSCs at each time point include the miR-320 paralogs (miR-320a/b/c) as well as other less commonly-reported miRNAs: miR-3175, miR-3960, miR-6089, miR-6090.
Table 1.
Top 30 most abundant mitomiRs expressed in MSCs at day 0, 3, 7 and 14 of osteogenic induction.
| Day 0 | Day 3 | Day 7 | Day 14 | ||||
|---|---|---|---|---|---|---|---|
| Transcript ID | Mean signal | Transcript ID | Mean signal | Transcript ID | Mean signal | Transcript ID | Mean signal |
| hsa-let-7b-5p | 14.19 | hsa-let-7b-5p | 13.71 | hsa-let-7b-5p | 12.54 | hsa-let-7b-5p | 13.91 |
| hsa-miR-24-3p | 12.95 | hsa-miR-24-3p | 12.59 | hsa-miR-3960 | 11.88 | hsa-let-7c-5p | 12.25 |
| hsa-let-7c-5p | 12.38 | hsa-let-7c-5p | 11.87 | hsa-miR-6090 | 11.49 | hsa-miR-4449 | 11.45 |
| hsa-miR-4449 | 11.80 | hsa-miR-221-3p | 11.71 | hsa-miR-6089 | 11.46 | hsa-miR-3960 | 11.34 |
| hsa-miR-221-3p | 11.64 | hsa-miR-4449 | 11.60 | hsa-miR-320a | 11.33 | hsa-miR-320a | 11.27 |
| hsa-let-7a-5p | 11.40 | hsa-miR-23a-3p | 11.37 | hsa-miR-320b | 11.03 | hsa-miR-221-3p | 11.27 |
| hsa-miR-320a | 11.19 | hsa-let-7a-5p | 11.09 | hsa-miR-3665 | 11.00 | hsa-miR-4497 | 11.16 |
| hsa-miR-23a-3p | 11.18 | hsa-miR-145-5p | 11.00 | hsa-miR-3196 | 10.87 | hsa-miR-24-3p | 11.14 |
| hsa-miR-3960 | 11.01 | hsa-miR-320a | 10.99 | hsa-miR-2115-5p | 10.81 | hsa-let-7a-5p | 11.03 |
| hsa-miR-320b | 10.97 | hsa-miR-125b-5p | 10.96 | hsa-miR-4497 | 10.78 | hsa-miR-320b | 11.00 |
| hsa-miR-3196 | 10.90 | hsa-miR-320b | 10.81 | hsa-miR-8069 | 10.77 | hsa-miR-6090 | 10.92 |
| hsa-miR-3651 | 10.88 | hsa-miR-100-5p | 10.78 | hsa-miR-4488 | 10.69 | hsa-miR-6089 | 10.91 |
| hsa-miR-125b-5p | 10.82 | hsa-miR-222-3p | 10.69 | hsa-miR-4466 | 10.63 | hsa-miR-145-5p | 10.64 |
| hsa-miR-6090 | 10.75 | hsa-miR-23b-3p | 10.68 | hsa-miR-3175 | 10.62 | hsa-miR-3196 | 10.62 |
| hsa-miR-100-5p | 10.73 | hsa-miR-3175 | 10.55 | hsa-let-7c-5p | 10.50 | hsa-miR-3665 | 10.56 |
| hsa-miR-6089 | 10.64 | hsa-let-7e-5p | 10.52 | hsa-miR-4508 | 10.42 | hsa-miR-320c | 10.48 |
| hsa-miR-3175 | 10.63 | hsa-miR-103a-3p | 10.44 | hsa-miR-320c | 10.30 | hsa-miR-7704 | 10.32 |
| hsa-let-7e-5p | 10.58 | hsa-miR-320c | 10.34 | hsa-miR-5787 | 10.28 | hsa-miR-125b-5p | 10.32 |
| hsa-miR-222-3p | 10.44 | hsa-miR-26a-5p | 10.29 | hsa-miR-1237-5p | 10.28 | hsa-miR-8069 | 10.22 |
| hsa-miR-23b-3p | 10.32 | hsa-miR-3960 | 10.13 | hsa-miR-762 | 10.25 | hsa-miR-4488 | 10.19 |
| hsa-miR-4497 | 10.31 | hsa-miR-3651 | 10.03 | hsa-miR-6088 | 10.24 | hsa-miR-3175 | 10.03 |
| hsa-miR-145-5p | 10.30 | hsa-miR-191-5p | 9.97 | hsa-miR-4787-5p | 10.18 | hsa-miR-23a-3p | 10.00 |
| hsa-let-7i-5p | 10.25 | hsa-miR-6090 | 9.94 | hsa-miR-7704 | 10.15 | hsa-miR-4466 | 9.99 |
| hsa-miR-103a-3p | 10.11 | hsa-miR-214-3p | 9.85 | hsa-miR-6125 | 10.14 | hsa-miR-100-5p | 9.94 |
| hsa-miR-320c | 10.09 | hsa-let-7d-5p | 9.83 | hsa-miR-6729-5p | 10.10 | hsa-miR-4508 | 9.94 |
| hsa-miR-26a-5p | 10.08 | hsa-miR-7704 | 9.79 | hsa-miR-4516 | 10.09 | hsa-miR-4787-5p | 9.93 |
| hsa-miR-3665 | 10.06 | hsa-miR-22-3p | 9.79 | hsa-miR-8072 | 10.04 | hsa-miR-5787 | 9.91 |
| hsa-miR-8069 | 9.95 | hsa-miR-6089 | 9.76 | hsa-miR-6775-5p | 9.96 | hsa-let-7e-5p | 9.87 |
| hsa-miR-214-3p | 9.94 | hsa-let-7i-5p | 9.71 | hsa-miR-6727-5p | 9.95 | hsa-miR-2115-5p | 9.83 |
| hsa-miR-4454 | 9.86 | hsa-miR-31-5p | 9.66 | hsa-miR-6803-5p | 9.75 | hsa-miR-1237-5p | 9.82 |
Note: miRNAs highlighted in bold font have been identified only in humans (according to the current miRBase database: www.mirbase.org).
Supplemental Table 2 shows the entire list of detectable mature miRNAs in whole cell extracts of MSCs at day 0 (595 miRNAs), day 3 (621 miRNAs), day 7 (570 miRNAs) and day 14 (734 miRNAs). Note that substantially more miRNAs were detected in whole cell extracts from MSCs at day 7 and 14 of osteogenic differentiation compared to MSC mitochondrial extracts at the same time points. Table 2 shows the top 30 most abundant mature miRNAs in MSC whole cell extracts prior to and during osteogenic induction. Similar to abundant miRNAs identified in mitochondria extracts (Table 1), members of the let-7 family were also among those most abundantly expressed in MSC whole cell extracts at each time point, with let-7b-5p again being the most abundant (Table 2). Other aforementioned miRNAs (except miR-3175) that were abundant in MSC mitochondrial extracts at each time point (i.e. miR-320 members, miR-3960, miR-6089, miR-6090) were also found in the top 30 list at each time point in the whole cell extracts. Table 2 shows that, compared to mitomiRs, there are many more abundantly-expressed miRNAs from whole cell extracts that are common to all time points (e.g. miR-23b-3p, miR-24-3p, miR-26a-5p, miR-125b-5p, miR-145-5p, miR-214-3p, miR-221-3p, miR-222-3p, miR-4497). As will be discussed more in the following section, there were more substantial changes in mitomiR expression profiles at day 7 or day 14 of osteogenic induction compared to day 0. This explains why the list of the top 30 highest expressed mitomiRs is quite different at these time points compared to that shown for day 0 non-induced MSCs (Table 1).
Table 2.
Top 30 most abundant miRNAs in MSC whole cell extracts at day 0, 3, 7 and 14 of osteogenic induction.
| Day 0 | Day 3 | Day 7 | Day 14 | ||||
|---|---|---|---|---|---|---|---|
| Transcript ID | Mean signal | Transcript ID | Mean signal | Transcript ID | Mean signal | Transcript ID | Mean signal |
| hsa-let-7b-5p | 13.92 | hsa-let-7b-5p | 14.13 | hsa-let-7b-5p | 14.19 | hsa-let-7b-5p | 14.52 |
| hsa-let-7c-5p | 12.83 | hsa-let-7c-5p | 13.25 | hsa-let-7c-5p | 13.30 | hsa-let-7c-5p | 13.48 |
| hsa-miR-24-3p | 12.61 | hsa-let-7a-5p | 12.38 | hsa-let-7a-5p | 12.66 | hsa-let-7a-5p | 13.19 |
| hsa-miR-23a-3p | 12.48 | hsa-miR-24-3p | 12.25 | hsa-miR-320a | 12.33 | hsa-miR-24-3p | 12.86 |
| hsa-let-7a-5p | 12.29 | hsa-miR-320a | 12.23 | hsa-miR-320b | 12.14 | hsa-miR-320a | 12.59 |
| hsa-miR-222-3p | 12.25 | hsa-miR-320b | 12.09 | hsa-miR-24-3p | 12.05 | hsa-miR-320b | 12.45 |
| hsa-miR-125b-5p | 12.18 | hsa-miR-3960 | 12.09 | hsa-miR-125b-5p | 12.01 | hsa-let-7e-5p | 12.25 |
| hsa-miR-320a | 11.98 | hsa-miR-125b-5p | 12.09 | hsa-miR-3960 | 11.93 | hsa-miR-23a-3p | 12.16 |
| hsa-miR-23b-3p | 11.85 | hsa-miR-23a-3p | 12.01 | hsa-miR-4497 | 11.88 | hsa-miR-320c | 12.04 |
| hsa-miR-320b | 11.82 | hsa-let-7e-5p | 11.84 | hsa-let-7e-5p | 11.83 | hsa-miR-221-3p | 11.84 |
| hsa-let-7e-5p | 11.82 | hsa-miR-320c | 11.75 | hsa-miR-145-5p | 11.81 | hsa-miR-26a-5p | 11.83 |
| hsa-miR-3960 | 11.77 | hsa-miR-4497 | 11.68 | hsa-miR-320c | 11.81 | hsa-let-7d-5p | 11.82 |
| hsa-miR-221-3p | 11.63 | hsa-miR-222-3p | 11.65 | hsa-miR-23a-3p | 11.63 | hsa-miR-125b-5p | 11.76 |
| hsa-miR-100-5p | 11.61 | hsa-miR-23b-3p | 11.64 | hsa-miR-23b-3p | 11.49 | hsa-miR-23b-3p | 11.67 |
| hsa-miR-320c | 11.50 | hsa-miR-6869-5p | 11.46 | hsa-miR-7704 | 11.42 | hsa-miR-222-3p | 11.53 |
| hsa-miR-6869-5p | 11.39 | hsa-miR-145-5p | 11.44 | hsa-miR-222-3p | 11.40 | hsa-miR-3960 | 11.42 |
| hsa-miR-4497 | 11.34 | hsa-miR-3665 | 11.30 | hsa-miR-6089 | 11.37 | hsa-miR-214-3p | 11.31 |
| hsa-miR-7704 | 11.28 | hsa-miR-221-3p | 11.27 | hsa-miR-221-3p | 11.29 | hsa-miR-6089 | 11.18 |
| hsa-miR-145-5p | 11.16 | hsa-miR-6090 | 11.21 | hsa-miR-6090 | 11.21 | hsa-miR-145-5p | 11.06 |
| hsa-miR-26a-5p | 11.14 | hsa-miR-26a-5p | 11.19 | hsa-miR-3665 | 11.21 | hsa-miR-6090 | 11.04 |
| hsa-miR-3665 | 11.13 | hsa-miR-7704 | 11.18 | hsa-miR-26a-5p | 11.12 | hsa-miR-103a-3p | 10.79 |
| hsa-miR-31-5p | 11.07 | hsa-miR-6089 | 11.17 | hsa-let-7d-5p | 11.10 | hsa-miR-3665 | 10.79 |
| hsa-miR-6090 | 11.05 | hsa-let-7d-5p | 10.80 | hsa-miR-193b-3p | 11.09 | hsa-miR-7704 | 10.72 |
| hsa-miR-6089 | 10.99 | hsa-miR-193a-5p | 10.71 | hsa-miR-6869-5p | 11.05 | hsa-miR-100-5p | 10.68 |
| hsa-miR-193a-5p | 10.76 | hsa-miR-4787-5p | 10.63 | hsa-miR-181a-5p | 10.81 | hsa-miR-191-5p | 10.64 |
| hsa-let-7d-5p | 10.72 | hsa-miR-125a-5p | 10.61 | hsa-miR-92a-3p | 10.69 | hsa-let-7i-5p | 10.61 |
| hsa-miR-125a-5p | 10.69 | hsa-miR-214-3p | 10.60 | hsa-miR-214-3p | 10.61 | hsa-miR-199a-3p | 10.58 |
| hsa-miR-92a-3p | 10.67 | hsa-miR-181a-5p | 10.59 | hsa-miR-4787-5p | 10.56 | hsa-miR-199b-3p | 10.58 |
| hsa-miR-214-3p | 10.44 | hsa-miR-100-5p | 10.58 | hsa-miR-574-3p | 10.33 | hsa-miR-4497 | 10.52 |
| hsa-miR-4787-5p | 10.42 | hsa-miR-193b-3p | 10.57 | hsa-miR-193a-5p | 10.21 | hsa-miR-92a-3p | 10.47 |
Note: miRNAs highlighted in bold font have been identified only in humans (according to the current miRBase database: www.mirbase.org).
Supplemental Table 3 lists all detectable stem-loop precursor miRNAs (pre-miRNAs) from mitochondrial extracts of MSCs at each time point (day 0: 149; day 3: 183; day 7: 97; day 14: 118). In general, the average signal intensity of detectable pre-miRNAs was lower than that observed for mature miRNAs. While mature let-7b-5p was highly expressed in all mitochondria extracts at each time point (Table 1), let-7b pre-miRNA was expressed at relatively low levels. In addition, while pre-miR-711 was found to be one of the most highly expressed pre-miRNAs in MSC mitochondria at each time point, expression of the mature miR-711 was only detected at low levels in mitochondria extracts from day 3 induced MSCs (Supplemental Table 1). However, some of the more highly expressed pre-miRNAs observed in all samples did appear to correlate with abundant expression of the corresponding mature miRNA in MSC mitochondria including pre-miR-320a, pre-miR-6089 and pre-miR-4449. Supplemental Table 4 shows all detectable pre-miRNAs in whole cell MSC extracts at each time point (day 0: 204; day 3: 187; day 7: 169; day 14: 265). Some of the most abundant stem-loop RNAs detected at each time point were also the most abundantly expressed at all time points in mitochondrial extracts as shown in Supplemental Table 3 (i.e. pre-miR-320a, 6089, 6800 and 6880). In addition, pre-miR-320e appeared to be highly expressed at all time points in both mitochondria and whole cell extracts, whereas mature miRNA levels of this miR-320 paralog was relatively low (Supplemental Tables 1 and 2). In terms of total numbers detected, substantially fewer mitochondria-derived pre-miRNAs were found at day 7 and day 14 when compared to numbers detected in whole cell extracts at the same time points. A similar trend was also found for the total number of mature mitochondria-associated miRNAs detected at day 7 or day 14 when compared to whole cell extracts.
3.4. Differentially expressed miRNAs in purified mitochondria extracts and whole cells extracts from MSCs during osteogenesis
Based on our cut-off parameters for differential expression (fold change ≥1.5; adjusted p value <0.05), (see Section 2.5), we did not detect any significant changes in mitomiR expression in MSCs at day 3 of osteogenic induction when compared to day 0 (Supplemental Fig. 6). However, compared to day 0, many mitomiRs were found to be expressed at significantly higher or lower levels at day 7 and day 14 time points, but particularly at day 7 (Supplemental Fig. 6). Hierarchical cluster analysis also revealed distinct subsets of mitomiRs displaying different expression profiles in MSCs at day 7 and 14 of osteogenesis when compared to day 0 or day 3 time points (Fig. 4). Differentially-expressed miRNAs from whole cell extracts were detected at days 3, 7 and 14 of osteogenic induction compared to day 0, but particularly at day 14 (Supplemental Fig. 7 and Fig. 5).
Fig. 4.
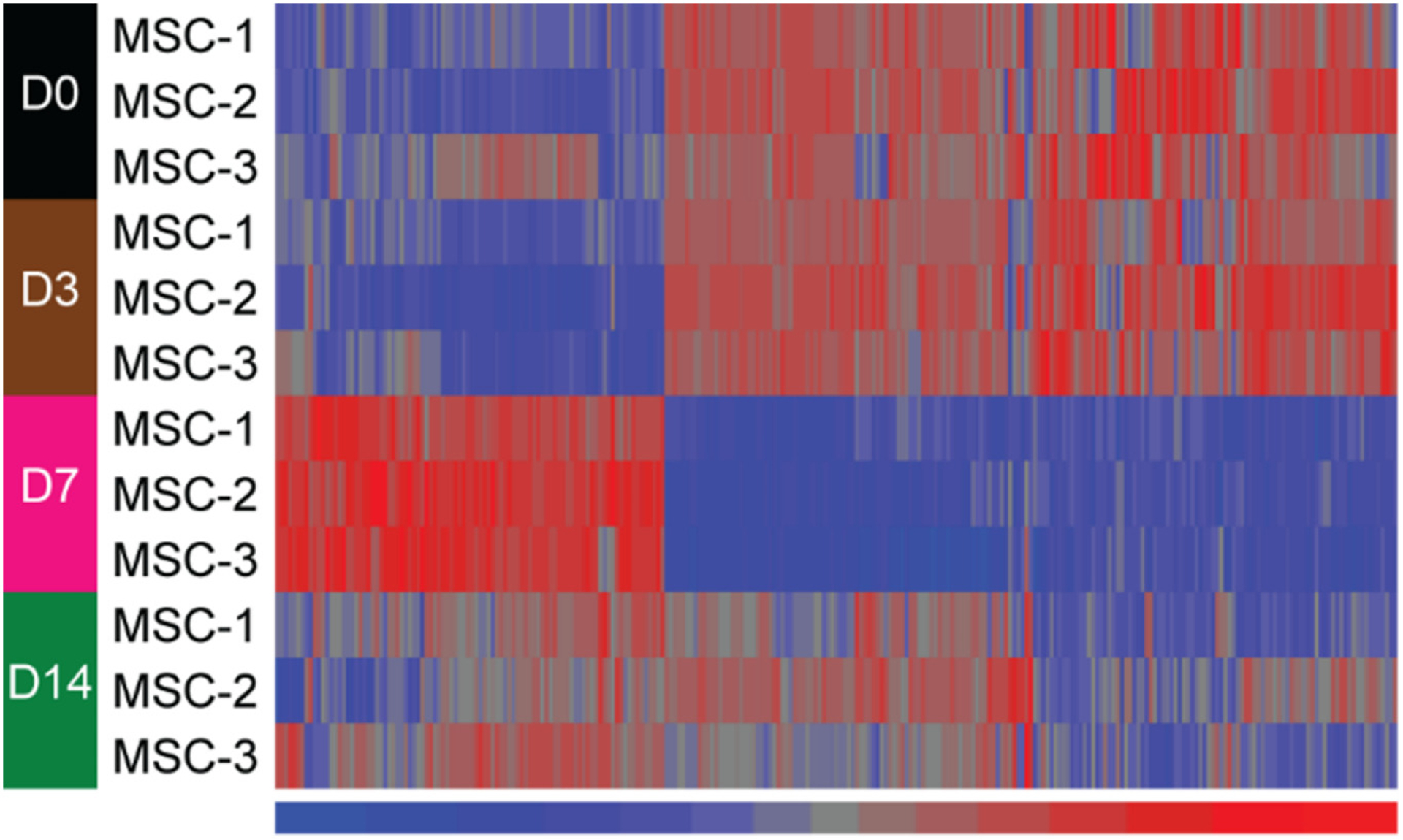
Heat map of 271 differentially-expressed mitomiRs from comparisons between day 0 and either day 7 or day 14. Significant differential expression was set at fold change absolute ≥ 1.5 and adjusted p value <0.05. These miRNAs were clustered in an unsupervised manner with Euclidean distance and Ward method. Signal values in the heat map were z-score normalized with a mean of 0 and a standard deviation of 1 across the miRNAs. Red color denotes higher signal and blue color represents lower signal. Note that there are more differentially expressed mitomiRs at the day 7 time point when compared to day 0.
Fig. 5.
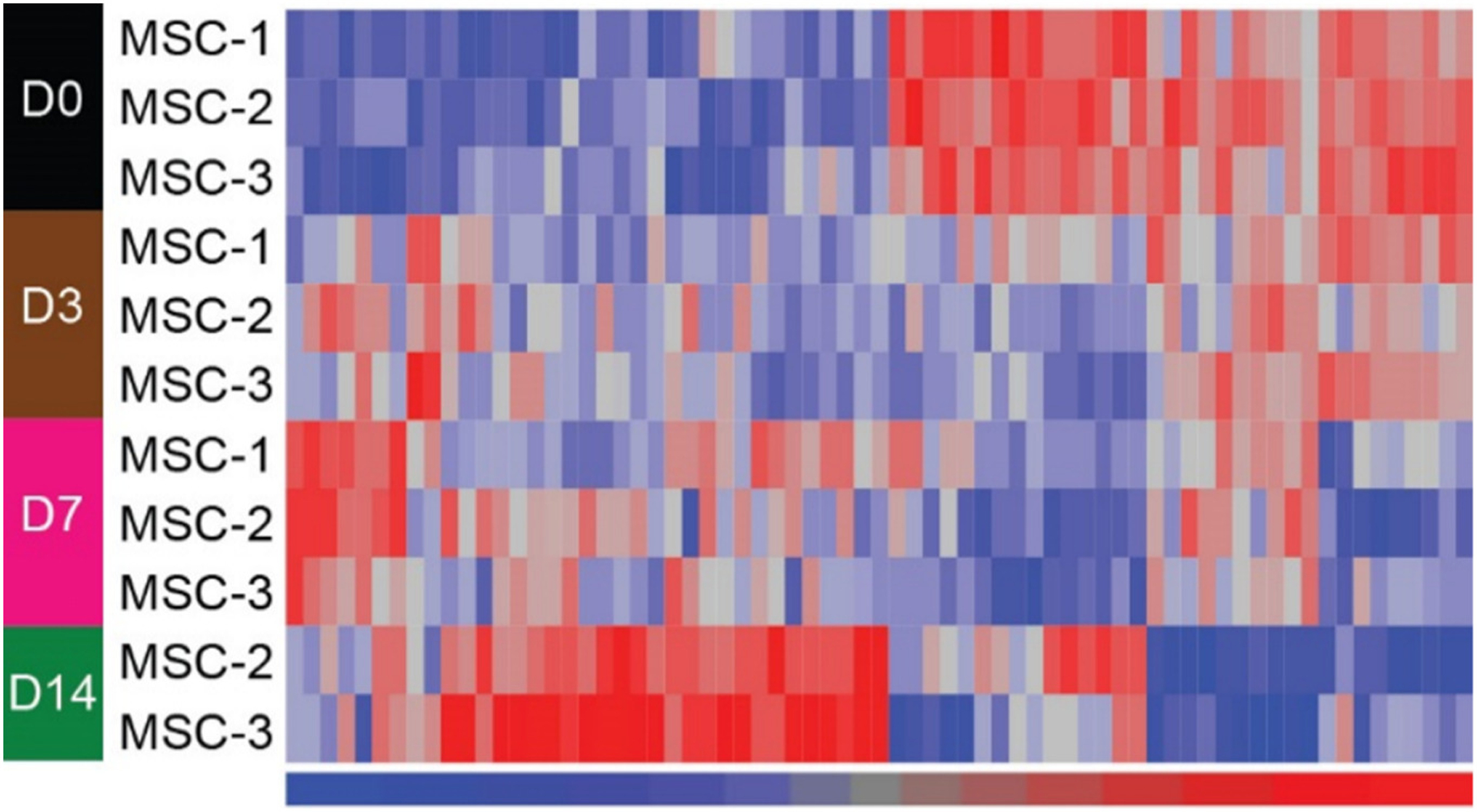
Heat map of differentially-expressed miRNAs from whole cell extracts following comparisons between day 0 and days 3, 7 or 14. Significant differential expression was set at fold change absolute ≥ 1.5 and adjusted p value <0.05. miRNAs were clustered in an unsupervised manner with Euclidean distance and Ward method. Signal values in the heat map were z-score normalized with a mean of 0 and a standard deviation of 1 across the miRNAs. Red color denotes higher signal and blue color represents lower signal. Note there are two samples for MSC day 14 since one was an outlier. Also note that there are more differentially expressed miRNAs identified at day 14 compared to day 0.
Supplemental Table 5 lists a total of 91 mitomiRs that were found to be increased at day 7, and 175 mitomiRs that were decreased at day 7 compared to day 0. From this data, Table 3 lists the top 30 most differentially-expressed mitomiRs at day 7. Table 4 shows that, overall, only 8 mitomiRs were expressed at significantly higher levels at day 14, and 30 mitomiRs were expressed at significantly lower levels at day 14 when compared to day 0. Common mitomiRs with increased expression at both the day 7 and day 14 time points were miR-4485, miR-483-5p, miR-4484, miR-4492, miR-4508 and miR-3620-5p (Tables 3, 4). Increased mitomiRs unique to the day 14 time point were miR-4646-5p and miR-615-3p. While the majority of decreased mitomiRs were unique to either the day 7 or day 14 time point, some were found to be reduced at both time points (i.e. miR-16-5p, miR-199a/b-3p, miR-31-5p, miR- 29a-3p, miR-20a-5p, miR-130a-3p, miR-21-5p) (Tables 3, 4).
Table 3.
Top 30 mitomiRs showing significantly increased or decreased expression levels in MSCs at day 7 of osteogenic differentiation. Fold change expression levels relative to day 0 are shown in addition to the Benjamini-Hochberg adjusted P values.
| Increased miRNA (day 7 vs day 0) | Decreased miRNA (day 7 vs day 0) | ||||
|---|---|---|---|---|---|
| Transcript ID | Fold change | P value | Transcript ID | Fold change | P value |
| hsa-miR-6893-5p | 21.76 | 6.91E-05 | hsa-miR-26a-5p | 343.26 | 1.28E-06 |
| hsa-miR-483-5p | 5.59 | 0.001 | hsa-miR-16-5p | 334.65 | 3.41E-07 |
| hsa-miR-4485 | 5.16 | 0.001 | hsa-miR-23b-3p | 314.70 | 4.62E-06 |
| hsa-miR-4484 | 4.48 | 0.001 | hsa-miR-199a-3p | 226.65 | 3.41E-07 |
| hsa-miR-4492 | 4.29 | 1.6E-04 | hsa-miR-199b-3p | 226.65 | 3.41E-07 |
| hsa-miR-6132 | 4.11 | 0.001 | hsa-miR-31-5p | 183.50 | 6.76E-08 |
| hsa-miR-378 h | 3.78 | 0.001 | hsa-miR-29a-3p | 135.70 | 1.42E-07 |
| hsa-miR-6753-5p | 3.73 | 0.001 | hsa-miR-151a-5p | 133.66 | 8.57E-07 |
| hsa-miR-3620-5p | 3.69 | 1.11E-04 | hsa-miR-361-5p | 112.31 | 1.42E-07 |
| hsa-miR-101-5p | 3.48 | 0.006 | hsa-miR-17-5p | 109.49 | 1.28E-06 |
| hsa-miR-4505 | 3.42 | 0.001 | hsa-miR-20a-5p | 97.79 | 2.37E-04 |
| hsa-miR-4668-5p | 3.39 | 0.002 | hsa-miR-34a-5p | 86.75 | 8.88E-08 |
| hsa-miR-3613-3p | 3.36 | 0.002 | hsa-miR-23a-3p | 86.05 | 3.69E-04 |
| hsa-miR-1268a | 3.21 | 0.004 | hsa-miR-152-3p | 82.80 | 1.34E-06 |
| hsa-miR-1268b | 3.07 | 0.007 | hsa-miR-107 | 81.11 | 1.22E-06 |
| hsa-miR-8075 | 3.06 | 3.05E-04 | hsa-let-7i-5p | 76.33 | 6.07E-04 |
| hsa-miR-4508 | 2.99 | 1.34E-04 | hsa-miR-24-3p | 74.40 | 3.43E-06 |
| hsa-miR-5001-5p | 2.98 | 0.037 | hsa-miR-708-5p | 65.13 | 1.41E-05 |
| hsa-miR-4463 | 2.96 | 0.006 | hsa-miR-143-3p | 63.12 | 2.36E-05 |
| hsa-miR-6787-5p | 2.94 | 0.001 | hsa-let-7e-5p | 62.27 | 8.72E-04 |
| hsa-miR-6126 | 2.82 | 0.027 | hsa-miR-106a-5p | 60.89 | 1.53E-06 |
| hsa-miR-6724-5p | 2.81 | 0.002 | hsa-miR-379-5p | 60.03 | 9.89E-07 |
| hsa-miR-4750-3p | 2.80 | 0.023 | hsa-miR-140-3p | 51.88 | 1.16E-06 |
| hsa-miR-4707-5p | 2.80 | 0.015 | hsa-let-7d-5p | 50.97 | 3.99E-05 |
| hsa-miR-7108-5p | 2.74 | 0.006 | hsa-miR-199a-5p | 49.72 | 9.19E-07 |
| hsa-miR-4763-3p | 2.67 | 0.001 | hsa-let-7f-5p | 46.50 | 1.156E-06 |
| hsa-miR-6803-5p | 2.65 | 3.85E-04 | hsa-miR-27b-3p | 40.86 | 2.155E-05 |
| hsa-miR-4649-5p | 2.64 | 0.004 | hsa-miR-130a-3p | 39.98 | 3.79E-07 |
| hsa-miR-7109-5p | 2.63 | 0.019 | hsa-miR-103a-3p | 39.94 | 7.79E-05 |
| hsa-miR-4487 | 2.61 | 0.003 | hsa-miR-21-5p | 39.17 | 4.05E-06 |
Note: miRNAs highlighted in bold font have been identified only in humans (according to the current miRBase database: www.mirbase.org).
Table 4.
Complete list of all significantly differentially expressed mitomiRs in MSCs at day 14 of osteogenesis. Fold change expression levels relative to day 0 are shown in addition to the Benjamini-Hochberg adjusted P values.
| Increased miRNA (day 14 vs day 0) | Decreased miRNA (day 14 vs day 0) | ||||
|---|---|---|---|---|---|
| Transcript ID | Fold change | P value | Transcript ID | Fold change | P value |
| hsa-miR-4485 | 10.21 | 0.003 | hsa-miR-210-3p | 30.24 | 0.003 |
| hsa-miR-4646-5p | 9.56 | 0.024 | hsa-miR-20a-5p | 20.03 | 0.031 |
| hsa-miR-483-5p | 8.55 | 0.004 | hsa-miR-629-5p | 12.29 | 0.001 |
| hsa-miR-4484 | 2.72 | 0.042 | hsa-miR-195-5p | 12.13 | 0.009 |
| hsa-miR-615-3p | 2.54 | 0.034 | hsa-miR-1271-5p | 11.64 | 0.004 |
| hsa-miR-4492 | 2.46 | 0.031 | hsa-miR-27a-3p | 11.40 | 0.032 |
| hsa-miR-4508 | 2.13 | 0.016 | hsa-miR-421 | 9.13 | 0.003 |
| hsa-miR-3620-5p | 2.06 | 0.038 | hsa-miR-16-5p | 8.67 | 0.006 |
| hsa-miR-494-3p | 7.73 | 0.045 | |||
| hsa-miR-29b-1-5p | 6.81 | 0.034 | |||
| hsa-miR-199a-3p | 6.48 | 0.009 | |||
| hsa-miR-199b-3p | 6.48 | 0.009 | |||
| hsa-miR-378a-3p | 6.21 | 0.003 | |||
| hsa-miR-29a-3p | 4.95 | 0.005 | |||
| hsa-miR-181a-2-3p | 4.74 | 0.004 | |||
| hsa-miR-214-5p | 4.34 | 0.047 | |||
| hsa-miR-31-5p | 4.26 | 0.004 | |||
| hsa-miR-3651 | 4.22 | 0.014 | |||
| hsa-miR-15b-5p | 4.19 | 0.047 | |||
| hsa-miR-15a-5p | 4.11 | 0.034 | |||
| hsa-miR-6819-5p | 3.65 | 0.029 | |||
| hsa-miR-21-5p | 3.60 | 0.050 | |||
| hsa-miR-424-3p | 3.51 | 0.006 | |||
| hsa-miR-6825-5p | 3.37 | 0.042 | |||
| hsa-miR-376c-3p | 3.28 | 0.004 | |||
| hsa-miR-130a-3p | 3.20 | 0.018 | |||
| hsa-miR-25-3p | 2.80 | 0.034 | |||
| hsa-miR-22-5p | 2.64 | 0.031 | |||
| hsa-miR-4732-5p | 2.50 | 0.026 | |||
| hsa-miR-128-3p | 2.34 | 0.009 | |||
Note: miRNAs highlighted in bold font have been identified only in humans (according to the current miRBase database: www.mirbase.org).
With respect to differentially-expressed miRNAs from whole cell extracts, two were identified as upregulated at day 3 vs day 0 (miR-4730: 2.80 fold, p = 0.027; miR-1237 h-5p: 2.49 fold, p = 0.027) and three were downregulated at day 3 vs day 0 (miR-132-3p: 4.08 fold, p = 0.027; miR-1271-5p: 2.11 fold, p = 0.043; miR-23a-5p: 1.93 fold, p = 0.043). Table 5 shows a total of seven miRNAs that were significantly increased and fourteen that were significantly decreased at day 7. Table 6 indicates a total of twenty four miRNAs showing increased expression at day 14 and twenty four miRNAs that were significantly decreased at day 14. Of these, only miR-652-3p was found to be increased at both the day 7 and day 14 time points whereas two miRNAs (miR-7162-3p and miR-23a-5p) were significantly reduced at both osteogenic time points. All the other differentially-expressed miRNAs from whole cell extracts were unique to the day 7 or day 14 time point further highlighting the dynamic expression and regulation of these non-coding RNAs at specific stages of osteoblast differentiation.
Table 5.
Complete list of all miRNAs from whole cell extracts showing significantly increased or decreased expression levels at day 7 of osteogenic induction compared to day 0. Fold change expression levels are shown in addition to the Benjamini-Hochberg adjusted P values.
| Increased miRNA (day 7 vs day 0) | Decreased miRNA (day 7 vs day 0) | ||||
|---|---|---|---|---|---|
| Transcript ID | Fold change | P value | Transcript ID | Fold change | P value |
| hsa-miR-4485 | 4.61 | 0.037 | hsa-miR-4521 | 9.02 | 0.025 |
| hsa-miR-328-3p | 4.32 | 0.024 | hsa-miR-29b-1-5p | 4.24 | 0.021 |
| hsa-miR-574-3p | 2.90 | 0.003 | hsa-miR-27a-5p | 4.17 | 0.029 |
| hsa-miR-652-3p | 2.75 | 0.040 | hsa-miR-132-3p | 3.90 | 0.023 |
| hsa-miR-425-3p | 2.31 | 0.029 | hsa-miR-7162-3p | 3.67 | 0.039 |
| hsa-miR-370-3p | 2.27 | 0.021 | hsa-miR-138-5p | 3.00 | 0.021 |
| hsa-miR-181a-5p | 1.91 | 0.043 | hsa-miR-23a-5p | 2.94 | 0.003 |
| hsa-miR-20a-5p | 2.58 | 0.038 | |||
| hsa-miR-3972 | 2.53 | 0.038 | |||
| hsa-miR-106a-5p | 2.50 | 0.029 | |||
| hsa-miR-17-5p | 2.50 | 0.029 | |||
| hsa-miR-1271-5p | 2.28 | 0.023 | |||
| hsa-miR-31-5p | 2.09 | 0.036 | |||
| hsa-miR-5006-5p | 2.04 | 0.038 | |||
Note: miRNAs highlighted in bold font have been identified only in humans (according to the current miRBase database: www.mirbase.org).
Table 6.
Complete list of all miRNAs from whole cell extracts showing significantly increased or decreased expression levels at day 14 of osteogenic induction compared to day 0. Fold change expression levels are shown in addition to the Benjamini-Hochberg adjusted P values.
| Increased miRNA (day 14 vs day 0) | Decreased miRNA (day 14 vs day 0) | ||||
|---|---|---|---|---|---|
| Transcript ID | Fold change | P value | Transcript ID | Fold change | P value |
| hsa-miR-34c-5p | 6.05 | 0.001 | hsa-miR-1263 | 5.22 | 0.011 |
| hsa-miR-34c-3p | 4.70 | 0.040 | hsa-miR-101-5p | 4.87 | 0.027 |
| hsa-miR-4649-5p | 4.18 | 0.027 | hsa-miR-548a-3p | 4.83 | 0.011 |
| hsa-miR-4484 | 3.91 | 0.001 | hsa-miR-92b-3p | 3.83 | 0.011 |
| hsa-miR-7150 | 3.82 | 0.006 | hsa-miR-629-5p | 3.80 | 0.035 |
| hsa-miR-664a-5p | 3.72 | 0.001 | hsa-miR-7162-3p | 3.79 | 0.028 |
| hsa-miR-652-3p | 3.22 | 0.018 | hsa-miR-5189-3p | 3.75 | 0.001 |
| hsa-miR-6782-5p | 3.22 | 0.012 | hsa-miR-197-3p | 3.65 | 0.001 |
| hsa-miR-6824-5p | 3.14 | 0.018 | hsa-miR-212-3p | 3.56 | 0.023 |
| hsa-miR-324-5p | 2.94 | 0.027 | hsa-miR-4304 | 3.56 | 0.012 |
| hsa-miR-198 | 2.85 | 0.024 | hsa-miR-4330 | 3.50 | 0.040 |
| hsa-miR-483-5p | 2.67 | 0.035 | hsa-miR-615-3p | 3.21 | 0.008 |
| hsa-miR-6716-5p | 2.59 | 0.016 | hsa-miR-138-1-3p | 3.06 | 0.014 |
| hsa-miR-6889-5p | 2.50 | 0.011 | hsa-miR-210-3p | 3.00 | 0.004 |
| hsa-miR-6787-5p | 2.18 | 0.029 | hsa-miR-3911 | 2.82 | 0.040 |
| hsa-miR-939-5p | 2.13 | 0.042 | hsa-miR-4669 | 2.54 | 0.048 |
| hsa-miR-4738-3p | 1.99 | 0.049 | hsa-miR-3972 | 2.44 | 0.035 |
| hsa-miR-6794-5p | 1.98 | 0.034 | hsa-miR-7847-3p | 2.40 | 0.013 |
| hsa-miR-6126 | 1.97 | 0.029 | hsa-miR-1271-5p | 2.15 | 0.023 |
| hsa-miR-505-5p | 1.95 | 0.035 | hsa-miR-6127 | 2.06 | 0.021 |
| hsa-miR-423-5p | 1.93 | 0.017 | hsa-miR-4298 | 2.04 | 0.030 |
| hsa-miR-134-5p | 1.87 | 0.022 | hsa-miR-5100 | 1.99 | 0.043 |
| hsa-miR-432-5p | 1.84 | 0.030 | hsa-miR-193a-5p | 1.96 | 0.040 |
| hsa-miR-214-3p | 1.83 | 0.012 | hsa-miR-23a-5p | 1.92 | 0.023 |
Note: miRNAs highlighted in bold font have been identified only in humans (according to the current miRBase database: www.mirbase.org).
When comparing differentially-expressed miRNA profiles obtained from MSC mitochondria and whole cell extracts, miR-4485 was the only miRNA found to be significantly increased at day 7 vs day 0. Common miRNAs that were all down-regulated at day 7 in both mitochondria and whole cell extracts were miR-31-5p, miR-17-5p, miR-20a-5p and miR-106a-5p. It should be noted that the fold change decrease in these miRNAs was substantially greater in the mitochondria samples when compared to whole cell extracts (Tables 3, 6). The only miRNA found to be significantly increased at day 14 in both mitochondria and whole cell extracts was miR-4484, while common miRNAs that were decreased at this time point were miR-629-5p and miR-1271-5p. Interestingly, the majority of significantly increased mitomiRs at day 7 and day 14 (Tables 3, 4) were found to be those that are less well-described in the published literature and, to date, have only been identified in humans (according to the miRBase website). As expected, these miRNAs were found to be expressed in whole cell extracts (Supplemental Table 2), but not differentially expressed during osteogenesis. These “human-specific” miRNAs have been highlighted in bold font in Tables 1–6. In comparison, the mitomiRs showing significantly lower levels of expression at day 7 and 14 are those more commonly reported in the published literature and have been identified in multiple species.
We then carried out RT-qPCRs using the same mitochondria extract samples analyzed in the array to attempt to correlate a few of the most significant expression changes. For the day 7 time points, we confirmed the significant increase in expression of miR-4485 and miR-483-5p (Fig. 6A, B) and the decreased expression of miR-26a-5p and miR-20a-5p (Fig. 6C, D). A significant decrease in expression of miR-20a-5p and miR-26a-5p was also confirmed at the day 14 time point (Fig. 6C, D). While a significant increase in miR-4485 was noted by qPCR at day 14, the fold change increase was not higher than the fold change increase at day 7 as found in the array data. Similarly, qPCR did not detect a significant increase in expression of miR-483-5p at day 14 as was found in the array. Of note, although miR-6893-5p displayed the highest fold change at day 7 compared to day 0, we could not demonstrate this by qPCR. This was due to low expression levels detected for this miRNA in all of the mitochondria extract samples (i.e. high Ct values). This may be due, in part, to the high GC content of the cDNA fragment amplified in the specific RT-qPCR miRNA assay.
Fig. 6.
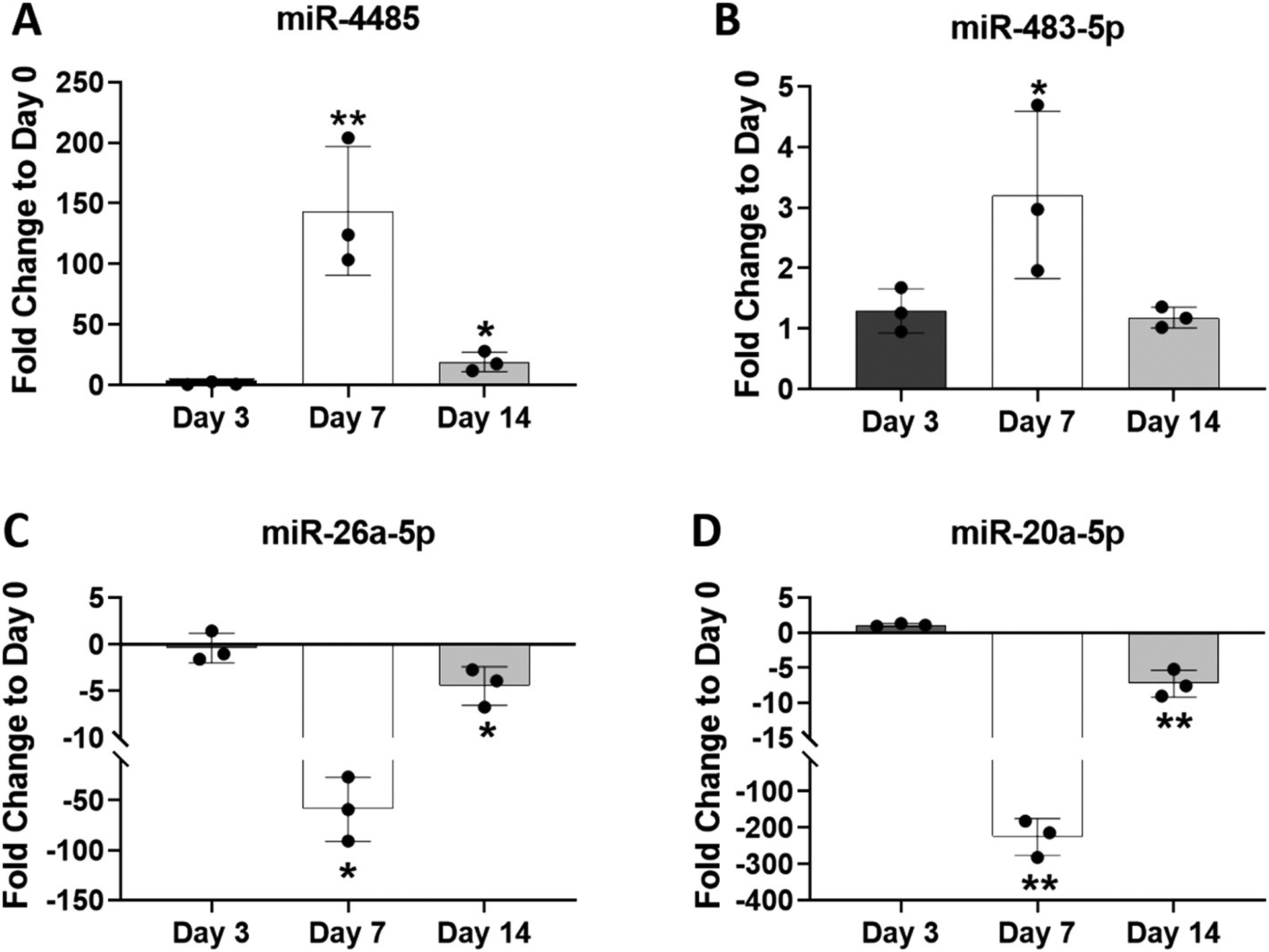
Analysis of four differentially-expressed mitomiRs by RT-qPCR. RNA from the same three mitochondrial extracts analyzed via microarray were also analyzed by RT-qPCR. Expression levels of miR-4485 (A), miR-483-5p (B), miR-26a-5p (C) and miR-20a-5p (D) were normalized to RNU44 and plotted as fold change expression compared to day 0. Data are expressed ± SD; n = 3. *p < 0.05; **p < 0.01.
In the whole cell extract samples, qPCR showed increased miR-4485 expression, the highest at day 7 in agreement with the array data (Fig. 7A), and a trend toward increased miR-181a-5p expression at day 7 (Fig. 7B). Significant increases in miR-483-5p expression were found at all time points, the highest being at day 14 in agreement with the array results (Fig. 7C). Significant decreases in expression of miR-138-5p and miR-20a-5p were confirmed at the day 7 time point as also found via miRNA microarray analysis (Fig. 7D, E).
Fig. 7.
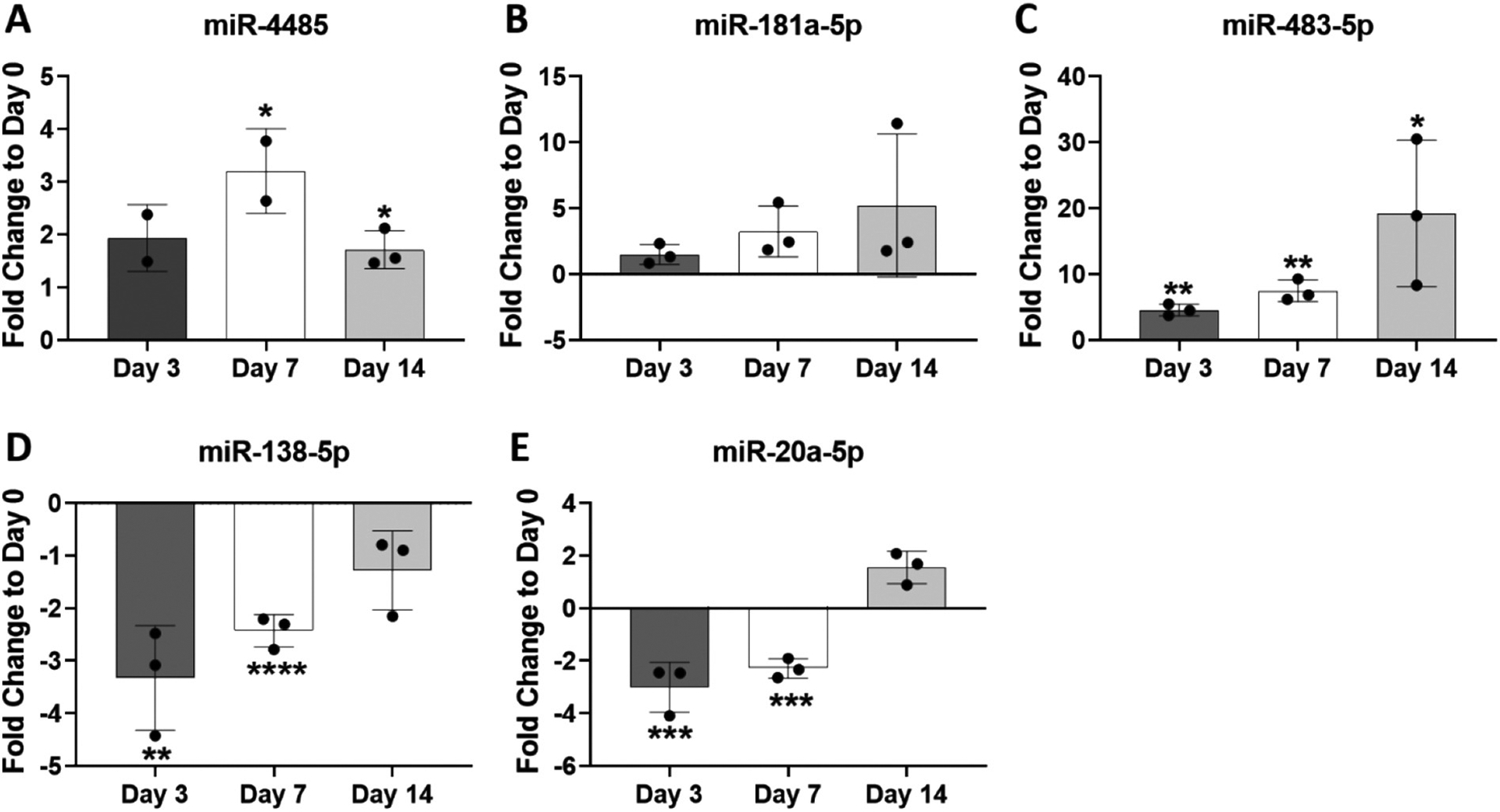
Analysis of four differentially-expressed miRNAs in whole cell extracts by RT-qPCR. RNA from the same three whole cell extracts analyzed via microarray were also analyzed by RT-qPCR. Expression levels of miR-4485 (A), miR-181a-5p (B), miR-483-5p (C), miR-138-5p (D) and miR-20a-5p (E) were normalized to RNU44 and plotted as fold change expression compared to day 0. Data are expressed ± SD; n = 3. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
4. Discussion
In this study, we present novel findings on expression profiles of miRNAs present in mitochondrial extracts of human MSCs at different time points during osteogenic differentiation. To date, there have been no reports in the field of bone biology on the expression levels of such mitochondria-associated miRNAs (mitomiRs) in skeletal progenitor cells or during osteoblast differentiation.
Overall, we were intrigued to detect so many miRNAs present in mitochondria extracts of MSCs. We followed the same protocol described in previous studies identifying miRNAs in purified mitochondria extracts of HeLa cells or human primary muscle cells [4,5]. We were confident that we had obtained highly purified mitochondria extracts given that we did not detect 28S and 18S RNA (Supplemental Fig. 1), and that our samples were enriched in mitochondria-specific mRNA and protein and devoid of an ER-specific peptide (Fig. 2). Also, by treating our extracts with RNAse A, we reduced the possibility of residual cytosolic RNA contamination [5–7].
From a growing number of published reports, it is now recognized that non-coding RNAs (including miRNA, siRNA, rRNA, tRNA, long non-coding RNAs) are present in or associated with mitochondria [28,29]. With respect to the term “mitomiR”, this describes miRNAs that may be associated with the outer mitochondrial membrane, the intermembrane space, the inner mitochondrial membrane or within the central mitochondrial matrix. While there is some evidence that miRNAs may be transcribed directly from the mitochondrial genome [9,30,31], the majority of mitomiRs identified in this study are derived from the nuclear genome, suggesting that mechanisms must exist to permit import or trafficking of miRNAs into the mitochondria.
In other published work identifying mitomiR expression profiles, the number of miRNAs associated with mitoplasts (i.e. mitochondria devoid of the outer membrane) was lower than that found in more crude extracts [8,9]. This suggests that many mitomiRs may be present within/associated with the outer membrane and/or within the intermembrane space of mitochondria. Our study involved analysis of intact mitochondria containing the outer membrane which may partly explain why we detected high numbers of mitomiRs in our array analyses. Also, we utilized a human specific Affymetrix GeneChip™ array containing 2578 human mature miRNA probe sets and 2025 human pre-miRNA (stem-loop) probe sets. Previous studies reporting fewer mitomiRs could also be due to the fact that their approaches utilized older arrays containing fewer human miRNA probe sets or other methods such as RT-qPCR arrays. Additionally, variations in cut-off parameters used to identify levels above background may likely also contribute to the differences in numbers of mitomiRs detected in our work compared to other studies. Interestingly, we also detected a number of precursor miRNAs in mitochondrial extracts, as has also been reported by Barrey et al [5]. In fact, this study also showed, via in situ hybridization and confocal microscopy, the presence of both mature miRNAs and pre-miRNAs in the mitochondria of human skeletal muscle myoblasts. The fact that Ago 2 and the enzyme Dicer has been identified in mitochondrial extracts [4,13,14] suggests the presence of active miRNA ribonucleoprotein complexes and the potential for processing of pre-miRNA to mature miRNA in mitochondria.
Many of the mitomiRs identified in previous reports were also found to be present in mitochondrial extracts of human bone marrow-derived MSCs in the present study. Of note, mitomiR profiling in other studies revealed expression of let-7 family members in HeLa cells, HEK-293 cells and human primary skeletal muscle cells [4,5,9]. Interestingly, let-7b and other let-7 family members were abundantly expressed in MSC mitochondria extracts, regardless of osteogenic induction (Table 1). In addition, let-7 paralogs were also found to be some of the most abundantly expressed miRNAs in whole cell extracts at each time point during osteogenesis, highlighting their widespread expression in different compartments of the cell (Table 2). Collectively, these findings support the data from many published reports demonstrating that the highly conserved let-7 family members play a number of critical roles in regulating proper cell function and health, including metabolic processes [32,33]. A similar pattern of abundant expression in mitochondria and whole cell extracts at all time points was found for miR-320 family members (miR-320a/b/c). There are hundreds of publications describing a number of functional roles for these conserved miRNAs including one describing miR-320a as a regulator of mitochondrial metabolism in skeletal muscle cells [34].
Other abundantly expressed miRNAs found in both mitochondria and whole cell extracts at each time point during osteogenesis include miR-3960, miR-6089 and miR-6090 (Tables 1 and 2). A literature search shows only fifteen publications on miR-3960 over the past ten years and, among these, there are two reports describing a potential function in regulating osteogenic differentiation [35,36]. There are very few published reports on miR-6089 and miR-6090, both of which are two examples of more recently-discovered miRNAs (note the high identification numbers) and, to date, have only been identified in humans as indicated in the miRBase database. In addition, another “human-specific” miRNA, miR-5787, is included in the top 30 abundant mitomiRs list at day 7 and 14 of osteogenesis, but not at day 0 or 3 (Table 1). This miRNA was detected in whole cell extracts (Supplemental Table 2) but was not among the most abundantly expressed in these samples (Table 2). While there are very few reports in the published literature on this mRNA, one study has identified miR-5785 as a mitomiR functioning in regulating metabolism in favor of oxidative phosphorylation [37].
Our data revealed that day 7 of MSC osteogenesis was the key time point where the highest amount of significantly differentially-expressed mitomiRs (relative to day 0) was found. A total of 91 mitomiRs were increased and 175 were decreased at day 7 (Supplemental Table 5). From Table 3, showing the top 30 differentially-expressed mitomiRs at day 7, those showing decreased expression are mostly conserved between species and changes in expression are all highly significant (based on the low adjusted P value) with substantial fold change decreases, some reaching over 300-fold. Also, a PubMed search shows that these reduced mitomiRs are well-described in the literature as playing functional roles in may cellular processes, including osteogenesis. The majority of mitomiRs found to be decreased at day 7 of osteogenesis were not found to be differentially-expressed in whole cell extracts at the same time point. A different set of miRNAs were apparently reduced in whole cell extracts including miR-138-5p, which is in agreement with our previous work describing this miRNA as an inhibitor of osteogenesis [38]. Such distinct expression profiles suggest that decreased mitomiR levels are not due to transcriptional suppression of their genes in the nucleus, but likely due to relocation/redistribution of these miRNAs from mitochondria to the cytoplasm or other organelles. Also, major decreases in mitomiR levels at day 7 compared to day 0 cannot be due to potential differences in the quality/integrity of mitochondria extracted from cells at these time points; our data shows similarities in mitochondria ultrastructure (Supplemental Fig. 1) and functionality (as assessed by uptake of TMRE; Fig. 3) from MSCs at day 0, 7 and 14 of osteogenic induction.
A possible explanation for these substantial changes in mitomiR expression levels may be related to processes controlling mitochondrial dynamics. It is known that mitochondria undergo fusion and fission as a means to control their function, morphology, quality and abundance within cells; such dynamic alterations are critical for controlling the cellular response to stress and maintaining proper cell function [39]. Mitochondrial fusion and fission are complex, highly organized processes regulated by a range of evolutionarily conserved proteins located at the outer or inner mitochondrial membrane resulting in a more fused, elongated network or a fragmented network, respectively. It is now apparent that mitochondrial fusion and fission are also important processes governing cell commitment and differentiation [40]. Mitochondria in MSCs predominantly undergo fusion during osteogenic commitment resulting in elongated networks [41,42]. Such changes have been observed at various time points during MSC osteogenesis, but particularly from day 7 onwards when oxidative phosphorylation is likely required to provide sufficient energy for cells to generate a functional, mineralized extracellular matrix [41]. With respect to the substantial decreases in mitomiR expression observed at day 7 of MSC osteogenesis in our study, this may be partly due to the extensive morphological rearrangements occurring within the outer and inner mitochondrial membranes, possibly resulting in relocation of miRNAs present at these sites to other compartments of the cell.
Regardless of these dynamic changes to mitochondria at day 7, the levels of many mitomiRs were also found to increase at this time point in mitochondrial extracts, albeit at much lower fold change differences (~2–5 fold) compared to those that were reduced (Table 3). One of these elevated miRNAs found at both the day 7 and day 14 time point was miR-483-5p (Tables 3 and 4). One report identified a role for this miRNA in targeting FIS1, a protein involved in regulating mitochondrial fission [43,44]. Interestingly, studies from our lab and others have reported negative effects of miR-483 over-expression on chondrogenesis and cartilage homeostasis [45,46]. This may be partly explained by the suppressive effects on mitochondrial fission given that chondrocytes are known to rely mostly on glycolysis and thus contain mostly fragmented mitochondria due to fission [40,42]. However, this cytoplasmic function of miR-483-5p in suppressing mitochondrial fission may have beneficial effects on osteoblast differentiation, but it remains determined if this miRNA performs a functional role within or associated with the mitochondria.
Another miRNA identified as significantly increased at day 7 was miR-378 h. Of note, miR-378 was also identified in mitochondria of murine cardiac cells and, in response to diabetic insult in mice, this miRNA was re-distributed to a specific subpopulation of mitochondria in the heart. It was also found to target the mitochondrial gene ATP6 to regulate mitochondrial function [10]. In addition, mice devoid of miR-378 exhibited elevated oxidative capacity [47] and, recently, miR-378 was shown to suppress MSC osteogenesis in vitro and bone formation in vivo [48]. Both of these studies analyzed the miR-378a paralog present within the PGC-1β gene, which encodes a transcriptional co-activator that regulates metabolism and mitochondrial biogenesis. Apparently miR-378 h has been identified only in humans and is located within the gene encoding fatty acid hydroxylase domain containing 2 (FAXDC2). The functional mature strand of miR-378a is almost identical to miR-378 h and they share the same seed sequence. It will be interesting to explore the role of these paralogs further in the context of regulating mitochondria function and osteoblast differentiation.
It is also important to highlight that the majority of increased mitomiRs at day 7 are those that have thus far only been identified in humans and have not been well-studied to date (see miRNAs highlighted in bold in Table 3). These human mitomiRs were also identified in whole cells extracts (Supplemental Table 2) but were not differentially-expressed during osteogenesis, except for miR-4485 (Table 5). There are a few published studies on miR-4485, which was also identified as increased at day 14 in mitochondrial extracts as well (Table 4). The gene encoding this miRNA is located on chromosome 11, but it is also predicted to be present within a long non-coding RNA transcribed from the mitochondrial genome [31]. Another two miRNAs also increased at day 7 in our study may also be derived from the mitochondria genome in addition to the nuclear genome (miR-4484 and miR-4463) [9]. Of interest, miR-4485 was found to be enriched in the mitochondria of a human breast cancer cell line and regulate mitochondrial function [49]. This study by Sripada et al predicted that miR-4485 was predominantly derived from the nuclear genome and transported into the mitochondria. Whether or not this miRNA plays a role in regulating mitochondrial metabolism in the context of osteoblast differentiation remains to be determined.
It is also worth noting that the stem-loop precursor sequence encoding miR-4485, as well as that of a number of other human-specific mitomiRs listed in Table 3, is unusually short. In addition, the pre-miRNAs encoding some of the others have small loops and a number of them contain a sequence encoding either a 3p or 5p mature miRNA strand, not both. A previous study also found that some mitomiRs identified in HeLa cells had some unusual features (e.g. differences in thermodynamic stability and length compared to well-described cytoplasmic miRNAs), but it should be noted that some of these mitomiRs have since been identified as mitochondrial tRNAs and are no longer defined as miRNAs [4]. With respect to the less well-described human-specific mitomiRs identified in our work, it will be important to keep track of updates to miRBase database on a regular basis to confirm that they are indeed bone fide miRNAs before pursuing their potential function in the future. It may be that these non-coding RNAs are indeed miRNAs, but perhaps are processed and function in a more non-canonical manner. It has already been reported that some miRNAs, including mitomiR-1, function to enhance expression of its target gene rather than canonical role of target gene suppression [22]. If indeed the human mitomiRs identified in our work are truly miRNAs, then the fact that they are increased during osteoblast differentiation suggests a potential role in regulating mitochondrial metabolism or dynamics, and this will be an avenue for future research. Interestingly, a recent study suggests that gene regulatory networks involving miRNAs are necessary to permit proper cell function in scenarios of high metabolic and anabolic activity [50]. It may be that a subset of miRNAs exist only in higher species with increased metabolic demands and play a role to control cellular processes in such scenarios to maintain healthy cellular differentiation and homeostasis.
In summary, this study provides more evidence supporting the existence of mitomiRs in human cells. A particularly novel aspect of this work is demonstrating how mitomiR expression profiles change in response to MSC differentiation toward the osteoblast lineage. The fact that hundreds of mitomiRs were detected suggests the possibility that many of them may simply be stored within the mitochondria (perhaps within P-bodies or endosomes associated with mitochondria) and are non-functional until they are released into the cytosol. However, this work also points to a number of mitomiR candidates to explore further to determine potential functional roles in regulating not only osteoblast differentiation, but also processes that are specifically controlled by mitochondria including energy metabolism, autophagy, cell viability and calcium trafficking. It will also be interesting to explore the location of specific mitomiRs of interest within the mitochondria. This could be addressed by comparing mitomiR expression levels in whole mitochondria extracts compared to purified mitoplasts devoid of the outer mitochondrial membrane. Other more involved and sophisticated approaches to determine mitomiR localization include fluorescence in situ hybridization combined with super-resolution microscopy as well as mitochondria-specific tagging of RNA [51]. It will also be interesting to determine if expression levels of specific mitomiR candidates are altered with age given the associations between aging, mitochondrial dysfunction and impaired bone healing [52,53].
Supplementary Material
Acknowledgements
We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with microarrays and genomic analysis. Thanks also to Erica Lantelme (FACS Core, Department of Pathology & Immunology at Washington University School of Medicine) for assistance with FACS analysis and data interpretation. We gratefully acknowledge Dr. Sanja Sviben, Gregory Strout and Dr. James Fitzpatrick for their assistance in electron microscopy studies conducted at the Washington University Center for Cellular Imaging (WUCCI), which is supported in part by Washington University School of Medicine, The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), the Foundation for Barnes-Jewish Hospital (3770), the Washington University Rheumatic Diseases Research Resource-based Center (NIH P30 AR073752) and the Washington University Musculoskeletal Research Center (MRC) (NIH P30 AR074992).
Funding sources
This work was supported by NIH R01 AR077203 and NIH R21 AR075730 (to AM), and NIH P30 AR057235 (Washington University Musculoskeletal Research Center).
Abbreviations
- miRNA
microRNA
- mitomiRs
mitochondria-associated microRNAs
- nt
nucleotide
- RISC
RNA-induced silencing complex
- ncRNA
non-coding RNA
- Ago 2
argonaute 2
- P bodies
processing bodies
- ER
endoplasmic reticulum
- pri-miRNA
primary miRNA
- pre-miRNA
precursor miRNA
- tRNA
transfer RNA
- rRNA
ribosomal RNA
- mt-COX1
mitochondria-encoded cytochrome c oxidase 1
- mt-ND1
mitochondria-encoded NADH-ubiquinone oxidoreductase chain 1
- KDEL
lysine-aspartic acid-glutamic acid-leucine peptide
- TOM22
translocase of the outer mitochondrial membrane 22
- FACS
flow cytometry and fluorescence activated cell sorting
- TMRE
tetramethylrhodamine, ethyl ester
- ALP
alkaline phosphatase
- OCN
osteocalcin
- TEM
transmission electron microscopy
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bone.2021.116058.
CRediT authorship contribution statement
Hongjun Zheng designed experiments and acquired, analyzed and interpreted data. He also generated figures and contributed to manuscript writing. Jin Liu acquired and analyzed data. Jinsheng Yu analyzed and interpreted microarray data and prepared figures. Audrey McAlinden designed research, interpreted data and wrote the manuscript. All authors approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no conflicts of interest with the contents of this article.
References
- [1].Bartel DP, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell 116 (2004) 281–297. [DOI] [PubMed] [Google Scholar]
- [2].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell 136 (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Leung AK, The whereabouts of microRNA actions: cytoplasm and beyond, Trends Cell Biol. 25 (2015) 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, Henrion-Caude A, Nuclear outsourcing of RNA interference components to human mitochondria, PLoS One 6 (2011), e20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X, Pre-microRNA and mature microRNA in human mitochondria, PLoS One 6 (2011), e20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bian Z, Li LM, Tang R, Hou DX, Chen X, Zhang CY, Zen K, Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions, Cell Res. 20 (2010) 1076–1078. [DOI] [PubMed] [Google Scholar]
- [7].Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ, MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis, RNA Biol. 6 (2009) 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS, The human mitochondrial transcriptome, Cell 146 (2011) 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R, Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA, PLoS One 7 (2012), e44873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Stricker JC, Croston TL, Baseler WA, Lewis SE, Martinez I, Hollander JM, Translational regulation of the mitochondrial genome following redistribution of mitochondrial microRNA in the diabetic heart, Circ. Cardiovasc. Genet 8 (2015) 785–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sieber F, Placido A, El Farouk-Ameqrane S, Duchene AM, Marechal-Drouard L, A protein shuttle system to target RNA into mitochondria, Nucleic Acids Res. 39 (2011), e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Calvo SE, Mootha VK, The mitochondrial proteome and human disease, Annu. Rev. Genomics Hum. Genet 11 (2010) 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, Steenbergen C, Nuclear miRNA regulates the mitochondrial genome in the heart, Circ. Res 110 (2012) 1596–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang WX, Visavadiya NP, Pandya JD, Nelson PT, Sullivan PG, Springer JE, Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury, Exp. Neurol 265 (2015) 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sripada L, Tomar D, Singh R, Mitochondria: one of the destinations of miRNAs, Mitochondrion 12 (2012) 593–599. [DOI] [PubMed] [Google Scholar]
- [16].Bandiera S, Mategot R, Girard M, Demongeot J, Henrion-Caude A, MitomiRs delineating the intracellular localization of microRNAs at mitochondria, Free Radic. Biol. Med 64 (2013) 12–19. [DOI] [PubMed] [Google Scholar]
- [17].Latronico MV, Condorelli G, The might of microRNA in mitochondria, Circ. Res 110 (2012) 1540–1542. [DOI] [PubMed] [Google Scholar]
- [18].Geiger J, Dalgaard LT, Interplay of mitochondrial metabolism and microRNAs, Cell. Mol. Life Sci 74 (2017) 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Macgregor-Das AM, Das S, A microRNA’s journey to the center of the mitochondria, Am. J. Physiol. Heart Circ. Physiol 315 (2018) H206–H215. [DOI] [PubMed] [Google Scholar]
- [20].Das S, Kohr M, Dunkerly-Eyring B, Lee DI, Bedja D, Kent OA, Leung AK, Henao-Mejia J, Flavell RA, Steenbergen C, Divergent effects of miR-181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets, J. Am. Heart Assoc 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li H, Zhang X, Wang F, Zhou L, Yin Z, Fan J, Nie X, Wang P, Fu XD, Chen C, Wang DW, MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation, Circulation 134 (2016) 734–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD, MicroRNA directly enhances mitochondrial translation during muscle differentiation, Cell 158 (2014) 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zheng H, Liu J, Tycksen E, Nunley R, McAlinden A, MicroRNA-181a/b-1 over-expression enhances osteogenesis by modulating PTEN/PI3K/AKT signaling and mitochondrial metabolism, Bone 123 (2019) 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Benjamini Y, Hochberg Y, Controlling the false discovery rate: a practical and powerful approach to multiple testing, J. R. Stat. Soc. Ser. B Methodol 57 (1995) 289–300. [Google Scholar]
- [26].Ye Y, Huang A, Huang C, Liu J, Wang B, Lin K, Chen Q, Zeng Y, Chen H, Tao X, Wei G, Wu Y, Comparative mitochondrial proteomic analysis of hepatocellular carcinoma from patients, Proteomics Clin Appl 7 (2013) 403–415. [DOI] [PubMed] [Google Scholar]
- [27].Kaushal PS, Sharma MR, Agrawal RK, The 55S mammalian mitochondrial ribosome and its tRNA-exit region, Biochimie 114 (2015) 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vendramin R, Marine JC, Leucci E, Non-coding RNAs: the dark side of nuclear-mitochondrial communication, EMBO J. 36 (2017) 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gusic M, and Prokisch H (2020) ncRNAs: new players in mitochondrial health and disease? Front Genet 11, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shinde S, Bhadra U, A complex genome-microRNA interplay in human mitochondria, Biomed. Res. Int 2015 (2015) 206382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bianchessi V, Badi I, Bertolotti M, Nigro P, D’Alessandra Y, Capogrossi MC, Zanobini M, Pompilio G, Raucci A, Lauri A, The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in endothelial cells, J. Mol. Cell. Cardiol 81 (2015) 62–70. [DOI] [PubMed] [Google Scholar]
- [32].Roush S, Slack FJ, The let-7 family of microRNAs, Trends Cell Biol. 18 (2008) 505–516. [DOI] [PubMed] [Google Scholar]
- [33].Jiang S, A regulator of metabolic reprogramming: MicroRNA Let-7, Transl. Oncol 12 (2019) 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dahlmans D, Houzelle A, Andreux P, Jorgensen JA, Wang X, de Windt LJ, Schrauwen P, Auwerx J, Hoeks J, An unbiased silencing screen in muscle cells identifies miR-320a, miR-150, miR-196b, and miR-34c as regulators of skeletal muscle mitochondrial metabolism, Mol Metab 6 (2017) 1429–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xia ZY, Hu Y, Xie PL, Tang SY, Luo XH, Liao EY, Chen F, Xie H, Runx2/miR-3960/miR-2861 positive feedback loop is responsible for osteogenic transdifferentiation of vascular smooth muscle cells, Biomed. Res. Int 2015 (2015) 624037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY, Guo LJ, Xie H, Zhou HD, Wu XP, Luo XH, A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation, J. Biol. Chem 286 (2011) 12328–12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen W, Wang P, Lu Y, Jin T, Lei X, Liu M, Zhuang P, Liao J, Lin Z, Li B, Peng Y, Pan G, Lv X, Zhang H, Ou Z, Xie S, Lin X, Sun S, Ferrone S, Tannous BA, Ruan Y, Li J, Fan S, Decreased expression of mitochondrial miR-5787 contributes to chemoresistance by reprogramming glucose metabolism and inhibiting MT-CO3 translation, Theranostics 9 (2019) 5739–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zheng H, Ramnaraign D, Anderson BA, Tycksen E, Nunley R, and McAlinden A (2018) MicroRNA-138 inhibits osteogenic differentiation and mineralization of human dedifferentiated chondrocytes by regulating RhoC and the actin cytoskeleton. JBMR Plus, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Westermann B, Mitochondrial fusion and fission in cell life and death, Nat Rev Mol Cell Biol 11 (2010) 872–884. [DOI] [PubMed] [Google Scholar]
- [40].Ren L, Chen X, Chen X, Li J, Cheng B, Xia J, Mitochondrial dynamics: fission and fusion in fate determination of mesenchymal stem cells, Front Cell Dev Biol 8 (2020) 580070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shum LC, White NS, Mills BN, Bentley KL, Eliseev RA, Energy metabolism in mesenchymal stem cells during osteogenic differentiation, Stem Cells Dev. 25 (2016) 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Forni MF, Peloggia J, Trudeau K, Shirihai O, Kowaltowski AJ, Murine mesenchymal stem cell commitment to differentiation is regulated by mitochondrial dynamics, Stem Cells 34 (2016) 743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fan S, Chen WX, Lv XB, Tang QL, Sun LJ, Liu BD, Zhong JL, Lin ZY, Wang YY, Li QX, Yu X, Zhang HQ, Li YL, Wen B, Zhang Z, Chen WL, Li JS, miR-483-5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1, Cancer Lett. 362 (2015) 183–191. [DOI] [PubMed] [Google Scholar]
- [44].Tian T, Lv X, Pan G, Lu Y, Chen W, He W, Lei X, Zhang H, Liu M, Sun S, Ou Z, Lin X, Cai L, He L, Tu Z, Wang X, Tannous BA, Ferrone S, Li J, Fan S, Long noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of pre-miRNA processing, Clin. Cancer Res 25 (2019) 3673–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Anderson BA, McAlinden A, miR-483 targets SMAD4 to suppress chondrogenic differentiation of human mesenchymal stem cells, J. Orthop. Res 35 (2017) 2369–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wang H, Zhang H, Sun Q, Wang Y, Yang J, Yang J, Zhang T, Luo S, Wang L, Jiang Y, Zeng C, Cai D, Bai X, Intra-articular delivery of antago-miR-483-5p inhibits osteoarthritis by modulating matrilin 3 and tissue inhibitor of metalloproteinase 2, Mol. Ther 25 (2017) 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [47].Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, Bassel-Duby R, Olson EN, Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 15330–15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Feng L, Zhang JF, Shi L, Yang ZM, Wu TY, Wang HX, Lin WP, Lu YF, Lo JHT, Zhu DH, Li G, MicroRNA-378 suppressed osteogenesis of MSCs and impaired bone formation via inactivating Wnt/beta-catenin signaling, Mol Ther Nucleic Acids 21 (2020) 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sripada L, Singh K, Lipatova AV, Singh A, Prajapati P, Tomar D, Bhatelia K, Roy M, Singh R, Godbole MM, Chumakov PM, Singh R, hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells, J Mol Med (Berl) 95 (2017) 641–651. [DOI] [PubMed] [Google Scholar]
- [50].Cassidy JJ, Bernasek SM, Bakker R, Giri R, Pelaez N, Eder B, Bobrowska A, Bagheri N, Nunes Amaral LA, Carthew RW, Repressive gene regulation synchronizes development with cellular metabolism, Cell 178 (980–992) (2019), e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jeandard D, Smirnova A, Tarassov I, Barrey E, Smirnov A, Entelis N, Import of non-coding RNAs into human mitochondria: a critical review and emerging approaches, Cells 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Haas RH, Mitochondrial dysfunction in aging and diseases of aging, Biology (Basel) 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Clark D, Nakamura M, Miclau T, Marcucio R, Effects of aging on fracture healing, Curr Osteoporos Rep 15 (2017) 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


