Abstract
The objective of this study was to investigate the presence and persistence of carbapenemase-producing Klebsiella spp. isolated from wastewater and treated wastewater from two tertiary hospitals in Mexico. We conducted a descriptive cross-sectional study in two hospital wastewater treatment plants, which were sampled in February 2020. We obtained 30 Klebsiella spp. isolates. Bacterial identification was carried out by the Matrix-Assisted Laser Desorption/Ionization-Time of Flight mass spectrometry (MALDI-TOF MS®) and antimicrobial susceptibility profiles were performed using the VITEK2® automated system. The presence of carbapenem resistance genes (CRGs) in Klebsiella spp. isolates was confirmed by PCR. Molecular typing was determined by pulsed-field gel electrophoresis (PFGE). High rates of Klebsiella spp. resistance to cephalosporins and carbapenems (80%) were observed in isolates from treated wastewater from both hospitals. The molecular screening by PCR showed the presence of blaKPC and blaOXA-48-like genes. The PFGE pattern separated the Klebsiella isolates into 19 patterns (A–R) with three subtypes (C1, D1, and I1). Microbiological surveillance and identification of resistance genes of clinically important pathogens in hospital wastewater can be a general screening method for early determination of under-detected antimicrobial resistance profiles in hospitals and early warning of outbreaks and difficult-to-treat infections.
Keywords: Klebsiella pneumoniae, carbapenem resistance, waste and treated water, public health
1. Introduction
The spread of pathogens of public health importance is increasingly rapid between communities and countries [1]. The emerging and re-emerging infectious disease outbreaks have been increasing in recent years, mainly through transmission by direct or indirect contact between humans, animals (including food) and the environment (water) [2,3]. Therefore, an epidemiological surveillance system is an important tool for the identification, monitoring and prediction of infectious diseases [4,5], not only in healthcare institutions, but also through the screening of wastewater environments to detect the presence of resistance bacteria from hospital sources into the community environment [6,7].
Carbapenem-resistant Enterobacteriaceae (CRE) are one of the important threats to global public health, generating significant morbidity and mortality as well as associated prolonged healthcare and increased cost [8,9]. CRE are currently defined by the Centers for Disease Control and Prevention (CDC) as: “an isolate of Enterobacteriaceae that is resistant to ertapenem, imipenem, meropenem or doripenem according to the Clinical Laboratory Standards Institute (CLSI) or documentation that the isolate produces a carbapenemase” [10,11,12]. There are different bacterial genus belonging to the Enterobacteriaceae family, which are present in the microbiota of humans, animals and the environment, among which Klebsiella pneumoniae stands out [13,14,15].
The worldwide spread of carbapenemase-producing Klebsiella pneumoniae (KPC) is a latent threat due to its capacity for dissemination in healthcare-associated infections (HAIs) and the accumulation of resistance mechanisms through the production of carbapenemase enzymes [16,17,18]. The first strain of KPC was reported in North Carolina in the United States [19], and the geographic distribution was limited to this country until 2005 when KPC was reported in a patient in France [20]. In Latin America, KPC was reported in Colombia in 2006 [21], followed by other countries such as Argentina, Brazil, Uruguay and Chile from 2011–2012 [16]. The prevalence of KPC in Mexico is less than 5%, according to the reports made by Plan Universitario de Control de la Resistencia Antimicrobiana (PUCRA) (2018) and Red Temática de Investigación y Vigilancia de la Farmacorresistencia (INVIFAR) (2020, 2021) [22,23,24]. In Mexico, the first report of KPC-3 (ST258) (n = 24) was made in 2010 as part of an outbreak with a case rate fatality of 55% [25].
KPC strains have been isolated mainly in HAIs in patients with prolonged hospital stays, contaminated medical devices and fomites in healthcare personnel, among others. However, being a ubiquitous bacterium, KPC strains have also been isolated from animals and aquatic environments such as surface water and sewage [13,26,27]. The selection of antibiotic-resistant KPC has increased due to horizontal gene transfer and exchange of mobile genetic elements, producing a great impact on public health [28]. These healthcare-acquired multidrug-resistant bacteria have spread to the environment due to the persistence of antibiotic resistance genes located in mobile genetic elements (MGEs) capable of spreading efficiently among bacteria in the sewage environment [29]. The identification of multidrug-resistance (MDR) or extensively drug-resistance (XDR) bacteria as well as antimicrobial resistant genes in freshwater and wastewater could be considered a good surveillance system for early detection of this event. In addition, the presence and persistence of these bacteria or their material genetics after water treatment could be considered a potentially important medium for the development and spread of antimicrobial resistance, especially in environments with inadequate sanitation [30]. This study explores the presence of carbapenem-resistant Klebsiella pneumoniae in hospital wastewater and even after treatment as a means of spread with gradual health risk.
2. Results
2.1. KPC Isolates
Among the 30 isolates, 26 (86.7%) were confirmed by MALDI-TOF MS® and VITEK 2® as K. pneumoniae and 4 (13.3%) as K. oxytoca. Half of the isolates were isolated from treated water and the other 50% from raw wastewater. Seven isolates of K. pneumoniae (three from raw wastewater/four from treated wastewater) and two of K. oxytoca (one from each site) were collected from the wastewater treatment plant (WWTP) of the Hospital Regional de Alta Especialidad de Ixtapaluca (HRAEI), while 19 isolates of K. pneumoniae (eleven from raw wastewater/eight from treated wastewater) and two of K. oxytoca (both from treated wastewater) were collected at the WWTP of Instituto Nacional de Cancerología (INCAN).
2.2. Antimicrobial Susceptibility
The antimicrobial susceptibility results showed that 18 isolates were resistant to ampicillin-sulbactam, ceftazidime and ceftriaxone with a MIC90 of >32, >64 and >64 mg/L, respectively, 16 isolates showed resistance to piperacillin-tazobactam, cefoxitin, doripenem, ertapenem and imipenem (MIC90 of >128, >64, >8, >8, >16 mg/L, respectively), 17 were resistant to cefepime and ciprofloxacin with a MIC90 of >64 and >4 mg/L, eight isolates showed resistance to gentamicin (MIC90 of 16 mg/L), only three isolates were resistant to amikacin with a MIC90 of 16 mg/L, and all isolates were sensitive to tigecycline with MIC90 of 1 mg/L. High rates of Klebsiella spp. resistance to cephalosporins and carbapenems (80%) were observed in isolates from treated wastewater from both hospitals.
The isolates collected in both WWTPs were grouped into nine antimicrobial susceptibility profiles. The most frequents profiles were: sensitive to all antibiotics (n = 12) and sensitive only to amikacin, gentamicin and tigecycline (n = 8), (Table 1). Thirteen (81.2%) carbapenem-producing Klebsiella spp. were ESBL producers.
Table 1.
Phenotyping properties of carbapenemase-producing Klebsiella spp. present in wastewater treatment plant (WWTP) of Hospital Regional de Alta Especialidad de Ixtapaluca (HRAEI) and Instituto Nacional de Cancerología (INCAN).
| Hospital | Sample Site | Code | Isolated | Genes | AMP | TZP | FOX | CAZ | CTX | CEF | DOR | ERT | IPM | MEM | AMK | GEN | CIP | TIG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HRAEI | Raw Wastewater | 1A-1 | K. pneumoniae | 16 | <4 | <4 | 4 | 16 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |
| 1A-2 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |||
| IXT02B | K. pneumoniae | blaKPC, blaOXA | >32 | >128 | 16 | >64 | 16 | 2 | >8 | >8 | 8 | >16 | 2 | 1 | 1 | <0.5 | ||
| IXT05B | K. oxytoca | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |||
| Treated Wastewater | 5A-1 | K. pneumoniae | blaKPC | >32 | >128 | 16 | >64 | >64 | >64 | >8 | >8 | >16 | >16 | <2 | >16 | 2 | 2 | |
| 5A-2 | K. pneumoniae | >32 | 8 | <4 | 4 | >64 | 2 | <0.12 | <0.5 | <0.25 | <0.25 | 4 | <1 | 2 | 2 | |||
| 5C-2 | K. pneumoniae | blaKPC | >32 | >128 | 16 | 64 | 16 | 2 | >8 | >8 | 8 | >16 | 2 | 1 | 2 | <0.5 | ||
| 5D-3 | K. pneumoniae | >32 | >128 | >64 | >64 | >64 | >64 | >8 | >8 | >16 | >16 | >64 | 16 | >4 | <0.5 | |||
| IXT09C | K. oxytoca | >32 | >128 | >64 | >64 | >64 | >64 | >8 | >8 | >16 | >16 | 16 | 1 | 1 | <0.5 | |||
| INCAN | Raw Wastewater | 9A-1 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | 1 | |
| 9A-2 | K. pneumoniae | blaKPC | >32 | >128 | 16 | 64 | 16 | 2 | >8 | >8 | 8 | >16 | >64 | 8 | 1 | 1 | ||
| 9D-3 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | 1 | |||
| 10B-2 | K. pneumoniae | 2 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | 1 | |||
| 10B-3 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | <1 | <0.025 | 1 | |||
| 17A-1 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |||
| 17A-2 | K. pneumoniae | blaKPC | >32 | >128 | 8 | >64 | 16 | 2 | >8 | >8 | >16 | >16 | 2 | 1 | 1 | <0.5 | ||
| 17C-1 | K. pneumoniae | blaKPC | >32 | >128 | 4 | >64 | 8 | 2 | >8 | >8 | >16 | >16 | 2 | 1 | 1 | <0.5 | ||
| 17D-3 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |||
| 20-Mar | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |||
| CAN14E | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | <0.5 | |||
| Treated Wastewater | 13A-1 | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | <1 | <0.025 | 1 | ||
| 13A-2 | K. pneumoniae | blaKPC | >32 | >128 | 16 | >64 | 16 | 2 | >8 | >8 | >16 | >16 | >64 | 8 | 0.5 | 1 | ||
| 13C-1 | K. oxytoca | blaKPC | >32 | >128 | 4 | >64 | 16 | 2 | >8 | >8 | >16 | >16 | 2 | 1 | >4 | <0.5 | ||
| 13C-2 | K. pneumoniae | blaKPC | >32 | >128 | >64 | >64 | 32 | 8 | >8 | >8 | 8 | >16 | <2 | 8 | 1 | 1 | ||
| 16-ene | K. pneumoniae | blaKPC | >32 | >128 | 8 | >64 | >64 | 2 | >8 | >8 | 8 | >16 | 2 | 1 | 1 | <0.5 | ||
| 16-Mar | K. oxytoca | blaKPC | >32 | >128 | >64 | >64 | >64 | 32 | >8 | >8 | >16 | >16 | 8 | 1 | 2 | 2 | ||
| 21A-1 | K. pneumoniae | blaKPC | >32 | >128 | 8 | >64 | 8 | 2 | >8 | 4 | 8 | >16 | 2 | 8 | 2 | 1 | ||
| 21D-1 | K. pneumoniae | blaKPC | >32 | >128 | 8 | >64 | 16 | 2 | >8 | >8 | >16 | >16 | 2 | >16 | >4 | 1 | ||
| CAN15F | K. pneumoniae | blaKPC | >32 | >128 | 8 | 64 | 8 | 2 | >8 | 4 | 8 | >16 | <2 | >16 | >4 | 1 | ||
| 13D-3-k | K. pneumoniae | 4 | <4 | <4 | <1 | <1 | <1 | <0.12 | <0.5 | <0.25 | <0.25 | <2 | 1 | <0.025 | 1 |
Ampicillin-sulbactam (AMP), piperacillin-tazobactam (TZP), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CTX), cefepime (CEF), doripenem (DOR), ertapenem (ERT), imipenem (IPM), meropenem (MEM), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP) and tigecycline (TIG).
2.3. Detection of Carbapenem-Resistance Genes
The molecular screening by PCR showed the presence of specific carbapenem-resistance genes (blaKPC and blaOXA-48-like); it was observed that 13 (81.25%) Klebsiella spp. isolated carried blaKPC while one isolate was detected to harbor both blaKPC and blaOXA-48-like. In only two isolated carbapenem-resistant, the genotype could not be determined; further sequencing analysis is being performed to fully characterized.
2.4. Molecular Typing of Pulsed-Field Gel Electrophoresis (PFGE)
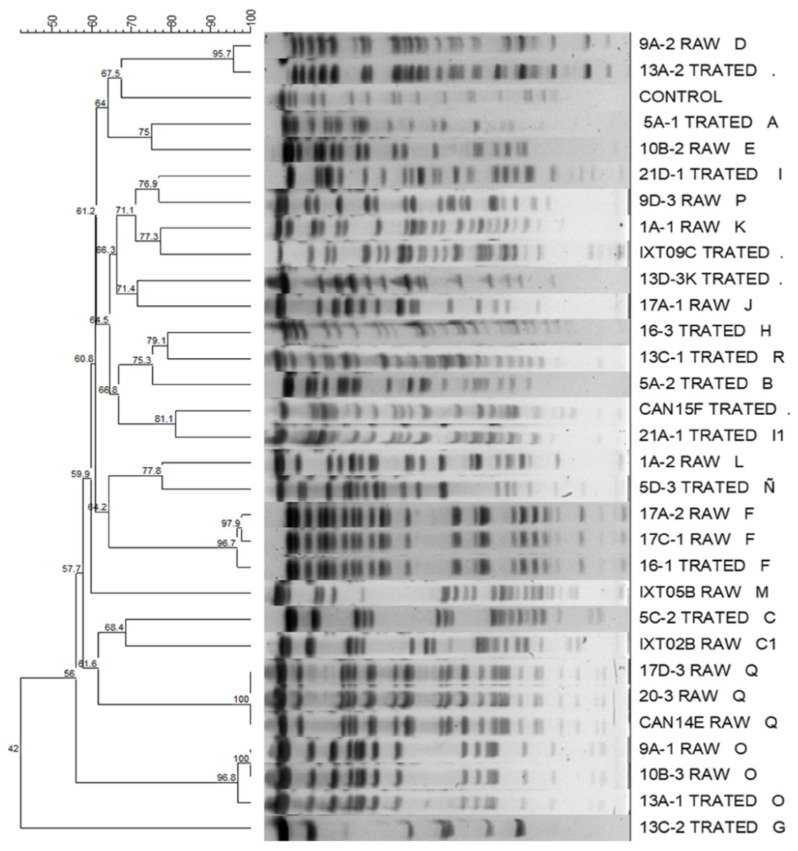
The PFGE separated the Klebsiella isolates into 19 patterns (A–R) with three subtypes (C1, D1, and I1). The patterns (A, B, C, D1, F, G, H, I, J, N, Ñ, O, R and I1) were present in the Klebsiella isolates from the treated wastewater, whereas the patterns (D, E, F, K, L, M, C1, O, P, J and Q) were in the isolates isolated from raw wastewater. The most represented patterns were F, I, O and Q, each with three isolates, and C, D and J, each with two isolates; the remaining patterns were represented by a single isolate. Klebsiella isolates with patterns C, D, F, J and O were isolated in samples of raw wastewater and treated wastewater. The clones found were related to the WWTP of origin. It was found that the majority patterns (D, F, I, J, O and Q) were only present in the WWTP of the INCAN, while in the HRAEI WWTP, the Klebsiella isolates did not show clonality, except for two isolates (C and C1). The results of the computer analysis of the banding patterns showed similarity percentages that were from 100% to 42% (Figure 1).
Figure 1.
Dendrogram based on pulsed-field gel electrophoresis (PFGE) patterns after digestion with enzyme Xba I of Klebsiella spp. isolates isolated from hospital wastewater.
When the majority PFGE patterns were compared with the antimicrobial susceptibility profile, it was observed that the isolates with patterns O, Q and J were sensitive to all the antibiotics tested. The isolates with patterns C and F were only sensitive to amikacin, gentamicin and tigecycline; the isolates with pattern I were only sensitive to amikacin and tigecycline, and isolates with pattern D were resistant to all the antibiotics tested, except tigecycline.
3. Discussion
HAIs caused by carbapenem-resistant K. pneumoniae strains have been reported in hospitals of several regions of the world, including Mexico [8]. However, the present study suggests that hospital wastewater becomes an important source of persistence and spread of multidrug-resistant bacteria, causing these bacteria to escape from WWTPs into the environment. Our findings are important from a public health viewpoint.
Among the total number (30) of Klebsiella spp. isolates, 26 were confirmed K. pneumoniae and only four as K. oxytoca; this is in accordance with a study by Samanta et al., and Sharma et al. also reported the presence of Klebsiella spp. in wastewater [31,32]. The present study shows the presence of carbapenemase-producing Klebsiella spp. in the studied WWTPs (16/30); some of these isolates were also resistant to beta-lactams (18/30), cephalosporins (17/30), fluoroquinolones (17/30) and aminoglycosides (8/30). In a study conducted in Bangladesh, it was observed that strains of Klebsiella spp. (n = 46) isolated from hospital wastewater were resistant to the same groups of antibiotics [26]. In another study carried out in South Africa, 60 Klebsiella strains were isolated, of which 28% were resistant to cephalosporins [33], while in Ethiopia in 2012, 30 Klebsiella strains were isolated in hospital wastewater, all strains were resistant to ampicillin, ten showed resistance to ceftazidime and six were resistant to gentamicin [27]. These results are similar to those found in our study. In the present study it was observed that 16 (53.3%) isolates of the Klebsiella spp. were resistant to carbapenems, similar results were observed in the study by Ebomah et al. [34], where high percentages of resistance to meropenem (69%) were observed. This is contrary to the study by Peneş et al. [35] that reported low resistance to meropenem and imipenem. Some other studies reported Klebsiella spp. with a high percentage of resistance against carbapenems as observed in our study for imipenem, carbapenem and meropenem [36,37]. The detection of K. pneumoniae strains resistant to carbapenems has also been reported by Perilli et al. [38] in wastewater samples collected throughout a year of study; these strains (n = 20) were also resistant to beta lactams and quinolones, as observed in our work. In addition, it was observed that 80% (12/15) of the Klebsiella isolates obtained from treated wastewater were resistant to carbapenems. This result has important implications for public health, since carbapenems are antibiotics of last choice whose resistance may be spreading to the environment through resistant bacteria present in treated wastewater, which is often reused. Interestingly, we found that all the Klebsiella spp. isolates analyzed were sensitive to tigecycline; this result is consistent with that reported by Islam et al. [26], who detected that the K. pneumoniae strains (n = 46) collected in their study were sensitive to tigecycline; this differs from that reported by Obasi et al. and Teban-Man et al., who detected strains of K. pneumoniae resistant to tigecycline isolated from wastewater 57.2 (4/7) and 64% (21/33), respectively [39,40]. The MIC90 results in carbapenemase-producing Klebsiella spp. isolates in this study ranged from >8 to >16 mg/L, being lower than those reported by Islam et al. [26] in carbapenemase-producing Klebsiella spp. isolates obtained from hospital wastewater.
This study detected the presence of CRG in Klebsiella isolates and found the genes blaKPC and blaOXA-48-like. The OXA-48 gene is one of the most blaOXA-48-like common variants belonging to class D carbapenemase. The first report of the blaOXA-48 gene was in Turkey in 2001 [18], and the prevalence of OXA-48 genes among Klebsiella strains was reported in different countries (Saudi Arabia, China, Turkey and France) [41,42,43,44]. We found that only one strain of K. pneumoniae carried blaOXA-48-like, and this differs from the findings by Gibbon, et al., who reported a high abundance of blaOXA-48 (29%) in strains of K. pneumoniae and K. ornithinolytica isolated of WWTP influent [45]. The blaKPC carbapenemases gene has worldwide prevalence [46,47,48]; in our study we detected that 81.25% of the Klebsiellas spp. isolates carried the blaKPC gene, and this is different from the study by Ebomah et al., who reported 10 isolates carried blaOXA-48-like; 17 carried blaKPC; and 73 isolates carried blaNDM-1 [34]. As observed in the studies conducted, the frequency of CRG varies in different geographical areas.
Molecular typing by PFGE was performed to investigate the clonal relationship between the K. pneumoniae isolates. In our study, we detected a remarkable diversity of patterns (n = 19), but seven patterns predominated; this heterogenicity of patterns and predominance of specific clones has been observed in other studies, such as those published by de Oliveira et al. and Daoud et al., who found three clonal groups and a unique group, respectively, in strains isolated from wastewater [49,50]. Another of our important findings was that the strains of Klebsiella spp. that carried the majority of PFGE patterns were found both in raw wastewater and treated wastewater, demonstrating its ability to persist in its passage through the WWTP. The results obtained showed that the majority clonal patterns were related to the WWTP of origin; this could be in relation to the Klebsiella clones that circulate in each hospital. The clonality of K. pneumoniae in hospitals has been widely documented. In a study carried in Tehran, Iran, PFGE revealed related patterns among K. pneumoniae isolates [51]. Another study in Shiraz, Iran, PFGE analysis showed 11 patterns of K. pneumoniae, observing important clonal relationships [52].
Although carbapenem-resistant and beta-lactam-resistant enterobacteria are widely explored in the hospital setting, little is known about this resistance phenotype in other environments. There are studies on Klebsiella spp. strains resistant to these groups of antimicrobials in different aquatic environments (rivers, sewage, among others) [53,54,55]. Resistance to other antibiotics such as colistin is a point that we propose to address in the carbapenemase-producing Klebsiella spp. isolates collected in this study.
These results highlight the need to carry out continuous monitoring programs for antibiotic-resistant bacteria in hospital WWTPs as part of the biosafety program of each institution. A greater number of studies need to be carried out in the hospital WWTPs of our country, to try to understand and reduce the routes of dissemination of resistant bacteria [56].
4. Materials and Methods
4.1. Study Sites and Sample Collection
The study was conducted at Mexico City Metropolitan Zone in February 2020. Wastewater samples from the Regional High Specialty Hospital of Ixtapaluca (Hospital Regional de Alta Especialidad de Ixtapaluca—HRAEI) and the National Institute of Oncology (Instituto Nacional de Cancerología—INCAN) were the primary sampling sources. These two tertiary-level hospitals were chosen based on several criteria, such as public management, bed capacity, location and the presence of WWTP in their facilities. Wastewater samples were collected in duplicate, before and after passing through each hospital’s wastewater treatment plant (WWTP); the average inlet flow comes from the hospital wards of each of the care centers. The plant configuration begins with a pretreatment process, which consists of removing coarse waste not degradable by conventional biological processes, then dosing the water flow to the aeration and extended aeration process mediated by regulating valves in several chambers. The treatment continues with a clarification and secondary sedimentation stage consisting of chambers, precipitating the thickened sludge. Finally, a disinfection process is carried out by contact with calcium hypochlorite tablets to conclude its passage through granular activated carbon filters, as well as by ultraviolet light (UV) germicidal equipment of the effluent. Selected samples were collected in sterile 1000 mL containers, transported at 4 °C to the Research Center of Infectious Diseases at the National Institute of Public Health (Instituto Nacional de Salud Pública—INSP) for processing.
4.2. Microbial Culturing and Identification
Microbial culturing was performed within four hours of sample collection. We used an aliquot of 100 mL of the raw wastewater and 200 mL of the treated wastewater to be centrifuged at 5000 rpm for 20 min; the pellet was dissolved in 10 mL of phosphate buffer solution (PBS). Prior to plating, the samples were diluted 1:1000 in PBS, and then were plating in HiCrome™ ECC Agar plates and then incubated at 37 °C for 18 h. Different presumptive isolates of Klebsiella spp. were selected for further identification. A total of 30 isolates of Klebsiella spp. were identified by mass spectrometry, using the Microflex LT MALDI-TOF MS® (Bruker Daltonics, Bremen, Germany) equipment.
4.3. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility profiles and detection of ESBLs of Klebsiella spp. isolates were performed by the VITEK 2® automated system (BioMérieux, Marcy l’Etoile, France). A panel of 14 antimicrobials were used: ampicillin-sulbactam (AMP), piperacillin-tazobactam (TZP), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CTX), cefepime (CEF), doripenem (DOR), ertapenem (ERT), imipenem (IPM), meropenem (MEM), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP) and tigecycline (TIG). The minimum inhibitory concentration (MIC) was interpreted according to the breakpoints established by the CLSI, 2021 document [12].
4.4. Detection of Carbapenem-Resistance Genes
The presence of carbapenem resistance genes (CRGs) in Klebsiella spp. isolates was confirmed by PCR. The assay included the following CRGs: blaKPC, blaOXA-48-like, blaNDM-1 and blaIMP [57,58,59,60]. The bacterial DNA was extracted by performing the boiling method, and the DNA templates were stored at −20 °C for PCR analysis. PCR products were visualized by electrophoresis on ethidium bromide stained 1% agarose gel. The primers used, sizes of the amplicons and reaction protocols are listed in Table 2.
Table 2.
Primer sequences of target carbapenems genes and their respective amplicon sizes and PCR cycling conditions.
| Target Gene | Primer Sequence (5’-3’) | Amplicon Size (bp) | PCR Cycling Condition | References |
|---|---|---|---|---|
| blaNDM-1 | F: GGTTTGGCGATCTGGTTTTC R: GGAATGGCTCATCACGATC |
621 | Initial denaturation 10 min at 94 °C. Denaturation at 94 °C for 30 s, annealing at 52 °C for 40 s, extension at 72 °C for 50 s, by 36 cycles, and final extension at 72 °C for 10 min. | Dogonchi AA, et al. [57] |
| blaIMP | F: ATAGCCATCCTTGTTTAGCTC R: TCTGCGATTACTTTATCCTC |
818 | Initial denaturation 10 min at 94 °C. Denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min by 30 cycles, and final extension at 72 °C for 10 min. | Aubron, CL, et al. [58] |
| blaKPC | F: ATGTCACTGTATCGCCGTCT R: TTTTCAGAGCCTTACTGCCC |
893 | Initial denaturation 15 min at 95 °C. Denaturation at 94 °C for 1 min, annealing at 62 °C for 1 min, extension at 72 °C for 1 min by 38 cycles, and final extension at 72 °C for 10 min. | Bradford, PA, et al. [59] |
| blaOXA-48-like | F: TTGGTGGCATCGATTATCGG R: GAGCACTTCTTTTGTGATGGC |
744 | Initial denaturation 5 min at 94 °C. Denaturation at 94 °C for 1 min, annealing at 60 °C for 1 min, extension at 72 °C for 1 min by 30 cycles, and final extension at 72 °C for 10 min. | Poirel, LC, et al. [60] |
4.5. Molecular Typing of PFGE
The genomic DNA was prepared as described above; after digestion with Xba I endonuclease, the DNA was separated using a CHEF-DR II system (BioRad, Birmingham, UK) [61]. The Salmonella serotype Braenderup strain (H9812) was included in each PFGE gel as control. The different clones were compared according to the criteria of Tenover [62].
4.6. Computer Fingerprint Analysis
Computer analysis of PFGE profiles was done using the Gel Compar II software, v.6.6.11 (Applied Maths, Inc.; Sint-Martens-Latem, Belgium) after visual inspection. The Lambda PFGE Ladder marker was included in each gel to normalize the PFGE profiles. The Dice coefficients were calculated and were then transformed into an agglomerative cluster by the unweighted pair group method with arithmetic average (UPGMA).
5. Conclusions
The results of this work showed the presence of carbapenemase-producing Klebsiella spp. isolates in the influents and effluents of the hospital wastewater treatment plants studied, evidencing the ability of these pathogens to survive WWTP treatment systems, as well as their potential ability to persist and spread through wastewater to other environments. The detection of carbapenemase-producing Klebsiella spp. isolates in hospital wastewater in our country highlights the need to establish antimicrobial resistance surveillance programs for this and other clinically important microorganisms.
The presence of Klebsiella isolates in hospital wastewater may create a real problem for public health, mainly due to the capacity for dissemination of these bacteria and their genes. Given the rapid spread of antimicrobial resistance in different ecological niches around the world, epidemiological surveillance of antimicrobial resistance raises the possibility of monitoring the persistence and spread of antimicrobial-resistant bacteria and their resistance genes not only in the hospital environment, but also in niches where their residues converge, such as wastewater.
Acknowledgments
We thank the administrative staff of the Research Center of Infectious Diseases at the National Institute of Public Health (Instituto Nacional de Salud Pública—INSP) for their efforts and valuable support, the nurse Bertha García Pineda of the Instituto Nacional de Cancerología for her support in obtaining the samples, QFB Yvonne Guadalupe Villalobos Zapata, QC Norma Irene López García and QFB Veronica Esteban Kenel of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán for their technical support in the use of MALDI-TOF MS® and PCR.
Author Contributions
Conceptualization, C.M.A.-A., M.G.-L., M.E.V.-M., B.A.C.-Q.; methodology, M.G.-L., M.E.V.-M., M.B.-d.-V., B.A.C.-Q., P.C.-J., A.S.-G.; writing—original draft preparation, M.G.-L., M.E.V.-M., C.M.A.-A.; writing—review and editing, C.M.A.-A., M.G.-L., M.E.V.-M., M.B.-d.-V., P.C.-J., A.P.-d.-L., A.S.-G.; funding acquisition, C.M.A.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed by FOSISS-CONACYT-2017, number 290618; M.G.-L. is a PhD student with a scholarship from CONACYT.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical approval was obtained from the Ethic Committee of the Instituto Nacional de Salud Pública (CI: 1164).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alpuche-Aranda C.M. Infecciones emergentes el gran reto de la salud global: COVID-19. Salud Publica Mex. 2020;62:123. doi: 10.21149/11284. [DOI] [PubMed] [Google Scholar]
- 2.McArthur D.B. Emerging Infectious Diseases. Nurs. Clin. N. Am. 2019;54:297–311. doi: 10.1016/j.cnur.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heymann D.L., Brilliant L. Surveillance in eradication and elimination of infectious diseases: A progression through the years. Vaccine. 2011;29:D141–D144. doi: 10.1016/j.vaccine.2011.12.135. [DOI] [PubMed] [Google Scholar]
- 5.Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daughton C.G. Wastewater surveillance for population-wide COVID-19: The present and future. Sci. Total Environ. 2020;736:139631. doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logan L.K., Weinstein R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartsch S.M., McKinnell J.A., Mueller L.E., Miller L.G., Gohil S.K., Huang S.S., Lee B.Y. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin. Microbiol. Infect. 2017;23:48.e9–48.e16. doi: 10.1016/j.cmi.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention CRE Technical Information. [(accessed on 5 May 2020)]; Available online: https://www.cdc.gov/hai/organisms/cre/technical-info.html#Definition.
- 11.Chea N., Bulens S.N., Kongphet-Tran T., Lynfield R., Shaw K.M., Vagnone P.S., Kainer M.A., Muleta D.B., Wilson L., Vaeth E., et al. Improved Phenotype-Based Definition for Identifying Carbapenemase Producers among Carbapenem-Resistant Enterobacteriaceae. Emerg. Infect. Dis. 2015;21:1611–1616. doi: 10.3201/eid2109.150198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute . M100: Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2021. [Google Scholar]
- 13.Bonardi S., Pitino R. Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: A challenge for human health. Ital. J. Food Saf. 2019;8:7956. doi: 10.4081/ijfs.2019.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köck R., Daniels-Haardt I., Becker K., Mellmann A., Friedrich A.W., Mevius D., Schwarz S., Jurke A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect. 2018;24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Von Wintersdorff C.J.H., Penders J., Van Niekerk J.M., Mills N.D., Majumder S., Van Alphen L.B., Savelkoul P.H.M., Wolffs P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vera-Leiva A., Barría-Loaiza C., Carrasco-Anabalón S., Lima C., Aguayo-Reyes A., Domínguez M., Bello-Toledo H., González-Rocha G. KPC: Klebsiella pneumoniae carbapenemasa, principal carbapenemasa en enterobacterias. Rev. Chil. Infectol. 2017;34:476–484. doi: 10.4067/S0716-10182017000500476. [DOI] [PubMed] [Google Scholar]
- 17.López V.J.A., Echeverri T.L.M. K. pneumoniae: ¿la nueva “superbacteria”? Patogenicidad, epidemiología y mecanismos de resistencia. Iatreia. 2010;23:157–165. [Google Scholar]
- 18.Queenan A.M., Bush K. Carbapenemases: The Versatile β-Lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yigit H., Queenan A.M., Anderson G.J., Domenech-Sanchez A., Biddle J.W., Steward C.D., Alberti S., Bush K., Tenover F.C. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naas T., Nordmann P., Vedel G., Poyart C. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 2005;49:4423–4424. doi: 10.1128/AAC.49.10.4423-4424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villegas M.V., Lolans K., Correa A., Suarez C.J., Lopez J.A., Vallejo M., Quinn J.P. First detection of the plasmid-mediated class a carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 2006;50:2880–2882. doi: 10.1128/AAC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de León Rosales S.P. Plan Universitario de Control de la Resistencia Antimicrobiana. Volume 53. Universidad Nacional Autónoma de México; Mexico City, Mexico: 2018. [Google Scholar]
- 23.Garza-González E., Franco-Cendejas R., Morfín-Otero R., Echaniz-Aviles G., Rojas-Larios F., Bocanegra-Ibarias P., Flores-Treviño S., Ponce-De-León A., Rodríguez-Noriega E., Alavez-Ramírez N., et al. The Evolution of Antimicrobial Resistance in Mexico during the Last Decade: Results from the INVIFAR Group. Microb. Drug Resist. 2020;26:1372–1382. doi: 10.1089/mdr.2019.0354. [DOI] [PubMed] [Google Scholar]
- 24.Garza-González E., Bocanegra-Ibarias P., Bobadilla-del-Valle M., Alfredo Ponce-de-León-Garduño L., Esteban-Kenel V., Silva-Sánchez J., Garza-Ramos U., Barrios-Camacho H., López-Jácome L.E., Colin-Castro C.A., et al. Drug resistance phenotypes and genotypes in Mexico in representative gram-negative species: Results from the infivar network. PLoS ONE. 2021;16:e0248614. doi: 10.1371/journal.pone.0248614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez-Zulueta P., Silva-Sánchez J., Barrios H., Reyes-Mar J., Vélez-Pérez F., Arroyo-Escalante S., Ochoa-Carrera L., Delgado-Sapien G., Morales-Espinoza M.D.R., Tamayo-Legorreta E., et al. First Outbreak of KPC-3-Producing Klebsiella pneumoniae (ST258) Clinical Isolates in a Mexican Medical Center. Antimicrob. Agents Chemother. 2013;57:4086–4088. doi: 10.1128/AAC.02530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam M.A., Islam M., Hasan R., Hossain M.I., Nabi A., Rahman M., Goessens W.H.F., Endtz H.P., Boehm A.B., Faruque S.M. Environmental Spread of New Delhi Metallo-β-Lactamase-1-Producing Multidrug-Resistant Bacteria in Dhaka, Bangladesh. Appl. Environ. Microbiol. 2017;83:e00793-17. doi: 10.1128/AEM.00793-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moges F., Endris M., Belyhun Y., Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Res. Notes. 2014;7:215. doi: 10.1186/1756-0500-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berendonk T.U., Manaia C.M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., Bürgmann H., Sørum H., Norström M., Pons M.-N., et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization (WHO) Food and Agricultura Organization of the United Nations (FAO) World Organisation or Animal Health (OIE) Technical Brief on Water, Sanitation, Hygiene and Wastewater Management to Prevent Infections and Reduce the Spread of Antimicrobial Resistance. WHO; Geneva, Switzerland: 2020. [Google Scholar]
- 31.Samanta A., Mahanti A., Chatterjee S., Joardar S.N., Bandyopadhyay S., Sar T.K., Mandal G.P., Dutta T.K., Samanta I. Pig farm environment as a source of beta-lactamase or AmpC-producing Klebsiella pneumoniae and Escherichia coli. Ann. Microbiol. 2018;68:781–791. doi: 10.1007/s13213-018-1387-2. [DOI] [Google Scholar]
- 32.Sharma M., Chetia P., Puzari M., Neog N., Borah A. Menace to the ultimate antimicrobials among common Enterobacteriaceae clinical isolates in part of North-East India. bioRxiv. 2019:610923. doi: 10.1101/610923. [DOI] [Google Scholar]
- 33.King T.L.B., Schmidt S., Essack S.Y. Antibiotic resistant Klebsiella spp. from a hospital, hospital effluents and wastewater treatment plants in the uMgungundlovu District, KwaZulu-Natal, South Africa. Sci. Total Environ. 2020;712:135550. doi: 10.1016/j.scitotenv.2019.135550. [DOI] [PubMed] [Google Scholar]
- 34.Ebomah K.E., Okoh A.I. Detection of carbapenem-resistance genes in Klebsiella species recovered from selected environmental niches in the Eastern Cape Province, South Africa. Antibiotics. 2020;9:425. doi: 10.3390/antibiotics9070425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peneş N.O., Muntean A.A., Moisoiu A., Muntean M.M., Chirca A., Bogdan M.A., Popa M.I. An overview of resistance profiles ESKAPE pathogens from 2010–2015 in a tertiary respiratory center in Romania. Rom. J. Morphol. Embryol. 2017;58:909–922. [PubMed] [Google Scholar]
- 36.Abderrahim A., Djahmi N., Pujol C., Nedjai S., Bentakouk M.C., Kirane-Gacemi D., Dekhil M., Sotto A., Lavigne J.P., Pantel A. First Case of NDM-1-Producing Klebsiella pneumoniae in Annaba University Hospital, Algeria. Microb. Drug Resist. 2017;23:895–900. doi: 10.1089/mdr.2016.0213. [DOI] [PubMed] [Google Scholar]
- 37.Al-Agamy M.H., Aljallal A., Radwan H.H., Shibl A.M. Characterization of carbapenemases, ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J. Infect. Public Health. 2018;11:64–68. doi: 10.1016/j.jiph.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Perilli M., Bottoni C., Pontieri E., Segatore B., Celenza G., Setacci D., Bellio P., Strom R., Amicosante G. Emergence of blaKPC-3-Tn4401a in Klebsiella pneumoniae ST512 in the municipal wastewater treatment plant and in the university hospital of a town in central Italy. J. Glob. Antimicrob. Resist. 2013;1:217–220. doi: 10.1016/j.jgar.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Obasi A., Nwachukwu S., Ugoji E., Kohler C., Göhler A., Balau V., Pfeifer Y., Steinmetz I. Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae from Pharmaceutical Wastewaters in South-Western Nigeria. Microb. Drug Resist. 2017;23:1013–1018. doi: 10.1089/mdr.2016.0269. [DOI] [PubMed] [Google Scholar]
- 40.Teban-Man A., Farkas A., Baricz A., Hegedus A., Szekeres E., Pârvu M., Coman C. Wastewaters, with or without hospital contribution, harbour MDR, carbapenemase-producing, but not hypervirulent Klebsiella pneumoniae. Antibiotics. 2021;10:361. doi: 10.3390/antibiotics10040361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo L., An J., Ma Y., Ye L., Luo Y., Tao C., Yang J. Nosocomial outbreak of oxa-48-producing Klebsiella pneumoniae in a Chinese hospital: Clonal transmission of st147 and st383. PLoS ONE. 2016;11:e0160754. doi: 10.1371/journal.pone.0160754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuzon G., Ouanich J., Gondret R., Naas T., Nordmann P. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob. Agents Chemother. 2011;55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nazik H., Ongen B., Ilktac M., Aydin S., Kuvat N., Sahin A., Yemisen M., Mete B., Durmus M.S., Balkan I.I., et al. Carbapenem resistance due to blaOXA-48 among ESBL-producing scherichia coli and Klebsiella pneumoniae isolates in a univesity hospital, Turkey. Southeast Asian J. Trop. Med. Public Health. 2012;43:1178–1185. [PubMed] [Google Scholar]
- 44.Shibl A., Al-Agamy M., Memish Z., Senok A., Khader S.A., Assiri A. The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2013;17:e1130–e1133. doi: 10.1016/j.ijid.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 45.Gibbon M.J., Couto N., David S., Barden R., Standerwick R., Jagadeesan K., Birkwood H., Dulyayangkul P., Avison M.B., Kannan A., et al. A high prevalence of blaOXA-48 in Klebsiella (Raoultella) ornithinolytica and related species in hospital wastewater in South West England. Microb. Genom. 2021;7:000509. doi: 10.1099/mgen.0.000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheppard A.E., Stoesser N., Wilson D.J., Sebra R., Kasarskis A., Anson L.W., Giess A., Pankhurst L.J., Vaughan A., Grim C.J., et al. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob. Agents Chemother. 2016;60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoesser N., Sheppard A.E., Peirano G., Anson L.W., Pankhurst L., Sebra R., Phan H.T.T., Kasarskis A., Mathers A.J., Peto T.E.A., et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci. Rep. 2017;7:5917. doi: 10.1038/s41598-017-06256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mesquita E., Ribeiro R., Silva C.J.C., Alves R., Baptista R., Condinho S., Rosa M.J., Perdigão J., Caneiras C., Duarte A. An update on wastewater multi-resistant bacteria: Identification of clinical pathogens such as Escherichia coli o25b:H4-b2-st131-producing ctx-m-15 esbl and kpc-3 carbapenemase-producing Klebsiella oxytoca. Microorganisms. 2021;9:576. doi: 10.3390/microorganisms9030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Oliveira D.V., Nunes L.S., Barth A.L., Van Der Sand S.T. Genetic Background of β-Lactamases in Enterobacteriaceae Isolates from Environmental Samples. Microb. Ecol. 2017;74:599–607. doi: 10.1007/s00248-017-0970-6. [DOI] [PubMed] [Google Scholar]
- 50.Daoud Z., Farah J., Sokhn E.S., El Kfoury K., Dahdouh E., Masri K., Afif C., Abdel-Massih R.M., Matar G.M. Multidrug-Resistant Enterobacteriaceae in Lebanese Hospital Wastewater: Implication in the One Health Concept. Microb. Drug Resist. 2018;24:166–174. doi: 10.1089/mdr.2017.0090. [DOI] [PubMed] [Google Scholar]
- 51.Shahcheraghi F., Aslani M.M., Mahmoudi H., Karimitabar Z., Solgi H., Bahador A., Alikhani M.Y. Molecular study of carbapenemase genes in clinical isolates of Enterobacteriaceae resistant to carbapenems and determining their clonal relationship using pulsed-field gel electrophoresis. J. Med. Microbiol. 2017;66:570–576. doi: 10.1099/jmm.0.000467. [DOI] [PubMed] [Google Scholar]
- 52.Hashemizadeh Z., Hosseinzadeh Z., Azimzadeh N., Motamedifar M. Dissemination pattern of multidrug resistant carbapenemase producing Klebsiella pneumoniae isolates using pulsed-field gel electrophoresis in southwestern Iran. Infect. Drug Resist. 2020;13:921–929. doi: 10.2147/IDR.S227955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siddiqui M.T., Mondal A.H., Sultan I., Ali A., Haq Q.M.R. Co-occurrence of ESBLs and silver resistance determinants among bacterial isolates inhabiting polluted stretch of river Yamuna, India. Int. J. Environ. Sci. Technol. 2019;16:5611–5622. doi: 10.1007/s13762-018-1939-9. [DOI] [Google Scholar]
- 54.Siddiqui M.T., Mondal A.H., Gogry F.A., Husain F.M., Alsalme A., Haq Q.M.R. Plasmid-mediated ampicillin, quinolone, and heavy metal co-resistance among esbl-producing isolates from the Yamuna River, New Delhi, India. Antibiotics. 2020;9:826. doi: 10.3390/antibiotics9110826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gogry F.A., Siddiqui M.T., Haq Q.M.R. Emergence of mcr-1 conferred colistin resistance among bacterial isolates from urban sewage water in India. Environ. Sci. Pollut. Res. 2019;26:33715–33717. doi: 10.1007/s11356-019-06561-5. [DOI] [PubMed] [Google Scholar]
- 56.Aarestrup F.M., Woolhouse M.E.J. Using sewage for surveillance of antimicrobial resistance. Science. 2020;367:630–632. doi: 10.1126/science.aba3432. [DOI] [PubMed] [Google Scholar]
- 57.Dogonchi A.A., Ghaemi E.A., Ardebili A., Yazdansetad S., Pournajaf A. Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: A potential threat to clinical therapeutics. Tzu Chi Med. J. 2018;30:90–96. doi: 10.4103/tcmj.tcmj_101_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aubron C., Poirel L., Ash R.J., Nordmann P. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 2005;11:260–264. doi: 10.3201/eid1102.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bradford P.A., Bratu S., Urban C., Visalli M., Mariano N., Landman D., Rahal J.J., Brooks S., Cebular S., Quale J. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 2004;39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 60.Poirel L., Héritier C., Tolün V., Nordmann P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han H., Zhou H., Li H., Gao Y., Lu Z., Hu K., Xu B. Optimization of pulse-field gel electrophoresis for subtyping of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health. 2013;10:2720–2731. doi: 10.3390/ijerph10072720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.