Significance
Lipid droplets (LDs) are ubiquitous organelles that play important roles in cellular energy homeostasis, tightly regulating the accumulation and release of lipids. In macrophages, lipids accumulate in LDs during inflammation. However, it is unclear how inflammatory activation promotes the accumulation of lipids in LDs, and how the dynamic between lipid accumulation and breakdown could drive or inhibit inflammation. Elucidating the role of lipid accumulation during inflammation may provide important knowledge to influence inflammatory processes during health and disease. We identify the importance of the hypoxia-inducible lipid droplet–associated protein and the intracellular adipose triglyceride lipase in the regulation of lipid accumulation and breakdown in inflammatory macrophages. Furthermore, we determine the regulatory effect of lipid breakdown from LDs in supporting inflammation.
Keywords: immunometabolism, lipid droplets, macrophages, HILPDA, ATGL
Abstract
In response to inflammatory activation by pathogens, macrophages accumulate triglycerides in intracellular lipid droplets. The mechanisms underlying triglyceride accumulation and its exact role in the inflammatory response of macrophages are not fully understood. Here, we aim to further elucidate the mechanism and function of triglyceride accumulation in the inflammatory response of activated macrophages. Lipopolysaccharide (LPS)-mediated activation markedly increased triglyceride accumulation in macrophages. This increase could be attributed to up-regulation of the hypoxia-inducible lipid droplet–associated (HILPDA) protein, which down-regulated adipose triglyceride lipase (ATGL) protein levels, in turn leading to decreased ATGL-mediated triglyceride hydrolysis. The reduction in ATGL-mediated lipolysis attenuated the inflammatory response in macrophages after ex vivo and in vitro activation, and was accompanied by decreased production of prostaglandin-E2 (PGE2) and interleukin-6 (IL-6). Overall, we provide evidence that LPS-mediated activation of macrophages suppresses lipolysis via induction of HILPDA, thereby reducing the availability of proinflammatory lipid precursors and suppressing the production of PGE2 and IL-6.
Lipid droplets (LDs) are ubiquitous organelles that serve as intracellular energy stores by compartmentalizing lipids, mainly in the form of triglycerides (TGs). While LDs were originally identified in adipocytes, their presence has been confirmed in the majority of mammalian cells. The surface of LDs is covered by numerous LD-related proteins, many of which are involved in the regulation of the storage and release of lipids (1). Besides mediating energy storage, LDs are increasingly recognized to be directly or indirectly involved in the regulation of other cellular processes, ranging from the storage of hydrophobic vitamins and signaling precursors to the management of endoplasmic reticulum (ER) and oxidative stress and the production of inflammatory mediators (2, 3).
Macrophages are innate immune cells that play distinct roles in tissue homeostasis and form the front line in host defense against pathogens. Inflammatory activation of macrophages induces the accumulation of TGs (4, 5). In particular, various pathogen-associated molecular patterns (PAMPs), including lipopolysaccharide (LPS), can specifically reprogram lipid metabolism in macrophages, leading to increased lipid storage (5). However, rather than being an unintentional consequence of lipid metabolic reprogramming, there is growing evidence that LDs are actively involved in affecting the inflammatory response of macrophages (5). Studies suggest that besides being hijacked by infectious pathogens as a source of energy (6, 7), LDs may also serve as a sink for proteins involved in antimicrobial defense mechanisms (8). Currently, how PAMPs promote the accumulation of LDs in macrophages is unclear.
Adipose triglyceride lipase (ATGL) is an LD-associated protein that is present in numerous cell types. It catalyzes the first step in the hydrolytic cleavage of intracellular TGs, generating nonesterified fatty acids (9–11). Intriguingly, in macrophages, ATGL is not only responsible for the lipolysis of stored TGs but also appears to be involved in the immune response. Indeed, deficiency of ATGL in lipid-laden macrophages was shown to attenuate the release of the proinflammatory cytokine interleukin-6 (IL-6) and enhance the release of antiinflammatory IL-10 (12). In addition, deficiency of ATGL was found to promote macrophage apoptosis (13). Although these studies have pointed to a role for intracellular lipolysis in the functional properties of macrophages, the precise role of TG accumulation and lipolysis in the immune function of macrophages remains undefined.
HILPDA (hypoxia-inducible lipid droplet–associated) is a small LD-associated protein (14) that was found to promote lipid storage in several cell types, including macrophages (15, 16). Previously, we and others showed that HILPDA functions as a direct physiological inhibitor of ATGL in macrophages (16, 17). Consistent with the role of HILPDA as an inhibitor of ATGL, HILPDA deficiency was accompanied by a marked reduction in lipid storage in fatty acid–treated macrophages (16). Whether HILPDA plays a role in the stimulation of TG storage by PAMPs is unknown. In principle, modification of ATGL-mediated lipolysis by HILPDA could be leveraged to further elucidate the role of TG accumulation in the inflammatory response of macrophages. Accordingly, here we set out to investigate the mechanism by which LPS enhances LD formation in macrophages, as well as investigate the role of TG accumulation in the inflammatory response of LPS-treated macrophages, using HILPDA-deficient macrophages as a model. We find that activation of macrophages by LPS reduces ATGL-mediated TG hydrolysis via up-regulation of HILPDA, leading to reduced production of prostaglandin-E2 (PGE2) and the proinflammatory cytokine IL-6.
Results
Toll-Like Receptor Stimulation Leads to LD Accumulation through Inhibition of ATGL by HILPDA.
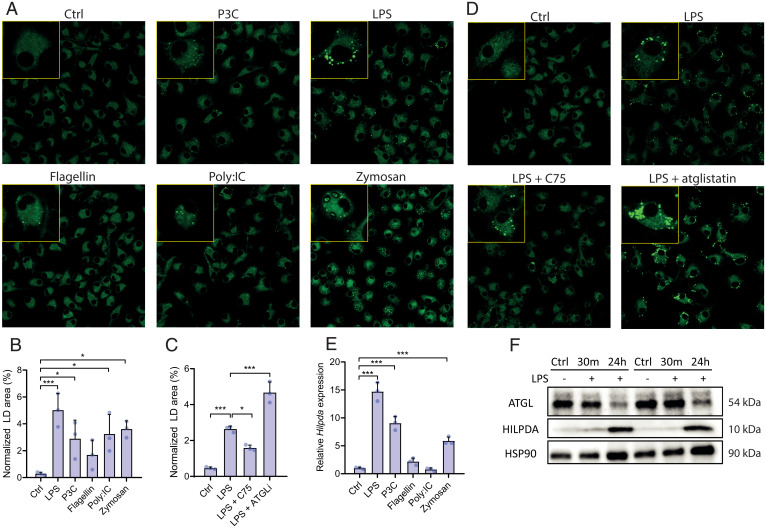
In order to investigate how inflammatory activation promotes lipid accumulation in macrophages, we first studied the stimulatory effect of LPS and various other Toll-like receptor (TLR) ligands on TG accumulation in bone marrow–derived macrophages (BMDMs), including Pam3Cysk (P3C; TLR-2), LPS (TLR-4), flagellin (TLR-5), polyinosinic-polycytidylic acid (poly:IC; TLR-3), and opsonized zymosan (TLR-2) (Fig. 1A). All TLR ligands increased TG accumulation in BMDMs, with LPS showing the largest effect (Fig. 1B). To gain insight into the mechanism underlying the increased TG storage upon LPS treatment, BMDMs were treated with the metabolic modulators C75, an inhibitor of fatty acid synthase (18), and atglistatin, an inhibitor of ATGL (19). Whereas inhibition of fatty acid synthesis by C75 decreased LD accumulation, inhibition of ATGL by atglistatin markedly increased accumulation of LDs in LPS-treated BMDMs (Fig. 1 C and D). Although de novo synthesis of fatty acids may be partly responsible for the increased accumulation of lipids, decreased lipolysis by ATGL inhibition may also play a role in LPS-induced LD accumulation. Previously, we showed that besides serving as a direct substrate for TG synthesis, fatty acids promote LD accumulation in BMDMs by raising protein levels of the endogenous ATGL inhibitor HILPDA (16). To investigate if HILPDA may play a role in LPS-induced LD accumulation, we first studied the effect of different TLR ligands on HILPDA levels. Various TLR ligands, especially LPS, markedly induced Hilpda messenger RNA (mRNA) levels (Fig. 1E). Concurrent with an increase in Hilpda mRNA, 24-h treatment with LPS dramatically increased HILPDA protein levels (Fig. 1F) while simultaneously reducing ATGL protein levels. These data hint at a potential role of HILPDA and ATGL in the augmentation of lipid storage after LPS treatment (Fig. 1F).
Fig. 1.
TLR stimulation leads to LD accumulation through HILPDA and ATGL. (A and B) BODIPY staining and LD quantification in wild-type BMDMs treated with vehicle (Ctrl), P3C, LPS, flagellin, poly:IC, or zymosan for 24 h. (C and D) BODIPY staining in wild-type BMDMs treated with vehicle (Ctrl), LPS, LPS and C75, or LPS and atglistatin (ATGLi). (E) mRNA levels of Hilpda in wild-type BMDMs treated with vehicle (Ctrl), LPS, P3C, flagellin, poly:IC, or zymosan for 24 h. (F) Protein levels of ATGL and HILPDA in wild-type BMDMs treated with vehicle (Ctrl) or LPS for 30 min or 24 h. HSP90 was used as loading control. Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
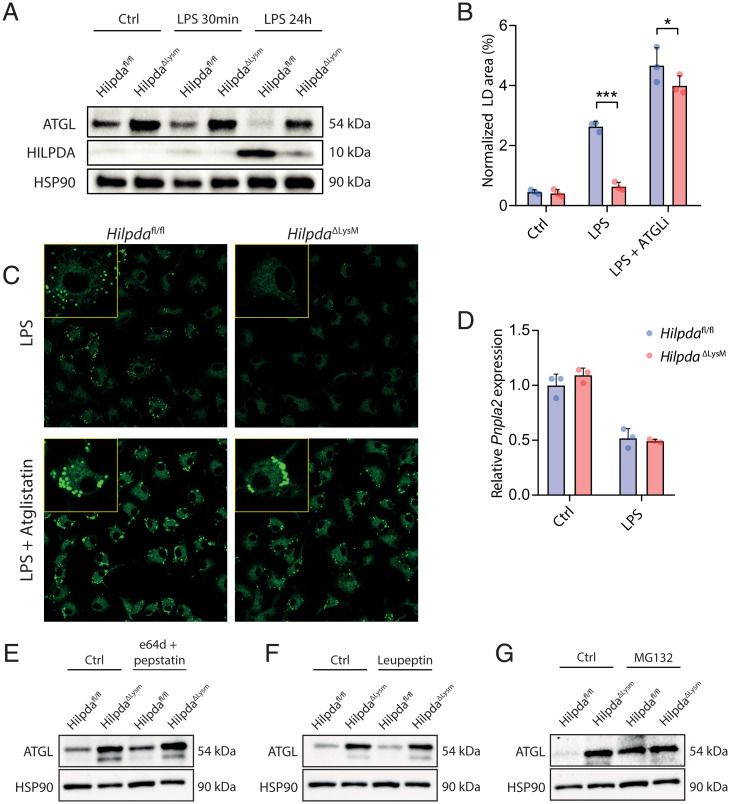
To study the role of HILPDA in LPS-induced lipid accumulation, we used a murine myeloid-specific knockout model for HILPDA (16). BMDMs derived from myeloid-specific HILPDA-deficient mice (HilpdaΔLysM) expressed little to no HILPDA protein after LPS stimulation, whereas BMDMs derived from their floxed littermates (Hilpdafl/fl) showed substantial up-regulation of HILPDA protein in response to LPS (Fig. 2A). Consistent with an important role of HILPDA in LPS-induced TG accumulation, the LD area was significantly lower in LPS-treated HilpdaΔLysM macrophages compared with Hilpdafl/fl macrophages (Fig. 2 B and C), whereas no differences in LD area were observed under control conditions (SI Appendix, Fig. S1). Intriguingly, the difference in TG accumulation between LPS-treated HilpdaΔLysM and Hilpdafl/fl macrophages was almost completely abolished by atglistatin, indicating that the reduced TG accumulation in LPS-treated HilpdaΔLysM macrophages is mostly due to enhanced ATGL activity (Fig. 2 B and C). To further investigate the connection between HILPDA and ATGL, we determined ATGL protein levels. In Hilpdafl/fl BMDMs, ATGL protein tended to be down-regulated already after 30 min of LPS stimulation, with an even stronger down-regulation after 24 h, again showing an inverse pattern compared with HILPDA protein (Fig. 2A). Strikingly, ATGL protein levels were much higher in HilpdaΔLysM compared with Hilpdafl/fl BMDMs, which was most evident after 24-h LPS treatment, indicating that HILPDA reduces ATGL protein levels (Fig. 2A). These data demonstrate that LPS treatment decreases ATGL protein levels by inducing HILPDA.
Fig. 2.
LPS promotes TG accumulation in macrophages by inducing HILPDA, which inhibits TG lipolysis by stimulating proteasomal degradation of ATGL. (A) Protein expression levels of ATGL and HILPDA in HilpdaΔLysM and Hilpdafl/fl BMDMs treated with vehicle (Ctrl) or LPS for 30 min or 24 h. HSP90 was used as loading control. (B and C) BODIPY staining and LD quantification of HilpdaΔLysM and Hilpdafl/fl BMDMs treated with LPS or LPS and atglistatin for 24 h. (D) mRNA levels of Pnpla2 (ATGL) in HilpdaΔLysM and Hilpdafl/fl BMDMs treated with vehicle (Ctrl) or LPS for 24 h. (E–G) ATGL protein expression in HilpdaΔLysM and Hilpdafl/fl BMDMs treated with vehicle (Ctrl) or e64d and pepstatin (E), vehicle (Ctrl) or leupeptin (F), and vehicle (Ctrl) or MG132 (G). HSP90 was used as loading control. Data are represented as mean ± SD. *P < 0.05, ***P < 0.001.
Subsequently, we tried to clarify the mechanism by which HILPDA decreases ATGL protein expression. HILPDA deficiency in BMDMs did not influence ATGL (Pnpla2) mRNA levels (Fig. 2D). Accordingly, we hypothesized that HILPDA might promote ATGL protein degradation. To verify this hypothesis, we treated HilpdaΔLysM and Hilpdafl/fl macrophages with specific lysosomal protease (e64d and leupeptin) or ubiquitin-proteasome (MG132) inhibitors. Treatment with e64d or leupeptin did not influence ATGL protein levels, suggesting that ATGL is not degraded via the lysosomes (Fig. 2 E and F). In contrast, treatment of Hilpdafl/fl macrophages with MG132 markedly induced ATGL protein levels, suggesting that ATGL is degraded via the ubiquitin-proteasome pathway (Fig. 2G). Consistent with a stimulatory effect of HILPDA on proteasomal degradation of ATGL, MG132 failed to increase ATGL protein levels in HilpdaΔLysM macrophages (Fig. 2G). Differences in ATGL protein levels between HilpdaΔLysM and Hilpdafl/fl macrophages were completely abolished after MG132 treatment, suggesting that the repressive effect of HILPDA on ATGL protein levels is entirely mediated by enhanced proteasomal degradation of ATGL. Overall, these data suggest that LPS promotes TG accumulation in macrophages by inducing HILPDA, which in turn decreases ATGL protein levels and consequent TG lipolysis by stimulating proteasomal degradation of ATGL.
HILPDA Deficiency in Myeloid Cells Facilitates the Inflammatory Phenotype In Vivo.
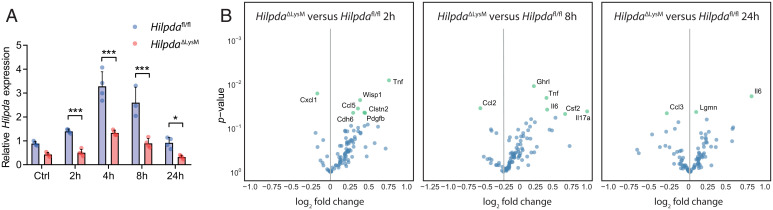
Based on the findings presented above, myeloid-specific HILPDA-deficient mice provide an excellent model to examine the impact of modulating macrophage TG accumulation on LPS-induced whole-body inflammation. To pursue this question, classical inflammation was induced by intraperitoneal injection of LPS in HilpdaΔLysM and Hilpdafl/fl mice for 2, 4, 8, or 24 h. Similar to the observation in BMDMs, LPS stimulated Hilpda expression in splenic Hilpdafl/fl macrophages, with a maximal effect observed after 4 h (Fig. 3A). Hilpda mRNA levels were reduced by 51 to 65% in LPS-treated splenic macrophages from HilpdaΔLysM mice compared with Hilpdafl/fl macrophages (Fig. 3A). To study the effect of macrophage-specific HILPDA deficiency on plasma inflammatory markers, plasma collected at different time points of LPS-induced inflammation was subjected to proteomic profiling, allowing for the semiquantification of a protein biomarker panel consisting of 92 biomarkers (Olink). Principal-component analysis (PCA) clearly separated the different time points (SI Appendix, Fig. S2A). Relative differences in the biomarkers between HilpdaΔLysM and Hilpdafl/fl mice were mapped in a volcano plot using a statistical threshold of P value < 0.05 (uncorrected). At baseline and at the 4-h time point, very few proteins were significantly different between the two sets of mice (SI Appendix, Fig. S2B). However, for the 2-, 8-, and 24-h time points, a higher number of proteins met the statistical threshold (Fig. 3B). Interestingly, proteins such as tumor necrosis factor (TNF) and IL-6 were higher in the HilpdaΔLysM mice compared with Hilpdafl/fl littermates at several time points, hinting at a higher degree of inflammation in the HilpdaΔLysM mice.
Fig. 3.
HILPDA deficiency in myeloid cells affects the inflammatory state in vivo. (A) Hilpda gene expression in isolated splenic macrophages from HilpdaΔLysM and Hilpdafl/fl mice 2, 4, 8, or 24 h after intraperitoneal injection of LPS or 24 h after intraperitoneal injection of saline (Ctrl). (B) Relative fold change and uncorrected P values in plasma proteomic profiles in HilpdaΔLysM and Hilpdafl/fl mice 2, 8, or 24 h after intraperitoneal injection of LPS. Data are represented as mean ± SD. *P < 0.05, ***P < 0.001.
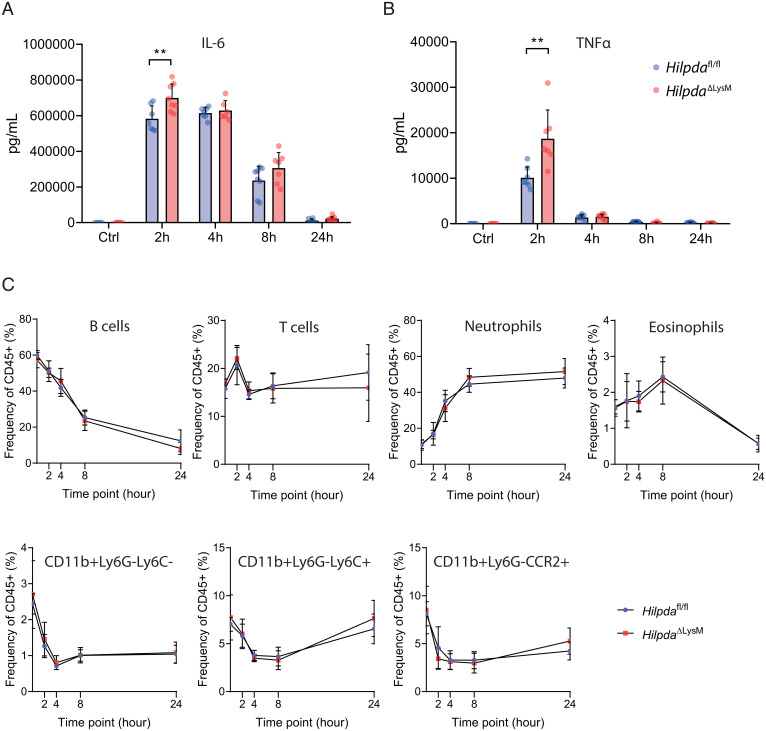
We next pursued a multiplex-based approach to measure actual protein levels of circulating inflammatory cytokines and chemokines. Each inflammatory marker followed a unique pattern following LPS injection over time. Interestingly, concentrations of IL27p28/IL30, IP-10, MCP-1, MIP-1α, MIP-2, IFNγ, IL-10, and IL-1β did not differ between the two genotypes (SI Appendix, Fig. S3 A–H). In contrast, both IL-6 (Fig. 4A) and TNFα (Fig. 4B) were significantly higher in HilpdaΔLysM than in Hilpdafl/fl mice at their measured peak concentrations, 2 h after LPS injection, confirming the biomarker-based data and further suggesting an increased inflammatory response in HilpdaΔLysM mice. To find out whether changes in immune cell populations drove the potential differences in inflammation, we assessed the relative abundance of different immune cell populations in the circulation by flow cytometry. Although the relative abundance of the measured immune cell populations in the circulation clearly changed over time upon LPS-induced inflammation, no differences were found between the two genotypes in populations of B cells, T cells, neutrophils, eosinophils, and three subsets of monocytes (CD11b+Ly6G−Ly6C−, CD11b+Ly6G−Ly6C+, and CD11b+Ly6G−CCR2+) (Fig. 4C). Additionally, no differences were found in the relative abundance of T cell subsets (SI Appendix, Fig. S3I).
Fig. 4.
Myeloid deficiency of HILPDA affects inflammatory parameters without altering relative immune cell populations in vivo. (A and B) Plasma concentration of IL-6 (A) and TNFα (B) in HilpdaΔLysM and Hilpdafl/fl mice 2, 4, 8, or 24 h after intraperitoneal injection of LPS or 24 h after intraperitoneal injection of saline (Ctrl). (C) Relative circulating immune cell populations in HilpdaΔLysM and Hilpdafl/fl mice 2, 4, 8, or 24 h after intraperitoneal injection of LPS or 24 h after intraperitoneal injection of saline (baseline). Data are represented as mean ± SD. **P < 0.01.
HILPDA-Deficient Macrophages Show an Increased Proinflammatory Phenotype Ex Vivo.
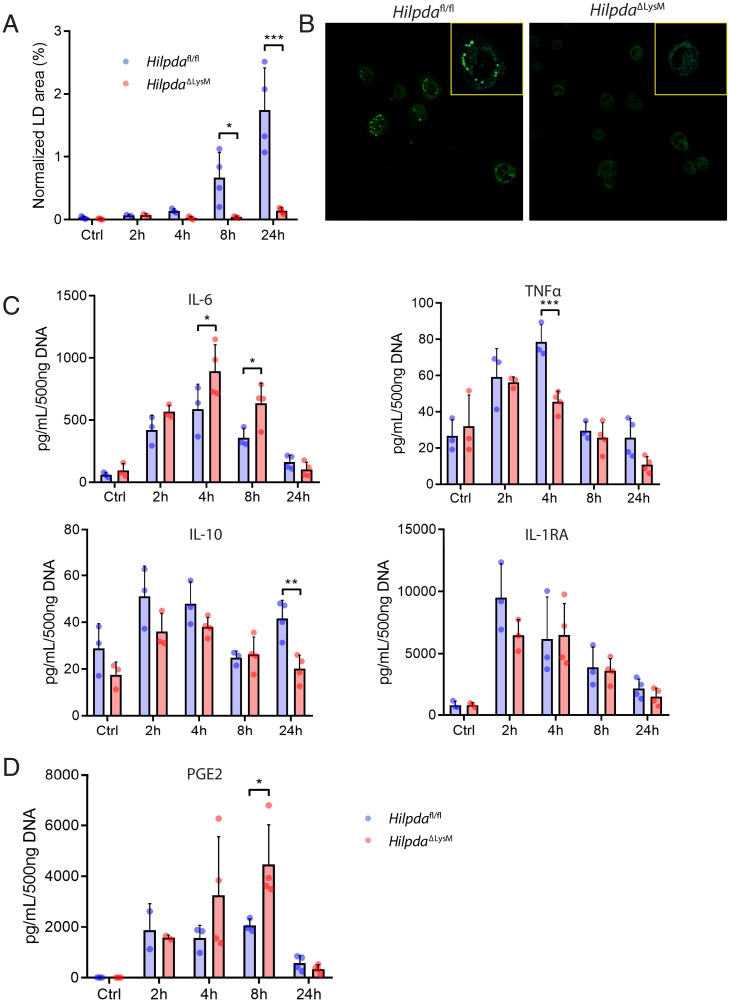
Next, we asked whether modulation of lipid storage by HILPDA could influence the secretion of inflammatory mediators from peritoneal macrophages. To that end, peritoneal macrophages were isolated from HilpdaΔLysM and Hilpdafl/fl mice at different time points after LPS injection, and the release of several inflammatory mediators was measured. In agreement with the data in BMDMs, LPS caused a clear induction in lipid storage in the Hilpdafl/fl macrophages but not in HilpdaΔLysM macrophages, becoming most evident 8 to 24 h after LPS injection (Fig. 5 A and B). Since we observed increased proinflammatory markers in vivo, we set out to investigate the proinflammatory phenotype of the peritoneal macrophages by measuring cytokine release (Fig. 5C). Interestingly, 4 and 8 h after LPS injection, IL-6 release was significantly higher in HilpdaΔLysM compared with Hilpdafl/fl macrophages, suggesting an increased inflammatory phenotype in peritoneal macrophages (Fig. 5C). By contrast, 4 h after LPS injection, the release of TNFα was significantly lower in HilpdaΔLysM compared with Hilpdafl/fl macrophages (Fig. 5C).
Fig. 5.
HILPDA-deficient macrophages show an increased proinflammatory phenotype after in vivo LPS treatment. (A and B) BODIPY staining and LD quantification in peritoneal macrophages isolated from HilpdaΔLysM and Hilpdafl/fl mice 2, 4, 8, or 24 h after intraperitoneal injection of LPS or 24 h after intraperitoneal injection of saline (Ctrl). (C and D) Concentrations of IL-6, TNFα, IL-10, and IL-1RA (C) or PGE2 (D) in ex vivo supernatant from peritoneal macrophages isolated from HilpdaΔLysM and Hilpdafl/fl mice 2, 4, 8, or 24 h after intraperitoneal injection of LPS or 24 h after intraperitoneal injection of saline (Ctrl). Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
IL-10 release showed a trend toward a reduction in HilpdaΔLysM compared with Hilpdafl/fl macrophages for most time points, which reached significance 24 h after LPS injection (Fig. 5C). No significant differences were observed for IL-1 receptor antagonist (IL-1RA) (Fig. 5C). Inasmuch as HILPDA deficiency reduces LD accumulation in LPS-treated macrophages by enhancing ATGL-mediated lipolysis, we wondered whether HILPDA deficiency may increase the release of lipid-derived inflammatory mediators. Indeed, 8 h after LPS injections, the release of PGE2 by peritoneal macrophages was significantly higher in HilpdaΔLysM macrophages than in Hilpdafl/fl macrophages (Fig. 5D). These data suggest that in HILPDA-deficient macrophages, lipids are directed from LDs toward the production of prostaglandins.
HILPDA Deficiency Does Not Affect Key Macrophage Effector Functions.
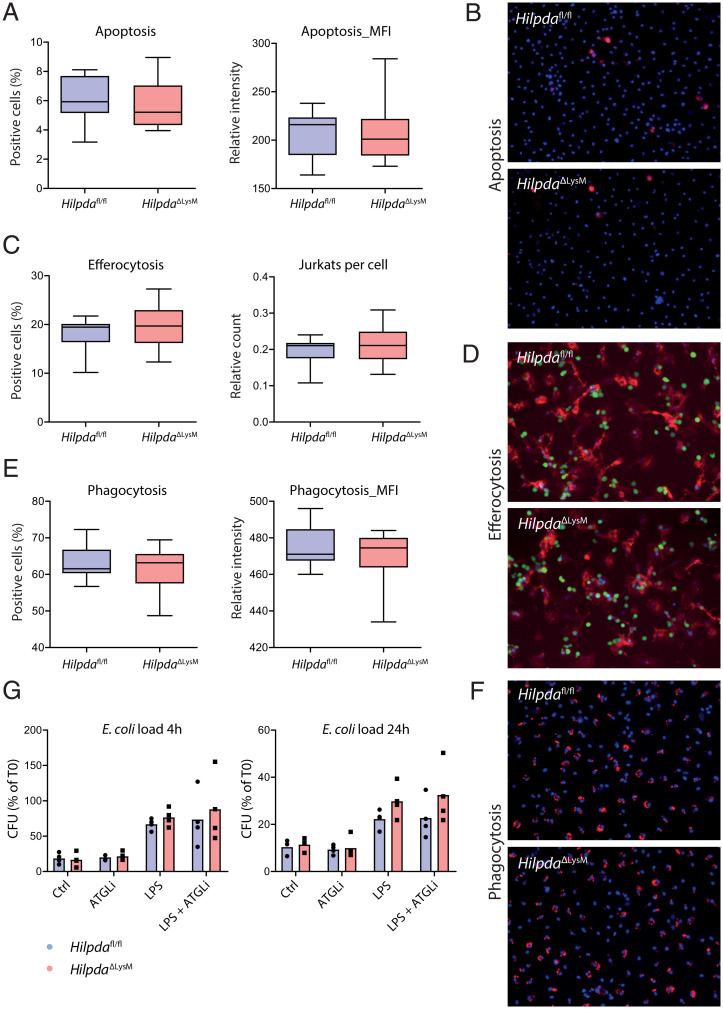
After observing clear differences in the inflammatory activation between Hilpdafl/fl and HilpdaΔLysM macrophages, we wondered whether similar differences could be observed in effector functions that are associated with inflammation. Interestingly, previous research highlighted the importance of TG hydrolysis by ATGL for phagocytosis (20) and linked ATGL deficiency to increased apoptosis in macrophages (13). Since HILPDA-deficient macrophages consistently show increased ATGL protein expression levels, we set out to study whether apoptosis may be down-regulated and whether efferocytosis and phagocytosis may be amplified in LPS-treated HilpdaΔLysM and Hilpdafl/fl macrophages. To study these characteristics, we used a high-content analysis approach based on live-cell fluorescence labeling assays, assessed using an automated confocal image reader and single-cell-based image analysis. To measure relative levels of apoptosis, BMDMs were stained with Annexin-V. Both the percentage of positive cells and the mean fluorescence intensity were similar in HilpdaΔLysM and Hilpdafl/fl macrophages, indicating that apoptosis is not affected by HILPDA deficiency (Fig. 6 A and B). To assess efferocytosis, HilpdaΔLysM or Hilpdafl/fl BMDMs were cocultured with apoptotic Jurkat E6.1 cells. Both the percentage of cells positive for the efferocytosis as well as the mean count of ingested Jurkat cells per macrophage did not differ between HilpdaΔLysM and Hilpdafl/fl BMDMs (Fig. 6 C and D). Lastly, to assess phagocytosis, BMDMs were coincubated with fluorescently labeled zymosan bioparticles. No differences in the percentage of positive cells or mean fluorescence intensity were found between HilpdaΔLysM and Hilpdafl/fl macrophages (Fig. 6 E and F). Taken together, our data suggest that deficiency of HILPDA does not affect apoptosis in HilpdaΔLysM versus Hilpdafl/fl macrophages, nor does it increase the capacity for efferocytosis or phagocytosis. Since recent research has highlighted the important role of LDs in the bacterial killing capacity of macrophages (8), we investigated whether the decreased retention of LDs after LPS treatment in HilpdaΔLysM macrophages could impact their killing capacity. The microbial killing of Escherichia coli was similar between atglistatin-treated HilpdaΔLysM and Hilpdafl/fl macrophages, and treatment with LPS did not lead to significant differences in remaining E. coli load after 4 or 24 h of LPS treatment (Fig. 6G).
Fig. 6.
HILPDA deficiency does not affect key macrophage effector functions. (A and B) Percentage of positive cells and mean fluorescence intensity (MFI) following Annexin-V and Hoechst staining in HilpdaΔLysM and Hilpdafl/fl BMDMs after treatment with LPS for 24 h. (C and D) Percentage of positive cells and amount of ingested Jurkat cells (green) following efferocytosis assay and staining in HilpdaΔLysM and Hilpdafl/fl BMDMs (actin, red; nuclei, blue) after treatment with LPS for 24 h. (E and F) Percentage of positive cells and MFI after phagocytosis assay and staining in HilpdaΔLysM and Hilpdafl/fl BMDMs (beads, red; nuclei, blue) after treatment with LPS for 24 h. (G) Relative E. coli load after 4 or 24 h of bacterial killing in HilpdaΔLysM and Hilpdafl/fl BMDMs after treatment with LPS for 24 h.
Increased Production of PGE2 in HILPDA-Deficient Macrophages Drives Increased Production of IL-6.
Next, we investigated the mechanism underlying the effect of HILPDA deficiency on PGE2 and IL-6 production using BMDMs. In line with peritoneal macrophages, IL-6 production was significantly higher in HilpdaΔLysM compared with Hilpdafl/fl BMDMs after 8 and 24 h of LPS treatment. Subsequently, we determined whether the observed differences in PGE2 and IL-6 might have been due to differential activation of transcriptional factors controlling the inflammatory response. However, phosphorylation of cJUN, STAT3, and NF-κB was not significantly different between HilpdaΔLysM and Hilpdafl/fl macrophages (SI Appendix, Fig. S4), indicating that the acute activation of inflammatory pathways is intact. Alternatively, since we previously showed that lipid loading leads to increased oxidation of excess fatty acids in HilpdaΔLysM compared with Hilpdafl/fl macrophages (16), we determined whether the oxidation of fatty acids or other metabolic parameters were differently regulated in HilpdaΔLysM macrophages. Comparing oxygen consumption rate (OCR) before and after the addition of etomoxir, an inhibitor of fatty acid oxidation (FAO), did not reveal consistent and significant differences in FAO capacity in LPS-treated BMDMs (24 h) between the two genotypes (SI Appendix, Fig. S5A). Additionally, no differences were observed in the extracellular acidification rate (ECAR) after 24 h of LPS treatment, suggesting the absence of long-term differences in glycolytic rate (SI Appendix, Fig. S5B). These results suggest that deficiency of HILPDA does not affect the metabolic capacity of macrophages.
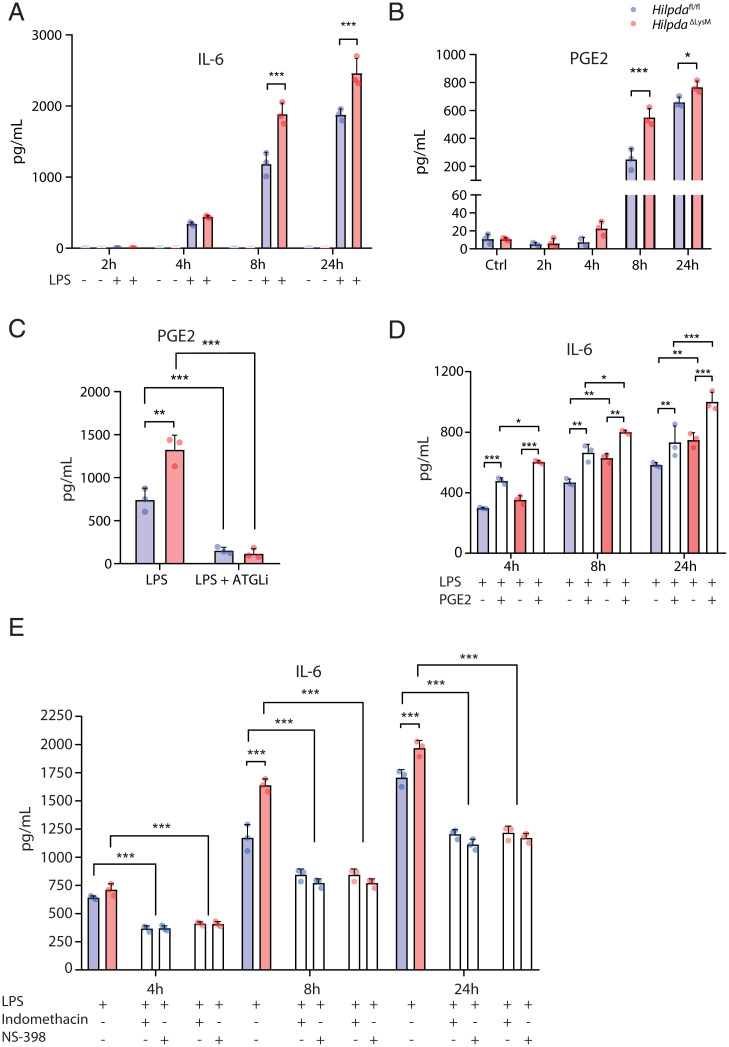
We subsequently hypothesized that the observed increase in PGE2 in HilpdaΔLysM macrophages could be driven by an increased conversion of excess fatty acids into lipid-related inflammatory mediators, which in turn may drive the increase in IL-6 release (21). To examine this hypothesis, we first measured whether the increased production of IL-6 and PGE2 in HilpdaΔLysM macrophages could be confirmed in vitro. Indeed, after 8 and 24 h of LPS treatment, the production of both IL-6 and PGE2 was significantly higher in HilpdaΔLysM than Hilpdafl/fl macrophages (Fig. 7 A and B). Next, we measured the release of PGE2 in BMDMs from HilpdaΔLysM and Hilpdafl/fl mice treated with LPS and vehicle or LPS and atglistatin for 24 h. Although the production of PGE2 was found to be consistently higher in BMDMs from HilpdaΔLysM versus Hilpdafl/fl mice, the production of PGE2 in both genotypes was similarly inhibited by the addition of atglistatin (Fig. 7C), validating the importance of lipolysis by ATGL for the endogenous production of PGE2. Subsequently, we tested whether the increased levels of PGE2 could drive the observed increase in IL-6 production. The addition of exogenous PGE2 to BMDMs from HilpdaΔLysM and Hilpdafl/fl mice treated with LPS significantly increased the production of IL-6 in both genotypes after 4, 8, and 24 h (Fig. 7D). Inhibiting cyclooxygenase 1 (COX1) and COX2, the alleged drivers of PGE2 synthesis, by a general COX1/2 inhibitor (indomethacin) or a specific COX2 inhibitor (NS-398), drastically decreased the production of PGE2 in both genotypes after 8 or 24 h of LPS stimulation (SI Appendix, Fig. S5C). Accordingly, these inhibitors also significantly decreased the production of IL-6 in macrophages from both genotypes, abolishing the difference between HilpdaΔLysM and Hilpdafl/fl macrophages (Fig. 7E) and verifying the association between increased PGE2 production and subsequent IL-6 production in HilpdaΔLysM macrophages.
Fig. 7.
Increased production of PGE2 in HILPDA-deficient macrophages drives increased production of IL-6. (A and B) Concentration of IL-6 (A) or PGE2 (B) from HilpdaΔLysM and Hilpdafl/fl BMDMs treated with vehicle (Ctrl) or LPS for 2, 4, 8, or 24 h. (C) Concentration of PGE2 from HilpdaΔLysM and Hilpdafl/fl BMDMs after treatment with LPS or LPS and atglistatin for 24 h. (D) Concentration of IL-6 from HilpdaΔLysM and Hilpdafl/fl BMDMs treated with LPS and/or PGE2 for 4, 8, or 24 h. (E) Concentration of IL-6 from HilpdaΔLysM and Hilpdafl/fl BMDMs treated with LPS and indomethacin or NS-398 for 4, 8, or 24 h. Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In the current study, we investigated the mechanism underlying the LPS-induced accumulation of TGs in macrophages, as well as the role of TG accumulation in LPS-induced inflammation. Our data demonstrate that LPS promotes TG accumulation in macrophages by inducing HILPDA, which, in turn, lowers ATGL protein levels by enhancing the proteasomal degradation of ATGL. The decrease in TGs in HILPDA-deficient myeloid cells is associated with a heightened inflammatory response, both in vivo and ex vivo, and is linked to an elevated production of PGE2. Our data suggest that the tight regulation of lipid efflux from LDs by ATGL and HILPDA is important for the regulation of the inflammatory response by controlling the production of PGE2, which in turn modulates the release of IL-6.
Although a decrease in ATGL-mediated lipolysis plays an important part in stimulating the accumulation of TGs in LDs during inflammation, this phenomenon is likely the result of an interplay between several variables. As observed, the expression of HILPDA increases during inflammation, decreasing ATGL protein levels and thereby limiting the breakdown of TGs from LDs. At the same time, oxidation of fatty acids is often limited in inflammatory macrophages (22, 23). Additionally, the synthesis or uptake of fatty acids (4) and subsequent synthesis of TGs (24) are also significantly enhanced after inflammatory activation. Together, these processes lead to the physical accumulation of LDs in macrophages. However, HILPDA deficiency prevents the inducing effects of LPS on TG accumulation in LDs, underlining the importance of ATGL inhibition in this process.
Lack of visible LDs in our HILPDA-deficient macrophages is distinct from the lack of LDs that results from the inhibition of TG synthesis, as seen by Castoldi and colleagues (24). In their DGAT1-deficient macrophages, the inhibition of TG synthesis caused a lack of lipid storage in LDs. Consequently, a decreased amount of precursors for the production of prostaglandins was available, leading to an attenuated inflammatory response in macrophages (24). In our study, HILPDA deficiency in myeloid cells increased ATGL-mediated lipolysis and thus likely enhanced the availability of fatty acids as precursors for inflammatory mediators. In line with this notion, we observed an increased inflammatory phenotype. The magnitude of the inflammatory phenotype logically differed between in vivo, ex vivo, and in vitro situations, and the effects of myeloid HILPDA deficiency on cytokine secretion were most pronounced in vitro. The relatively modest in vivo effects of HILPDA deficiency could partly be explained by the relatively modest reduction in Hilpda expression in splenic macrophages of HilpdaΔLysM mice. Overall, the modest in vivo effects emphasize the complexity of studying the effects of a myeloid-specific gene deficiency on markers of systemic inflammation. However, the enhanced inflammatory phenotype in macrophages, both in vitro and ex vivo, was consistently exemplified by an increased production of IL-6, likely effectuated by increased levels of PGE2. Hence, our data confirm the importance of the availability of TG pools in the inflammatory process in macrophages. Moreover, the results of this study pinpoint ATGL-mediated lipolysis as an important factor in the inflammatory process by tightly controlling the flow of lipids from LDs toward the synthesis of inflammatory mediators, thereby precisely determining the magnitude of the inflammatory response.
Interestingly, the timing of the in vivo and ex vivo differences in cytokine levels between LPS-treated wild-type and myeloid-specific HILPDA-deficient mice preceded the differences in visible LD content in peritoneal macrophages, as differences in the accumulation of visible LDs only became significant 8 h after LPS treatment. A potential explanation for the apparent discrepancy is that already at baseline, LD degradation exceeds LD synthesis in HILPDA-deficient macrophages as a result of a reduction in HILPDA-mediated ATGL inhibition, leading to a higher flux of lipids. This higher flux of lipids in HilpdaΔLysM macrophages is not dependent on LPS-induced regulation of HILPDA or ATGL and may be the driving force of differences in cytokine secretion in vivo and ex vivo between genotypes at earlier time points prior to LPS-induced LD accumulation.
The question remains how the mechanism that connects ATGL overactivation to IL-6 secretion may translate toward the systemic level. There is ample evidence that increased systemic IL-6 levels can enhance lipolysis and FAO in various tissues and cell types (25, 26), whereas blocking endogenous IL-6 signaling can prevent the mobilization of free fatty acids (27). These findings identify IL-6 as an important systemic lipolytic factor. Although the mechanism by which IL-6 influences ATGL-mediated lipolysis in macrophages is still unknown, it can be hypothesized that the increased secretion of IL-6 resulting from amplified ATGL-mediated lipolysis in HilpdaΔLysM macrophages may lead to the formation of a positive feedback loop, further stimulating lipolytic and inflammatory processes. However, IL-6 is also thought to play a key role in alternative macrophage polarization and was suggested to limit the inflammatory effects of LPS (28). This apparent incongruence emphasizes the complex nature of this cytokine and warrants further study on the interplay of IL-6 in macrophage lipid metabolism and inflammation.
While the link between PGE2, IL-6, and lipid accumulation is clear, the connection between LDs and TNFα is less straightforward. In the plasma of HilpdaΔLysM mice treated with LPS, the concentration of TNFα is significantly higher after 2 h of LPS treatment. However, in peritoneal macrophages lacking HILPDA, significantly lower TNFα levels were released ex vivo after a 4-h LPS treatment in vivo. It seems that TNFα release could be inhibited by PGE2 in some cases (29, 30), which may partly explain this observation. However, reduced TNFα levels were not visible in peritoneal macrophages after an 8-h LPS treatment, whereas the PGE2 release of peritoneal macrophages was still high at that time point. Thus, our model cannot provide conclusive results on the connection between TNFα and lipid metabolism. In nonimmune cells, TNFα is thought to stimulate the accumulation of lipids in LDs via acyl-CoA synthetases in (31) and to stimulate lipogenesis (32). At the same time, TNFα may also stimulate lipolysis (33), possibly through ATGL (34). Since these pathways counteract each other, it would be interesting to investigate which lipid-related metabolic effects of TNFα are replicable in macrophages and how this may enhance or inhibit the inflammatory signature of macrophages.
In both neutrophils and mast cells lacking ATGL, decreased release of inflammatory lipid mediators can be seen (35, 36). These results are in agreement with our results using the ATGL inhibitor atglistatin, which directly inverted the increased levels of PGE2 in HilpdaΔLysM macrophages. Eicosanoids, including PGE2, are signaling lipids synthesized from arachidonic acid by highly organized COX enzymes that can be directly recruited to LD membranes. Synthesis of eicosanoids during LPS-induced inflammation is heavily dependent on the liberation of esterified arachidonic acid from intracellular phospholipid or neutral lipid pools (37). Whereas specific hydrolysis of membrane glycerophospholipids or neutral lipids by phospholipase A2 enzymes was conventionally regarded as the main source of fatty acids for eicosanoid production (38, 39), the current study builds upon the increasing evidence that ATGL also controls the release of fatty acid precursors from LDs for the production of eicosanoids in immune cells (35, 36).
It is well-established that prostaglandins, including PGE2, can directly regulate the inflammatory response by influencing the release of cytokines. The regulation of different cytokines by PGE2 is complex and determined by separate molecular mechanisms (29, 40). Whether PGE2 exerts pro- or antiinflammatory effects is likely also dependent on the timing of experiments and the use of different in vitro or in vivo systems. Endogenous production of PGE2 in macrophages has led to inhibition of TNFα release, but also to the induction of either transcription or release of IL-10, IL-1β, and IL-6 (21, 29, 30, 40, 41). Especially the direct role between PGE2 and IL-6 is often highlighted to be specific (21) and formed the basis of the observed phenotypes within the current study.
The fact that careful regulation of ATGL is crucial for an adequate immune response in macrophages, besides providing fatty acid precursors for inflammatory mediators, becomes further apparent in studies using ATGL−/− macrophages. In macrophages lacking ATGL, TGs accumulate, leading to mitochondrial dysfunction, defects in macrophage polarization, ER stress, decreased ability to migrate, and decreased ability to use phagocytosis, accompanied by increased induction of mitochondrial apoptosis (12, 13, 20). A decreased activation of ATGL in macrophages, for instance by deleting its coactivator CGI-58, also diminishes phagocytosis, although this phenotype is not accompanied by mitochondrial dysfunction (42). Intriguingly, in our results, which in many ways show the exact opposite of the processes found in macrophages with decreased ATGL activity, no evidence for increased phagocytosis or efferocytosis ability was found. Thus, whereas active ATGL was identified as a crucial factor to enable phagocytosis, a further increase in ATGL expression does not seem to increase the phagocytic ability of macrophages.
To enable precise control of ATGL in macrophages, ATGL expression is regulated by several inhibitors, including HILPDA. We have shown that HILPDA is induced by lipid influx (16) and, as observed in this study, inflammatory activation by LPS. Previous studies have also shown that HILPDA inhibits ATGL via direct physical interaction (16, 17). Here we present evidence that HILPDA promotes the proteasomal degradation of ATGL in BMDMs, explaining the direct effects of HILPDA on LD homeostasis in macrophages (15, 16). The exact mechanism by which HILPDA enhances ATGL degradation is unclear. It can be hypothesized that by binding to ATGL, HILPDA may cause a conformational change in ATGL, leading to the recruitment of a particular ubiquitin ligase. Further studies are necessary to carefully dissect the underlying mechanism.
In conclusion, our study demonstrates that HILPDA is directly involved in regulating the breakdown of ATGL during inflammatory activation in macrophages, thereby controlling ATGL-mediated lipolysis and mediating the increase in TG accumulation upon LPS treatment. Furthermore, we establish an important role for ATGL-mediated lipolysis in the production of PGE2 and subsequent modulation of IL-6 release.
Materials and Methods
Animal Studies.
All animal experiments were approved by the animal welfare committee of Wageningen University (2016.W-0093.015). HilpdaΔLysM and Hilpdafl/fl mice were bred as described before (16). Mice had unlimited access to food (standard chow) and water and were housed under normal light–dark cycles in humidity- and temperature-controlled specific pathogen-free conditions. Both male and female mice were used for the isolation of primary cell cultures. For the in vivo study, male HilpdaΔLysM and Hilpdafl/fl mice, aged 9 to 12 wk, were housed with two or three littermates per cage and randomly allocated to the different time points. Mice were weighed prior to injection to calculate specific concentrations of LPS. At each time point, eight mice per genotype received an intraperitoneal injection of LPS (from E. coli O55:B5; L6529; Sigma-Aldrich) at a concentration of 2 mg/kg body weight in an end volume of 200 µL sterile saline solution. Control groups were injected with 200 µL sterile saline solution. After the indicated time points, mice were anesthetized with isoflurane and blood was collected via orbital puncture in ethylenediaminetetraacetate (EDTA)-containing tubes (Sarstedt). After blood collection, mice were immediately killed by cervical dislocation. Subsequently, peritoneal macrophages and tissues were harvested.
Flow Cytometry.
Before collection of blood plasma, 25 µL blood was aliquoted from each sample and stained with antibodies against CD45-Alexa700 (103127), CD11b-FITC (101205), F4/80-PE (123109), Ly6C-Brilliant Violet 421 (128031), CCR2-PE/Cy7 (150611), CD3-APC (100235), CD4-PerCP/Cy5.5 (100539), CD8-Brilliant Violet 785 (100749), CD19-PE/Dazzle 594 (115553), and Ly6G-APC/Fire 750 (127651) (all purchased from BioLegend). Red blood cells (RBCs) were lysed with RBC lysis buffer (00-4333-57; eBioscience, Thermo Fisher Scientific), and samples were measured on the CytoFLEX Flow Cytometry System (Beckman Coulter). Results were analyzed using CytExpert acquisition and analysis software, version 2.4 (Beckman Coulter) and FlowJo analysis software (BD Biosciences).
Plasma Measurements.
EDTA tubes containing blood samples were centrifuged for 15 min at 5,000 rpm at 4 °C. Plasma was aliquoted and stored at −80 °C until further measurements. The V-PLEX Mouse Cytokine 19-Plex Kit (K15255D-1; Meso Scale Diagnostics) and MESO QuickPlex SQ 120 (Meso Scale Diagnostics) were used to determine the concentrations of IL27p28/IL30, IP-10, MCP-1, MIP-1α, MIP-2, IFNγ, IL-10, IL-1β, IL-6, and TNFα in the plasma. A high-multiplex biomarker panel (Olink Target Mouse Exploratory) was used to explore 92 biomarkers in the mouse plasma using the proprietary Proximity Extension Assay technology (Olink Proteomics).
Primary Cell Isolation.
Peritoneal macrophages, splenic cells, and BMDMs were isolated. All techniques that were used to isolate, grow and treat the cells are described in the SI Appendix.
In Vitro Experiments.
BMDMs were treated with LPS from E. coli (O55:B5; L6529; Sigma-Aldrich) and flagellin from Salmonella typhimurium (Sigma-Aldrich) at a concentration of 100 ng/mL, Pam3CysK4 (L2000; EMC Microcollections) at a concentration of 5 µg/mL, poly:IC (Sigma-Aldrich) at a concentration of 20 µg/mL, and zymosan from Saccharomyces cerevisiae (Sigma-Aldrich) at a concentration of 100 µg/mL. Zymosan was opsonized to increase uptake by phagocytosis and sonicated just before use. Atglistatin (SML1075; Sigma-Aldrich) was used at a concentration of 20 μM, and cells were pretreated for 1 h before further treatment. C75 (C5490; Sigma-Aldrich) was used at a concentration of 5 µg/mL, and cells were pretreated for 1 h before further treatment. Indomethacin (I7378; Sigma-Aldrich) was used at 10 µM and NS-398 (N194; Sigma-Aldrich) was used at 1 µM, and cells were pretreated for 1 h before further treatment. PGE2 (2296; Tocris, Bio-Techne) was added at 0.1 µM 1 h after treatment with LPS.
Confocal Imaging.
The visualization of LD accumulation was studied by plating BMDMs on 8- or 18-well glass-bottom µ-slides (80807 and 81817; Ibidi) coated with collagen type I (50201; Ibidi). After treatment, cells were washed with PBS and fixed with 3.7% paraformaldehyde. Next, cells were stained with 2 μg/mL BODIPY 493/503 (D3922; Invitrogen, Thermo Fisher Scientific) and ActinRed555 (R37112; Thermo Fisher Scientific) and mounted with Vectashield Antifade Mounting Medium (H-1000-10; Vector Laboratories). Imaging was performed on the TCS SP8 X System (Leica Microsystems) using the 63 × 1.20 numerical aperture water-immersion objective lens. The pinhole was set to 1 Airy unit, and fluorescent probes were excited using a white-light laser (50% laser output) with laser power set to 1.5%. Fluorescence emission was detected using internal hybrid detectors. Images were processed and analyzed for LDs with CellProfiler software (43).
Immunoblotting.
Cells were lysed in RIPA lysis buffer (89900; Thermo Fisher Scientific) supplemented with phosphatase and protease inhibitors. Precast 4 to 15% polyacrylamide gels were used to separate protein lysates, and proteins were transferred onto nitrocellulose membranes using a Trans-Blot Turbo Semi-Dry Transfer Cell with Trans-Blot Turbo PVDF Transfer Packs (Bio-Rad Laboratories). After blocking in nonfat milk, membranes were incubated overnight at 4 °C with primary antibody for HILPDA, a kind gift of Christina Warnecke, Department of Nephrology and Hypertension, University Hospital Erlangen, Friedrich-Alexander-University Erlangen-Nürnberg, Erlangen, Germany; ATGL (2138S; Cell Signaling Technology); phospho–NF-κB p65 (Ser536; 3033T; Cell Signaling Technology); phospho-Stat3 (Tyr705; 9145T; Cell Signaling Technology); phospho–c-Jun (Ser63; 9261S; Cell Signaling Technology); ACTIN (5057; RRID: AB_10694076; Cell Signaling Technology); or HSP90 (4874S; RRID: AB_2121214; Cell Signaling Technology). Membranes were subsequently incubated with secondary antibody (anti-rabbit immunoglobulin G, horseradish peroxidase–linked antibody; 7074; Cell Signaling Technology) for 1 h at 4 °C. Membranes were developed using Clarity ECL substrate (Bio-Rad Laboratories) and images were acquired with the ChemiDoc MP System (Bio-Rad Laboratories).
Enzyme-Linked Immunosorbent Assay.
IL-6, TNFα, IL-10, and IL-1RA concentrations were measured in cell supernatants with DuoSet Sandwich Enzyme-Linked Immunosorbent Assay (ELISA) Kits (DY406/DY410/DY417/DY490; R&D Systems, Bio-Techne) according to the manufacturer’s instructions. PGE2 was measured with the PGE2 ELISA Kit Monoclonal (514010; Cayman Chemicals). Normalization for peritoneal macrophages was performed by determining the DNA concentration per well (Quant-iT dsDNA Assay Kit High Sensitivity; Q33120; Thermo Fisher Scientific).
Functional Assays.
BMDMs were treated with LPS (100 ng/mL) for 24 h to induce LD accumulation and used for various functional assays. Technical details can be found in the SI Appendix.
Bacterial Killing Assay.
E. coli were grown to an optical density of 1 at 600 nm and diluted toward 1 × 105 colony-forming units (CFUs) per milliliter. BMDMs from HilpdaΔLysM and Hilpdafl/fl mice were seeded at a density of 50,000 cells per well in 96-well plates and treated overnight with atglistatin, LPS, or LPS + atglistatin for 24 h. BMDMs were coincubated with E. coli (total multiplicity of infection 100) for 30 min and subsequently washed with 200 µg/mL gentamycin. Cells were lysed directly after 30 min (t = 0), or after 4 (t = 4) or 24 h (t = 24) of incubation at 37 °C and 10% CO2. BMDMs were lysed and bacteria were plated in serial dilutions on Lysogeny broth-agar plates overnight at 30 °C, after which the CFUs were counted.
Extracellular Flux Assay.
The extracellular flux of HilpdaΔLysM and Hilpdafl/fl BMDMs was measured after 24-h treatment with LPS (100 ng/mL) using the Seahorse XF-96 Analyzer (Agilent Technologies). BMDMs were seeded in XF-96 plates (Agilent Technologies) and cultured at 37 °C and 5% CO2. Before measurement, BMDMs were washed and cultured in Seahorse XF base medium (Agilent Technologies) without sodium bicarbonate, supplemented with 2 mM l-glutamine for the glycolytic stress test or with 2 mM l-glutamine and 25 mM glucose for the measurement of oxidative phosphorylation, for 1 h at 37 °C in a non-CO2 incubator. For the glycolytic stress test, glucose (25 mM), oligomycin (1.5 μM), and 2-DG (50 mM) were injected, respectively, and the ECAR was measured at baseline and following injections. For the measurement of OXPHOS and FAO, etomoxir (50 μM) was injected and the OCR was measured at baseline and following the injection. Calculations were made using the Seahorse XF-96 software Wave Desktop 2.6 (RRID: SCR_014526; Agilent Technologies).
qPCR.
Total RNA was isolated with TRIzol reagent (Invitrogen, Thermo Fisher Scientific), and from spleen macrophages using the RNeasy Micro Kit (QIAGEN). Complementary DNA (cDNA) was synthesized from 500 ng RNA with the iScript cDNA Kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. The CFX384 Touch Real-Time Detection System (Bio-Rad Laboratories) was used to assess amplification of Pnpla2 templates (forward primer 5′-CAACGCCACTCACATCTACGG-3′; reverse primer 5′-GGACACCTCAATAATGTTGGCAC-3′) or Hilpda templates (forward primer 5′-TCGTGCAGGATCTAGCAGCAG-3′; reverse primer 5′-GCCCAGCACATAGAGGTTCA-3′) with the SensiMix Kit for SYBR Green reactions (QT650-05; BioLine). Mouse 36b4 expression was used to normalize the quantification (forward primer 5′-ATGGGTACAAGCGCGTCCTG-3′; reverse primer 5′-GCCTTGACCTTTTCAGTAAG-3′).
Power Calculation Animal Study.
From a pilot study, we concluded that macrophages from HilpdaΔLysM mice after stimulation with LPS showed a 1.4-fold up-regulation in the mRNA expression of Il6, compared with Hilpdafl/fl mice. SDs between several experiments were tested between 0.1 and 0.2. The group size was estimated with a power calculation assuming use of a one-way ANOVA with a significance level of 0.05 and a power of 80%, including at least 10 pairwise comparisons between groups, leading to an estimation of around n = 7 mice needed per group. To allow compensation for unforeseen circumstances and account for potential loss of mice during the study, we included 8 mice per genotype, per group.
Statistical Analysis.
Analysis and visualization for the Olink data were performed with the R language (CRAN; RRID: SCR_003005; https://www.r-project.org) using the packages ggbiplot (PCA) and ggplot2 (volcano plots). Details on statistical analyses can be found in the figure legends. N represents either the number of animals used or the number of in vitro replications performed. Data are represented as mean ± SD or as indicated. Statistical analyses were performed using the unpaired Student’s t test, Mann–Whitney U test, or two-way ANOVA followed by either Bonferroni’s or Sidak’s post hoc multiple-comparisons test, if both genotype and treatment were found to be significant. A value of P < 0.05 was considered statistically significant. All data were visualized and analyzed using Prism version 5.0 or 8.0 (GraphPad Software) or R Studio (PBC; https://www.rstudio.com/).
Supplementary Material
Acknowledgments
We thank Merel Defour, Benthe van der Lugt, Julia van Heck, Jenny Jansen, Anneke Hijmans, Gregorio Fazzi, and Ricky Siebeler for their practical assistance and Dr. Christina Warnecke for providing the HILPDA antibody. This work was financed by grants from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (2014/12393/ALW), Diabetes Fonds (2015.82.1824), and Hartstichting (ENERGISE CVON2014-02).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114739119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Gross D. A., Silver D. L., Cytosolic lipid droplets: From mechanisms of fat storage to disease. Crit. Rev. Biochem. Mol. Biol. 49, 304–326 (2014). [DOI] [PubMed] [Google Scholar]
- 2.den Brok M. H., Raaijmakers T. K., Collado-Camps E., Adema G. J., Lipid droplets as immune modulators in myeloid cells. Trends Immunol. 39, 380–392 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Welte M. A., Gould A. P., Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1260–1272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feingold K. R., et al. , Mechanisms of triglyceride accumulation in activated macrophages. J. Leukoc. Biol. 92, 829–839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh W. Y., et al. , Toll-like receptors induce signal-specific reprogramming of the macrophage lipidome. Cell Metab. 32, 128–143.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Péan C. B., et al. , Regulation of phagocyte triglyceride by a STAT-ATG2 pathway controls mycobacterial infection. Nat. Commun. 8, 14642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocchiaro J. L., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H., Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U.S.A. 105, 9379–9384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch M., et al. , Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science 370, eaay8085 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Zimmermann R., et al. , Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S., Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: Ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279, 47066–47075 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Jenkins C. M., et al. , Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279, 48968–48975 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Aflaki E., et al. , Impaired Rho GTPase activation abrogates cell polarization and migration in macrophages with defective lipolysis. Cell. Mol. Life Sci. 68, 3933–3947 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aflaki E., et al. , Triacylglycerol accumulation activates the mitochondrial apoptosis pathway in macrophages. J. Biol. Chem. 286, 7418–7428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattijssen F., et al. , Hypoxia-inducible lipid droplet-associated (HILPDA) is a novel peroxisome proliferator-activated receptor (PPAR) target involved in hepatic triglyceride secretion. J. Biol. Chem. 289, 19279–19293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier A., et al. , Hypoxia-inducible protein 2 Hig2/Hilpda mediates neutral lipid accumulation in macrophages and contributes to atherosclerosis in apolipoprotein E-deficient mice. FASEB J. 31, 4971–4984 (2017). [DOI] [PubMed] [Google Scholar]
- 16.van Dierendonck X. A. M. H., et al. , HILPDA uncouples lipid droplet accumulation in adipose tissue macrophages from inflammation and metabolic dysregulation. Cell Rep. 30, 1811–1822.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Padmanabha Das K. M., et al. , Hypoxia-inducible lipid droplet-associated protein inhibits adipose triglyceride lipase. J. Lipid Res. 59, 531–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhajda F. P., et al. , Synthesis and antitumor activity of an inhibitor of fatty acid synthase. Proc. Natl. Acad. Sci. U.S.A. 97, 3450–3454 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer N., et al. , Development of small-molecule inhibitors targeting adipose triglyceride lipase. Nat. Chem. Biol. 9, 785–787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandak P. G., et al. , Efficient phagocytosis requires triacylglycerol hydrolysis by adipose triglyceride lipase. J. Biol. Chem. 285, 20192–20201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams J. A., Shacter E., Regulation of macrophage cytokine production by prostaglandin E2. Distinct roles of cyclooxygenase-1 and -2. J. Biol. Chem. 272, 25693–25699 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Vats D., et al. , Oxidative metabolism and PGC-1β attenuate macrophage-mediated inflammation. Cell Metab. 4, 13–24 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Prados J.-C., et al. , Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 185, 605–614 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Castoldi A., et al. , Triacylglycerol synthesis enhances macrophage inflammatory function. Nat. Commun. 11, 4107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carey A. L., et al. , Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes 55, 2688–2697 (2006). [DOI] [PubMed] [Google Scholar]
- 26.van Hall G., et al. , Interleukin-6 stimulates lipolysis and fat oxidation in humans. J. Clin. Endocrinol. Metab. 88, 3005–3010 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Trinh B., et al. , Blocking endogenous IL-6 impairs mobilization of free fatty acids during rest and exercise in lean and obese men. Cell Rep. Med. 2, 100396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauer J., et al. , Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat. Immunol. 15, 423–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacKenzie K. F., et al. , PGE(2) induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A-SIK-CRTC3 pathway. J. Immunol. 190, 565–577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S. H., et al. , Distinct protein kinase A anchoring proteins direct prostaglandin E2 modulation of Toll-like receptor signaling in alveolar macrophages. J. Biol. Chem. 286, 8875–8883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung H. S., et al. , TNF-α induces acyl-CoA synthetase 3 to promote lipid droplet formation in human endothelial cells. J. Lipid Res. 61, 33–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feingold K. R., Grunfeld C., Tumor necrosis factor-alpha stimulates hepatic lipogenesis in the rat in vivo. J. Clin. Invest. 80, 184–190 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauner H., Petruschke T., Russ M., Röhrig K., Eckel J., Effects of tumour necrosis factor alpha (TNFα) on glucose transport and lipid metabolism of newly-differentiated human fat cells in cell culture. Diabetologia 38, 764–771 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Jin D., et al. , TNF-α reduces g0s2 expression and stimulates lipolysis through PPAR-γ inhibition in 3T3-L1 adipocytes. Cytokine 69, 196–205 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Schlager S., et al. , Adipose triglyceride lipase acts on neutrophil lipid droplets to regulate substrate availability for lipid mediator synthesis. J. Leukoc. Biol. 98, 837–850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dichlberger A., Schlager S., Maaninka K., Schneider W. J., Kovanen P. T., Adipose triglyceride lipase regulates eicosanoid production in activated human mast cells. J. Lipid Res. 55, 2471–2478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pacheco P., et al. , Lipopolysaccharide-induced leukocyte lipid body formation in vivo: Innate immunity elicited intracellular loci involved in eicosanoid metabolism. J. Immunol. 169, 6498–6506 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Leslie C. C., Cytosolic phospholipase A2: Physiological function and role in disease. J. Lipid Res. 56, 1386–1402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jarc E., Petan T., A twist of FATe: Lipid droplets and inflammatory lipid mediators. Biochimie 169, 69–87 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Williams J. A., Pontzer C. H., Shacter E., Regulation of macrophage interleukin-6 (IL-6) and IL-10 expression by prostaglandin E2: The role of p38 mitogen-activated protein kinase. J. Interferon Cytokine Res. 20, 291–298 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Hinson R. M., Williams J. A., Shacter E., Elevated interleukin 6 is induced by prostaglandin E2 in a murine model of inflammation: Possible role of cyclooxygenase-2. Proc. Natl. Acad. Sci. U.S.A. 93, 4885–4890 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goeritzer M., et al. , Deletion of CGI-58 or adipose triglyceride lipase differently affects macrophage function and atherosclerosis. J. Lipid Res. 55, 2562–2575 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpenter A. E., et al. , CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.