Significance
Functional plasticity of the nociceptive circuit is a remarkable feature and is of clinical relevance. As an example, nociceptors lower their threshold upon tissue injury, a process known as allodynia that would facilitate healing by guarding the injured areas. However, long-lasting hypersensitivity could lead to chronic pain, a debilitating disease not effectively treated. Therefore, it is crucial to dissect the mechanisms underlying basal nociception and nociceptive hypersensitivity. In both vertebrate and invertebrate species, conserved transient receptor potential (Trp) channels are the primary transducers of noxious stimuli. Here, we provide a precedent that in Drosophila larvae, heat sensing in the nociception and hypersensitivity states is mediated by distinct heat-sensitive neurons and TrpA1 alternative isoforms.
Keywords: nociception, nociceptive hypersensitivity, Drosophila, transient receptor potential (Trp), alternative splicing
Abstract
Acute nociception is essential for survival by warning organisms against potential dangers, whereas tissue injury results in a nociceptive hypersensitivity state that is closely associated with debilitating disease conditions, such as chronic pain. Transient receptor potential (Trp) ion channels expressed in nociceptors detect noxious thermal and chemical stimuli to initiate acute nociception. The existing hypersensitivity model suggests that under tissue injury and inflammation, the same Trp channels in nociceptors are sensitized through transcriptional and posttranslational modulation, leading to nociceptive hypersensitivity. Unexpectedly and different from this model, we find that in Drosophila larvae, acute heat nociception and tissue injury-induced hypersensitivity involve distinct cellular and molecular mechanisms. Specifically, TrpA1-D in peripheral sensory neurons mediates acute heat nociception, whereas TrpA1-C in a cluster of larval brain neurons transduces the heat stimulus under the allodynia state. As a result, interfering with synaptic transmission of these brain neurons or genetic targeting of TrpA1-C blocks heat allodynia but not acute heat nociception. TrpA1-C and TrpA1-D are two splicing variants of TrpA1 channels and are coexpressed in these brain neurons. We further show that Gq-phospholipase C signaling, downstream of the proalgesic neuropeptide Tachykinin, differentially modulates these two TrpA1 isoforms in the brain neurons by selectively sensitizing heat responses of TrpA1-C but not TrpA1-D. Together, our studies provide evidence that nociception and noncaptive sensitization could be mediated by distinct sensory neurons and molecular sensors.
As an ancient property of the nervous system, nociception is essential for survival by protecting organisms from potential dangers, such as high-intensity thermal and mechanical stimulation (1–4). Remarkably, tissue injury causes hypersensitivity of the nociceptive responses, manifested as lowered activation threshold (i.e., allodynia) and increased response magnitude to suprathreshold stimulation (i.e., hyperalgesia) (5–8). Although short-term hypersensitivity is beneficial by enhanced protection of the injured area during tissue healing, hypersensitivity may persist after healing. This prolonged hypersensitivity no longer serves protective functions but is rather associated with debilitating diseases, including chronic pain. Thus, acute nociception and hypersensitivity reflect two different functional states of the nociceptive circuit and elucidating their differences is likely to shed light on the mechanisms of chronic pain.
Drosophila larvae have emerged as an important genetically tractable model for studying acute nociception and nociceptive hypersensitivity (9–12). Despite a simpler nervous system, larvae exhibit robust nocifensive withdrawal behavior to noxious heat, mechanical and chemical stimulation (12–15). Moreover, a well-controlled and robust tissue injury model has been established (9). In this model, brief exposure of larvae to UVC light elicits injury and apoptosis of epidermal cells, resulting in behavioral hypersensitivity to heat stimulation, including allodynia and hyperalgesia. Genetic studies have identified several proalgesic factors, including Tachykinin, tumor necrosis factor-α, and Hedgehog in regulation of nociceptive hypersensitivity (9–11). Importantly, these proalgesic agents also contribute to pain hypersensitivity in mammals (10, 16), suggesting the signaling mechanisms driving transition from acute nociception to nociceptive hypersensitivity are conserved.
From arthropods to mammals, transient receptor potential (Trp) ion channel family members are the primary molecular detectors of thermal and chemical stimuli that activate nociceptors to produce acute nociception (17–19). In addition to their essential roles in nociception, genetic and pharmacological studies have implicated Trp channels, most notably TrpV1 and TrpA1, as critical regulators of nociceptive hypersensitivity (7). For example, through transcriptional and posttranslational modulation, proalgesic agents and inflammatory factors released after tissue injury sensitize TrpV1 and TrpA1 responses to noxious stimuli, resulting in hypersensitivity of cellular and behavioral responses (7, 20). These findings have stimulated enormous interest in developing new analgesics by antagonizing Trp channel activity (18). However, as a foreseeable adverse effect of this strategy, not only hypersensitivity but also nociception will be affected. Indeed, animal and clinical studies indicate that treatment with TrpV1 antagonists impairs heat nociception, thereby causing unwanted burning injury; moreover, it produces hyperthemia due to a critical role of TrpV1 in controlling core body temperature (21–23). Hence, it would be important to develop strategies that can block Trp channel sensitization while leaving intact their beneficial functions in acute nociception and homeostatic regulation (24, 25).
TrpA1 is activated by environmental irritant chemicals and proalgesic agents (26). Strikingly, its chemosensory functions are conserved across 500 million y of animal evolution (3). In addition, heat and related infrared sensing by TrpA1 have been discovered in insects, such as Drosophila and mosquitoes (27, 28), as well as frogs (29, 30) and snakes (31), although TrpA1 thermal sensitivity in mammals remains controversial (26, 32–34). In Drosophila, alternative splicing produces five TrpA1 isoforms that differ in the N-terminal cap, as well as the linker domain connecting ankyrin repeats to the transmembrane domain (28, 35). Previous reports indicate that Drosophila TrpA1 isoforms are functional different (27, 35). To further characterize in vivo expression and functions of individual TrpA1 isoforms, we have modified the Drosophila TrpA1 genomic locus to generate knockin (KI) flies that only express a single isoform, and knockout (KO) flies that lack select isoforms (28). We find that TrpA1-C and TrpA1-D are two functional isoforms expressed in class 4 dendritic arborization (C4da) neurons (28), the primary nociceptors in the larval peripheral nervous system (PNS) (13). Ca2+ imaging and behavioral analyses indicate that acute heat nociception is primarily mediated by a single isoform TrpA1-D in C4da neurons, with negligible contribution from TrpA1-C (28). However, TrpA1-C bears intrinsic heat sensitivity as heat activates TrpA1-C in heterologous cells (28). Therefore, while TrpA1-D mediates heat nociception, TrpA1-C is normally a dormant heat sensor in vivo.
Inspired by the diverse functions of TrpA1 isoforms in acute heat nociception, herein, we aim to determine how individual TrpA1 isoforms contribute to heat allodynia after tissue injury. Contrary to our expectation that heat allodynia results from sensitization of TrpA1-D in C4da neurons, we find that heat allodynia involves TrpA1-C and a cluster of heat-sensing neurons in larval brain. Heat responses of these brain neurons are sensitized after tissue injury and blocking their synaptic transmission impairs behavioral allodynia. We further show that Gq-phospholipase C (PLC) specifically sensitizes heat responses of TrpA1-C but not TrpA1-D in larval brain neurons. Taken together, our studies provide evidence that acute nociception and hypersensitivity could exploit distinct neurons and molecular sensors.
Results
Heat Is Transduced by Distinct TrpA1 Isoforms in Nociception and Allodynia States.
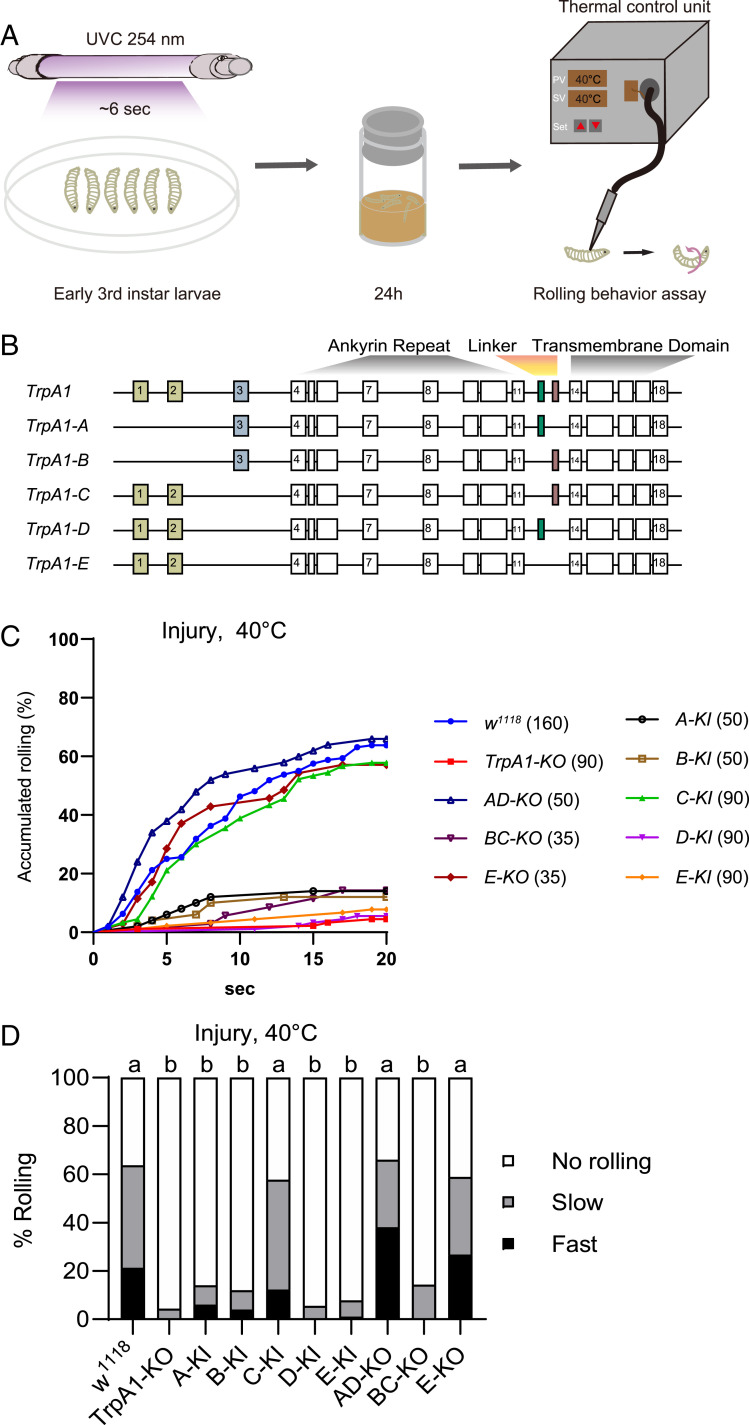
By stimulating a third-instar Drosophila larva using a custom-built heat probe with constant set-point temperature, we and others have shown that larvae exhibit nocifensive rolling behavior only when the probe temperature is set at 42 °C or higher (9, 28). This acute nociception requires TrpA1 activity in larval C4da neurons (28, 35). To induce thermal hypersensitivity, we adopted an established tissue injury paradigm by brief exposure of larvae to UVC (Fig. 1A), which elicited epidermal cell apoptosis, as previously reported (SI Appendix, Fig. S1I) (9). We found that ∼60% of UVC but not mock-treated larvae rolled in response to otherwise subthreshold stimulation of 40 °C, starting at ∼4 h after UVC treatment (SI Appendix, Fig. S1 C and D). Moreover, UVC-treated larvae responded to subthreshold temperature as low as 32 °C (SI Appendix, Fig. S1 E and F). These results indicate that UVC elicits heat allodynia in larvae, with a magnitude of behavioral hypersensitivity consistent with a previous report (9). To investigate the heat transduction mechanisms underlying allodynia, we assessed the role of TrpA1, which has been implicated in heat hypersensitivity in larvae (10). We found that heat allodynia was almost completely abolished in TrpA1-KO larvae (Fig. 1 C and D), an allele we have generated by removing the entire TrpA1 genomic region, including exons and introns (28). These data indicate that TrpA1 is responsible for transducing heat stimuli in the allodynia state.
Fig. 1.
TrpA1-C controls heat allodynia. (A) Schematic diagram of heat allodynia of Drosophila larvae. The nocifensive rolling behavioral assay was performed on third-instar larvae, at 24 h after brief exposure to UVC light (UVC, ∼6 s, 20 mJ/cm2). (B) Diagram of TrpA1 alternative splicing, with alternative exons marked in colors. (C) Accumulated percentage of larvae showing rolling behavioral responses to a 40 °C-heat probe at 24 h after UVC treatment. Larvae with knockout (KO) or knockin (KI) of select TrpA1 isoforms were analyzed, including TrpA1-KO, TrpA1-AD-KO (AD-KO), TrpA1-BC-KO (BC-KO), TrpA1-E-KO (E-KO), TrpA1-A-KI (A-KI), TrpA1-B-KI (B-KI), TrpA1-C-KI (C-KI), TrpA1-D-KI (D-KI), TrpA1-E-KI (E-KI). The sample numbers are indicated with brackets. (D) Rolling responses in C are categorized into three groups: no rolling (white bar, no rolling recorded in less than 20 s), slow rolling (gray bar, rolling occurred after 5 s but within 20 s), and fast rolling (black bar, rolling occurred in no longer than 5 s). This category scheme was applied to all heat probe behavioral assay in this study unless noted otherwise. Log-rank tests, corrected by total comparing group numbers, were performed to compare datasets displayed in accumulated response curves in C. For clarity, results of statistical analysis are instead marked in the accompanying bar graph in D. Columns with different superscripts (a and b) are significantly different from each other (P < 0.05).
Alternative splicing generates five TrpA1 isoforms (Fig. 1B). To dissect their functions in vivo, we have previously generated an extensive collection of TrpA1 isoform-specific alleles, including knockin flies each expressing a single isoform (e.g., TrpA1-C-KI, in which TrpA1-C is intact but TrpA1-A/B/D/E isoforms are depleted), as well as knockout flies lacking select isoforms (e.g., TrpA1-BC-KO allele depletes TrpA1-B and -C but expresses TrpA1-A/D/E isoforms; see SI Appendix, Fig. S1A for details) (28). We have previously shown that a single isoform, TrpA1-D, expressed in C4da neurons is necessary and sufficient for acute heat nociception, whereas the other four isoforms including TrpA1-C, which is coexpressed in C4da neurons, are not required (28). Based on the prevailing hypersensitivity model, we hypothesized that heat allodynia would involve sensitization of TrpA1-D in C4da neurons, whereas TrpA1-A/B/C/E isoforms are not involved. To test this hypothesis, we assessed how TrpA1 isoform alleles respond to subthreshold stimulation of 40 °C after UVC treatment. We found that whenever TrpA1-C is depleted (e.g., in TrpA1-BC-KO and in TrpA1-A/B/D/E-KI larvae), heat allodynia was severely impaired with extent indistinguishable from that of TrpA1-KO (Fig. 1 C and D). Conversely, heat allodynia was normal and indistinguishable from that of wild-type larvae whenever TrpA1-C is expressed (e.g., in TrpA1-C-KI, TrpA1-AD-KO, and TrpA1-E-KO larvae) (Fig. 1 C and D). We further showed that UVC-treated TrpA1-C-KI larvae responded to subthreshold temperature as low as 32 °C (SI Appendix, Fig. S1 G and H), similar to wild-type (SI Appendix, Fig. S1 E and F). These results are unexpected, as they suggest that a single isoform TrpA1-C appears to mediate heat allodynia, whereas other TrpA1 isoforms including TrpA1-D contribute minimally.
Together with our published results (28), these data further demonstrate that heat nociception and allodynia exploit different transduction mechanisms; a single isoform, TrpA1-D, is the heat transducer for acute nociception, whereas another isoform, TrpA1-C, transduces heat stimuli in the allodynia state. As a result, TrpA1-D depletion abolishes acute heat nociception but not allodynia; conversely, TrpA1-C depletion abolishes heat allodynia without affecting nociception. Because in the current behavioral paradigm heat nociception and allodynia involve TrpA1-D and TrpA1-C but no other TrpA1 isoforms, we will focus on these two isoforms in the rest of report.
Nociception and Allodynia Require TrpA1 Activity in the PNS and Central Nervous System, Respectively.
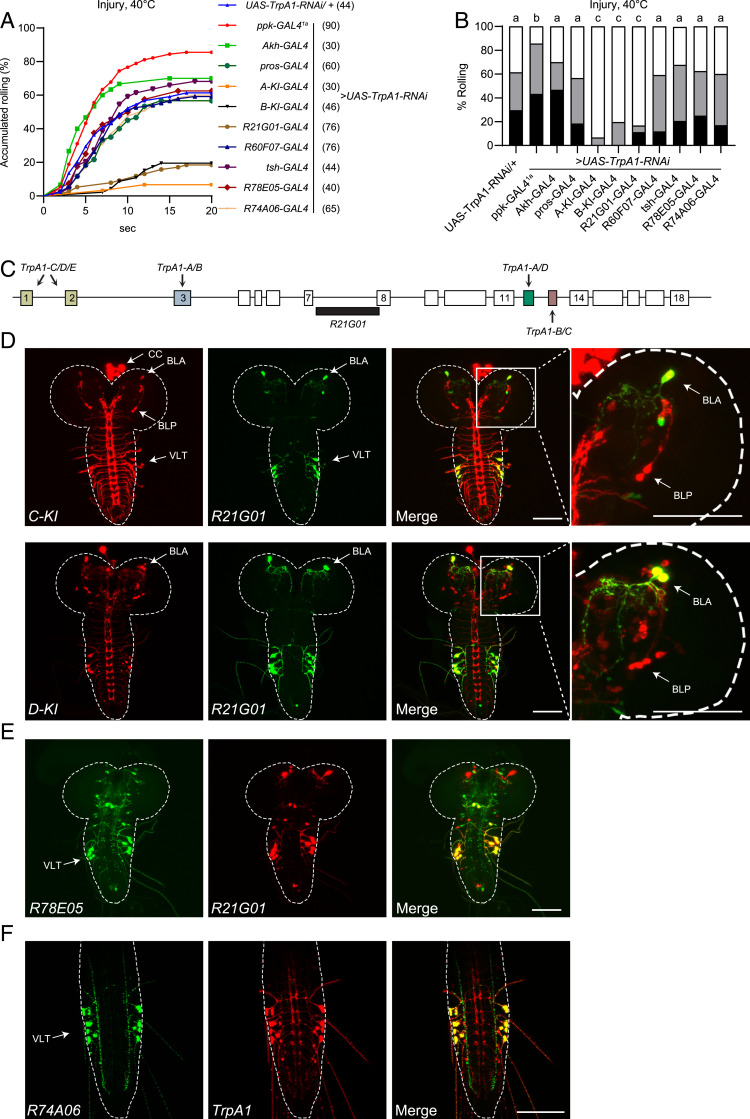
Examining the TrpA1-C-KI-T2A-GAL4 knockin flies that report endogenous TrpA1-C expression, we have shown TrpA1-C expression in multiple larval tissues, including the central nervous system (CNS), corpus cardiacum (CC) cells, the gut, and C4da neurons in the PNS (28). The genomic organization of TrpA1 does not allow specific knockdown of TrpA1-C by RNA interference (RNAi) without affecting other isoforms. Therefore, to determine the cellular loci of TrpA1 action in heat allodynia, we expressed the RNAi construct that has been shown to effectively knock down all TrpA1 isoforms by targeting the common region (28, 36). By tissue-specific knockdown of TrpA1 expression, we have previously shown that heat nociception requires TrpA1 in C4da neurons in the PNS, but not in the CNS (28). Here, we exploited the same approach to determine where TrpA1 is required for heat allodynia, by expressing UAS-TrpA1-RNAi under control of GAL4 drivers that overlapped with TrpA1-C expression. Unexpected and contrary to our initial prediction, we found heat allodynia was unaffected when TrpA1 was knocked down by the C4da-neuron–specific driver ppk-GAL41a (Fig. 2 A and B; see also SI Appendix, Fig. S5F for ppk-GAL41a expression) (37). Similarly, TrpA1 knockdown under control of ppk1.9-GAL4, another C4da neuron driver (9–11, 38), did not affect heat allodynia (SI Appendix, Fig. S5 D and E; see also SI Appendix, Fig. S5F for ppk1.9-Gal4 expression). The lack of effect was not due to RNAi efficacy as larvae bearing ppk-GAL41a and UAS-TrpA1-RNAi exhibit drastically reduced acute nociceptive responses to a 44 °C probe (28). These data suggest that heat allodynia does not require TrpA1 activity in C4da neurons.
Fig. 2.
TrpA1 in larval brain BLA neurons is required for heat allodynia. (A and B) Behavioral responses to a 40 °C heat probe of larvae in which UAS-TrpA1-RNAi was expressed in various tissues under control of different GAL4 drivers. A copy of UAS-Dcr2 was included to increase RNAi efficiency. Behavioral assay was performed 24 h after UVC treatment. The sample numbers are indicated with brackets in A. Log-rank tests, corrected by total comparing group numbers, were performed to compare dataset displayed in accumulated response curves in A. Results of statistical analysis are marked in the bar graphs in B for clarity, and columns with different superscripts (a, b, and c) are significantly different from each other (P < 0.05). (C) Diagram of the cis-regulatory element R21G01 (black bar), located at the TrpA1 intron between exons 7 and 8. Arrows indicate alternative exons. (D) Expression of TrpA1-C, TrpA1-D, and R21G01 in larval CNS in transheterozygotes bearing TrpA1-(C or -D)-KI-T2A-lexA > lexAop2-6XmCherry and R21G01-GAL4 > UAS-6XGFP. BLA, BLP, VLT, and CC cells, are indicated by arrows. Enlarged view of a brain lobe is shown in the right-most panels. (E) R78E05 and R21G01 expression in larval CNS in transheterozygotes bearing R78E05-GAL4 > UAS-6XGFP and R21G01-lexA > lexAop2-6XmCherry. (F) R74A06 and TrpA1 expression in larval ventral nerve cord in transheterozygotes bearing R74A06-GAL4 > UAS-6XGFP and TrpA1-T2A-lexA > lexAop2-6XmCherry. (Scale bars in D–F, 100 μm.)
Next, we knocked down TrpA1 with TrpA1-B-KI-T2A-GAL4, a driver that reports endogenous expression of the TrpA1-B isoform (28). Notably, larvae express TrpA1-C and TrpA1-B in identical cells, with the exception that TrpA1-C but not TrpA1-B is expressed in C4da neurons (28). As a result, TrpA1-B-KI-T2A-GAL4 marks all TrpA1-C+ cells except for C4da neurons (28). We found that heat allodynia was severely impaired when TrpA1 was knocked down under control of TrpA1-B-KI-T2A-GAL4 (Fig. 2 A and B), indicating that heat allodynia requires TrpA1 in cells other than C4da neurons. We substantiated this conclusion by showing that TrpA1 knockdown by TrpA1-A-KI-T2A-GAL4, which marks all TrpA1+ cells except for C4da neurons (28), significantly impaired heat allodynia (Fig. 2 A and B). To determine whether allodynia requires TrpA1 in nonneuronal tissues, we knocked down TrpA1 in CC cells by Akh-GAL4 (39), or in the gut by pros-GAL4 (40, 41), and found that heat allodynia was not affected (Fig. 2 A and B). These data suggest that TrpA1 activity in C4da neurons, CC cells, or the gut does not play a major role in heat allodynia, further raising a possibility that heat allodynia involves TrpA1 activity in larval CNS. By crossing TrpA1-C-KI-T2A-GAL4 with a reporter line, we showed TrpA1-C expression in three anatomically distinct neuronal clusters in larval CNS (Fig. 2D). They included two clusters in larval brain: that is, BLP (brain lateral posterior) neurons and BLA (brain lateral anterior) neurons, and a third cluster VLT (VNC lateral and TrpA1+) neurons in the ventral nerve cord (VNC) (28, 42).
Consistent with our previous observation that TrpA1-C is expressed in a subgroup of TrpA1-D+ neurons (28), our analysis of the TrpA1-D-KI-T2A-GAL4 allele revealed TrpA1-D expression in BLA, BLP, VLT, and other neurons in larval CNS (Fig. 2D). Therefore, TrpA1-C and TrpA1-D isoforms are coexpressed in BLA, BLP, and VLT neurons. In an effort to identify drivers that can distinguish BLA, BLP, and VLT neurons, we found R21G01-GAL4, driven by the TrpA1 cis-regulatory sequence (Fig. 2C), was specifically expressed in BLA and VLT, but not in BLP or other PNS neurons (Fig. 2D and SI Appendix, Fig. S1K). Moreover, TrpA1 knockdown by R21G01-GAL4 severely impaired heat allodynia (Fig. 2 A and B). To assess the role of VLT neurons, we knocked down TrpA1 with the pan-VNC driver tsh-GAL4 (43), or with two additional drivers R78E05-GAL4 and R74A06-GAL4 that were expressed in VLT but not BLA neurons (Fig. 2 E and F), and found that heat allodynia was unaffected (Fig. 2 A and B). Therefore, TrpA1 activity in BLA but not VLT neurons appears to be critical for heat allodynia. Finally, we showed that TrpA1 activity in BLP was not required for heat allodynia, as knocking down TrpA1 under control of R60F07-GAL4, a driver labeling BLP but not BLA or VLT neurons (42), did not affect heat allodynia (Fig. 2 A and B).
In summary, these cell-specific RNAi analyses suggest that heat allodynia requires TrpA1 activity in BLA neurons in larval brain. Altogether, our findings support the notion that heat nociception and allodynia involve distinct neurons and molecular sensors; nociception is mediated by TrpA1-D in PNS C4da neurons, whereas in the allodynia state heat is transduced by TrpA1-C in brain BLA neurons.
BLA Neurons Control Heat Allodynia and Are Sensitized after Epidermal Injury.
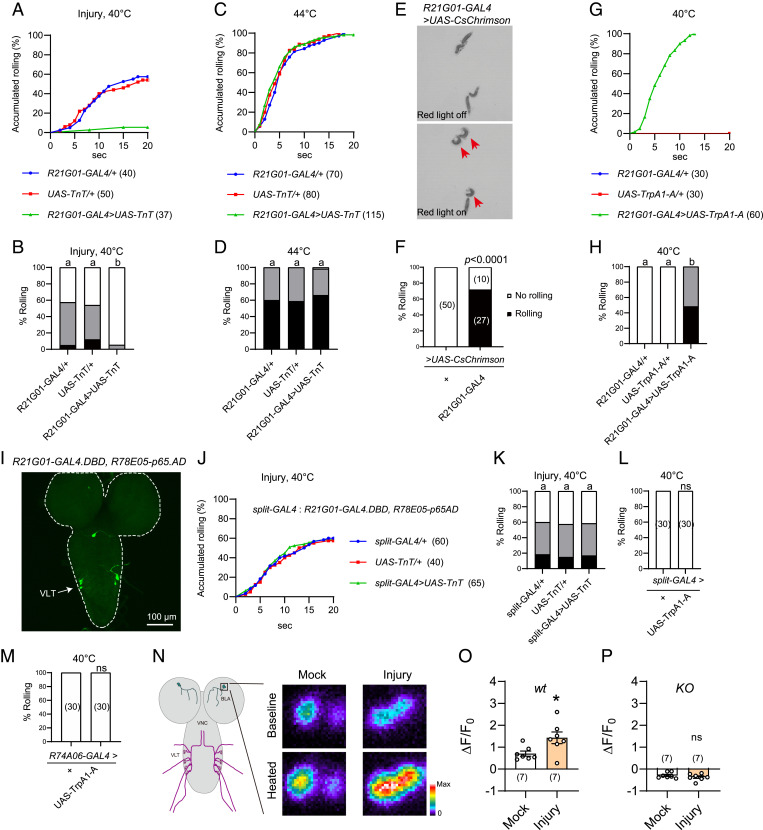
Next, we expressed tetanus toxin (TnT), a neurotoxin that blocks synaptic transmission by cleaving synaptobrevin (44), under R21G01-GAL4 control. We found that TnT expression almost completely abolished heat allodynia (Fig. 3 A and B). In contrast, heat nociception was unaffected as these TnT expressing larvae responded normally to suprathreshold stimulation of 44 °C (Fig. 3 C and D). These results indicate that larval CNS neurons marked by R21G01-GAL4 are specifically required for heat allodynia. Next, we activated these CNS neurons by expressing the light-sensitive CsChrimson under R21G01-GAL4 control and stimulating larvae with 620-nm red light (45). We found light stimulation elicited rolling of over 70% larvae (Fig. 3 E and F, and Movies S1 and S2). To substantiate this finding, we activated neurons by another approach. Specifically, we expressed heat-sensitive TrpA1-A (i.e., the A isoform of TrpA1), which is activated by warm temperature as low as ∼29 °C (46), under R21G01-GAL4 control. To ensure robust neuronal activation, we stimulated larvae with a 40 °C probe and found that 100% larvae rolled (Fig. 3 G and H). In contrast, control larvae bearing R21G01-GAL4, or UAS-TrpA1-A alone failed to roll (Fig. 3 G and H). These optogenetics and thermogenetics results together indicated that activation of R21G01-GAL4+ CNS neurons is sufficient to elicit rolling behavior. R21G01-GAL4 was expressed in BLA and VLT neurons (Fig. 2D); we therefore attempted to determine behavioral outcome after perturbation of VLT neurons. There is no available GAL4 driver that specifically labels VLT neurons. As such, we exploited the split-GAL4 system, in which the DNA binding domain (GAL4.DBD) and the activation domain (p65AD) of GAL4 are expressed in distinct cell groups, allowing functional reconstitution of GAL4 activity only in cells that express both GAL4.DBD and p65AD (47, 48). Intriguingly, we found combining R21G01-GAL4.DBD and R78E05-p65AD reconstituted GAL4 activity specifically in VLT neurons (Fig. 3I). We further showed that expression of TnT under split-GAL4 control did not affect heat allodynia (Fig. 3 J and K). Moreover, thermogenetic activation of VLT neurons by split-GAL4 or R74A06-GAL4 failed to induce rolling (Fig. 3 L and M). Taken together, our results suggest that BLA but not VLT neurons are important for heat allodynia.
Fig. 3.
BLA neurons mediate heat responses in the allodynia state. (A and B) Behavioral responses of larvae bearing R21G01-GAL4 > UAS-TnT to a 40 °C heat probe, at 24 h after UVC treatment. (C and D) Behavioral responses of larvae bearing R21G01-GAL4 > UAS-TnT to a 44 °C heat probe, without UVC treatment. (E and F) Optogenetics-induced rolling behavior of larvae bearing R21G01-GAL4 > UAS-CsChrimson. 2.1 × 2.1 cm2 test fields are shown. Larvae displaying rolling are indicated with arrows in E. Here, behavioral responses are categorized into two groups, no rolling (white bar, if no rolling was observed during 20 s of light stimulation), and rolling (black bar, rolling occurred in 20 s). (G and H) Rolling behavior of larvae bearing R21G01-GAL4 > UAS-TrpA1-A, in response to a 40 °C-heat probe. (I) VLT neurons in larval ventral nerve cord were specifically labeled by the split-GAL4 (R21G01-GAL4.DBD, R78E05-p65AD). (J and K) Rolling behavior of larvae bearing split-GAL4 > UAS-TnT to a 40 °C-heat probe, at 24 h after UVC. (L and M) Thermogenetics stimulation of larvae bearing split-GAL4 > UAS-TrpA1-A (L) or R74A06-GAL4 > UAS-TrpA1-A (M) by a 40 °C heat probe did not induce rolling behavior. (N) Representative heat responses of BLA neurons from larvae bearing R21G01-GAL4 > UAS-GCaMP6s, at 24 h after mock or UVC treatment. A diagram for BLA and VLT neurons in larval CNS, labeled by R21G01-GAL4 is displayed, Left. Squared area (25 μm × 25 μm) including the cell body of BLA neurons is enlarged (Right) to show somatic heat responses. Dissected larval brains were stimulated using a program-controlled heat block. (O and P) Summary results of peak heat responses of BLA neurons from wild-type (O) and TrpA1-KO (P) larvae. Larvae were mock or UVC-treated 24 h before imaging. Genotypes used for GCaMP imaging are: wt (w; UAS-GCaMP6s/+; R21G01-GAL4/+). KO (w; UAS-GCaMP6s/+; R21G01-GAL4, TrpA1-KO/TrpA1-KO). For A–D, G and H, and J–M, the sample numbers are indicated with brackets. Log-rank tests were performed and corrected by total comparing group numbers. Results of statistical analysis are marked in the corresponding bar graphs for clarity, and columns with different superscripts (a and b) are significantly different from each other (P < 0.05). ns, no significant difference (P > 0.05). For F, Fisher’s exact test was performed. For O and P, unpaired Student’s t tests were performed. *P < 0.05, ns, no significant difference.
To measure heat responses of BLA neurons, we performed GCaMP imaging in dissected larval brain. We found that BLA neurons from control larvae were activated by heat, a response that was abolished in TrpA1-KO larvae (Fig. 3 N–P and SI Appendix, Fig. S2 A and B). Next, we bath-applied the Na+ channel blocker tetrodotoxin (TTX). TTX blocks action potentials in Drosophila brain (49); thus, it should block synaptic inputs to BLA neurons. We found TTX did not alter heat responses of BLA neurons (SI Appendix, Fig. S2C), suggesting that BLA are intrinsically heat-sensitive neurons that detect heat through TrpA1. Furthermore, heat responses of BLA neurons were potentiated after UVC treatment, and this potentiation was not observed in TrpA1-KO larvae (Fig. 3 N–P). Together, our results suggest that UVC sensitizes TrpA1-mediated heat responses of BLA neurons, leading to allodynia.
Gq-PLC Sensitizes Heat Responses of TrpA1-C but Not TrpA1-D.
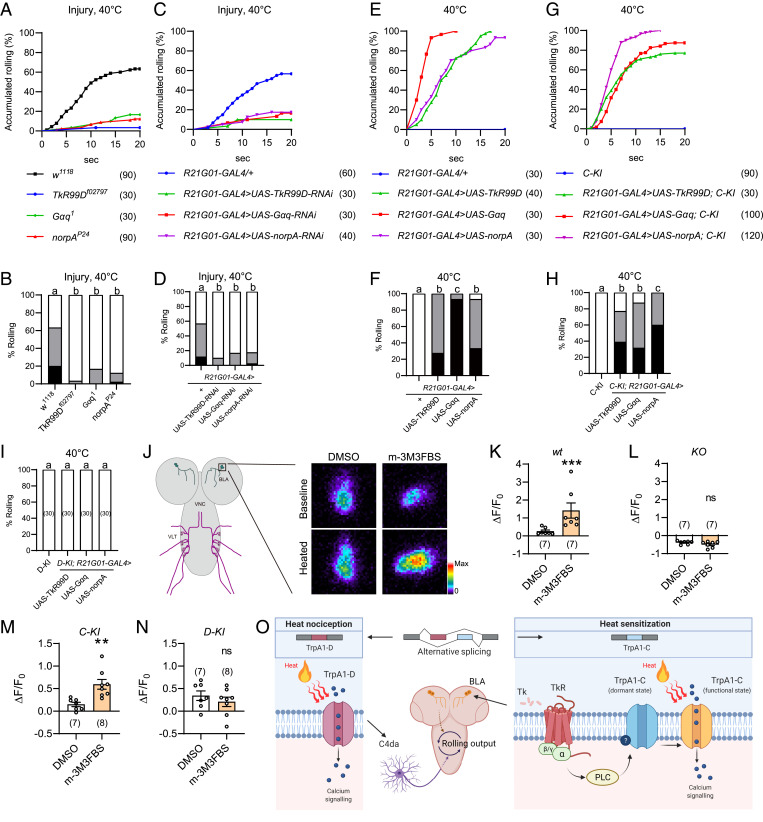
A previous study has shown the sensitization agent Tachykinin, a neuropeptide expressed in larval brain, critically controls heat allodynia of Drosophila larvae (11). Consistently, we found that heat allodynia was severely impaired in larvae bearing the null allele of TkR99D (Tachykinin receptor 99D) (Fig. 4 A and B). TkR99D is a G protein-coupled receptor linked to Gq-PLC signaling (11); we therefore assessed the role of Gq and PLC and found that heat allodynia was severely impaired in larvae bearing the null allele of Gαq (Gα49B) and the hypomorphic allele of PLCβ (norpA) (Fig. 4 A and B). Furthermore, heat allodynia was markedly reduced after RNAi knockdown of TkR99D, Gαq, or norpA under control of R21G01-GAL4 (Fig. 4 C and D). In contrast, TkR99D knockdown in C4da neurons under control of either ppk-GAL41a or another C4da-neuron-specific driver ppk-GAL4vk37 did not affect heat allodynia (SI Appendix, Fig. S5 A and B; see SI Appendix, Fig. S5F for ppk-GAL4vk37 expression) (37). These data indicate that heat allodynia requires TkR99D in larval brain neurons, but not C4da neurons. Because BLA but not VLT neurons were critically involved in heat allodynia, these results further suggest that heat allodynia requires TkR99D-Gq-PLC signaling in BLA neurons. Previous studies in Drosophila have shown that activity of TkR99D, Gαq, and PLC can be increased by overexpression (11, 50–54). Here, we found overexpression of TkR99D, Gαq, or norpA under control of R21G01-GAL4 was sufficient to induce heat allodynia (Fig. 4 E and F). Together, these loss-of-function and gain-of-function analyses suggest that TkR99D-Gq-PLC signaling in BLA neurons critically controls heat allodynia.
Fig. 4.
Gq-PLC signaling modulates heat responses of TrpA1-C but not TrpA1-D. (A and B) Injury-induced heat allodynia was impaired in larvae bearing genetic mutation of TkR, Gq, or PLC. Behavioral assay was performed 24 h after UVC treatment. (C and D) Injury-induced heat allodynia was impaired in larvae in which TkR, Gq, or PLC was knocked down in BLA neurons. Behavioral assay was performed 24 h after UVC treatment. (E and F) Heat allodynia was induced in wild-type larvae in which TkR, Gq or PLC was overexpressed in BLA neurons, in the absence of tissue injury. (G and H) Heat allodynia was induced in TrpA1-C-KI larvae in which TkR, Gq, or PLC was overexpressed in BLA neurons, in the absence of tissue injury. (I) No heat allodynia was induced in TrpA1-D-KI larvae in which TkR, Gq, or PLC was overexpressed in BLA neurons, in the absence of tissue injury. (J) Representative somatic heat responses of wild-type BLA neurons, in the presence of DMSO or the PLC agonist m-3M3FBS (20 μM). Larvae bearing R21G01-GAL4 > UAS-GCaMP6s were used, in the absence of tissue injury. A diagram for BLA and VLT neurons in CNS is displayed (Left), and BLA cell body is enlarged (Right) to show somatic heat responses. (K–N) Summary results of peak heat responses of BLA neurons from wild-type (K), TrpA1-KO (L), TrpA1-C-KI (M), and TrpA1-D-KI (N) larvae. Their genotypes are: wt (w; UAS-GCaMP6s/+; R21G01-GAL4/+), KO (w; UAS-GCaMP6s/+; R21G01-GAL4, TrpA1-KO/TrpA1-KO), C-KI (w; R21G01-lexA/lexAop2-myr-GCaMP6s; TrpA1-C-KI/TrpA1-C-KI), and D-KI (w; R21G01-lexA/lexAop2-myr-GCaMP6s; TrpA1-D-KI/TrpA1-D-KI). (O) A working model showing two distinct transduction mechanisms regulate acute heat nociception and allodynia. Nociception is mediated by TrpA1-D in peripheral C4da neurons, while allodynia by tissue injury is mediated by TrpA1-C in brain BLA neurons. Tachykinin receptor/G protein/PLC cascades specifically sensitize TrpA1-C by switching it from a dormant to a functional state. For A to I, the sample numbers are indicated with brackets. Log-rank test was performed and corrected by total comparing group numbers. Results of statistical analysis are marked in the corresponding bar graphs, and columns with different superscripts (a, b, and c) are significantly different from each other (P < 0.05). ns, no significant difference (P > 0.05). For K, Mann–Whitney U test was performed. ***P < 0.001. For L–N, unpaired Student’s t tests were performed. *P < 0.05, **P < 0.01.
Next, we determined whether Gq-PLC activity could differentially regulate TrpA1-C versus TrpA1-D. We found that overexpression of TkR99D, Gαq, or norpA under control of R21G01-GAL4 was sufficient to elicit robust heat allodynia in TrpA1-C-KI larvae, with ∼80% larvae rolled at 40 °C (Fig. 4 G and H). In contrast, no heat allodynia was induced in TrpA1-D-KI larvae (Fig. 4I). These behavioral data suggest that Gq-PLC activation in BLA neurons sensitizes heat responses of TrpA1-C but not TrpA1-D. To substantiate this conclusion, we performed GCaMP imaging to determine how acute pharmacological activation of PLC regulates heat responses of BLA neurons. We found that brief treatment of dissected larval brains with the PLC agonist m-3M3FBS, which activates PLC in Drosophila (55), significantly potentiated heat responses of BLA neurons in wild-type larvae (Fig. 4 J and K and SI Appendix, Fig. S3). On the other hand, heat responses were abolished, and no potentiation was observed in TrpA1-KO larvae (Fig. 4L and SI Appendix, Fig. S3). These data indicate that PLC sensitizes heat responses of TrpA1 in BLA neurons. We further found that m-3M3FBS potentiated heat responses of BLA neurons in TrpA1-C-KI but not TrpA1-D-KI larvae (Fig. 4 M and N and SI Appendix, Fig. S3), providing cellular evidence that PLC sensitizes TrpA1-C but not TrpA1-D. Based on these behavioral and Ca2+ imaging results, we propose that activation of Gq-PLC in BLA neurons specifically sensitizes TrpA1-C but not TrpA1-D, leading to cellular and behavioral hypersensitivity.
Finally, we determined whether Gq-PLC could differentially modulate TrpA1-C and TrpA1-D in C4da neurons, which express TrpA1-C and TrpA1-D at comparable levels (28). Consistent with our previous report (28), we found that TrpA1-C-KI larvae exhibited severe defects of acute heat nociception as only ∼20% rolled in response to a 44 °C probe (SI Appendix, Fig. S4 A and B). However, the response ratio was increased to 100% after Gαq or norpA overexpression in C4da neurons under ppk-GAL4vk37 control (SI Appendix, Fig. S4 A and B). In contrast, Gαq or norpA overexpression did not further increase rolling responses of TrpA1-D-KI larvae (SI Appendix, Fig. S4 C and D). Together, these results strengthen the conclusion that Gq-PLC specifically sensitizes TrpA1-C but not TrpA1-D.
Discussion
Acute nociception and hypersensitivity reflect two functional states of the nociceptive circuit. Elucidating the differences between these two states is important as it will likely shed light on chronic pain mechanisms. Based on results from the current and prior studies (28), we propose a working model (Fig. 4O). In this model, acute heat nociception and heat allodynia in Drosophila larvae involve distinct transduction mechanisms, including different TrpA1 ion channel isoforms as heat transducers (i.e., TrpA1-D vs. TrpA1-C) and different heat-sensitive neurons (i.e., C4da neurons in the PNS vs. BLA neurons in the CNS). These conclusions are supported by the following observations. First, TrpA1-D mediates acute heat nociception (28), whereas TrpA1-C mediates heat allodynia (Fig. 1 C and D). Second, RNAi knockdown of TrpA1 in BLA neurons impairs heat allodynia (Fig. 2 A and B), but not heat nociception (28). On the other hand, TrpA1 knockdown in C4da neurons abolishes acute nociception (28), but has little effect on heat allodynia (Fig. 2 A and B and SI Appendix, Fig. S5 D and E). Third, perturbing signaling of the sensitization agent Tachykinin by knocking down TKR99D in BLA neurons impaired heat allodynia (Fig. 4 C and D). On the other hand, TKR99D knockdown in C4da neurons did not affect heat allodynia (SI Appendix, Fig. S5 A and B). Fourth, synaptic transmission of BLA neurons is required for heat allodynia (Fig. 3 A and B), but not for acute nociception (Fig. 3 C and D). Fifth, artificial activation of BLA neurons by optogenetics or thermogenetics is sufficient to elicit rolling behavior (Fig. 3 E–H). Sixth, heat responses of BLA neurons are potentiated after epidermal injury (Fig. 3 N–P). Therefore, multiple heat-sensitive neurons are distributed in larval PNS and CNS, to regulate nociception and allodynia. Such anatomical and molecular segregation suggests a possibility to counteract allodynia while leaving nociception intact, by targeting allodynia-specific neurons or molecular sensors.
A previous study has shown that RNAi knockdown of TKR99D by ppk1.9-GAL4 impairs heat allodynia, and TkR99D expression by ppk1.9-GAL4 rescues the allodynia defect of TkR99D mutants (11). These results suggest that Tachykinin acts on C4da neurons to elicit allodynia, a conclusion that appears inconsistent with our model (Fig. 4O). To investigate this discrepancy, we expressed UAS-TkR99D-RNAi under control of three different C4da neuron drivers: ppk-GAL41a and ppk-GAL4vk37 used in the present study (37), and ppk1.9-GAL4 used in the previous study (11). We found TkR99D knockdown by ppk1.9-GAL4 significantly reduced heat allodynia (SI Appendix, Fig. S5 A and B), consistent with the previous report (11), In contrast, TkR99D knockdown by ppk-GAL41a or ppk-GAL4vk37 had no effect on heat allodynia (SI Appendix, Fig. S5 A and B). These results raised possibilities that GAL4 drivers could have different strength in C4da neurons, or alternatively could differ in their expression patterns. To test these possibilities, we expressed GFP under control of ppk-GAL41a, ppk-GAL4vk37, or ppk1.9-GAL4. We found GFP intensity in C4da neurons was comparable between ppk1.9-GAL4 and ppk-GAL41a, arguing against driver strength differences (SI Appendix, Fig. S5C). We next examined the expression patterns. Although ppk-GAL41a and ppk-GAL4vk37 were barely detected in larval tissues other than C4da neurons, ppk1.9-GAL4 was expressed in ring gland, lymph gland, fat body and hemocytes (SI Appendix, Fig. S5F). Therefore, the discrepancy between the present study and the previous one might be caused by different GAL4 drivers used; the defective heat allodynia observed with ppk1.9-GAL4 could result from TkR99D knockdown in tissues other than C4da neurons.
To further determine the cellular loci for allodynia, we performed genetic rescue studies by expressing TkR99D in UVC-treated TkR99D mutant larvae, or by expressing TrpA1 in UVC-treated TrpA1 mutant larvae. We found the expression of TkR99D or TrpA1 in either BLA or C4da neurons was sufficient to rescue allodynia phenotype of the respective mutant larvae (SI Appendix, Fig. S6 A, B, E, and F). To assess whether the rescue effects could result from ectopic TkR99D or TrpA1 activity, we repeated the genetic rescue studies but this time in larvae not treated with UVC. We expect that if TKR99D and TrpA1 were restored to their endogenous expression levels in either BLA or C4da neurons, mutant larvae not treated with UVC should not exhibit allodynia. Contrary to our expectation, we found that TkR99D or TrpA1 expression in either BLA or C4da neurons was sufficient to induce rolling responses to 40 °C in the respective mutant larvae (SI Appendix, Fig. S6 C, D, G, and H). These results suggest the rescue effects we observed in UVC-treated mutant larvae could result from ectopic activity of TkR99D and TrpA1 in BLA or C4da neurons, likely due to Gal4-UAS mediated overexpression.
Activation of BLA or C4da neurons is sufficient to elicit rolling, prompting us assess whether they could act in the same neural pathway. To address this, we first determined whether BLA and C4da neurons could directly interact. We conducted GFP reconstitution across synaptic partners (GRASP) studies but found no reconstituted GFP signals (SI Appendix, Fig. S7 A and B). The Drosophila genome encodes no ATP receptors. To assess whether C4da neurons could activate BLA neurons, we expressed the ionotropic ATP receptor P2X2 in C4da neurons (56). We validated that ATP application strongly activated C4da neurons (SI Appendix, Fig. S7 C and D); however, this treatment failed to activate BLA neurons (SI Appendix, Fig. S7 E and F). Together, our results suggest that C4da and BLA neurons are two independent groups of heat-sensitive neurons (Fig. 4O). Several interneurons in the rolling circuit have been identified to act downstream of C4da neurons (57); it will be of interest to investigate whether rolling induced by BLA neurons is also mediated by the same set of interneurons.
Larvae are small, suggesting that heat applied to the body wall, as in the behavioral study, could alter brain temperature. To estimate temperature changes in brain, we inserted a tiny thermal probe inside the larval body into the brain lobe location. Touching a larva with a 40 °C heat probe elicited a rapid temperature rise with a plateau at ∼34 °C (SI Appendix, Fig. S1J). These results suggest that external heat could effectively stimulate larval brain neurons.
TrpA1-C exhibits higher sensitivity to reactive oxygen species (ROS) than other TrpA1 isoforms (28). As heat has been implicated in ROS production (58), one possible mechanism underlying heat allodynia would be sensitization of TrpA1-C by ROS. To determine whether ROS could play a role in heat allodynia, we expressed an ROS scavenger, catalase (59), in BLA neurons under R21G01-GAL4 control. We found catalase expression did not affect allodynia (SI Appendix, Fig. S8 A and B). Although the human catalase used here has been shown to be secreted from cells to counteract ROS in the extracellular space (59), it may not effectively reduce intracellular ROS levels. Therefore, a role of intracellular ROS in heat hypersensitivity cannot be completely ruled out.
The only difference between TrpA1-C and TrpA1-D is the linker connecting ankyrin repeats to transmembrane domains (Fig. 1B and SI Appendix, Fig. S1B). Our results suggest TrpA1-C and TrpA1-D differentially responded to Gq-PLC activity (Fig. 4 G–I, M, and N), suggesting the linker domains could differentially interact with Gq-PLC downstream effectors. One such effector could be phosphatidylinositol 4,5-bisphosphate (PIP2), the lipid in membrane inner leaflet that has been shown to interact with ion channels to regulate their functions (18, 20, 60). Indeed, PIP2 has also been proposed to inhibit TrpA1, as antibody sequestration of PIP2 potentiates human TrpA1 responses in heterologous systems (20). One known mechanism by which PIP2 interacts with ion channels is via electrostatic interactions between positively charged residues and negatively charged PIP2 (61). Based on the published TrpA1 structure (62), the linker domain is in close proximity to inner membrane leaflet thus could potentially interact with PIP2. When analyzing the linker domain, we found no obvious differences, such as phosphorylation sites, but noticed more positively charged Lysine and Arginine in TrpA1-C than TrpA1-D (seven in TrpA1-C vs. three in TrpA1-D) (SI Appendix, Fig. S1B), raising a possibility that PIP2 could differentially interacts with TrpA1-C and TrpA1-D through electrostatic interaction. Therefore, one hypothesis would be that PIP2 inhibits TrpA1-C in the normal condition, and this inhibition is released by sensitization agents such as Tachykinin through PLC-mediated PIP2 depletion, leading to TrpA1-C sensitization (Fig. 4 G, H, and M). On the other hand, PIP2 does not appear to inhibit TrpA1-D as PLC activation did not alter TrpA1-D heat responses (Fig. 4 I and N). This hypothesis predicts that increasing the ratio of TrpA1-C over PIP2 would sensitize TrpA1-C. Indeed, we have shown that although TrpA1-C is normally heat-insensitive in C4da neurons in vivo, overexpression of TrpA1-C in C4da neurons renders robust heat responses of TrpA1-C (28), consistent with the notion that increasing TrpA1-C levels allows escaping from PIP2 inhibition.
Alternative splicing is enriched in the nervous system and is thought to expand the functional genome capacity (63, 64). By revealing TrpA1-D and TrpA1-C as heat transducers exclusively for nociception and allodynia, respectively, our study suggested alternative splicing as a previously underappreciated mechanism that regulates nociception and hypersensitivity. Although splicing variants of mammalian TrpA1 with different linker domains, like those of Drosophila TrpA1-C and -D, has not been identified, other splicing variants of human and mouse TrpA1 have been identified and many of these variants exhibit functional diversity in vitro (65, 66). It will be of interest to determine expression of TrpA1 splicing variants and their contribution to mammalian nociception and hypersensitivity. In summary, our study highlights the importance of investigating distinct in vivo functions of alternative isoforms of sensory ion channels to advance our understandings of nociception and hypersensitivity.
Finally, UVC was used in the present study as an experimental paradigm to induce tissue injury and heat allodynia. It is worth noting that UVC is an unnatural stimulus as it does not reach the surface of earth. On the other hand, UVA is natural and can cause skin damage to humans. It will be interesting to know whether exposure of larvae to UVA could elicit heat hypersensitivity by employing the similar cellular and transduction mechanisms described here.
Materials and Methods
Fly Stocks.
Flies were raised in cornmeal/yeast-based food and maintained in incubators with the temperature set at 25 °C and humidity set at 60% in a 12:12-h light:dark cycle. The fly stocks were used as follows. w1118, TrpA1 isoform knockout and knockin stocks (28), isoform-T2A-GAL4/lexA drivers (28), UAS-TrpA1-A (28), 386Y-GAL4 (BL#25410), R21G01-GAL4 (BL#48951), R21G01-lexA (BL#61521), pros-GAL4 (a gift from Wei Zhang, Tsinghua University, Beijing, China), tsh-GAL4 (a gift from Yulong Li, Peking University School of Life Sciences, Beijing, China), Akh-GAL4 (a gift from Rebecca Yang, Duke University Medical School, Durham, NC), UAS-TnT (BL#28837), R60F07-GAL4 (BL#45358), R78E05-GAL4 (BL#39998), R74A06-GAL4 (BL#47398), R21G01-GAL4.DBD (BL#69471), R78E05-p65.AD (BL#68630); UAS-Dcr2 (BL#24650), UAS-TrpA1-RNAi (BL#36780), UAS-TkR99D-RNAi (BL#27513), UAS-Gαq-RNAi (BL#33765), UAS-norpA-RNAi (BL#31113); UAS-6XGFP (BL#52261, BL#52262), lexAop2-6XmCherry (BL#52271, BL#52272), UAS-EGFP (from Wei Xie), UAS-GCaMP6s (BL#42746), lexAop2-myr-GCaMP6s (28), UAS-GCaMP6m (BL#42748), lexAop2-GCaMP6f (BL#44277), UAS-CsChrimson (BL#55135), Gαq1 (BL#42257), norpAP24 (BL#9048), UAS-Gαq (BL#30734), UAS-norpA (BL#35529), nsyb-GRASP (BL#64314), ppk-Gal41a (37), ppk-Gal4vk37 (37), UAS-P2X2 (67), and TkR99Df02797, UAS-TkR99D, and ppk1.9-Gal4 (gifts from Michael J. Galko, The University of Texas MD Anderson Cancer Center, Houston, Texas).
Epidermal Injury by UVC Treatment.
Early third-instar larvae were briefly cleaned by distilled water and then anesthetized by ether for about 2 min until immobilized. Larvae were then transferred to a SYLGARD silicone (Dow Corning) plate with the dorsal side facing up. The plate was then placed inside a Spectolinker XL-1000 UV cross-linker (254 nm, Spectronics Corporation) and exposed to a brief UVC radiation (∼6 s, ∼20 mJ/cm2, measured by UV spectrophotometer AccuMAX XS-254, Spectroline). After treatment, larvae were returned to mashed fly food and recovered for 24 h before behavioral assay was performed.
Thermal Nociception Assay.
Thermal nociception assay was performed as previously described (9, 28). Briefly, the heat stimulus was applied by a custom-made heat probe with a PID (Proportional Integral Derivative) controller. Early third-instar larvae were cleaned with distilled water and stimulated by the heat probe tip at abdominal segments 4 to 6 in the dorsal side. Rolling was defined as at least one 360° roll. One larva was stimulated only once. The latency between stimulation onset and rolling was recorded with up to a 20-s cutoff. According to the latency, responses were divided into three categories, fast rolling (≤5 s), slow rolling (>5 but ≤20 s), and no rolling (>20 s). The assays were performed at room temperature (about 23 °C) under a dissection scope (Nikon SMZ800) and a light source (Fostec ACE).
Confocal Microscopy and Immunofluorescence.
Drosophila larval tissues were dissected in ice-cold phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde (PFA/PBS) for 1 h at room temperature. After fixation, samples were washed by 0.3% Triton X-100/PBS for 4 × 15 min. Then the samples continue to be washed by PBS for 3 × 15 min. Samples were then mounted on glass slides with mounting media (Vectashield H-1000, Vector Lab). Images were acquired with a confocal microscope (Zeiss LSM700). For immunostaining, fixed tissues were incubated in blocking buffer (5% normal donkey serum, 0.3% Triton X-100/PBS) for 1 h at room temperature. Samples were then incubated with anti–Fasciclin-3 for labeling epidermal membranes (Developmental Studies Hybridoma Bank, 7G10; 1:50) overnight at 4 °C. After washing, the samples were incubated with secondary antibody goat-anti-mouse Cy3 (Jackson ImmunoResearch; 1:1,000) for 1 h at room temperature. Samples were washed by PBS for 3 × 15 min and mounted on glass slides for imaging. For HRP staining, dissected larval fillets were incubated with anti-HRP-Cy3 (Jackson ImmunoResearch, 123-165-021; 1:500) for 1.5 h at room temperature.
Program-Controlled Peltier Plate.
To apply heat stimulation to dissected larvae brain, we built a program-controlled Peltier device. Briefly, a Peltier plate (12706, 4 × 4 cm) was attached on a copper block (4 cm × 4 cm) with circulated cooling water. The Peltier plate and copper block were then mounted on the sample stage of the confocal microscope. The power controller for the Peltier device was an FTC200 TEC board (Accuthermo Technology) connected with an FTX700D H-bridge amplifier and a 300 W 12 V DC power. The real-time temperature of samples was monitored by a T-type thermal coupler (Omega 5SRTC-TT-T-36-36). The device was controlled by the FTC200 controller software. The PID parameters were optimized to achieve fast and smooth temperature holding and ramping curves.
GCaMP Imaging.
The larval brain was dissected in HL3 (NaCl 70 mM, KCl 5 mM, CaCl2 1.5 mM, MgCl2 20 mM, NaHCO3 10 mM, Trehalose 5 mM, Sucrose 115 mM, Hepes 5 mM, pH 7.2) and transferred to program-controlled Peltier plate containing ∼10 μL HL3 solution with a cover glass. The brain sample was placed between the Peltier plate and the cover glass, allowing it to rest for 10 min at 20 °C before imaging. The temperature program was controlled by FTC200 software (Accuthermo Technology) as follows; holding at 20 °C for 60 s, ramping up to 40 °C at about 0.5 °C/s, and holding at 40 °C for 80 s. R21G01-GAL4 > UAS-GCaMP6s or R21G01-lexA > lexAop2-myr-GCaMP6s larvae were used to monitor Ca2+ responses of BLA neurons upon heat stimulation. Data were recorded on a Zeiss LSM700 confocal microscope with a Zeiss 20×/1.0 NA water-immersion objective lens (421452-9800). GCaMP6s fluorescence was excited by a 488 nm laser. Images were acquired at 512 × 512 pixels, 12-bit dynamic range in a time-lapse mode. Region-of-interest (ROI) was selected from the somatic region of BLA neurons. Changes of GCaMP6s fluorescence were calculated by ΔF/F0 = (Ft − F0)/F0, where Ft was the fluorescent value of an ROI of a given frame. F0 was defined as the value of the same ROI in the 60-s frame. Maximal ΔF/F0 was the maximum value during the imaging period from 60 to 120 s. For TrpA1-KO, the ΔF/F0 was calculated from the frame when temperature reached 40 °C. For m-3M3FBS treatment, larval brains were incubated with m-3M3FBS (20 μM) on the 20 °C heat plate for 10 min before imaging. As negative control m-3M3FBS, equal amounts of DMSO vehicle were applied. We noted that BLA neurons showed reduced heat responses when DMSO was present.
For GCaMP imaging of P2X2 responses to ATP, larvae were carefully dissected and mounted on SYLGARD silicone plate in HL3 saline with VNC connected to the peripheral nervous system. After 5-min rest, ATP solution (Sigma, A2383-5G) was gently added with a final ATP concentration of 1 mM. Thirty seconds of averaged fluorescence intensities before and after ATP application were used to calculate baseline and response values.
Optogenetics.
To stimulate BLA neurons, we expressed CsChrimson under control of R21G01-GAL4. Two copies of UAS-CsChrimson were used to enhance the expression levels. The larvae were fed on food containing all-trans-retinal (400 μM) and maintained in dark throughout the entire development and the experimental procedure. Third-instar larvae were used for behavioral assay in a 10-cm Petri dish. To deliver red light, we connected a 10 × 10 cm LED back-light plate (620 nm, 12 Volts, 4.8 Watts) to an LED power driver (ONI-P12R). The behavioral videos were recorded with an industry camera (CGimagetech, CGU2-130M) adapted with an infrared long-pass filter (Zomei, >760 nm).
Brain Temperature Measurement.
A tiny thermocouple (IT-24P, Physitemp) was inserted into the brain lobe location (segment T3-A1) of the third-instar larvae through an excision at the posterior end. The thermocouple was connected with the temperature logging device to monitor temperature changes. The heat probe contacted larval dorsal side around segment A4, the same way as we performed rolling behavioral assay, for a duration of 20 s.
Statistical Analyses.
Statistical analysis was performed by Graphpad Prism software. The latency of rolling behavior was analyzed using the Log-rank (Mantel-Cox) test. The P values were corrected by multiplying total comparing numbers. Whenever t test was used, the dataset passed normality test. A dataset of Fig. 4K did not satisfy normal distribution, therefore, the nonparametric Mann–Whitney U test was used instead. For optogenetic stimulation assay, Fisher’s exact test was performed. In all figures, ns, P > 0.05, *P < 0.05, **P < 0.01, and ***P < 0.001. Columns with different superscripts (a, b, and c) are significantly different from each other (P < 0.05). Error bars are SEMs.
Supplementary Material
Acknowledgments
We thank Drs. Michael J. Galko and Seol Hee Im for technical support for behavioral hypersensitivity assay; Drs. Fan Wang, Michael J. Galko, and Rebecca Yang for critical comments on the manuscript; Drs. Wei Zhang, Yulong Li, Rebecca Yang, and the Bloomington Drosophila Stock Center for the fly stocks; and Biorender.com for model figure illustration. Y.X. is supported by the NIH Awards 5R01NS089787 and 1R21NS107924 and Human Frontier Science Program Young Investigator Award RGY0090/2014.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. P.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113645119/-/DCSupplemental.
Data Availability
All study data are included in the main text and supporting information.
References
- 1.Baliki M. N., Apkarian A. V., Nociception, pain, negative moods, and behavior selection. Neuron 87, 474–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Julius D., Basbaum A. I., Molecular mechanisms of nociception. Nature 413, 203–210 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Kang K., et al. , Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464, 597–600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walters E. T., Nociceptive biology of Molluscs and Arthropods: Evolutionary clues about functions and mechanisms potentially related to pain. Front. Physiol. 9, 1049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hucho T., Levine J. D., Signaling pathways in sensitization: Toward a nociceptor cell biology. Neuron 55, 365–376 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Ji R. R., Woolf C. J., Neuronal plasticity and signal transduction in nociceptive neurons: Implications for the initiation and maintenance of pathological pain. Neurobiol. Dis. 8, 1–10 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Basbaum A. I., Bautista D. M., Scherrer G., Julius D., Cellular and molecular mechanisms of pain. Cell 139, 267–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costigan M., Scholz J., Woolf C. J., Neuropathic pain: A maladaptive response of the nervous system to damage. Annu. Rev. Neurosci. 32, 1–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babcock D. T., Landry C., Galko M. J., Cytokine signaling mediates UV-induced nociceptive sensitization in Drosophila larvae. Curr. Biol. 19, 799–806 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babcock D. T., et al. , Hedgehog signaling regulates nociceptive sensitization. Curr. Biol. 21, 1525–1533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im S. H., et al. , Tachykinin acts upstream of autocrine Hedgehog signaling during nociceptive sensitization in Drosophila. eLife 4, e10735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.W. D. Tracey, Jr, Wilson R. I., Laurent G., Benzer S., Painless, a Drosophila gene essential for nociception. Cell 113, 261–273 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Hwang R. Y., et al. , Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 17, 2105–2116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im S. H., Galko M. J., Pokes, sunburn, and hot sauce: Drosophila as an emerging model for the biology of nociception. Dev. Dyn. 241, 16–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko T., et al. , Serotonergic modulation enables pathway-specific plasticity in a developing sensory circuit in Drosophila. Neuron 95, 623–638.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woolf C. J., Allchorne A., Safieh-Garabedian B., Poole S., Cytokines, nerve growth factor and inflammatory hyperalgesia: The contribution of tumour necrosis factor alpha. Br. J. Pharmacol. 121, 417–424 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montell C., Drosophila sensory receptors-a set of molecular Swiss army knives. Genetics 217, 1–34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julius D., TRP channels and pain. Annu. Rev. Cell Dev. Biol. 29, 355–384 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Montell C., The TRP superfamily of cation channels. Sci. STKE 2005, re3 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Dai Y., et al. , Sensitization of TRPA1 by PAR2 contributes to the sensation of inflammatory pain. J. Clin. Invest. 117, 1979–1987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kort M. E., Kym P. R., TRPV1 antagonists: Clinical setbacks and prospects for future development. Prog. Med. Chem. 51, 57–70 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Garami A., et al. , Contributions of different modes of TRPV1 activation to TRPV1 antagonist-induced hyperthermia. J. Neurosci. 30, 1435–1440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavva N. R., et al. , Pharmacological blockade of the vanilloid receptor TRPV1 elicits marked hyperthermia in humans. Pain 136, 202–210 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Vay L., Gu C., McNaughton P. A., The thermo-TRP ion channel family: Properties and therapeutic implications. Br. J. Pharmacol. 165, 787–801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanack C., et al. , GABA blocks pathological but not acute TRPV1 pain signals. Cell 160, 759–770 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Bautista D. M., et al. , TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell 124, 1269–1282 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Kang K., et al. , Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature 481, 76–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu P., et al. , Polymodal nociception in Drosophila requires alternative splicing of TrpA1. Curr. Biol. 29, 3961–3973.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito S., Saito C. T., Nozawa M., Tominaga M., Elucidating the functional evolution of heat sensors among Xenopus species adapted to different thermal niches by ancestral sequence reconstruction. Mol. Ecol. 28, 3561–3571 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Saito S., et al. , Analysis of transient receptor potential ankyrin 1 (TRPA1) in frogs and lizards illuminates both nociceptive heat and chemical sensitivities and coexpression with TRP vanilloid 1 (TRPV1) in ancestral vertebrates. J. Biol. Chem. 287, 30743–30754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gracheva E. O., et al. , Molecular basis of infrared detection by snakes. Nature 464, 1006–1011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandewauw I., et al. , A TRP channel trio mediates acute noxious heat sensing. Nature 555, 662–666 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Story G. M., et al. , ANKTM1, a TRP-like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 112, 819–829 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Viswanath V., et al. , Opposite thermosensor in fruitfly and mouse. Nature 423, 822–823 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Zhong L., et al. , Thermosensory and nonthermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat-sensor domains of a thermoTRP Channel. Cell Rep. 1, 43–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu C., Luo J., He L., Montell C., Perrimon N., Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca(2+) signaling in the Drosophila midgut. eLife 6, e22441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han C., et al. , Integrins regulate repulsion-mediated dendritic patterning of drosophila sensory neurons by restricting dendrites in a 2D space. Neuron 73, 64–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ainsley J. A., et al. , Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 13, 1557–1563 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Guntur A. R., et al. , Drosophila TRPA1 isoforms detect UV light via photochemical production of H2O2. Proc. Natl. Acad. Sci. U.S.A. 112, E5753–E5761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du E. J., et al. , TrpA1 regulates defecation of food-borne pathogens under the control of the duox pathway. PLoS Genet. 12, e1005773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amcheslavsky A., Jiang J., Ip Y. T., Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J., Shen W. L., Montell C., TRPA1 mediates sensation of the rate of temperature change in Drosophila larvae. Nat. Neurosci. 20, 34–41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berni J., Pulver S. R., Griffith L. C., Bate M., Autonomous circuitry for substrate exploration in freely moving Drosophila larvae. Curr. Biol. 22, 1861–1870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sweeney S. T., Broadie K., Keane J., Niemann H., O’Kane C. J., Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14, 341–351 (1995). [DOI] [PubMed] [Google Scholar]
- 45.Klapoetke N. C., et al. , Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamada F. N., et al. , An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfeiffer B. D., et al. , Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luan H., Peabody N. C., Vinson C. R., White B. H., Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehnert B. P., Baker A. E., Gaudry Q., Chiang A. S., Wilson R. I., Distinct roles of TRP channels in auditory transduction and amplification in Drosophila. Neuron 77, 115–128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asahina K., et al. , Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell 156, 221–235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian N. X., Winitz S., Johnson G. L., Epitope-tagged Gq alpha subunits: Expression of GTPase-deficient alpha subunits persistently stimulates phosphatidylinositol-specific phospholipase C but not mitogen-activated protein kinase activity regulated by the M1 muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. U.S.A. 90, 4077–4081 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley G. G., Ondrako J. M., Reks S. E., Fuel and hormone regulation of phospholipase C beta 1 and delta 1 overexpressed in RINm5F pancreatic beta cells. Mol. Cell. Endocrinol. 177, 107–115 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Ignell R., et al. , Presynaptic peptidergic modulation of olfactory receptor neurons in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 106, 13070–13075 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon Y., Shim H. S., Wang X., Montell C., Control of thermotactic behavior via coupling of a TRP channel to a phospholipase C signaling cascade. Nat. Neurosci. 11, 871–873 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Delgado R., et al. , Light-induced opening of the TRP channel in isolated membrane patches excised from photosensitive microvilli from Drosophila photoreceptors. Neuroscience 396, 66–72 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Lima S. Q., Miesenböck G., Remote control of behavior through genetically targeted photostimulation of neurons. Cell 121, 141–152 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Ohyama T., et al. , A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Arenas O. M., et al. , Activation of planarian TRPA1 by reactive oxygen species reveals a conserved mechanism for animal nociception. Nat. Neurosci. 20, 1686–1693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fogarty C. E., et al. , Extracellular Reactive Oxygen Species Drive Apoptosis-Induced Proliferation via Drosophila Macrophages. Curr. Biol. 26, 575–584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopes C. M., et al. , Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron 34, 933–944 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Suh B. C., Hille B., PIP2 is a necessary cofactor for ion channel function: How and why? Annu. Rev. Biophys. 37, 175–195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paulsen C. E., Armache J. P., Gao Y., Cheng Y., Julius D., Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 525, 552 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Dillman A. A., et al. , mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat. Neurosci. 16, 499–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X., et al. , Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166, 1147–1162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Y., Suzuki Y., Uchida K., Tominaga M., Identification of a splice variant of mouse TRPA1 that regulates TRPA1 activity. Nat. Commun. 4, 2399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang H., Tay S. H., Ng W., Ng S. Y., Soong T. W., Targeting novel human transient receptor potential ankyrin 1 splice variation with splice-switching antisense oligonucleotides. Pain 162, 2097–2109. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin X., et al. , A subset of DN1p neurons integrates thermosensory inputs to promote wakefulness via CNMa signaling. Curr. Biol. 31, 2075–2087 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and supporting information.