Abstract
Microcin H47 is a bactericidal antibiotic produced by a naturally occurring Escherichia coli strain isolated in Uruguay. The microcin genetic system is located in the chromosome and extends over a 10-kb DNA segment containing the genes required for microcin synthesis, secretion, and immunity. The smallest microcin synthesis gene, mchB, was sequenced and shown to encode a highly hydrophobic peptide. An mchB-phoA gene fusion, which directed the synthesis of a hybrid bifunctional protein with both PhoA and microcin H47-like activities, was isolated. The results presented herein lead us to propose that microcin H47 is indeed a ribosomally synthesized peptide antibiotic and that its peptide precursor already has antibiotic activity of the same specificity as that of mature microcin.
The production of ribosomally synthesized peptide antibiotics can be considered a successful strategy for mediating bacterial antagonistic relationships, as judged by its widespread occurrence among bacteria. These antibiotics are produced by gram-positive bacteria as well as by gram-negative bacteria and appear to share some features, such as their relatively small size (1.18 to 9.00 kDa) and their mode of secretion into the extracellular medium, which is dependent on dedicated export mechanisms (9, 16, 18). Many of these antibiotics are synthesized as peptide precursors which are subsequently modified by other proteins whose determinants are linked to the antibiotic structural gene. Examples of modified peptide antibiotics are the lantibiotics and most microcins (3, 8, 14, 18, 22, 23). The secretion step can also involve the processing of an N-terminal leader peptide (16, 18). Most of the peptide antibiotics produced by gram-positive organisms are proposed to act as ionophores at the level of the cytoplasmic membrane, dissipating the electrochemical gradient across this membrane (6, 28, 31). In contrast, microcins, which are produced by gram-negative bacteria, exhibit a wide range of cellular targets; e.g., microcin B17 has been shown to be an inhibitor of DNA gyrase (32), microcin C7 inhibits protein synthesis (14), and colicin V disrupts the membrane potential (35).
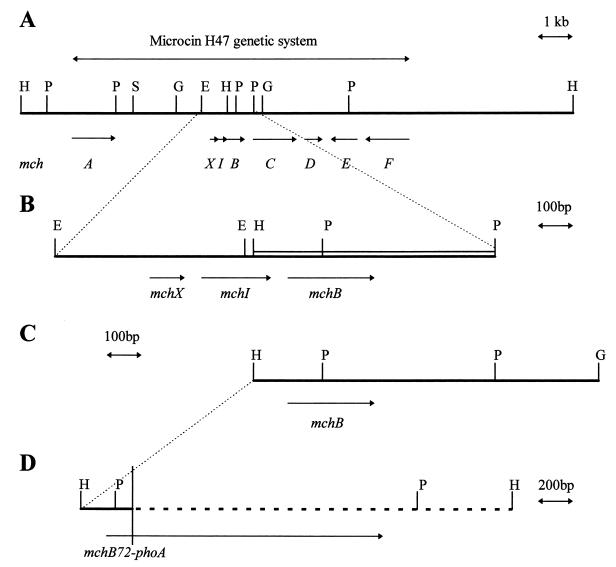
Microcin H47 (MccH47) is a bactericidal antibiotic produced by a naturally occurring Escherichia coli strain. Due to its size, it shares with other microcins the ability to pass through cellophane membranes. The MccH47 genetic determinants are clustered in a 10-kb DNA segment located in the chromosome (20, 21). Saturation mutagenesis of the MccH47 system revealed the presence of four genes, mchA, mchB, mchC, and mchD, devoted to MccH47 synthesis; an immunity gene, mchI, encoding a small, 69-residue integral membrane peptide; and two further genes, mchE and mchF, required for the secretion of the antibiotic into the extracellular medium (10, 27) (Fig. 1A). The expression of the unlinked tolC gene was also shown to be necessary for the production of MccH47 in normal amounts (10). A small gene, mchX, was found upstream of the immunity determinant; preliminary results point to its involvement in the activation of its own expression and probably in that of downstream immunity and production genes (27). The complexity of the MccH47 antibiotic system parallels that of other microcin systems, such as those of microcins B17 and C7 (13, 18). In addition, the MccH47 system contains a ca. 3-kb DNA segment located between the genes mchA and mchX which could not be clearly related to any antibiotic function and was thus provisionally called the “silent region” (10, 27) (Fig. 1A).
FIG. 1.
(A) DNA region containing the MccH47 genetic system cloned in the recombinant plasmid pEX2000. The physical map as well as the extension of the antibiotic system is shown. mch genes, with their directions of transcription, are represented by the italic letters and arrows underneath the map. (B) Enlargement of the mchXIB cluster. The double line represents the DNA segment whose nucleotide sequence is presented in Fig. 2. Arrows indicate directions of transcription for mch genes. (C) DNA segment containing the mchB gene cloned in plasmid pUY100. (D) DNA segment cloned in plasmid pMVD10. Continuous line, mch DNA; dashed line, TnphoA DNA. The mchB72-phoA gene fusion is shown underneath. The vertical line indicates the junction site between mchB and phoA. E, EcoRI; G, BglII; H, HindIII; P, PstI; S, SalI. The restriction maps are complete for the enzymes listed above, except for EcoRI in panel A, where only one site is indicated, and except in panel D, where they are not shown.
In the central region of the MccH47 genetic system, three small genes have been identified. They are mchX; the immunity determinant, mchI; and the smallest microcin production gene, mchB (Fig. 1A and B). mchX and mchI are known to be transcribed in the same direction, towards mchB. We have previously proposed that these three genes would be cotranscribed in the rightwards direction, based on the polar effects of insertion mutations located in mchX (10, 27).
In the present work, we deal with MccH47 production and provide evidence that this antibiotic is ribosomally synthesized as a peptide precursor. The smallest of the four genes required for microcin synthesis, mchB, is proposed to be the structural gene for MccH47. Its peptide product would exert an antibiotic action with the same specificity as that of mature MccH47.
MATERIALS AND METHODS
Culture media.
Luria-Bertani (LB) rich medium and M63 minimal medium were used (25). M63 minimal medium was supplemented with a carbon source at a final concentration of 0.2% and with thiamine at 1 mg/liter. The antibiotics were added at the following final concentrations (in micrograms per milliliter): ampicillin (AP), 50; chloramphenicol (CM), 30; kanamycin (KM), 30. The chromogenic indicator 5-bromo-4-chloro-3-indolylphosphate (XP) for detecting alkaline phosphatase activity was added to solid media at 40 μg/ml.
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed and described in Table 1.
TABLE 1.
Bacterial strains, plasmids, and phage used for this study
| Strain, plasmid, or phage | Genotype or phenotype | Reference |
|---|---|---|
| E. coli K-12 | ||
| CC118 | araD139 Δ(ara-leu)7697 argE(Am) galE galK ΔlacX74 ΔphoA20 recA1 rpoB rpsE thi | 24 |
| RYC1000 | araD139 gyrA ΔlacU169 rbsΔ7 recA56 relA rpsL thiA | 10 |
| CM1470 recA | asnB32 atp-706(ΔIBEFHA) recA relA1 spoT1 thi-1 | 34 |
| Plasmids | ||
| pEX2000 | pBR322 carrying the MccH47 system | 10 |
| pEX4 | pUC13 carrying the MccH47 system | 21 |
| pEX4::Tn5 76 | pEX4 with Tn5 in mchB | 10 |
| pUY69 | pUC13 carrying mchI | 27 |
| pMVD10 | pACYC184 carrying the mchB72-phoA gene fusion | This work |
| pUY100 | pUC13 carrying mchB | This work |
| Phage | ||
| λ-TnphoA | b221 cI857 Pam3 with TnphoA in or near rex | 15 |
Microcin production and immunity or resistance assays.
Microcin production was assayed by patch test, which consisted of picking each strain under study with a sterile toothpick and stabbing it onto a lawn of MccH47-sensitive cells (e.g., RYC1000). After incubation of the plates, halos of growth inhibition appeared around MccH47-producing stabs. Microcin immunity or resistance was evaluated by the cross-streaking method, and detection of intracellular MccH47 was performed by cell lysis (10). The antibiotic activity in the cell lysates was assayed on a lawn of microcin-sensitive cells.
Manipulation and sequencing of DNA.
Plasmid DNA purification, transformation of competent E. coli cells, restriction enzyme digestions, and other routine DNA manipulations were performed as described previously (29). Sequencing reactions were carried out by the chain termination method (30), applied to double-stranded DNA, with synthetic oligonucleotides and the Sequenase 2.0 kit from United States Biochemicals (Cleveland, Ohio).
TnphoA mutagenesis.
To generate gene fusions with phoA, strain CC118(pEX2000) was mutagenized with λTnphoA as described previously (15). Separate experiments were carried out to obtain independent mutants. Insertion mutants were selected on LB-AP-KM-XP plates. The chromogenic indicator XP allowed a distinction to be made between PhoA+ clones (blue) and PhoA− clones (white). Blue colonies were purified and their plasmid DNA was extracted and used to transform CC118 cells; transformants were selected on LB-AP-KM-XP plates. In principle, blue transformant colonies had received plasmid DNA with a TnphoA insertion, generating a gene fusion whose product was being exported.
Alkaline phosphatase assay.
Cultures were grown overnight in LB medium and then diluted 1/50 into fresh medium and grown further to bring them into exponential-phase growth by the time of assay. The assay was done twice for each strain. The alkaline phosphatase activities were determined as described previously (5).
Nucleotide sequence accession number.
The sequence of the DNA fragment containing mchB has been deposited in the EMBL database under accession no. AJ009631.
RESULTS
Nucleotide sequence of the mchB gene and analysis of its predicted product.
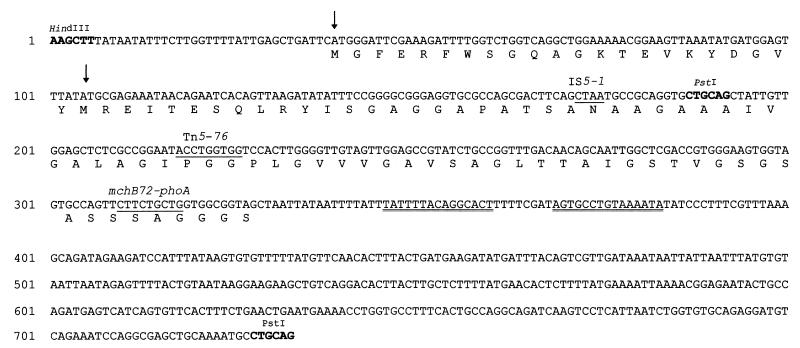
A DNA segment containing mchB was completely sequenced. As expected, mchB was found downstream of mchI, being transcribed in the same direction (Fig. 1B and 2). Two possible in-frame ATG translation start sites were found at positions 39 and 105, initiating two open reading frames (ORFs) of 97 and 75 codons, respectively. The first ATG is preceded by a GAG at a distance of 8 bp, and the corresponding ORF overlaps the end of the immunity gene by 16 codons. Six nucleotides upstream of the second translation initiation codon there is a potential ribosome binding site, GGAG. This second ORF begins 20 bp after the end of the immunity gene and appears to be the best candidate for mchB; it would encode a 6,600-Da peptide (Fig. 2). In this communication, we operationally define the 75-codon ORF as the mchB gene. Downstream of mchB, there is a 15-bp inverted repeat, with 8 bp between the repeated sequences (Fig. 2).
FIG. 2.
DNA sequence of the 733-bp HindIII-PstI segment containing mchB. The numbers on the left refer to nucleotide positions. HindIII and PstI sites are indicated. The predicted amino acid sequence of MchB is shown. Two potential start codons were found (arrows). The sequences of the direct repeats created by transposon insertions are underlined. The TnphoA insertion is indicated with the fusion name. Downstream of mchB, a 15-bp inverted repeat is indicated by double underlining.
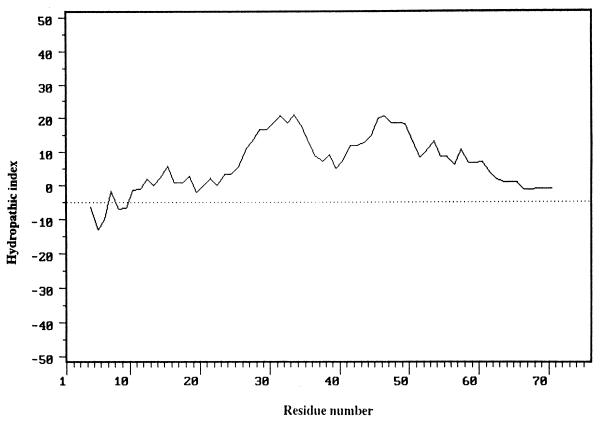
The predicted MchB protein product presents an extensive hydrophobic region extending over its 65 carboxy-terminal residues, where no charged amino acids were found (Fig. 2 and 3). Computer-aided analyses indicated that MchB would be an integral membrane peptide (7, 17). Moreover, a Sec signal peptide at its amino terminus, including four possible leader peptidase cleavage consensus sites located in the stretch between residues 22 and 29, is also recognized (33). This observation suggests that MchB could be exported via the Sec pathway, including the signal peptide processing step. However, a double-glycine motif, present in many peptides secreted via dedicated ATP-binding cassette (ABC) export systems (16), was also identified in the MchB amino terminus (Fig. 4). An ABC export mechanism would imply a concomitant cleavage of MchB with the release of a 60-residue peptide into the extracellular medium. In sum, the data from sequencing indicate that the mchB gene product must be exported and, most probably, is also processed during the course of its export. However, both types of export pathways, Sec and ABC, appear to be compatible with the deduced MchB sequence.
FIG. 3.
Hydropathic profile of the predicted MchB peptide. The sequence was analyzed by using a 9-residue window and the hydropathic index computation described in reference 19.
FIG. 4.
Comparison of the putative double-glycine signal peptide of MchB with the amino acid sequences of the double-glycine signal peptides belonging to the following peptide antibiotics: colicin V (CvaC), lactococcin Gα (LagA), pediocin PA-1 (PedA), lactococcin Gβ (LagB), plantaricin A (PlnA), Enterococcus faecalis cytolysin (CylL1 and CylL2), lactococcin A (LcnA), lactococcin B (LcnB), lactococcin M (LcnM and LcnN), and lactococcin DR (LcnDR1). Except for MchB, all sequences were extracted from reference 16. The asterisks indicate the MchB residues found in the same position in four or more of the other antibiotic peptides. The arrow indicates the consensus cleavage site.
The MchB amino acid sequence was compared with sequences in databases by using BLASTP 1.4.11 (1). No sequence similarities were found.
Construction and analysis of an mchB-phoA gene fusion.
Strain CC118(pEX2000) was mutagenized with TnphoA as described in Materials and Methods. Plasmid pEX2000 is a pBR322 derivative bearing the DNA segment represented in Fig. 1A. Cells carrying pEX2000 produce and are immune to MccH47. A PhoA+ plasmid mutant clone was analyzed, and its TnphoA insertion was sequenced and shown to originate an in-frame mchB-phoA gene fusion. This fusion was called mchB72-phoA, in reference to the mchB codon where the junction site was located (Fig. 2). The alkaline phosphatase activity of fusion-bearing cells was measured as described in Materials and Methods, and a mean value of 147 U was obtained, indicating that MchB must contain an export signal in its amino-terminal portion. As expected for a clone mutated in one of the antibiotic synthesis genes, this mutant strain was impaired for microcin production as analyzed by patch test and remained immune to MccH47.
For several purposes, we attempted to subclone and express the mchB72-phoA fusion in the absence of the remaining mch genes. A HindIII fragment containing the gene fusion (Fig. 1D) was ligated to HindIII-digested pACYC184, and the ligation mixture was used to transform strain CC118(pEX4::Tn5 76). Plasmid pEX4::Tn5 76 carries the entire MccH47 system with a Tn5 insertion in mchB (Fig. 2). As we considered the hypothesis that mchB could be the structural gene for MccH47, it was thought that the PhoA+ phenotype conferred by the fusion would depend on the presence of the mchE and mchF genes, whose products we knew were involved in MccH47 export. Therefore, plasmid pEX4::Tn5 76 would provide the remaining mch genes in trans. AP-resistant (Apr), CM-resistant (Cmr), PhoA+ transformant clones were selected. One of them harbored the desired construction, and this plasmid was called pMVD10 (Fig. 1D). The plasmid DNA from this strain was employed to transform strain CC118, with selection of the Cmr clones, in order to obtain transformants with pMVD10 and not with pEX4::Tn5 76. Surprisingly, all transformants were also Apr, i.e., they carried both plasmids.
This result suggested that cells bearing plasmid pMVD10 could not grow unless another mch gene(s) was also present. Several different strains were then transformed with the plasmid DNA mixture of pMVD10 and pEX4::Tn5 76 to ascertain which additional gene(s) was needed for pMVD10 transformants to be viable. The DNA mixture employed was previously digested with BglII, which cuts pEX4::Tn5 76 and has no target on pMVD10. By this procedure, the probability of appearance of pEX4::Tn5 76 transformants was enormously decreased, since linear DNA is not able to transform normal E. coli cells. The strains employed for these experiments contained different DNA fragments from the MccH47 system cloned into plasmids compatible with pMVD10. In all cases, Cmr transformants were obtained in the context of strains immune to MccH47, and no growth appeared when strains lacking immunity were used. The most conclusive result was reached by transforming cells with plasmid pUY69, which only contains the mchI gene: abundant Cmr colonies appeared, indicating that pMVD10 gave rise to a toxic effect which was specifically overcome by the presence of the immunity gene, mchI. It was concluded that pMVD10 encodes a deleterious activity with MccH47 specificity which impairs cell growth in the absence of its cognate immunity peptide.
However, cells carrying pMVD10 together with pEX4::Tn5 76 or pUY69 did not produce inhibition halos on a lawn of sensitive cells. To assay their intracellular microcin activity, they were lysed and the cell lysates were tested as described in Materials and Methods. No inhibition halos were detected. Therefore, strains with pMVD10 are capable of producing an MccH47-like activity, detected by its toxic effect on the producing cell itself, but are unable to produce inhibition halos when assayed on a lawn of MccH47-sensitive cells.
With regard to the MchB72-PhoA mode of export, it was important to discern if it was dependent on the presence of the remaining mch genes and, particularly, on mchE and mchF. If this were so, strains carrying both plasmids pMVD10 and pUY69 (without any other mch gene) would lack alkaline phosphatase activity. This was not the case, and CC118(pMVD10, pUY69) exhibited a level of alkaline phosphatase activity (120 U) similar to that of CC118(pEX2000) mchB72-phoA. It should be noted that the gene fusion is expressed in pMVD10, as judged by the enzymatic activity of its product, even when the mchB promoter is most probably absent in the cloned fragment.
Is mchB the structural gene for MccH47?
At this stage of the work, it was not evident which genetic determinant in pMVD10 was responsible for the toxic MccH47-like activity detected. Was it the mchB72-phoA fusion, or was it another small mch gene, as yet unidentified? The following experiments shed light on this dilemma.
When RYC1000 cells were transformed with pMVD10, no transformant clones appeared on the plates after 24 h of incubation. The plates were incubated for an additional 2 days, and a few Cmr clones grew. Five clones proved to grow stably in LB medium with CM and were analyzed.
Three of these clones turned out to be PhoA−. RYC1000 cells were transformed with their plasmid DNA, and abundant Cmr colonies appeared. It was concluded that these three clones were plasmid mutants which had been affected in the MccH47 structural genetic determinant. Their plasmids were called pMVD10-1, pMVD10-2, and pMVD10-3 and had molecular sizes ca. 1.5 kb greater than that of pMVD10, most probably due to transposition of insertion sequences (IS). This was in fact shown to be the case, and the sites of the insertions and their nature were determined by DNA sequencing for the first two plasmids. In pMVD10-1, the junction site with the left end of IS5 was located in codon 23 of the mchB portion of the gene fusion (Fig. 2). pMVD10-2 bore IS10, whose O end was sequenced. The site of this insertion was precisely mapped to codon 31 of the phoA moiety of the gene fusion (counting included the linker residues). The aberrant fusion protein produced would have 103 residues, the last one being directed by the phoA-IS10 junction. In pMVD10-3, the site of the putative IS insertion was determined by restriction analysis and was found to be located in the phoA region of pMVD10, far away from the mchB-phoA junction. In the three pMVD10 mutant derivatives, the sequence analysis was extended to confirm that no second mutation had occurred upstream of the IS insertions, in the mchB portion of the gene fusion. No alteration of the mchB sequence was found.
The information provided by these mutants indicated that the mchB-phoA gene fusion codes for a bifunctional hybrid protein involved in both toxic MccH47-like and alkaline phosphatase activities. The insertion disruption of either of its constituent portions, mchB or phoA, simultaneously abolished both activities. These results strongly indicated that the mchB gene itself must code for a peptide with the MccH47-like toxic activity.
The remaining two Cmr clones that appeared when RYC1000 cells were transformed with pMVD10 retained the PhoA+ phenotype. Their plasmids were used to transform RYC1000 cells, and no growth appeared, as happened with pMVD10, suggesting that these two clones were chromosomal mutants. Both exhibited poor growth, and when analyzed for their MccH47 sensitivity, they proved to be completely resistant, contrasting with the three plasmid mutant clones presented above, which remained microcin sensitive. As we had previously isolated MccH47-resistant mutants with a similar phenotype whose mutations mapped to the atp operon in the chromosome (our unpublished results), the two clones under study were assayed for their Atp phenotype. It is known that atp mutants, i.e., those affected in the ATP synthase complex, cannot grow on minimal succinate medium (4). The clones under study were thus assayed on this medium: none of these mutants grew, suggesting that they were also affected in the atp locus. Moreover, CM1470 recA, a Δatp strain (34), as well as previously selected MccH47-resistant atp mutants, was able to grow when transformed with pMVD10.
The wild-type mchB gene was subcloned in a 1-kb HindIII-BglII DNA fragment under the control of the lac promoter in the pUC13 vector: the recombinant plasmid pUY100 in principle contained no other mch gene (Fig. 1C). It was deduced to encode MccH47-like activity, as did pMVD10, since it could be propagated in cells of atp mutants such as CM1470 recA, but not in those of atp+ nonimmune strains. Paralleling pMVD10-containing cells, those carrying pUY100 could not inhibit the growth of an MccH47-sensitive strain, as assayed either by performing a patch test or by checking the activity of cell lysates (see Materials and Methods).
DISCUSSION
The results presented here describe the mchB gene, which had previously been identified as one of the four mch genes required for the synthesis of MccH47 (10). This small ORF is now proposed to be the structural gene for the antibiotic, and this would therefore lead us to conclude that MccH47 is indeed a ribosomally synthesized peptide antibiotic.
The mchB gene is located in the center of the microcin genetic system, downstream of mchX and mchI, which are contiguous and are expressed in the same direction. These three genes were proposed to constitute an operon (27). A 15-bp inverted repeat found downstream of mchB could well accomplish transcription termination functions at the end of this operon.
The deduced amino acid sequence of the MchB product reveals a highly hydrophobic structure. Its analysis pointed to the possible export of this peptide and revealed that, from the structural point of view, two export pathways, the Sec general pathway and an ABC dedicated secretion system, could be employed. Indeed, at the amino terminus of the MchB peptide, both a Sec export signal sequence (33) and a double-glycine signal sequence were found. This double-glycine motif has been detected in several secreted peptide antibiotics and was shown to signal a site where the peptide is cleaved during the export process. This cleavage is carried out by an ABC transporter, which is always specific for its substrate (16). In the case of MccH47, the mchE and mchF gene products were shown to be required for its secretion into the extracellular medium. The TolC outer membrane protein, not linked to the microcin system, is also involved in the production mechanism (10). These requirements for MccH47 secretion are reminiscent of those for colicin V, whose dedicated exporter belongs to the ABC superfamily (9, 11). Accordingly, the mchB72-phoA gene fusion directs the synthesis of a hybrid protein with alkaline phosphatase activity, indicating that it is in fact exported. However, this enzymatic activity is present in all clones bearing the fusion, independently of their genetic context relative to the remaining mch genes, i.e., at least the PhoA moiety of the hybrid protein was able to be exported to the periplasm even in the absence of mchE and mchF. In view of all these data, we propose that MchB, besides apparently conforming to the structural requirements for its ABC secretion, would reach the periplasm, at least to some extent, even in the absence of a dedicated export system. The study of microcin molecular structure will certainly shed light on its mode of secretion.
Cells with pMVD10 or pUY100 cannot grow unless they also contain the mchI gene. Both plasmids must therefore encode a toxic activity, specifically overcome by MccH47 immunity. It is well known that the immunity protein cognate to a peptide or protein antibiotic is specific and unique to that particular antibacterial molecule, a fact upon which the cross-immunity criterion for the classification of ribosomally synthesized antibiotics was based (2, 26). We therefore concluded that the toxic activity detected had the same specificity as MccH47 and that both pMVD10 and pUY100 must code for an antibiotically active MccH47 precursor. Cross-resistance results also support this conclusion: atp mutants were isolated as resistant to mature MccH47 (our unpublished results), and spontaneous chromosomal mutants with an Atp− phenotype were isolated by their ability to grow when transformed with pMVD10. All these Atp mutants, isolated by different procedures, plus CM1470 recA (34), a strain with an atp deletion, turned out to be resistant to exogenous MccH47 as well as to the endogenous toxic microcin-like activity. These results also indicate that the ATP synthase complex would play an essential role in the MccH47 mode of action.
The smallest microcin so far described is C7, a heptapeptide encoded by a 7-codon ORF (12). The 318-bp mch DNA segment cloned in pMVD10 and responsible for the MccH47-like toxicity seemed large enough to bear several small ORFs besides mchB, each one of which could have been the microcin structural gene. The analysis of the mchB-phoA fusion oriented our thinking regarding this dilemma: is mchB or another small ORF in pMVD10 the structural gene? Three spontaneous mutants were obtained from cells carrying pMVD10 which could grow normally, i.e., they did not suffer from the toxic MccH47-like effect. They carried IS insertions on pMVD10. One of them was located on the mchB portion of the gene fusion, so it did not help us in the elucidation process: the expression of another small gene could be affected by this insertion. However, the remaining two IS insertions were found in the phoA moiety of the fusion, and each one abolished the PhoA and toxic phenotypes at the same time, while the mchB portion remained intact. This clearly meant that the hybrid MchB-PhoA protein was involved in both functions, i.e., that it was bifunctional, and therefore that the mchB portion of the fusion was directly involved in the MccH47-like toxicity. The toxic phenotype conferred by pUY100 coincides with the results obtained with pMVD10. Together, these results support our working hypothesis that mchB is the structural gene for MccH47, although other possibilities cannot be completely ruled out.
Cells carrying the mchB gene but lacking the remaining mch genes previously known to be required for MccH47 synthesis (10) did not produce inhibition halos on a lawn of sensitive cells. This was not due to a defect in microcin secretion, since lysates of these cells were also unable to kill other cells. Therefore, the toxic MccH47-like activity detected in these cells appears to be unable to act exogenously, i.e., the MchB peptide would not enter other cells and reach its target. Therefore, we propose that the remaining microcin synthesis genes, mchA, C, and D, could be involved in the maturation of MchB, endowing it with the ability to enter other cells. A certain parallelism is found with microcin B17 maturation, which depends on the action of a three-protein enzyme complex, the microcin B17 synthase. In this case, posttranslational modifications confer antibiotic activity to the molecule (22). However, results presented in this work indicate that the MchB peptide already possesses antibiotic activity and is able to interact with its target in the same way as mature MccH47. A similar conclusion was reached for microcin C7, whose peptide precursor inhibits protein synthesis in vitro but cannot affect the growth of sensitive cells (14).
The results presented in this work strongly support the hypothesis that the mchB gene, already known to be involved in MccH47 synthesis, is the structural gene for this antibiotic. Its peptide product, the microcin precursor, would already possess antibiotic activity of the same specificity as mature microcin, based on cross-immunity and cross-resistance criteria. Its size is appropriate for a gene encoding a microcin peptide, since microcins have molecular masses under 9,000 Da. The corresponding MchB product exhibits a double-glycine motif, characteristic of many peptide antibiotic precursors secreted through ABC exporters. MccH47 purification and N-terminal sequencing will give the definitive answer to these concerns.
ACKNOWLEDGMENTS
This work was supported by International Foundation for Science grant F/1884-1, by European Community grant CI1*-CT92-0011, by Comisión Sectorial de Investigación Científica de la Universidad de la República (Montevideo, Uruguay), by Consejo Nacional de Investigaciones Científicas y Tecnológicas grant 82, and by Programa de Desarrollo de las Ciencias Básicas, Montevideo, Uruguay.
We thank Colin Manoil for kindly supplying λTnphoA and the bacterial strains required for the TnphoA mutagenesis and Ole Michelsen for strain CM1470 recA. We are also indebted to María Parente for excellent technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Baquero F, Moreno F. The microcins. FEMS Microbiol Lett. 1984;23:117–124. [Google Scholar]
- 3.Bayer A, Freund S, Nicholson G, Jung G. Posttranslational backbone modifications in the ribosomal biosynthesis of the glycine-rich antibiotic microcin B17. Angew Chem Int Ed Engl. 1993;32:1336–1339. [Google Scholar]
- 4.Boogerd F C, Boe L, Michelsen O, Jensen P R. atp mutants of Escherichia coli fail to grow on succinate due to a transport deficiency. J Bacteriol. 1998;180:5855–5859. doi: 10.1128/jb.180.22.5855-5859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and Φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 6.Bruno M E C, Montville T J. Common mechanistic action of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1993;59:3003–3010. doi: 10.1128/aem.59.9.3003-3010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claros M G, Von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 8.De Vos W M, Kuipers O P, Van Der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 9.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaggero C, Moreno F, Laviña M. Genetic analysis of microcin H47 antibiotic system. J Bacteriol. 1993;175:5420–5427. doi: 10.1128/jb.175.17.5420-5427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilson L, Mahanty H K, Kolter R. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 1990;9:3875–3884. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González-Pastor J E, San Millán J L, Moreno F. The smallest known gene. Nature. 1994;369:281. doi: 10.1038/369281a0. [DOI] [PubMed] [Google Scholar]
- 13.González-Pastor J E, San Millán J L, Castilla M A, Moreno F. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J Bacteriol. 1995;177:7131–7140. doi: 10.1128/jb.177.24.7131-7140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guijarro J I, González-Pastor J E, Baleux F, San Millán J L, Castilla M A, Rico M, Moreno F, Delepierre M. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J Biol Chem. 1995;270:23520–23532. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 15.Gutiérrez C, Barondess J, Manoil C, Beckwith J. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. J Mol Biol. 1987;195:289–297. doi: 10.1016/0022-2836(87)90650-4. [DOI] [PubMed] [Google Scholar]
- 16.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 17.Klein P, Kanehisa M, De Lisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 18.Kolter R, Moreno F. Genetics of ribosomally synthesized peptide antibiotics. Annu Rev Microbiol. 1992;46:141–164. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- 19.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 20.Laviña M, Gaggero C. Genetic determinants for microcin H47, an Escherichia coli chromosome-encoded antibiotic. In: James R, Lazdunski F, Pattus F, editors. Bacteriocins, microcins, and lantibiotics. Heidelberg, Germany: Springer-Verlag KG; 1992. pp. 413–416. [Google Scholar]
- 21.Laviña M, Gaggero C, Moreno F. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J Bacteriol. 1990;172:6585–6588. doi: 10.1128/jb.172.11.6585-6588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y-M, Milne J C, Madison L L, Kolter R, Walsh C T. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 23.Madison L L, Vivas E I, Li Y-M, Walsh C T, Kolter R. The leader peptide is essential for the post-translational modification of the DNA-gyrase inhibitor microcin B17. Mol Microbiol. 1997;23:161–168. doi: 10.1046/j.1365-2958.1997.2041565.x. [DOI] [PubMed] [Google Scholar]
- 24.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 26.Pugsley A P. Escherichia coli K12 strains for use in the identification and characterization of colicins. J Gen Microbiol. 1985;131:369–376. doi: 10.1099/00221287-131-2-369. [DOI] [PubMed] [Google Scholar]
- 27.Rodríguez E, Laviña M. Genetic analysis of microcin H47 immunity. Can J Microbiol. 1998;44:692–697. doi: 10.1139/cjm-44-7-692. [DOI] [PubMed] [Google Scholar]
- 28.Ruhr E, Sahl H-G. Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob Agents Chemother. 1985;27:841–845. doi: 10.1128/aac.27.5.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venema K, Abee T, Haandrikman A J, Leenhouts K J, Kok J, Konings W N, Venema G. Mode of action of lactococcin B, a thiol-activated bacteriocin from Lactococcus lactis. Appl Environ Microbiol. 1993;59:1041–1048. doi: 10.1128/aem.59.4.1041-1048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizán J L, Hernández-Chico C, Del Castillo I, Moreno F. The peptide antibiotic microcin B17 induces double strand cleavage of DNA mediated by gyrase. EMBO J. 1991;10:467–476. doi: 10.1002/j.1460-2075.1991.tb07969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Von Meyenburg K, Jorgensen B B, Michelsen O, Sorensen L, McCarthy J E G. Proton conduction by subunit a of the membrane-bound ATP synthase of Escherichia coli revealed after induced overproduction. EMBO J. 1985;4:2357–2363. doi: 10.1002/j.1460-2075.1985.tb03939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C-C, Konisky J. Colicin V-treated Escherichia coli does not generate membrane potential. J Bacteriol. 1984;158:757–759. doi: 10.1128/jb.158.2.757-759.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]