Significance
Streamers, filamentous bacterial biofilms formed in flowing systems, are ubiquitous in natural and artificial environments, where they cause clogging of devices and spreading of infections. Despite their impact, little is known about the nature and properties of streamers and their response to fluid flow. Here, we uncover the specific contribution of bacterial secreted extracellular DNA and exopolysaccharide Pel, two important components in Pseudomonas aeruginosa biofilms, to the formation and the mechanical properties of the streamers. We then show how this knowledge can be used to control biofilm streamer formation, both to inhibit or to promote it.
Keywords: bacterial biofilms, Pseudomonas aeruginosa, extracellular DNA, biofilm rheology, fluid flow
Abstract
Across diverse habitats, bacteria are mainly found as biofilms, surface-attached communities embedded in a self-secreted matrix of extracellular polymeric substances (EPS), which enhance bacterial recalcitrance to antimicrobial treatment and mechanical stresses. In the presence of flow and geometric constraints such as corners or constrictions, biofilms can take the form of long, suspended filaments (streamers), which bear important consequences in industrial and clinical settings by causing clogging and fouling. The formation of streamers is thought to be driven by the viscoelastic nature of the biofilm matrix. Yet, little is known about the structural composition of streamers and how it affects their mechanical properties. Here, using a microfluidic platform that allows growing and precisely examining biofilm streamers, we show that extracellular DNA (eDNA) constitutes the backbone and is essential for the mechanical stability of Pseudomonas aeruginosa streamers. This finding is supported by the observations that DNA-degrading enzymes prevent the formation of streamers and clear already formed ones and that the antibiotic ciprofloxacin promotes their formation by increasing the release of eDNA. Furthermore, using mutants for the production of the exopolysaccharide Pel, an important component of P. aeruginosa EPS, we reveal an concurring role of Pel in tuning the mechanical properties of the streamers. Taken together, these results highlight the importance of eDNA and of its interplay with Pel in determining the mechanical properties of P. aeruginosa streamers and suggest that targeting the composition of streamers can be an effective approach to control the formation of these biofilm structures.
The extracellular matrix confers stability to biofilms (1, 2) and protects the bacterial community against chemical and mechanical insults (3, 4). This protected environment makes biofilm bacteria a major cause of chronic infections in clinical environments and of clogging in industrial flow systems (5–8). The diverse biopolymers composing the extracellular matrix form the three-dimensional scaffold of the biofilm and are responsible for both adhesion to the surface and cohesion within the biofilm. The extracellular polymeric substances (EPS) include polysaccharides, proteins, extracellular DNA (eDNA), and lipids; however, the composition can vary greatly depending on the microorganisms present, the shear forces experienced, temperature, and nutrient availability (3). The chemical composition and the resulting intermolecular interactions between EPS components drive the self-assembly of the components and determine the structure and the mechanical properties of the biofilm network (9–12). The rheological signature of biofilms is a viscoelastic response to external forces, characterized by an instantaneous elastic deformation followed by a viscous flow, which allows dissipation of long-lasting stresses without structural failure (13). This rheological behavior is key to biofilm persistence in flow (14, 15) and allows biofilms to adapt their architecture to colonize very diverse environments (16–18).
Streamers are perhaps the most profound example of biofilm structural specialization in response to environmental conditions (19–21). When the viscoelastic biofilm matrix is subjected to the hydrodynamic forces generated by flows, biofilms can take the form of long filamentous suspended structures, the streamers, which increase the carrying capacity of biofilms in natural ecosystems (22). Indeed, their localization within the bulk fluid flow, in contrast to the thin-film morphology of surface-associated biofilms, confers streamers a greater impact on the uptake of microorganisms and debris flowing by and clogging (23, 24). Structures morphologically similar to streamers can also be formed by abiotic materials in flow, as shown by flowing a suspension of polyacrylamide and polystyrene particles through a porous structure (25). Aggregation processes and the viscous nature of the polymeric matrix play a crucial role in the formation of these filamentous structures (26, 27). These observations suggest that streamer formation is driven by multiple factors, including the local three-dimensional hydrodynamic profile and EPS mechanical properties, in addition to bacterial physiology. Yet, to date, a mechanistic link between extracellular matrix composition, the interaction between components, and the streamers’ morphology and rheology is still not established. This is in part due to the suspended nature of streamers, which makes it particularly challenging to measure their physicochemical properties without adequate in situ measurement techniques (21).
The opportunistic pathogen Pseudomonas aeruginosa is one of the best-characterized model organisms in the study of biofilm development (28–32). It forms biofilms in diverse habitats, including soil, rivers, medical devices, and human organs (6, 33, 34), with different architectures: In the surface-attached morphology, P. aeruginosa biofilms often show a characteristic mushroom-like shape (35, 36), whereas in a flowing fluid they can form streamers (19, 20, 23). P. aeruginosa can synthesize three exopolysaccharides: alginate, Pel, and Psl (37). Alginate production is typical of mucoidal strains isolated from patients with chronic pulmonary infections and is usually lost upon domestication (28). Most environmental and clinical isolates produce the polysaccharide Pel and some also produce the polysaccharide Psl (17, 37–40). Psl promotes early colony formation, increases elasticity, and strengthens the scaffold of biofilms, whereas Pel promotes adhesion among cells and to surfaces, decreases biofilm viscosity, thereby increasing the ability to spread on surfaces (17), and provides protection against antibiotics (39). In strains deficient in the Psl gene cluster, such as PA14, Pel is the primary exopolysaccharide and forms the scaffold of the entire biofilm (29, 30, 39, 41). In addition to polysaccharides, a protein, the extracellular adhesin CdrA, contributes to the structural stability of the biofilm matrix (42, 43).
eDNA is a functional component of P. aeruginosa biofilms (44–46). eDNA was initially considered a minor component, primarily relevant as a gene pool to be exploited for horizontal gene transfer (47), but its contribution to biofilm formation has been recently reevaluated (45, 46, 48). The presence of eDNA has also been documented in single- and multispecies biofilms formed in different environments (35, 49–58) by Staphylococcus aureus (49, 51, 52, 59), Myxococcus xanthus (53), Burkholderia cenocepacia (54), Staphylococcus epidermidis (51, 55), and Bacillus subtilis (56) and in mixed environmental samples (50, 57, 58). eDNA is released by cell death and lysis, which mainly occur in the interior of biofilms (e.g., the stalk of the mushrooms) (44, 60, 61). Explosive cell lysis is triggered by the SOS response that is induced by genotoxic stress, such as ultraviolet radiation or treatment with sub-MIC (minimum inhibitory concentration) of fluoroquinolone antibiotics, including ciprofloxacin (60). Thanks to the binding capability of the highly charged DNA molecule, eDNA protects the community from antimicrobials and antibiotics (62, 63). Additionally, eDNA promotes cell–cell adhesion (44) and biofilm self-organization (64) and enables the formation of a stable biofilm structure, as indirectly demonstrated by the degradation of biofilms by treatment with DNase I (45, 65), a nonspecific nuclease that cleaves DNA. Nevertheless, a quantification of the impact of eDNA and its interaction with the other components of the biofilm matrix on the rheological properties of biofilms is still missing.
Lectin staining has shown that Pel, composed of positively charged amino sugars, binds eDNA in the stalk of the mushroom structures in mature biofilms, conferring structural stability (35, 66). Biochemical analysis has recently shown that Pel also binds to CdrA, protecting it from proteolysis (42). Due to their opposite charges, Pel and eDNA cross-link by ionic bonding. The ionic-binding mechanism between Pel and eDNA is pH-dependent, since Pel is cationic until the isoelectric point at pH 6.7 to 6.9, where Pel carries no charge (35). This finding suggests that chemical heterogeneity within a biofilm (67) can influence Pel’s ability to bind to eDNA and, consequently, its localization within the biofilm structure. To date, eDNA–Pel colocalization and cross-linkage have been investigated in the mushroom architecture of surface-attached biofilms (35), but the influence of the eDNA–Pel interaction on streamer formation and rheological properties has remained unknown.
In this work, we analyze the distinct role of eDNA and Pel in the formation of P. aeruginosa streamers and show how their interaction tunes their rheological properties. We show that eDNA is the constitutive element of the backbone of streamers, essential for their formation and stability. This finding allows controlling the formation of streamers. Treatment with DNase I, a nonspecific nuclease that cleaves DNA, completely suppresses their formation and clears already formed streamers in the wild-type strain. The efficacy of the treatment is reduced in Pel-overproducing strains, due to the protecting action of Pel. On the other hand, sublethal concentrations of ciprofloxacin, an antibiotic known to induce cell lysis (60), can paradoxically stimulate the formation of streamers. This effect is due to the release of bacterial eDNA in the flowing solution that is captured by the streamers. By varying the composition of the EPS using Pel-mutant strains, we show that an increase in the Pel content determines an increase in the elastic modulus and the viscosity of the matrix, indicating that eDNA–Pel interaction results in stiffer streamers. Taken together, these findings provide a mechanistic understanding of the structural role of different matrix components in the formation of P. aeruginosa biofilm streamers.
Results
eDNA Represents the Backbone of Biofilm Streamers.
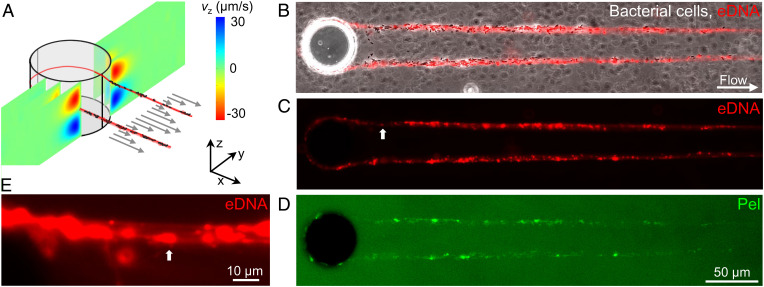
Obstacles in a flow field act as a tethering point for the formation of biofilm streamers. This scenario, which occurs, for example, in porous media, in the human body and filters, can be recapitulated in its simplest form by isolated pillars in a microfluidic channel. In our microfluidic platform, reproducible formation of streamers could be achieved on 50-µm-diameter, isolated pillars (Fig. 1A and Materials and Methods) exposed to a constant flow of a diluted suspension of P. aeruginosa PA14. Fluorescence and phase-contrast video microscopy were used to characterize the dynamics and composition of streamers during their formation (Fig. 1 B–D). The first strands formed after 3 to 5 h, and after about 10 to 15 h they had become millimeter-long stable filaments (Fig. 1B). Around each pillar, we observed the formation of two symmetric streamers, tethered on the side of the pillar and suspended at channel middepth (Fig. 1A). At the flow velocities used in our experiments, bacterial cells, in addition to colonizing the inner walls of the microfluidic channel, attached to the side of the pillar facing the flow (68) and EPS accumulated around the curved pillar surface (19) before being extruded by downstream flow (27). Biomass accumulation is recognized as the dominant mechanism for streamer initiation (22) and is promoted by the secondary flow around the curved surfaces (19), while the extrusion process is favored by the extensional component of the flow downstream of the pillar.
Fig. 1.
eDNA is a major component of PA14 biofilm streamers. (A) Schematic of a biofilm streamer formed around a pillar, due to the combined action of the main flow (gray arrows) and the secondary flow (color scale). The x (gray arrows) and the z component (color scale) of the flow velocity around a 50-µm pillar were numerically computed (COMSOL Multiphysics) for a mean flow velocity of 500 µm/s. (B–D) Representative phase-contrast and fluorescence images of a portion of the biofilm streamers formed by P. aeruginosa PA14 WT cells attached to a 50-µm pillar within a microfluidic channel after 20 h of continuous flow of a dilute bacterial suspension at U = 2 mm/s. (E) Magnified view of C. The white arrow indicates corresponding points in the figures. Images were taken at channel middepth. Bacterial cells were imaged in phase contrast (B), eDNA was stained using red-fluorescent PI (2 μg/mL; B, C, and E) and Pel was stained using green-fluorescent WFL (50 μg/mL; D).
This experimental system allowed us to identify the spatial localization of different components of streamers: Bacterial cells were visualized with phase-contrast microscopy (Fig. 1B), while eDNA and Pel were visualized by epifluorescence microscopy after staining with propidium iodide (PI) (Fig. 1C) and Wisteria floribunda lectin (WFL) (Fig. 1D), respectively (Materials and Methods). Cell aggregates were mainly located in the first 200 µm downstream from the pillar, whereas further downstream we mainly observed single bacteria embedded in the streamers (Fig. 1B). Both Pel and eDNA were present on the filament: eDNA was homogeneously distributed along the streamer (Fig. 1 B and C), whereas Pel was mainly colocalized with cell aggregates, resulting in a higher abundance in the region of the filament closer to the pillar (Fig. 1D). We observed a continuous red PI-stained thread in all the streamers, suggesting that bundles of eDNA connect the bacterial cells within the streamers (Fig. 1 C and E). The continuous distribution of eDNA within streamer filaments indicates that eDNA formed the backbone structure of the streamers (Fig. 1C), whereas Pel was localized in micrometer-sized patches, mainly located in the cells’ clusters (SI Appendix, Fig. S1).
The Morphology and Mechanical Properties of Streamers Depend on Pel Abundance.
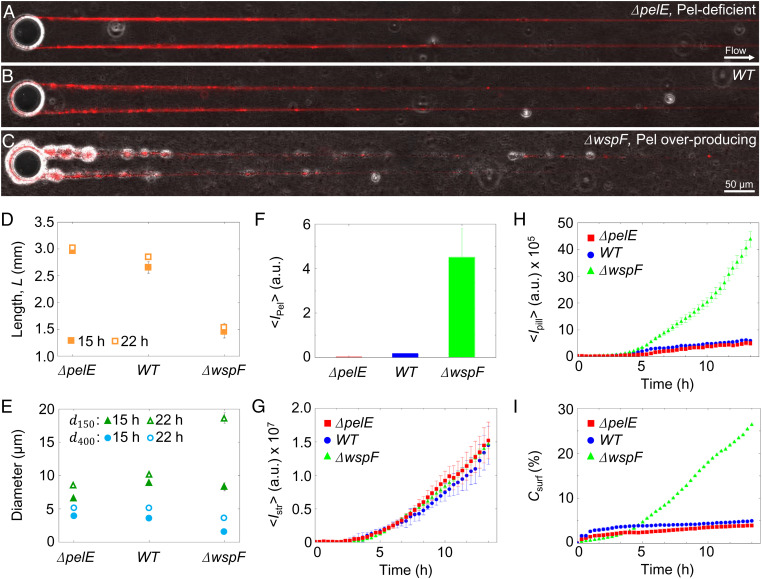
Pel is not essential for the formation of streamers, but it has an impact on their morphology. The role of Pel was determined by comparing the streamers formed by the wild-type strain (PA14 WT), a mutant strain lacking the ability to produce Pel (PA14 ΔpelE), and a Pel-overproducer strain (PA14 ΔwspF). All three strains produced millimeter-long streamers, on the same timescale (Fig. 2 A–C). WFL staining confirmed that PA14 ΔpelE streamers lacked Pel (Fig. 2F and SI Appendix, Fig. S1A), while Pel-overproducer streamers showed a 10-fold increase in the quantity of Pel compared to WT (Fig. 2F and SI Appendix, Fig. S1B). Pel-deficient streamers were on average 3.03 mm 0.03 mm long after 22 h, marginally (∼10%) longer than WT streamers and approximately twice as long as Pel-overproducer streamers, which reached an average length of 1.47 mm 0.28 mm (Fig. 2D). Differences in Pel content also affected the average diameter of streamers, which we measured at different distances from the pillar (Materials and Methods). After 22 h, WT streamers display a diameter d150 = 10 µm 0.2 µm in the vicinity of the pillar and d400 = 5 µm 0.3 µm in the downstream region. Compared to WT streamers, after 22 h Pel-deficient streamers showed a diameter d150 = 8.4 µm 0.2 µm (∼15%) in the vicinity of the pillar, which is 15% smaller compared to WT (Fig. 2E). Instead, the diameter of Pel-overproducer streamers near the pillar was comparable to the WT one after 15 h (d150 = 8.9 µm 0.3 µm for WT and d150 = 8.3 µm 1.2 µm for ΔwspF) and double that of WT streamers after 22 h (d150 = 10 µm 0.2 for WT and d150 = 18.6 µm 1.6 µm for ΔwspF). The trend is inverted in the downstream portion of the filament, where the diameter of the Pel-overproducer strain was approximately half that of WT streamers (Fig. 2E). Data on streamers’ geometry were quantified over 25 replicates. Cell aggregates were also mainly located in the vicinity of the pillar (Fig. 2 A–C). Pel-deficient streamers (Fig. 2A) had substantially fewer and smaller cell clusters than WT streamers (Fig. 2B), while in Pel-overproducer streamers bacterial aggregates were larger and more abundant (Fig. 2C).
Fig. 2.
Pel affects the morphology of PA14 biofilm streamers. (A–C) Representative overlaid phase-contrast (bacterial cells) and red fluorescence (eDNA) images of the biofilm streamers formed by P. aeruginosa PA14 Pel-deficient (ΔpelE) (A), WT (B), and Pel-overproducing (ΔwspF) (C) cells attached to a 50-µm pillar after 15 h of continuous flow of a dilute bacterial suspension at U = 2 mm/s. The images are representative of the streamer morphology obtained with the different mutant strains. (D) Length of streamers after 15 h (filled symbols) and 22 h (open symbols) of continuous flow, measured from fluorescence images for ΔpelE, WT, and ΔwspF strains. (E) Diameter of streamers in the vicinity of the pillar (150 µm from the pillar, (green triangles) and in the downstream region (400 µm from the pillar, (blue circles) after 15 h (filled symbols) and 22 h (open symbols) of continuous flow, measured from fluorescence images for the three strains. (F) Green fluorescence intensity of streamers, , corresponding to the Pel content, for the three strains after 24 h. (G) Red fluorescence intensity of streamers, , corresponding to the eDNA content, as a function of time (red squares, ΔpelE; blue circles, WT; green triangles, ΔwspF). (H) Red fluorescence intensity measured on the pillar surface, , as a function of time for the same suspensions as G. (I) Coverage of the bottom surface of the channel, , as a function of time for the same suspensions as G and H. Error bars represent the SEM.
Despite the differences in morphology associated with the different amounts of Pel, the quantity of eDNA per unit length of the streamer, , as assessed by the red fluorescence intensity of PI staining (Materials and Methods and SI Appendix, Fig. S2), was comparable in the three strains and showed the same, increasing trend with time (Fig. 2G). This trend is linear from 5 h onward (Fig. 2G) and is consistent with the previously proposed filtration model (22, 23, 27), which attributes the streamer’s growth to the addition of mass captured by the streamer from the flowing fluid. This dynamics contrasts with the differences in biofilm growth on the surface among the three strains, which was determined by analyzing the colonization of the horizontal surface of the microfluidic channel and the vertical surface of the pillar. In terms of surface coverage, quantified as the fraction Csurf of the surface covered by bacteria, the Pel-deficient strain shows the lowest surface growth, with Csurf < 4% after 14 h (Fig. 2I), closely followed by the WT (Csurf 5%). In line with this observation, WT and ΔpelE showed limited colonization of the pillar surface compared to the ΔwspF mutant, as quantified by the red fluorescence intensity on the pillar, (Fig. 2H). The Pel-overproducing strain displayed greater surface growth, leading to five times higher surface coverage (Fig. 2I) and seven times higher biomass accumulation on the pillar (Fig. 2H) in comparison with the WT strain.
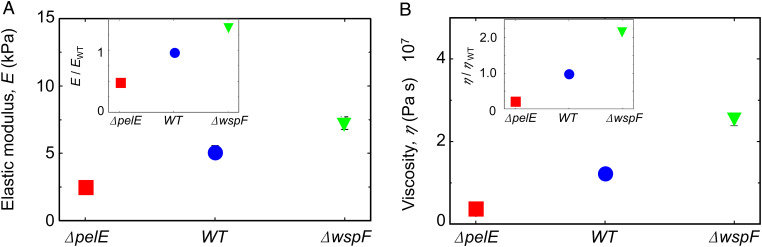
Pel not only affects the morphology of the streamers but also their viscoelastic behavior. We studied streamer rheology in situ using a newly developed technique that relies on measuring the deformation of the filaments in response to a controlled variation of fluid shear (Materials and Methods) (69). Specifically, streamers grown at a flow velocity of 2 mm/s were deformed by suddenly exposing them to a flow velocity of 4 mm/s for 5 min. We observed that streamers underwent a deformation typical of viscoelastic materials: On a timescale of seconds, they displayed an elastic deformation, which was recovered once flow velocity was lowered, followed by a slow viscous deformation on a timescale of minutes (SI Appendix, Fig. S3). All three strains exhibited this viscoelastic deformation; however, the elastic modulus E and the effective viscosity η associated with the deformation depended on the Pel content. The elastic modulus of WT streamers (E = 5.1 kPa 0.9 kPa) was double that of Pel-deficient streamers (E = 2.5 kPa 0.5 kPa) and 30% smaller than that of Pel-overproducer streamers (E = 7.2 kPa 0.9 kPa) (Fig. 3A, Inset). The impact of Pel abundance on the effective viscosity was even more pronounced: WT streamers had approximately a fivefold higher effective viscosity (η = 11.6 MPa s 1.7 MPa s) than Pel-deficient streamers (η = 2.7 MPa s 0.5 MPa s), while Pel-overproducing streamers had a viscosity 2.5-fold greater than WT streamers (η = 25.3 MPa s 2.9 MPa s) (Fig. 3B, Inset). Therefore, since both E and η increased with increasing Pel abundance, we conclude that Pel causes stiffening in the viscoelastic response of biofilm streamers (Fig. 3).
Fig. 3.
Pel stiffens biofilm streamers. (A) Elastic modulus, E, of biofilm streamers formed by P. aeruginosa PA14 ΔpelE (red), WT (blue), and ΔwspF (green) cells attached to a 50-µm pillar. Points show the average of the five independent average values associated to each bacterial batch. The data were collected for five independent bacterial batches, each one prepared on a different day. The error bar is the SEM. Values were obtained by analyzing the deformation in situ of a portion of filament between 400 µm and 1 mm from the pillar. (Inset) The elastic modulus is rescaled by the elastic modulus of PA14 WT, E/EWT. (B) Effective viscosity, η, measured in the same experiments as A. (Inset) The effective viscosity is rescaled by the viscosity of PA14 WT, η/ηWT. Error bar represents the SEM.
The formation of streamers by P. aeruginosa was also observed under more acidic and more alkaline conditions. The variation in pH did not affect streamer morphology; however, variation in pH altered the rheological properties of the streamers (SI Appendix, Fig. S4). On the other hand, the matrix protein CdrA, which has been shown to make a critical contribution to robust biofilm formation in flow cells by PA14 due to its capacity to bind to Pel (42), does not affect the formation or the mechanical properties of PA14 streamers (SI Appendix, Fig. S5).
eDNA Degradation Prevents Streamer Formation.
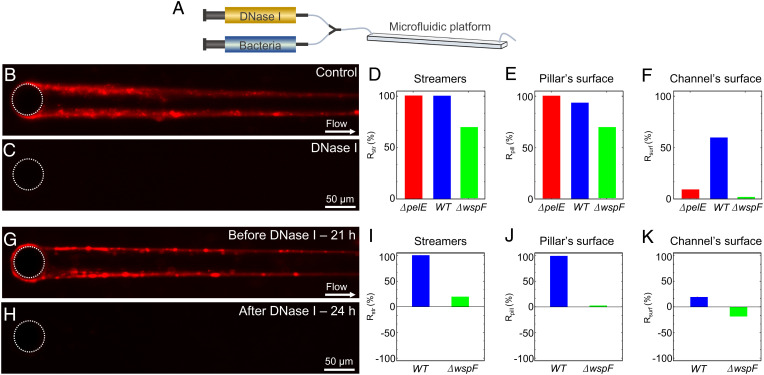
DNA degradation caused by treatment with the enzyme DNase I completely prevented the formation of streamers by PA14 WT, demonstrating that eDNA is an essential structural component of streamers in PA14. PA14 WT did not form any streamers over 24 h when treated with DNase I solution, together with its activators CaCl2 and MgCl2, from the start of the experiment (Fig. 4 A, C, and D and SI Appendix, Fig. S6I). DNase treatment also prevented the accumulation of biomass around the pillar (Fig. 4 C and E). Phase-contrast imaging confirmed that streamers were not formed under DNase I treatment (SI Appendix, Fig. S6 B, D, F, and H). Control experiments with only CaCl2 and MgCl2 revealed streamer formation (SI Appendix, Fig. S6 A, C, E, and G), confirming that DNase I was responsible for preventing streamer formation. In contrast to its effect on streamers and on the pillar surface, DNase I treatment only modestly hampered biofilm presence on the channel’s bottom surface, reducing bacterial surface-coverage Csurf by about 50% (Fig. 4F): this supports the conclusion that eDNA is more important (in fact, essential) in the formation of streamers than in the formation of surface-attached biofilms. By focusing on early-stage biofilm formation (t < 24 h), we did not investigate the maturation of the biofilm, which according to the literature is prevented by DNase I (45). In the Pel-deficient case, DNase I also completely prevented streamer formation and biofilm accumulation around the pillar surface, but its effect on the channel’s surface was even weaker than for the WT (SI Appendix, Fig. S7 and Fig. 4 D–F).
Fig. 4.
DNase I prevents biofilm streamer formation. (A) Schematic of the flow configuration during the experiment in which DNase I flow is injected at the beginning of the experiment. (B and C) Representative fluorescence images of biofilm streamers formed by P. aeruginosa PA14 WT cells attached to a 50-µm pillar after 24 h of continuous flow of a dilute bacterial suspension at U = 2 mm/s untreated (B) or treated (C) with 1 mg/mL DNase I. (D–F) Reduction in comparison to a control channel with no DNase treatment, Rstr, after 24 h of DNase I treatment calculated for the fluorescence intensity of the streamers, , (D), reduction, Rpill, in the fluorescence intensity of the biofilm around the pillar surface, (E), and reduction, Rsurf, in the surface coverage, (F), for P. aeruginosa PA14 ΔpelE (red), WT (blue), and ΔwspF (green). (G and H) Representative fluorescence images of the biofilm streamers formed by P. aeruginosa PA14 WT cells attached to a 50-µm pillar after 21 h of continuous flow of an untreated diluted bacterial suspension at U = 2 mm/s (G) and of the same streamer after 3 h of treatment with 1 mg/mL DNase I (H). (I–K) Reduction in comparison to a control channel with no DNase treatment, Rstr, for the signal after 3 h of DNase I treatment of streamer formed after 21 h of continuous flow of an untreated dilute bacterial suspension for the fluorescence intensity of the streamers, (I), reduction, Rpill, in the fluorescence intensity of the biofilm around the pillar surface, , (J), and reduction, Rsurf, for the surface coverage, (K) for P. aeruginosa PA14 WT (blue) and ΔwspF (green).
An overproduction of Pel in the biofilm formed by PA14 ΔwspF displayed a reduced effect of the DNase I treatment, represented by an ∼60% reduction in streamer formation and colonization of the pillar surface (Fig. 4 D and F). Additionally, the thickness of the biofilm on the pillar measured using phase-contrast microscopy seemed to increase during the treatment with DNase I and had an opposite trend compared to the PI fluorescent signal (Fig. 4E and SI Appendix, Fig. S8 A–E). The biofilm structures that survive the DNase I treatment showed a high Pel content, as observed by staining Pel with WFL in the biofilm remaining after the treatment (SI Appendix, Fig. S9 C and G). eDNA is colocalized with Pel within these structures, as shown by staining (SI Appendix, Fig. S9 D and H). This suggests that Pel protects eDNA from the DNase I treatment. In line with the higher surface colonization ability of PA14 ΔwspF (Fig. 2I), surface coverage was not affected by DNase I treatment (SI Appendix, Fig. S8G and Fig. 4F), confirming that Pel plays a prominent role in the formation of a surface-attached biofilm.
To understand whether DNase I treatment could also effectively remove streamers after their formation, we exposed established streamers of PA14 WT and PA14 ΔwspF to DNase I. This treatment completely removed a 21-h PA14 WT streamer, along with the biomass on the pillars, within 30 min (Fig. 4 I and J and SI Appendix, Fig. S6 P and Q), whereas it only reduced biofilm presence on the channel’s surface by 17% (Fig. 4L). Phase-contrast microscopy confirmed that DNase I treatment completely removed streamers (SI Appendix, Fig. S6). In contrast, streamers formed by PA14 ΔwspF were reduced by 20% (Fig. 4I and SI Appendix, Fig. S8H) and almost unaffected observing the Pel-rich biofilm formed around pillars (Fig. 4J and SI Appendix, Fig. S8O). The biofilm on the channel’s surface was also unaffected and continued to increase during the treatment, showing a 15% increase during the 3-h treatment (Fig. 4K and SI Appendix, Fig. S8P).
To explore and highlight the consequences of DNase I treatment on streamer formation in topographically more complex environments, we performed experiments with PA14 WT in a microfluidic model of a porous medium composed of thousands of identical pillars. When a dilute suspension of PA14 WT was flown through the porous medium, two different biofilm morphologies could be identified: In the first millimeter, a dense biofilm clog formed, whereas the downstream pillars supported a dense network of streamers, each akin to those observed on isolated pillars (SI Appendix, Fig. S10A). DNase I treatment prevented clogging when started before streamers formation (SI Appendix, Fig. S10 and Movie S1) and reduced clogging when started after a mature streamer network had formed (SI Appendix, Fig. S11 and Movie S2). When DNase I was added to the flow from the beginning of the experiment, no streamers formed for the 23 h of the experiment (SI Appendix, Fig. S10 B and C and Movie S1). This observation further strengthens the conclusion that eDNA filaments are the structural elements of streamers, which in turn cause clogging. Thus, preventing the formation of streamers by removing eDNA could avoid or delay clogging. In experiments in which a 20-h WT streamer network was treated with DNase I, the efficacy of the treatment depended on the biofilm morphology. While eDNA in the clog was removed, resulting in shrinkage of the biofilm (SI Appendix, Fig. S11 A and B), some biomass remained, which most likely was composed of Pel (SI Appendix, Movie S2). In contrast, the network of streamers was efficiently removed by DNase I (SI Appendix, Fig. S11 C and D), confirming that in PA14 WT eDNA is essential for the integrity of streamers (SI Appendix, Movie S2). The formation of biofilm streamers and the inhibition of their formation with DNase I was found to be a consistent feature across other P. aeruginosa strains and mutants with different polysaccharide composition of the extracellular matrix (SI Appendix, Fig. S12).
Antibiotic-Induced eDNA Release Promotes Streamer Formation.
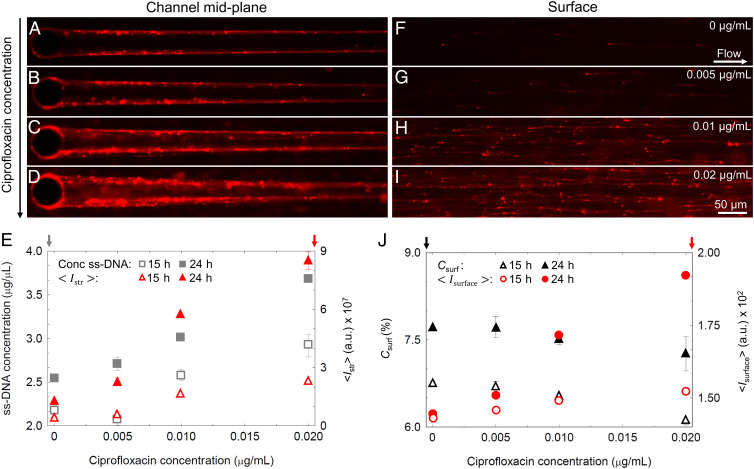
In further experiments, we observed that a higher abundance of eDNA resulted in thicker streamers. The eDNA concentration in the system can be increased by exposing bacteria to ciprofloxacin, an antibiotic that induces cell lysis and consequently the release of cytosolic contents, including DNA (60, 70). We performed experiments in which PA14 WT cells were exposed to three ciprofloxacin concentrations below the MIC, namely 0.005 μg/mL, 0.01 μg/mL, and 0.02 μg/mL, as they were flown through the microfluidic device with isolated pillars (Materials and Methods and SI Appendix, Fig. S13). We observed that increasing concentrations of ciprofloxacin caused an increase in the diameter of the streamer (Fig. 5 A–D) and in the eDNA concentration in the streamer, measured as the average red fluorescent intensity, (SI Appendix, Fig. S13B). The difference between the different ciprofloxacin concentrations in was initially small (t < 15 h; dotted line in SI Appendix, Fig. S13B and red open triangles in Fig. 5E) and then rapidly increased (SI Appendix, Fig. S13B). After 24 h, was 10-fold higher in streamers exposed to 0.02 μg/mL of ciprofloxacin than in the untreated control (red filled triangles in Fig. 5E). was linearly correlated with ciprofloxacin concentration at both 15 h and 24 h (Fig. 5E). The same trend was displayed by the concentration of single-stranded DNA (ssDNA) measured in the effluent of the devices (Materials and Methods and gray squares in Fig. 5E). Taken together, these data suggest that the increase in diameter of streamers is determined by an increased concentration of eDNA.
Fig. 5.
Ciprofloxacin increases biofilm streamer formation. (A–D) Representative fluorescence images of the biofilm streamers formed by P. aeruginosa PA14 WT attached to a 50-µm pillar after 24 h of continuous flow at U = 2 mm/s of a dilute bacterial suspension containing ciprofloxacin at concentrations of 0 μg/mL (A), 0.005 μg/mL (B), 0.01 μg/mL (C), and 0.02 μg/mL (D). Images were taken at channel middepth. (E) Concentration of ssDNA in solution (gray squares, left axis) and average red fluorescence intensity of the streamers, (red triangles, right axis) as a function of ciprofloxacin concentration, measured after 15 h (open symbols) and 24 h (filled symbols) of continuous flow of a dilute PA14 WT suspension. Error bar represents the SEM. (F–I) Fluorescence images of the eDNA network formed on the surface in the same channels as A–D, with ciprofloxacin at concentrations of 0 μg/mL (F), 0.005 μg/mL (G), 0.01 μg/mL (H), and 0.02 μg/mL (I). Images were taken on the glass surface of the channel 3 mm upstream from the pillar. (J) Surface coverage (percentage), (black triangles, left axis) and red fluorescence intensity, , measured on the horizontal surface of the channel (red circles, right axis) as a function of ciprofloxacin concentration, measured after 15 h (open symbols) and 24 h (filled symbols) in the same conditions as A–D. Error bar represents the SEM.
Ciprofloxacin caused a slight decrease in bacterial surface coverage, Csurf, which showed a maximal 10% reduction at the highest ciprofloxacin concentration tested (SI Appendix, Fig. S13C and Fig. 5J). However, the treatment increased the abundance of eDNA on the surface and promoted the development of a network of eDNA filaments, originating from the lysis of single cells and then spread downstream by the flow (Fig. 5 F–I). We quantified the eDNA increase on the surface by measuring the average red fluorescent intensity of the surface, (red circles in Fig. 5J and Materials and Methods). increases linearly with the antibiotic concentration and therefore ssDNA in solution. The highest ciprofloxacin concentration tested showed a 10-fold increase in compared to the control. This observation strengthens the hypothesis that the increased growth of streamers is due to higher eDNA concentration in solution rather than cell capture, since the numbers of cells on the surface of the channel and in the effluent solution actually slightly decreased with increasing antibiotic concentration, whereas the concentration of eDNA in the effluent solution and on the surface notably increased (Fig. 5 E–I). The decrease in the number of cells can be attributed to stress-induced explosive lysis causing the death of a limited portion of the population.
A similar trend was observed with the Pel-deficient PA14 ΔpelE mutant (SI Appendix, Fig. S14 A, B, and E), which showed a slight increase in streamer size with increasing ciprofloxacin concentration, and in the Pel-overproducing PA14 ΔwspF mutant (SI Appendix, Fig. S14 C–E), which showed a more pronounced increase in streamer diameter. We thus speculate that the absence of Pel reduces the eDNA binding sites available on the streamers and consequently its absorption, thus reducing the size of the streamers. Similarly, Pel overproduction enhances eDNA absorption and streamer growth.
Discussion
We have shown that eDNA is the fundamental structural component of P. aeruginosa biofilm streamers. Our observations demonstrated that the backbone of biofilm streamers consists of eDNA. The exopolysaccharide Pel is a further component of streamers, yet not an essential one, since a Pel-deficient mutant was still able to form streamers. In previous work (19, 20, 23), a Pel-deficient mutant did not form streamers in geometries where the filament had to cross the streamlines of the flow, as is the case in a channel with a series of corners. In contrast, our results in a geometry in which streamer growth occurred along the direction of streamlines show that a Pel-deficient mutant did form millimeter-long streamers. We hypothesize that the extensional component of the flow around the pillar may have favored the formation of streamers. Indeed, as previously shown using λ-phage DNA, a shear flow can induce and control the assembly of DNA, when the condition γ τ > 1 is satisfied, where γ is the flow shear rate and τ is the relaxation time of the DNA molecule (71, 72). In our configuration (SI Appendix) we are indeed in the regime where the flow can favor DNA assembly and consequently the creation of streamers.
Our data indicate that the quantity of Pel secreted by bacteria affects the morphology of streamers. The Pel-deficient strain was found to have limited coverage of the flat surface, as reported in previous work (30, 38, 39), in which deficiency in Pel production was found to reduce the surface colonization ability and prevent the formation of a mature surface-attached biofilm. We found that Pel also promotes the formation of cell aggregates along the streamer and is mainly localized within these aggregates, whereas eDNA forms the millimeter-long continuum thread that supports the streamer structure. Interestingly, an increase in Pel corresponds to an increase of bacterial aggregates, supporting the idea that Pel promotes cell–cell adhesion not only in the surface-attached biofilm configuration (32) but also within suspended streamer filaments. Comparison of results using Pel-mutants showed that Pel has an important effect on the streamers’ rheological behavior: The higher the concentration of Pel, the higher the elastic modulus and effective viscosity of the streamers, and thus their stiffness.
eDNA is an abundant and structurally important component of P. aeruginosa surface-attached biofilms (44, 45). Evidence has emerged that eDNA in surface-attached biofilms is localized in distinct patterns that depend on the age of the biofilm and that this, in turn, determines the biofilm structure: Young colonies present eDNA mostly on the upper surface, old colonies near the substratum (44). Here we have demonstrated that, for streamers, eDNA constitutes the skeleton of the streamer filaments. This observation is compatible with a previous report (20), where staining of streamers formed by P. aeruginosa PAO1 with Triticum vulgaris (WGA) and Canavalia ensiformis (ConA) lectins allowed the visualization of the exopolysaccharide aggregates in the proximity of corners, but not of the EPS filaments spanning the channel, suggesting that the filaments were formed by eDNA. Moreover, while mutants deficient in the production of Pel, Psl, and CdrA have been shown to be defective in biofilm formation on flat surfaces (19, 20, 30, 42, 43), in our system they develop streamers that are structurally similar to those formed by the WT strains. Due to the prominent role of eDNA, the impact of the other components of the matrix is likely to be less critical for the structural stability of streamers than of surface-attached biofilms. This observation can have significant implications in flow networks since bacteria with limited biofilm-forming capability on a surface can still develop streamers and thus contribute to the systems clogging.
In addition to our direct visualization of eDNA distribution within streamers (Fig. 2 A–C), the role of eDNA is further supported by our experiments with DNA-degrading enzymes. In the Pel-deficient and WT strains, DNase I treatment completely prevented the formation of streamers, in both single-pillar (Fig. 4) and porous media configurations (SI Appendix, Fig. S10), and even established streamers. In Pel-overproducer streamers, treatment with DNase I was less effective and did not lead to complete streamer disruption (SI Appendix, Figs. S8 and S9). When Pel is abundant in the filament, eDNA degradation leads only to a partial disruption of the biofilm matrix. This suggests that the eDNA–Pel ionic interaction shields the eDNA from the enzymatic action of DNase I or helps to maintain the degraded eDNA fragments within the biofilm matrix, confirming the observation reported in ref. 73. Conversely, the presence of Psl does not restrain DNase I-induced degradation of the eDNA forming the streamers (SI Appendix, Fig. S12), further highlighting the protective role of the eDNA–Pel ionic interaction against enzymatic degradation.
The key characteristic of eDNA in the context of biofilm formation is its binding affinity (35, 74). It is known that in mature mushroom structures formed by P. aeruginosa PA14 eDNA is found in the stalk, where it binds to the positively charged exopolysaccharide Pel, resulting in increased structural stability (35). It has recently been shown that in surface-attached biofilms eDNA can be stabilized by proteins, resulting in the formation of lattice structures (74). Here, we showed that eDNA–Pel cross-links are not essential for the formation of biofilm streamers but significantly contribute to shaping their morphology and mechanical properties. In fact, we have shown that the Pel-overproducer strain generates shorter streamers with a larger diameter and large cell aggregates close to the pillar and that, conversely, the Pel-deficient strain forms long and slender filaments, easily shaped by the flow. Based on these observations, we hypothesize that the magnitude of the flow at which streamers are formed will have a significant impact on their morphology. Hence, the final morphology appears to be determined by the interplay between the force exerted by the flow and the biofilm matrix composition.
Using mutant strains in Pel production, we were also able to quantify the structural and mechanical properties of the eDNA–Pel interaction by directly measuring the elastic and viscous response to controlled mechanical stress. Interestingly, in our experiments the ratio of elastic modulus and effective viscosity, which represents the elastic relaxation time, scales according to an almost universal relation previously proposed for biofilms (14) (SI Appendix, Fig. S15). Moreover, we show that by causing an increase in both the elastic modulus and the viscosity, Pel enhances the stiffness of the streamer. Pel is a very small molecule (0.5 kDa) (28), even compared to fragments of eDNA (one base pair of double-stranded DNA is 0.65 kDa). Given that the opposite charge of eDNA and Pel may favor the creation of ionic bonds (28), we hypothesize that Pel molecules interconnect eDNA chains, thus reducing the slip between them and consequently affecting the response to deformations of the whole biofilm structure. The pH-induced change in the mechanical properties confirms the importance (and the still unexplored complexity) of this interaction in determining the biofilm mechanical response. These findings depict the streamers formed by P. aeruginosa PA14 as a bicomponent network, formed by a combination of a short-chain (Pel) and a long-chain (eDNA) compound, connected by ionic interactions; networks with these characteristics, in which the shorter chain acts as an energy-dissipating element and improves the mechanical properties of the material, are known as double-network gels (75). The analogy between biofilm streamers and synthetic double-network gels—which have been recently developed to create hydrogels more resistant to deformations—suggests that the key element for biofilm mechanical resistance could reside in the interaction between the different components of EPS, with an important role played by eDNA.
Our results show that induction of lysis of P. aeruginosa PA14 by treatment with a sublethal concentration of ciprofloxacin promotes streamer formation by increasing the concentration of eDNA, leading to an increase of the streamer biomass. A recent study demonstrated that sublethal antibiotic concentrations promoted bacterial aggregation and resulted in elevated susceptibility to intestinal expulsion (76). Our experiments provide evidence that, by increasing the amount of eDNA, some antibiotic treatments could enhance the formation of biofilms by promoting the occurrence of streamers which, given their spatial localization, eventually lead to clogging in medical devices such as stents or catheters and to the spreading of infection due to biofilm detachment. Moreover, since it is becoming clear that bacterial eDNA plays an important role in potentiating inflammation (77, 78), these findings are paving the way for future studies on the effects of antibiotic treatments on biofilm-forming pathogens and the host immune response.
Materials and Methods
Bacterial Cultures.
Experiments were performed using P. aeruginosa strain PA14 WT, the pel deletion mutant PA14 ΔpelE, the Pel-overproducer strain PA14 ΔwspF, and the cdrA deletion mutant PA14 ΔCdrA. P. aeruginosa PAO1 WT, the pel deletion mutant PAO1 ΔpelA, the psl deletion mutant PAO1 ΔpslB, and the pel and psl double mutant PAO1 ΔpelA ΔpslB (D) were kindly provided by Tim Tolker-Nielsen, Costerton Biofilm Center, University of Copenhagen, Copenhagen, Denmark. Single colonies were grown from frozen stocks on Luria broth agar plates at 37 °C for 24 h. P. aeruginosa suspensions were prepared by inoculating 3 mL tryptone broth (TB; 10 g/L tryptone) with cells from a single colony and incubating for 3 h at 37 °C, while shaking at 200 rpm. The suspensions were then diluted in fresh TB to final optical density at 600 nm (OD600) < 0.01.
eDNA and Pel quantities were visualized and measured using fluorescence staining methods: for eDNA visualization, PI (Sigma-Aldrich) was added to the medium to a final concentration of 2 μg/mL, and for Pel visualization WFL (bioWorld) was dissolved in phosphate-buffered saline to a final concentration of 50 μg/mL. When Pel and eDNA were visualized simultaneously on the same streamer, WFL was flown for 30 min, images were captured, and only thereafter staining with PI was performed. This protocol (35, 66) allowed visualization of the two components on the same streamer, while avoiding possible interactions between the two stains. For experiments involving different pH values, we adjusted pH to 5.8 by adding HCl and to 7.8 by adding NaOH to the culture media. For experiments involving degradation of eDNA, DNase I (Sigma-Aldrich) was dissolved in TB to a final concentration of 1 mg/mL and CaCl2 and MgCl2 (final concentrations 0.12 mM) were added as activators. For antibiotic treatment experiments, ciprofloxacin (Sigma-Aldrich) was first dissolved in 0.1 N HCl to a final concentration of 20 mg/mL and then further diluted in TB to obtain solutions of final concentration 0.005 μg/mL, 0.01 μg/mL, and 0.02 μg/mL. All samples of the antibiotic treatment experiment were assessed by high-sensitivity double-stranded DNA assay and high-sensitivity ssDNA assay according to established protocols using Quibit 3.0 (Thermo Fisher Scientific).
Microfluidic Assays.
To analyze streamer formation in flow, we fabricated a polydimethylsiloxane (PDMS) microfluidic device with four channels on the same chip, each containing six cylindrical pillars of diameter 50 μm. The channel was 40 μm high and 1 mm wide. Pillars were located at the center of the channel and the distance between pillars was 5 mm. The flow was driven by a syringe pump (neMESYS 290N; CETONI). Prior to use, all microfluidic channels were washed with 2 mL of TB medium. A diluted PA14 bacteria suspension (OD600 < 0.01; cell concentration < 106 cells/mL) was flown for 24 h. All experiments were performed at room temperature (T = 20 ± 0.5 °C). The viability of the cell suspension after incubation in the syringe was assessed by flow cytometry (SI Appendix) following live/dead staining. The impact of different growth regimes on streamer formation was assessed in an experiment described in SI Appendix, Fig. S16.
In the experiments in which DNase I was used, the DNase I solution was flown in a Y connector (P-514; IDEX) located before the inlet to avoid contact between the cells and the enzyme before the channel inlet (Fig. 4A). To perform DNase I treatment on mature streamers, a shut-off valve (P-782; IDEX) was inserted between the syringe containing the DNase I solution and the Y connector . The shut-off valve was kept closed during streamer growth and then opened to expose the mature streamer to DNase I treatment. In the experiments in which ciprofloxacin was used, the antibiotic solution was flown in a Y connector (P-514; IDEX) located before the inlet to avoid contact between the cells and ciprofloxacin before the channel inlet, using the same configuration described for the DNase I treatments (Fig. 4A). For the experiment with the model porous medium containing 75-µm pillars, the device was fabricated using PDMS and flow was driven by a syringe pump (neMESYS 290N; CETONI).
Cell Imaging and Tracking.
All imaging was performed on an inverted microscope (Ti-Eclipse; Nikon) using a digital camera (ORCA-Flash4.0 V3 Digital CMOS camera; Hamamatsu Photonics). Bacterial cells were imaged using phase-contrast microscopy (20× magnification). Biofilm composition was quantified using epifluorescence microscopy (20× magnification). During biofilm streamer growth, images were taken every 15 min both in phase contrast and epifluorescence, unless specified otherwise in the figure legends. During mechanical tests on biofilm streamers, images were taken before the tests in epifluorescence and once per second in phase-contrast during the tests. All images of biofilm streamers were obtained at channel middepth, while surface coverage was evaluated on the glass wall of the microfluidic channel in a region located 3 mm upstream of the pillars.
Statistics and Derivations.
All image analysis was performed in Fiji-Image J (79). All images of streamers are examples from experiments that were repeated three times with consistent results. A complete description of the statistics and derivation is reported in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Sam Charlton, Steffen Geisel, and Ela Burmeister for the insightful discussions; Prof. Tim Tolker-Nielsen for providing the P. aeruginosa PAO1 mutant strains; and support from Swiss National Science Foundation PRIMA Grant 179834 (to E.S.), Gordon and Betty Moore Foundation Marine Microbial Initiative Investigator Award GBMF3783 (to R.S.), and Simons Foundation Grant 542395 (to R.S.) as part of the Principles of Microbial Ecosystems Collaborative (PriME).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113723119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Flemming H.-C., et al. , Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Flemming H. C., Wuertz S., Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Flemming H. C., Wingender J., The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Peterson B. W., et al. , Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol. Rev. 39, 234–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall-Stoodley L., et al. , Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 65, 127–145 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Donlan R. M., Costerton J. W., Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemming H. C., Biofouling in water systems--cases, causes and countermeasures. Appl. Microbiol. Biotechnol. 59, 629–640 (2002). [DOI] [PubMed] [Google Scholar]
- 8.M. Caldara, C. Belgiovine, E. Secchi, R. Rusconi, Environmental, microbiological, and immunological features of bacterial biofilms associated with implanted medical devices. Clin. Microbiol. Infect. 35, e00221-20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desmond P., Best J. P., Morgenroth E., Derlon N., Linking composition of extracellular polymeric substances (EPS) to the physical structure and hydraulic resistance of membrane biofilms. Water Res. 132, 211–221 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Pandit S., et al. , The exopolysaccharide component of extracellular matrix is essential for the viscoelastic properties of Bacillus subtilis biofilms. Int. J. Mol. Sci. 21, 6755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kundukad B., et al. , Mechanical properties of the superficial biofilm layer determine the architecture of biofilms. Soft Matter 12, 5718–5726 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Stewart E. J., Ganesan M., Younger J. G., Solomon M. J., Artificial biofilms establish the role of matrix interactions in staphylococcal biofilm assembly and disassembly. Sci. Rep. 5, 13081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlton S. G. V., et al. , Regulating, measuring, and modeling the viscoelasticity of bacterial biofilms. J. Bacteriol. 201, e00101–e00119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw T., Winston M., Rupp C. J., Klapper I., Stoodley P., Commonality of elastic relaxation times in biofilms. Phys. Rev. Lett. 93, 098102 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Gloag E. S., Fabbri S., Wozniak D. J., Stoodley P., Biofilm mechanics: Implications in infection and survival. Biofilm 2, 100017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall-Stoodley L., Costerton J. W., Stoodley P., Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Chew S. C., et al. , Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. MBio 5, e01536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadell C. D., Ricaurte D., Yan J., Drescher K., Bassler B. L., Flow environment and matrix structure interact to determine spatial competition in Pseudomonas aeruginosa biofilms. eLife 6, e21855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rusconi R., Lecuyer S., Autrusson N., Guglielmini L., Stone H. A., Secondary flow as a mechanism for the formation of biofilm streamers. Biophys. J. 100, 1392–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusconi R., Lecuyer S., Guglielmini L., Stone H. A., Laminar flow around corners triggers the formation of biofilm streamers. J. R. Soc. Interface 7, 1293–1299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh U. U., Ali H., Ghosh R., Kumar A., Bacterial streamers as colloidal systems: Five grand challenges. J. Colloid Interface Sci. 594, 265–278 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Scheidweiler D., Peter H., Pramateftaki P., de Anna P., Battin T. J., Unraveling the biophysical underpinnings to the success of multispecies biofilms in porous environments. ISME J. 13, 1700–1710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drescher K., Shen Y., Bassler B. L., Stone H. A., Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl. Acad. Sci. U.S.A. 110, 4345–4350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas I., Sadrzadeh M., Kumar A., Impact of bacterial streamers on biofouling of microfluidic filtration systems. Biomicrofluidics 12, 044116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debnath N., et al. , Abiotic streamers in a microfluidic system. Soft Matter 13, 8698–8705 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Hassanpourfard M., et al. , Bacterial floc mediated rapid streamer formation in creeping flows. Sci. Rep. 5, 13070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S., Kumar A., Formation and post-formation dynamics of bacterial biofilm streamers as highly viscous liquid jets. Sci. Rep. 4, 7126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wozniak D. J., et al. , Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U.S.A. 100, 7907–7912 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman L., Kolter R., Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Friedman L., Kolter R., Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann E. E., Wozniak D. J., Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36, 893–916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazli M., et al. , Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 16, 1961–1981 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Konrad B., Gerd D., “Ecology and epidemiology of Pseudomonas aeruginosa” in Pseudomonas aeruginosa as an Opportunistic Pathogen (Springer Science+Business Media, 1993), pp. 1–18. [Google Scholar]
- 34.Driscoll J. A., Brody S. L., Kollef M. H., The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Jennings L. K., et al. , Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 112, 11353–11358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klausen M., Aaes-Jørgensen A., Molin S., Tolker-Nielsen T., Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50, 61–68 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Franklin M. J., Nivens D. E., Weadge J. T., Howell P. L., Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2, 1–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colvin K. M., et al. , The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colvin K. M., et al. , The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7, e1001264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billings N., et al. , The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 9, e1003526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooley B. J., et al. , The extracellular polysaccharide Pel makes the attachment of P. aeruginosa to surfaces symmetric and short-ranged. Soft Matter 9, 3871–3876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichhardt C., et al. , The versatile Pseudomonas aeruginosa biofilm matrix protein CdrA promotes aggregation through different extracellular exopolysaccharide interactions. J. Bacteriol. 202, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reichhardt C., Wong C., Passos da Silva D., Wozniak D. J., Parsek M. R., CdrA interactions within the Pseudomonas aeruginosa biofilm matrix safeguard it from proteolysis and promote cellular packing. MBio 9, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allesen-Holm M., et al. , A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59, 1114–1128 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Whitchurch C. B., Tolker-Nielsen T., Ragas P. C., Mattick J. S., Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Seviour T., et al. , The biofilm matrix scaffold of Pseudomonas aeruginosa contains G-quadruplex extracellular DNA structures. NPJ Biofilms Microbiomes 7, 27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ibáñez de Aldecoa A. L., Zafra O., González-Pastor J. E., Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 8, 1390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montanaro L., et al. , Extracellular DNA in biofilms. Int. J. Artif. Organs 34, 824–831 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Rice K. C., et al. , The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8113–8118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Böckelmann U., et al. , Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 262, 31–38 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Izano E. A., Amarante M. A., Kher W. B., Kaplan J. B., Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dengler V., Foulston L., DeFrancesco A. S., Losick R., An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 197, 3779–3787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu W., et al. , DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS One 7, e51905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novotny L. A., Amer A. O., Brockson M. E., Goodman S. D., Bakaletz L. O., Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One 8, e67629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Z., et al. , Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology (Reading) 153, 2083–2092 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Peng N., et al. , The exopolysaccharide-eDNA interaction modulates 3D architecture of Bacillus subtilis biofilm. BMC Microbiol. 20, 115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang L., Schramm A., Neu T. R., Revsbech N. P., Meyer R. L., Extracellular DNA in adhesion and biofilm formation of four environmental isolates: A quantitative study. FEMS Microbiol. Ecol. 86, 394–403 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Dominiak D. M., Nielsen J. L., Nielsen P. H., Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 13, 710–721 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Zheng Y., Joo H. S., Nair V., Le K. Y., Otto M., Do amyloid structures formed by Staphylococcus aureus phenol-soluble modulins have a biological function? Int. J. Med. Microbiol. 308, 675–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turnbull L., et al. , Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7, 11220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Webb J. S., et al. , Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiang W. C., et al. , Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulcahy H., Charron-Mazenod L., Lewenza S., Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 4, e1000213 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gloag E. S., et al. , Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc. Natl. Acad. Sci. U.S.A. 110, 11541–11546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eckhart L., Fischer H., Barken K. B., Tolker-Nielsen T., Tschachler E., DNase1L2 suppresses biofilm formation by Pseudomonas aeruginosa and Staphylococcus aureus. Br. J. Dermatol. 156, 1342–1345 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Reichhardt C., Parsek M. R., Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 10, 677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart P. S., Franklin M. J., Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Secchi E., et al. , The effect of flow on swimming bacteria controls the initial colonization of curved surfaces. Nat. Commun. 11, 2851 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.G. Savorana, J. Słomka, R. Stocker, R. Rusconi, E. Secchi, A microfluidic platform for characterizing the structure and rheology of biofilm streamers. 10.1101/2022.02.22.481486. Accessed 9 March 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brazas M. D., Hancock R. E. W., Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49, 3222–3227 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haber C., Wirtz D., Shear-induced assembly of λ-phage DNA. Biophys. J. 79, 1530–1536 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeom M. S., Lee J., The mechanism of the self-assembly of associating DNA molecules under shear flow: Brownian dynamics simulation. J. Chem. Phys. 122, 184905 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Jennings L. K., et al. , Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 34, 108782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Devaraj A., et al. , The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc. Natl. Acad. Sci. U.S.A. 116, 25068–25077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun J. Y., et al. , Highly stretchable and tough hydrogels. Nature 489, 133–136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlomann B. H., Wiles T. J., Wall E. S., Guillemin K., Parthasarathy R., Sublethal antibiotics collapse gut bacterial populations by enhancing aggregation and expulsion. Proc. Natl. Acad. Sci. U.S.A. 116, 21392–21400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watters C., Fleming D., Bishop D., Rumbaugh K. P., Host responses to biofilm. Prog. Mol. Biol. Transl. Sci. 142, 193–239 (2016). [DOI] [PubMed] [Google Scholar]
- 78.Serrage H. J., Jepson M. A., Rostami N., Jakubovics N. S., Nobbs A. H., Understanding the matrix: The role of extracellular DNA in oral biofilms. Front Oral Health 2, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.