Abstract
Alcohol use disorder (AUD) has a complex pathogenesis, making it a difficult disorder to treat. Identifying relevant signaling pathways in the brain may be useful for finding new pharmacological targets to treat AUD. The receptor tyrosine kinase ALK activates the transcription factor STAT3 in response to ethanol in cell lines. Here, we demonstrate ALK activation and upregulation of known STAT3 target genes (Socs3, Gfap, and Tnfrsf1a) in the prefrontal cortex (PFC) and ventral hippocampus (HPC) of mice after 4 days of binge-like ethanol drinking. Mice treated with the STAT3 inhibitor stattic drank less ethanol than vehicle-treated mice, demonstrating the behavioral importance of STAT3. To identify novel ethanol-induced target genes downstream of the ALK and STAT3 pathway, we analyzed the NIH LINCS L1000 database for gene signature overlap between ALK inhibitor (alectinib and NVP-TAE684) and STAT3 inhibitor (niclosamide) treatments on cell lines. These genes were then compared to differentially expressed genes in the PFC of mice after binge-like drinking. We found 95 unique gene candidates, out of which 57 had STAT3 binding motifs in their promoters. We further demonstrated by qPCR that expression of the putative STAT3 genes Nr1h2, Smarcc1, Smarca4, and Gpnmb were increased in either the PFC or HPC after binge-like drinking. Together, these results indicate activation of the ALK-STAT3 signaling pathway in the brain after binge-like ethanol consumption, identify putative novel ethanol-responsive STAT3 target genes, and suggest that STAT3 inhibition may be a potential method to reduce binge drinking in humans.
Keywords: alcohol use disorder, neuroimmune, anaplastic lymphoma kinase, LINCS, signal transducer and activator of transcription 3
1. INTRODUCTION
Alcohol use disorder (AUD) is a psychiatric illness characterized by excessive alcohol consumption, an irresistible compulsion to drink alcohol, an inability to abstain from drinking despite negative consequences, and a negative affective state during withdrawal from alcohol use. In 2017, an estimated 173.3 million people consumed alcohol in the United States, corresponding to over 70% of the United States population over the age of 181. Alcohol use is linked to increased risk of cancers, cardiovascular diseases, self-harm, and many other negative consequences2. In addition to the human cost, the financial burden associated with AUD was estimated at $249 billion in 2010, of which over 75% was attributed to binge drinking3. Binge drinking is defined by the National Institute on Alcohol Abuse and Alcoholism as a pattern of drinking that brings the blood alcohol concentration to 0.08 g/dL, generally after 4 drinks for women and 5 drinks for men, in about 2 hours4. According to the 2018 National Survey on Drug Use and Health, approximately 26% of adults age 18 and over reported binge drinking in the past month5. Binge drinking is a strong predictor of both acute and long-term alcohol-related problems, and has increased 7.7% between 2000–2016, particularly in the female, Black, and over 50 population6. These statistics highlight the need to further understand the neurobiological pathways involved in binge drinking.
Anaplastic lymphoma kinase (ALK) is a receptor tyrosine kinase that is expressed in the brain and whose overactivity has been implicated in many different types of cancers7–10. Ligands of ALK include pleiotrophin, midkine, heparin, ALK and LTK ligand 1 (ALKAL1 or FAM150a), and ALKAL2 (or FAM150b)11–14. One of the first indications that ALK might be involved in AUD came from Alk gene knockout (Alk −/−) mice. Alk −/− mice had increased ethanol-induced sedation, consumed more ethanol in several drinking procedures, and, interestingly, did not escalate their drinking after chronic ethanol exposure compared to Alk +/+ mice15–17. In contrast to the drinking phenotype observed in Alk −/− mice, systemic inhibition of ALK using the small molecule inhibitors, NVP-TAE684 and alectinib, and locally reducing ALK expression in the ventral tegmental area using RNA interference, resulted in decreased binge-like ethanol drinking18. The opposing phenotypes between the Alk −/− mice and manipulations of Alk in adult mice on ethanol drinking may be explained by compensatory mechanisms occurring during development, such as constitutive activation of the MAPK/ERK pathway observed in Alk −/− mice15. Mice treated with NVP-TAE684 also had decreased ethanol conditioned place preference, suggesting that ALK modulates alcohol consumption by facilitating its rewarding properties. Finally, polymorphisms in the human ALK gene have been associated with subjective responses to alcohol and alcohol dependence15,19, demonstrating a potential role for ALK in AUD.
ALK signals through several downstream effectors, including signal transducer and activator of transcription 3 (STAT3)8. STAT3 is a transcription factor whose target genes regulate diverse biological processes such as development, inflammation, and proliferation20. STAT3 is activated by phosphorylation of tyrosine 705 (Y705), allowing for STAT3 dimerization and translocation into the nucleus, where it binds to STAT-responsive elements20. Cell culture experiments demonstrated that ethanol treatment of the human neuroblastoma cell line, IMR-32, activated ALK and STAT3, as measured by tyrosine phosphorylation, and that STAT3 activation in response to ethanol was blocked by treatment with NVP-TAE684, suggesting that STAT3 acts downstream of ALK in response to ethanol in these cells21. The role of STAT3 in alcohol-related disease has been well-studied in the liver22, but its function in the brain in the context of alcohol use disorder is less understood. STAT3 is activated in the hippocampus (HPC)23 and Edinger-Westphal nucleus24 after an acute ethanol injection, and in the cerebellum and hippocampus after chronic intermittent alcohol exposure25,26.
Based on these findings, we hypothesized that the ALK-STAT3 pathway would be activated after binge-like drinking in mice. We tested this hypothesis by examining the phosphorylation of ALK and STAT3, and the expression of known STAT3 target genes, Gfap27 Socs328, Tnfrsf1a29, and Lcn230,31, in the ventral HPC and prefrontal cortex (PFC), after mice underwent 4 days of the drinking in the dark (DID) procedure, which models binge-like ethanol drinking32. We also tested whether inhibition of STAT3, similarly to ALK inhibition18, would reduce binge-like ethanol intake in mice. Finally, in order to discover novel genes downstream of the ALK-STAT3 signaling pathway that are altered by ethanol drinking, we interrogated the National Institutes of Health Library of Integrated Network-Based Cellular Signatures (LINCS) L100033 database for overlapping differentially expressed genes in cells treated with ALK and STAT3 inhibitors, and compared these to differentially expressed genes in the PFC of mice that had undergone long-term ethanol binge-like drinking34. Our results demonstrate that ALK is activated and that select STAT3 target gene expression increases in the ventral HPC and PFC after 4 sessions of binge-like drinking. We also provide evidence of ethanol-induced expression of putative novel STAT3 target genes in the brain that encode proteins involved in chromatin remodeling and innate immunity.
2. MATERIALS AND METHODS
2.1. Animals
Male and female C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) at 8 weeks old and used for experiments beginning at 10 weeks old. Mice had access to food and water ad libitum and were singly housed in climate-controlled rooms with a 12 h light/dark cycle (lights off at 10 am and on at 10 pm) and were tested during the dark cycle. Mice were fed Teklad 7912 diet (Envigo, Indianapolis, IN, USA). All animals were cared for in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and all procedures were approved by the UIC Animal Care and Use Committee.
2.2. Drinking in the dark (DID)
DID was conducted as previously described32. Briefly, mice were individually housed in the reverse dark/light cycle room for two weeks prior to experiments. After this acclimation period, each mouse was given access to a single 10 ml sipper tube containing 20% ethanol (v/v) or water (as a control in western blot and qPCR experiments only), 3 h into the dark cycle, for 2 h on the first three days and 4 h on the 4th day. The volume of fluid consumed was measured at the end of each session. On the 4th day, volume of fluid consumed was measured at both 2 and 4 h. To test sucrose consumption, a separate cohort of mice underwent DID as described for ethanol, except the sipper tubes contained 2% sucrose dissolved in water instead of ethanol. A total of 136 mice underwent DID with ethanol or water for measuring gene expression and protein levels, 44 mice underwent DID with ethanol in the stattic experiments and 24 mice underwent DID with sucrose in the stattic experiments. The specific numbers of mice used for each experiment are indicated in the results section.
2.3. Tissue collection
Mice were euthanized by rapid decapitation either immediately after the final drinking session (defined as the “0 h” timepoint) or 24 hours after sipper tubes were removed. Trunk blood (20 μl) was collected in heparinized capillary tubes, transferred to 1.5 ml centrifuge tubes, and snap frozen and stored at −80°C for subsequent determination of blood ethanol concentrations (BECs). Brains were rapidly removed from the cranium after decapitation and cut into 1 mm-thick sections on ice using a stainless-steel brain matrix (Zivic Instruments Inc., Pittsburgh, PA, USA). The medial PFC (containing infralimibic and prelimbic regions, ~1.9 mm anterior to bregma) and ventral HPC (~4.7 mm posterior to bregma) were dissected out of each section using a razor blade, transferred to 1.5 ml centrifuge tubes, snap frozen, and stored at −80°C until processing for protein analysis by western blots and RNA levels by quantitative real-time PCR (qPCR).
2.4. Stattic administration
Mice underwent the DID protocol as indicated above. On days 2, 3, and 4, mice were injected subcutaneously (s.c.) with 50 μl stattic (~20 mg/kg35,36, Selleck Chemicals, Houston, TX USA) dissolved in 100% dimethylsulfoxide (DMSO), or DMSO as a vehicle control, 30 minutes before sipper tubes were placed on the home cage. Due to the low injection volume, the dose was not precisely 20 mg/kg in each mouse. The concentration of stattic solution injected was adjusted based on the average weight of the males and females separately to approximate a 20 mg/kg dose. For determination of blood ethanol levels, in this experiment only, blood was collected from the tail vein.
2.5. Blood ethanol concentration (BEC) measurement
BECs were measured using the nicotinamide adenine dinucleotide-alcohol dehydrogenase (NAD-ADH) enzymatic assay as previously described37. β-NAD and yeast ADH were purchased from Sigma Aldrich (St. Louis, MO, USA).
2.6. Western blot
Frozen tissue was lysed and manually homogenized using a pestle in ice cold 1X RIPA buffer (Cell Signaling Technology, Danvers, MA, USA) with 1X Halt protease inhibitor (Thermo Fisher Scientific, Waltham, MA, USA), 1X Halt phosphatase inhibitor (Thermo Fisher Scientific), 1 mM sodium fluoride, and 1 mM sodium orthovanadate. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein (25 μg) were separated by gel electrophoresis on precast 4–12% Tris-glycine gels (Novex, Thermo Fisher Scientific) and transferred to nitrocellulose membranes using the Trans-Blot Turbo system (Bio-Rad, Hercules, CA, USA). Membranes were blocked with Odyssey Blocking TBS Buffer (LI-COR, Lincoln, NE, USA). Membranes were incubated with primary (4°C, overnight) and secondary (1 hour, room temperature) antibodies in 5% BSA in TBST (20 mM Tris, 150 mM NaCl, pH 7.4, 0.1% Tween-20). Primary antibodies were the following: STAT3, Cell Signaling Technology #9139, RRID: AB_331757, 1:1000 dilution; phosphorylated STAT3 (pSTAT3, Y705), Cell Signaling Technology #9145, RRID: AB_2491009, 1:1000 dilution; ALK #381518, 1:500 dilution; phosphorylated ALK (pALK, Y1278), Cell Signaling Technology #6941, RRID: AB_10860598, 1:500 dilution; and β-actin, Sigma-Aldrich #A5441, RRID: AB_476744, 1:10,000 dilution. Secondary antibodies were the following: IRDye 680RD Donkey anti-Mouse IgG, LI-COR Biosciences #925–68072, RRID: AB_2814912, 1:10,000 dilution and IRDye 800CW Donkey anti-Rabbit IgG, LI-COR Biosciences #925–32213, RRID: AB_2715510, 1:5000 dilution). Blots were imaged on Odyssey Fc Dual-Mode Imaging system (LI-COR) and analyzed using Image Studio Lite (LI-COR).
2.7. Bioinformatics
To identify ethanol-responsive genes that might be downstream of ALK-STAT3 signaling, we mined the NIH LINCS L1000 database to identify genes differentially expressed after treatment of cell lines with ALK and STAT3 inhibitors and compared these with differentially expressed genes in the PFC of female C57BL/6J mice that had consumed ethanol in the DID procedure for 36 days34. We downloaded the differential gene expression signatures for over 20,000 compounds (L1000 level 5 data) from Gene Expression Omnibus (GEO) (Phase I: GSE92742, Phase II: GSE7013), where each signature is the result of a different drug, dose, cell line, and time point for which differential expression was assessed. A subset of the L1000 data was generated for niclosamide (STAT3 inhibitor), alectinib (ALK inhibitor), and NVP-TAE684 (ALK inhibitor) using the signature id field from the metadata file available on GEO. We defined each signature as genes with |z| > 2 as recommended by Broad Institute (personal communication). We determined an ALK inhibitor signature as the overlapping genes between alectinib and NVP-TAE684 treatments across all cell lines. We next identified the genes common between the ALK and niclosamide treatments across cell lines and defined these as the ALK-STAT3 signature. Finally, we compared the ALK-STAT3 signature with genes that were differentially expressed in the PFC after DID to find the overlap. To assess the statistical significance of the overlaps we used the hypergeometric distribution. Analyses were implemented in R version 3.4.3. Pathway analysis of genes that overlapped between the ALK-STAT3 and DID signatures was performed using Ingenuity Pathway Analysis (IPA, Qiagen).
2.8. RNA isolation, reverse transcription, and qPCR
Total RNA was extracted from brain tissues using the RNeasy Mini kit (QIAGEN, Germantown, MD, USA) according to manufacturer’s instructions. RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific) and qPCR was performed using SYBR green PCR master mix (Bio‐Rad, Hercules, CA). Primers are listed in Table 1. Relative mRNA levels were determined using the 2-ΔΔCt method using the reference genes Hprt and Rpl13a. Gene expression data are shown normalized to Rpl13a, as the results did not differ when normalization was done with either Hprt or Rpl13a.
Table 1.
Primer sequences for qPCR
| Gene | Forward primer (5’–3’) | Reverse primer (5’–3’) |
|---|---|---|
| Alk | TGCCGCTCTCATTGATCCTC | TGCTTCCGACGGTACACAAT |
| Gfap | ATCGAGATCGCCACCTACAG | CTCACATCACCACGTCCTTG |
| Gpnmb | CATGGAAAGTCTCTGCGGGG | TGCTCTCTCATGTGATCGGG |
| Hprt | GTTGGGCTTACCTCACTGCT | TCATCGCTAATCACGACGCT |
| Lcn2 | TGAGTGTCATGTGTCTGGGC | AACTGATCGCTCCGGAAGTC |
| Rpl13a | AGGGGCAGGTTCTGGTATTG | GGGGTTGGTATTCATCCGCT |
| Smarcc1 | AACGGCATCAGGGGACATTT | GCTGTCTGGGTAGAAACCCC |
| Smarca4 | TGACAGAGAAGCAGTGGCTCAAG | TCGCTGTCTCGCTTACGCTT |
| Stat3 | CCCGTACCTGAAGACCAAGTTC | ACACTCCGAGGTCAGATCCA |
| Socs3 | TAGACTTCACGGCTGCCAAC | CGGGGAGCTAGTCCCGAA |
| Tnfrsf1a | TGTAACTGCCATGCAGGGTT | GTGACATTTGCAAGCGGAGG |
2.9. Statistical analysis
Data are presented as mean ± SEM. All statistical analyses were performed using Prism software (version 8, GraphPad, San Diego, CA USA). For DID experiments, 2-way repeated measures (RM) ANOVA was used with day as the within-subject factor and treatment as the between-subject factor. For western blot and qPCR result, a 2-way ANOVA was used with the factors of sex and treatment, followed by post-hoc Tukey’s multiple comparisons tests when a significant interaction was observed. A Student’s t-test was used to compare 4 h ethanol consumption and BECs.
3. RESULTS
3.1. DID
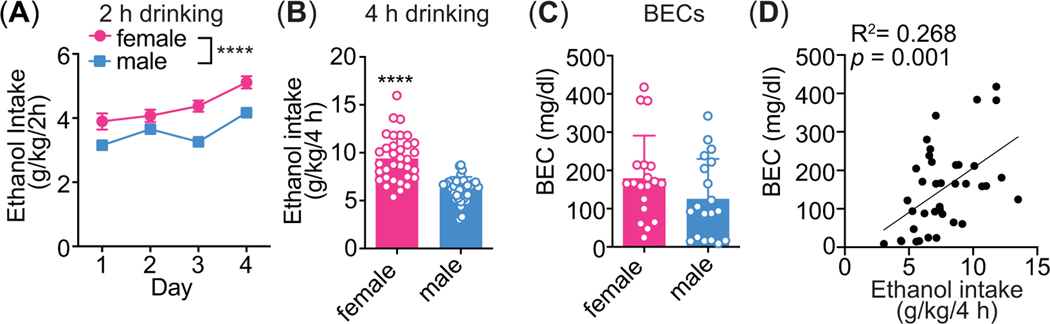
Male and female mice underwent the 4 day DID protocol with 20% ethanol or water (as a control) in order to determine the effects of binge-like ethanol drinking on ALK and STAT3 phosphorylation by western blot and on gene expression by qPCR in the ventral HPC and medial PFC. Female mice consumed more ethanol than males during the 2 h drinking sessions (Figure 1A, n = 34 per sex, sex: F1, 66 = 20.93, p < 0.0001, time: F3, 198 = 13.84, p < 0.0001, interaction: F3, 198 = 1.36, p = 0.26) and during the 4 h drinking session on the 4th day (Figure 1B, n = 34 per sex, female mean: 9.4 ± 2.4 g/kg/4 h, male mean: 6.2 ± 1.27 g/kg/4 h; t66 = 6.82, p < 0.0001). BECs were measured in the mice that were sacrificed immediately after the drinking session (0 h group) and were over the NIAAA-defined binge level of 80 mg/dl, although BECs were not significantly different between females and males (Figure 1C, n = 19 per sex per group, females: 179.6 ± 111.2 mg/dl, males: 126.1 ± 104.0 mg/dl, t36 = 1.53, p = 0.13). Finally, BECs and 4 h ethanol intake on the 4th day were significantly positively correlated (Figure 1D, n = 38, R2 = 0.262, p = 0.001).
Figure 1.
Drinking in the dark (DID). Mice consumed 20% ethanol or water in the DID protocol for 2 h on the first 3 days and for 4 h on the 4th day. Ethanol intake was measured at 2 and 4 h on the 4th day. (A) 2 h ethanol intake (g/kg) by female (n = 34) and male mice (n = 34), ****p < 0.0001. (B) 4 h ethanol intake (g/kg) by female and male mice. (C) Blood ethanol concentrations (BECs) in female (n = 19) and male mice (n = 19), taken at the end of the 4 h drinking session on the 4th day for the 0 h group only. (D) Correlation between day 4 ethanol intake (g/kg/4 h) and BECs (n = 38).
3.2. Activation of ALK in mouse PFC and HPC after binge-like ethanol drinking
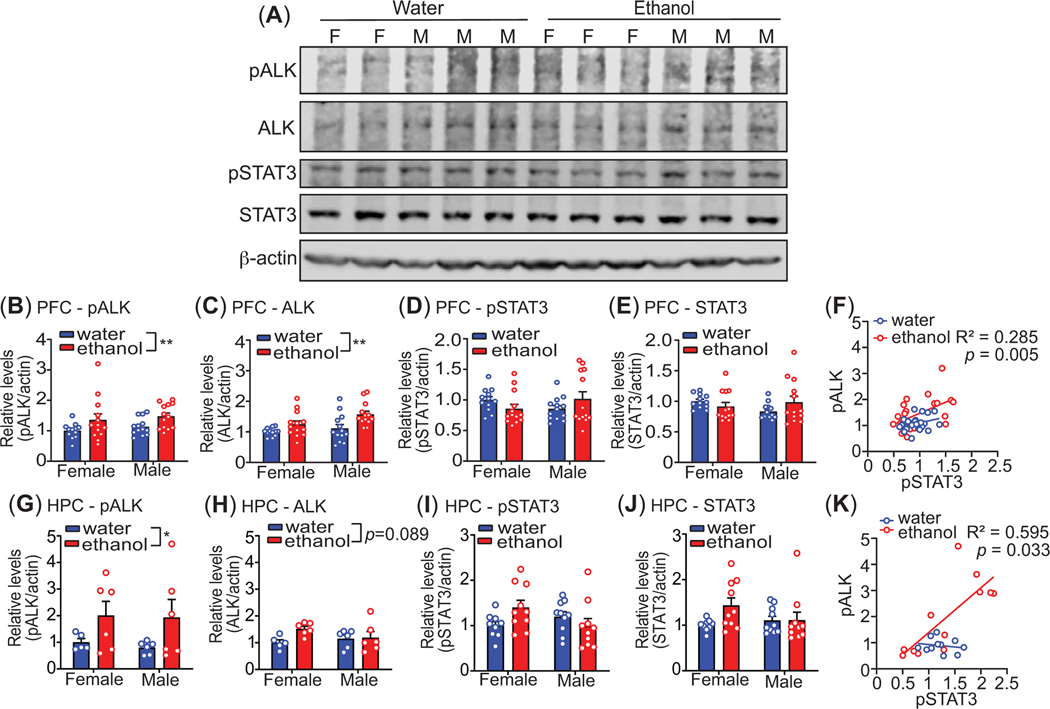
To investigate whether ALK and STAT3 are activated in the brain after binge-like ethanol intake, we measured ALK and STAT3 tyrosine phosphorylation in the mouse PFC and HPC immediately after the 4 h drinking session on day 4 of the DID protocol. Tissue lysates were subjected to western blotting with antibodies to pALK, ALK, pSTAT3, STAT3, and β-actin (as a loading control). Levels of pALK, relative to β-actin, were significantly higher in the PFC after binge-like ethanol drinking in both sexes (Figure 2A-B, n = 12–13 per sex per group, treatment: F1, 47 = 7.40, p = 0.0091, sex: F1, 47 = 0.99, p = 0.32, interaction: F1, 47 = 0.0044, p = 0.95). Total ALK protein was also increased in the PFC of both sexes after ethanol consumption (Figure 2C, n = 12–13 per sex per group, treatment: F1, 47 = 11.75, p = 0.0013, sex: F1, 47 = 4.59, p = 0.037, interaction: F1, 47 = 1.05, p = 0.31). Similar results were obtained in the HPC, with a significant increase in pALK observed after ethanol drinking in both sexes (Figure 2G, n = 6 per sex per group, treatment: F1, 19 = 5.50, p = 0.03, sex: F1, 19 = 0.092, p = 0.77, interaction: F1, 19 = 0.019, p = 0.89). Total ALK protein appeared to be elevated in the HPC after ethanol drinking, but this increase was not statistically significant (Figure 2H, n = 6 per sex per group, treatment: F1, 19 = 3.22, p = 0.089, sex: F1, 47 = 1.05, p = 0.31, interaction: F1, 19 = 2.37, p = 0.14). These results indicate that ALK is activated in the mouse PFC and HPC after binge-like ethanol drinking.
Figure 2.
Activation of ALK in mouse medial prefrontal cortex (PFC) and ventral hippocampus (HPC) after binge-like ethanol drinking. Mice drank ethanol or water in the DID protocol for 4 days and the prefrontal cortex (PFC, n = 12 – 13 per sex per group) and ventral hippocampus (HPC, n = 6 per sex per group) were collected immediately after the 4th drinking session. Protein lysates were subjected to western blotting with antibodies to phosphorylated ALK (pALK), ALK, phosphorylated STAT3 (pSTAT3), STAT3, and β-actin as a protein loading reference on the gels. (A) Representative western blots from PFC samples. M, male; F, female. (B-E, G-J) Quantification of relative band intensity from western blots of the PFC (B-E) and the HPC (G-J) for pALK (B, G), ALK (C, H), pSTAT3 (D, I), and STAT3 (E, J). (F, K) Correlations between pSTAT3 and pALK in the PFC (F) and HPC (K). The R2 and p-values indicate a significant association between pSTAT3 and pALK in mice that consumed ethanol. Data are shown as the mean ± S.E.M. *p<0.05 and **p< 0.01, main effect of ethanol by 2-way ANOVA.
Levels of pSTAT3 and STAT3 in the PFC were not significantly altered by ethanol drinking, although there was a trend towards a sex by treatment interaction for pSTAT3 levels. (Figure 2D-E, n = 12–13 per sex per group, pSTAT3, treatment: F1, 47 = 0.011, p = 0.92, sex: F1, 47 = 0.012, p = 0.91, interaction: F1, 47 = 3.66, p = 0.062; STAT3, treatment: F1, 47 = 0.24, p = 0.63, sex: F1, 47 = 0.54, p = 0.47, interaction: F1, 47 = 3.27, p = 0.077). In the HPC, there was a significant sex by treatment interaction in pSTAT3 levels (Fig. 3I, n = 9–10 per sex per group, treatment: F1, 35 = 0.52, p = 0.48, sex: F1, 35 = 0.62, p = 0.44, interaction: F1, 35 = 4.93, p = 0.033); however, Tukey’s post-hoc multiple comparisons testing did not reveal any significant differences between the groups. Total STAT3 protein was unaltered in the HPC after ethanol drinking (Figure 3J, n = 9–10 per sex per group, treatment: F1, 35 = 2.72, p = 0.11, sex: F1, 35 = 0.68, p = 0.42, interaction: F1, 35 = 2.6, p = 0.12). To further explore a potential relationship between activation of ALK and STAT3 after ethanol drinking, we examined the association between levels of pALK and pSTAT3 in both the PFC and HPC. Interestingly, there was a significant positive correlation between levels of pALK and pSTAT3 in both the PFC and HPC, but only in mice that consumed ethanol (Figure 1F, PFC, n = 25–26 per group, R2 = 0.285, p = 0.005; Figure 1K, HPC, n = 11–12 per group, R2 = 0.595, p = 0.0033). Neither ALK nor STAT3 protein levels correlated with BECs or the amount of ethanol consumed (data not shown). These results suggest that although we did not observe significant activation of STAT3 after ethanol drinking, ALK and STAT3 are coregulated in these brain regions after ethanol drinking.
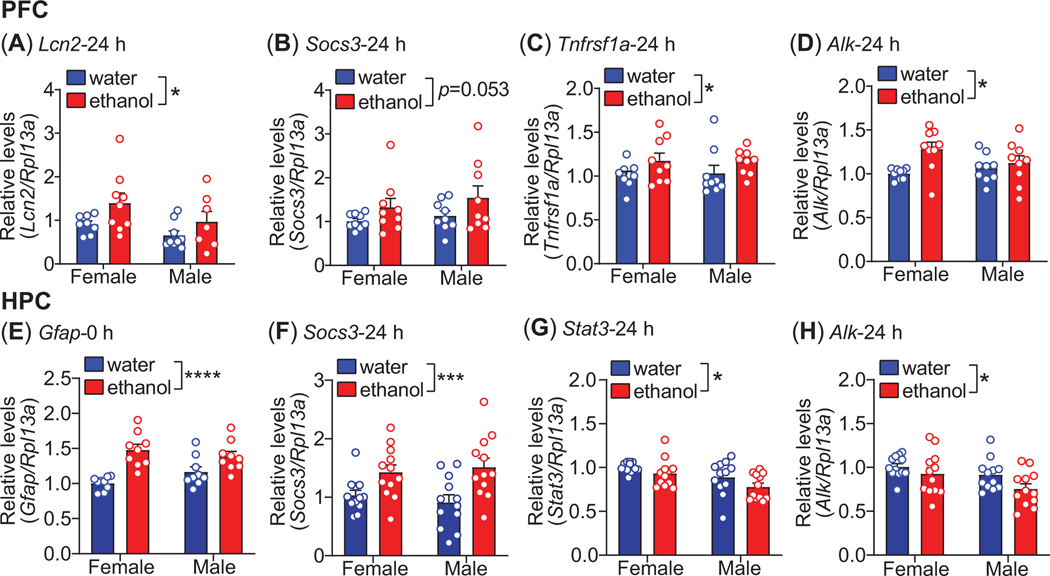
Figure 3.
Increased expression of STAT3 target genes after binge-like drinking. Mice drank ethanol or water in the DID protocol for 4 days and ventral hippocampus (HPC, n=9 per sex per group) and medial prefrontal cortex (PFC, n=7–9 per sex per group) were dissected from the brain immediately (0 h) or 24 h after the 4th drinking session as indicated. Total RNA was isolated and gene expression measured by quantitative real-time PCR. (A-D) Gene expression in the PFC for Lcn2 (A), Socs3 (B), Tnfrsf1a (C), and Alk (D). (E-H) Gene expression in the HPC for Gfap (E), Socs3 (F) Stat3 (G), and Alk (H). All data are shown as the mean ± S.E.M. *p < 0.05, ***p<0.001, ****p < 0.0001, main effect of ethanol by 2-way ANOVA.
3.3. Increased expression of STAT3 target genes after binge-like drinking
We next examined the effect of binge-like drinking on the expression of known STAT3 target genes (Socs328, Lcn230,31, Tnfrsf1a29, and Gfap27) in the PFC and HPC with the rationale that activation of ALK-STAT3 signaling by ethanol drinking will lead to increased expression of STAT3 transcriptional targets. Mice underwent the DID protocol with either ethanol or water as described above, and PFC and HPC were collected either immediately after the 4th drinking session (0 h timepoint) or 24 h after the last drinking session. The 24 h timepoint was chosen because there could be a delay between the activation of STAT3 by ethanol and the accumulation of STAT3 transcriptional target gene mRNA.
In the PFC, Lcn2 and Tnfrsf1a mRNA levels did not differ between the ethanol and water drinking groups at the 0 h timepoint (data not shown). However, both Lcn2 and Tnfrsf1a gene expression were significantly higher in the PFC of the ethanol-drinking group 24 h later (Figure 3A, C, n = 7–9 per sex per group, Lcn2, treatment: F1, 29 = 5.25, p = 0.029, sex: F1, 29 = 3.81, p = 0.061, interaction: F1, 29 = 0.24, p = 0.63; Tnfrsf1a, treatment: F1, 32 = 4.48, p = 0.042, sex: F1, 32 = 0.0041, p = 0.95, interaction: F1, 32 = 0.070, p = 0.79). Similar to Lcn2 and Tnfrsf1a, expression of Socs3 in the PFC was not altered between the ethanol and water drinking groups immediately after the drinking session (data not shown). Although Socs3 appeared to be elevated in the ethanol drinking group compared to the water drinking group 24 h later, this did not quite reach statistical significance (Figure 3B, n = 9 per sex per group, treatment: F1, 32 = 4.05, p = 0.053, sex: F1, 32 = 0.86, p = 0.36, interaction: F1, 32 = 0.072, p = 0.79). We observed a significant treatment by sex interaction in Gfap expression in the PFC at the 0 h timepoint that appeared to be due to increased Gfap expression in females that drank ethanol compared to the other groups (Supplementary Figure 1A, n = 9 per sex per group, treatment: F1, 32 = 0.92, p = 0.35, sex: F1, 32 = 0.79, p = 0.38, interaction: F1, 32 = 4.69, p = 0.038), although none of the post-hoc comparisons were statistically significant. Similar results were obtained for Gfap expression in the PFC at the 24 h timepoint (Supplementary Figure 1B, n = 9 per sex per group, treatment: F1, 32 = 0.15, p = 0.70, sex: F1, 32 = 0.73, p = 0.40, interaction: F1, 32 = 7.03, p = 0.012). Finally, we examined Alk and Stat3 gene expression in the PFC after binge-like ethanol drinking. Stat3 expression was not significantly altered by ethanol drinking at either of the timepoints (data not shown). At the 0 h timepoint, Alk expression was higher in females that drank ethanol compared with the water drinking females and in males that drank ethanol compared with water drinking females (Supplementary Figure 1C, n = 9 per sex per group, treatment: F1, 32 = 14.51, p = 0.0006, sex: F1, 32 = 0.28, p = 0.60, interaction, F1, 32 = 4.39, p = 0.044; Tukey’s multiple comparisons test: female water vs. female ethanol, p = 0.0012 and female water vs. male ethanol, p = 0.022). The increase in Alk gene expression in the PFC persisted 24 hours after the final drinking session (Figure 3D, n = 9 per sex per group, treatment: F1, 32 = 7.08, p = 0.012, sex: F1, 32 = 0.58, p = 0.45, interaction: F1, 32 = 2.92, p = 0.097). The increase in Alk gene expression in the PFC is consistent with higher levels of ALK protein after binge-like ethanol drinking (Figure 2C).
In the HPC, Gfap expression was increased at the 0 h timepoint in mice that drank ethanol (Figure 3E, n = 9 per sex per group, treatment: F1, 32 = 27.57, p < 0.0001, sex: F1, 32 = 0.30, p = 0.59, interaction: F1, 32 = 3.73, p = 0.062). In contrast to Gfap, Socs3 expression in the HPC was not altered by ethanol drinking at 0 h (data not shown) but was higher in the ethanol drinking group compared to the water drinking group at 24 h (Figure 3F, n=12 per sex per group, treatment: F1, 44 = 15.65, p = 0.0003, sex: F1, 44 = 0.022, p = 0.88, interaction: F1, 44 = 0.67, p = 0.42). Interestingly, Stat3 expression in the HPC was not altered by ethanol drinking at the 0 h timepoint but was reduced in ethanol drinking mice compared to water drinking mice 24 h after the last drinking session (Figure 3G, n = 12 per sex per group, treatment: F1, 44 = 4.27, p = 0.044; sex: F1, 44 = 9.58, p = 0.0034, interaction: F1, 44 = 0.23, p = 0.63). The decrease in Stat3 in the HPC after ethanol drinking might be explained by the robust increase in Socs3 at 24 h because SOCS3 is a negative feedback regulator of STAT3 signaling28. Like Stat3, Alk expression did not change at 0 h (data not shown) but was decreased 24 h after the last drinking session in the HPC of mice that drank ethanol (Figure 3H, n=12 per sex per group, treatment: F1, 44 = 4.74, p = 0.035, sex: F1, 44 = 5.77, p = 0.021, interaction: F1, 44 = 0.52, p = 0.48). Notably, Stat3 and Alk expression levels were significantly positively correlated at the 24 h timepoint in mice that consumed ethanol but not in mice that drank water (Supplementary Figure 1D, R2 = 0.726, p < 0.0001), providing additional evidence of coregulation of ALK and STAT3 after ethanol drinking. Together, these results demonstrate that the expression of known STAT3 transcriptional target genes increase in the brain after ethanol drinking, and that these changes in gene expression are temporally and regionally distinct. Moreover, alterations in Alk and Stat3 gene expression in response to ethanol are different between the PFC and HPC.
3.4. Inhibition of STAT3 in mice decreases binge-like drinking
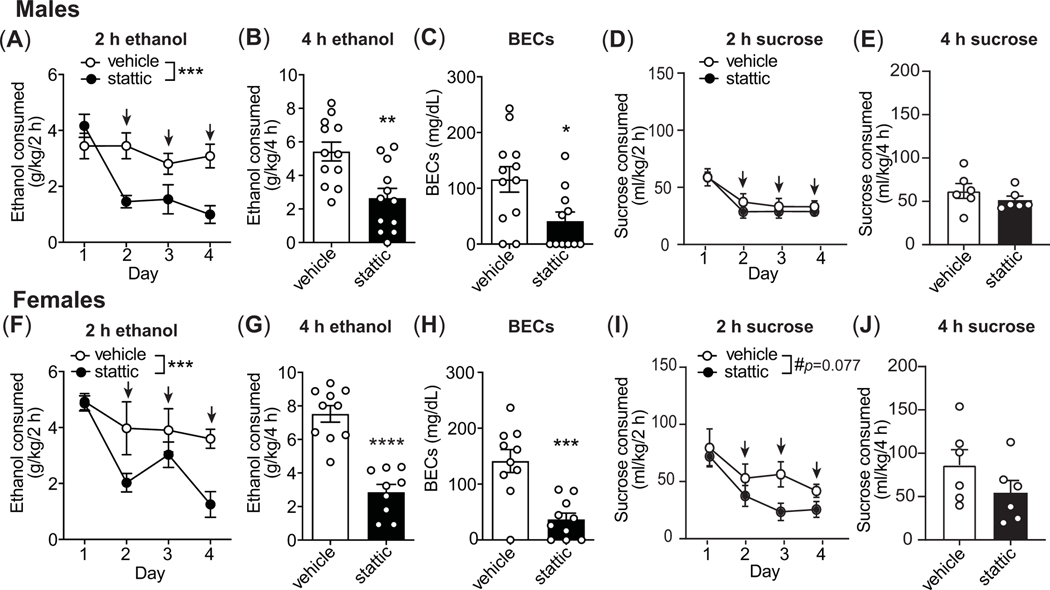
To determine if STAT3 plays a role in binge-like drinking, we next tested the effect of STAT3 inhibition on DID using stattic, which is a STAT3-selective non-peptide inhibitor. Stattic (20 mg/kg, s.c.) or vehicle was administered 30 min before the start of the 2nd, 3rd, and 4th drinking session. No injections were given on the first drinking day. Treatment with stattic dramatically decreased binge-like drinking in both female and male mice during the 2 h ethanol drinking sessions when compared to vehicle-treated mice (Figure 4A, males, n = 12 per group, two-way RM ANOVA on days 2–4, treatment: F1, 22 = 21.23, p = 0.0001, day: F2, 43 = 0.62, p = 0.54, interaction: F2, 43 = 1.30, p = 0.28; Figure 4F, females, n = 10 per group, treatment: F1, 18 = 15.53, p = 0.001, day: F2, 36 = 0.88, p = 0.42, interaction: F2, 36 = 0.47, p = 0.63). Stattic was also effective in reducing ethanol intake during 4 h on the 4th day in both sexes when compared to vehicle treatment (Figure 4B, males, n = 12 per group, t22 = 3.45, p = 0.0023; Figure 4G, females, n = 10 per group, t18 = 6.26, p < 0.0001). BECs were correspondingly reduced in mice treated with stattic compared to vehicle (Figure 4C, males, n = 12 per group, vehicle, 116.1 ± 79.1 mg/dl, stattic, 41.16 ± 54.54 mg/dl, t21 = 2.62, p = 0.016; Figure 4H, females, n = 10 per group, vehicle, 141.2 ± 65.3 mg/dl, stattic, 36.89 ± 34.85 mg/dl, t18 = 4.46, p = 0.0003).
Figure 4.
STAT3 inhibition in mice decreases binge-like drinking. Mice drank ethanol (males, n=12 per group; females, n=10 per group) or sucrose (n=6 per sex per group) in the DID protocol for 4 days and were injected with 20 mg/kg of stattic subcutaneously 30 min before the drinking sessions on days 2, 3, and 4 (indicated by arrows). (A-B) Ethanol consumed in g/kg body weight by males during the 2 h sessions (A) and the 4 h session on day 4 (B). (C) Blood ethanol concentrations (BECs) in males after the 4 h session on day 4. (D-E) Sucrose consumed in ml/kg body weight by males during the 2 h sessions (D) and the 4 h session on day 4 (E). (F-G) Ethanol consumed by females during the 2 h sessions (F) and the 4 h session on day 4 (G). (H) BECs in females after the 4 h session on day 4. (I-J) Sucrose consumed by females during the 2 h sessions (I) and the 4 h session on day 4 (J). All data are presented as the mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, and ****p < 0.0001.
We also compared baseline drinking on the first day (no injections) with drinking on treatment days 2–4. Vehicle injections on days 2–4 did not change ethanol consumption when compared to day 1 in either male or female mice, while stattic treatment significantly decreased ethanol consumption when comparing day 1 to 2 (p = 0.0003), 1 to 3 (p = 0.0025), and 1 to 4 (p = 0.0032) in male mice, and day 1 to 2 (p = 0.0019) and 1 to 4 (p < 0.0001) in female mice. In total, these results demonstrate that inhibition of STAT3 decreases binge-like ethanol intake in male and female mice.
We next tested for the effect of stattic on sucrose consumption to determine if the inhibitory effect of stattic on ethanol drinking could be generalized to another rewarding substance. Stattic treatment did not significantly alter sucrose drinking when compared with vehicle treatment during the 2 h drinking sessions in either sex (Figure 4D, males, n = 6 per group, two-way RM ANOVA on days 2–4, treatment: F1, 10 = 0.70, p = 0.42, day: F2, 20 = 0.14, p = 0.87, interaction: F2, 20 = 0.16, p = 0.85; Figure 4I, females, n = 6 per group, treatment: F1, 10 = 3.88, p = 0.077, day: F2, 20 = 1.90, p = 0.18, interaction: F2, 20 = 1.38, p = 0.27). Stattic also did not significantly affect sucrose drinking during the 4 h session on day 4 (Figure 4E, males, n = 6 per group, t10 = 1.08, p = 0.30; Figure 4J: females, n = 6 per group, t10 = 1.39, p = 0.19) relative to the vehicle-treated animals. However, when compared to baseline sucrose drinking on the first day, within group analysis using Tukey’s multiple comparisons test showed that stattic treatment decreased 2 h sucrose intake between days 1 and 2 (p = 0.029), 1 and 3 (p = 0.008), and 1 and 4 (p = 0.0008) in male mice, and between days 1 and 2 (p = 0.027), 1 and 3 (p = 0.0053), and 1 and 4 (p = 0.016) in female mice. However, this result is somewhat confounded by a suppressive effect of vehicle treatment on sucrose intake when comparing day 1 with days 2–4 (day 1 vs. 2, p = 0.045) in male mice. We also measured water intake in mice treated with stattic in a single 2 h drinking session. Stattic treatment did not alter water consumption in male or female mice (males, n = 9 per goup, vehicle: 25.73 ± 4.46 ml/kg/2 h, stattic: 27.79 ± 12.82 ml/kg/2 h, t16 = 0.33, p = 0.74; females, n = 3 per group, vehicle: 17.21 ± 2.171 ml/kg/2 h, stattic: 22.09 ± 3.828 ml/kg/2h, t4 = 1.92, p = 0.13).
We also tested if another STAT3 inhibitor, niclosamide, would have the same suppressive effect as stattic on ethanol and sucrose drinking. Similar to stattic, niclosamide reduced both ethanol and sucrose intake in male and female mice (Supplementary Figure 2). Together, these results indicate that STAT3 inhibition reduces both ethanol and sucrose intake, with a greater effect on ethanol intake, but does not affect water drinking. STAT3 may therefore be generally be involved in motivation to consume rewarding substances.
3.5. Identification of novel ethanol-responsive genes downstream of ALK-STAT3 signaling
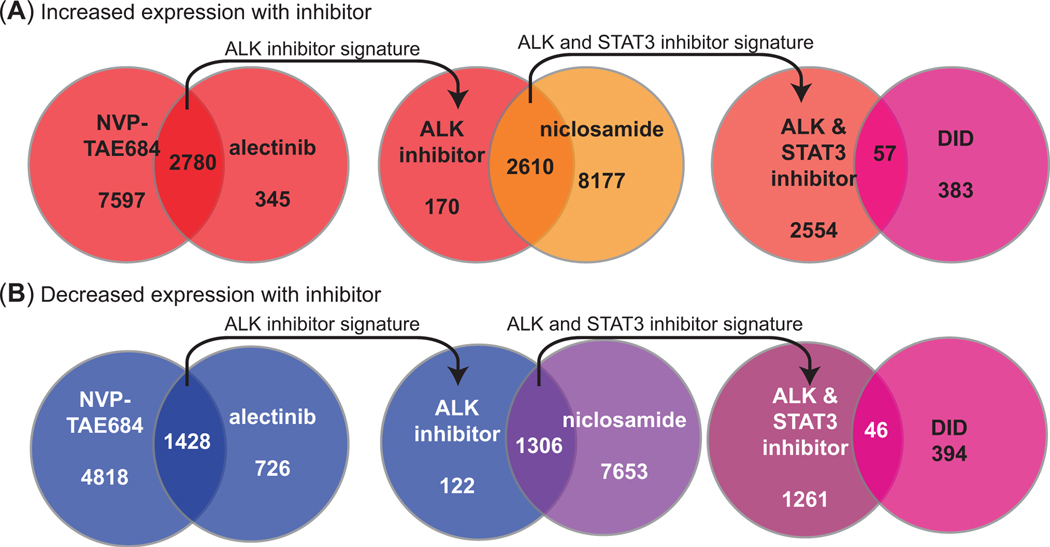
Given that the expression of known STAT3 target genes increase in the PFC and HPC after binge-like ethanol drinking, we reasoned that there could be novel ethanol-responsive genes downstream of ALK-STAT3 signaling. To begin to identify these genes, we mined the NIH LINCS L1000 database. LINCS L1000 catalogues perturbations in 1000 genes (described as “transcriptomic signatures”) in cell lines after treatment with small molecules. Two ALK inhibitors (alectinib and NVP-TAE684) and one STAT3 inhibitor (niclosamide) have been tested on multiple cell lines each. We identified the transcriptomic signatures of the five cell lines (A375, MCF7, PC3, HA1E, and HT29) that had been tested with each of the inhibitors. First, the overlap between the gene signatures of the two ALK inhibitors was identified (Figure 5). This yielded 2780 genes that increased and 1428 genes that decreased in response to both ALK inhibitors. The number of genes was significantly greater than expected by chance (genes that increased, 1.06 fold enrichment, hypergeometric p = 1.32 × 10−18; genes that decreased, 1.31 fold enrichment, hypergeometric p = 1.82 × 10−58), as would be predicted for two inhibitors selective for ALK. The ALK inhibitor signature was then compared with the gene signature of the STAT3 inhibitor to identify genes that are regulated by the ALK-STAT3 pathway. This yielded 2610 genes that increased and 1306 genes that decreased in response to the inhibitors. Interestingly, the overlap in the number of genes that were altered by both ALK and STAT3 inhibitors were also greater than those expected by chance (genes that increased, 1.07 fold enrichment, hypergeometric p = 2.52 × 10−38; genes that decreased, 1.26 fold enrichment, hypergeometric p = 6.54 × 10−78), suggesting a significant overlap in genes downstream of both ALK and STAT3 signaling. Finally, the genes from ALK-STAT3 inhibitors were compared to genes that were differentially expressed in the PFC of female C57BL/6J mice after 36 days of DID34. By using this approach, we discovered 57 and 46 genes whose expression increased and decreased, respectively, after treatment with ALK and STAT3 inhibitors and were differentially expressed after DID (Figure 5). Of the total of 103 genes identified, 8 were found in both groups (increasing and decreasing), leaving 95 unique genes (Supplementary Table 1).
Figure 5.
Overlapping gene expression changes between ALK and STAT3 inhibitors from LINCS L1000 and comparison with differentially expressed genes in the mouse prefrontal cortex after ethanol drinking. (A-B) Diagrams showing the number of genes whose expression increased (A) or decreased (B) after ALK (NVP-TAE684 and alectinib) and STAT3 (niclosamide) inhibitor treatments of cell lines and those that overlapped with differentially expressed genes in the prefrontal cortex of female mice after ethanol drinking in the drinking in the dark (DID) protocol.
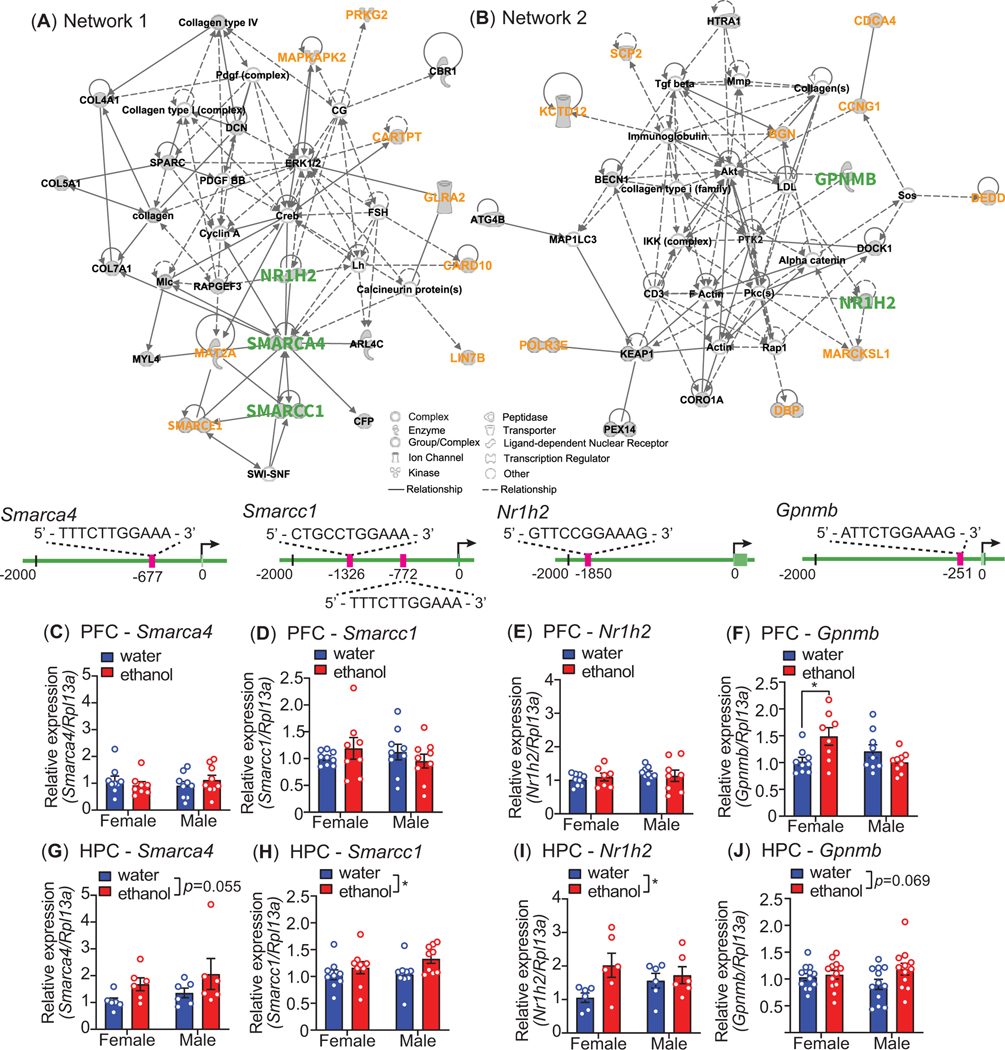
We next performed IPA on the 95 genes to determine if they fit into any defined pathways or cellular networks. IPA yielded 7 networks, with the top 2 networks (Figure 6) having connectivity scores of 45 (“Cell Death and Survival, Cellular Compromise, Tissue Development”) and 36 (“Cardiovascular System Development and Function, Cell Morphology, Cellular Assembly and Organization”). This demonstrated that within the candidate gene list, there are several interconnected clusters of genes with related functions. To further narrow down STAT3 regulated genes in this list, we performed a STAT3 binding motif analysis in the 2 kb upstream promoter region of the 95 genes identified in the LINCS analysis. Gene symbols from the LINCS1000 analysis were first mapped to Ensembl IDs. The transcription start site (TSS) for each gene was identified and the genomic sequence for the 2 kb upstream promotor region was extracted from the mouse Mm10 genome using SAMtools38. The STAT3 consensus motif was downloaded from JASPAR39 and motif search was done using the FIMO tool40.This analysis identified 91 motifs present in 57 of the 95 genes (Supplementary Table 2), showing that more than half of the genes identified through the LINCS analysis contained putative STAT3 binding motifs in their 2 kb promoter region and could be direct targets of STAT3.
Figure 6.
Ingenuity Pathway Analysis (IPA) and expression of genes from IPA networks after ethanol drinking. Overlapping differentially expressed genes from LINCS and the prefrontal cortex (PFC) of mice that drank ethanol were analyzed by IPA. (A) Network 1 and (B) network 2 from IPA. Genes indicated with green and orange text had predicted STAT3 binding motifs in their promoters. Diagrams below each network show the predicted STAT3 motifs of genes in green text that were selected for experimental validation by qPCR in mouse PFC (7–10 per sex per group) and ventral hippocampus (HPC, n=6–12 per sex per group) after ethanol or water drinking in the 4 day drinking in the dark procedure. (C-J) Relative mRNA expression of Smarca4 in the PFC (C) and HPC (G), Smarcc1 in the PFC (D) and HPC (H), Nr1h2 in the PFC (E) and HPC (I), and Gpnmb in the PFC (I) and HPC (J). All data are shown as the mean ± S.E.M, *p < 0.05.
Finally, we measured the expression of several genes in the IPA networks that had predicted STAT3 binding motifs. All of the following analyses were performed on PFC and HPC collected immediately (0 h timepoint) after the 4th DID session with water or ethanol as described above. From network 1 (Figure 6), we measured the expression of Smarca4, Smarcc1, and Nr1h2 (which was found in both network 1 and 2). All three genes possess putative STAT3 binding motifs (Figure 6). Four sessions of ethanol drinking did not alter Smarca4, Smarcc1, or Nr1h2 expression in the PFC (Figure 6C-E). However, Smarca4 expression nearly significantly increased in the HPC after ethanol drinking (Figure 6G, n = 6 per sex per group, treatment: F1, 20 = 4.14, p = 0.055, sex: F1, 20 = 1.13, p = 0.30, interaction: F1, 20 = 0.009, p = 0.93), and Smarcc1 and Nr1h2 expression levels were significantly increased in the HPC of mice that consumed ethanol (Smarcc1, Figure 6H, n = 9 per sex per group, treatment: F1, 32 = 4.91, p = 0.034, sex: F1, 32 = 0.79, p = 0.38, interaction: F1, 32 = 0.70, p = 0.41; Nr1h2, Figure 6I, n = 6, treatment: F1, 20 = 4.89, p = 0.039, sex: F1, 20 = 0.17, p = 0.68, interaction: F1, 20 = 2.48, p = 0.13). From network 2, we measured Gpnmb expression after DID. Gpnmb also contains a putative STAT3 binding motif in its proximal upstream promoter region. In the PFC, we did not observe a significant main effect of ethanol or sex, but there was a significant ethanol-by-sex interaction on Gpnmb expression. Post hoc Tukey’s multiple comparisons test demonstrated that there was a significant increase in Gpnmb expression only in female mice that drank ethanol (Figure 6F, n = 9 per sex per group, treatment: F1, 31 = 1.4, p = 0.25, sex: F1, 31 = 1.78, p = 0.19, interaction: F1, 31 = 9.36, p = 0.0046; post hoc Tukey’s multiple comparisons, control vs ethanol, female: p = 0.029, male: p = 0.54). In the HPC, we observed a nearly significant increase in Gpnmb expression in mice that drank ethanol, although this effect appears to be driven largely by an increase in the male mice (Figure 6J, n = 12 per sex per group, treatment: F1, 44 = 3.48, p = 0.069, sex: F1, 44 = 0.027, p = 0.87, interaction: F1, 44 = 1.8, p = 0.17). These results suggest that there are brain-region and sex-specific increases in the expression of genes after ethanol binge-like drinking that are potentially regulated by the ALK-STAT3 pathway.
4. DISCUSSION
This study demonstrates that ethanol activates ALK signaling and increases the expression of known STAT3 target genes in the medial PFC and ventral HPC of mice after 4 days of binge levels of ethanol drinking. Although STAT3 was not significantly activated after ethanol drinking, pALK and pSTAT3 levels were significantly correlated in both the PFC and HPC after ethanol consumption, suggesting that STAT3 signaling is activated by ethanol. Additional evidence is provided by experiments with the STAT3 inhibitors (stattic and niclosamide), which were effective in decreasing binge-like ethanol drinking after systemic administration. Finally, we identified novel putative ethanol-responsive target genes downstream of ALK-STAT3 signaling using the LINCS L1000 database. To our knowledge, this is the first study to provide evidence that ethanol consumption can activate the ALK-STAT3 pathway in vivo. We previously found that that ethanol treatment of a neuroblastoma cell line activated ALK and STAT3 in an ALK-dependent manner21, but it was not known if ethanol drinking could activate this particular pathway in the brain.
Another group found modest but statistically significant increases in pSTAT3 in the cerebellum and HPC of C57BL/6J mice during withdrawal from 3 cycles of 3 days each of chronic intermittent ethanol vapor exposure25,26 and in the HPC after a single 4 g/kg injection of ethanol in mice23. Because of this, we expected to see increased pSTAT3 at least in the HPC after ethanol drinking. There are several possible explanations for this discrepancy. The first possibility is that STAT3 is not activated by only 4 bouts of binge-like ethanol drinking and that STAT3 activation may require higher doses or chronic exposure to ethanol. The second is timing; STAT3 activation may either be more transient than ALK activation or occur only during ethanol withdrawal. The third possible explanation is that STAT3 activation only happens in a subset of cells in the PFC and/or HPC, and that in total tissue homogenates, cell-type specific changes were masked41. Based on our demonstration that Gfap is increased after ethanol drinking, we suspect that STAT3 may be activated in astrocytes (see below). Gfap is a well-known target of STAT3 and activated STAT3 is a characteristic of reactive astrocytes27,42. We also only measured ethanol-induced changes in the ventral HPC and it is possible that activation of STAT3 after binge-like ethanol drinking may only occur in the dorsal HPC. However, we did find a significant positive correlation between pALK and pSTAT3 in both the PFC and HPC, and only in mice that drank ethanol. This suggests that pSTAT3 and pALK are coregulated by ethanol and provides some evidence that STAT3 may be activated during ethanol drinking in response to ALK activation.
Although we provide some evidence that STAT3 is activated in response to ethanol downstream of ALK, this does not mean that STAT3 could only be activated in an ALK-dependent manner by ethanol. STAT3 is also activated by a number of other cytokines, chemokines, and interferons20. Specifically, ethanol increases the transcription of the Il6 gene43. STAT3 is constitutively activated in the brains of transgenic mice that overexpress Il6 in astrocytes, demonstrating a clear role for IL-6 in STAT3 activation in the brain23,25,26.
Given that STAT3 is a transcription factor, we hypothesized that STAT3 activation by ethanol would lead to an increase in the expression of known STAT3 transcriptional targets, so we measured the expression of the documented STAT3 target genes Gfap44, Socs345, Lcn230, and Tnfrsf1a46. Gene expression increased in either the PFC or HPC after 4 days of DID. The increase in Gfap and Lcn2, which are two markers of reactive astrocytes42, suggest that even a short duration of binge-like drinking is sufficient to induce astrocyte reactivity in the mouse brain. This corroborates increases in GFAP observed by other investigators after varying lengths of ethanol exposure23,47,48. Increased GFAP immunoreactivity has also been observed after long-term ethanol consumption in rodents after intermittent ethanol access, but not after continuous access to ethanol49. In fact, multiple factors, including the drinking model and sex, appear to play a role in the regulation of GFAP49,50. Our observation of increased Gfap after ethanol drinking only in female mice is consistent with other researchers that have also seen a larger induction of GFAP in female mice after ethanol consumption50,51.
To our knowledge, this is the first report of Lcn2 upregulation in the CNS after ethanol exposure. LCN2 is a small, secreted glycoprotein involved in anti-microbial defense through the sequestering of iron52. In an transcriptomic study of reactive astrocytes comparing two different in vivo models of neuroimmune activation (ischemic stroke and lipopolysaccharide injection), Lcn2 was found to be one of the strongest markers of reactive astrocytes53. Interestingly, pathway analysis of the two reactive astrocyte transcriptomes identified “acute phase response signaling” as the top pathway53, highlighting the role of STAT3 in regulating astrocyte reactivity. The exact roles of LCN2 in the CNS are not clear, but several studies indicate that Lcn2 is upregulated after innate immune challenge53–55, and both pro-inflammatory56,57 and anti-inflammatory55 roles of LCN2 have been reported. Notably, LCN2 plays a detrimental role in alcohol-induced liver disease58,59. Further studies are necessary to understand the function of LCN2 in the CNS the context of alcohol use.
Tumor necrosis factor alpha (TNFα) is a potent pro-inflammatory cytokine that signals through TNF receptor 1 and TNF receptor 2 to activate the activation protein-1 (AP-1) and nuclear factor-kappa B (NF-κB) pathways. Increased Tnf expression occurs after acute and chronic ethanol exposure in rodents48,60. Tnfrsf1a, which encodes TNF receptor 1, the major mediator of TNFα signaling, is a transcriptional target of STAT346. Of note, Tnfrsf1a knockout mice showed a modest, non-significant decrease in voluntary 2-bottle ethanol consumption at high ethanol concentrations (15% and 17%) compared with wild-type controls, and mice lacking both Tnrsf1a and Il1r1 drank significantly less ethanol compared to wild-type mice and did not escalate their drinking in an every-other-day 2-bottle choice protocol with increasing ethanol concentrations61.
STAT3 activity is tightly regulated by SOCS362. SOCS3 is potent inhibitor of JAK/STAT3 signaling and its expression is primarily regulated by STAT3. We observed a robust increase in Socs3 gene expression 24 h after the ethanol drinking session, suggesting that Socs3 is induced in order to attenuate increased STAT3 activity in response to ethanol. Notably, Alk and Stat3 gene expression were reduced in the HPC 24 h after the ethanol drinking session and it is possible that this is due to negative feedback via induction of SOCS3. In addition to negatively regulating STAT3 activity, SOCS3 also dampens other immune-response pathways63. For example, SOCS3 can inhibit NF-κB signaling after stimulation of toll-like receptors64. This has implications for AUD as toll-like receptor activation of NF-κB is a critical mediator of alcohol-induced immune response48,65. As such, an increase in Socs3 may be seen as an anti-inflammatory response to alcohol use. Whether this response is protective or harmful in the context of AUD requires further investigation.
Many of the known STAT3 target genes have been identified in relation to cancer progression and tumorigenesis66. To discover novel ethanol-responsive genes that act within the ALK-STAT3 pathway, we used a bioinformatics approach to discover genes that were altered in cell lines after treatment with ALK and STAT3 inhibitors and compared these genes with those that were changed in the PFC of female C57BL/6J mice that had undergone 36 DID sessions34. IPA and STAT3 binding motif analysis were performed on this gene set and the expression of candidate genes Smarca4, Smarcc1, Nr1h2, and Gpnmb were increased in either the HPC or PFC after 4 DID sessions.
The Nr1h2 gene encodes liver X receptor β (LXRβ), one of the two nuclear oxysterol receptors and a member of the nuclear receptor superfamily. LXRα and LXRβ are endogenous cholesterol sensors and transcription factors for genes involved in cholesterol metabolism67. Despite the name, LXRβ is not highly expressed in the liver, but is found in abundance in the brain, immune cells, and in the gut epithelium68. Cholesterol is a major component of the brain. Approximately a quarter of the body’s cholesterol is found in the brain in the form of myelin69. Cholesterol is metabolized de novo in the CNS, primarily by astrocytes, because the BBB does not allow the passage of lipoproteins70. In the context of AUD, Alsebaaly et al found brain region specific changes in genes involved in cholesterol metabolism71. In male Long-Evans rats during protracted withdrawal 3 weeks after 47 days of every other day 2 bottle choice ethanol consumption, Abca1, an LXR target gene, was decreased in the PFC and increased in the NAc71, although no changes in Nr1h2 were found in the aforementioned study. In addition to their role in cholesterol metabolism, LXRs have been shown to have an anti-inflammatory effect67, which adds another layer of intrigue in defining the role of neuroimmune activation in AUD.
Glycoprotein nonmelanoma protein B (GPNMB) is a type I transmembrane glycoprotein that is widely expressed throughout the body and brain72. The elevation of GPNMB or alterations in GPNMB gene have been linked to neurological disorders including Parkinson’s disease73,74, Alzheimer’s disease75, Gaucher’s disease76 and amyotrophic lateral sclerosis77. Studies indicate that GPNMB acts in a neuroprotective manner by attenuating astrocyte mediated pro-inflammatory responses78,79. The increase in Gpnmb in mouse PFC after binge-like drinking suggests that there may be anti-inflammatory events occurring in the brain to counterbalance the pro-inflammatory responses to ethanol. To date, there are no studies showing direct transcriptional regulation of Gpnmb by STAT3, but we found that the Gpnmb upstream promoter region contains a STAT3 binding site. Overall, we observed increased expression of both pro- (Lcn2, Gfap, Tnfrsf1a) and anti- (Socs3, Nr1h2, Gpnmb) inflammatory genes that are known to be or putative targets of STAT3 in the PFC and HPC after 4 days of binge-like drinking, suggesting a complex and dynamic neuroimmune response to ethanol drinking.
We also observed a cluster of genes encoding chromatin remodelers (Smarce1, Smarca4, and Smarcc1) in our bioinformatics analysis and measured the expression of two genes (Smarca4 and Smarcc1) that are part of the SWI/SNF complex, also known as the BRG1/BRM-associated factor (BAF) complex. The SWI/SNF complexes remodel chromatin by regulating nucleosome positioning, using energy generated through ATP hydrolysis. Substantial research has highlighted the role of epigenetics in AUD80 and in addiction at large81. Of note, various genes within the SWI/SNF complexes have been implicated in alcohol-related phenotypes and in human alcohol dependence. A study in Caenorhabditis elegans found that most of the SWI/SNF genes (12 out of 13) were required for either initial sensitivity to ethanol or for acute functional recovery from ethanol exposure82, highlighting the connection between ethanol and the SWI/SNF complex. In vitro studies have also found ethanol-induced changes in gene expression or DNA methylation of SWI/SNF complex genes83,84. Finally, a transcriptomic study of mouse hippocampus after chronic intermittent ethanol vapor exposure found increased expression of several Smarc genes, including Smarce1, Smarca4, and Smarcc185. In the cancer field, the emerging role for STAT3 in epigenetic regulation is a recent area of focus86. Our results suggest that ethanol induced STAT3 activation and regulation of genes encoding chromatin remodeling enzymes may be contributing to the epigenetic landscape of AUD.
Finally, the upregulation of both pro-inflammatory cytokine receptor Tnfrsf1a and of anti-inflammatory mediators Gpnmb and Socs3 demonstrates that binge-like ethanol drinking activates a brain neuroimmune response that cannot be easily categorized into merely pro- or anti-inflammatory. Substantial research has been conducted on the role of neuroimmune response87,88 and astrocytes89–91 on the pathophysiology of AUD. Our results add to the existing neuroimmune literature and suggest that STAT3 may play a role in activation of the neuroimmune response and astrocytes after ethanol consumption. It is also important to note that while we have focused this discussion on the potential role of STAT3 in mediating an astrocytic immune response in the brain, STAT3 is expressed in microglia, neurons, and oligodendrocytes, and the contribution of STAT3 in these cells should not be ignored. Future studies should determine the cell types and brain regions in which STAT3 acts to regulate gene expression and behavioral responses to ethanol.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Subhash C. Pandey for helpful discussions of the data. This work was funded by grants from the National Institute on Alcohol Abuse and Alcoholism (U01 AA020912 to AWL, P50 AA022538 to AWL, T32 AA026577 to KH, U01 AA020926 to RDM, F32 AA028148 to LBF, and T32 AA 007471 to LBF) and the National Center for Advancing Translational Science (UL1 TR002003 to MMC). The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Funding Information: National Institute on Alcohol Abuse and Alcoholism, Grant/Award Numbers: U01 AA020912 (to AWL), P50 AA022538 (to AWL), T32 AA026577 (to KH), U01 AA020926 (to RDM), F32 AA028148 (to LBF), and T32 AA007471 (to LBF). National Center for Advancing Translational Science Grant/Award Number UL1 TR002003 to MMC.
Footnotes
SUPPORTING INFORMATION
The data that support the findings of this study are available as Supplementary files associated with this paper.
CONFLICT OF INTEREST
The authors declare no competing conflicts of interest.
REFERENCES
- 1.White AM, Castle IP, Hingson RW, Powell PA. Using Death Certificates to Explore Changes in Alcohol-Related Mortality in the United States, 1999 to 2017. Alcohol Clin Exp Res. 2020;44(1):178–187. [DOI] [PubMed] [Google Scholar]
- 2.Alcohol use and burden for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet (London, England). 2018;392(10152):1015–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 National and State Costs of Excessive Alcohol Consumption. American journal of preventive medicine. 2015;49(5):e73–e79. [DOI] [PubMed] [Google Scholar]
- 4.Alcoholism NIoAAa. NIAAA Newsletter. In. Vol 32004. [Google Scholar]
- 5.(SAMSA) SAaMHSA. 2018 National Survey on Drug Use and Health. In: Table 2.1B Tobacco Product and Alcohol Use in Lifetime PY, and Past Month among Persons Aged 12 or Older, by Age Group: Percentages, 2017 and 2018, ed2019. [Google Scholar]
- 6.Grucza RA, Sher KJ, Kerr WC, et al. Trends in Adult Alcohol Use and Binge Drinking in the Early 21st-Century United States: A Meta-Analysis of 6 National Survey Series. Alcohol Clin Exp Res. 2018;42(10):1939–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yau NK, Fong AY, Leung HF, et al. A Pan-Cancer Review of ALK Mutations: Implications for Carcinogenesis and Therapy. Current cancer drug targets. 2015;15(4):327–336. [DOI] [PubMed] [Google Scholar]
- 8.Vernersson E, Khoo NK, Henriksson ML, Roos G, Palmer RH, Hallberg B. Characterization of the expression of the ALK receptor tyrosine kinase in mice. Gene expression patterns : GEP. 2006;6(5):448–461. [DOI] [PubMed] [Google Scholar]
- 9.Bilsland JG, Wheeldon A, Mead A, et al. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology. 2008;33(3):685–700. [DOI] [PubMed] [Google Scholar]
- 10.Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14(4):439–449. [DOI] [PubMed] [Google Scholar]
- 11.Murray PB, Lax I, Reshetnyak A, et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Science signaling. 2015;8(360):ra6. [DOI] [PubMed] [Google Scholar]
- 12.Reshetnyak AV, Murray PB, Shi X, et al. Augmentor alpha and beta (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc Natl Acad Sci U S A. 2015;112(52):15862–15867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoica GE, Kuo A, Aigner A, et al. Identification of anaplastic lymphoma kinase as a receptor for the growth factor pleiotrophin. J Biol Chem. 2001;276(20):16772–16779. [DOI] [PubMed] [Google Scholar]
- 14.Stoica GE, Kuo A, Powers C, et al. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J Biol Chem. 2002;277(39):35990–35998. [DOI] [PubMed] [Google Scholar]
- 15.Lasek AW, Lim J, Kliethermes CL, et al. An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PLoS One. 2011;6(7):e22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangieri RA, Maier EY, Buske TR, Lasek AW, Morrisett RA. Anaplastic Lymphoma Kinase Is a Regulator of Alcohol Consumption and Excitatory Synaptic Plasticity in the Nucleus Accumbens Shell. Frontiers in pharmacology. 2017;8:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweitzer P, Cates-Gatto C, Varodayan FP, et al. Dependence-induced ethanol drinking and GABA neurotransmission are altered in Alk deficient mice. Neuropharmacology. 2016;107:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutton JW 3rd, Chen H, You C, Brodie MS, Lasek AW. Anaplastic lymphoma kinase regulates binge-like drinking and dopamine receptor sensitivity in the ventral tegmental area. Addict Biol. 2017;22(3):665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res. 2011;45(11):1419–1425. [DOI] [PubMed] [Google Scholar]
- 20.Bharadwaj U, Kasembeli MM, Robinson P, Tweardy DJ. Targeting Janus Kinases and Signal Transducer and Activator of Transcription 3 to Treat Inflammation, Fibrosis, and Cancer: Rationale, Progress, and Caution. Pharmacol Rev. 2020;72(2):486–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He D, Chen H, Muramatsu H, Lasek AW. Ethanol activates midkine and anaplastic lymphoma kinase signaling in neuroblastoma cells and in the brain. J Neurochem. 2015;135(3):508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hepatoprotective Gao B. and anti-inflammatory cytokines in alcoholic liver disease. J Gastroenterol Hepatol. 2012;27 Suppl 2:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts AJ, Khom S, Bajo M, et al. Increased IL-6 expression in astrocytes is associated with emotionality, alterations in central amygdala GABAergic transmission, and excitability during alcohol withdrawal. Brain Behav Immun. 2019;82:188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachtell RK, Tsivkovskaia NO, Ryabinin AE. Alcohol-induced c-Fos expression in the Edinger-Westphal nucleus: pharmacological and signal transduction mechanisms. J Pharmacol Exp Ther. 2002;302(2):516–524. [DOI] [PubMed] [Google Scholar]
- 25.Gruol DL, Huitron-Resendiz S, Roberts AJ. Altered brain activity during withdrawal from chronic alcohol is associated with changes in IL-6 signal transduction and GABAergic mechanisms in transgenic mice with increased astrocyte expression of IL-6. Neuropharmacology. 2018;138:32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruol DL, Melkonian C, Huitron-Resendiz S, Roberts AJ. Alcohol alters IL-6 Signal Transduction in the CNS of Transgenic Mice with Increased Astrocyte Expression of IL-6. Cellular and molecular neurobiology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagisawa M, Nakashima K, Arakawa H, et al. Astrocyte differentiation of fetal neuroepithelial cells by interleukin-11 via activation of a common cytokine signal transducer, gp130, and a transcription factor, STAT3. J Neurochem. 2000;74(4):1498–1504. [DOI] [PubMed] [Google Scholar]
- 28.Auernhammer CJ, Bousquet C, Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc Natl Acad Sci U S A. 1999;96(12):6964–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egusquiaguirre SP, Yeh JE, Walker SR, Liu S, Frank DA. The STAT3 Target Gene TNFRSF1A Modulates the NF-kappaB Pathway in Breast Cancer Cells. Neoplasia. 2018;20(5):489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shiratori-Hayashi M, Koga K, Tozaki-Saitoh H, et al. STAT3-dependent reactive astrogliosis in the spinal dorsal horn underlies chronic itch. Nat Med. 2015;21(8):927–931. [DOI] [PubMed] [Google Scholar]
- 31.Jung M, Oren B, Mora J, et al. Lipocalin 2 from macrophages stimulated by tumor cell-derived sphingosine 1-phosphate promotes lymphangiogenesis and tumor metastasis. Science signaling. 2016;9(434):ra64. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiology & behavior. 2005;84(1):53–63. [DOI] [PubMed] [Google Scholar]
- 33.Subramanian A, Narayan R, Corsello SM, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171(6):1437–1452 e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One. 2013;8(3):e59870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das A, Salloum FN, Durrant D, Ockaili R, Kukreja RC. Rapamycin protects against myocardial ischemia-reperfusion injury through JAK2-STAT3 signaling pathway. J Mol Cell Cardiol. 2012;53(6):858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Z, Wang X, Ma L, et al. Inhibition of STAT3 signaling targets both tumor-initiating and differentiated cell populations in prostate cancer. Oncotarget. 2014;5(18):8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zapata A, Gonzales RA, Shippenberg TS. Repeated ethanol intoxication induces behavioral sensitization in the absence of a sensitized accumbens dopamine response in C57BL/6J and DBA/2J mice. Neuropsychopharmacology. 2006;31(2):396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England). 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornes O, Castro-Mondragon JA, Khan A, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48(D1):D87–d92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics (Oxford, England). 2011;27(7):1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erickson EK, Farris SP, Blednov YA, Mayfield RD, Harris RA. Astrocyte-specific transcriptome responses to chronic ethanol consumption. Pharmacogenomics J. 2018;18(4):578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceyzeriat K, Abjean L, Carrillo-de Sauvage MA, Ben Haim L, Escartin C. The complex STATes of astrocyte reactivity: How are they controlled by the JAK-STAT3 pathway? Neuroscience. 2016;330:205–218. [DOI] [PubMed] [Google Scholar]
- 43.Gano A, Doremus-Fitzwater TL, Deak T. Sustained alterations in neuroimmune gene expression after daily, but not intermittent, alcohol exposure. Brain Res. 2016;1646:62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hol EM, Pekny M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Current opinion in cell biology. 2015;32:121–130. [DOI] [PubMed] [Google Scholar]
- 45.Croker BA, Krebs DL, Zhang JG, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4(6):540–545. [DOI] [PubMed] [Google Scholar]
- 46.Egusquiaguirre SP, Yeh JE, Walker SR, Liu S, Frank DA. The STAT3 Target Gene TNFRSF1A Modulates the NF-κB Pathway in Breast Cancer Cells. Neoplasia. 2018;20(5):489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Satriotomo I, Miki T, Itoh M, Ameno K, Ijiri I, Takeuchi Y. Short-term ethanol exposure alters calbindin D28k and glial fibrillary acidic protein immunoreactivity in hippocampus of mice. Brain Res. 2000;879(1–2):55–64. [DOI] [PubMed] [Google Scholar]
- 48.Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J Neurosci. 2010;30(24):8285–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bull C, Syed WA, Minter SC, Bowers MS. Differential response of glial fibrillary acidic protein-positive astrocytes in the rat prefrontal cortex following ethanol self-administration. Alcohol Clin Exp Res. 2015;39(4):650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alfonso-Loeches S, Pascual M, Guerri C. Gender differences in alcohol-induced neurotoxicity and brain damage. Toxicology. 2013;311(1–2):27–34. [DOI] [PubMed] [Google Scholar]
- 51.Wilhelm CJ, Hashimoto JG, Roberts ML, Bloom SH, Andrew MR, Wiren KM. Astrocyte Dysfunction Induced by Alcohol in Females but Not Males. Brain pathology (Zurich, Switzerland). 2016;26(4):433–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Molecular cell. 2002;10(5):1033–1043. [DOI] [PubMed] [Google Scholar]
- 53.Zamanian JL, Xu L, Foo LC, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32(18):6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ip JP, Noçon AL, Hofer MJ, Lim SL, Müller M, Campbell IL. Lipocalin 2 in the central nervous system host response to systemic lipopolysaccharide administration. J Neuroinflammation. 2011;8:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang SS, Ren Y, Liu CC, et al. Lipocalin-2 protects the brain during inflammatory conditions. Mol Psychiatry. 2018;23(2):344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naudé PJ, Nyakas C, Eiden LE, et al. Lipocalin 2: novel component of proinflammatory signaling in Alzheimer’s disease. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26(7):2811–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bi F, Huang C, Tong J, et al. Reactive astrocytes secrete lcn2 to promote neuron death. Proc Natl Acad Sci U S A. 2013;110(10):4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai Y, Jogasuria A, Yin H, et al. The Detrimental Role Played by Lipocalin-2 in Alcoholic Fatty Liver in Mice. Am J Pathol. 2016;186(9):2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wieser V, Tymoszuk P, Adolph TE, et al. Lipocalin 2 drives neutrophilic inflammation in alcoholic liver disease. J Hepatol. 2016;64(4):872–880. [DOI] [PubMed] [Google Scholar]
- 60.Baxter-Potter LN, Henricks AM, Berger AL, Bieniasz KV, Lugo JM, McLaughlin RJ. Alcohol vapor exposure differentially impacts mesocorticolimbic cytokine expression in a sex-, region-, and duration-specific manner. Neuroscience. 2017;346:238–246. [DOI] [PubMed] [Google Scholar]
- 61.Karlsson C, Schank JR, Rehman F, et al. Proinflammatory signaling regulates voluntary alcohol intake and stress-induced consumption after exposure to social defeat stress in mice. Addict Biol. 2017;22(5):1279–1288. [DOI] [PubMed] [Google Scholar]
- 62.Carow B, Rottenberg ME. SOCS3, a Major Regulator of Infection and Inflammation. Front Immunol. 2014;5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker BJ, Akhtar LN, Benveniste EN. SOCS1 and SOCS3 in the control of CNS immunity. Trends Immunol. 2009;30(8):392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baetz A, Frey M, Heeg K, Dalpke AH. Suppressor of cytokine signaling (SOCS) proteins indirectly regulate toll-like receptor signaling in innate immune cells. J Biol Chem. 2004;279(52):54708–54715. [DOI] [PubMed] [Google Scholar]
- 65.Pandey SC. TLR4-MyD88 signalling: a molecular target for alcohol actions. British journal of pharmacology. 2012;165(5):1316–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carpenter RL, Lo HW. STAT3 Target Genes Relevant to Human Cancers. Cancers. 2014;6(2):897–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mouzat K, Chudinova A, Polge A, et al. Regulation of Brain Cholesterol: What Role Do Liver X Receptors Play in Neurodegenerative Diseases? International journal of molecular sciences. 2019;20(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Warner M, Gustafsson JA. Estrogen receptor β and Liver X receptor β: biology and therapeutic potential in CNS diseases. Mol Psychiatry. 2015;20(1):18–22. [DOI] [PubMed] [Google Scholar]
- 69.Hussain G, Wang J, Rasul A, et al. Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids in health and disease. 2019;18(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pfrieger FW. Outsourcing in the brain: do neurons depend on cholesterol delivery by astrocytes? BioEssays : news and reviews in molecular, cellular and developmental biology. 2003;25(1):72–78. [DOI] [PubMed] [Google Scholar]
- 71.Alsebaaly J, Dugast E, Favot L, Rabbaa Khabbaz L, Solinas M, Thiriet N. Persistent Neuroadaptations in the Expression of Genes Involved in Cholesterol Homeostasis Induced by Chronic, Voluntary Alcohol Intake in Rats. Frontiers in molecular neuroscience. 2018;11:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang JJ, Ma WJ, Yokoyama S. Expression and immunolocalization of Gpnmb, a glioma-associated glycoprotein, in normal and inflamed central nervous systems of adult rats. Brain and behavior. 2012;2(2):85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moloney EB, Moskites A, Ferrari EJ, Isacson O, Hallett PJ. The glycoprotein GPNMB is selectively elevated in the substantia nigra of Parkinson’s disease patients and increases after lysosomal stress. Neurobiology of disease. 2018;120:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murthy MN, Blauwendraat C, Guelfi S, Hardy J, Lewis PA, Trabzuni D. Increased brain expression of GPNMB is associated with genome wide significant risk for Parkinson’s disease on chromosome 7p15.3. Neurogenetics. 2017;18(3):121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H, Dey KK, Chen PC, et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer’s disease. Molecular neurodegeneration. 2020;15(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murugesan V, Liu J, Yang R, et al. Validating glycoprotein non-metastatic melanoma B (gpNMB, osteoactivin), a new biomarker of Gaucher disease. Blood cells, molecules & diseases. 2018;68:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu S, Wuolikainen A, Wu J, et al. Targeted Multiple Reaction Monitoring Analysis of CSF Identifies UCHL1 and GPNMB as Candidate Biomarkers for ALS. Journal of molecular neuroscience : MN. 2019;69(4):643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Neal ML, Boyle AM, Budge KM, Safadi FF, Richardson JR. The glycoprotein GPNMB attenuates astrocyte inflammatory responses through the CD44 receptor. J Neuroinflammation. 2018;15(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Budge KM, Neal ML, Richardson JR, Safadi FF. Glycoprotein NMB: an Emerging Role in Neurodegenerative Disease. Molecular neurobiology. 2018;55(6):5167–5176. [DOI] [PubMed] [Google Scholar]
- 80.Pandey SC, Kyzar EJ, Zhang H. Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology. 2017;122:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hamilton PJ, Nestler EJ. Epigenetics and addiction. Current opinion in neurobiology. 2019;59:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mathies LD, Blackwell GG, Austin MK, et al. SWI/SNF chromatin remodeling regulates alcohol response behaviors in Caenorhabditis elegans and is associated with alcohol dependence in humans. Proc Natl Acad Sci U S A. 2015;112(10):3032–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin Exp Res. 2011;35(4):735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burrowes SG, Salem NA, Tseng AM, et al. The BAF (BRG1/BRM-Associated Factor) chromatin-remodeling complex exhibits ethanol sensitivity in fetal neural progenitor cells and regulates transcription at the miR-9–2 encoding gene locus. Alcohol. 2017;60:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith ML, Lopez MF, Archer KJ, Wolen AR, Becker HC, Miles MF. Time-Course Analysis of Brain Regional Expression Network Responses to Chronic Intermittent Ethanol and Withdrawal: Implications for Mechanisms Underlying Excessive Ethanol Consumption. PLoS One. 2016;11(1):e0146257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wingelhofer B, Neubauer HA, Valent P, et al. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia. 2018;32(8):1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roberto M, Patel RR, Bajo M. Ethanol and Cytokines in the Central Nervous System. Handbook of experimental pharmacology. 2018;248:397–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Erickson EK, Grantham EK, Warden AS, Harris RA. Neuroimmune signaling in alcohol use disorder. Pharmacol Biochem Behav. 2019;177:34–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adermark L, Bowers MS. Disentangling the Role of Astrocytes in Alcohol Use Disorder. Alcohol Clin Exp Res. 2016;40(9):1802–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bull C, Freitas KC, Zou S, et al. Rat nucleus accumbens core astrocytes modulate reward and the motivation to self-administer ethanol after abstinence. Neuropsychopharmacology. 2014;39(12):2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erickson EK, DaCosta AJ, Mason SC, Blednov YA, Mayfield RD, Harris RA. Cortical astrocytes regulate ethanol consumption and intoxication in mice. Neuropsychopharmacology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.