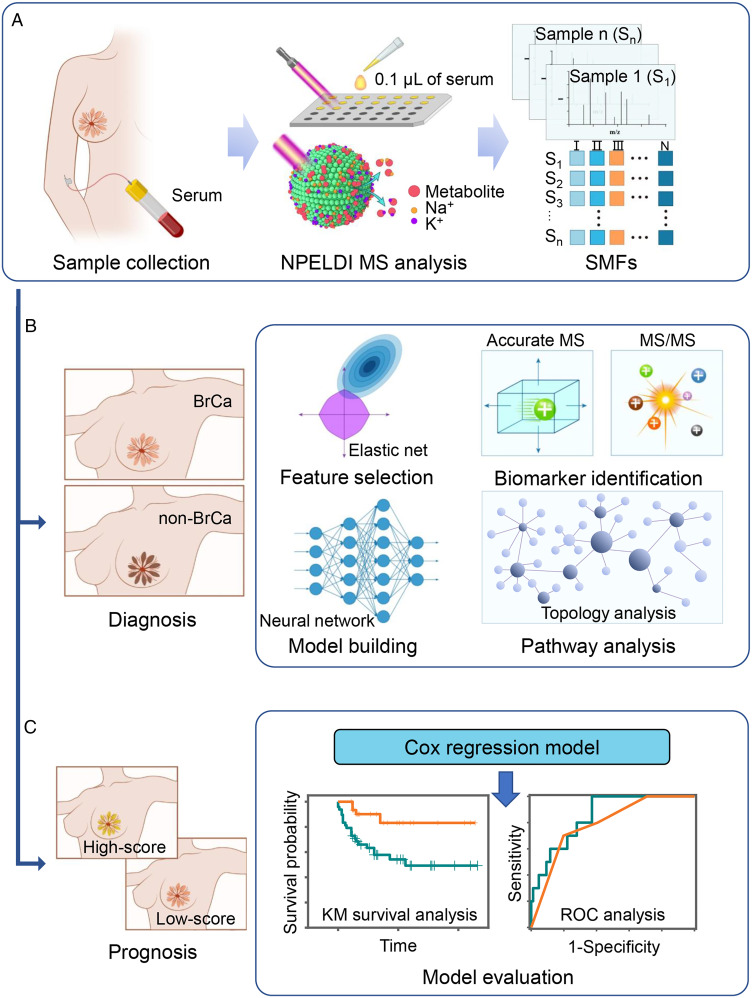
Significance
Breast cancer (BrCa) is the most common cancer worldwide, and high-performance metabolic analysis is emerging in diagnosis and prognosis of BrCa. Here, we used nanoparticle-enhanced laser desorption/ionization mass spectrometry to record serum metabolic fingerprints of BrCa in seconds, achieving high reproducibility and low consumption of direct serum detection. Our analytical method, combined with the aid of machine learning algorithms, was demonstrated to provide high diagnostic efficiency with accuracy of 88.8% and desirable prognostic prediction (P < 0.005). Furthermore, seven metabolic biomarkers differentially enriched in BrCa serum and their related pathways were identified. Together, our findings provide a tool to characterize BrCa and highlight certain metabolic signatures as potential diagnostic and prognostic factors of diseases including but not limited to BrCa.
Keywords: diagnosis, prognosis, serum metabolic fingerprints, mass spectrometry, breast cancer
Abstract
High-performance metabolic analysis is emerging in the diagnosis and prognosis of breast cancer (BrCa). Still, advanced tools are in demand to deliver the application potentials of metabolic analysis. Here, we used fast nanoparticle-enhanced laser desorption/ionization mass spectrometry (NPELDI-MS) to record serum metabolic fingerprints (SMFs) of BrCa in seconds, achieving high reproducibility and low consumption of direct serum detection without treatment. Subsequently, machine learning of SMFs generated by NPELDI-MS functioned as an efficient readout to distinguish BrCa from non-BrCa with an area under the curve of 0.948. Furthermore, a metabolic prognosis scoring system was constructed using SMFs with effective prediction performance toward BrCa (P < 0.005). Finally, we identified a biomarker panel of seven metabolites that were differentially enriched in BrCa serum and their related pathways. Together, our findings provide an efficient serum metabolic tool to characterize BrCa and highlight certain metabolic signatures as potential diagnostic and prognostic factors of diseases including but not limited to BrCa.
Breast cancer (BrCa) is the most common cancer with more than 1,300,000 new cases and 450,000 deaths each year worldwide (1). High-performance analytical tools are critical in the early and efficient diagnosis and treatment of BrCa (2). Currently, the clinical practice still relies on conventional methods of histopathological classification and imaging tools, such as mammography, magnetic resonance imaging, and ultrasound. However, these approaches need large instruments or rigorous operations, which are time consuming and not always reliable, especially in early diagnosis (3, 4). However, liquid biopsy using blood samples is advantageous in its ease to apply in a noninvasive and high-throughput manner and can serve as a promising tool to facilitate early detection, prediction of metastatic potential, and selection of therapy. In this regard, a growing body of molecular biomarkers, such as serum circulating extracellular vesicles, nucleic acid methylation, circulating tumor cells, and autoantibodies, are being intensively pursued (5–7).
Malignant cells acquire distinct metabolic reprogramming in response to a variety of extrinsic and intrinsic cell stressors to initiate transformation and growth programs (8–11). Analysis of metabolites (molecular weight (MW) < 1,000 Da) can help to reveal the real-time status of living systems. Increasing studies on BrCa have highlighted the value of metabolomics in early diagnosis and in therapeutic and prognostic predictions (12, 13). Over the past decade, analytical techniques, such as NMR spectroscopy and mass spectrometry (MS), have been developed for the comprehensive screening of cancer metabolomes (14, 15). MS is emerging through precisely measuring the mass-to-charge ratio (m/z) of metabolites with molecular identification capability. However, the application of MS in liquid- and gas-phase detection relies on chromatography for sample purification and metabolite enrichment, hindering the analytical speed and capacity. In contrast, laser desorption/ionization (LDI)-MS uses nanoparticle matrices as an alternative of chromatography for solid-phase detection (16–18). It offers a new high-performance technique that helps with advanced metabolic analysis of BrCa.
Herein, we used nanoparticle-enhanced LDI-MS (NPELDI-MS) to record global serum metabolic fingerprints (SMFs) of BrCa patients (n = 169), benign breast disease (BBD) patients (n = 21), and healthy donors (HDs; n = 135). We first demonstrated high detection reproducibility (∼95% of features with intensity coefficients of variance (CVs) < 30% in serum samples), fast analytical speed (∼30 s per sample), and minimal sample consumption (∼100 nL per sample) in a label-free manner (Scheme 1A). We achieved desirable diagnostic performance with an area under the curve (AUC) of 0.948 (95% confidence interval (CI) of 0.922 to 0.973) of BrCa by machine learning on SMFs (Scheme 1B). Further, a metabolic prognosis scoring system (MP-score) built by SMFs effectively predicted the prognosis and survival of patients (P < 0.005; Scheme 1C). Of note, a diagnostic model was generated based on a biomarker panel of seven metabolites, affording robust discrimination efficiency with an AUC of 0.865 (95% CI of 0.820 to 0.911). Therefore, our work will facilitate the development of advanced metabolic analysis of BrCa and the screening of metabolic alterations toward therapeutic intervention.
Scheme 1.
SMFs to decode BrCa. (A) Serum samples were collected from enrolled subjects and microarrayed on chips for NPELDI-MS analysis in metabolic fingerprinting. (B) Machine learning of SMFs was conducted by feature selection and model building to diagnose the BrCa group from the non-BrCa group, and a biomarker panel was identified for pathway analysis. (C) The Cox regression model was applied to build a prognosis prediction model based on SMFs. The Kaplan–Meier (KM) curve, log-rank testing, and time-dependent ROC curve analysis were conducted for survival analysis of low- and high-score groups.
Results
High-Performance Serum Metabolite Characterization by NPELDI-MS.
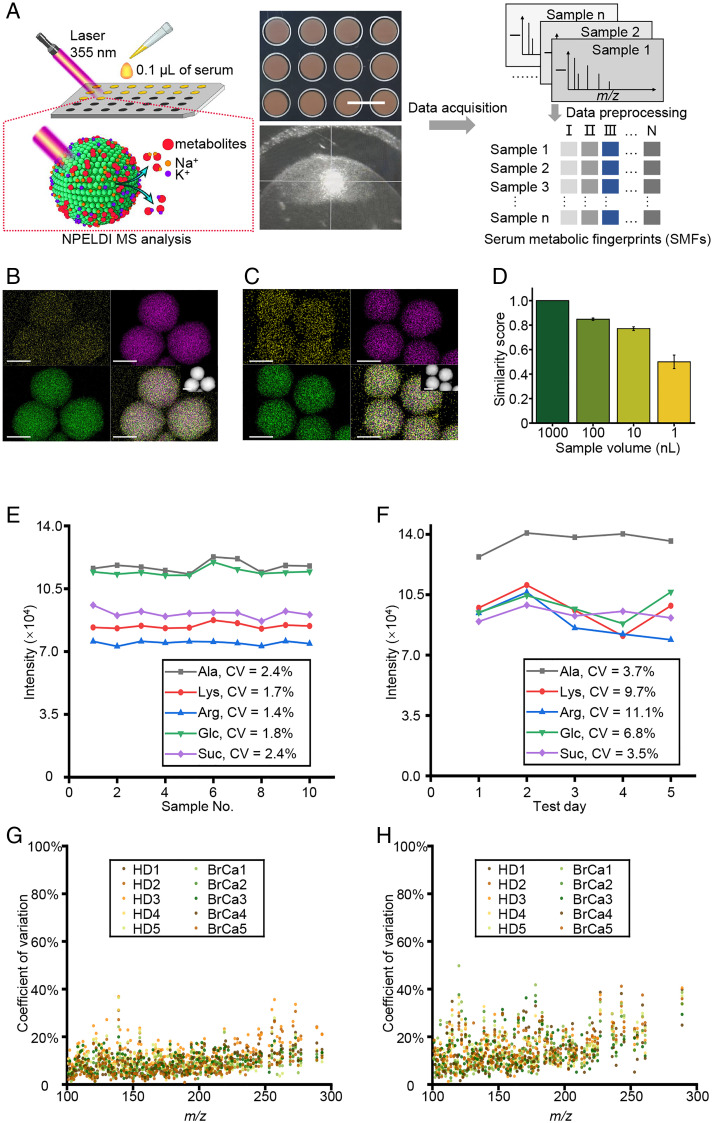
To improve the analytical speed, sample consumption, and reproducibility of LDI-MS, we conducted high-performance serum metabolic fingerprinting by NPELDI-MS (Fig. 1A). The ferric nanoparticles were prepared using a modified low-cost solve-thermal method (Materials and Methods), showing the designed surface roughness structure as an ideal matrix for NPELDI-MS. Due to the size-selective trapping and affinity-based cationization of metabolites by the surface nanostructures of nanoparticles (SI Appendix, Fig. S1), NPELDI-MS allowed direct detection of metabolites from the interference of proteins and salts in serum (SI Appendix, Fig. S2) with minimum sample treatment at high speed (∼30 s per sample). To validate the size-selective trapping of nanoparticles, histamine (His) and bovine serum albumin (BSA), as representatives for metabolites (MW < 1,000 Da) and macromolecules (MW > 1,000 Da), respectively, were mixed with nanoparticles to form nanoparticle–analyte hybrids. Consequently, the elemental mapping analysis of the nanoparticle–analyte hybrids showed a significantly higher molecular size–selective trapping rate (defined as the ratio of carbon signal intensity on the nanoparticles to the background) for His than for BSA (P < 0.001; Fig. 1 B and C and SI Appendix, Table S1). Importantly, the NPELDI-MS process afforded fast analytical speed with 2 s per sample (with 2,000 laser shots at a pulse frequency of 1,000 Hz; Materials and Methods), which could be coupled with on-chip microarray (384 samples per chip) to achieve automatic large-scale sample screening (Fig. 1A).
Fig. 1.
High-performance serum metabolite characterization by NPELDI-MS. (A) Illustration of NPELDI-MS. The Upper digital image shows the MS chips after microarray printing, and the Bottom image was recorded during NPELDI-MS. (Scale bar, 5 mm.) (B and C) Elemental mappings of the nanoparticle–analyte (His in B and BSA in C) hybrids are shown with O in yellow, Fe in purple, and C in green, respectively. The insert images were high-angle annular dark-field images of nanoparticle–analyte hybrids. (Scale bars, 200 nm.) (D) The similarity scores between 1,000 nL of pristine serum and its dilutions by 10- to 1,000-fold using 100 to 1 nL of pristine serum. The error bars were calculated as SD of five repeated experiments. (E and F) Intensities of five molecular peaks (gray line for [Ala + Na]+ at an m/z of 112.04, red line for [Lys + Na]+ at an m/z of 169.09, blue line for [Arg + Na]+ at an m/z of 197.19, green line for [Glc + Na]+ at an m/z of 203.05, and purple line for [Suc + Na]+ at an m/z of 365.11) for intrachip (ten replicates per sample) in E and interchip (five chips for 5 d, one chip per day) detection in F, respectively. (G and H) CV distribution of intensities for the apparent molecular peaks in ten serum samples (five HDs denoted as HD1 to HD5 and five BrCa patients denoted as BrCa1 to BrCa5) for intrachip (ten replicates per sample) in G and interchip detection (five chips for 5 d, one chip per day) in H, respectively.
Of note, we calculated the cosine similarity score of apparent molecular peaks (with average intensity of >500) between native serum and its dilutions using 1 to 100 nL of serum to identify the minimum sample volume for detection. We obtained qualified similarity scores above 0.771 using 10 to 100 nL of serum (Fig. 1D) due to the efficient absorption and transfer of laser energy for enhanced detection sensitivity by two to six orders (compared with organic matrices; SI Appendix, Fig. S3 and Table S2). Specifically, 108 apparent molecular peaks of metabolites were observed, likely owing to the high sensitivity of NPELDI-MS, which helped to form a reservoir for SMFs. Further, to avoid sampling heterogeneity, the pristine and diluted serum samples were all homogenized by a vortex mixer before dilution and loading to the plate. Additionally, we also conducted an experiment to illustrate this issue by sampling and diluting n = 5 serum samples five times and testing these samples by NPELDI-MS. Consequently, the average similarity scores were higher than 0.97 with SEs lower than 0.02 (SI Appendix, Fig. S4) in each sample, showing the homogeneity of our analytical method. Additionally, the effect of the nanoparticle size on the LDI efficiency was studied by changing the time of hydrothermal reaction according to a previous study (19). Consequently, the results showed that in the range of 250 to 400 nm, the number of signals with a signal-to-noise ratio > 3 kept stable and had no significant changes (P > 0.05) in serum samples, illustrating that nanoparticle size would not influence the signal enhancement in the range of 250 to 400 nm, and the optimal reaction time was set as 10 h considering the size uniformity of nanoparticles (SI Appendix, Fig. S5).
We also developed a protocol to determine the detection reproducibility of NPELDI-MS in both the intrachip (ten replicates per sample) and interchip (five chips for 5 d, one chip per day) contexts. We included one standard sample that was a mixture of five standard metabolites, including alanine (Ala), lysine (Lys), arginine (Arg), glucose (Glc), and sucrose (Suc), and ten serum samples (five HDs and five BrCa patients). The intensity CVs of the five molecular peaks ([Ala + Na]+ at an m/z of 112.04, [Lys + Na]+ at an m/z of 169.09, [Arg + Na]+ at an m/z of 197.19, [Glc + Na]+ at an m/z of 203.05, and [Suc + Na]+ at an m/z of 365.11) in the standard sample were 1.36 to 2.45% and 3.50 to 11.08% for intrachip and interchip detection, respectively (Fig. 1 E and F and SI Appendix, Tables S3 and S4). The desirable reproducibility of NPELDI-MS was attributed to the homogeneous morphology of nanoparticle–analyte cocrystallization, distinct from the random sample crystallization with organic matrices (SI Appendix, Fig. S3). Consistently, for features (apparent molecular peaks with average intensity of >500) in serum samples, 97.37 to 100.00% and 93.39 to 98.35% of these peaks displayed intensity CVs of <30% regarding intrachip and interchip detection, respectively (SI Appendix, Table S5). These results demonstrated that NPELDI-MS could achieve high performance in profiling SMFs with desirable speed, less sample consumption, and high reproducibility.
Characterization of BrCa-Specific SMFs.
To determine BrCa-specific SMFs, we collected serum samples prospectively from stage I to IV treatment-naive BrCa patients (n = 169, with 43 individuals in stage I, 52 individuals in stage II, 36 individuals in stage III, and 38 individuals in stage IV), BBD patients (n = 21), and HDs (n = 135) (Fig. 2A). We performed the histopathological evaluation of all samples by two independent pathologists. Clinical characteristics, including age at pathological diagnosis, subtype, and tumor, node, metastasis (TNM) stage are summarized (SI Appendix, Table S6).
Fig. 2.
Characterization of BrCa-specific SMFs. (A) The age and stage distribution of 169 BrCa patients and age distribution of 21 BBD patients as well as 135 HDs. (B) Three typical mass spectra at the m/z range of 100 to 400 are shown for serum samples of BrCa, BBD, and HD on the Left, while the frequency distribution of similarity scores on the Right was calculated for each group by SMFs within the same group. (C) A heat map of independent metabolic fingerprints for 325 serum samples was plotted using 301 m/z signals through data preprocessing. The color scale was processed by logarithmic correction. (D) The OPLS-DA classification for the BrCa (red points), BBD (yellow points), and HD (green points) group.
We constructed a serum metabolic database based on the high-throughput NPELDI-MS analysis. In total, there were ∼124,000 data points in the origin MS result acquired per sample, where strong m/z signals were obtained with total ion counts (calculated as the summation of the MS intensity of each serum sample) of ∼1.19 to 1.44 × 108 at the low mass range of 100 to 400 Da (Fig. 2B and SI Appendix, Fig. S6) due to the high resolution of 0.005 Da and high LDI sensitivity of small metabolites with the detection limit of femtomole (SI Appendix, Table S2) (20). Typically, over 95% of samples shared high similarity scores over 0.8 with typical mass spectra in each group, indicating the reliability of SMFs and the potency for further diagnostic and prognostic applications. Specifically, we also analyzed the ferric nanoparticles directly without adding any other analyte by NPELDI-MS, and several peaks observed could be indexed as the fragments of matrix ([FexOy]+; SI Appendix, Fig. S7A). However, those peaks were suppressed and not detected in the mass spectra of serum samples and thus would not interfere with further analysis. Although ferric nanoparticles possess intrinsic nanoenzymatic activity, no iron-adducted peaks could be observed for metabolites (such as the ferrocene-like ion formed by N-heterocyclic species; SI Appendix, Fig. S7 B–D) (21). Additionally, to evaluate the system variation during the long-term sample test, we have applied standard samples that consisted of five standard metabolites (including Ala, Lys, Arg, Glc, and Suc) as quality control (QC) samples. The QC samples were tested during the whole sample test procedure at regular intervals. Consequently, the QC samples can be gathered into a separate cluster (R2X(cum) = 0.860, R2Y(cum) = 0.746, and Q2(cum) = 0.664; SI Appendix, Fig. S8) by orthogonal partial least squares discriminant analysis (OPLS-DA; SI Appendix, Fig. S6). Further, the intensity CVs of the five molecular peaks ([Ala + Na]+ at an m/z of 112.04, [Lys + Na]+ at an m/z of 169.09, [Arg + Na]+ at an m/z of 197.19, [Glc + Na]+ at an m/z of 203.05, and [Suc + Na]+ at an m/z of 365.11) in the standard sample were 3.74 to 4.59%, which demonstrated desirable system variation during the long-term and large-scale sample test (SI Appendix, Table S7). Notably, the SMF referred to 301 m/z signals was extracted from origin MS results by data preprocessing (Fig. 2C), including binning, smoothing, baseline correction, peak detection, and alignment (Materials and Methods), which was the basis for feature selection and biomarker screening toward diagnostic and prognostic use.
Particularly, OPLS-DA was performed to visualize the distribution of samples based on SMFs, showing that BrCa and HD samples can be roughly separated into two clusters in the OPLS-DA score plot (R2X(cum) = 0.805, R2Y(cum) = 0.432, and Q2(cum) = 0.372; Fig. 2D). However, the separation of these two groups was not clear enough, which indicated the necessity of introducing an advanced machine learning algorithm to help interpret data and improve the diagnostic efficiency. These findings demonstrated that SMFs generated by NPELDI-MS could be used as a readout to distinguish BrCa patients from non-BrCa (control group, including HD and BBD) individuals.
Construction of an SMF-Based Diagnostic Model for BrCa.
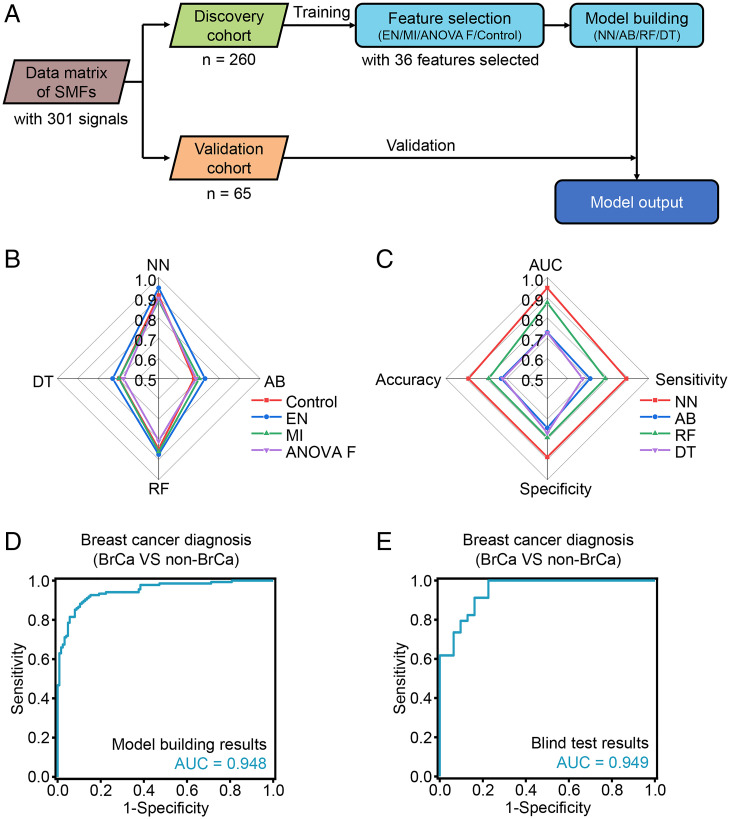
To complement the histopathology-based diagnosis, we determined whether SMF-based metabolomic information (consisting of 301 signals) could be used as a liquid diagnostic tool to distinguish cancers from noncancers. We constructed shared and unique BrCa SMFs by performing NPELDI-MS and data preprocessing and randomly split these serum samples into a discovery cohort of n = 260 (n = 135 BrCa and n = 125 non-BrCa) and an independent validation cohort of n = 65 (n = 34 BrCa and n = 31 non-BrCa) with well-matched age (P > 0.05).
Machine learning using SMFs from the cohorts included feature selection and model building (Fig. 3A). Typically, univariant methods (analysis of variance F value [ANOVA F] and mutual information [MI]) and a model-based method (elastic net [EN]) were applied for feature selection, and SMFs without any selection were set as the control for comparison. Subsequently, four algorithms (neural network [NN], AdaBoost [AB], random forest [RF], and decision tree [DT]) were performed for model building. All the models achieved an AUC of >0.673 for discriminating BrCa compartments from non-BrCa compartments in the discovery cohort. Specifically, EN in feature selection showed optimized performance with an AUC of 0.727 to 0.948 among all four algorithms with an optimized cutoff frequency of 95% and 36 features selected (Materials and Methods and SI Appendix, Fig. S9) compared with ANOVA F (AUC of 0.673 to 0.897, P < 0.005 by paired t test), MI (AUC of 0.696 to 0.884, P < 0.05), and control (AUC of 0.674 to 0.911, P < 0.01; Fig. 3B and SI Appendix, Table S8). Furthermore, in the model building, NN afforded the best performance with an AUC of 0.948 (95% CI of 0.922 to 0.973) compared with AB (AUC of 0.728, 95% CI of 0.665 to 0.790, P < 0.0001 by Delong test), RF (AUC of 0.875, 95% CI of 0.833 to 0.917, P < 0.0001), and DT (AUC of 0.727, 95% CI 0.664 to 0.791, P < 0.0001), given 36 features as selected by EN (Fig. 3C). Importantly, we obtained consistent results (with an AUC of 0.949, 95% CI of 0.901 to 0.996, accuracy of 83.1%, sensitivity of 82.4%, and specificity of 83.9%; Fig. 3E) for the independent validation cohort in the blind test with EN for feature selection and NN for model building compared with that in the discovery cohort (AUC of 0.948, 95% CI of 0.922 to 0.973, accuracy of 88.8%, sensitivity of 88.9%, and specificity of 88.8%; Fig. 3D), validating the diagnostic value of SMFs for BrCa.
Fig. 3.
The machine learning model for BrCa diagnosis. (A) Workflow for building the diagnostic model. (B) The AUCs in the differentiation of BrCa compartments from non-BrCa compartments in the discovery cohort, using three typical algorithms (red line for control, blue line for EN, green line for MI, and purple line for ANOVA F) in feature selection and four typical algorithms in model building (NN, AB, RF, and DT). (C) Model evaluation of four typical machine learning algorithms (NN, AB, RF, and DT) based on 36 features selected by EN, including AUC, sensitivity, specificity, precision, and accuracy. (D and E) The ROC curves for NN modeled by features to diagnose BrCa compartments from non-BrCa compartments of the discovery cohort (D) and validation cohort (E).
These results suggested that SMF-based metabolomic information derived from BrCa exhibited excellent performance distinguishing BrCa patients from non-BrCa individuals. Specifically, the best diagnostic AUC could be obtained by applying EN for feature selection and NN for model building.
Construction of an SMF-Based Prognostic Model for BrCa Survival Prediction.
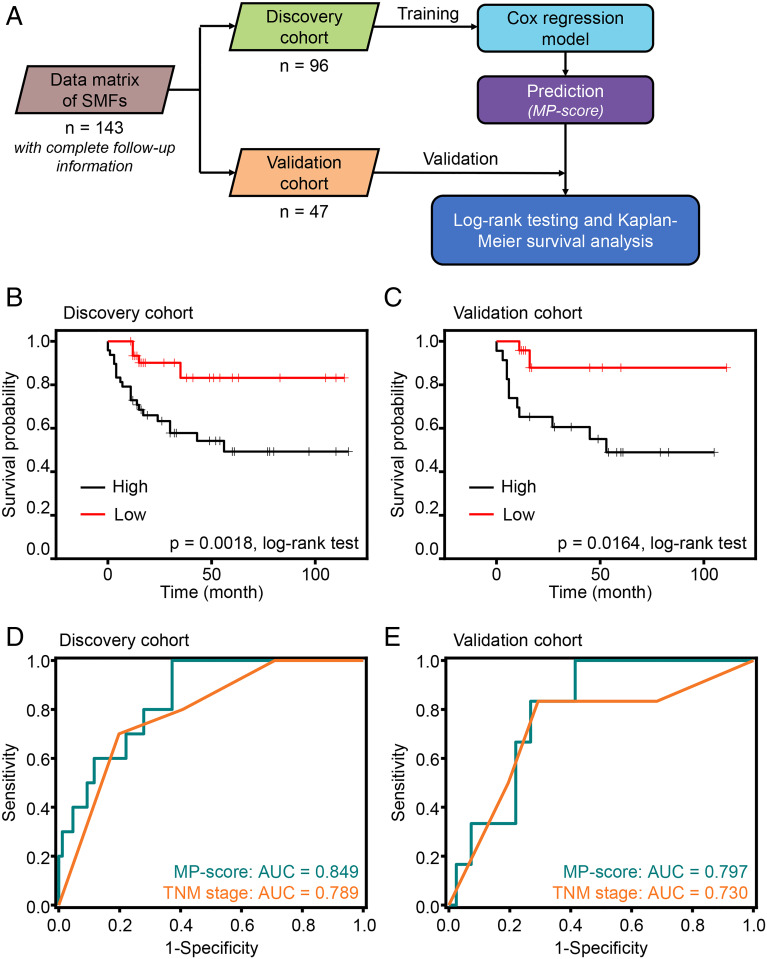
Importantly, we analyzed the SMF dataset from patients with complete follow-up information. These patients were randomly split into a discovery cohort (containing 96 observations with 26 events) and a validation cohort (containing 47 observations with 13 events), with a median follow-up time of 16 mo (ranging from less than 1 mo to 116 mo). The Cox regression model was implemented to construct an MP-score with a four-metabolite panel (Fig. 4A and SI Appendix, Table S9). Kaplan–Meier curves were generated with a dichotomized label, which divided the patients into high-score and low-score groups according to the median MP-score. As a result, the median survival time in the low-score group was significantly more prolonged than that in the high-score group (P = 0.0018 in the discovery cohort and P = 0.0164 in the validation cohort by log-rank test; Fig. 4 B and C), illustrating the desirable prognosis efficiency.
Fig. 4.
Prognostic prediction of BrCa based on SMFs. (A) Workflow for building the prognostic model for calculating MP-score. (B and C) Overall survival curves of patients with BrCa with low risk (red line) or high risk (black line) of death according to the MP-score (constructed via Cox regression model) in the discovery (B) and validation cohorts (C). (D and E) Time-dependent ROC and corresponding AUCs for 9-mo survival predicted by MP-score (green line) and TNM stage (orange line) in the discovery (D) and validation cohorts (E).
To further determine the predictive ability of MP-score, we performed time-dependent receiver operating characteristic curve (ROC) analysis to characterize the prediction performance of the four-metabolite MP-score, with the comparison of conventional TNM (as defined by the American Joint Committee on Cancer guidelines) staging system (22, 23). The ROC analysis demonstrated that the MP-score displayed a comparable prediction performance with an AUC of 0.849 (95% CI of 0.749 to 0.949) in the discovery cohort and 0.797 (95% CI of 0.657 to 0.936) in the validation cohort compared with the TNM staging system (with an AUC of 0.789 and 95% CI of 0.664 to 0.914 in the discovery cohort and an AUC of 0.730 and 95% CI of 0.503 to 0.956, in the validation cohort; Fig. 4 D and E).
Taken together, we proposed an efficient model based on SMFs for prognostic prediction, illustrating the broad application of our detection method in guiding treatment planning clinically.
Identification of a Metabolic Biomarker Panel and Pathway Analysis.
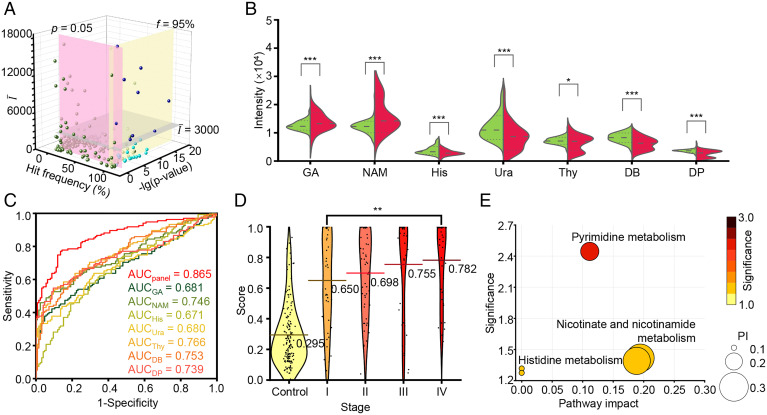
To further determine specific BrCa serum metabolic biomarkers, we filtered the metabolites from the data matrix of SMFs based on the stepwise statistical screening criteria (mean intensity > 3,000, hit frequency > 95%, and P < 0.05) by comparing BrCa compartments with non-BrCa compartments (Fig. 5A). Using the screening strategy, we identified seven metabolites, of which amounts were significantly different in serum samples of BrCa patients compared with those in non-BrCa samples. Subsequently, we validated these seven metabolites as l-glyceric acid (GA), nicotinamide (NAM), His, uracil (Ura), thymine (Thy), 3,4-dihydroxybenzylamine (DB), and dehydro-phenylalanine (DP) (24, 25), respectively, through accurate MS using Fourier transform ion cyclotron resonance (FT-ICR)-MS or MS/MS using time-of-flight (TOF)-MS (SI Appendix, Fig. S10 and Table S10). Among them, His, Ura, Thy, DB, and DP were down-regulated (P < 0.05), while GA and NAM were up-regulated (P < 0.001) in BrCa compartments compared with in BBD and HD compartments (Fig. 5B).
Fig. 5.
Biomarker panel construction and pathway analysis. (A) Selection of biomarker candidates according to mean intensity (), hit frequency (f), and P value (p). (B) The violin plot illustrated the differential expression of seven metabolites between the BrCa group (red) and non-BrCa group (green), and P values (***, P < 0.001; *, P < 0.05) are indicated on the Top of each violin plot. (C) The ROC curves showed a higher AUC of 0.865 using the metabolic biomarker panel than a single metabolic biomarker (AUC of 0.680 to 0.766). (D) Score distribution of individuals from the non-BrCa group (labeled as control) and BrCa patients in stage I/II/III/IV. The lines show the average scores of each group. (E) The potential pathways that were differentially regulated in the BrCa group and non-BrCa group; each circle’s color and size were correlated to the P value and pathway impact (PI). A total of three pathways were considered with a pathway impact of >0.1 and hit number ≥1, including 1) pyrimidine metabolism, 2) nicotinate and NAM metabolism, and 3) histidine metabolism.
In multibiomarker analysis, the NN model built by seven metabolites exhibited an enhanced diagnostic AUC of 0.865 (95% CI of 0.820 to 0.911), which was superior to the analysis of a single metabolic biomarker with limited AUCs of 0.680 to 0.766 (P < 0.05; Fig. 5C and SI Appendix, Table S10). Importantly, we observed an increased diagnostic score (defined as the probability of being diagnosed as BrCa patients using the NN model built by these seven metabolites) of stage IV BrCa compartments (average score of 0.782) compared with stage I, II, and III compartments (average scores of 0.650, 0.698, and 0.755, respectively; Fig. 5D). Additionally, stage IV BrCa cases showed the highest average score of 0.782, which was significantly higher than that of stage I cases (average score of 0.650, P < 0.05). In the early-stage diagnosis related to subtypes of BrCa, the sensitivity for BrCa at stage I was highest for the triple-negative/basal-like BrCa (85.71%) and lowest for HER2-enriched BrCa (20%). Due to the complexity of multiclassification machine learning (26), more samples were required to build models toward direct BrCa staging.
We further performed pathway enrichment analysis to determine the biological relevance and metabolic signaling of the seven metabolites. In the pathway topology analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway library, there were three metabolic pathways related to BrCa with a pathway impact > 0.1 and hit number (the number of matched metabolites in the pathway) ≥ 1, including 1) pyrimidine metabolism, 2) nicotinate and NAM metabolism, and 3) histidine metabolism (Fig. 5E and SI Appendix, Table S11). Pyrimidine metabolism reflected adaptive metabolic reprogramming to the up-regulated transcriptional activity due to cancer cell proliferation (27, 28). Nicotinate and NAM metabolism represented a high rate of NAM adenine dinucleotide turnover in cancer cells corresponding to the high rate of proliferation and DNA synthesis (29–32). Histidine metabolism can be associated with the modulation of inflammatory and hypersensitivity responses owing to the vital immunomodulatory role of His (33, 34).
Discussion
Recently, the advancement of analytical techniques to characterize the complexity of tumor metabolism has shed light on understanding multiple types of cancers (7). Most liquid/gas-phase MS techniques need rigorous sample treatment of ∼0.5 to 1 h and a sample volume of ∼10 to 500 µL per sample by chromatography to remove unwanted biomolecules (e.g., salts and proteins) at high concentrations of ∼milligrams/milliliter to overcome the sample complexity and enrich the target metabolites (35–37). By comparison, our approach offers a high analytical speed (∼30 s per sample) with simple sample treatment and low sample volume (∼10 to 100 nL), both of which were improved by orders of magnitudes owing to the on-chip nanoparticle-assisted selective and sensitive LDI for small metabolites in complex biofluids and microarray-based automatic detection.
Notably, the reproducibility of metabolic fingerprinting is critical in large-scale applications. In NMR-based metabolic fingerprinting, ∼95% of features displayed intensity CVs < 30% due to the stable magnetic field with the frequency locked by isotope-labeled additives (35, 38). In metabolic fingerprinting based on conventional MS, ∼70% of features displayed intensity CVs < 30% owing to the multiprocedures during sample treatment by chromatography methods and distinct ionization efficiency of various metabolites (37). In contrast, our approach displayed ∼95% of features with intensity CVs < 30% due to simple sample pretreatment and enhanced ionization efficiency toward small metabolites. Therefore, we have achieved high-performance serum metabolic fingerprinting with desirable analytical speed, sample volume, and detection reproducibility, which can be fundamental for feature selection and machine learning in the next stage. The size-selective trapping effect of nanoparticles could be attributed to the nanoscale surface roughness of nanoparticles, making the small metabolites trapped by nanocrevices on the surface and facilitating efficient ionization of small metabolites compared with macromolecule (39–41). The high detection sensitivity of ferric nanoparticles could be mainly concluded into two aspects: 1) For photo-thermal properties, the ferric nanoparticles exhibited strong ultraviolet (UV) laser absorption and low thermal conductivity and could be heated to a high temperature by the laser irradiation toward the efficient molecular desorption (20, 42), and 2) for the unique ionization mechanism of NPELDI, a positive ion layer (including H+, Na+, and K+) was formed on the negatively charged surface of ferric nanoparticles (39), and the molecules containing polar functional groups (such as a hydroxyl group) can be ionized on the surface of ferric nanoparticles through the dipole–dipole interaction between the molecule and the nanoparticles (43), which would facilitate the production of cation-adducted species.
In a case–control design, we enrolled 325 pathologically defined subjects in this work (Materials and Methods), comparable to previous studies (5, 12, 44–47). To determine the minimum sample number to conduct machine learning, we used a power analysis of ten samples (five/five BrCa/non-BrCa compartments) as a pilot study and obtained a power of > 0.8 with the sample number of 200 (100/100 BrCa/non-BrCa compartments) at a false discovery rate of 0.10 (SI Appendix, Fig. S11), validating that the machine learning results were at a sufficient confidence level.
Classical diagnostic strategies, such as physical examination, mammography, and biopsy, are still limited for large-scale screening due to their inherent characteristics, calling for medical professionals with rich experience of years, large instruments of high cost in purchase/maintenance, or inevitable invasiveness with low population compliance (4, 48, 49). Previously, other MS techniques, including liquid chromatography-MS (LC-MS), gas chromatography-MS (GC-MS), and matrix-assisted laser desorption ionization-MS, have been explored in previous studies for BrCa diagnosis (12, 47, 50–52). Compared with those studies (SI Appendix, Table S12), our work was conducted based on a well-designed cohort and showed desirable diagnostic performance with an AUC of 0.948 (95% CI of 0.922 to 0.973), which is promising for large-scale screening use in clinical practice.
Moreover, an MP-score was constructed using a four-metabolite panel by analyzing the SMF dataset from patients with complete follow-up information. Although the TNM staging system has been regarded as the main prognostic predictor, the four–metabolite-based MP-score model showed comparable prediction efficiency, achieving high prognosis efficiency of P = 0.0018 with an AUC of 0.849 (95% CI of 0.749 to 0.949). Of note, the NPELDI–MS-based SMF analysis only required a blood test, which is promising for universal and large-scale point-of-care applications.
To further determine specific BrCa serum metabolic biomarkers, we successfully identified a biomarker panel of seven BrCa-specific serum small metabolites and validated them through accurate mass measurements or MS/MS. Among them, His, Ura, Thy, DB, and DP were down-regulated (P < 0.05), while GA and NAM were up-regulated (P < 0.001) in BrCa compartments. Then, we constructed a seven-metabolite NN model that achieved high diagnosis efficiency with an AUC of 0.865 (95% CI of 0.820 to 0.911). KEGG pathway analysis revealed three metabolic pathways related to the seven metabolites: pyrimidine metabolism, nicotinate, NAM metabolism, and histidine metabolism. Further study needs to be conducted to determine the underlying mechanism of how they precisely contribute to BrCa development and progression.
In general, we have established a high-performance serum metabolic fingerprinting platform based on NPELDI-MS and have identified BrCa-specific metabolites as potential diagnostic/prognostic markers. Our work would contribute to the metabolic analysis of BrCa and provide intervention targets toward cancer treatment.
Materials and Methods
Clinical Subjects and Sample Harvesting.
This study was conducted in accordance with the principles of the Declaration of Helsinki, and the study protocol was approved by the ethical committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine (Shanghai, China, permit number [2017]KY053). Serum samples were collected from Shanghai General Hospital, and signed informed consents were obtained from participants or their authorized representatives prior to all study procedures.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Funds (Grant 81971771, Grant 81772802, Grant 82073269, and Grant M-0349), the Ministry of Science and Technology of China (Grant 2017YFE0124400 and Grant 2018YFC1312800), the Shanghai Institutions of Higher Learning (Grant 2021-01-07-00-02-E00083), the Shanghai Science and Technology Innovation Action Plan (Grant 20XD1402800), the Clinical Research Plan of Shanghai Shen-kang Hospital Development Centre (Grant SHDC2020CR2065B), the Shanghai Rising-Star Program (Grant 19QA1404800), the Innovation Research Plan by the Shanghai Municipal Education Commission (Grant ZXWF082101), the Clinical Research Innovation Plan of Shanghai General Hospital (Grant CTCCR-2016B05), the Natural Science Foundation of Jiangsu Province (Grant BK20181186), the Innovation Research Plan by the Shanghai Municipal Education Commission (Grant ZXWF082101), the National Research Center for Translational Medicine Shanghai (Grant TMSK-2021-124, NRCTM(SH)-2021-06), the Medical-Engineering Joint Funds of Shanghai Jiao Tong University (Grant YG2019QNA44, Grant YG2021ZD09, Grant YG2022QN107), and the Shanghai Science and Technology Commission (Grant 20JC1410100).
Footnotes
Competing interest statement: The authors have filed patents for both the technology and the use of the technology to detect biosamples.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122245119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Cancer Genome Atlas Network, Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabár L., et al. , The incidence of fatal breast cancer measures the increased effectiveness of therapy in women participating in mammography screening. Cancer 125, 515–523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffy S. W., et al. , Mammography screening reduces rates of advanced and fatal breast cancers: Results in 549,091 women. Cancer 126, 2971–2979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reisenauer C., Fazzio R. T., Hesley G., JOURNAL CLUB: Ultrasound-guided breast interventions: Low incidence of infectious complications with use of an uncovered probe. AJR Am. J. Roentgenol. 208, 1147–1153 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Vinik Y., et al. , Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci. Adv. 6, eaba5714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong C.-Q., et al. , A panel of tumor-associated autoantibodies for the detection of early-stage breast cancer. J. Cancer 12, 2747–2755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ignatiadis M., Sledge G. W., Jeffrey S. S., Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 18, 297–312 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Hart C. D., et al. , Serum metabolomic profiles identify ER-positive early breast cancer patients at increased risk of disease recurrence in a multicenter population. Clin. Cancer Res. 23, 1422–1431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B., The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Faubert B., Solmonson A., DeBerardinis R. J., Metabolic reprogramming and cancer progression. Science 368, eaaw5473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahendralingam M. J., et al. , Mammary epithelial cells have lineage-rooted metabolic identities. Nat. Metab. 3, 665–681 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Hadi N. I., et al. , Serum metabolomic profiles for breast cancer diagnosis, grading and staging by gas chromatography-mass spectrometry. Sci. Rep. 7, 1715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suman S., Sharma R. K., Kumar V., Sinha N., Shukla Y., Metabolic fingerprinting in breast cancer stages through 1H NMR spectroscopy-based metabolomic analysis of plasma. J. Pharm. Biomed. Anal. 160, 38–45 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Gao H., et al. , Application of ex vivo 1H NMR metabonomics to the characterization and possible detection of renal cell carcinoma metastases. J. Cancer Res. Clin. Oncol. 138, 753–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin L., et al. , LC-MS-based serum metabolic profiling for genitourinary cancer classification and cancer type-specific biomarker discovery. Proteomics 12, 2238–2246 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Su H., et al. , Plasmonic alloys reveal a distinct metabolic phenotype of early gastric cancer. Adv. Mater. 33, e2007978 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Cao J., et al. , Metabolic fingerprinting on synthetic alloys for medulloblastoma diagnosis and radiotherapy evaluation. Adv. Mater. 32, e2000906 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Zhang M., et al. , Ultra‐fast label‐free serum metabolic diagnosis of coronary heart disease via a deep stabilizer. Adv. Sci. (Weinh.) 8, e2101333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng H., et al. , Monodisperse magnetic single-crystal ferrite microspheres. Angew. Chem. Int. Ed. Engl. 44, 2782–2785 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Yagnik G. B., et al. , Large scale nanoparticle screening for small molecule analysis in laser desorption ionization mass spectrometry. Anal. Chem. 88, 8926–8930 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Gao L., et al. , Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2, 577–583 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Giuliano A. E., et al. , Breast cancer—Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67, 290–303 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Weiss A., et al. , Validation study of the American Joint Committee on Cancer Eighth Edition prognostic stage compared with the anatomic stage in breast cancer. JAMA Oncol. 4, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee C., Paul M., Xie L., van der Donk W. A., Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–684 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Malek K., Królikowska A., Bukowska J., pH and substrate effect on adsorption of peptides containing Z and E dehydrophenylalanine. surface-enhanced Raman spectroscopy studies on Ag nanocolloids and electrodes. J. Phys. Chem. B 118, 4025–4036 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Mohan S., et al. , Multi-modal prediction of breast cancer using particle swarm optimization with non-dominating sorting. Int. J. Distrib. Sens. Netw. 16, 155014772097150 (2020). [Google Scholar]
- 27.Gong Y., et al. , Metabolic-pathway-based subtyping of triple-negative breast cancer reveals potential therapeutic targets. Cell Metab. 33, 51–64 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Rabinovich S., et al. , Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv H., et al. , NAD+ metabolism maintains inducible PD-L1 expression to drive tumor immune evasion. Cell Metab. 33, 110–127 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Chowdhry S., et al. , NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 569, 570–575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiarugi A., Dölle C., Felici R., Ziegler M., The NAD metabolome—A key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Hasmann M., Schemainda I., FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63, 7436–7442 (2003). [PubMed] [Google Scholar]
- 33.Nicoud M. B., et al. , Study of the antitumour effects and the modulation of immune response by histamine in breast cancer. Br. J. Cancer 122, 348–360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X. D., et al. , Histamine deficiency promotes inflammation-associated carcinogenesis through reduced myeloid maturation and accumulation of CD11b+Ly6G+ immature myeloid cells. Nat. Med. 17, 87–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckonert O., et al. , Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2, 2692–2703 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Dunn W. B., et al. ; Human Serum Metabolome (HUSERMET) Consortium, Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Want E. J., et al. , Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 5, 1005–1018 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Dumas M.-E., et al. , Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: The INTERMAP Study. Anal. Chem. 78, 2199–2208 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang L., et al. , Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 11, 3556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang L., et al. , Plasmonic silver nanoshells for drug and metabolite detection. Nat. Commun. 8, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun X., et al. , Metabolic fingerprinting on a plasmonic gold chip for mass spectrometry based in vitro diagnostics. ACS Cent. Sci. 4, 223–229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu H.-W., Unnikrishnan B., Anand A., Mao J.-Y., Huang C.-C., Nanoparticle-based laser desorption/ionization mass spectrometric analysis of drugs and metabolites. J. Food Drug Anal. 26, 1215–1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., et al. , Magnetic solid phase extraction of brominated flame retardants and pentachlorophenol from environmental waters with carbon doped Fe3O4 nanoparticles. Appl. Surf. Sci. 321, 126–135 (2014). [Google Scholar]
- 44.Tan B., et al. , Identifying potential serum biomarkers of breast cancer through targeted free fatty acid profiles screening based on a GC-MS platform. Biomed. Chromatogr. 34, e4922 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Dowling P., et al. , Metabolomic and proteomic analysis of breast cancer patient samples suggests that glutamate and 12-HETE in combination with CA15-3 may be useful biomarkers reflecting tumour burden. Metabolomics 11, 620–635 (2015). [Google Scholar]
- 46.Louis E., et al. , Phenotyping human blood plasma by 1H-NMR: A robust protocol based on metabolite spiking and its evaluation in breast cancer. Metabolomics 11, 225–236 (2015). [Google Scholar]
- 47.Lee S. B., et al. , Breast cancer diagnosis by analysis of serum N-glycans using MALDI-TOF mass spectroscopy. PLoS One 15, e0231004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehman C. D., et al. ; Breast Cancer Surveillance Consortium, Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern. Med. 175, 1828–1837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verkooijen H. M., et al. , Diagnostic accuracy of large-core needle biopsy for nonpalpable breast disease: A meta-analysis. Br. J. Cancer 82, 1017–1021 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozar N., et al. , Identification of novel diagnostic biomarkers in breast cancer using targeted metabolomic profiling. Clin. Breast Cancer 21, e204–e211 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Silva A. A. R., et al. , Multiplatform investigation of plasma and tissue lipid signatures of breast cancer using mass spectrometry tools. Int. J. Mol. Sci. 21, 3611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jasbi P., et al. , Breast cancer detection using targeted plasma metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1105, 26–37 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.