Abstract
In addition to the BlaB metallo-β-lactamase, Chryseobacterium (Flavobacterium) meningosepticum CCUG 4310 (NCTC 10585) constitutively produces a 31-kDa active-site serine β-lactamase, named CME-1, with an alkaline isoelectric pH. The blaACME gene that encodes the latter enzyme was isolated from a genomic library constructed in the Escherichia coli plasmid vector pACYC184 by screening for cefuroxime-resistant clones. Sequence analysis revealed that the CME-1 enzyme is a new class A β-lactamase structurally divergent from the other members of this class, being most closely related to the VEB-1 (also named CEF-1) and PER β-lactamases and the Bacteroides chromosomal cephalosporinases. The blaACME determinant is located on the chromosome and exhibits features typical of those of C. meningosepticum resident genes. The CME-1 protein was purified from an E. coli strain that overexpresses the cloned gene via a T7-based expression system by means of an anion-exchange chromatography step followed by a gel permeation chromatography step. Kinetic parameters for several substrates were determined. CME-1 is a clavulanic acid-susceptible extended-spectrum β-lactamase that hydrolyzes most cephalosporins, penicillins, and monobactams but that does not hydrolyze cephamycins and carbapenems. The enzyme exhibits strikingly different kinetic parameters for different classes of β-lactams, with both Km and kcat values much higher for cephalosporins than for penicillins and monobactams. However, the variability of both kinetic parameters resulted in overall similar acylation rates (kcat/Km ratios) for all types of β-lactam substrates.
Production of β-lactamases is the most prevalent mechanism of bacterial resistance to β-lactam antibiotics. Three molecular families of active-site serine β-lactamases (classes A, C, and D) and one of metallo-β-lactamases (class B) have evolved in the bacterial kingdom (2, 10, 17).
Molecular class A β-lactamases are the most widespread β-lactam-degrading enzymes in clinical isolates, in which they can occur either as chromosomally encoded enzymes resident in the species or as acquired enzymes encoded by genetic determinants carried on mobile elements (10, 23). Class A enzymes are remarkably versatile from the functional standpoint. Some of them show relatively narrow substrate profiles, while others exhibit broader substrate specificities. They are usually susceptible to mechanism-based β-lactamase inhibitors, such as clavulanic acid, sulbactam, and tazobactam, but inhibitor-resistant variants also exist (10, 26). From the structural standpoint, although all class A β-lactamases share conserved sequence motifs that are the landmarks for classification, a considerable heterogeneity occurs among members of this group and various evolutionary lineages have been identified (10, 12, 25). The enzymes within each lineage often exhibit a consistent functional behavior. However, under the strong selective pressure generated by intense β-lactam usage, fine allelic variants of certain enzymes (e.g., TEM and SHV) that show a significant modification of the substrate specificity and/or susceptibility to inhibitors have been selected (10, 21, 23, 26).
In this paper we report on the cloning and characterization of a Chryseobacterium (formerly Flavobacterium) meningosepticum chromosomal gene (blaACME) that encodes a class A β-lactamase which is structurally rather divergent from the other class A enzymes, being most closely related to members of the class A lineage including the VEB-1 (also named CEF-1) (34, 46) and PER (6, 29, 30) β-lactamases and the Bacteroides chromosomal cephalosporinases (32, 36, 42). CME-1 is a clavulanic acid-susceptible extended-spectrum β-lactamase active on narrow- to expanded-spectrum cephalosporins (except for cephamycins), penicillins, and monobactams, and it exhibits strikingly different kinetic parameters with different groups of β-lactam substrates.
MATERIALS AND METHODS
Bacterial strains and genetic vectors.
C. meningosepticum CCUG 4310 (NCTC 10585) was used as the source of DNA for construction of the genomic library. This reference strain was selected since it has been reported to be highly related to most C. meningosepticum clinical isolates (31). Escherichia coli DH5α (GIBCO-BRL, Gaithersburg, Md.) and BL21(DE3) (Novagen, Inc., Madison, Wis.) were used as the hosts for recombinant plasmids. Bacterial strains were always grown aerobically at 37°C. Plasmid pACYC184 (11) was used as the vector for construction of the C. meningosepticum genomic library. Plasmid pBC-SK (Stratagene, La Jolla, Calif.) was used for some subcloning steps.
Antibiotics.
Antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise specified. Nitrocefin was from Unipath (Milan, Italy), imipenem was from Merck Research Laboratories (Rahway, N.J.), ceftazidime was from Glaxo-Wellcome (Verona, Italy), cefepime and aztreonam were from Bristol-Myers Squibb Co. (Wallingford, Conn.), carumonam was from Hoffmann-La Roche (Basel, Switzerland), and clavulanic acid was from SmithKline Beecham Pharmaceuticals (Brentford, United Kingdom). All antibiotic solutions were prepared immediately before use.
β-Lactamase assays.
β-Lactamase activity in crude cell extracts was assayed spectrophotometrically. Reactions were always performed in 50 mM sodium phosphate buffer (PB; pH 7.0) at 25°C in a total volume of 0.75 ml. Imipenem hydrolysis was monitored at a λ of 299 nm with a substrate concentration of 0.12 mM. Nitrocefin hydrolysis was monitored at a λ of 482 nm with a substrate concentration of 0.075 mM. Inhibition of enzymatic activity by EDTA was determined by measuring the residual activity after incubation of the crude extract for 20 min at 25°C in the presence of 20 mM EDTA. A control without EDTA was always run in parallel. Crude extracts were prepared as follows. Cells were grown in Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) aerobically at 37°C, collected by centrifugation, washed once in 50 mM PB, resuspended in the same buffer (1/10 of the original culture volume), and disrupted by sonication (six times for 15 s each time at 50 W). The supernatant obtained after centrifugation at 10,000 × g for 10 min to remove cell debris represented the crude extract. The protein concentration in solution was assayed by the method of Bradford (9) with a commercial kit (Bio-Rad protein assay; Bio-Rad, Richmond, Calif.), with bovine serum albumin used as a standard.
Protein electrophoretic techniques.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of protein preparations was performed as described by Laemmli (22) with final acrylamide concentrations of 15 and 5% (wt/vol) for the separating and the stacking gels, respectively. After electrophoresis the protein bands were stained with Coomassie brilliant blue R-250. Zymogram detection of β-lactamase activities after SDS-PAGE was performed essentially as described previously (24). After the renaturation treatment, the bands of β-lactamase activity were revealed by the appearance of purple-stained bands after overlaying the gel with filter paper previously soaked in a 0.25 mM nitrocefin solution in PB. Analytical isoelectric focusing (IEF) was performed on precast 6% acrylamide gels containing an ampholine gradient in the pH range of 3.5 to 9.5 (Pharmacia Biotech, Uppsala, Sweden) with a Multiphor II flat-bed apparatus (Pharmacia). Proteins were focused at a constant temperature (6°C) for 3 h at 1 W/cm. After focusing, the β-lactamase activity was revealed as described above for renaturing SDS-PAGE.
Recombinant DNA methodology.
Basic recombinant DNA procedures were performed as described by Sambrook et al. (39). Construction of the genomic library from C. meningosepticum CCUG 4310 has been described previously (37). Southern blot analysis was performed on nylon membranes (Hybond-N; Amersham, Little Chalfont, United Kingdom), according to the manufacturer’s instructions, with randomly primed 32P-labeled probes. The blaB-specific probe used to recognize clones carrying the blaB gene was made of a 0.21-kb HindIII fragment internal to the C. meningosepticum blaB gene (37). Restriction endonucleases and DNA modification enzymes were from Boehringer (Mannheim, Germany).
DNA sequencing and computer analysis of sequence data.
DNA sequencing was performed by the dideoxy-chain termination method with a Sequenase, version 2.0, DNA sequencing kit (Amersham) and custom sequencing primers. The sequences of both strands were determined with denatured double-stranded DNA templates. Computer analysis of the sequence data was performed with an updated version (version 8.1) of the University of Wisconsin Genetics Computer Group (UWGCG) package (15). Similarity searches against sequence databases were performed with an updated version of the BLAST program (1) at the BLAST network service of the Swiss Institute for Experimental Cancer Research. Comparison of codon usage tables was performed with the CORRESPOND program of the UWGCG package, as described by Grantham et al. (19). Multiple sequence alignments were generated with the help of the CLUSTAL W program (44). Phylogenetic analysis was performed by the neighbor-joining method (38) with the bootstrap tree option of the CLUSTAL W program and by allowing for 1,000 bootstrap trials.
Purification of the CME-1 enzyme.
The CME-1 enzyme was purified from E. coli BL21(DE3)(pBlaA-CNB) as follows. The strain was grown in 6 liters of brain heart infusion broth containing chloramphenicol (85 μg/ml) for 16 h at 37°C. The cells were harvested by centrifugation, washed twice with 50 mM Tris-HCl (Tris buffer; pH 8.5), resuspended in 300 ml of TB, and disrupted by sonication (five times for 30 s each time at 60 W). Cell debris was removed by high-speed centrifugation (105,000 × g for 60 min at 4°C), and the clarified supernatant was loaded onto an S-Sepharose FF column (2.5 by 30 cm; Pharmacia) equilibrated with TB. After washing of the column with the same buffer, the bound proteins were eluted with a linear NaCl gradient (0 to 1 M) in TB. The fractions that showed β-lactamase activity (nitrocefin was used as the substrate) were pooled, dialyzed against PB (pH 7.0), concentrated 10-fold by ultrafiltration, and loaded onto a Superdex-75 column (1.6 by 75 cm; Pharmacia) that had been equilibrated and eluted with the same buffer. The β-lactamase-containing elution peak was concentrated at 0.5 mg/ml and was stored at −80°C until use.
N terminus sequencing and electrospray mass spectrometry.
The N-terminal sequence of the purified CME-1 protein was determined with a gas-phase sequencer (Procise-492; Applied Biosystems, Foster City, Calif.) after resuspension of the protein (50 pmol) in a 0.1% (vol/vol) trifluoroacetic acid solution and loading of the sample onto a polyvinylidene difluoride membrane (Millipore Corp., Bedford, Mass.). Electrospray mass spectrometry was performed with a VG Platform (Micromass, Manchester, United Kingdom). The purified protein was in acetronitrile-water (50:50 [vol/vol]; pH 6.5). The source temperature was kept at 80°C. The sample was directly introduced into the ionization chamber (at atmospheric pressure) through a steel capillary with a flow rate of 40 μl/min. The sampling cone voltage was maintained at 40 V.
Determination of kinetic parameters.
Substrate hydrolysis by the purified enzyme was monitored by following the absorbance variation with a lambda 2 spectrophotometer (Perkin-Elmer, Rahway, N.J.) equipped with thermostatically controlled cells and connected to an International Business Machines-compatible personal computer via an RS232C serial interface. The wavelengths and changes in extinction coefficient were as follows: penicillin G, 235 nm and −775 M−1 cm−1; ampicillin and piperacillin, 235 nm and −820 M−1 cm−1; carbenicillin, 235 nm and −780 M−1 cm−1; nitrocefin, 482 nm and +15,000 M−1 cm−1; cephaloridine and cefepime, 260 nm and −10,000 M−1 cm−1; cephalothin, 260 nm and −6,500 M−1 cm−1; cefuroxime, 260 nm and −7,600 M−1 cm−1; cefoxitin, 260 nm and −7,700 M−1 cm−1; cefotaxime, 260 nm and −7,500 M−1 cm−1; ceftazidime, 260 nm and −9,000 M−1 cm−1; imipenem, 299 nm and −9,000 M−1 cm−1; aztreonam, 320 nm and −700 M−1 cm−1; and carumonam, 310 nm and −810 M−1 cm−1. Km and kcat values were determined by analyzing either the complete hydrolysis time courses (14) when the reaction velocity was sufficiently high to allow complete substrate hydrolysis within a few minutes or under initial-rate conditions by using the Hanes-Woolf plot (40). The low Km values for penicillins and aztreonam were measured as Ki with 100 μM nitrocefin as the reporter substrate. The Ki value was determined by the plot of V0/Vi versus I, yielding a line whose slope is Kms/(Kms + S) · Ki, where V0 and Vi are the initial rates of nitrocefin hydrolysis in the absence and presence of the inhibitor, respectively, I is the inhibitor concentration, S is the reporter substrate concentration, and Kms is the Michaelis constant of the enzyme for the reporter substrate. Inactivation by clavulanic acid was monitored with 100 μM nitrocefin as the reporter substrate. All the determinations were performed at 30°C in PB with bovine serum albumin (50 μg/ml). The total reaction volume was 0.6 ml in all cases. The enzyme concentration in the reaction was in the range of 20 to 200 nM.
RESULTS
Production of an active-site serine β-lactamase by C. meningosepticum CCUG 4310.
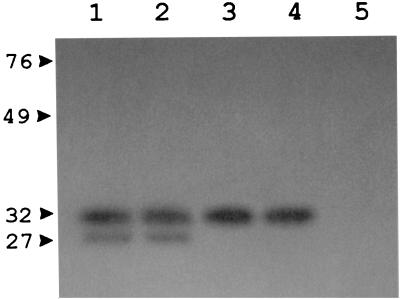
C. meningosepticum CCUG 4310 produces a molecular class B metallo-β-lactamase that is active against several substrates including carbapenems and that is susceptible to inhibition by chelating agents (37). Measurement of the β-lactamase activity of crude extracts prepared from this strain showed that, after treatment with EDTA, the imipenem-hydrolyzing activity was nearly completely inhibited, while a consistent nitrocefin-hydrolyzing activity was still detectable (Table 1), suggesting the additional presence of one or more active-site serine enzymes. Production of this residual EDTA-resistant activity was apparently constitutive (Table 1). A zymogram analysis of the crude extract, performed after renaturing SDS-PAGE with the nitrocefin chromogenic substrate, yielded a major band of activity at approximately 31 kDa and a minor band of activity at approximately 27 kDa; both of these bands appeared to be produced constitutively (Fig. 1). Considering that the 27-kDa band likely corresponds to the BlaB metalloenzyme (37), zymogram results suggested that the EDTA-resistant activity present in the crude extract was contributed by a serine β-lactamase consisting of a 31-kDa polypeptide. IEF analysis of the crude extract, which was developed with nitrocefin, yielded two bands of β-lactamase activity that focused at pH 8.5 and >9, respectively; both of these bands appeared to be produced constitutively (data not shown). Considering that the pI 8.5 band likely corresponds to the BlaB metalloenzyme (37), IEF results suggested that the EDTA-resistant activity present in the crude extract was contributed by a serine enzyme with an alkaline isoelectric pH.
TABLE 1.
β-Lactamase activities of crude extracts of C. meningosepticum CCUG 4310 and E. coli DH5α(pBlaA-4c)a
| Sampleb | Sp act (μmol/min/mg of protein) against:
|
|||
|---|---|---|---|---|
| Nitrocefin | Nitrocefin EDTA | Imipenem | Imipenem EDTA | |
| CCUG 4310, not induced | 0.182 | 0.127 (69)c | 0.078 | <0.005 |
| CCUG 4310, induced | 0.197 | 0.134 (68) | 0.083 | <0.005 |
| DH5α(pBlaA-4c) | 0.465 | 0.458 (98) | <0.005 | NAd |
| DH5α(pACYC184) | <0.010 | NA | <0.005 | NA |
The basal activity of E. coli DH5α(pACYC184) is also shown for comparison. Data represent mean values for three measurements. The standard deviations were always lower than 10%.
The crude extracts of CCUG 4310 were prepared from exponentially growing cultures, either without antibiotic (not induced) or with ampicillin (25 μg/ml) added 2 h before collection (induced). The crude extracts of E. coli strains were prepared from early-stationary-phase cultures.
The values in parentheses are the percentage of the activity measured with nitrocefin alone.
NA, not assayed.
FIG. 1.
Results of zymogram analysis performed after renaturing SDS-PAGE with the chromogenic cephalosporin nitrocefin as the substrate for detection of β-lactamase activity. Lanes: 1, crude extract from CCUG 4310, not induced; 2, crude extract from CCUG 4310 induced with ampicillin; 3, purified CME-1 enzyme; 4, crude extract from E. coli DH5α(pBlaA-4c); 5, crude extract from E. coli DH5α(pACYC184). The crude extracts were prepared as described in Table 1. Protein size standards are indicated in kilodaltons on the left.
Cloning of the C. meningosepticum genetic determinant encoding the 31-kDa active-site serine β-lactamase.
A genomic library of C. meningosepticum CCUG 4310, constructed in the E. coli multicopy plasmid vector pACYC184 and transformed into E. coli DH5α, was replica plated on a medium containing cefuroxime (50 μg/ml). Three cefuroxime-resistant clones were obtained from approximately 7 × 103 screened transformants. A Southern hybridization analysis of the plasmids carried by these clones with a blaB-specific probe showed that two of them contained a cloned copy of the previously characterized blaB gene (37), while the remaining one, named pBlaA-4c, did not contain any blaB-related sequences (data not shown). This clone was able to produce an EDTA-resistant β-lactamase that was unable to hydrolyze imipenem (Table 1) and that, in zymograms performed after renaturing SDS-PAGE, appeared to be contributed by a 31-kDa polypeptide (Fig. 1).
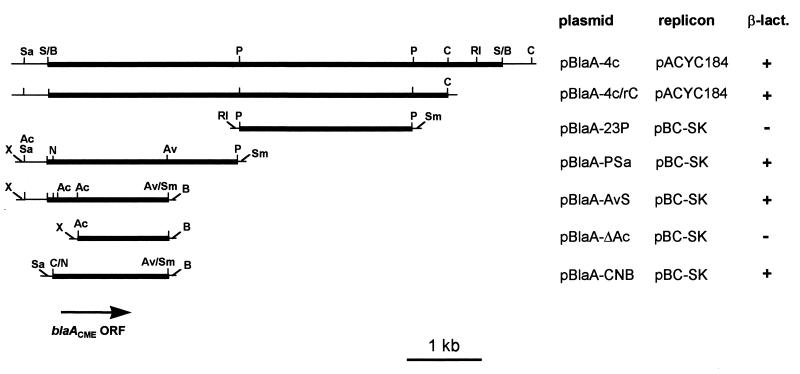
The β-lactamase-encoding determinant carried by clone pBlaA-4c was mapped within a 1.7-kb NspV-AvaII fragment by subcloning analysis (Fig. 2). The origin of the cloned fragment from a single chromosomal region of the donor strain was confirmed by a Southern hybridization analysis performed with the genomic DNA of C. meningosepticum CCUG 4310 by using the 1.7-kb NspV-AvaII fragment as a probe. The probe hybridized to the band of undigested chromosomal DNA and recognized single restriction fragments of 4.3 and 5 kb after digestion with NspV and PstI, respectively (data not shown).
FIG. 2.
Restriction map of the DNA insert of plasmid pBlaA-4c and subcloning strategy. Thick lines represent insert sequences, while thin lines represent vector sequences. The location and orientation of the blaACME ORF is indicated. Crude extracts prepared from early-stationary-phase cultures of E. coli clones carrying each recombinant plasmid were assayed for production of β-lactamase activity (β-lact.) as described in the Materials and Methods section. Abbreviations: Ac, AccI; Av, AvaII; Av/Sm, AvaII-SmaI junction; B, BamHI; C, ClaI; C/N, ClaI-NspV junction; N, NspV; P, PstI; RI, EcoRI; S/B, Sau3AI-BamHI junction; Sa, SalI; Sm, SmaI; X, XhoI.
Sequence analysis of the β-lactamase-encoding determinant.
The nucleotide sequence of the DNA insert of plasmid pBlaA-AvS (Fig. 2) was determined. An 888-bp open reading frame (ORF) (Fig. 3) which encoded a polypeptide that showed, in a BLAST search, the highest similarity scores with other class A β-lactamases was identified. Results of subcloning experiments were consistent with the identification of this ORF, named blaACME, as the β-lactamase-encoding determinant (Fig. 2).
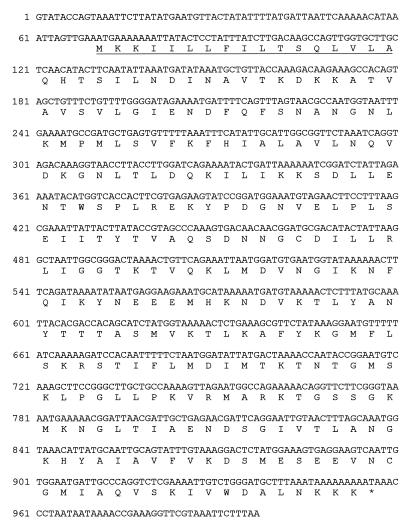
FIG. 3.
Nucleotide sequence of the blaACME gene and flanking regions. Nucleotide 1 corresponds to the first base of the AccI restriction site located upstream of the gene. The deduced amino acid sequence of the CME-1 protein is reported below the nucleotide sequence. The underlined region corresponds to the experimentally determined signal peptide for secretion.
The blaACME ORF encodes a 295-amino acid polypeptide whose amino-terminal sequence exhibits features typical of those of bacterial signal peptides that target secretion into the periplasmic space via the general secretory pathway (Fig. 3). According to the results of sequencing of the N terminus of the purified CME-1 protein (see below), the cleavage site is located after the Ala-17 residue. This would yield a mature protein with a calculated molecular mass and a pI of 30,878 Da and 9.38, respectively, which are in good agreement with the experimental results obtained with the purified protein (see below).
The G+C content of the blaACME ORF is 34.2%, being similar to those of the other sequenced C. meningosepticum genes recorded in release 56 of the EMBL sequence database (range, 36.1 to 41.6%). The codon usage of blaACME was not significantly different from that of the other sequenced C. meningosepticum genes (D squared value = 1.33).
Comparison of the CME-1 enzyme with other class A β-lactamases at primary structure level.
The BLAST search performed with the CME-1 protein as a query returned the highest similarity scores (scores, >300) with the group of class A β-lactamases that included the VEB-1 (also named CEF-1) (34, 46), PER-1 (30), and PER-2 (6) enzymes and the Bacteroides chromosomal cephalosporinases (32, 36, 42). Lower similarity scores were returned for the other class A β-lactamases (Table 2 and data not shown).
TABLE 2.
Class A β-lactamases included in phylogenetic analysis
| Enzyme | Organism | Accession no. (databasea) | % Identityb | BLAST scoresc | Reference or source |
|---|---|---|---|---|---|
| CME-1 | Chryseobacterium meningosepticum CCUG 4310 | AJ006275 (EM) | This study | ||
| VEB-1d | Escherichia coli MG-1 | AF078527 (EM) | 46 | 587 | 34 |
| CblA | Bacteroides uniformis WAL-7088 | P30898 (SW) | 40 | 547 | 42 |
| CepA | Bacteroides fragilis CS30 | L13472 (EM) | 36 | 492 | 36 |
| PER-1 | Pseudomonas aeruginosa RNL-1 | P37321 (SW) | 39 | 486 | 30 |
| PER-2 | Salmonella enterica serotype Typhimurium JMC | X93314 (EM) | 38 | 486 | 6 |
| CfxA | Bacteroides vulgatus CLA341 | P30899 (SW) | 29 | 328 | 32 |
| BRO-1 | Moraxella catarrhalis ATCC 53879 | Q59514 (SW) | 27 | 236 | 8 |
| PSE-1 | Pseudomonas aeruginosa RPL11 | Q03170 (SW) | 23 | 198 | 20 |
| ULA27 | Citrobacter diversus ULA27 | P22390 (SW) | 25 | 184 | 33 |
| L2 | Stenotrophomonas maltophilia 1275 IID | P96465 (SW) | 24 | 176 | 49 |
| NMC-A | Enterobacter cloacae NOR-1 | P52663 (SW) | 24 | 169 | 27 |
| SHV-1 | Escherichia coli P453 | P14557 (SW) | 23 | 164 | 5 |
| TEM-1 | Salmonella enterica serotype Paratyphi R7268 | P00810 (SW) | 23 | 162 | 43 |
| Sme-1 | Serratia marcescens S6 | P52682 (SW) | 21 | 145 | 28 |
| MEN-1 | Escherichia coli MEN | P28585 (SW) | 22 | 144 | 4 |
| OXY-2 | Klebsiella oxytoca D488 | P23954 (SW) | 21 | 138 | 35 |
| PenA | Burkholderia cepacia 249 | U85041 (EM) | 22 | 135 | 45 |
| BlaI | Yersinia enterocolitica Y-56 | Q01166 (SW) | 22 | 133 | 41 |
EM, EMBL/GenBank nucleotide sequence database; SW SwissProt protein sequence database.
Compared to the sequence of the mature CME-1 protein and calculated on the basis of the multiple alignment used for phylogenetic analysis.
These scores were returned after submission of the 295-amino-acid polypeptide encoded by the blaACME ORF as a query at the BLAST network service using WU-BLAST server version 2.0a13.
The sequence of VEB-1 is identical to that of CEF-1, which is encoded by a gene found in a P. aeruginosa integron (46).
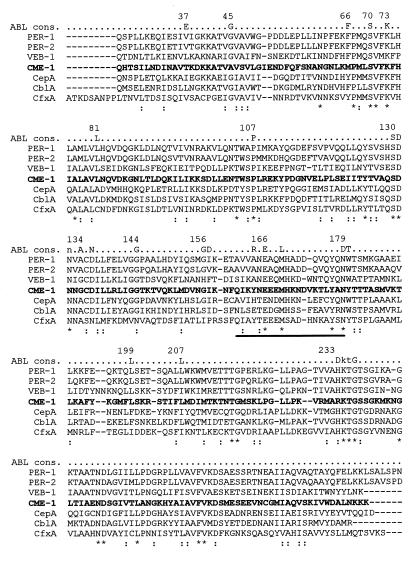
A multiple sequence alignment analysis of the CME-1 enzyme with its closest neighbors is shown in Fig. 4, together with the previously defined consensus sequence for class A β-lactamases (26). Of the nine invariant residues reported as typical of all class A enzymes (Gly-45, Ser-70, Lys-73, Pro-107, Ser-130, Asp-131, Ala-134, Glu-166, and Gly-236) (26), seven are retained in the CME-1 protein, while an alanine residue is substituted for Gly-45 and a glycine residue is substituted for Ala-134 (Fig. 4). Concerning the other conserved residues of the ABL consensus sequence, substitutions never reported in other class A enzymes were found at positions 37 (Thr), 66 (Met), and 233 (Arg). Compared with its closest neighbors and with most other class A proteins, the CME-1 enzyme contains an extra residue within the Ω-loop region (Fig. 4).
FIG. 4.
Sequence alignment of the CME-1 protein (in boldface) with its closest class A neighbors. Identical residues are indicated by an asterisk; conservative substitutions are indicated by a colon. The enzyme names and corresponding sequence references are the same as those in Table 2. The conserved residues of the ABL consensus sequence (ABL cons.) (26) are reported above the alignment, and some relevant amino acid positions, according to the ABL numbering scheme (3), are also indicated. The Ω-loop region is indicated by a horizontal bar below the sequences.
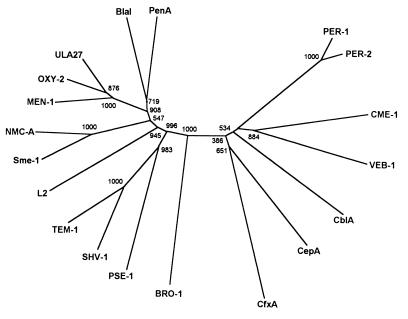
Phylogenetic relationships among the CME-1 enzyme, its closest neighbors, and 12 additional proteins representative of the major lineages of class A β-lactamases of gram-negative bacteria (Table 2) were analyzed by construction of an unrooted tree. Results of this analysis indicated that the CME-1 enzyme is rather divergent from the other class A β-lactamases and confirmed its closest overall evolutionary relatedness with members of the lineage that includes the PER and VEB-1 β-lactamases and the Bacteroides cephalosporinases. In particular, CME-1 and VEB-1 appear to have diverged early from a common ancestor that originated during the initial phases of class A β-lactamase evolution (Fig. 5).
FIG. 5.
Unrooted tree showing the phyletic relationships among 19 different class A β-lactamases, including the CME-1 enzyme and its closest neighbors. Sequence names are the same as those in Table 2. Numbers at each branching point indicate the number/1,000 bootstrap trials returned for that point.
Purification and characterization of CME-1 enzyme.
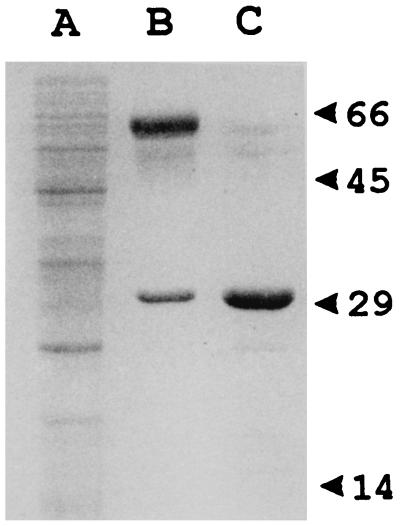
Overexpression of the blaACME gene was obtained by introducing recombinant plasmid pBlaA-CNB in which the blaACME ORF is located downstream of the T7 promoter flanking the polylinker of pBC-SK (Fig. 2), in the T7 RNA polymerase-producing E. coli host BL21(DE3). The CME-1 enzyme was purified from a crude lysate of E. coli BL21(DE3)(pBlaA-CNB) by means of an anion-exchange chromatography step followed by a gel permeation chromatography step. By SDS-PAGE the purified protein appeared as a single 31-kDa band and was estimated to be >95% pure (Fig. 6). The isolectric pH of the purified protein was >9 (data not shown). The amino-terminal sequence of the purified protein was determined to be NH2-QHTSI. The Mr of the purified protein, as determined by electrospray mass spectrometry, was 30,870 ± 12.
FIG. 6.
SDS-PAGE analysis of the purification steps of the CME-1 protein. Lanes: A, clarified extract of E. coli BL21(DE3)(pBlaA-CNB); B, pooled fractions with β-lactamase activity eluted from the S-Sepharose column; C, pooled fractions with β-lactamase activity eluted from the Superdex-75 column. Protein size standards are indicated (in kilodaltons) on the right.
The purified CME-1 protein appeared to be active against several β-lactam substrates including narrow- to expanded-spectrum cephalosporins, penicillins, and monobactams. No hydrolysis of cefoxitin and imipenem was detected (Table 3). Kinetic parameters were markedly different for members of different β-lactam families. The enzyme showed higher kcat values for cephalosporins than for penicillins (10- to 500-fold) and monobactams (100- to 700-fold). On the other hand, the affinities of CME-1 for penicillins and monobactams were much higher than those for cephalosporins, resulting in overall similar acylation rates (kcat/Km ratios) with the various substrates (Table 3). The enzyme was completely inactivated after 60 s of exposure to clavulanic acid at a 1:50 (enzyme:inhibitor) molar ratio. After 24 h of incubation, a small recovery of activity was observed under these conditions, while no recovery of activity was observed when a 1:10,000 (enzyme:inhibitor) molar ratio was used. In short competitive assays a Ki of 0.36 μM was calculated for clavulanic acid. No significant modifications of the kinetic parameters measured with nitrocefin were detectable after exposure of the enzyme to EDTA concentrations of up to 20 mM for 20 min or with phosphate ion concentrations ranging from 20 to 250 mM in the assay buffer.
TABLE 3.
Kinetic parameters of purified CME-1 enzyme
| Substrate | Km (μM)ab | kcat (s−1)a | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| Penicillin G | 3.1 ± 0.2 | 1.2 ± 0.07 | 0.39 |
| Ampicillin | 5.1 ± 0.4 | 2.9 ± 0.18 | 0.57 |
| Carbenicillin | 7.0 ± 0.4 | 2.6 ± 0.17 | 0.37 |
| Piperacillin | 0.12 ± 0.01 | 0.23 ± 0.02 | 1.92 |
| Nitrocefin | 25 ± 1.2 | 134 ± 8.0 | 5.36 |
| Cephaloridine | 87 ± 5.7 | 74 ± 4.3 | 0.85 |
| Cephalothin | 105 ± 4.4 | 118 ± 6.0 | 1.12 |
| Cefuroxime | 106 ± 4.6 | 102 ± 6.2 | 0.96 |
| Cefoxitin | NDc | NHd | |
| Cefotaxime | 50 ± 3.0 | 27 ± 1.6 | 0.54 |
| Ceftazidime | 511 ± 36 | 68 ± 4.4 | 0.13 |
| Cefepime | 487 ± 39 | 48 ± 2.7 | 0.099 |
| Imipenem | ND | NH | |
| Aztreonam | 5.0 ± 0.3 | 0.23 ± 0.02 | 0.046 |
| Carumonam | 3.6 ± 0.3 | 0.17 ± 0.01 | 0.047 |
Values are means ± standard deviations of three measurements.
Determined as Ki when Km was lower than 10 μM.
not determined.
NH, no hydrolysis detected.
DISCUSSION
In addition to the BlaB metallo-β-lactamase (37), C. meningosepticum CCUG 4310 also produces a class A serine β-lactamase named CME-1. Similarly to BlaB (37), CME-1 is encoded by a chromosomal gene which, according to the G+C content and codon usage, appears to be resident in the species. Production of either enzyme appears to be independent of the presence of β-lactam inducers, with relatively high basal levels of activity. Although β-lactam susceptibility always depends on the interplay of several factors, the constitutive production of these two β-lactamases, whose combined substrate profiles include virtually all the major β-lactam families (37; this study), likely provides a relevant contribution to the natural high-level β-lactam resistance shown by C. meningosepticum (7, 16).
CME-1 is a new class A β-lactamase whose primary structure is rather divergent from those of other class A enzymes. Its closest structural neighbors are the recently described VEB-1 enzyme (also named CEF-1) encoded by a gene found in E. coli and Pseudomonas aeruginosa integrons (34, 46), the PER-1 extended-spectrum β-lactamase detected among clinical isolates of P. aeruginosa, Acinetobacter, and Salmonella enterica serotype Typhimurium (29, 47, 48), the PER-2 extended-spectrum β-lactamase detected among clinical isolates of the family Enterobacteriaceae (6), and the chromosomally encoded cephalosporinases of various Bacteroides species (32, 36, 42). Results of a phylogenetic analysis performed with representative enzymes of all major class A lineages of gram-negative bacteria, which were in overall agreement with those of previous studies (10, 12, 25), confirmed that CME-1 is most closely related to the former group of enzymes and represents a new member that diverged rather early during the evolutionary history of that lineage. Members of this lineage constitute a distinct molecular subfamily among the class A β-lactamases encountered in gram-negative bacteria, including both resident and mobile enzymes with common ancestries. Identification of additional enzymes that belong to this subfamily would help provide a better understanding of the evolutionary history of class A β-lactamases.
According to its functional properties, CME-1 could be included in group 2e of the Bush-Jacoby-Medeiros classification scheme (10). In fact, CME-1 exhibits good catalytic efficiencies toward most cephalosporin substrates, including the expanded-spectrum cephalosporins (such as cefotaxime, ceftazidime, and cefepime), with kcat values in the range of 25 to 100 s−1 and kcat/Km ratios in the range of 105 to 106 M−1 · s−1. The enzyme is also able to hydrolyze penicillins and monobactams, although with lower efficiencies. Interestingly, with penicillins and monobactams both the kcat and the Km values are considerably lower than those observed with cephalosporins, eventually resulting in overall similar acylation efficiencies (kcat/Km ratios, 5 × 104 M−1 · s−1 for monobactams and in the range of 1 × 105 to 1 × 106 M−1 · s−1 for penicillins). Owing to these kinetic properties, the CME-1 enzyme appears to be an interesting model for further investigation of (i) the structure-function relationships of extended-spectrum class A β-lactamases and (ii) the correspondence between kinetic parameters and the impact of enzyme production on microbial susceptibility to various β-lactams.
The overall functional behavior of CME-1 resembles those of the Bacteroides cephalosporinases (32, 36, 42) and the VEB-1 (34) and PER β-lactamases (6, 29). However, compared to PER-1 and VEB-1, which are the enzymes of this group for which some kinetic data are available (29, 34), CME-1 exhibits a more pronounced diversification of kinetic parameters toward cephalosporins, penicillins, and monobactams, suggesting the existence of functional heterogeneities among members of this lineage. A detailed evaluation of the kinetic parameters of the enzymes that belong to this subfamily would provide interesting comparative data. Although the kinetic parameters of CME-1 with nitrocefin were not affected by the phosphate ion concentration in the assay buffer, it might be interesting to further investigate whether the phosphate ion concentration differentially affects the kinetic parameters of CME-1 toward certain substrates, as reported for PER-1 (13).
The molecular size and overall functional properties of the CME-1 enzyme from C. meningosepticum CCUG 4310 appeared to be quite similar to those of a serine β-lactamase previously purified from a C. meningosepticum clinical isolate (isolate GN14059) (18). However, the isoelectric pH values of the two enzymes are strikingly different (>9.0 for CME-1 versus 5.1 reported for the enzyme from isolate GN14059 [18]). Since in extracts of CCUG 4310 we were unable to detect any band of β-lactamase activity that focused in the acidic pH range, the β-lactamase purified from GN14059 could be an acquired active-site serine enzyme structurally different from the resident CME-1 enzyme. It will be interesting to investigate this point further by analyzing β-lactamase production among several different C. meningosepticum isolates.
ACKNOWLEDGMENTS
This work was supported by grants 97.04260.CT04 and 98.00510.CT04 from the Italian National Research Council and by a grant from the Belgian Government (PAI P4/03).
We thank Tiziana di Maggio for technical support and Francesco Lissi and Elena Sestini for secretarial assistance.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc London Biol. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Ambler R P, Coulson A F W, Frère J-M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthélémy M, Péduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochem Biophys Acta. 1992;1122:15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- 5.Barthélémy M, Péduzzi J, Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1) Biochem J. 1988;251:73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauernfeind A, Stemplinger L, Jungwirth R, Mangold P, Amann S, Akalin E, Ang Ö, Bal C, Casellas J M. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob Agents Chemother. 1996;40:616–620. doi: 10.1128/aac.40.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloch K C, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine (Baltimore) 1997;76:30–41. doi: 10.1097/00005792-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Bootsma H J, van Dijk H, Verhoef J, Fleer A, Mooi F. Molecular characterization of the BRO β-lactamase of Moraxella (Branhamella) catarrhalis. Antimicrob Agents Chemother. 1996;40:966–972. doi: 10.1128/aac.40.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couture F, Lachapelle J, Levesque R C. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol. 1992;6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 13.Danels F, Hall L M C, Gur D, Akalin H E, Livermore D M. Transferable production of PER-1 β-lactamase in Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;35:281–294. doi: 10.1093/jac/35.2.281. [DOI] [PubMed] [Google Scholar]
- 14.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frère J-M, Waley S G. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and dd-peptidases. Biochem Pharmacol. 1987;36:2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fass R J, Barnishan J. In vitro susceptibility of nonfermentative gram-negative bacilli other than Pseudomonas aeruginosa to 32 antimicrobial agents. Rev Infect Dis. 1980;2:841–853. doi: 10.1093/clinids/2.6.841. [DOI] [PubMed] [Google Scholar]
- 17.Frère J M. Beta-lactamases and bacterial resistance to antibiotics. Mol Microbiol. 1995;16:385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 18.Fujii T, Sato K, Yokota E, Maejima T, Inoue M, Mitsuhashi S. Properties of a broad spectrum β-lactamase isolated from Flavobacterium meningosepticum GN14059. J Antibiot. 1988;41:81–85. doi: 10.7164/antibiotics.41.81. [DOI] [PubMed] [Google Scholar]
- 19.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:43–74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huovinen P, Jacoby G A. Sequence of the PSE-1 β-lactamase gene. Antimicrob Agents Chemother. 1991;35:2428–2430. doi: 10.1128/aac.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Livermore D M. β-lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massidda O, Rossolini G M, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matagne A, Lamotte-Brasseur J, Frère J-M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naas T, Nordmann P. Analysis of a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc Natl Acad Sci USA. 1994;91:7693–7697. doi: 10.1073/pnas.91.16.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naas T, Vandel L, Sougakoff W, Livermore D M, Nordmann P. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob Agents Chemother. 1994;38:1262–1270. doi: 10.1128/aac.38.6.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:962–969. doi: 10.1128/aac.37.5.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owen R J, Snell J J S. Deoxyribonucleic acid reassociation in the classification of flavobacteria. J Gen Microbiol. 1976;93:89–102. doi: 10.1099/00221287-93-1-89. [DOI] [PubMed] [Google Scholar]
- 32.Parker A C, Smith C J. Genetic and biochemical analysis of a novel Ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob Agents Chemother. 1993;37:1028–1036. doi: 10.1128/aac.37.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perilli M, Franceschini N, Segatore B, Amicosante G, Oratore A, Duez C, Joris B, Frère J-M. Cloning and nucleotide sequencing of the gene encoding the β-lactamase from Citrobacter diversus. FEMS Microbiol Lett. 1991;67:79–84. doi: 10.1016/0378-1097(91)90448-j. [DOI] [PubMed] [Google Scholar]
- 34.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynaud A, Péduzzi J, Barthélémy M, Labia R. Cefotaxime-hydrolysing activity of the β-lactamase of Klebsiella oxytoca D488 could be related to a threonine residue at position 140. FEMS Microbiol Lett. 1991;65:185–192. doi: 10.1016/0378-1097(91)90301-p. [DOI] [PubMed] [Google Scholar]
- 36.Rogers M B, Parker A C, Smith C J. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob Agents Chemother. 1993;37:2391–2400. doi: 10.1128/aac.37.11.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossolini G M, Franceschini N, Riccio M L, Mercuri P S, Perilli M, Galleni M, Frère J-M, Amicosante G. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem J. 1998;332:145–152. doi: 10.1042/bj3320145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Segel I H. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1976. pp. 236–241. [Google Scholar]
- 41.Seoane A, Garcia Lobo J M. Nucleotide sequence of a new class A β-lactamase gene from the chromosome of Yersinia enterocolitica: implications for the evolution of class A β-lactamases. Mol Gen Genet. 1991;228:215–220. doi: 10.1007/BF00282468. [DOI] [PubMed] [Google Scholar]
- 42.Smith C J, Bennett T K, Parker A C. Molecular and genetic analysis of the Bacteroides uniformis cephalosporinase gene cblA, encoding the species-specific β-lactamase. Antimicrob Agents Chemother. 1994;38:1711–1715. doi: 10.1128/aac.38.8.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trepanier S, Prince A, Huletsky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother. 1997;41:2399–2405. doi: 10.1128/aac.41.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tribuddharat C, Fennewald M A. EMBL/GenBank database entry no. AF078527. Hinxton, United Kingdom: EMBL European Bioinformatics Institute; 1998. [Google Scholar]
- 47.Vahaboglu H, Hall L M C, Mulazimoglu L, Dodanli S, Yildirim I, Livermore D M. Resistance to extended-spectrum cephalosporins, caused by PER-1 β-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J Med Microbiol. 1995;43:294–299. doi: 10.1099/00222615-43-4-294. [DOI] [PubMed] [Google Scholar]
- 48.Vahaboglu H, Ötztürk R, Aigün G, Coskunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamase among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh T R, MacGowan A P, Bennett P M. Sequence analysis and enzyme kinetics of the L2 serine β-lactamase from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1460–1464. doi: 10.1128/aac.41.7.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]