Significance
Our results demonstrate the existence of early cellular pathways and network alterations in oligodendrocytes in the alpha-synucleinopathies Parkinson’s disease and multiple system atrophy. They further reveal the involvement of an immune component triggered by alpha-synuclein protein, as well as a connection between (epi)genetic changes and immune reactivity in multiple system atrophy. The knowledge generated in this study could be used to devise novel therapeutic approaches to treat synucleinopathies.
Keywords: oligodendrocytes, transcriptome, alpha-synuclein, maturation, HLA
Abstract
Limited evidence has shed light on how aSYN proteins affect the oligodendrocyte phenotype and pathogenesis in synucleinopathies that include Parkinson’s disease (PD) and multiple system atrophy (MSA). Here, we investigated early transcriptomic changes within PD and MSA O4+ oligodendrocyte lineage cells (OLCs) generated from patient-induced pluripotent stem cells (iPSCs). We found impaired maturation of PD and MSA O4+ OLCs compared to controls. This phenotype was associated with changes in the human leukocyte antigen (HLA) genes, the immunoproteasome subunit PSMB9, and the complement component C4b for aSYN p.A53T and MSA O4+ OLCs, but not in SNCAtrip O4+ OLCs despite high levels of aSYN assembly formation. Moreover, SNCA overexpression resulted in the development of O4+ OLCs, whereas exogenous treatment with aSYN species led to significant toxicity. Notably, transcriptome profiling of genes encoding proteins forming Lewy bodies and glial cytoplasmic inclusions revealed clustering of PD aSYN p.A53T O4+ OLCs with MSA O4+ OLCs. Our work identifies early phenotypic and pathogenic changes within human PD and MSA O4+ OLCs.
The synucleinopathies are a group of neurodegenerative diseases typically characterized by the presence of alpha-synuclein (aSYN) protein-rich aggregates termed Lewy bodies (LBs) within neurons in both Parkinson’s disease (PD) and dementia with Lewy bodies (DLB). Alternatively, aSYN-positive glial cytoplasmic inclusions (GCIs) within oligodendrocytes uniquely characterize multiple system atrophy (MSA) (1). Recent case reports, however, revealed the presence of GCI-like structures in oligodendrocytes and/or astrocytes in postmortem brains of PD patients carrying genetic variations in LRRK2 (p.I1371V), SNCA (p.G51D and p.A53E), and DJ1 (p.L172Q) genes (2–5). Interestingly, inclusions could not be identified in oligodendrocytes in PD brains carrying the aSYN variation p.H50Q or the duplication of the SNCA locus, despite high levels of phosphorylated aSYN (2). These data suggest the existence of pathological differences between familial PD cases. However, the reason(s) for the development of aSYN-positive GCI-like structures in certain forms of PD but not others remains unknown.
We and others have previously shown that rodent and human oligodendrocyte lineage cells (OLCs) exhibit different levels of aSYN during development (6–8). Interestingly, increased expression of the SNCA gene encoding for aSYN was found higher within OLCs in the MSA brain (9). Moreover, OLCs have been shown to take up different aSYN assemblies in vitro (10–12). Thus, identifying the changes induced by the presence of pathogenic aSYN as a result of SNCA expression coupled with the uptake of aSYN assemblies capable of inducing de novo aggregation could allow for the identification of therapeutic pathways that could be modulated to treat synucleinopathies. Through transcriptomic analysis of patient-induced pluripotent stem cell (iPSC)-derived O4+ OLCs, we identified signaling pathways that induced OLCs to adopt an antigen-presenting phenotype, impairing their maturation into myelinating oligodendrocytes. We further demonstrated that the antigen-presenting phenotype can be triggered by a fibrillar aSYN variant via activation of inflammatory pathways including activation of the immunoproteosome in PD (p.A53T aSYN) and MSA OLCs.
Results
Transcriptomic Analysis Identifies Increased Expression of Genes Encoding Major Histocompatibility Complex (MHC)–Class Proteins and Decreased Expression of Genes Involved in Differentiation and Maturation in Human aSYN p.A53T O4+ OLCs.
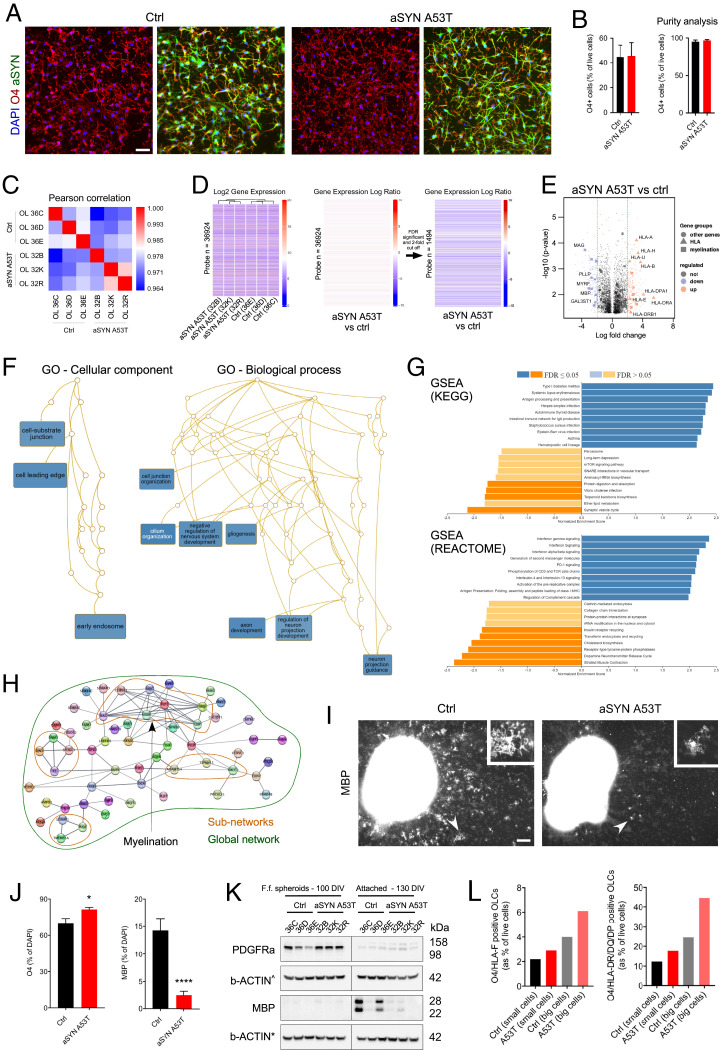
To investigate whether human oligodendrocytes display cellular alterations in PD, we used control (13, 14) and PD-derived iPSC lines harboring the heterozygous aSYN variation p.A53T (15) (SI Appendix, Fig. S1). Familial aSYN p.A53T leads to early PD onset (16) and robust cellular pathology in iPSC-derived aSYN p.A53T neurons (17). In this study, we generated OLCs from aSYN p.A53T iPSCs using our published protocol (6, 18). Free-floating 100-d-old spheroids expressed OLC canonical markers SOX10, OLIG2, and NKX2.2 (SI Appendix, Fig. S2) but mostly lacked O4, a marker of the premyelinating oligodendrocyte stage. To stimulate the maturation of oligodendrocyte precursor cells (OPCs) into O4+ OLCs, we seeded the spheroids onto coated surfaces and then confirmed that OLCs naturally expressed the SNCA gene [Fig. 1 A and B (6)]. We next performed Affymetrix GeneChip microarray to examine transcriptomic changes in fluorescence-activated cell sorter (FACS)–purified O4+ OLCs (SI Appendix, Fig. S3 and Dataset S1). Pearson’s correlation revealed a segregation between the samples, classifying control and aSYN p.A53T O4+ OLCs separately (Fig. 1C). Out of the 135,750 probes analyzed, we identified 1,494 annotated genes significantly up- or down-regulated (Fig. 1D) with at least a twofold change. Thirty-three of the 50 most significantly down-regulated genes encoded for proteins involved in oligodendrocyte differentiation and maturation, whereas human leukocyte antigen (HLA) genes encoding for MHC-class I and II proteins were highly up-regulated (Fig. 1E). Gene ontology (GO) annotation enrichment with hypergeometric distribution indicated significant enrichment of cellular component and biological process terms (Fig. 1F). The cellular component terms included cell-substrate junction and early endosome, whereas the biological process terms included gliogenesis and negative regulation of the nervous system development.
Fig. 1.
Transcriptomic analysis of FACS-purified O4+ OLCs revealed decreased expression of genes necessary for oligodendrocyte differentiation and maturation and increased expression of genes encoding MHC-Class I and II proteins. (A) Abundance of O4+ OLCs coexpressing WT and aSYN p.A53T in adherent cultures. (Scale bar 50 μm.) (B) O4+ OLCs isolated by FACS from 130-d-old adherent cultures. The bar diagrams represent the percentages of healthy WT (control) and aSYN p.A53T O4+ OLCs following sorting (based on n = 3 differentiated iPSC clones per genotype) and purity of the samples after reanalysis. Results are presented as mean ± SEM. (C) Pearson correlation analysis for control and aSYN p.A53T samples. (D) Gene expression patterns clustered by genotype. In all, 1,494 annotated genes were differentially expressed in control and aSYN p.A53T O4+ OLCs using a twofold cutoff. (E) Volcano plots showing distribution of significantly up- and down-regulated genes in aSYN p.A53T O4+ OLCs. Vertical dashed lines indicate twofold cutoff. (F) GO analysis of biological process and cellular component terms revealed changes in aSYN p.A53T O4+ OLCs. (G) GSEA reveals groups of genes enriched or depleted in aSYN p.A53T O4+ OLCs. (H) Network analysis using Cytoscape for genes down-regulated in aSYN p.A53T O4+ OLCs allows identification of subnetworks, including the subnetwork “myelination.” (I) Representative images of adherent cultures stained for myelin binding protein (MBP). Arrowheads indicate MBP+ cells. (Scale bar, 100 μm.) (J) Quantification of O4+ and MBP+ cells in adherent cultures. Data are mean ± SEM; n = 3. P values: ****P < 0.0001; *P < 0.05. Statistical analysis performed using two-tailed t test. (K) Western blot analysis reveals different levels of PDGFRα and MBP in aSYN p.A53T and control free-floating and adherent cultures enriched in OPCs and OLCs, respectively (Fig. 3A). (L) FACS-based quantification of O4+/HLA-F+ and O4+/HLA-DR/DQ/DP+ OLCs (small and big cells) in cultures aged 130 DIV. Ctrl, control; DAPI, 6′-diamidino-2-phenylindole; FDR, false discovery rate; F.f., free-floating; IgA, immunoglobulin A; rRNA, ribosomal RNA; TCR, T cell receptor for antigen; tRNA, transfer RNA.
To gain further insight into the genetic and cellular changes, we performed gene set enrichment analysis (GSEA) (19). We identified positive and negative gene enrichment for several pathways, including the immune system (antigen processing and presentation) and oligodendrocyte differentiation and maturation (lipid metabolism; Fig. 1G). Importantly, the transcriptomic changes were specific for oligodendrocytes since the analysis of the transcriptome of aSYN p.A53T and control iPSC-derived midbrain spheroids containing TH+/FOXA2+ neurons revealed alterations in different categories, including the catecholamine metabolic process defined by six genes, namely DDC, AGTR2, TH, EPAS1, NR4A2 (known as NURR1), and DBH (SI Appendix, Fig. S4 and Dataset S2).
Next, we used Stringdb via Cytoscape (20) to identify interconnections between the genes significantly up- or down-regulated by at least twofold. Several genes were connected, especially those coding for the subnetwork MAG-CNPase-MBP-MOG-PLP1-MOBP-CLDN11 (Fig. 1H), components of the myelin proteome (21). We also found a decrease in the expression of peptidylarginine deiminase type II, an enzyme required for oligodendrocyte differentiation and myelination (22). MicroRNAs, which regulate the translation of messenger RNAs into proteins, were also altered (Dataset S1). Among them, we identified microRNA (miR)-9, miR-138, and miR-219, which are associated with oligodendrocyte differentiation and maturation (23). Significant changes in lipid-related gene expression were also identified (Dataset S4). These data suggest that terminal differentiation of aSYN p.A53T oligodendrocytes was impaired. Indeed, using fluorescent image analysis, we found a greater number of O4+ OLCs but a lower number of MBP+ mature oligodendrocytes (24, 25) in aSYN p.A53T cultures compared to controls (Fig. 1 I and J). These data were confirmed by Western blot analysis (Fig. 1K). Our analysis further revealed increased levels of the OPC marker PDGFRα (24, 25) in free-floating aSYN p.A53T spheroid cultures aged 100 d in vitro (DIV) when compared to controls (Fig. 1K and SI Appendix, Table S1 and Dataset S1). To validate these findings, we differentiated OLCs from mouse embryonic stem cells generated from wild-type (WT) and the M83 mouse model of PD, which expresses the human aSYN p.A53T transgene under the prion promoter (18, 26, 27). In line with our human OLC data, we observed a lower number of CNPase+ OLCs compared to controls, indicating terminal differentiation was affected by aSYN p.A53T (SI Appendix, Fig. S5). Finally, flow cytometry analysis of human OLCs revealed ∼1.5-fold increase in the number of double-positive O4+/HLA-F+ and O4+/HLA-DR/DQ/DP+ OLCs in aSYN p.A53T cultures compared to controls (Fig. 1I), a result in line with our gene expression profiles.
Taken together, our data indicate that human O4+ aSYN p.A53T OLCs have increased expression of HLA genes and presumably a delayed maturation, since OLCs are capable of myelinating axons in the M83 PD model (28–30). Our data further suggest that during development, O4+ aSYN p.A53T OLCs may exhibit an immune reactive phenotype, similar to that observed in multiple sclerosis (31).
Transcriptomic Analysis Revealed Different Cellular Alterations Linked to Oligodendrocyte Differentiation and Immune Reactivity in PD and MSA Patients’ O4+ OLCs.
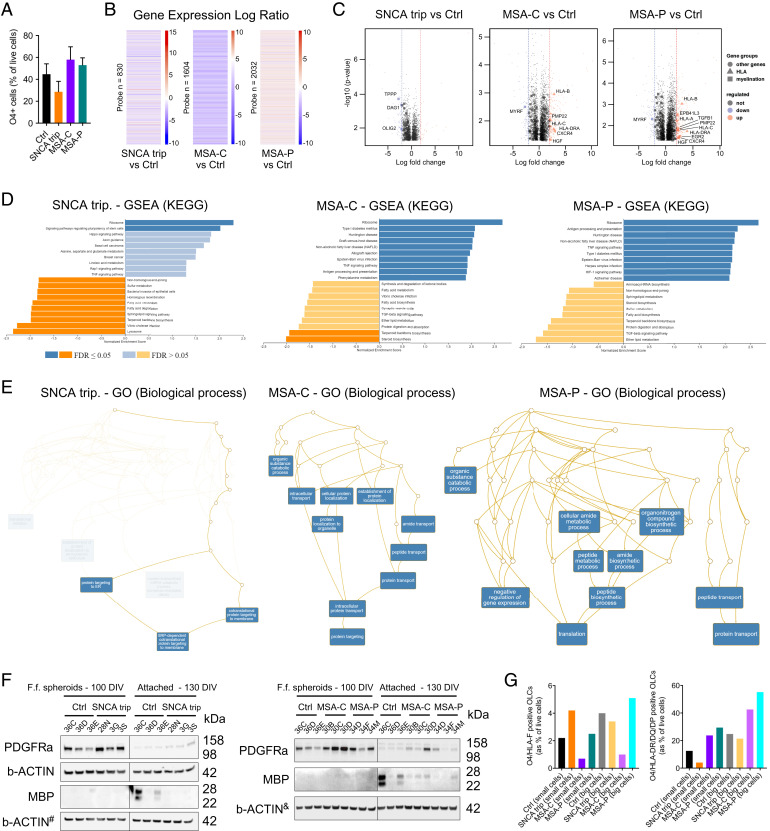
We next examined whether transcriptomic changes also appeared in O4+ OLCs generated from an individual carrying a triplication of the SNCA locus (referred to as SNCAtrip) and two MSA patients—one diagnosed with progressive ataxia and the other with predominant PD-like symptoms. Thus, we generated OLCs using several disease-specific iPSC lines from PD (SNCAtrip: CSC-3G, CSC-3S, and CSC-28N) (6, 14, 32) and MSA (MSA-C: CSC-30B, CSC-30C, and CSC-30D; and MSA-P: CSC34D, CSC-34F, and CSC-34M; SI Appendix, Fig. S1). We identified significant changes in the expression of many genes in FACS-purified O4+ OLCs derived from SNCAtrip (830 genes), MSA-C (1,604 genes), and MSA-P (2,032 genes) (Fig. 2 A and B and SI Appendix, Fig. S3 and Datasets S6 and S7). Interestingly, similar to aSYN p.A53T O4+ OLCs, genes encoding for MHC-class molecules were significantly up-regulated within MSA-C and MSA-P O4+ OLCs but not in SNCAtrip O4+ OLCs (Fig. 2C). However, genes encoding for proteins involved in oligodendrocyte differentiation and maturation were down-regulated for all genotypes (Fig. 2C and SI Appendix, Table S1 and Datasets S4 and S5). These findings are in line with the GSEA data, which revealed positive enrichment of antigen presenting and processing within MSA-C and MSA-P O4+ OLCs but negative enrichment for lipid metabolism in all three genotypes (Fig. 2D).
Fig. 2.
Transcriptomic profiling of SNCAtrip, MSA-P, and MSA-C O4+ OLCs revealed alterations in genes linked to oligodendrocyte maturation and in MHC Class I and II proteins. (A) Abundance of O4+ OLCs in SNCAtrip, MSA-C, and MSA-P adherent cultures aged 130 DIV. The bar diagram represents the percentage of living healthy and synucleinopathies O4+ OLCs measured by flow cytometry (based on n = 3 differentiated iPSC clones per genotype); results are presented as mean ± SEM. (B) Heatmaps showing 830; 1,604; and 2,032 annotated genes differentially expressed in controls and SNCAtrip, MSA-C, and MSA-P O4+ OLCs using a twofold cutoff. (C) Volcano plots showing significant up- and down-regulated genes in SNCAtrip, MSA-C, and MSA-P O4+ OLCs. Vertical dashed lines indicate twofold cutoff. (D) GSEA revealed groups of genes enriched and depleted in SNCAtrip, MSA-C, and MSA-P O4+ OLCs and compared to control. (E) GO analysis of biological process revealed changes in SNCAtrip, MSA-C, and MSA-P O4+ OLCs. (F) Western blot indicated different levels of PDGFRα and MBP in 100- and 130-d-old SNCAtrip, MSA-C, MSA-P, and control cultures. The symbols # and & indicate that the membranes were also used to evaluate aSYN levels (Fig. 3A) (G) Quantification by flow cytometry of O4+/HLA-F+ and O4+/HLA-DR/DQ/DP+ OLCs in adherent cultures. Ctrl, control; ER, endoplasmic reticulum; FDR, false discovery rate; F.f., free-floating; SRP, signal-recognition particle; TGF, transforming growth factor; tRNA, transfer RNA.
The significantly enriched GO terms for SNCAtrip O4+ OLCs included signal recognition particle-dependent cotranslational protein targeting to membrane, for MSA-C: intracellular protein transport and protein targeting, and for MSA-P: peptide biosynthetic process and translation (Fig. 2E). Similar to aSYN p.A53T O4+ OLCs, we observed high levels of PDGFRα and low levels of MBP in the SNCAtrip, MSA-C, and MSA-P O4+ OLCs (Fig. 2F). We also confirmed changes in the percentage of double-positive O4+/HLA-F+ and O4+/HLA-DR/DQ/DP+ OLCs in SNCAtrip as well as MSA cultures (Fig. 2G). Overall, the changes identified in MSA-C and MSA-P O4+ OLCs were more similar to those identified for aSYN p.A53T O4+ OLCs than those for SNCAtrip O4+ OLCs. This suggests that PD aSYN p.A53T OLC pathology could be considered an intermediate between PD SNCAtrip and MSA OLC pathologies in terms of disease severity, onset, and progression (33, 34).
Transcriptomic Changes within O4+ OLCs Overexpressing or Exposed to aSYN Species Differently Influence the Cellular Phenotypes of WT OLCs.
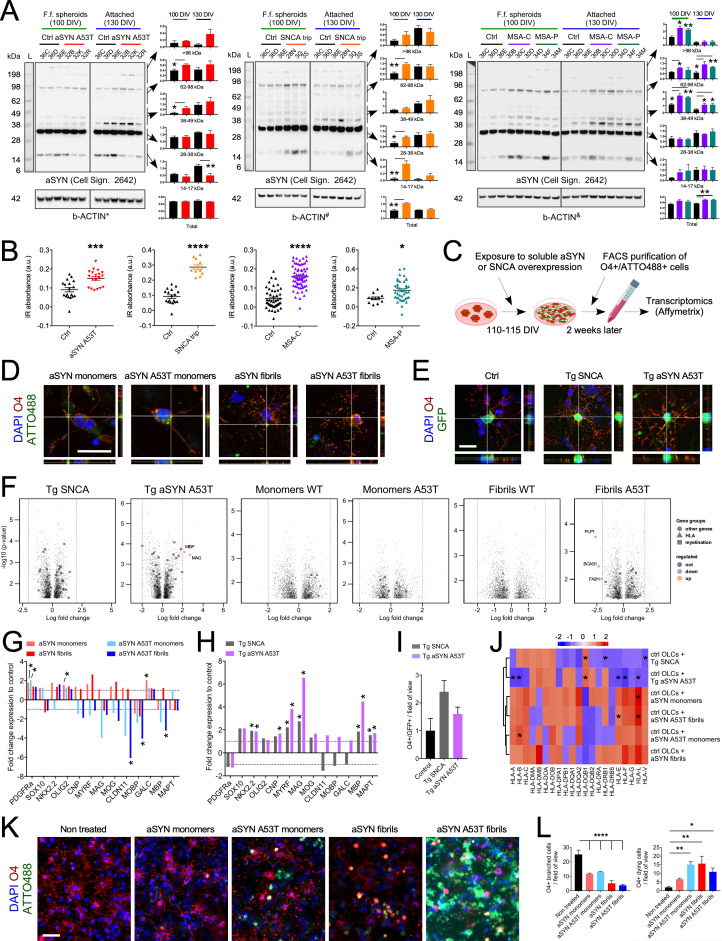
Because the studied genotypes are linked to aSYN pathology, we examined the levels of aSYN in attached or spheroid OLCs enriched from synucleinopathies versus control cultures. Immunoblot analysis for aSYN within the aSYN p.A53T OLC cultures revealed lower levels of monomeric aSYN (14 kDa) but higher levels of high–molecular weight (MW) aSYN at 100 and 130 DIV (Fig. 3A, Left). For SNCAtrip OLCs attached or spheroid cultures, we observed an increase in monomeric and high-MW aSYN at 100 and 130 DIV, respectively (Fig. 3A, Middle). For MSA-C and MSA-P cultures, we observed a significant increase of high-MW aSYN species both in free-floating spheroids and in attached OLC cultures (Fig. 3A, Right). Intriguingly, we also observed significant changes in phosphorylation of aSYN at serine 129 in 100 DIV spheroids (SI Appendix, Fig. S6). Additionally, we found that total aSYN levels were changed in SNCAtrip, MSA-C, and MSA-P but not in aSYN p.A53T spheroids or attached OLC cultures (Fig. 3A). Fourier-transform infrared (FTIR) analysis further revealed an increase in β-sheets structural content, reminiscent of protein aggregation in FACS-purified O4+ OLCs (Fig. 3B). Interestingly, aSYN was also present in the culture medium, but its levels did not correlate with cell death (SI Appendix, Fig. S7). Together, these data suggest that 1) aSYN could induce cellular dysfunction due to its endogenous expression and/or following uptake from the culture medium, 2) the structure and properties of the aSYN species present in OLCs may vary between the different genotypes studied, and 3) the changes in aSYN levels observed in MSA OLCs may relate to an underlying (epi)genetic pathological process.
Fig. 3.
O4+ OLCs overexpressing WT or p.A53T aSYN or exposed to WT or p.A53T aSYN assemblies exhibit cellular alterations. (A) Western blot analysis revealed changes in total aSYN levels and levels of 14 kDa and high-MW aSYN species in synucleinopathy spheroids and attached cultures. Data are presented as mean ± SEM; n = 3. P values: **P < 0.01; *P < 0.05. Statistical analysis was performed using a two-tailed t test. Membranes are the same as those presented in Figs. 1K and 2F; hence, identical b-ACTIN controls are shown (referred to as *, #, and &). (B) Scatter plots show FTIR analysis of β-sheet structural content (the ratio between 1,628 cm−1 and 1,665 cm−1) within FACS-purified healthy, aSYN p.A53T, SNCAtrip, MSA-C, and MSA-P O4+ OLCs (based on n = 15 to 17 spectra measurements for the n = two iPSC clones tested for SNCAtrip and n = 3 iPSC clones tested for healthy, aSYN p.A53T, MSA-C, and MSA-P). Results are presented as mean ± SEM. P values: ****P < 0.0001; ***P < 0.001; *P < 0.05. Statistical analysis was performed using two-tailed t test. (C) Experimental approach used for assessing transcriptomic changes within O4+ control OLCs induced by overexpression of SNCA or treatment with aSYN monomers and fibrils. (D) Confocal images show human aSYN-ATTO488+ aggregates within O4+ OLCs following a 2-wk treatment with WT and p.A53T human aSYN monomers or fibrils. (Scale bar, 25 μm.) (E) Confocal images show GFP+/O4+ OLCs a week after aSYN transduction. (Scale bar, 25 μm.) (F) Volcano plots show significantly up- and down-regulated genes in FACS-purified GFP+/O4+ and ATTO488+/O4+ OLCs following viral transduction or exposure to aSYN species, respectively. Vertical dashed lines indicate twofold cutoff. (G) Bar diagram for changes in expression of genes associated with oligodendrocyte differentiation in FACS-purified ATTO488+/O4+ OLCs, following treatment with aSYN assemblies. Data are mean ± SEM; n = 3. Horizontal dashed lines indicate onefold change. *P < 0.05. Statistical analysis by two-tailed t test. (H) Bar diagram shows changes in expression of genes associated with oligodendrocyte differentiation in FACS-purified GFP+/O4+ OLCs following gene transduction. Data are mean ± SEM; n = 3. *P < 0.05. Statistical analysis by two-tailed t test. (I) Quantification of the number of GFP+/O4+ OLCs in transduced cultures. (J) Heatmap shows changes in expression of HLA genes in treated or transduced FACS-purified OLCs. n = 3. *P < 0.05. (K) Images of healthy OLC cultures 1 wk after treatment with WT and p.A53T human aSYN monomers or aSYN fibrils. (L) Bar diagrams show the number of O4+ OLCs in cultures treated with ATTO-labeled WT and p.A53T human aSYN monomers or aSYN fibrils. Results are presented as mean ± SEM; n = 3. P values: ****P < 0.0001; **P < 0.01; *P < 0.05. Statistical analysis performed using one-way ANOVA. (Scale bar, 50 μm.) a.u., arbitrary units; Cell Sign., cell signaling; Ctrl, control; DAPI, 6′-diamidino-2-phenylindole; F.f., free-floating; L, ladder; Tg., transgenic.
To examine whether the transcriptomic changes identified within PD and MSA O4+ OLCs above (i.e., down-regulation of oligodendrocyte differentiation genes and/or up-regulation of HLA genes) were triggered by endogenous expression or uptake of aSYN, we either overexpressed WT or p.A53T aSYN in OLCs or exposed them to monomeric or fibrillar assemblies (WT or p.A53T aSYN). We employed a retroviral vector-based approach to overexpress aSYN transgenes in dividing iPSC-derived neural progenitors and OPCs present in control cultures aged 110 to 115 DIV; this allowed us to capture only the changes induced by aSYN in immature OLCs. The viral constructs contained a green fluorescent protein (GFP) reporter, whereas exogenous aSYN assemblies were coupled to ATTO-488, thus enabling purification of O4+/GFP+ and O4+/ATTO488+ OLCs by FACS (Fig. 3 D–F and SI Appendix, Figs. S8 and S9). Transcriptomic analysis (Datasets S6 and S7) revealed that treatment of O4+ OLCs with aSYN p.A53T fibrils led to a decrease in expression of CLDN11, MOBP, and MBP genes, common markers of myelination; this was not observed for WT fibrils (Fig. 3 G and H). In contrast, WT and p.A53T aSYN overexpression triggered a significant increase in the expression of NKX2.2, CNPase, MYRF, MAG, MBP, and MAPT genes, markers expressed during oligodendrocyte differentiation (Fig. 3 G and I), but not associated with an increase in OLC numbers (Fig. 3J). While overexpression of WT and p.A53T aSYN triggered a low increase in HLA-DQb1 gene expression, treatment with aSYN species mostly increased expression of HLA genes, in particular HLA-L (Fig. 3K). The up-regulation of HLA genes, however, was not as strong as those derived from the PD p.A53T aSYN or MSA iPSC-derived O4+ OLCs, probably due to ongoing cellular toxicity. Indeed, we observed cell death for O4+ OLCs induced by aSYN monomers, which was even more evident with aSYN fibrils (Fig. 3 K and L). Altogether, these data may suggest that aSYN is important during OLC differentiation and that the aSYN assemblies that build over time in them or that are taken up by them may be responsible for the immune reactive phenotype of the OLCs.
Transcriptomic Profiling of Genes Encoding Proteins That Compose GCIs and LBs Indicated Changes Linked to the Immunoproteasome in Human O4+ OLCs, Revealing a Stratification of Synucleinopathy Cases.
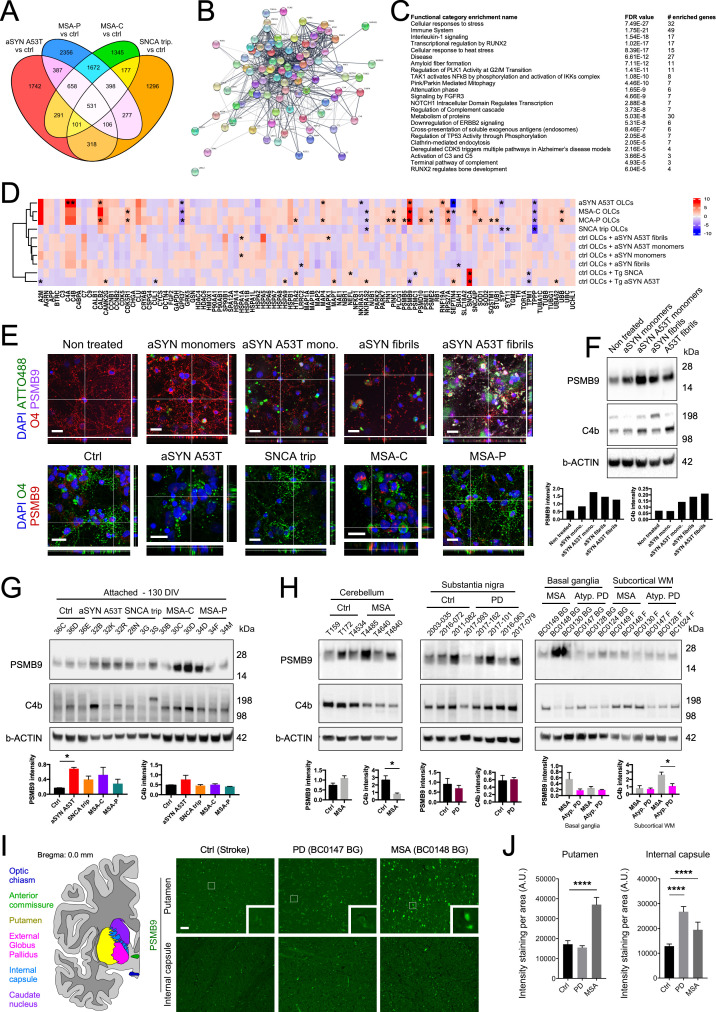
Next, we examined whether transcriptomic changes within O4+ OLCs could provide insights into the pathological hallmarks identified within postmortem PD and MSA brains. We employed a targeted approach to identify changes in the expression of genes encoding for proteins composing GCIs and LBs. We reasoned that if early cellular changes existed, they would be reflected in pathways engaged early in oligodendrocyte pathogenesis. This rationale was based on the fact that within the studied genotypes (Fig. 4A and Dataset S8), some gene expression changes in O4+ OLCs were shared but others were not, suggesting common and genotype-specific alterations. Thus, we examined the expression of 99 genes encoding proteins previously identified in GCIs and LBs (35–46). We classified the genes into interconnected pathways based on the Reactome database (Fig. 4B). Notably, the greatest enriched functional gene category for 99 targeted genes was related to immune response (Fig. 4C). Interestingly, PD aSYN p.A53T O4+ OLCs exhibited changes in the expression of several genes that closely resembled those whose expression was altered within MSA-C and MSA-P O4+ OLCs, resulting in the clustering with MSA-C and MSA-P O4+ OLCs but not with PD SNCAtrip OLCs (Fig. 4D and Dataset S8). PSMB9 expression was significantly increased in PD aSYN p.A53T O4+ OLCs, suggesting involvement of the immunoproteasome (ref. 47; Fig. 4D). Increased expression of C4a and C4b genes involved in the solubilization of immune aggregates and protein antigens (48, 49) was also observed (Dataset S9). We further confirmed the increased levels of PSMB9 and C4b proteins by immunocytochemistry and Western blot analyses both in synucleinopathies and aSYN assembly-treated cultures (Fig. 4 E–G).
Fig. 4.
Transcriptomic profiling of genes encoding proteins composing GCIs and LBs stratifies aSYN p.A53T O4+ OLCs with MSA-C and MSA-P O4+ OLCs, but not with aSYN SNCAtrip O4+ OLCs. (A) Venn diagram for genes whose expression is higher than twofold and was significantly changed between the different genetic backgrounds studied. (B) Network analysis with Cytoscape showed the relationships between the genes associated with GCIs and LBs. (C) GO term enrichment analysis identified 23 significantly affected pathways within the network identified in B. (D) Heatmap for the expression of genes encoding proteins identified in GCIs and LBs identified in aSYN p.A53T, SNCAtrip, MSA-P, MSA-C O4+ OLCs, control O4+ OLCs treated with aSYN species, or control O4+ OLCs overexpressing the WT or p.A53T SNCA transgene. Cluster analysis revealed stratification of the samples into three categories: class 1: aSYN p.A53T, MSA-P, and MSA-C O4+ OLCs; class 2: SNCAtrip, WT aSYN fibrils-, p.A53T aSYN fibrils-, monomeric WT aSYN-, and monomeric p.A53T aSYN-treated WT OLCs; class 3: WT OLCs overexpressing tg WT SNCA and Tg. p.A53T aSYN transgenes. n = 3; * denotes significant changes. (E) Confocal images for PSMB9 in synucleinopathy OLCs and OLCs treated with WT and p.A53T haSYN monomers or aSYN fibrils. (Scale bars, 10 µm.) (F) Western blot analysis showed increased levels of immunoproteasome subunit PSMB9 and C4b in treated cultures compared to control samples. Bar diagrams show quantification for the blots presented. (G) Western blot analysis confirmed increased levels of immunoproteasome subunit PSMB9 in MSA and p.A53T aSYN OLCs. C4b protein level was elevated in MSA OLCs compared to control samples. Data are presented as mean ± SEM; n = 3. P values: *P < 0.05. Statistical analysis was performed using one-way ANOVA. (H) Western blot for PSMB9 and C4b present in GCIs and LBs in the RIPA-soluble fraction from the cerebellum, the substantia nigra, the basal ganglia, and the subcortical white matter (WM) regions of the human postmortem brain of MSA, PD, healthy, and atypical PD (Atyp. PD) patients. Data are presented as mean ± SEM; n = 3. P values: *P < 0.05. Statistical analysis was performed by two-tailed t test. (I) Schematic for the analyzed human brain regions (the different brains regions are color coded). Immunohistochemistry shows increased PSMB9 intensity staining in the putamen of MSA samples and internal capsule in both PD and MSA samples compared to controls. Insets represent corresponding zoomed areas. (J) Quantification of PSMB9 intensity staining per area in the putamen and internal capsule. P values: ****P < 0.0001. (Scale bar, 50 μm.) A.U., arbitrary units; Ctrl, control; DAPI, 6′-diamidino-2-phenylindole; FDR, false discovery rate; mono., IKK, inhibitor of nuclear factor-κB kinase; monomers; NFκb, nuclear factor κB; Tg, transgenic.
To validate our results, we performed Western blot analysis of PD and MSA postmortem brain regions (SI Appendix, Table S2). We found trends toward increased PSMB9 levels within the cerebellum and basal ganglia of MSA brains and within the substantia nigra of PD cases compared to controls (Fig. 4H). The level of C4b, however, varied depending on the brain regions analyzed. C4b level was low in the cerebellum of MSA patients compared to controls, whereas it was high in the substantia nigra and subcortical white matter in PD and MSA samples, respectively. These differences may reflect patient heterogeneity and region-specific neuropathological differences at the end stage of the disease (50), which may correlate or vary from early-stage cellular pathogenesis identified in patient iPSC-derived OLCs. Importantly, using immunohistochemical techniques we identified PSMB9 levels as being more elevated in MSA and PD compared to controls, not only in OLIG2+ and GCI+ cells but also in MAP2+ and GFAP+ cell types (Dataset S9). Taken together, our data suggest that these diseases present a strong immunologic component that appears early during the disease process and continues to the end stage of the disease (Fig. 4 I and J).
Discussion
Alterations in the transcriptome of OLCs in PD and MSA postmortem brains have only been recently described (51, 52). Here, we present evidence for early OLC pathology and/or change in function in PD and MSA. However, these phenotypes may not appear for all PD patients due to the natural lowering of SNCA expression during OLCs maturation (6). Nonetheless, they provide information on possible early cellular dysfunctions that occur in MSA, where SNCA expression is higher than in healthy brain (9), or in PD forms, where GCI-like structures are identified in OLCs following postmortem examination (2–5). Importantly and unexpectedly, our work suggests a connection between (epi)genetic changes and immune reactivity in MSA.
The first major observation emerging from our study is that the maturation of iPSC-derived O4+ OLCs from PD and MSA patients was substantially impaired. The transcription of genes and levels of MBP protein associated with OLC differentiation and myelination were significantly decreased in PD and MSA. Because mice overexpressing transgenic human aSYN p.A53T have myelinated axons (27), we speculate that the impaired maturation of PD and MSA iPSC-derived OLCs is rather due to a delay than a blockage toward becoming myelinating OLCs. An alternative explanation could be that more immune reactive OLCs are produced in PD and MSA, therefore shifting the proportions of OLC subtypes that are produced during development (53–55). Indeed, one can speculate that in the disease state, the proportion of immunological OLCs increases due to a change in function toward becoming immune reactive, an effective response, for example, against an early buildup of aSYN assemblies. Our data also indicate substantial changes in cholesterol, galactosyl ceramide, and ethanolamine plasminogen lipids important for myelin formation (56). Changes in lipid composition in PD brain cell types were recently described (57), and the interplay between lipids in PD GBA gene variants in the process of aSYN aggregation is currently under scrutiny (58).
The overexpression of WT and p.A53T SNCA transgenes in dividing neural progenitors and OPCs induced expression of oligodendroglial genes NKX2.2, CNPase, MAG, MOG, and MBP coupled by down-regulation of early ventral forebrain lineage gene DLX5 and astroglial genes GFAP and CD44 in FACS-purified GFP+/O4+ OLCs. Interestingly, the maturation of PDGFRα+ OPCs into MBP+ oligodendrocytes was previously shown to be delayed, and demyelination was reported following overexpression of SNCA in mature oligodendrocytes (29, 30). These data suggest that SNCA expression is required for neural progenitors to commit toward oligodendrocyte lineage or OPCs to mature into O4+ OLCs and that SNCA down-regulation is necessary for OLCs to further mature into myelinating lipid-rich oligodendrocytes. As recently suggested, aberrant reexpression of SNCA in diseases could trigger myelination defects (59). Further, we found changes in levels of monomeric and high-MW aSYN in the studied genotypes. In addition, p.A53T aSYN fibrils led to a down-regulation of late-stage oligodendrocyte differentiation genes. Previous work suggests that naturally occurring high-MW aSYN species have a physiological role (60) and targeted mutagenesis of SNCA prevents the formation of aggregation-prone monomers in mice (61, 62). We speculate that this may apply to oligodendrocyte fate and maturation. Increased levels of high-MW or monomeric aSYN may be required for oligodendrocyte lineage specification, whereas low levels of high-MW aSYN or monomers may be necessary for the maturation and maintenance of OPCs into MBP+ oligodendrocytes. Hence, high aSYN levels in oligodendrocytes may delay maturation or change their function toward becoming antigen-presenting cells; the presence of p.A53T aSYN may have a similar effect due to its genetic variation and conformational change of the protein (63, 64). Our finding that diseased OLCs have impaired maturation concurs with data suggesting that in PD there is widespread pathology of cerebral white matter (65) associated with cognitive decline (66). These findings are also interesting in light of Windrem and colleagues (67) showing that iPSC-derived glia generated from patients suffering schizophrenia exhibit a cell autonomous pathology associated with delayed maturation.
The second major observation in this study is the involvement of an innate inflammatory component, revealed by increased expression of complement component C4b, HLA, and immunoproteasome subunit genes in PD p.A53T aSYN and in MSA O4+ OLCs. A recent study by Kirby and colleagues (31) showed that oligodendroglia become immune reactive instead of maturing into myelinating oligodendroglia in an experimental model of multiple sclerosis. Our data demonstrate that this applies also to MSA and PD p.A53T aSYN iPSC-derived O4+ OLCs and may be due to both structures and properties of the high-MW aSYN as well as the p.A53T variation. There are major differences between aSYN fibrils generated in vitro under controlled conditions compared to those obtained from PD and MSA brains, as revealed by seeding activity and cryogenic electron microscopy (68–72). Indeed, Peng and colleagues (70) identified GCI-derived aSYN seeds to be 1,000-fold more potent than LB-derived seeds following de novo aggregation. Additionally, MSA patient brain extracts greatly triggered aSYN aggregation in an engineered human embryonic kidney cell line and a mouse model of familial PD (M83 mice overexpressing pA53T aSYN) compared to PD patient brain extracts (71). This may explain why O4+ OLCs pathogenesis in MSA-C and MSA-P was much more pronounced than in SNCAtrip O4+ OLCs or following treatment of control O4+ OLCs with human WT aSYN fibrils.
In both MSA pathologies (MSA-P and MSA-C) and the PD SNCAtrip, protein aggregates contain WT aSYN, whereas in patients carrying the heterozygous genetic variation p.A53T aSYN, mutant aSYN is also incorporated within the aggregates. Hence, it was surprising to see that gene expression changes within the p.A53T aSYN O4+ OLCs resembled those present within MSA O4+ OLCs more than those present within the SNCAtrip O4+ OLCs. Gene expression analysis further revealed that aSYN p.A53T O4+ OLCs follow a transcriptomic pattern more similar to that of MSA-P and MSA-C O4+ OLCs than that of SNCAtrip O4+ OLCs, despite both aSYN p.A53T and SNCAtrip leading to PD. Based on previous work (70, 71, 73), we speculated that the structural properties, seeding propensity, and pathogenicity of p.A53T aSYN assemblies are closer to MSA assemblies than aSYN SNCAtrip ones and that these characteristics were induced by cell autonomous changes that are closer between p.A53T aSYN and MSA genotypes than the PD SNCAtrip genotype (based on our gene expression data). Thus, PD aSYN p.A53T OLC pathological severity, disease onset, and progression could be considered an intermediate between PD SNCAtrip and MSA pathologies.
It is important to mention that MSA is more lethal and that the disease has a more severe phenotype than idiopathic PD. Since posttranscriptional protein modifications in a disease-specific intracellular milieu might lead to structural heterogeneity of protein fibrils (74) and likely make them more or less harmful and/or prone to aggregation, future efforts should be directed toward unraveling the structure and properties of aSYN fibrillar polymorphs in different brain regions and cell types of patients diagnosed with different synucleinopathies (SNCA variants, idiopathic PD, DLB, MSA-P, MSA-C, atypical PD, etc.). This would allow definition of the pathogenicity of these aggregates by performing large-scale characterization and side-by-side prion-like seeding experiments or cryoEM. Such work could help clarify the aggregation state of aSYN in patients with synucleinopathies. Unveiling the structure of pathogenic fibrillar polymorphs will be extremely important to 1) develop personalized treatments, for example, via patient-specific anti-aSYN antibodies that do not interfere with the natural function of monomeric and nonpathogenic aSYN oligomers but could stimulate an early immune response to neutralize pathogenic species, thus preventing toxicity to OLCs, and 2) to help predict phenoconversion from the prodromal stage, for example, from individuals affected by idiopathic rapid eye movement sleep behavior disorder (75), to establish accurate diagnosis and prognosis and implement preventive care.
Materials and Methods
Lead Contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Laurent Roybon (Laurent.roybon@med.lu.se).
Material Availability.
This study generated iPSC lines that are subject to material transfer agreement and general data protection regulation. Brain samples were obtained from The Netherlands Brain Bank (https://www.brainbank.nl/), The Portuguese Brain Bank (http://www.bancodecerebros.chporto.pt/), and The New York Brain Bank at Columbia University (http://columbianeuroresearch.org/taub/res-investigation.html#NewYork) (SI Appendix, Table S2).
Patient Identification, Protection, and Sampling.
All procedures were conducted in accordance with national and European Union directives. The patient biopsies utilized to generate the iPSCs were obtained with informed consent and after ethical committee approval at the Parkinson institute in Milan, Italy: Ethics Committee “Milano Area C” (https://comitatoeticoareac.ospedaleniguarda.it/) and registered under the number: 370–062015. The permit for reprogramming was delivered by the Swedish work environment authority to L.R. and registered under the number 20200–3211.
Human iPSC Lines.
The generation of iPSC lines CSC-3G, CSC-3S, CSC-32B, CSC-32K, and CSC-32R was previously published (6, 15, 32). Lines CSC-28N, CSC-36C, CSC-36D, and CSC-36E were recently described (13, 14). A similar approach was used to generate iPSCs from an MSA-C patient (iPSC lines CSC-30B; CSC-30C and CSC-30D) and an MSA-P patient (iPSC lines CSC-34D; CSC-34F and CSC-34M; SI Appendix, Fig. S1).
Human iPSC Lines Generation.
Human iPSCs were generated as previously described (32). The presence of the SNCAtrip (32) and the presence of the aSYN p.A53T variation (15) were previously confirmed. To assess the differentiation of the generated iPSCs into the three germ layers, cells were cultured as embryoid bodies in WiCell medium supplemented with 20 ng/mL FGF2 and plated on a 96-well plate for spontaneous differentiation, which was confirmed using immunofluorescent staining with antibodies against β-III-tubulin (Sigma-Aldrich, T8660, 1:200), smooth muscle actin (Sigma-Aldrich, A2547, 1:200), and α-fetoprotein (Sigma-Aldrich, A8542, 1:200) (SI Appendix, Fig. S1).
Oligodendrocyte Differentiation.
Human iPSC-derived OLCs were generated using a detailed protocol that was previously described (6, 18).
Generation of Midbrain Spheroids Containing FOXA2+/TH+ Neurons.
To differentiate the iPSC into midbrain dopaminergic neurons, previously established protocol was followed (76).
Differentiation of CNPase-Positive OLCs from Mouse Embryonic Stem Cells Derived from WT and M83 Mouse Model of PD.
The differentiation of mouse embryonic stem cells into CNPase+ oligodendrocytes was performed as previously described (18, 26).
FACS-Based Purification of O4+ OLCs.
For each sample, cells were dissociated using prewarmed Accutase, incubated for 20 min, resuspended in GDM medium without phenol red, and transferred to a FACS tube. After centrifugation, the pellet was resuspended in Dulbecco’s modified Eagle’s medium/F12 without phenol red containing 5% normal serum and O4 antibody. For exclusion of dead cells, 7-aminoactinomycin D (1:1,000) was added and incubated on ice for 5 min before sorting. Samples were analyzed using a BD FACSAria III (BD Biosciences) with FACSDiva v8.0 software (BD Biosciences) at the MultiPark Cellomics and Flow Cytometry Core technical platform, Lund University. The cytometer was set up using a 100-μm nozzle at a standard pressure of 20 psi and a frequency of 30.0 kHz and was calibrated daily using BD FACSDiva Cytometer Setup and Tracking (CS&T) software and CS&T Research Beads (BD Biosciences).
Immunocytochemistry, Immunohistochemistry, and Image Acquisition.
Immunocytochemistry (ICC) and immunohistochemistry were performed using standard protocols. Primary antibodies were diluted in blocking solution and incubated overnight at 4 °C: goat anti-FOXA2 (Santa Cruz, sc-6554, 1:250), mouse anti-TH (Millipore, MAB1318, 1:500), goat anti–SOX-10 (R&D, AF2867, 1:200), rabbit anti-OLIG2 (Millipore, AB9610, 1:200), mouse anti-NKX2.2 (DSHB, 74.5A5, 1:50), chicken anti-MAP2 (Abcam, ab92434, 1:2,500), mouse anti-O4 (Kindly shared by J. Goldman at Columbia University, in the City of New York, 1:50), rabbit anti-aSYN (Santa Cruz, SC7011-R, 1:200), chicken anti-MBP (Millipore, AB9348, 1:100), rabbit anti-PSMB9 (Abcam, ab3328, 1:500), mouse anti-GFAP (Sigma-Aldrich, G3893, 1:400), mouse anti–phospho-α-Synuclein (Ser129) clone 81A (Millipore, MABN826, 1:1,000), and goat anti-Olig2 (R&D, AF2418, 1:100). On the next day, appropriate Alexa Fluor 488-, 555-, or 647-conjugated secondary antibodies were used at 1:400 in phosphate-buffered saline (PBS) or PBS-Tween 20 for 1 h at room temperature in the dark. Additionally, cell nuclei were stained by 6′-diamidino-2-phenylindole (Sigma-Aldrich, D9542, 1:10,000). All fluorescent photomicrographs were taken using an inverted epifluorescence microscope (LRI – Olympus IX-73) or confocal microscope (Leica TCS SP8 confocal microscope [Leica Microsystems] equipped with Diode 405/405-nm and Argon (405-, 488-, 552-, 638-nm) lasers with an HP PL APO 63x/NA1.2 water immersion objective).
Western Blotting.
Spheroids aged 100 d and adherent cultures aged 130 d were lysed with RIPA buffer complemented with Halt Protease Inhibitor and Halt Phosphatase Inhibitor Mixtures. Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23227) according to the manufacturer’s instructions. Samples were combined with loading buffer, heated at 70 °C for 10 min, and loaded into Bolt 4 to 12% Bis-Tris Plus Gels. After electrophoresis, the samples were transferred to polyvinylidene difluoride membranes. Membranes were blocked in either 5% skim milk or 5% bovine serum albumin (for phosphorylated proteins) diluted in 0.1% PBS-Tween 20 and incubated overnight with target primary antibodies diluted in blocking solution at 4 °C: mouse anti–β-actin (Sigma-Aldrich, A5441, 1:20,000), rabbit anti-aSYN (Cell Signaling, 2642S, 1:500), rabbit anti-phosphoS129 (Abcam, ab1683811, 1:500), rabbit anti-PDGFRα (Santa Cruz, sc-338, 1:200), mouse anti-MBP (Abcam, ab62631, 1:2,000), rabbit anti-PSMB9 (Abcam, ab3328, 1:500), and rabbit anti-C4b (Abcam, ab66791, 1:500). The following day, the membranes were incubated with horseradish peroxidase–conjugated secondary antibodies (R&D Systems, 1:5,000) for 1 h. The blots were then visualized using Pierce Enhanced Chemo-Luminescence solution and imaged in a Chemi-Doc chemo-luminescence system (Bio-Rad Laboratories). Bands were quantified using Image Lab (Bio-Rad Laboratories). To restain membranes for different markers, Restor PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific, 46430) was used.
FTIR Microspectroscopy.
FTIR microspectroscopy analyses were performed on FACS-sorted O4+ OLCs aged 130 DIV and spheroids containing dopaminergic neurons aged 75 DIV. Cell pellets were washed and spread (1 μL) on the 1-mm-thick CaF2 spectrophotometric window and dried under nitrogen flow. For reproducibility, infrared (IR) spectra were taken from different areas of the cell pellet deposited on CaF2. The background spectra were measured from a clean area of the same CaF2 window, close to the cell pellet. Spectra were recorded on a Hyperion 3000 IR microscope (Bruker Scientific Instruments, Billerica, MA) coupled to a Tensor 27, which was used as the IR light source with 15Å∼ IR objective and mercury cadmium telluride detector. The measuring range was 900 to 4,000 cm−1, and the spectra collection was done in transmission mode at 4 cm−1 resolution from 250 to 500 coadded scans. All measurements were made at room temperature. For analysis of FTIR spectra, OPUS software (Bruker) and Orange (University of Ljubljana) were used after atmospheric compensation. Derivation of the spectra to the second-order using Savitzky-Golay of third polynomial order 3 with 9 smoothing points was used to unmask the peak corresponding to β-sheet structures (1,628 cm−1) and to eliminate the baseline contribution (77). To analyze β-sheet structural content, the 1,628 cm−1/1,656 cm−1 band ratio was studied (78).
Analysis of aSYN in Culture Media.
aSYN present in the growth media of OLC cultures aged 130 DIV was quantified using the U-PLEX Human α-Synuclein Kit (Meso Scale Discovery).
SNCA Overexpression.
Day 110 to 115–enriched oligodendrocyte cultures were transduced independently with retrovirus pCMMP-haSYNWT-IRES2-eGFP-WPRE, retrovirus pCMMP-haSYNA53T-IRES2-eGFP-WPRE, and retrovirus pCMMP-IRES2-eGFP-WPRE in GDM medium supplemented with protamine-sulfate (4 mg/mL) for 1 to 2 wk prior to sample collection for immunocytochemistry and FACS purification/analysis. Cells started to express GFP within 24 h (SI Appendix, Fig. S6). According to gene expression data, transgene expression was 30- to 60-fold greater than endogenous SNCA expression.
Generation of ATTO-Labeled Human Recombinant aSYN, Characterization of Fibrils, and Treatment.
Recombinant WT aSYN or p.A53T aSYN was purchased from Alexotech in a lyophilized form. To covalently label aSYN with ATTO-488 fluorescent tag, we incubated monomeric proteins with reactive ATTO dye freshly dissolved in dimethyl sulfoxide (Sigma-Aldrich) as previously reported (11). Unreactive dye was removed by gel filtration chromatography using GE Sephadex G-25 according to manufacturer’s instructions (GE Healthcare). Covalent labeling was confirmed using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SI Appendix, Fig. S6), and labeled fractions were further analyzed by Western blot and size exclusion chromatography as previously reported (12). Labeled fractions were combined and concentrated to 5 mg/mL using centrifugal filter units with a 3,000-MW cutoff (Millipore). To generate aSYN fibrils, we incubated aSYN monomers (5 mg/mL) at 37 °C for 5 d in an Eppendorf SS minishaker with constant shaking at 1,000 rpm. Assembled proteins were then aliquoted and stored at −80 °C for further analysis. To confirm aSYN fibrillar formation, we performed Thioflavin T (ThT) analysis by incubation of ThT (10 mM) with monomeric and protein assemblies prior to measuring fluorescence intensity. To further visualize protein assemblies, we prepared samples for transmission electron microscopy (TEM). Briefly, 5 μL of aSYN fibrils solution was deposited and adsorbed on a 400-mesh carbon-coated copper grid. The specimen was stained with 2% uranyl acetate. The images were acquired in a 120-kv Tecnai bioTWIN microscope equipped with an Olympus Veleta camera. TEM was performed at the Lund University bioimaging center (https://www.lbic.lu.se/explore-our-infrastructure/microscopy). For treatment, 110- to 115-DIV–enriched OLC cultures were treated with 10 µM monomers or fibrils in GDM medium supplemented 1 to 2 wk prior to sample collection for Western blot, ICC, or FACS purification for transcriptomic analysis.
Expression Data Analysis.
A total of 240,000 FACS-sorted O4+ OLCs were washed twice with PBS, and pellets were snap-frozen in liquid nitrogen and stored at −80 °C before shipping to Kompetenzzentrum Fluoreszente Bioanalytik (Germany) for gene expression analysis using the human Clariom D array (Affymetrix). RNA extraction and sample preparation were performed at an Affymetrix Service Provider and Core Facility, “KFB—Center of Excellence for Fluorescent Bioanalytics” (Resensburg, Germany), and carried out as described in the Affymetrix GeneChip WT PLUS Reagent Kit User Manual (Affymetrix, Inc., Santa Clara, CA).
Quantification and Statistical Analysis.
Statistical analyses.
Statistical analyses were performed either with GraphPad Prism 7 software or with R (https://www.r-project.org/). Data were presented as mean ± SEM. A P value <0.05 was considered significant: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Sample groups were subjected to unpaired t test or ANOVA, as indicated in the figure legends.
Gene expression analysis.
Summarized probe set signals in log2 scale were calculated by using the GCCN-SST-RMA algorithm with the Affymetrix GeneChip Expression Console v1.4 Software. Average signal values, comparison of fold changes, and significance P values were calculated with R. The P value was adjusted for multiple comparison by performing the Benjamini-Hochberg false discovery rate calculation in R. We flagged as significant genes for which the raw P values (sig._p-value) were less than 0.05 and less than the corresponding adjusted P value, as previously employed (14, 67). Heatmaps of gene expression and fold change between condition versus control were made with ComplexHeatmap version 2.4.3 (79). Volcano plots of log P value against log fold change were done using ggplot2 version 3.3.0 (80). KEGG pathway analysis was performed with WebGestalt version 0.4.4 (81). GO term enrichment was performed with hypergeometric distribution and GSEA using WebGestalt (http://www.webgestalt.org/) with Reactome and KEGG databases as background. To assess possible functional protein association networks, we used Stringdb via Cytoscape for genes significantly up- or down-regulated by at least twofold. The lists of lipid-associated genes whose expression was changed in the OLCs were generated by filtering the genes whose description contained the term “lipid” from each of the expression datasets. From the filtered lists, a subset of genes associated with lipid metabolism was generated by retaining only genes that were significant after adjusting for multiple comparisons.
Supplementary Material
Acknowledgments
We thank Marianne Juhlin for her outstanding technical assistance, the staff at the cytogenetic units at Lund University Hospital for preparing the samples for karyotyping, and Dr. Sara Linse at Lund University for helping with the expression and purification of aSYN, as well as ATTO488 labeling, postlabeling purification, and fibril formation of both proteins for some of the preparations we used. We are also thankful to Per Persson and Ferenc Borondics for providing access to FTIR equipment, Dr. Stefano Goldwurm from the Parkinson Institute, ASST PINI-CTO in Milan (https://www.asst-pini-cto.it/), Italy, and the members of the Cell and Stem Cell laboratory for their help with the generation of tools and reagents, Poul Henning Jensen (Aarhus University, Denmark) and Hilal Lashuel (Swiss Federal Institute of Technology, Lausanne, Switzerland), who provided WT SNCA and p.A53T aSYN complementary DNAs to generate the viral constructs, and James E. Goldman (Columbia University, New York) for his generous gift of O4 antibody. We acknowledge the Cell Line and DNA Biobank From Patients Affected by Genetic Diseases (Istituto G. Gaslini, Genova, Italy) and the Parkinson Institute Biobank and members of the Telethon Network of Genetic Biobanks funded by Telethon Italy (project no. GTB12001, http://biobanknetwork.telethon.it) for providing fibroblast samples. We are also thankful to the New York Brain Bank at Columbia University (82), The Netherlands Brain Bank, and the Portuguese Brain Bank of Centro Hospitalar Universitário do Porto (Portugal) for providing us with the patient postmortem tissue samples. This work was supported by Swedish private foundations (the Holger Crafoord Foundation, the Shaking Generation Foundation, the Petrus and Augusta Hedlunds Foundation, the Åke Wibergs Foundation, the Greta och Johan Kocks Foundation, donations for science, medicine, and technology at Fysiografen in Lund to L.R.), the Swedish Research Council (Grants VR-2015-03684 and VR-2121-02284 to L.R.), Brainstem–Stem Cell Center for Excellence in Neurology funded by the Innovation Fund Denmark (to L.R.), and the Olav Thon Foundation in Norway (to L.R.). M.V.’s laboratory is supported by the Swedish Research Council (Grant VR-2015-02510). L.R. and G.K.G. received funding for the project from the European Union’s Horizon 2020 research and innovation program via the Swedish Research Council (2015-06798) under the aegis of the Joint Programme for Neurodegeneration, to develop spheroid models. L.R.’s laboratory is part of the Lund Stem Cell Center and the Strategic Research Area MultiPark at Lund University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111405119/-/DCSupplemental.
Data Availability
The datasets generated during this study are available in the supporting information.
Change History
March 28, 2022: Figure 3b has been updated from a bar graph to a scatter plot for clarity and the labels for Figure 3f have been updated for consistency. Figure 4 has also been updated.
References
- 1.Burn D. J., Jaros E., Multiple system atrophy: Cellular and molecular pathology. Mol. Pathol. 54, 419–426 (2001). [PMC free article] [PubMed] [Google Scholar]
- 2.Kiely A. P., et al. , Distinct clinical and neuropathological features of G51D SNCA mutation cases compared with SNCA duplication and H50Q mutation. Mol. Neurodegener. 10, 41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee K., et al. , LRRK2 p.Ile1371Val mutation in a case with neuropathologically confirmed multi-system atrophy. J. Parkinsons Dis. 8, 93–100 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Pasanen P., et al. , Novel alpha-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol. Aging 35, 2180.e1-5 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Taipa R., et al. , DJ-1 linked parkinsonism (PARK7) is associated with Lewy body pathology. Brain 139, 1680–1687 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Djelloul M., et al. , Alpha-synuclein expression in the oligodendrocyte lineage: An in vitro and in vivo study using rodent and human models. Stem Cell Reports 5, 174–184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richter-Landsberg C., Gorath M., Trojanowski J. Q., Lee V. M., Alpha-synuclein is developmentally expressed in cultured rat brain oligodendrocytes. J. Neurosci. Res. 62, 9–14 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asi Y. T., et al. , Alpha-synuclein mRNA expression in oligodendrocytes in MSA. Glia 62, 964–970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaji S., et al. , Pathological endogenous α-synuclein accumulation in oligodendrocyte precursor cells potentially induces inclusions in multiple system atrophy. Stem Cell Reports 10, 356–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes J. F., et al. , Alpha-synuclein transfers from neurons to oligodendrocytes. Glia 62, 387–398 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Reyes J. F., et al. , Binding of α-synuclein oligomers to Cx32 facilitates protein uptake and transfer in neurons and oligodendrocytes. Acta Neuropathol. 138, 23–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pomeshchik Y., et al. , Human iPSC-derived hippocampal spheroids: An innovative tool for stratifying Alzheimer disease patient-specific cellular phenotypes and developing therapies. Stem Cell Reports 15, 256–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russ K., et al. , TNF-α and α-synuclein fibrils differently regulate human astrocyte immune reactivity and impair mitochondrial respiration. Cell Rep. 34, 108895 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Azevedo C., et al. , Generation of an induced pluripotent stem cell line (CSC-32) from a patient with Parkinson’s disease carrying a heterozygous variation p.A53T in the SNCA gene. Stem Cell Res. (Amst.) 43, 101694 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Polymeropoulos M. H., et al. , Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Ryan S. D., et al. , Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell 155, 1351–1364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djelloul M., Azevedo C., Pomeshchik Y., Hammarberg A., Roybon L., Reporting on methods to generate and purify rodent and human oligodendrocytes from different sources. Stem Cell Res. (Amst.) 20, 58–66 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Liao Y., Wang J., Jaehnig E. J., Shi Z., Zhang B., WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47 (W1), W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahn O., Tenzer S., Werner H. B., Myelin proteomics: Molecular anatomy of an insulating sheath. Mol. Neurobiol. 40, 55–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcão A. M., et al. , PAD2-mediated citrullination contributes to efficient oligodendrocyte differentiation and myelination. Cell Rep. 27, 1090–1102.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galloway D. A., Moore C. S., miRNAs as emerging regulators of oligodendrocyte development and differentiation. Front. Cell Dev. Biol. 4, 59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emery B., Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Goldman S. A., Kuypers N. J., How to make an oligodendrocyte. Development 142, 3983–3995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chumarina M., et al. , Derivation of mouse embryonic stem cell lines from tyrosine hydroxylase reporter mice crossed with a human SNCA transgenic mouse model of Parkinson’s disease. Stem Cell Res. (Amst.) 19, 17–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giasson B. I., et al. , Neuronal alpha-synucleinopathy with severe movement disorder in mice expressing A53T human alpha-synuclein. Neuron 34, 521–533 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Ettle B., et al. , α-Synuclein-induced myelination deficit defines a novel interventional target for multiple system atrophy. Acta Neuropathol. 132, 59–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ettle B., et al. , Intracellular alpha-synuclein affects early maturation of primary oligodendrocyte progenitor cells. Mol. Cell. Neurosci. 62, 68–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.May V. E., et al. , α-Synuclein impairs oligodendrocyte progenitor maturation in multiple system atrophy. Neurobiol. Aging 35, 2357–2368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirby L., et al. , Oligodendrocyte precursor cells present antigen and are cytotoxic targets in inflammatory demyelination. Nat. Commun. 10, 3887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmqvist S., et al. , Creation of a library of induced pluripotent stem cells from Parkinsonian patients. NPJ Parkinsons Dis. 2, 16009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittaker H. T., Qui Y., Bettencourt C., Houlden H., Multiple system atrophy: Genetic risks and alpha-synuclein mutations. F1000 Res. 6, 2072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spira P. J., Sharpe D. M., Halliday G., Cavanagh J., Nicholson G. A., Clinical and pathological features of a Parkinsonian syndrome in a family with an Ala53Thr alpha-synuclein mutation. Ann. Neurol. 49, 313–319 (2001). [PubMed] [Google Scholar]
- 35.Beyer K., Domingo-Sàbat M., Ariza A., Molecular pathology of Lewy body diseases. Int. J. Mol. Sci. 10, 724–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiba Y., et al. , Immunohistochemical localization of aggresomal proteins in glial cytoplasmic inclusions in multiple system atrophy. Neuropathol. Appl. Neurobiol. 38, 559–571 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa T., et al. , Role of TPPP/p25 on α-synuclein-mediated oligodendroglial degeneration and the protective effect of SIRT2 inhibition in a cellular model of multiple system atrophy. Neurochem. Int. 57, 857–866 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Langerveld A. J., Mihalko D., DeLong C., Walburn J., Ide C. F., Gene expression changes in postmortem tissue from the rostral pons of multiple system atrophy patients. Mov. Disord. 22, 766–777 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Nakamura S., Kawamoto Y., Nakano S., Akiguchi I., Kimura J., Cyclin-dependent kinase 5 and mitogen-activated protein kinase in glial cytoplasmic inclusions in multiple system atrophy. J. Neuropathol. Exp. Neurol. 57, 690–698 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Odagiri S., et al. , Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in α-synucleinopathy. Acta Neuropathol. 124, 173–186 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Papp M. I., Kahn J. E., Lantos P. L., Glial cytoplasmic inclusions in the CNS of patients with multiple system atrophy (striatonigral degeneration, olivopontocerebellar atrophy and Shy-Drager syndrome). J. Neurol. Sci. 94, 79–100 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Pountney D. L., et al. , Alpha B-crystallin is a major component of glial cytoplasmic inclusions in multiple system atrophy. Neurotox. Res. 7, 77–85 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Uversky V. N., Alpha-synuclein misfolding and neurodegenerative diseases. Curr. Protein Pept. Sci. 9, 507–540 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Wakabayashi K., et al. , The Lewy body in Parkinson’s disease and related neurodegenerative disorders. Mol. Neurobiol. 47, 495–508 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Xu G., Stevens S. M. Jr., Moore B. D., McClung S., Borchelt D. R., Cytosolic proteins lose solubility as amyloid deposits in a transgenic mouse model of Alzheimer-type amyloidosis. Hum. Mol. Genet. 22, 2765–2774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zatloukal K., et al. , p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol. 160, 255–263 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murata S., Takahama Y., Kasahara M., Tanaka K., The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 19, 923–931 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Flachsbart F., et al. , Investigation of complement component C4 copy number variation in human longevity. PLoS One 9, e86188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y., et al. , Diversity in intrinsic strengths of the human complement system: Serum C4 protein concentrations correlate with C4 gene size and polygenic variations, hemolytic activities, and body mass index. J. Immunol. 171, 2734–2745 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Halliday G. M., Holton J. L., Revesz T., Dickson D. W., Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 122, 187–204 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Bryois J., et al. ; Eating Disorders Working Group of the Psychiatric Genomics Consortium; International Headache Genetics Consortium; 23andMe Research Team, Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat. Genet. 52, 482–493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teeple E., et al. , Single nuclei sequencing of human putamen oligodendrocytes reveals altered heterogeneity and disease-associated changes in Parkinson’s disease and multiple system atrophy. bioRxiv [Preprint] (2021). 10.1101/2021.05.06.442967 (Accessed 7 May 2021). [DOI]
- 53.Chamling X., et al. , Single-cell transcriptomic reveals molecular diversity and developmental heterogeneity of human stem cell-derived oligodendrocyte lineage cells. Nat. Commun. 12, 652 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falcão A. M., et al. , Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 24, 1837–1844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jäkel S., et al. , Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Brien J. S., Stability of the myelin membrane. Science 147, 1099–1107 (1965). [DOI] [PubMed] [Google Scholar]
- 57.Brekk O. R., Honey J. R., Lee S., Hallett P. J., Isacson O., Cell type-specific lipid storage changes in Parkinson’s disease patient brains are recapitulated by experimental glycolipid disturbance. Proc. Natl. Acad. Sci. U.S.A. 117, 27646–27654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muñoz S. S., Petersen D., Marlet F. R., Kücükköse E., Galvagnion C., The interplay between glucocerebrosidase, α-synuclein and lipids in human models of Parkinson’s disease. Biophys. Chem. 273, 106534 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Marmion D. J., Peelaerts W., Kordower J. H., A historical review of multiple system atrophy with a critical appraisal of cellular and animal models. J. Neural Transm. (Vienna) 128, 1507–1527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bartels T., Choi J. G., Selkoe D. J., α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dettmer U., et al. , Parkinson-causing α-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nat. Commun. 6, 7314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nuber S., et al. , Abrogating native α-synuclein tetramers in mice causes a L-DOPA-responsive motor syndrome closely resembling Parkinson’s disease. Neuron 100, 75–90.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coskuner O., Wise-Scira O., Structures and free energy landscapes of the A53T mutant-type α-synuclein protein and impact of A53T mutation on the structures of the wild-type α-synuclein protein with dynamics. ACS Chem. Neurosci. 4, 1101–1113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Y., et al. , Cryo-EM structure of full-length α-synuclein amyloid fibril with Parkinson’s disease familial A53T mutation. Cell Res. 30, 360–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rae C. L., et al. , White matter pathology in Parkinson’s disease: The effect of imaging protocol differences and relevance to executive function. Neuroimage 62, 1675–1684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dadar M., Gee M., Shuaib A., Duchesne S., Camicioli R., Cognitive and motor correlates of grey and white matter pathology in Parkinson’s disease. Neuroimage Clin. 27, 102353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Windrem M. S., et al. , Human iPSC glial mouse chimeras reveal glial contributions to schizophrenia. Cell Stem Cell 21, 195–208.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bousset L., et al. , Structural and functional characterization of two alpha-synuclein strains. Nat. Commun. 4, 2575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guerrero-Ferreira R., et al. , Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8, e48907 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng C., et al. , Cellular milieu imparts distinct pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prusiner S. B., et al. , Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. U.S.A. 112, E5308–E5317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schweighauser M., et al. , Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van der Perren A., et al. , The structural differences between patient-derived α-synuclein strains dictate characteristics of Parkinson’s disease, multiple system atrophy and dementia with Lewy bodies. Acta Neuropathol. 139, 977–1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arakhamia T., et al. , Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180, 633–644.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye G., et al. , Predictors of conversion to α-synucleinopathy diseases in idiopathic rapid eye movement sleep behavior disorder. J. Parkinsons Dis. 10, 1443–1455 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Chumarina M., et al. , Cellular alterations identified in pluripotent stem cell-derived midbrain spheroids generated from a female patient with progressive external ophthalmoplegia and parkinsonism who carries a novel variation (p.Q811R) in the POLG1 gene. Acta Neuropathol. Commun. 7, 208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong J., Yu S., Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. (Shanghai) 39, 549–559 (2007). [DOI] [PubMed] [Google Scholar]
- 78.André W., Sandt C., Dumas P., Djian P., Hoffner G., Structure of inclusions of Huntington’s disease brain revealed by synchrotron infrared microspectroscopy: Polymorphism and relevance to cytotoxicity. Anal. Chem. 85, 3765–3773 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, New York, 2009). [Google Scholar]
- 81.Wang J., Vasaikar S., Shi Z., Greer M., Zhang B., WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45 (W1), W130–W137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramirez E. P. C., Keller C. E., Vonsattel J. P., The New York Brain Bank of Columbia University: Practical highlights of 35 years of experience. Handb. Clin. Neurol. 150, 105–118 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available in the supporting information.