Significance
Prenatal exposure to maternal infection increases the risk of developing mental health disorders, such as schizophrenia and autism spectrum disorder. Exposure to maternal immune activation has been associated with a number of neuroanatomical deficits in adolescent and adult offspring, with differing effects based on the gestational timing of infection. However, little is known about how the embryo brain is affected. We show, using whole-brain MRI, that maternal immune activation significantly affects brain anatomy. When the exposure occurs early in pregnancy, volume reductions are mainly observed, while the opposite is true for exposure later in pregnancy. Furthermore, we identify alterations to the density of certain classes of neurons and glia, which have been associated with stress and inflammation in the brain.
Keywords: maternal immune activation, embryo brain development, structural MRI, electron microscopy, brain development
Abstract
Exposure to maternal immune activation (MIA) in utero is a risk factor for neurodevelopmental and psychiatric disorders. MIA-induced deficits in adolescent and adult offspring have been well characterized; however, less is known about the effects of MIA exposure on embryo development. To address this gap, we performed high-resolution ex vivo MRI to investigate the effects of early (gestational day [GD]9) and late (GD17) MIA exposure on embryo (GD18) brain structure. We identify striking neuroanatomical changes in the embryo brain, particularly in the late-exposed offspring. We further examined the putative neuroanatomical underpinnings of MIA timing in the hippocampus using electron microscopy and identified differential effects due to MIA timing. An increase in apoptotic cell density was observed in the GD9-exposed offspring, while an increase in the density of neurons and glia with ultrastructural features reflective of increased neuroinflammation and oxidative stress was observed in GD17-exposed offspring, particularly in females. Overall, our findings integrate imaging techniques across different scales to identify differential impact of MIA timing on the earliest stages of neurodevelopment.
1. Introduction
Brain development is a remarkable and complex set of processes under the organizational control of genetic, environmental, and immune regulation. The tightly regulated nature and interdependence of these processes make them vulnerable to a variety of risk factors. Converging lines of evidence suggest an association between prenatal exposure to maternal infection and increased risk for a host of neurodevelopmental disorders in offspring, including schizophrenia and autism spectrum disorder (ASD) (1–3). Indeed, exposure to maternal immune activation (MIA) in animal models has been shown to induce neuroanatomical and behavioral changes relevant to many neurodevelopmental disorders (4). MIA leads to an increase in maternal proinflammatory cytokines and chemokines, which are thought to interfere with fetal brain development by disturbing its delicate ecosystem, potentially as a consequence of the microglial response (5–9). Exposure to MIA during the sensitive window of in utero brain development may alter neurodevelopmental trajectories, thereby increasing risk for neuropsychiatric disorders later in life (10, 11). Identifying these sensitive windows and their impact on later development is critical to our understanding of the effects of MIA exposure. Previous work from our group has demonstrated that the gestational timing of MIA exposure has a differential impact on offspring brain and behavioral development, with a greater variation observed following early MIA exposure (gestational day [GD]9) relative to exposure later in gestation (GD17) or to saline exposure (11). These differences may be attributable to variation in maternal immune responsiveness and fetal brain development at different gestational timings and to MIA-induced neurodevelopmental variation throughout development (12).
Although there is significant evidence that MIA exposure in utero alters brain-development trajectories in both human (13) and animal models (14, 15), it is unclear how soon after MIA exposure these changes can be detected. MIA-induced outcomes have been better characterized in adolescent and adult rodent offspring (12). However, to better understand the initiation and progression of MIA-induced variation in brain development, it is critical to study its impact at the earliest stages of life. Human neuroimaging studies have identified alterations in functional and structural connectivity in the infant brain following exposure to chronic, low-grade inflammation (as measured by interleukin-6 levels and/or C-reactive protein in the maternal plasma) (16–18). Even though some observations in early phases of life have been made, there is less information available regarding how MIA exposure impacts the morphogenesis of the fetus. Recent work on this topic suggests that MIA exposure induces acute up-regulation of genes involved in immune signaling, hypoxia, and angiogenesis in the fetal mouse brain (19). Further, alterations in neuronal proliferation, neuronal and glial specification, cortical lamination (19, 20), global messenger RNA translation, and altered nascent proteome synthesis have been reported (21).
These findings suggest that effects of MIA exposure may be detectable in the fetal and neonatal period across mouse and human studies. However, it is unclear whether the transcriptional and histological variation observed in rodents translates to the neuroanatomical changes detected by whole-brain imaging observed in human studies. Furthermore, although gestational timing has been shown to have differential effects in adolescent and adult offspring (11, 22), it is unclear how it affects neurodevelopment in its early phase. A better understanding of the neurodevelopmental sequelae of MIA exposure on very early brain development is of importance in the context of the current COVID-19 pandemic, as mothers who contracted the virus during pregnancy were more likely to have obstetric complications leading to poor fetal health outcomes that may impact downstream brain development, such as low birth weight, intrauterine growth restriction, and preterm birth (23, 24).
To build upon our previous investigations, in which we characterized brain development from adolescence to adulthood (11), we aimed to develop a chronology of how the timing of MIA exposure may impact brain development in utero, using the same gestational exposures as in our previous work. We leveraged structural MRI, an inherently three-dimensional (3D) imaging technique applicable for mouse phenotyping (25). This technique allows for a comparable assay across species, providing a potential avenue for establishing cross-species homology (26). We examine the effects of in utero exposure to early (GD9) or late (GD17) MIA with a viral mimetic, polyinosinic:polycytidylic acid (poly I:C), on embryo brain morphology at GD18 using high-resolution ex vivo whole-brain MRI. To better understand the cellular underpinnings of the volumetric changes identified by MRI, we utilized high-resolution electron microscopy (EM) to examine the density of certain cells, including apoptotic cells, dark neurons, and dark glial cells. These dark cells are unique from other neurons and glia, as they have distinct ultrastructural characteristics reflective of oxidative stress and are only identifiable with EM (27). We focused on dark and apoptotic cells, as they have been identified as a putative marker for neuroinflammation, cellular stress, apoptosis, and disease in the brain parenchyma (27–29). The dorsal hippocampus was selected as the region of interest, as it was differentially affected by GD9 and GD17 MIA timing. Further, alterations in this region have been consistently associated with neuropsychiatric disorders (30, 31), as well as in response to MIA exposure in our previous work (11) and that of others (32). Our results demonstrate neuroanatomical alterations in the GD18 embryo brain following MIA, with differential effects due to timing in many regions, including the dentate gyrus of the hippocampus. Here, we observed a significant increase in the density of apoptotic cells in the GD9-exposed embryos (but not GD17) relative to the control group, while an increase in the density of dark cells (both neurons and glia) was observed in the GD17-exposed offspring. These findings suggest that morphological changes due to MIA exposure are already detectable in the fetal brain and that the timing of MIA exposure may differentially impact the brain both at anatomical and cellular levels.
2. Results
2.1. Differential Effects due to MIA Timing on Embryo Brain Volume.
To investigate the effects of early or late prenatal MIA exposure on embryo brain development, pregnant dams were exposed to either poly I:C (P1530-25MG poly I:C sodium salt TLR ligand tested; Sigma-Aldrich) (5 mg/kg, intraperitoneally [i.p.]) or saline solution (NaCl) at GD9 or GD17 (Section 4.1). Embryos from each of these four groups were harvested at GD18 and fixed in order to acquire ex vivo high-resolution structural MRIs at the same timepoint (Section 4.2). Once we confirmed that there were no statistically significant differences between our two control groups, they were combined, leaving us with three treatment groups: saline (SAL), GD9-poly I:C (POL E), and GD17-poly I:C (POL L). We used deformation morphometry to create a consensus average of the group, and the Jacobian determinants were analyzed by using a mixed-effects model to examine relative volumetric differences between groups.
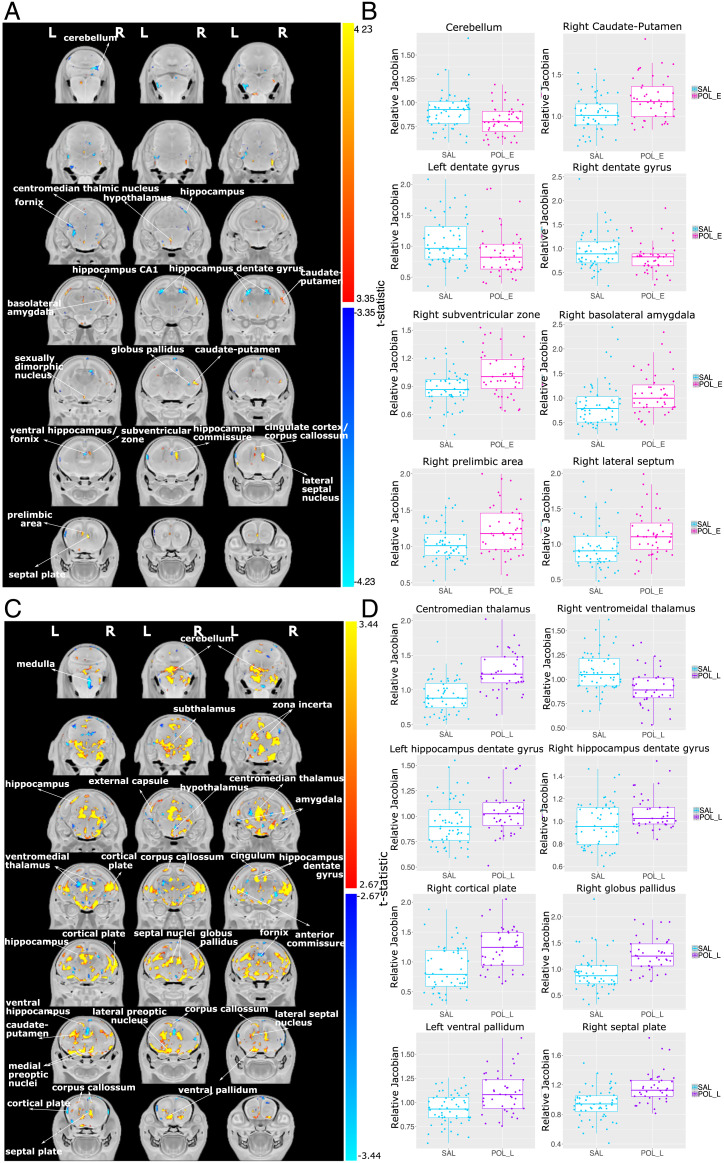
Overall, we observed differing patterns of whole-brain morphology dependent on MIA timing; decreases in brain volume were more prevalent following GD9 exposure, while striking brain-volume increases were observed following GD17 exposure. We observed significant neuroanatomical differences due to MIA timing. The POL E embryos exhibited smaller volumes relative to SAL in the globus pallidus, hippocampus (including the dentate gyrus, as well as more posterior regions), fornix, centromedian thalamic nucleus, and cerebellum. Larger volume was observed in the prelimbic area, lateral septum, subventricular zone, caudate-putamen, sexually dimorphic nucleus of the hypothalamus, basolateral amygdala, and CA1 region of the hippocampus in the POL E group relative to SAL (t = 4.242, <1% false discovery rate [FDR]) (Fig. 1).
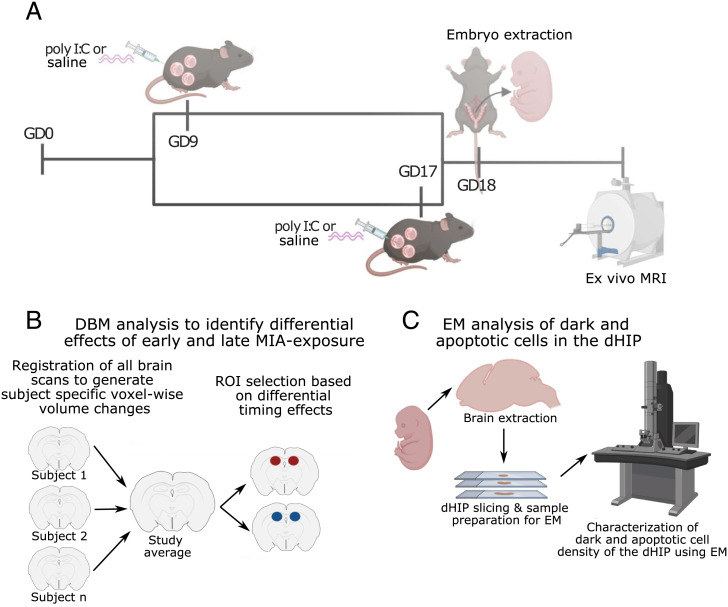
Fig. 1.
Experimental timeline. (A) Pregnant dams were injected (i.p.) with poly I:C (5 mg/kg) or vehicle (0.9% sterile NaCl) solution on GD9 or GD17. On GD18, pregnant dams were euthanized, and embryos were extracted and prepared for high-resolution ex vivo MRI. (B) Analysis flow of deformation-based morphometry analysis used to detect voxel-wise brain-volume differences due to early or late MIA exposure. The dorsal hippocampus was selected as a region of interest (ROI) for cellular investigation using EM due to differential effects of timing on bilateral volume. (C) Embryo brains were extracted from scanned samples, sliced, and prepared for EM investigation of dark glial cell, dark neuron, and apoptotic cell density. DBM, deformation-based morphometry; dHIP, dorsal hippocampus. Figure was made by using biorender (https://biorender.com).
GD17 MIA exposure induced very striking volumetric alterations, particularly volumetric increases in the brain of POL L offspring at GD18 (t = 3.234, <1% FDR) relative to SAL offspring. Regions of volume increase included the ventral pallidum, septal plate and lateral septal nucleus, medial and lateral preoptic nuclei, caudate-putamen, globus pallidus, hippocampus (both dentate gyrus and CA1 regions), cingulum, anterior commissure, cortical plate, corpus callosum, external capsule, centromedian thalamus, and cerebellum. Decreases in volume were observed in the ventral hippocampus, more anterior subregions of the cortical plate, bilateral amygdala, fornix, and ventromedial thalamus (Fig. 1).
Interestingly, POL E and POL L MIA exposures were observed to have opposite effects on brain volume in some regions implicated in neurodevelopmental disorders and identified in previous MIA studies (11, 14), such as the dorsal hippocampus, wherein GD9 exposure decreased volume and GD17 exposure increased volume. Similar observations were made for the centromedian thalamic nucleus. The septal nucleus and caudate-putamen were increased in both MIA-exposed groups. A significant difference between POL E and POL L embryo brain anatomy was also observed (t = 3.590, <1% FDR). These results are fully described in SI Appendix, section 2.2.1 and Fig. 4. Post hoc investigation of sex differences revealed no significant sex-by-group interactions.
2.2. EM of Embryo Dorsal Hippocampus.
Given the differential effects of MIA timing on the dorsal hippocampus, and that this region has been shown to be affected by MIA exposure in our own work (11) and in various neuropsychiatric disorders (31), it was selected for EM investigation of cell density. Specifically, we focused on apoptotic cells, dark neurons, and dark glia, defined by ultrastructural properties only identifiable with EM and indicative of potential oxidative stress and neuroinflammation in the brain parenchyma (27–29); this may suggest a specific vulnerability of these cells during this early neurodevelopmental time frame.
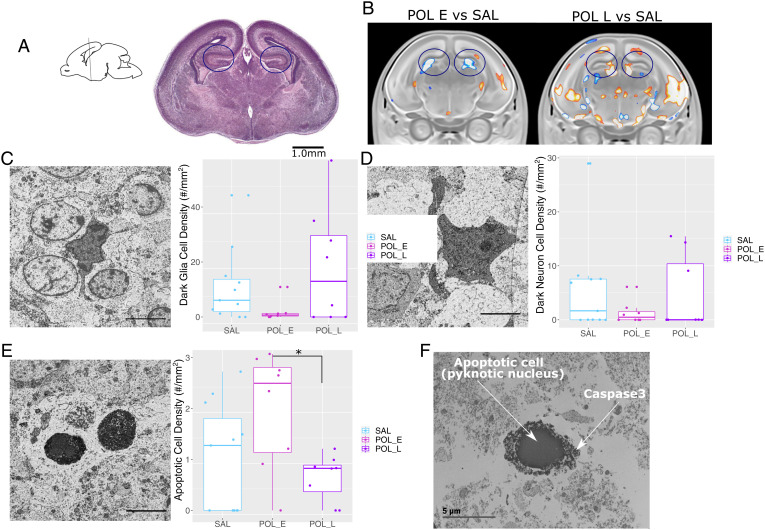
In keeping with norms in the field (due to sample size and distribution) (33), rather than using parametric statistics, we chose to use a nonparametric Kruskall–Wallis test, followed by a pairwise Wilcoxon-rank test to assess group differences in the average density of dark glial, dark neuronal, and apoptotic cells across slices (i.e., average per embryo). For aggregate density measure per group, there were no group differences in total cell density (χ2 = 0.680, df = 2, P = 0.712; SI Appendix, Fig. 6). Although the density of dark glial and dark neuron cells appeared to be higher in the POL L offspring, there were no differences in dark glial cell density (χ2 = 3.322, df = 2, P = 0.190) or dark neuron cell density (χ2 = 0.758, df = 2, P = 0.685). A significant group effect was observed for apoptotic cell density (χ2 = 6.349, df = 2, P = 0.042), wherein the POL E offspring appeared to have greater density than the POL L offspring (P = 0.053), as well as the SAL offspring, although not significantly so (P = 0.117) (Fig. 2; see SI Appendix, Fig. 7 for more representative images of dark glia, dark neurons, and apoptotic cells).
Fig. 2.
Neuroanatomical changes in the GD18 embryo brain following GD9 MIA or GD17 MIA exposure. (A) A t-statistic map of group (POL E vs. SAL) thresholded at 5% (Bottom; t = 3.35) and 1% FDR (Top; t = 4.23) overlaid on the study average. (B) Boxplots of peak voxels (voxels within a region of volume change showing largest effect) selected from regions of interest highlighted in white text in A. For all boxplots, the relative Jacobian determinants are plotted on the y axis. Here, a value of one means the voxel is no different from the average; anything above one is relatively larger, and below one is relatively smaller. For all boxplots, the midline represents the median of the data, the box represents the first and third quartiles, and the vertical lines represent 1.5× interquartile range of the data. Dots on the plot represent individual data points for each subject. (C) A t-statistic map of the group (POL L vs. SAL) thresholded at 5% (Bottom; t = 2.67) and 1% FDR (Top; t = 3.44) overlaid on the study average. (D) Boxplots of peak voxels (voxels within a region of volume change showing largest effect) selected from regions of interest highlighted in white text in C. For all boxplots, the relative Jacobian determinants are plotted on the y axis as in B.
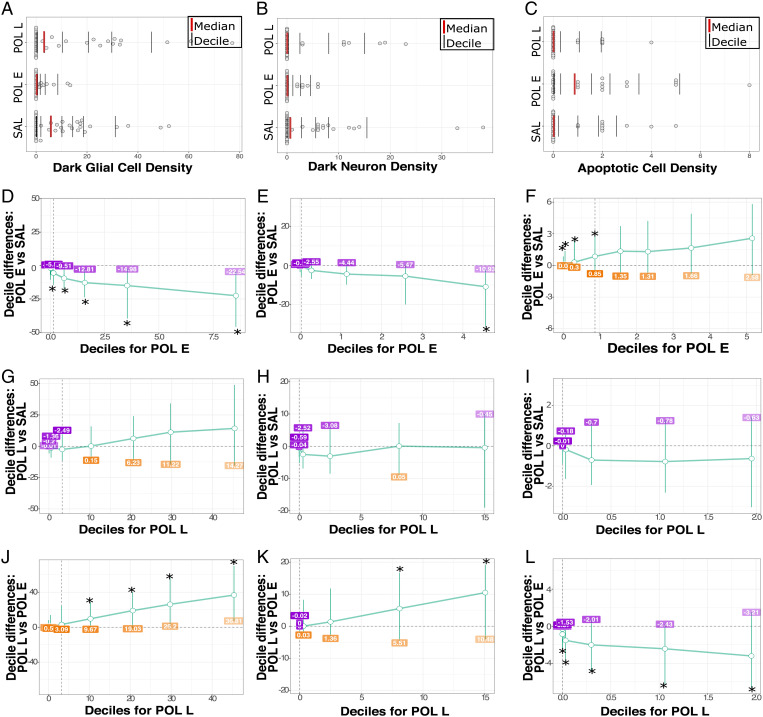
Given that the variance in cell density differed quite drastically between groups, we sought to examine if the distributions in cell density differed between groups using the most granular data we had. Rather than using the pooled data per mouse (as above), we applied the shift function to the density measures acquired from each slice per mouse (3) to maximize variance. The shift function (34) quantifies how two distributions differ based on deciles of the distributions, i.e., it describes how one distribution should be transformed to match the other and estimates how and by how much one distribution must be shifted.
Comparison of cell-density distributions across groups confirmed that there were no overall differences in total cell density between groups (apart from a significant difference between POL E and SAL distributions only at the seventh decile of distribution [P = 0.045]; SI Appendix, Tables 2–4 4).
Interestingly, dark glial density was significantly lower for POL E offspring relative to SAL at higher deciles of the distribution (5th through 9th decile, P < 0.020; SI Appendix, Table 5; Fig. 3). Distribution for POL L offspring was not different from SAL; however, they also had significantly more dark glia than POL E at higher deciles of the distribution (6th through 9th decile, P < 0.040; SI Appendix, Tables 6 and 7), suggesting that POL L have higher dark glial cell density than POL E at higher deciles of the distribution. Comparison of distributions for dark neurons revealed only subtle differences, with significantly higher density at higher deciles of the distribution for POL E relative to SAL (decile 9, P = 0.016; SI Appendix, Table 8) and POL L (deciles 8 and 9, P < 0.010; SI Appendix, Table 10), with no differences between POL L and SAL (SI Appendix, Table 9; Fig. 3). Finally, for apoptotic cell density, POL E offspring had significantly higher density across lower deciles of distribution relative to SAL (deciles 1 to 4, P = 0.039; SI Appendix, Table 11) and across all deciles relative to POL L (P < 0.020; SI Appendix, Table 12). No differences between POL L and SAL were observed (SI Appendix, Table 13; Fig. 3).
Fig. 3.
Differences in dark and apoptotic cell density with representative images captured using EM. (A) Sagittal slice orienting to the region of the hippocampus selected, with the corresponding brain slice stained with Cresyl Violet from the GD18 mouse brain atlas, coronal slice 14 (75). (Scale bar: 1 mm.) The region of interest is highlighted in the circles. (B) Representative slices of the hippocampus from the MRI results for POL E relative to SAL and POL L relative to SAL. The region of interest is highlighted in the circles. (C) Image acquired by scanning EM (25-nm resolution) in the dorsal hippocampus from representative offspring (equivalent to coronal slice 14 from A) highlighting dark glial cells. Boxplot showing dark glial cell density (per mouse) per group (n = 6 to 8 per group). (D) Representative dark neuron image with boxplot for dark neuron cell density per mouse. (E) Representative apoptotic cell image with boxplot for apoptotic cell density per mouse. (F) Representative apoptotic cell (with pyknotic nucleus), with positive active Caspase 3 staining surrounding the nucleus (black matter surrounding the nucleus). (Scale bars: 5 µm.) *P = 0.053.
Sex differences in distribution were also observed for all cell types (total, dark glia, dark neurons, and apoptotic cells). Of interest, increased density of dark glial cells was observed in the POL L females relative to SAL, while decreased density was observed for the POL E females, further described in SI Appendix, section 2.3.2 and Fig. 8, with the summary of results per decile provided in SI Appendix, Tables 14–37.
Finally, using immunocytochemical transmission EM, we confirmed that the cells we identified to be apoptotic [those with a pyknotic nucleus (28, 35–37)] also had positive active Caspase 3 staining (Fig. 2), given the role of active Caspase 3 in morphological changes and DNA fragmentation leading to apoptosis (38). Furthermore, we sought to determine whether the dark glial cells identified were colocalized with markers of active microglia using CD11b immunostaining, previously shown to be present in adult contexts of pathology (27). However, despite multiple efforts, no positive staining was identified with transmission EM, potentially due to the difficulty in establishing an EM-compatible immunostaining protocol.
3. Discussion
There is a well-established link between MIA exposure in utero and latent neuroanatomical and behavioral abnormalities that emerge in adolescence or adulthood, with relevance to schizophrenia and ASD pathology (39–41). However, limited work has been conducted in the early neurodevelopmental period (12). We leveraged high-resolution ex vivo MRI and EM to characterize the effects of MIA exposure at two gestational timepoints on the embryo brain at GD18. Our results suggest that the embryo mouse brain undergoes significant remodeling in response to MIA, particularly due to late gestational exposure, coupled with changes in the presence of dark and apoptotic cells in the hippocampus. Elucidating the neurodevelopmental changes across the embryonic periods following MIA exposure is an important step toward our understanding of MIA exposure as a primer of downstream psychopathology and as a risk factor for an array of neuropsychiatric disorders.
Interestingly, we see volume reductions due to early MIA exposure in several brain regions, whereas we see striking volume expansions following late exposure. Since the late-exposed embryo brains were harvested 24 h after immune exposure, we are likely capturing an acute neuroinflammatory or stress response, or an acceleration of brain development in response to the immune stimulus. However, given that there was no difference based on SAL timing, one can assume that any acute effects are attributable to the MIA itself. Interestingly, there is homology between regions affected in the embryo brains and those in which we observed altered neurodevelopmental trajectories from childhood to adulthood in our published work (11). Some of these regions include the striatum/caudate-putamen, hippocampus, lateral septum, cingulate cortex, and cerebellum, many of which have been implicated in neuroimaging studies of humans with schizophrenia or ASD (30, 42, 43). Previous animal studies also report that MIA in late gestation increases neuroinflammation in the embryo rat brain and decreases placental function, as measured by T2-signal intensity (44). These findings suggest that increased neuroinflammation and decreased placental function could, in part, be driving some of the volumetric increases in the late exposed embryo brain, which may provide some mechanisms underlying disease pathology.
To gain more insight into the putative cellular underpinnings of the volumetric changes, we performed EM experiments in the dorsal hippocampus, a region highly implicated in neurodevelopmental pathology (31). In addition to the current findings, we previously observed interesting transcriptional changes to genes involved in early neurodevelopmental processes in the dorsal hippocampus of adolescent MIA-exposed mice—although following a different gestational exposure (11). A similar study conducted in nonhuman primates also found that the largest transcriptional changes due to MIA exposure were found in the offspring hippocampus, relative to the prefrontal and anterior cingulate cortex (45), in line with our observations in the mouse (11). Alterations to hippocampal morphology, both at the gross-anatomical and molecular level, and function have been implicated in numerous MIA studies (46, 47). Importantly, dysfunction of this region has also been proposed as a central player in the pathophysiology of schizophrenia, as well as other neuropsychiatric illnesses, wherein dysfunction begins during subclinical stages and worsens as symptoms progress (31). Of course, other brain regions, which may or may not be connected to the hippocampus, play a role as well (30, 42, 43).
Identifying the cellular processes triggered by MIA exposure is critical to our understanding of how this risk factor may alter offspring neurodevelopmental trajectories. By leveraging high-resolution EM techniques, we have an unprecedented opportunity to investigate the brain parenchyma at nanoscale resolution (28). This allows for identification of changes in different cell types and their unique features. We identified differential effects due to gestational MIA timing. In GD9-exposed offspring, where decreased dorsal hippocampal volume was detected, we observed an increase in apoptotic cell density (confirmed via Caspase 3 staining) and a decrease in dark glial cell density compared to SAL offspring. Conversely, in GD17-exposed offspring, which had enlarged dorsal hippocampal volumes, there was a tendency for greater dark-cell density (although not significantly), particularly for dark glial cells in the late-exposed females. These cellular results align well with the volume-based MRI results, wherein increased apoptosis could be linked to the decreased volume in GD9-exposed offspring, while increased dark-cell density could be linked to an acute inflammatory response and increased volume in the GD17-exposed offspring. Importantly, this may point to differences in neuropathological mechanisms in the fetal brain associated with MIA exposure and to some putative sex differences that require further investigation.
We focused our analyses on dark cells, both neurons and glia, as well as apoptotic cells, as these have been frequently detected in response to stress (29), aging (48), and neurodegeneration (49). They are thought to play a role in both normal and pathological synapse and neuronal network formation (49). Dark neurons are typically defined by a darker appearance, with an accumulation of mitochondria and nuclear indentations, associated with structural plasticity (49–51), as well as markers of cellular stress (dilated endoplasmic reticulum and Golgi apparatus) (28). Dark glial cells, particularly microglia, also display cellular markers of stress and have been shown to have hyperramified processes, which often lead to increased physiologically relevant contents, such as synaptic contacts and increased phagocytic capacity (27, 52). Further, reports of microglial reactivity and density in the brains of prenatally immune-challenged animals early in life are mixed, with observations of increased density and motility, as well as no differences (53). Although we were not able to identify colocalization between dark glial and CD11b staining, this does not rule out the possibility that these cells are, in fact, dark microglia, but it necessitates future, cell-specific research. By focusing specifically on dark microglial cells in the future, we may gain better insight into the phenotypic variability of these reactive cells and parse some of the heterogeneity in the literature.
At GD9, corresponding with our first MIA exposure, microglia colonize the brain, initiating their development toward maturity. Interestingly, sex differences in the number and morphology of microglia have been observed, with males showing greater numbers of these cells earlier in development (postnatal day [PND]4) than females, who have more microglia later in development (PND30 to PND60) (54). GD9 also occurs at a time at which the fetal brain is undergoing extensive neural proliferation and migration, which transitions more toward circuit refinement and cortical organization by GD17 (our late-MIA timepoint) (55). Importantly, microglia may play important roles in the regulation of apoptosis (56), as well as the permeability and formation of the blood–brain barrier, which typically takes shape between GD13.5 and GD15.5 in the mouse. Apoptosis is a critical cellular process in early brain and placental development in utero (57). The process of apoptosis has been detected as early as GD5, but increases significantly toward the end of gestation, peaking in early postnatal life before dropping off (58). In contrast with our findings, previous rodent studies using immunohistochemical techniques have actually observed an increase in apoptosis following lipopolysaccharide exposure in late pregnancy in rats (59) and poly I:C exposure at GD17, but not GD9, in mice (22). The discrepancy between these findings and ours may be due to differences in techniques or features used to identify these cells; immunohistochemistry may be less sensitive than EM to detection of ongoing apoptosis.
Exposure to poly I:C at the dose selected (5 mg/kg) typically elicits an acute immune response peaking 3 to 4 h postinjection, resolving, typically, within 24 to 48 h (60). We chose this dose and administration to model a more acute inflammatory exposure and to match our previous work (11, 61). Some of the differences in MIA-exposed offspring may be due to differences in developmental stage of the fetus (discussed in greater detail below) or to the maternal milieu. The maternal immune and endocrine systems undergo numerous changes during gestation. During early pregnancy (corresponding with GD9 exposure), the mother is in a more proinflammatory state due to implantation and placentation, the need to repair uterine epithelium and cycle between cell death and repair. In contrast, the later phase of pregnancy (other than immediately preceding labor and delivery) is characterized by a more anti-inflammatory state, wherein the mother, placenta, and fetus have achieved a symbiotic relationship (62, 63). These differences in maternal environment may influence the effects observed by MIA challenge and may, in part, explain some of the differences in offspring effects observed in our study.
The results presented here should be considered in light of their limitations. The design of our embryo study would be more complete with an assessment of neuroanatomy acutely following the GD9 exposure at GD10; this would allow us to detect whether volume increases, as those detected in the GD17-exposed offspring, are a response to acute inflammation or specific to that gestational time point. Additionally, examining the brain of GD17-exposed offspring 9 d following MIA exposure, at PND8, could provide a similar delay after MIA before collection for imaging; however, the comparison of embryonic vs. postnatal brain development may have its own confounds. Unfortunately, the embryo is too small for MRI acquisition at GD9 or GD10; however, imaging could be performed by using other techniques, such as optical projection tomography (64). Regarding the EM analysis, we did not look at the extracellular space, which could contribute to the changes in volume detected in the MRI. In future work, cryofixation methods for preserving the EM samples would allow for more in-depth analyses of the extracellular space volume and composition (65).
The cytokine response elicited by our first and second batches of poly I:C differed, in that the second batch (to which a small subset of samples was exposed) did not elicit as strong of an immune response, particularly for the GD17 offspring. Although both batches of poly I:C were used within the 1-y mark for sample collection, the immunostimulatory potential of our second batch was tested when the product was older (2 y) than the first batch (1 y), leading to potential degradation of the material when immunostimulatory potential was tested in a satellite group of animals. An alternative approach to using satellite animals would be to measure cytokine response to poly I:C challenge in the experimental dams; this would allow for a more nuanced investigation of the relationship between the strength of the immune response in the mother and the degree of alteration observed in the offspring. Recent work by Mueller et al. (66) has shown that there may be resilience of susceptibility to MIA, depending on the magnitude of maternal inflammatory response. We opted for the use of satellite animals, as we were concerned about introducing an additional intervention and stressor to the experimental dams. Important work from the human neuroimaging literature has shown that effects of maternal stress can be detected at the structural level in the human neonatal brain, both across structure and function. Thus, we wanted to ensure that we were isolating the effects of MIA without an additional confound of stress (67–71). Here, we provide evidence for the presence of neuroanatomical alterations in the embryo brain, differentially affected by timing; however, in future work, it would be of high interest to associate the relationship between maternal cytokine responsiveness and offspring outcomes as a means to parse some of the heterogeneity observed. In addition to investigating the maternal immune response as a variance modulator, investigating the variance within and between litters is of high interest. While we do include litter as a random intercept in our mixed-effects modeling, investigating this more closely may be of high interest for future work.
We comprehensively examined the effects of prenatal MIA exposure, a known risk factor for neuropsychiatric disorders, at two gestational timepoints on embryo brain anatomy at the gross morphological and cellular levels. We identified striking neuroanatomical remodeling in the embryo brain, particularly following exposure in late gestation; we also observed sex-dependent alterations in the density of dark neuronal and glial cells in the dorsal hippocampus, with greater cell density in female offspring. This may reflect the initiation of pathological circuit remodeling. These findings show that MIA exposure induces striking neurodevelopmental changes in embryonic development, which may further our understanding of how this risk factor increases the likelihood of developing neuropsychiatric illnesses later in life.
4. Materials and Methods
4.1. Animals, Prenatal Immune Activation, and Sample Preparation.
C57BL/6J female and male mice of breeding age (8 to 12 wk old) were subject to timed mating procedures (described in SI Appendix, section 1.1) to generate pregnant dams. Pregnant dams were randomly assigned to one of four treatment groups (see Fig. 4 for experimental design): 1) poly I:C (P1530-25MG poly I:C sodium salt TLR ligand tested; Sigma-Aldrich) (5 mg/kg, i.p.) at GD9 (POL E; seven embryo dams); 2) 0.9% sterile NaCl solution (saline) at GD9 (SAL E; six embryo dams); 3) poly I:C at GD17 (5 mg/kg, i.p.) (POL L; seven embryo dams); or 4) saline at GD17 (SAL L; four embryo dams). Additionally, immunostimulatory potential of poly I:C was confirmed in a separate group of dams (see SI Appendix, section 1.1 for methods and SI Appendix, section 2.1, Table 1, and Figs. 1–3 for results).
Fig. 4.
Differences in distribution of dark glial, dark neuron, and apoptotic cell density per group. Distribution of dark glial cell density (A), dark neuron density (B), and apoptotic cell density (C) for all hippocampal slices per animal. The red line identifies the median of the data, while each black bar denotes a decile of distribution. A percentile bootstrapping technique applied to identify the difference in decile between the POL E and SAL for dark glial cell density (D), dark neuron density (E), and apoptotic cell density (F) showing decreased density of dark glia and increased density of apoptotic cells for POL E offspring. Next, POL L and SAL comparisons are shown for dark glia (G), dark neurons (H), and apoptotic cells (I). Finally, POL E vs. POL L density is shown for dark glia (J), dark neurons (K), and apoptotic cells (L), showing that POL L had higher density than POL E for dark glia and neurons, while POL L has lower apoptotic cell density. *P < 0.05.
4.1.1. Sample preparation for MRI.
On GD18, pregnant dams were euthanized; embryos were extracted and postfixed in 4% paraformaldehyde with 2% gadolinium (MRI contrast agent; Bracco Imaging S.p.A) in phosphate-buffered saline (PBS) for 1 wk. A piece of the yolk sac was collected for genotyping of each embryo to identify the sex of the mouse via presence of the SRY gene (performed by Transnetyx). Collections were performed in two separate cohorts (with two different poly I:C batches from the same supplier outlined in SI Appendix, section 1.2 and Table 1).
Table 1.
Final sample size for embryo MRI data following quality control
| Group | Males MRI(cohort 1/cohort 2) | Females MRI(cohort 1/cohort 2) | Litters(cohort 1/cohort 2) |
|---|---|---|---|
| SAL E | 14 (13/1) | 15 (13/2) | 7 (6/1) |
| SAL L | 12 (10/2) | 12 (6/6) | 4 (2/2) |
| POL E | 11 (6/5) | 17 (9/8) | 7 (4/3) |
| POL L | 14 (11/3) | 17 (15/2) | 7 (6/1) |
Collection was performed in two rounds with two different batches of poly I:C (same supplier and product). Number indicates total sample size following quality control. The number of samples coming from each collection cohort is noted in parentheses.
4.2. Magnetic Resonance Image Acquisition and Processing.
All samples were shipped to the Mouse Imaging Centre (Toronto) for scanning. A multichannel 7.0-T MRI scanner with a 40-cm-diameter bore (Varian Inc.) was used to acquire anatomical images of the entire embryo (whole body). A custom-built 16-coil solenoid array was used to acquire 40-μm3-resolution images from 16 samples concurrently (72) (see SI Appendix, section 1.3.1 for details).
Preprocessed embryo brain images (https://github.com/CoBrALab/documentation/wiki/Embryo-scan-preprocessing) of all subjects in the study were aligned by unbiased deformation-based morphometry using the antsMultivariateTemplateConstruction2.sh tool (https://github.com/CoBrALab/twolevel_ants_dbm) (73). The output of this iterative group-wise registration procedure generates a group average from all the scans in the study, as well as the minimum deformation field that is required to accurately map each individual subject to the average at the voxel level (see 3D representation of group average in SI Appendix, Fig. 5). Relative Jacobian determinants (74), which explicitly model only the nonlinear deformations and remove global linear transformation (attributable to differences in total brain size), were blurred by using a Gaussian kernel at 160-µm full-width-at-half-maximum to better conform to Gaussian assumptions for downstream statistical testing (see SI Appendix, section 1.3.2 for details).
4.3. EM.
After MRI scanning, embryo brains (SAL E, n = 4 [two males/two females]; SAL L, n = 7 [three males/four females]; POL E, n = 8 [four males/four females]; and POL L, n = 8 [four males/four females]) were extracted from the fixed samples and further postfixed with 3.5% acrolein in phosphate buffer (100 mM) (pH 7.4) overnight at 4 °C. Postfixed brains were sectioned to 50-μm sagittal slices by using a VT1200S vibratome (Leica Biosystem) and stored at −20 °C in cryoprotectant (30% glycerol and 30% ethylene glycol in PBS [50 mM] [pH 7.4]). Three brain sections in which the dorsal hippocampus was present [coronal section 12-15 (75), roughly equivalent to lateral 2.0 to 2.8 mm (76)], were processed for EM by using osmium-thiocarbohydrazide-osmium postfixation (77) (SI Appendix, section 1.4.1). Samples were sectioned into ∼70- to 75-nm ultrathin sections by using an Ultracut UC7 ultramicrotome (Leica Biosystems). Three levels of section-rubans were collected at an interval of 10 µm, glued on a specimen mount with conductive carbon adhesive tabs (Electron Microscopy Sciences), and imaged by array tomography at 25-nm resolution with an acceleration voltage of 1.4 kV and current of 1.2 nA using a Zeiss Crossbeam 540 Gemini scanning EM (Zeiss) (three images per embryo).
Images from the all four treatment groups were analyzed blind to the experimental conditions by using QuPath (v0.2.0-m3) software (78). For each picture, region areas were traced and measured to calculate cell density. Cell type and apoptotic state were determined by ultrastructural features. Total cell numbers, dark cells (neuronal and glial cells), and apoptotic cells were then counted within the dorsal hippocampus (CA1, CA3, and dentate gyrus). The percentage of dark-cell population or apoptotic cell number was calculated as a ratio over the total cell population (details in SI Appendix, section 1.4.1).
Finally, immunocytochemical transmission EM was used to determine whether cells identified as apoptotic, those with a pyknotic nucleus, were also positive for active Caspase 3 staining. Additionally, to investigate whether dark cells identified were specifically microglia, immunocytochemical staining against CD11b was performed (see SI Appendix, section 1.4.2 for details).
4.4. Statistical Analyses.
4.4.1. Neuroimaging data analysis.
Statistical analyses were performed by using the R software package (R version 3.5.1, RMINC version 1.5.2.2; https://www.r-project.org/). To assess the effects of poly I:C exposure at different gestational timepoints on embryo neuroanatomy, we ran a whole-brain voxel-wise linear mixed-effects model [“mincLmer”; lme4_1.1-21 package (79)] on the relative Jacobian determinant files using group and sex as fixed effects and number of pups per litter and cohort collection batch as random intercepts. A Satterthwaite approximation was used to compute degrees of freedom for every voxel (using the “mincLmerEstimateDF” function) . The FDR correction (using “mincFDR”) was applied to correct for multiple testing (80, 81) (see SI Appendix, section 1.5.1 for details). This analysis was run again with the POL L group as the reference in order to directly compare POL E to POL L differences (SI Appendix, section 2.2.1). Sex differences were explored as a follow-up analysis (SI Appendix, section 1.5.1). Putative differences in organ volume (i.e., lungs, heart, liver, etc.) for the embryos were also investigated by applying deformation-based morphometry (as described above) to the body cavity (SI Appendix, section 1.5.1); however, no differences were observed (SI Appendix, section 2.2.2).
4.4.2. EM.
A nonparametric Kruskall–Wallis test followed by a pairwise Wilcoxon test was used to assess group differences in the density of dark glial, dark neuronal, and apoptotic cells in the combined SAL, POL E, and POL L groups averaged per subject across slices (the SAL group was combined to maintain consistency with the MRI findings).
To maximize variance, the shift function was used to compare differences in the distribution of density measures acquired from each slice per mouse (3), rather than using the pooled data per mouse (34). This allows us to quantify how two distributions differ based on deciles of the distributions; i.e., it describes how one distribution should be transformed to match the other and estimates how and by how much one distribution must be shifted. When a significant difference is observed between deciles, it suggests that there is a specific difference in the density of the cell type investigated (i.e., dark glia) between groups; this allows us to determine whether differences are consistent across the entire distribution or more localized to one or both tails or the center. In the context of the cell-density data acquired, this technique allows us to compare groups beyond means or medians by accounting for the variance across the entire distribution of the data; this may provide us with a more nuanced understanding of differences between groups. Three pairwise comparisons were made (SAL–POL E, SAL–POL L, and POL L–POL E) on the distributions for dark glia, dark neuron, and apoptotic cell density. A percentile bootstrap technique was used to derive CIs based on differences in distribution at each decile of the distribution with a corresponding P value. This was then repeated to assess sex differences, so the same comparisons were made in only males and only females, followed by the same percentile bootstrap procedure.
Supplementary Material
Acknowledgments
We are grateful to Roulin Gao for providing training in how to harvest embryos. We thank Drs. Bruno Giros and Salah El Mestikawy for lending us their centrifuge. A preprint of this work is available on BioRxiv (https://doi.org/10.1101/2021.07.14.452084), and all the data (preprocessed embryo brain MRIs and EM data from the dorsal hippocampus) have been made publicly available on the Zenodo platform (https://zenodo.org/record/5164621), as has the code used to perform statistical analyses. Finally, we acknowledge our funding bodies, including the Canadian Institute of Health Research and Healthy Brains for Healthy Lives, for providing support for this research. Additionally, we thank the Fonds de Recherche du Québec en Santé for providing salary support for E.G., K.P., and M.M.C., as well as the Kappa Gamma Foundation of Canada for supporting the salary of E.G. M.-E.T. is a Tier 2 Canada Research Chair in Neurobiology of Aging and Cognition.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114545119/-/DCSupplemental.
Data Availability
Structural MRI and EM cell counts and cell-density data have been deposited in Zenodo, as is the code used for statistical analyses (https://doi.org/10.5281/zenodo.5164621). Instructions for running the ANTs DBM analysis can be found on GitHub (https://github.com/CoBrALab/twolevel_ants_dbm), as can information on MRI preprocessing steps (https://github.com/CoBrALab/documentation/wiki/Embryo-scan-preprocessing).
References
- 1.Brown A. S., et al. , A.E. Bennett Research Award. Prenatal rubella, premorbid abnormalities, and adult schizophrenia. Biol. Psychiatry 49, 473–486 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Brown A. S., et al. , Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch. Gen. Psychiatry 61, 774–780 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Selten J. P., van Duursen R., van der Graaf Y., Gispen-de Wied C., Kahn R. S., 736—Second-trimester exposure to maternal stress is a possible risk factor for psychotic illness in the child. Schizophr. Res. 24, 258 (1997). [Google Scholar]
- 4.Canetta S. E., Brown A. S., Prenatal infection, maternal immune activation, and risk for schizophrenia. Transl. Neurosci. 3, 320–327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi G. B., et al. , The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumusoglu S. B., Stevens H. E., Maternal inflammation and neurodevelopmental programming: A review of preclinical outcomes and implications for translational psychiatry. Biol. Psychiatry 85, 107–121 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Solek C. M., Farooqi N., Verly M., Lim T. K., Ruthazer E. S., Maternal immune activation in neurodevelopmental disorders. Dev. Dyn. 247, 588–619 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Boksa P., Effects of prenatal infection on brain development and behavior: A review of findings from animal models. Brain Behav. Immun. 24, 881–897 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Thion M. S., Ginhoux F., Garel S., Microglia and early brain development: An intimate journey. Science 362, 185–189 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Reisinger S., et al. , The poly(I:C)-induced maternal immune activation model in preclinical neuropsychiatric drug discovery. Pharmacol. Ther. 149, 213–226 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Guma E., et al. , Early or late gestational exposure to maternal immune activation alters neurodevelopmental trajectories in mice: An integrated neuroimaging, behavioural, and transcriptional study. Biol. Psychiatry 90, 328–341 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Guma E., Plitman E., Chakravarty M. M., The role of maternal immune activation in altering the neurodevelopmental trajectories of offspring: A translational review of neuroimaging studies with implications for autism spectrum disorder and schizophrenia. Neurosci. Biobehav. Rev. 104, 141–157 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Ellman L. M., et al. , Structural brain alterations in schizophrenia following fetal exposure to the inflammatory cytokine interleukin-8. Schizophr. Res. 121, 46–54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crum W. R., et al. , Evolution of structural abnormalities in the rat brain following in utero exposure to maternal immune activation: A longitudinal in vivo MRI study. Brain Behav. Immun. 63, 50–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piontkewitz Y., Arad M., Weiner I., Abnormal trajectories of neurodevelopment and behavior following in utero insult in the rat. Biol. Psychiatry 70, 842–851 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Graham A. M., et al. , Maternal systemic Interleukin-6 during pregnancy is associated with newborn amygdala phenotypes and subsequent behavior at 2 years of age. Biol. Psychiatry 83, 109–119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph M. D., et al. , Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat. Neurosci. 21, 765–772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spann M. N., Monk C., Scheinost D., Peterson B. S., Maternal immune activation during the third trimester is associated with neonatal functional connectivity of the salience network and fetal to toddler behavior. J. Neurosci. 38, 2877–2886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canales C. P., et al. , Sequential perturbations to mouse corticogenesis following in utero maternal immune activation. eLife 10, e60100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombardo M. V., et al. , Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol. Psychiatry 23, 1001–1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalish B. T., et al. , Maternal immune activation in mice disrupts proteostasis in the fetal brain. Nat. Neurosci. 24, 204–213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer U., et al. , The time of prenatal immune challenge determines the specificity of inflammation-mediated brain and behavioral pathology. J. Neurosci. 26, 4752–4762 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavalcante M. B., Cavalcante C. T. M. B., Sarno M., Barini R., Kwak-Kim J., Maternal immune responses and obstetrical outcomes of pregnant women with COVID-19 and possible health risks of offspring. J. Reprod. Immunol. 143, 103250 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes-Lagos J. J., et al. , A translational perspective of maternal immune activation by SARS-CoV-2 on the potential prenatal origin of neurodevelopmental disorders: The role of the cholinergic anti-inflammatory pathway. Front. Psychol. 12, 614451 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D., Zhang J., Recent progress in magnetic resonance imaging of the embryonic and neonatal mouse brain. Front. Neuroanat. 10, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barron H. C., Mars R. B., Dupret D., Lerch J. P., Sampaio-Baptista C., Cross-species neuroscience: Closing the explanatory gap. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 4, 376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bisht K., et al. , Dark microglia: A new phenotype predominantly associated with pathological states. Glia 64, 826–839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nahirney P. C., Tremblay M.-E., Brain ultrastructure: Putting the pieces together. Front. Cell Dev. Biol. 9, 629503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry M. S., et al. , Delta opioid receptor signaling promotes resilience to stress under the repeated social defeat paradigm in mice. Front. Mol. Neurosci. 11, 100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Erp T. G. M., et al. , Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry 21, 547–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman J. A., et al. , Hippocampal dysfunction in the pathophysiology of schizophrenia: A selective review and hypothesis for early detection and intervention. Mol. Psychiatry 23, 1764–1772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hui C. W., et al. , Prenatal immune challenge in mice leads to partly sex-dependent behavioral, microglial, and molecular abnormalities associated with schizophrenia. Front. Mol. Neurosci. 11, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fagerland M. W., t-tests, non-parametric tests, and large studies—a paradox of statistical practice? BMC Med. Res. Methodol. 12, 78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rousselet G. A., Pernet C. R., Wilcox R. R., Beyond differences in means: Robust graphical methods to compare two groups in neuroscience. Eur. J. Neurosci. 46, 1738–1748 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Colbourne F., Sutherland G. R., Auer R. N., Electron microscopic evidence against apoptosis as the mechanism of neuronal death in global ischemia. J. Neurosci. 19, 4200–4210 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q., et al. , Ultrastructural characteristics of neuronal death and white matter injury in mouse brain tissues after intracerebral hemorrhage: Coexistence of ferroptosis, autophagy, and necrosis. Front. Neurol. 9, 581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taatjes D. J., Sobel B. E., Budd R. C., Morphological and cytochemical determination of cell death by apoptosis. Histochem. Cell Biol. 129, 33–43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McIlwain D. R., Berger T., Mak T. W., Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 5, a008656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knuesel I., et al. , Maternal immune activation and abnormal brain development across CNS disorders. Nat. Rev. Neurol. 10, 643–660 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Estes M. L., McAllister A. K., Maternal immune activation: Implications for neuropsychiatric disorders. Science 353, 772–777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown A. S., Meyer U., Maternal immune activation and neuropsychiatric illness: A translational research perspective. Am. J. Psychiatry 175, 1073–1083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Erp T. G. M., et al. ; Karolinska Schizophrenia Project, Cortical brain abnormalities in 4474 individuals with schizophrenia and 5098 control subjects via the enhancing neuro imaging genetics through meta analysis (ENIGMA) consortium. Biol. Psychiatry 84, 644–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Rooij D., et al. , Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: Results from the ENIGMA ASD Working Group. Am. J. Psychiatry 175, 359–369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girard S., Tremblay L., Lepage M., Sébire G., IL-1 receptor antagonist protects against placental and neurodevelopmental defects induced by maternal inflammation. J. Immunol. 184, 3997–4005 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Page N. F., et al. , Alterations in retrotransposition, synaptic connectivity, and myelination implicated by transcriptomic changes following maternal immune activation in nonhuman primates. Biol. Psychiatry 89, 896–910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piontkewitz Y., Arad M., Weiner I., Tracing the development of psychosis and its prevention: What can be learned from animal models. Neuropharmacology 62, 1273–1289 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Couch A. C. M., et al. , Maternal immune activation primes deficiencies in adult hippocampal neurogenesis. Brain Behav. Immun. 97, 410–422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tremblay M.-È., Zettel M. L., Ison J. R., Allen P. D., Majewska A. K., Effects of aging and sensory loss on glial cells in mouse visual and auditory cortices. Glia 60, 541–558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bisht K., Sharma K., Lacoste B., Tremblay M.-È., Dark microglia: Why are they dark? Commun. Integr. Biol. 9, e1230575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Versaevel M., et al. , Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci. Rep. 4, 7362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimatkin S. M., Bon’ E. I., Dark neurons of the brain. Neurosci. Behav. Physiol. 48, 908–912 (2018). [Google Scholar]
- 52.St-Pierre M.-K., Šimončičová E., Bögi E., Tremblay M.-È., Shedding light on the dark side of the microglia. ASN Neuro 12, 1759091420925335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smolders S., Notter T., Smolders S. M. T., Rigo J.-M., Brône B., Controversies and prospects about microglia in maternal immune activation models for neurodevelopmental disorders. Brain Behav. Immun. 73, 51–65 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Schwarz J. M., Sholar P. W., Bilbo S. D., Sex differences in microglial colonization of the developing rat brain. J. Neurochem. 120, 948–963 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selemon L. D., Zecevic N., Schizophrenia: A tale of two critical periods for prefrontal cortical development. Transl. Psychiatry 5, e623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ozaki K., et al. , Maternal immune activation induces sustained changes in fetal microglia motility. Sci. Rep. 10, 21378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mor G., Abrahams V. M., Potential role of macrophages as immunoregulators of pregnancy. Reprod. Biol. Endocrinol. 1, 119 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mosley M., et al. , Patterns of cell death in the perinatal mouse forebrain. J. Comp. Neurol. 525, 47–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai Z., Pan Z. L., Pang Y., Evans O. B., Rhodes P. G., Cytokine induction in fetal rat brains and brain injury in neonatal rats after maternal lipopolysaccharide administration. Pediatr. Res. 47, 64–72 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Cunningham C., Campion S., Teeling J., Felton L., Perry V. H., The sickness behaviour and CNS inflammatory mediator profile induced by systemic challenge of mice with synthetic double-stranded RNA (poly I:C). Brain Behav. Immun. 21, 490–502 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Guma E., et al. , Subtle alterations in neonatal neurodevelopment following early or late exposure to prenatal maternal immune activation in mice. Neuroimage Clin. 32, 102868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dekel N., Gnainsky Y., Granot I., Mor G., Inflammation and implantation. Am. J. Reprod. Immunol. 63, 17–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mor G., Cardenas I., The immune system in pregnancy: A unique complexity. Am. J. Reprod. Immunol. 63, 425–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharpe J., Optical projection tomography as a new tool for studying embryo anatomy. J. Anat. 202, 175–181 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korogod N., Petersen C. C. H., Knott G. W., Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. eLife 4, e05793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mueller F. S., et al. , Behavioral, neuroanatomical, and molecular correlates of resilience and susceptibility to maternal immune activation. Mol. Psychiatry 26, 396–410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buss C., Entringer S., Swanson J. M., Wadhwa P. D., The role of stress in brain development: The gestational environment’s long-term effects on the brain. Cerebrum 2012, 4 (2012). [PMC free article] [PubMed] [Google Scholar]
- 68.Lautarescu A., Craig M. C., Glover V., Prenatal stress: Effects on fetal and child brain development. Int. Rev. Neurobiol. 150, 17–40 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Lenniger C., Espinoza-Heredia C., Trentacosta C., Thomason M. E., Associations between prenatal maternal cortisol levels and the developing human brain. Biol. Psychiatry 87 (9, suppl.), S126 (2020). [Google Scholar]
- 70.Graham A., et al. , Sex specific effects of maternal cortisol concentrations during pregnancy on the functional connectivity of the newborn limbic system. Psychoneuroendocrinology 83, 6 (2017). [Google Scholar]
- 71.Moog N. K., et al. , Prospective association of maternal psychosocial stress in pregnancy with newborn hippocampal volume and implications for infant social-emotional development. Neurobiol. Stress 15, 100368 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spencer Noakes T. L., Henkelman R. M., Nieman B. J., Partitioning k-space for cylindrical three-dimensional rapid acquisition with relaxation enhancement imaging in the mouse brain. NMR Biomed., 10.1002/nbm.3802 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Avants B. B., et al. , A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung M. K., et al. , A unified statistical approach to deformation-based morphometry. Neuroimage 14, 595–606 (2001). [DOI] [PubMed] [Google Scholar]
- 75.Schambra U., Prenatal Mouse Brain Atlas: Color Images and Annotated Diagrams of: Gestational Days 12, 14, 16 and 18 Sagittal, Coronal and Horizontal Section (Springer, Boston, 2008). [Google Scholar]
- 76.Franklin K. B., Paxinos G., The Mouse Brain in Stereotaxic Coordinates, Compact. The Coronal Plates and Diagrams (Elsevier Academic Press, Amsterdam, 2008). [Google Scholar]
- 77.Ellisman M., et al. , Bridging gaps in imaging by applying EM tomography and serial block face SEM, including a new genetically encoded tag for correlated light and 3D electron microscopy of intact cells, tissues and organisms: Integrating the resulting correlated image data using the Whole Brain Catalog. Microsc. Microanal. 17 (Suppl. 2), 2–3 (2011). [Google Scholar]
- 78.Bankhead P., et al. , QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models Usinglme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 80.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 81.Genovese C. R., Lazar N. A., Nichols T., Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15, 870–878 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural MRI and EM cell counts and cell-density data have been deposited in Zenodo, as is the code used for statistical analyses (https://doi.org/10.5281/zenodo.5164621). Instructions for running the ANTs DBM analysis can be found on GitHub (https://github.com/CoBrALab/twolevel_ants_dbm), as can information on MRI preprocessing steps (https://github.com/CoBrALab/documentation/wiki/Embryo-scan-preprocessing).