Significance
Shigella flexneri, a deleterious bacterium, causes massive human infection cases and deaths worldwide. To facilitate survival and replication in infected host cells, S. flexneri can secrete two highly similar E3 ligase effectors, IpaH1.4 and IpaH2.5, to subvert the linear ubiquitin chain assembly complex (LUBAC), a key player involved in numerous antibacterial signaling pathways of host cells but with poorly understood mechanisms. In this study, through systematic biochemical and structural characterization, we elucidate the multiple tactics adopted by IpaH1.4/2.5 to disarm the human LUBAC and provide mechanistic insights into the subversion of host LUBAC by IpaH1.4/2.5 of S. flexneri.
Keywords: LUBAC, HOIP, HOIL-1L, IpaH1.4, ubiquitination
Abstract
Shigella flexneri, a gram-negative bacterium, is the major culprit of bacterial shigellosis and causes a large number of human infection cases and deaths worldwide annually. For evading the host immune response during infection, S. flexneri secrets two highly similar E3 ligases, IpaH1.4 and IpaH2.5, to subvert the linear ubiquitin chain assembly complex (LUBAC) of host cells, which is composed of HOIP, HOIL-1L, and SHARPIN. However, the detailed molecular mechanism underpinning the subversion of the LUBAC by IpaH1.4/2.5 remains elusive. Here, we demonstrated that IpaH1.4 can specifically recognize HOIP and HOIL-1L through its leucine-rich repeat (LRR) domain by binding to the HOIP RING1 domain and HOIL-1L ubiquitin-like (UBL) domain, respectively. The determined crystal structures of IpaH1.4 LRR/HOIP RING1, IpaH1.4 LRR/HOIL-1L UBL, and HOIP RING1/UBE2L3 complexes not only elucidate the binding mechanisms of IpaH1.4 with HOIP and HOIL-1L but also unveil that the recognition of HOIP by IpaH1.4 can inhibit the E2 binding of HOIP. Furthermore, we demonstrated that the interaction of IpaH1.4 LRR with HOIP RING1 or HOIL-1L UBL is essential for the ubiquitination of HOIP or HOIL-1L in vitro as well as the suppression of NF-κB activation by IpaH1.4 in cells. In summary, our work elucidated that in addition to inducing the proteasomal degradation of LUBAC, IpaH1.4 can also inhibit the E3 activity of LUBAC by blocking its E2 loading and/or disturbing its stability, thereby providing a paradigm showing how a bacterial E3 ligase adopts multiple tactics to subvert the key LUBAC of host cells.
Ubiquitination plays pivotal roles in almost every cellular process in mammals, such as protein degradation, gene transcription, autophagy, and immune signaling (1–5). The ubiquitination event involves the covalent conjugation of the target substrate with one or multiple ubiquitin (Ub) catalyzed by a cascade of three enzymes: an E1 Ub-activating enzyme, an E2 Ub-conjugating enzyme, and an E3 Ub ligase (3, 6). In addition to decorating other substrates, Ub itself can be also ubiquitinated to form eight distinct types of poly-Ub chains, which are linked through the extreme N-terminal methionine residue (M1) or any one of the seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) within Ub (4, 7). Different linkage types of poly-Ub chains encode distinct signals and conduct specific cellular functions in mammalian cells (7–9). For instance, the well-known K48-linked poly-Ub chain can serve as a signal for proteasomal degradation, while the M1-linked poly-Ub chain (also named linear poly-Ub chain), in which the backbone carboxyl group of the extreme C-terminal G76 of one Ub is conjugated to the backbone amino group of the M1 residue in the preceding Ub thereby linking multiple Ub molecules together in a “head-to-tail” manner, is widely involved in immune-related signaling pathways (10–14).
The linear Ub chain assembly complex (LUBAC), which consists of a catalytic subunit HOIP and two regulatory subunits HOIL-1L and SHARPIN, is the only currently identified E3 ligase complex capable of catalyzing linear poly-Ub chains on protein substrates (10, 12, 15–19). HOIP is a multidomain-containing RING-between-RING (RBR)–type E3 ligase. It contains an N-terminal region: N-glycanase/Ub-associated domain (UBA) or UBX-containing protein domain for interacting with two crucial deubiquitinases, OTULIN and CYLD, both of which can trim M1-linked Ub chains (20–22), and a B-box–type zinc finger (ZF) followed by a canonical ZF (Fig. 1A). In addition, the middle region of HOIP contains two Nlp4-like ZF (NZF) domains, an atypical UBA, and a following hinge-like domain (Fig. 1A). The C-terminal part of HOIP has a unique linear Ub chain-determining domain (LDD) and a preceding characteristic RBR region, which is composed of a RING1 domain, an in-between-RING motif (IBR), and a RING2 domain (Fig. 1A). Notably, the RBR region together with the LDD domain forms the catalytic core of HOIP for assembling linear Ub chains (23–25), while the RING1 domain is responsible for recruiting Ub-conjugated E2 enzymes, such as UBE2L3 (10, 25, 26). Intriguingly, HOIL-1L is also an RBR-type E3 ligase (Fig. 1A). However, the catalytic activity of HOIL-1L is not directly involved in the linear Ub chain assembly (10). Instead, HOIL-1L can conjugate monoubiquitin onto all LUBAC subunits to attenuate the function of LUBAC by providing preferred monoubiquitinated substrates for HOIP-mediated auto linear ubiquitination of LUBAC (27). In addition, HOIL-1L has an N-terminal LUBAC-tethering motif (LTM), a Ub-like (UBL) domain, and a middle NZF domain that can selectively recognize linear Ub chains (28) (Fig. 1A). As a unique adaptor protein in LUBAC, SHARPIN has an N-terminal pleckstrin homology domain that can mediate the dimerization of SHARPIN (29), an LTM motif, and a UBL domain followed by a Ub-binding NZF domain (16) (Fig. 1A). Previously, studies from our group and other groups revealed that the UBL domains of HOIL-1L and SHARPIN can simultaneously interact with the HOIP UBA domain to assemble the ternary LUBAC (15, 30, 31), which is further stabilized by a cooperative interaction between the two LTM motifs of HOIL-1L and SHARPIN (31). Notably, these mutual interactions among the three subunits of LUBAC are demonstrated to be essential for the stability of the trimeric LUBAC in cells (15–17, 31, 32). Importantly, previous functional studies established that LUBAC plays crucial roles in NF-κB signaling and antibacterial selective autophagy (xenophagy) (12, 32–35). Not surprisingly, as a key player in immune signaling pathways, LUBAC is targeted by many pathogenic bacteria or viruses to subvert host responses during infections (18), such as S. flexneri (36, 37), Salmonella typhimurium (38), hepatitis B virus (39), hepatitis C virus (40), porcine reproductive and respiratory syndrome virus (41), and Epstein–Barr virus (42). However, until now, many of the detailed molecular mechanisms underlying the subversion of LUBAC by these pathogens are still largely unknown.
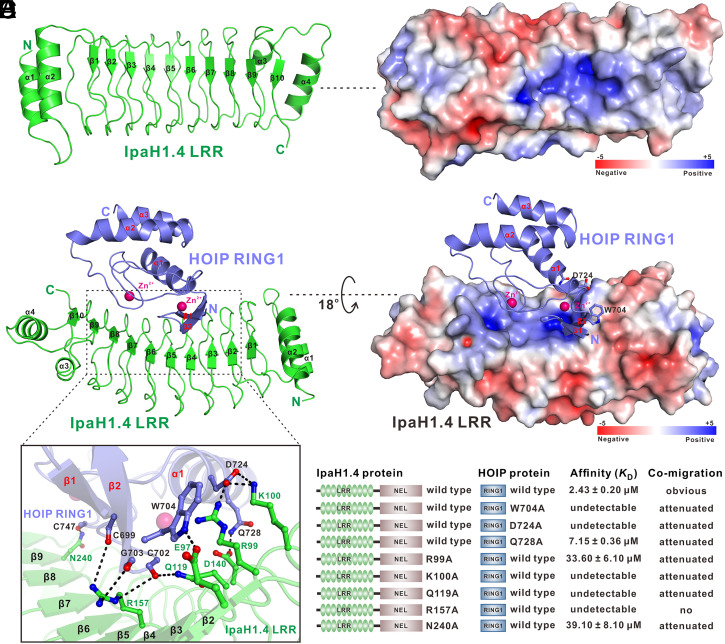
Fig. 1.
Biochemical characterization of the interaction between HOIP and IpaH1.4. (A) A schematic diagram showing the domain organizations of HOIP, HOIL-1L, SHARPIN, IpaH1.4/2.5, and UBE2L3. In this drawing, the intermolecular interactions of HOIP, HOIL-1L, and SHARPIN are indicated by black two-way arrows, while the HOIP/IpaH1.4, HOIL-1L/IpaH1.4, and HOIP/UBE2L3 interactions characterized in this study are further highlighted by red two-way arrows. (B and C) Analytical gel filtration chromatography analysis of the interaction between HOIP RING1 domain and full-length IpaH1.4 (B) or IpaH1.4 LRR domain (C). (D) The summarized mapping results of the interacting regions between IpaH1.4 and HOIP by analytical gel filtration chromatography; aa, amino acids. (E) Superposition plots of the 1H-15N HSQC spectra of 15N-labeled HOIP RING1 domain titrated with increasing molar ratios of unlabeled IpaH1.4 LRR domain. (F) ITC-based measurement of the binding affinity of full-length IpaH1.4 with the HOIP RING1 domain. The KD error is the fitted error obtained from the data analysis software when using the one-site binding model to fit the ITC data; DP, differential power measured by the ITC machine; ΔH, heat change measured by the ITC machine. (G) Overlay plot of the sedimentation velocity data of the HOIP RING1 domain (red), the IpaH1.4 LRR domain (blue), and the HOIP RING1/IpaH1.4 LRR complex (black); c(s), continuous sedimentation coefficient distribution; MW, molecular weight.
S. flexneri is a type of gram-negative enteric pathogen that is capable of invading the intestinal epithelium and replicating rapidly in the cytosol of host cells. It is the leading cause of shigellosis, an acute bloody diarrhea in humans (43). To suppress host defense during infection, S. flexneri secretes dozens of virulent factors into the cytoplasm of infected host cells (44). Several effector proteins have been shown to suppress the innate immune signaling or gene transcription of host cells (36, 45). Among these effector proteins, two highly similar E3 Ub ligases IpaH1.4 and IpaH2.5 were recently uncovered to suppress NF-κB activation by targeting LUBAC (37). Both IpaH1.4 and IpaH2.5 contain an N-terminal substrate-binding leucine-rich repeat (LRR) domain and a C-terminal catalytic E3 ligase (NEL) domain (Fig. 1A). They belong to the bacterial NEL E3 ligase family that can catalyze the K48-linked ubiquitination reaction by forming a Ub-thioester intermediate via a catalytic Cys residue in a conserved CXD motif of the NEL domain (46, 47). Strikingly, IpaH1.4/2.5 can directly interact with the HOIP and HOIL-1L subunits of LUBAC and mediate K48-linked ubiquitination of HOIP for proteasomal degradation (37). However, how IpaH1.4/2.5 recognizes HOIP and HOIL-1L as well as the downstream consequence of the recognition of HOIL-1L by IpaH1.4/2.5 remain elusive.
In this study, we biochemically and structurally characterized the interaction between IpaH1.4 and LUBAC and uncovered that the LRR domain of IpaH1.4 can specifically recognize the RING1 domain of HOIP. We determined the crystal structures of the apo form IpaH1.4 LRR, the IpaH1.4 LRR/HOIP RING1 complex, and the related HOIP RING1/UBE2L3 complex, which not only elucidates the detailed binding mechanisms of HOIP with IpaH1.4 and UBE2L3 but also unveils that the interaction of HOIP RING1 with IpaH1.4 can impede the UBE2L3 binding of HOIP. Furthermore, we also uncovered the binding mechanism of IpaH1.4 with HOIL-1L by solving the IpaH1.4 LRR/HOIL-1L UBL complex structure. Finally, we showed that IpaH1.4 can directly mediate the ubiquitination of HOIL-1L in vitro, albeit to a much less extent than that of HOIP, and, importantly, the specific interaction of IpaH1.4 LRR with HOIP RING1 or HOIL-1L UBL is essential for the in vitro ubiquitination of HOIP or HOIL-1L as well as the IpaH1.4-mediated suppression of NF-κB activation in cells. In all, our work elucidates the mechanistic basis underpinning the subversion of LUBAC by the E3 ligase IpaH1.4 of S. flexneri.
Results
Biochemical Characterization of the Interaction between IpaH1.4 and HOIP.
To elucidate how IpaH1.4/2.5 targets LUBAC, we first focused on the IpaH1.4/2.5 and HOIP interaction. Given that IpaH1.4 and IpaH2.5 share highly similar amino acid sequences, especially in the target-binding LRR domain (∼98.83% sequence identity) and the catalytic NEL domain region (SI Appendix, Fig. S1A), we chose IpaH1.4 for further biochemical and structural characterization. Using analytical gel filtration chromatography–based comigration assays with purified relevant HOIP and IpaH1.4 fragment proteins, we revealed that the full-length IpaH1.4 can specifically recognize the RING1 domain rather than the previously reported hinge region between the UBA and RING1 domains of HOIP (Fig. 1 B and D and SI Appendix, Fig. S2). Further reciprocal in vitro binding assays revealed that the LRR domain of IpaH1.4 alone is sufficient for the recognition of HOIP RING1 (Fig. 1 C and D and SI Appendix, Fig. S3). We further utilized NMR spectroscopy to characterize the interaction between HOIP RING1 and IpaH1.4 LRR. Titrations of 15N-labeled HOIP RING1 with unlabeled IpaH1.4 LRR proteins showed that many peaks in the 1H-15N heteronuclear single quantum coherence (HSQC) spectra of HOIP RING1 undergo significant peak broadenings or chemical shift changes (Fig. 1E), indicating that HOIP RING1 can directly bind to the LRR domain of IpaH1.4, and the interaction between these two proteins is in an equilibrium with an intermediate exchange rate. Further quantitative isothermal titration calorimetry (ITC) analysis uncovered that IpaH1.4 binds to HOIP RING1 with a binding stoichiometry of 1:1 (N value is about 1.0) and a dissociation constant (KD) value of ∼1.0 μM (Fig. 1F). Finally, using analytical ultracentrifugation-based assays, we elucidated that HOIP RING1 and IpaH1.4 LRR both are monomers in solution and can associate with each other to form a stable 1:1 stoichiometric complex (Fig. 1G), which is in line with our ITC data. Taken together, these biochemical results clearly demonstrate that the recognition of HOIP RING1 by IpaH1.4 LRR is responsible for the specific interaction between HOIP and IpaH1.4.
The Crystal Structure of the IpaH1.4 LRR/HOIP RING1 Complex.
To further gain mechanistic insights into the recognition of HOIP RING1 by IpaH1.4, we solved the atomic structures of the apo form IpaH1.4 LRR and IpaH1.4 LRR in complex with HOIP RING1 using X-ray crystallographic methods (SI Appendix, Table S1). The apo form structure of IpaH1.4 LRR reveals that IpaH1.4 LRR has a canonical horseshoe shape, which is composed of nine units of LRR capped by two N-terminal α-helices (α1 and α2) and a C-terminal α4-helix together with a short β10-strand that directly augments the β-sheet of LRR in a parallel manner (Fig. 2A). Further structural analyses revealed that IpaH1.4 LRR contains a positively charged groove located at its potential substrate-binding concave side (Fig. 2B). The overall architecture of IpaH1.4 LRR is highly similar to that of the LRR domain of IpaH3 from S. flexneri (SI Appendix, Fig. S4A). As expected, in the IpaH1.4 LRR/HOIP RING1 complex, IpaH1.4 LRR binds to HOIP RING1 through its concave side (Fig. 2C). No significant structural changes in the IpaH1.4 LRR domain can be observed upon its binding to HOIP (SI Appendix, Fig. S4B). Concurrently, HOIP RING1 is composed of two short N-terminal antiparallel β-strands together with three α-helices and forms a unique architecture coordinating two Zn2+ ions (Fig. 2C). In the complex structure, HOIP RING1 packs extensively with the solvent-exposed and highly positively charged groove of IpaH1.4 LRR mainly through its β-sheet and two Zn2+-binding regions, burying a total surface area of ∼614 Å2 (Fig. 2 C and D). Detailed structural analysis of the binding interface in the IpaH1.4 LRR/HOIP RING1 complex revealed that the specific interaction between IpaH1.4 LRR and HOIP RING1 is mainly mediated by polar interactions (Fig. 2E). Particularly, the positively charged side chains of R99 and K100 in IpaH1.4 LRR form two salt bridges with the negatively charged side chain of D724 in HOIP RING1 (Fig. 2E). In parallel, the side chains of E97 and D140 in IpaH1.4 LRR form two hydrogen bonds with the side chains of W704 and Q728 in HOIP RING1 (Fig. 2E). Meanwhile, the side chains of Q119, R157, and N240 in IpaH1.4 LRR directly couple with the backbone carbonyl groups of C699, C702, G703, and C747 in HOIP RING1 to form five specific hydrogen bonds (Fig. 2E). In addition, the IpaH1.4 LRR/HOIP RING1 complex is further stabilized by a cation–π interaction between the positively charged guanidyl moiety of IpaH1.4 R99 and the aromatic side chain of HOIP W704 (Fig. 2E). Notably, the key interface residues in IpaH1.4 for interacting with HOIP RING1 also can be found in IpaH2.5 but are quite divergent in the equivalent positions of other IpaH family proteins, such as IpaH3, IpaH7.8, and IpaH9.8 (SI Appendix, Fig. S1B), suggesting that HOIP is likely a specific target for IpaH1.4/2.5. Importantly, in accordance with their important structural roles, all the key interface residues of HOIP RING1 involved in the interaction with IpaH1.4 LRR are highly conserved across different eukaryotic species (SI Appendix, Fig. S5). Using site-directed mutagenesis together with comigration and ITC-based assays (SI Appendix, Figs. S6–S8), we further verified the interaction between IpaH1.4 LRR and HOIP RING1 observed in the complex structure. In agreement with our structural data, point mutations of key interface residues, such as the W704A, D724A, and Q728A mutations in HOIP RING1 or the R99A, K100A, Q119A, R157A, and N240A mutations in IpaH1.4 LRR, all largely attenuate or essentially disrupt the specific interaction between HOIP RING1 and IpaH1.4 LRR (Fig. 2F and SI Appendix, Figs. S6–S8).
Fig. 2.
Structural analyses of the apo form IpaH1.4 LRR domain and the HOIP RING1/IpaH1.4 LRR complex. (A) Ribbon diagram showing the overall structure of the IpaH1.4 LRR domain in the apo form. (B) Surface charge potential representation (contoured at ±5 kT/eV; blue/red) of the apo form IpaH1.4 LRR domain. (C) Ribbon diagram showing the overall structure of the HOIP RING1/IpaH1.4 LRR complex with a 1:1 binding stoichiometry. In this drawing, the two coordinated Zn2+ ions of HOIP RING1 are further indicated in the ball model. (D) The combined surface charge potential representation (contoured at ±5 kT/eV; blue/red) and the ribbon–stick–ball model showing the charge–charge interactions between HOIP RING1 and IpaH1.4 LRR in the complex structure. (E) The ribbon–stick–ball model showing the detailed interface between HOIP RING1 and IpaH1.4 LRR in the complex structure. In this drawing, the relevant side chains as well as backbone groups of the key binding interface residues are shown in the stick–ball mode, and the related hydrogen bonds and salt bridges involved in HOIP RING1/IpaH1.4 LRR binding are shown as dotted lines. (F) Summary of the ITC-measured binding affinities and the analytical gel filtration chromatography–based comigration results between full-length IpaH1.4 and the HOIP RING1 domain or their mutants.
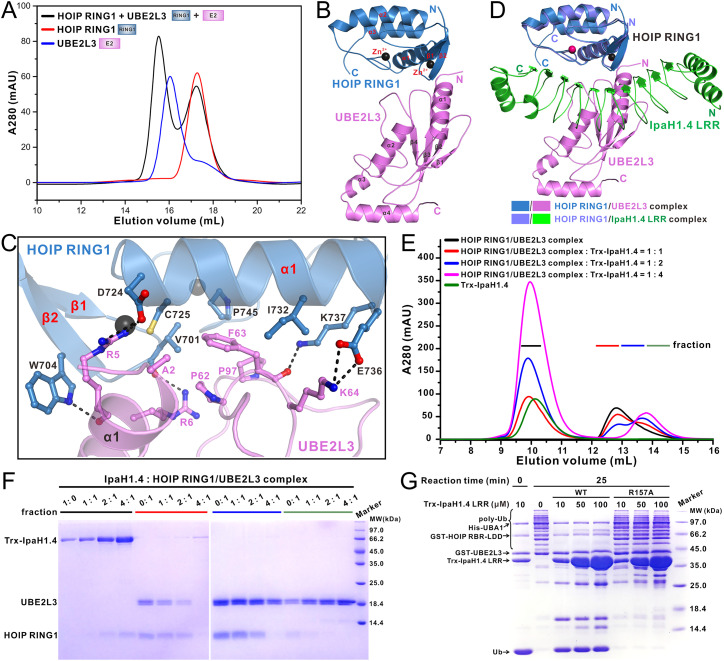
The Structure of the UBE2L3/HOIP RING1 Complex.
As an RBR-type E3 protein, HOIP relies on its RING1 domain to specifically recognize E2 proteins for catalyzing the linear Ub chain assembly. Previous studies have systematically characterized the efficiency of different E2 proteins in HOIP-catalyzed linear ubiquitination (10, 26), and it is well demonstrated that UBE2L3 is the cognate E2 species for HOIP with the highest catalytic efficiency (26). Indeed, our analytical gel filtration chromatography–based assays confirmed that UBE2L3 can effectively bind to the HOIP RING1 domain (Fig. 3A). Given that both UBE2L3 and IpaH1.4 can directly bind to HOIP RING1, it is intriguing to know what the relationship is between IpaH1.4 and UBE2L3 in binding to HOIP. Therefore, we solved the crystal structure of the UBE2L3/HOIP RING1 complex (SI Appendix, Table S2). In the complex structure, UBE2L3 features an architecture assembled by a four-stranded antiparallel β-sheet packing with four α-helices (Fig. 3B) and specifically binds to HOIP RING1 with an overall binding mode like that of the UBE2D2/HOIP RING1 interaction in the crystal structure of HOIP RBR-LDD in complex with UBE2D2∼Ub conjugate and monoubiquitin (Protein Data Bank [PDB] ID: 5EDV) (SI Appendix, Fig. S9A). Further detailed structural analysis revealed that the binding interface between UBE2L3 and HOIP RING1 is mainly formed by residues located in the N-terminal part of α1 as well as the β3–β4 and β4–α2 connecting loops of UBE2L3 and accommodates residues from the β-sheet and α1 regions of HOIP RING1 through both hydrophobic and polar interactions (Fig. 3 B and C). In particular, the hydrophobic side chains of UBE2L3 A2, P62, F63, and P97 residues patch against a hydrophobic pocket formed by the V701, C725, I732, and P745 residues of HOIP RING1 (Fig. 3C and SI Appendix, Fig. S9B). Meanwhile, the hydrophobic portion of the side chain of UBE2L3 R5 sits on the aromatic ring of W704 from HOIP RING1 (Fig. 3C and SI Appendix, Fig. S9B). Furthermore, the backbone carboxyl groups of R5 and F63 together with the polar side chain group of R6 from UBE2L3 interact with the W704, V701, and K737 residues of HOIP RING1 to form three highly specific hydrogen bonds (Fig. 3C). In addition, an Arg–Asp pair (Arg5UBE2L3–Asp724HOIP) and a Lys–Glu pair (Lys64UBE2L3–Glu736HOIP) of salt bridges further strengthen the UBE2L3 and HOIP interaction (Fig. 3C).
Fig. 3.
IpaH1.4 can inhibit the E3 catalytic activity of HOIP through blocking its E2 binding. (A) Analytical gel filtration chromatography analysis of the interaction between the HOIP RING1 domain and UBE2L3. (B) Ribbon diagram showing the overall structure of the HOIP RING1/UBE2L3 complex. (C) The ribbon–stick–ball representation showing the detailed binding interface in the HOIP RING1/UBE2L3 complex structure. In this drawing, the side chains as well as relevant backbone groups of the key binding interface residues are shown in the stick–ball mode, and the hydrogen bonds involved in HOIP RING1/UBE2L3 binding are shown as dotted lines. (D) Ribbon representations showing the structural comparison of the HOIP RING1/IpaH1.4 LRR complex (slate/green) with the HOIP RING1/UBE2L3 complex (blue/magenta). In this drawing, these two structures are overlaid by aligning the HOIP RING1 domain in these two complex structures. (E) Analytical gel filtration chromatography analyses of the purified HOIP RING1/UBE2L3 complex titrated with increasing molar ratios of full-length IpaH1.4 proteins. (F) SDS-PAGE combined with Coomassie blue staining analyses showing the protein components of the corresponding fractions collected from the analytic gel filtration chromatography experiments of the HOIP RING1/UBE2L3 complex titrated with different molar ratios of full-length IpaH1.4 in E. (G) In vitro linear Ub chain assembly assays showing that the catalytic activity of HOIP (RBR-LDD region) can be efficiently blocked by the wild-type IpaH1.4 LRR, while the R157A mutant of IpaH1.4 LRR, which cannot interact with HOIP RING1, loses the ability to inhibit the activity of HOIP.
IpaH1.4 LRR Can Inhibit the E3 Activity of HOIP through Blocking its E2 Binding.
Structural comparison analysis showed that some key interfacial residues of HOIP RING1, such as W704 and D724, are both involved in the interactions with IpaH1.4 LRR and UBE2L3 (Figs. 2E and 3C and SI Appendix, Fig. S5), and the UBE2L3-binding and IpaH1.4-binding sites on HOIP RING1 are heavily overlapped (Fig. 3D), suggesting a direct competition between E2 UBE2L3 and IpaH1.4 in binding to HOIP. We sought to measure the binding affinity between UBE2L3 and HOIP RING1 using the ITC method. However, the enthalpy change of the ITC titration assay of UBE2L3 with HOIP RING1 is too small to calculate a reliable KD value for the UBE2L3/HOIP RING1 interaction (SI Appendix, Fig. S10 A and B). Fortunately, as an alternative, we successfully measured the binding affinities of the HOIP UBA-RBR-LDD/HOIL-1L UBL complex with UBE2L3 and IpaH1.4 LRR (SI Appendix, Fig. S10 C and D). The ITC results revealed that IpaH1.4 LRR has a higher affinity (KD = 2.3 ± 0.4 μM) than UBE2L3 (KD = 5.8 ± 0.6 μM) for binding to HOIP RING1 (SI Appendix, Fig. S10 C and D). Consistent with our structural and ITC data, the analytical gel filtration chromatography coupled with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) assays showed that IpaH1.4 can readily compete with UBE2L3 for interacting with HOIP RING1 (Fig. 3 E and F). Since the recognition of E2 by HOIP RING1 is essential for the E3 activity of HOIP, we wondered whether IpaH1.4 LRR alone could inhibit the catalytic activity of HOIP for assembling linear Ub chains by binding to the HOIP RING1 domain. As expected, in our reconstituted in vitro linear Ub chain assembly assay, the isolated IpaH1.4 LRR domain can efficiently block the enzymatic activity of the HOIP RBR-LDD catalytic fragment to assemble linear Ub chains (Fig. 3G). In contrast, the R157A mutant of IpaH1.4 LRR, which loses the ability to interact with HOIP RING1 (Fig. 2F), has negligible inhibitory effects on the assembly of linear Ub chains mediated by HOIP RBR-LDD (Fig. 3G). Collectively, all these biochemical observations together with our structural analyses clearly demonstrated that IpaH1.4 can efficiently suppress the E3 activity of HOIP by disturbing its E2 binding in vitro.
Biochemical Characterization of the Interaction between IpaH1.4 and HOIL-1L.
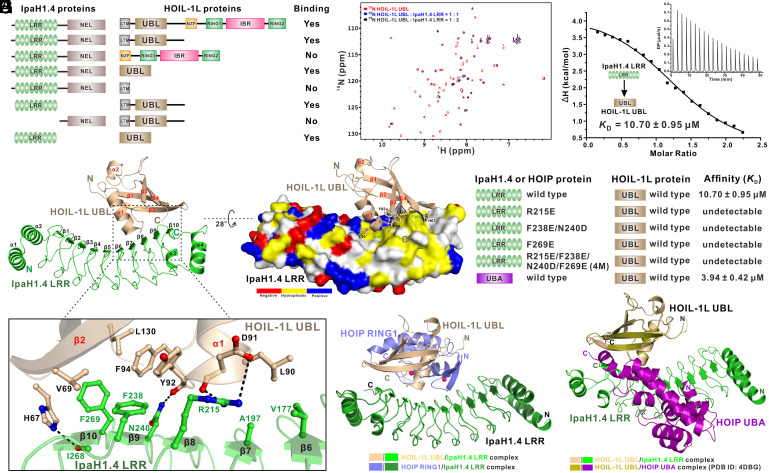
A previous study also showed that IpaH1.4 can directly target the HOIL-1L subunit of LUBAC (37). Using comigration assays with purified recombinant proteins from Escherichia coli, we first confirmed that IpaH1.4 can directly but weakly bind to HOIL-1L (Fig. 4A and SI Appendix, Fig. S11A). Subsequently, we carefully mapped the binding regions between HOIL-1L and IpaH1.4 and uncovered that the IpaH1.4/HOIL-1L interaction is mainly mediated by the specific binding between IpaH1.4 LRR and HOIL-1L UBL (Fig. 4A and SI Appendix, Fig. S11). Further NMR titration analyses using 15N-labeled HOIL-1L UBL titrated with unlabeled IpaH1.4 LRR confirmed that HOIL-1L UBL can directly interact with IpaH1.4 LRR (Fig. 4B). Intriguingly, an ITC-based quantitative analysis revealed that the interaction between IpaH1.4 LRR and HOIL-1L UBL has a KD value of ∼10.7 μM (Fig. 4C), which is much weaker than that of the IpaH1.4 LRR and HOIP RING1 interaction (Fig. 1F).
Fig. 4.
Biochemical and structural characterization of the interaction between the HOIL-1L UBL domain and IpaH1.4 LRR regions. (A) The summarized mapping results of the interacting regions between IpaH1.4 and HOIL-1L by analytical gel filtration chromatography–based assays. (B) Superposition plots of the 1H-15N HSQC spectra of the 15N-labeled HOIL-1L UBL domain titrated with increasing molar ratios of unlabeled IpaH1.4 LRR proteins. (C) ITC-based measurement of the binding affinity of IpaH1.4 LRR with HOIL-1L UBL. The KD error is the fitted error obtained from the data analysis software when using the one-site binding model to fit the ITC data. (D) Ribbon diagram showing the overall structure of the HOIL-1L UBL/IpaH1.4 LRR complex. (E) The combined surface representation and the ribbon–stick model showing the hydrophobic binding surface between IpaH1.4 LRR and HOIL-1L UBL. In this drawing, HOIL-1L UBL is displayed in the ribbon–stick model, and IpaH1.4 LRR is shown in surface representation colored by amino acid types. Specifically, the hydrophobic amino acid residues in the surface model of IpaH1.4 LRR are drawn in yellow, the positively charged residues are in blue, the negatively charged residues are in red, and the uncharged polar residues are in gray. (F) The ribbon–stick model showing the binding interface in the HOIL-1L UBL/IpaH1.4 LRR complex structure. The relevant hydrogen bonds and salt bridges involved in the HOIL-1L UBL/IpaH1.4 LRR interaction are shown as dotted lines. (G) The measured binding affinities between HOIL-1L UBL and IpaH1.4 LRR or their mutants by ITC-based assays. (H) Ribbon representations showing the structural comparisons of the HOIP RING1/IpaH1.4 LRR complex (slate/forest) with the HOIL-1L UBL/IpaH1.4 LRR complex (wheat/green). In this drawing, the two structures are overlaid by aligning IpaH1.4 LRR in these two complex structures. (I) Ribbon representations showing the structural comparisons of the HOIL-1L UBL/IpaH1.4 LRR complex (wheat/green) with the HOIL-1L UBL/HOIP UBA complex (olive/purple) (PDB ID: 4DBG). Here, the two structures are overlaid by aligning HOIL-1L UBL in these two complex structures.
Structure of the IpaH1.4 LRR/HOIL-1L UBL Complex.
To reveal the mechanistic basis for the recognition of HOIL-1L by IpaH1.4, we determined the crystal structure of the HOIL-1L UBL/IpaH1.4 LRR complex (SI Appendix, Table S2). In the final complex structure model, an asymmetric unit contains two HOIL-1L UBL and two IpaH1.4 LRR molecules (SI Appendix, Fig. S12A), and two distinct contacting sites (site 1 and site 2) between IpaH1.4 LRR and HOIL-1L UBL can be identified (SI Appendix, Fig. S12B). Particularly, the second interface (site 2) is mainly maintained by polar interactions, including salt bridges and hydrogen bonds (SI Appendix, Fig. S12C). However, point mutations of key interface residues of IpaH1.4 involved in site 2 do not affect the specific interaction between IpaH1.4 and HOIL-1L in solution (SI Appendix, Fig. S12 D–H). Therefore, the site 2 interface is merely induced by crystal packing, and only the site 1 interface is a real binding interface between IpaH1.4 and HOIL-1L in solution.
Further structural analyses of the interaction between IpaH1.4 LRR and HOIL-1L UBL in site 1 showed that HOIL-1L UBL adopts a Ub-like fold and specifically binds to the concave side of IpaH1.4 LRR via its α1 and β2 regions (Fig. 4D). Unsurprisingly, the binding of HOIL-1L UBL to IpaH1.4 LRR does not induce large conformational changes in IpaH1.4 LRR (SI Appendix, Fig. S13A), and the overall structure of HOIL-1L UBL in the IpaH1.4 LRR/HOIL-1L UBL complex is highly similar to that of HOIL-1L UBL in complex with HOIP UBA (SI Appendix, Fig. S13B). Detailed structural analyses of the binding interface between IpaH1.4 LRR and HOIL-1L UBL revealed that the side chains of V69, Y92, F94, and L130 from HOIL-1L UBL make hydrophobic contacts with the hydrophobic side chains of the F238 and F269 residues of IpaH1.4 LRR (Fig. 4 E and F). Concurrently, the hydrophobic side chain of HOIL-1L L90 packs against a hydrophobic pitch formed by the side chains of V177 and A197 residues of IpaH1.4 LRR (Fig. 4 E and F). Furthermore, the imidazole ring of H67 and the backbone carbonyl group of the Y92 residue of HOIL-1L respectively interact with the I268 and N240 residues of IpaH1.4 LRR to form two hydrogen bonds, and the negatively charged HOIL-1L D91 forms specific charge–charge and hydrogen-bonding interactions with the positively charged R215 residue of IpaH1.4 LRR (Fig. 4F). In line with their critical structural roles, all these key binding interface residues of HOIL-1L are highly conserved during evolution (SI Appendix, Fig. S14). Notably, the key interfacial residues of IpaH1.4 LRR for interacting with HOIL-1L UBL can also be found in IpaH2.5 but are missing in other S. flexneri IpaH family members (SI Appendix, Fig. S1B), implying that HOIL-1L is also likely a specific target of IpaH1.4/2.5. Using comigration and ITC analyses, we further validated the specific interactions between IpaH1.4 LRR and HOIL-1L UBL observed in the complex structure. Consistent with our structural results, mutations of key interfacial residues from IpaH1.4, including the R215E, F238E/N240D, F269E, and R215E/F238E/N240D/F269E (hereafter referred to as 4M) mutations of IpaH1.4 LRR, all essentially abolish the specific interaction between IpaH1.4 and HOIL-1L in solution (SI Appendix, Figs. S15 and S16 and Fig. 4G).
Relationship between IpaH1.4, HOIP, and HOIL-1L in Binding to Each Other.
Based on our study, both HOIL-1L UBL and HOIP RING1 can directly interact with IpaH1.4 LRR (Figs. 2E and 4F). However, further structural comparison analysis showed that HOIL-1L UBL and HOIP RING1 are unable to simultaneously bind IpaH1.4 LRR due to potential steric exclusion (Fig. 4H). Considering that the binding affinity between IpaH1.4 LRR and HOIP RING1 is roughly tenfold stronger than that of the IpaH1.4 LRR/HOIL-1L UBL interaction (Figs. 1F and 4C), IpaH1.4 should preferentially bind to HOIP rather than HOIL-1L when targeting LUBAC. In addition to interacting with IpaH1.4 LRR, HOIL-1L UBL can also directly bind to HOIP UBA through its α1 and β2 regions (SI Appendix, Fig. S17A), and the IpaH1.4-binding and HOIP-binding sites on HOIL-1L UBL are highly overlapped (Fig. 4I and SI Appendix, Fig. S14). Therefore, IpaH1.4 and HOIP should be competitive in binding to HOIL-1L. Interestingly, ITC-based analyses revealed that IpaH1.4 LRR has a weaker binding affinity (KD = 10.7 ± 0.1 μM) than HOIP UBA (KD = 3.9 ± 0.4 μM) for interacting with HOIL-1L UBL (Fig. 4 C and G and SI Appendix, Fig. S17C). Consistent with our structural and ITC results, further analytical gel filtration chromatography coupled with SDS-PAGE assays showed that IpaH1.4 is unable to form a ternary complex with the purified HOIP UBA/HOIL-1L UBL complex and is ineffective in destabilizing the HOIP UBA/HOIL-1L UBL interaction (SI Appendix, Fig. S17 D and E).
In Vitro Ubiquitination of HOIP and HOIL-1L by IpaH1.4 Relies on the Specific Interactions of IpaH1.4 LRR with HOIP RING1 and HOIL-1L UBL.
Our biochemical and structural data clearly demonstrated that IpaH1.4 can directly recognize HOIP and HOIL-1L through its LRR domain (Figs. 2 and 4). It is intriguing to know whether HOIP and HOIL-1L in the context of LUBAC can be recognized and ubiquitinated by IpaH1.4. Using a reconstituted in vitro ubiquitination assay with purified LUBAC core fragments together with relevant E1, E2, and Ub proteins, we revealed that IpaH1.4 can readily and preferentially catalyze the poly-ubiquitination of HOIP and very weakly ubiquitinates HOIL-1L (Fig. 5A and SI Appendix, Fig. S18A). In contrast, the SHARPIN component in the mini-LUBAC complex cannot be ubiquitinated by IpaH1.4 (Fig. 5A and SI Appendix, Fig. S18A), suggesting that IpaH1.4 only ubiquitinates directly bound substrate but cannot modify indirectly associated proteins, which might be out of the accessibility of the IpaH1.4 catalytic site. Importantly, the R157A mutation of IpaH1.4 that specifically disrupts the IpaH1.4/HOIP RING1 interaction but does not affect the IpaH1.4/HOIL-1L UBL binding (Fig. 2F and SI Appendix, Fig. S19A) essentially abolishes the poly-ubiquitination of HOIP mediated by IpaH1.4 (Fig. 5B). Interestingly, likely due to the elimination of the competitive binding from HOIP RING1, HOIL-1L can be more easily modified by the IpaH1.4 R157A mutant than the wild-type IpaH1.4 (Fig. 5 A and B). Furthermore, consistent with our aforementioned biochemical and structural results (Fig. 4), IpaH1.4 mutations that specifically disrupt the interaction between IpaH1.4 LRR and HOIL-1L UBL, such as the R215E and F238E/N240D mutations of IpaH1.4 (Fig. 4G), completely eliminate the ubiquitination of HOIL-1L conducted by IpaH1.4 (Fig. 5C).
Fig. 5.
The interaction of IpaH1.4 LRR with HOIP RING1 or HOIL-1L UBL is essential for the in vitro ubiquitination of HOIP or HOIL-1L as well as the suppression of NF-κB activation in cells. (A and B) In vitro ubiquitination assays showing the abilities of the wild-type IpaH1.4 (A) and the IpaH1.4 R157A mutant (B) to assemble K48-type Ub chains on the three subunits of LUBAC. To eliminate the potential ubiquitination events induced by HOIP or HOIL-1L, the catalytic-dead HOIP (480 to 1,072) C885A and HOIL-1L C460A mutants are used in these assays. Notably, the IpaH1.4 R157A mutant, which is unable to interact with HOIP RING1, almost loses the ability to assemble K48-type Ub chains on HOIP; IB, immunoblot; HA, hemagglutinin; WT, wild-type. (C) In vitro ubiquitination assays showing the abilities of the wild-type IpaH1.4 (Left), the IpaH1.4 R215E mutant (Middle), and the IpaH1.4 F238E/N240D mutant (Right) to mediate the poly-ubiquitination of HOIL-1L. Notably, the R215E and F238E/N240D mutants of IpaH1.4, both of which are unable to interact with HOIL-1L UBL, cannot mediate the poly-ubiquitination of HOIL-1L. (D) NF-κB reporter dual-luciferase assay using overexpressed LUBAC together with the wild-type IpaH1.4 or different IpaH1.4 variants. All luciferase activities are normalized to that of the control cells. Error bars denote the SD between three replicates. An unpaired Student's t test analysis was used to define a statistically significant difference, and the stars indicate the significant differences between the indicated bars (**, P ≤ 0.01; ***, P ≤ 0.001). (E) Representative fluorescent microscopy images of cultured HeLa cells infected with the wild-type S. flexneri M90T strain, the ipaH1.4/ipaH2.5 double-knockout S. flexneri (ΔipaH1.4/ΔipaH2.5) strain, or a S. flexneri (ΔipaH1.4/ΔipaH2.5) strain recomplemented with the wild-type IpaH1.4 or different IpaH1.4 mutant (R175A, C368A, R175A/C368A, 4M, and 4M/C368A) and stained with a specific antibody to the p65 subunit of NF-κB (red) and DAPI (blue) to show bacteria and nuclei of cells. For comparison, DAPI and p65 fluorescence intensity values in a cross-section of a selected cell (indicated with a white line) are shown in graphs on the Right. (F) Statistical results related to the nuclear translocation of p65 in HeLa cells infected with different S. flexneri strains in E. Bar graphs represent results from three independent experiments presented as the mean ± SD of >70 analyzed cells. An unpaired Student's t test analysis was used to define a statistically significant difference, and the stars indicate the significant differences between the indicated bars (**, P ≤ 0.01; ***, P ≤ 0.001); ns, not significant.
Recognition of HOIP and HOIL-1L by IpaH1.4 Is Required for the IpaH1.4-Mediated Suppression of NF-κB Activation in Cells.
Previous studies demonstrated that the overexpression of LUBAC in HEK293T cells can induce a strong activation of the NF-κB pathway that depends on the linear Ub assembly activity of LUBAC (15–17, 35), and IpaH1.4 can directly target LUBAC for proteasomal degradation, especially the catalytic HOIP subunit (37). To further evaluate the recognition of HOIP and HOIL-1L by IpaH1.4 on the NF-κB activation induced by LUBAC in cells, we established a reliable NF-κB reporter dual-luciferase assay to measure the activities of LUBAC in the presence of wild-type IpaH1.4 or different IpaH1.4 variants. As expected, overexpression of LUBAC alone in HEK293T cells can promote the NF-κB reporter luciferase level over 200-fold compared with the control, while the coexpression of LUBAC with the wild-type IpaH1.4 can effectively abolish the activation effect induced by LUBAC (Fig. 5D). Interestingly, the catalytic-dead C368A mutant of IpaH1.4 does not completely lose the inhibitory activity toward LUBAC but still retains about half of the inhibitory activity of the wild-type IpaH1.4 (Fig. 5D), suggesting that even without its E3 ubiquitination activity, the sole interaction of IpaH1.4 with LUBAC should contribute to the IpaH1.4-mediated suppression of NF-κB activation induced by LUBAC. Meanwhile, the IpaH1.4 R157A mutant and 4M mutant, which lose the HOIP-binding ability and the HOIL-1L-binding ability, respectively (Figs. 2F and 4G and SI Appendix, Fig. S19 A and B), both retain similar inhibitory activity as the IpaH1.4 C368A mutant (Fig. 5D), indicating that the recognition of HOIP and HOIL-1L by IpaH1.4 is required for the effective IpaH1.4-mediated inhibition of NF-κB activation induced by LUBAC. In line with our aforementioned biochemical results (Fig. 3 E–G), the IpaH1.4 4M/C368A mutant, which loses ubiquitination activity as well as HOIL-1L binding ability but retains the ability to interact with the HOIP RING1 for interfering with the E2 binding of HOIP, still has weak inhibitory effects on NF-κB activation induced by LUBAC (Fig. 5D). These observations demonstrated that the abilities for interacting with HOIP and HOIL-1L as well as the E3 ubiquitination activity of IpaH1.4 are required for the IpaH1.4-mediated suppression of NF-κB activation induced by overexpressed LUBAC in cells.
To further validate our structural conclusions under more physiological conditions, we generated an ipaH1.4/ipaH2.5 double-knockout (ΔipaH1.4/ΔipaH2.5) S. flexneri strain and recomplemented it with a wild-type ipaH1.4 gene or different ipaH1.4 mutant genes (R175A, C368A, R175A/C368A, 4M, and 4M/C368A) in the original ipaH1.4 locus by the flippase-mediated recombination method. Notably, all the wild-type ipaH1.4 and different ipaH1.4 mutant genes that were used to recomplement the ipaH1.4/ipaH2.5 double-knockout S. flexneri strain are C-terminally fused with a His tag gene sequence, and the proper protein expression levels of these ipaH1.4 variants in mutant strains were further confirmed by Western blotting using a specific anti-His antibody (SI Appendix, Fig. S20). Then, we used these engineered S. flexneri strains and the wild-type S. flexneri M90T strain to infect cultured HeLa cells. Subsequently, the nuclear localization of p65 in infected HeLa cells was quantified by immunofluorescence. The results showed that the wild-type S. flexneri M90T strain can effectively block p65 nuclear translocation in ∼70% of cells with bacterial infection, while the ipaH1.4/ipaH2.5 double-knockout S. flexneri (ΔipaH1.4/ΔipaH2.5) strain has a poor ability to block the nuclear translocation of p65 (Fig. 5 E and F). In line with a previous study (37), the recomplementation of the double-knockout strain with the wild-type ipaH1.4 confers the bacteria with a comparable ability as that of the wild-type strain to inhibit p65 nuclear translocation in infected cells (Fig. 5 E and F). In contrast, the S. flexneri strain recomplemented with the ipaH1.4 R157A mutant or the ipaH1.4 4M mutant is unable to inhibit p65 nuclear translocation (Fig. 5 E and F), suggesting that the binding abilities of IpaH1.4 to HOIP and HOIL-1L are critical for S. flexneri to suppress the nuclear translocation of p65. Interestingly, the strain recomplemented with the ipaH1.4 C368A (catalytic-dead) mutant can still inhibit p65 nuclear translocation (Fig. 5 E and F). As expected, the S. flexneri strain recomplemented with the ipaH1.4 R157A/C368A or 4M/C368A mutant has little inhibition on the nuclear translocation of p65, similar to that of the double-knockout strain (ΔipaH1.4/ΔipaH2.5) (Fig. 5 E and F). In all, these data clearly indicated that S. flexneri strains with mutants of IpaH1.4 defective in binding to HOIP RING1 or HOIL-1L UBL are attenuated in suppression of host immunity.
Discussion
The secretion of special E3 Ub ligases to target key proteins of infected host cells for proteasomal degradation is an efficient and smart strategy adopted by some deleterious bacteria to subvert the host antibacterial immune response for facilitating their survival and replication in host cells. For successful proliferation in infected human cells, S. flexneri uses two highly similar E3 effector proteins, IpaH1.4 and IpaH2.5, to subvert LUBAC through proteasomal degradation (37). In this study, we uncovered the molecular mechanism as well as the related downstream consequences for the recognition between IpaH1.4 and LUBAC through systematic biochemical and structural characterization. Particularly, we have elucidated that IpaH1.4 directly binds to the RING1 domain of HOIP and the UBL domain of HOIL-1L by different but partially overlapped surfaces of its N-terminal LRR domain (Figs. 2 and 4). Importantly, the interaction between IpaH1.4 and HOIP or HOIL-1L is indispensable for the poly-ubiquitination of HOIP or HOIL-1L by IpaH1.4 in vitro (Fig. 5). Interestingly, our structural and biochemical assays also revealed that the binding of IpaH1.4 to HOIP masks the E2-interacting surface in the RING1 domain of HOIP (Fig. 3), thus inhibiting the linear Ub chain assembly activity of LUBAC by blocking the E2 binding to HOIP in addition to inducing Ub–proteasome-dependent HOIP degradation. Notably, blocking the RING domain of a targeting E3 ligase is a common strategy adopted by many other E3 ligase inhibitors, such as glomulin (48), Emi1 (49), and Salmonella effector SopA (50). Intriguingly, the determined IpaH1.4/HOIL-1L complex structure uncovered that IpaH1.4 binds to a hydrophobic surface in the HOIL-1L UBL domain (Fig. 4 D–F), which also mediates the HOIL-1L/HOIP interaction (SI Appendix, Fig. S17 A and B). Therefore, in addition to mediating the poly-ubiquitination of HOIL-1L, IpaH1.4 might compete with HOIP for binding to HOIL-1L, thereby destabilizing the HOIL-1L/HOIP interaction in the LUBAC that is crucial for the stability of HOIP in cells (15, 16, 32). However, based on our competition binding assay and quantitative ITC results (SI Appendix, Fig. S17 C–E and Fig. 4C), under a condition with limited IpaH1.4 molecules, IpaH1.4 is unlikely to effectively impair the integrity of LUBAC by binding to HOIL-1L UBL. Therefore, when targeting LUBAC, IpaH1.4 should preferentially attack HOIP rather than HOIL-1L. However, when the HOIP level drops by proteasomal degradation as infection time progresses, it is likely that the exposed HOIL-1L becomes a follow-up substrate for IpaH1.4. Furthermore, considering that HOIL-1L alone has specific cellular functions (51), the recognition of HOIL-1L by IpaH1.4 might also affect the function of HOIL-1L in a LUBAC-independent scenario. Therefore, the crystal structures of the IpaH1.4/HOIP and IpaH1.4/HOIL-1L complexes solved in this study may provide potential drug targets for the future development of innovative therapeutic strategies against S. flexneri infection.
IpaH1.4 and IpaH2.5 share almost identical amino acid sequences in their LRR and NEL domains (SI Appendix, Fig. S1A); therefore, the binding mechanism of IpaH1.4 with LUBAC uncovered in this study should be also applicable to IpaH2.5. Indeed, in the NF-κB reporter dual-luciferase assays, similar phenomena were observed for IpaH1.4 and IpaH2.5 (Fig. 5D and SI Appendix, Fig. S18B). Interestingly, besides IpaH1.4/2.5, S. flexneri also secretes other E3 ligases of the NEL family, such as IpaH3, IpaH4.5, IpaH7.8, and IpaH9.8, which share almost identical amino sequences in their catalytic NEL domains but have distinct N-terminal substrate-binding LRR domains (SI Appendix, Fig. S21) (52). Sequence alignment analyses of these homologs uncovered that the critical residues for HOIP or HOIL-1L interaction are conserved in IpaH1.4 and IpaH2.5 but are absent in other IpaH members (SI Appendix, Fig. S1B). For example, the key HOIP-binding R157 residue of IpaH1.4/2.5 is replaced by an Asp, Ser, or Phe residue in other IpaH members; the hydrophobic HOIL-1L-binding F238 residue is substituted by a polar Ser, Thr, Asp, or Arg residue (SI Appendix, Fig. S1B). Therefore, only IpaH1.4/2.5 can specifically target HOIP and HOIL-1L, in line with a previous study (37). Apparently, the sequence diversities of the LRR domains confer NEL family proteins with different spectra of substrate specificities. Through the modular combination of a highly variable N-terminal substrate-binding domain and a highly conserved C-terminal NEL domain for catalyzing K48-conjugated Ub chain formation, the NEL family proteins evolve to become a wide-spread type of effector protein delivered by many bacterial pathogens, including Shigella, Salmonella, Yersinia, and Pseudomonas, to modulate essential host cell processes by targeting relevant host proteins for proteasomal degradation. Intriguingly, we elucidated that in addition to mediating K48-type ubiquitination as well as the subsequent proteasomal degradation of the substrate, IpaH1.4/2.5 can disturb the function of LUBAC by solely interacting with HOIP and/or HOIL-1L through its substrate-binding LRR domain. The ability of the IpaH1.4 LRR domain to directly inhibit the function of host LUBAC implies that the LRR domains of IpaH family proteins may have evolved to disrupt host signaling pathways before they fuse with the NEL domain. Apparently, the later addition of an LRR domain with a catalytic NEL domain that has an E3 ligase activity can further augment the inhibitory function on host client proteins of the LRR domain through proteasomal degradation.
Notably, our observations on the suppression of NF-κB signaling by IpaH1.4 mutants in the S. flexneri infection assay are somewhat different from that of the IpaH1.4 overexpression luciferase assay (Fig. 5 D–F). In the luciferase assay, the single disruption of IpaH1.4’s binding to either HOIP or HOIL-1L cannot completely abolish IpaH1.4’s inhibition of NF-κB signaling (Fig. 5D), likely due to the high concentration of overexpressed IpaH1.4 protein. While both assays agree that the catalytic-dead (C368A) IpaH1.4 mutant can still inhibit NF-κB activation, the binding ability to either HOIP or HOIL-1L is crucial for IpaH1.4 to suppress host NF-κB signaling (Fig. 5 D–F). Importantly, the fact that the catalytic-dead IpaH1.4 C368A mutant can still effectively suppress host NF-κB signaling underlines the direct interference on its host client protein of an IpaH E3 ligase through its LRR binding and reminds us that functional disruption of an IpaH E3 ligase by solely mutating its catalytic residue is likely to be insufficient. Thus, functional assays with a catalytic-dead mutant of IpaH E3 ligase might not give strong signals, and the relevant experimental data should be interpreted with caution.
Finally, based on our study together with other groups’ reports (30, 31, 37), we proposed a working model to depict the subversion of host LUBAC by the E3 effector IpaH1.4/2.5 of S. flexneri (SI Appendix, Fig. S22). In this model, HOIP associates with HOIL-1L and SHARPIN through the specific interactions between the HOIP UBA domain and the UBL domains of HOIL-1L and SHARPIN, forming the ternary LUBAC complex that can recognize the E2∼Ub conjugate through the HOIP RING1 domain to mediate linear ubiquitination of relevant substrates for antibacterial cellular processes (SI Appendix, Fig. S22). Upon S. flexneri infection, LUBAC is directly targeted and subverted by the bacterial E3 ligase IpaH1.4/2.5 secreted by S. flexneri. In particular, IpaH1.4/2.5 can specifically and preferentially recognize the RING1 domain of HOIP through its N-terminal LRR domain, thereby inhibiting the E3 activity of HOIP by blocking its E2∼Ub loading (SI Appendix, Fig. S22). Subsequently, IpaH1.4/2.5 subverts LUBAC in a two-step fashion. First, IpaH1.4/2.5 catalyzes the formation of K48-linked poly-Ub chains on HOIP and subsequently leads to the proteasomal degradation of HOIP (SI Appendix, Fig. S22). Second, in the absence of HOIP, HOIL-1L exposes its UBL domain to IpaH1.4/2.5, which further induces the K48-linked poly-ubiquitination of HOIL-1L and promotes its proteasomal degradation, thereby eventually resulting in the subversion of LUBAC.
Materials and Methods
Plasmids and Mutagenesis.
For recombinant protein expression in E. coli, the DNA fragments encoding desired proteins were cloned into a modified pET32M (Novagen) or pGEX-4T-1 (GE Healthcare) vector, which encodes a Trx-6×His or glutathione-S-transferase (GST) tag followed by an HRV 3C protease cutting sequence before the multiple cloning sites. We also constructed a modified pET32M vector in which the Trx-6×His tag is replaced by a maltose-binding protein (MBP)-6×His tag to produce MBP fusion protein. All the point mutations in constructs were introduced by standard PCR protocol. For overexpression in HEK293T cells, the coding sequences for full-length HOIL-1L, HOIP, and SHARPIN were inserted into pFLAG-CMV-1 (Sigma), resulting in a fusion protein with an N-terminal FLAG tag. The IpaH1.4 wild type or mutants were cloned into a modified version of pFLAG-CMV-1, in which a FLAG-mCherry tag was fused to the N terminus of IpaH1.4 or IpaH2.5. All point mutations were introduced by standard PCR methods and verified by DNA sequencing.
Protein Expression and Purification.
All the proteins used in structural studies and biochemical assays were expressed in E. coli BL21(DE3) as N-terminal Trx-6×His, MBP-6×His, or GST fusion proteins. If desired, the tag was cut by homemade HRV 3C protease, which recognizes a cutting site in the fusion protein following the tag. Protein expression in E. coli was induced by 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the optical density at 600 nm reached 0.6∼0.8, and E. coli was further cultured at 16 °C for 16 h. For the ZF-containing proteins, 0.1 mM ZnCl2 was added into culture to improve protein folding before IPTG induction. Trx/MBP-6×His or GST fusion proteins were affinity purified by Ni2+-nitriloacetic acid or glutathione resin (GE Healthcare), respectively, and further polished by size-exclusion column HiLoad Superdex 200 26/60 (GE Healthcare) in buffer A (20 mM Tris-HCl pH 7.5, 100 mM NaCl, and 1 mM dithiothreitol [DTT]). Proteins were concentrated to the desired concentration by ultrafiltration. Uniformly 15N-labeled HOIP RING1 or IpaH1.4 LRR protein was expressed in E. coli BL21(DE3) cells cultured with M9 minimal medium using 15NH4Cl (Cambridge Isotope Laboratories Inc.) as the sole nitrogen source and was purified similar to unlabeled proteins.
Size-Exclusion Chromatography and Comigration Assay.
Individual and mixed protein samples (500 μL) were loaded onto a HiLoad Superdex 200 Increase 10/300 column (GE Healthcare) and eluted in buffer A; absorbance was monitored at 280 nm.
ITC Assay.
Generally, proteins for titration were concentrated to about 500 μM (in syringe) or 50 μM (in cell) in buffer A. Titration was conducted at 25 °C with a PEAQ-ITC machine (Malvern Instrument). The dissociation constant and number of binding sites were deduced by fitting titration curves with a model for one set of binding sites in the PEAQ-ITC analysis software (Malvern Instrument).
Analytic Ultracentrifugation Assay.
Protein for analysis was collected from the peak fraction of a size-exclusion chromatography separation. The absorbance at a wavelength of 280 nm (A280 value) was adjusted to ∼0.5 with buffer A. The same batch of buffer A was used as a reference for absorbance monitoring during ultracentrifugation. Sedimentation velocity experiments were conducted at 142,250 × g with a Beckman XL-I analytical ultracentrifuge operated at 20 °C. The sedimentation velocity data were analyzed in the software SEDFIT (53) and fitted with a continuous sedimentation coefficient distribution c(s) model with parameters such as partial specific volume of protein samples and buffer density calculated in the program SEDNTERP (http://www.rasmb.org/).
NMR Spectroscopy.
The 15N-labeled protein for NMR studies was concentrated to ∼0.2 mM in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 6.5), 50 mM NaCl, and 1 mM DTT. All 15N-HSQC spectra were collected at 25 °C with an 800-MHz spectrometer (Agilent Technologies) equipped with an actively z gradient–shielded triple resonance probe.
Crystal Growth.
Protein in buffer A was concentrated to about 10∼20 mg/mL for crystallization condition screening with commercial buffer kits. Briefly, 1 μL of protein and an equal volume of reservoir buffer were mixed for a crystallization trial in a sitting drop manner. IpaH1.4 LRR was crystallized in a buffer containing 0.1 M sodium citrate (pH 4.0) and 0.8 M (NH4)2SO4. The IpaH1.4 LRR/HOIP RING1 complex crystal grew in a buffer containing 2% (vol/vol) 1, 4-dioxane, 0.1 M Tris-HCl (pH 8.0), and 15% (vol/vol) PEG3350. The UBE2L3/HOIP RING1 complex was crystallized in a buffer containing 0.06 M MgCl2, 0.06 M CaCl2, 0.1 M Tris-bicine (pH 8.1), 12% (vol/vol) MPD, 12% (wt/vol) PEG1000, and 12% (wt/vol) PEG3350. The IpaH1.4 LRR/HOIL-1L UBL complex crystal grew in a buffer containing 0.2 M (NH4)2SO4, 0.1 M Tris-HCl (pH 8.5), and 25% (wt/vol) PEG3350.
X-ray Diffraction and Structure Determination.
Single crystals were equilibrated with mother liquor containing 20% (vol/vol) glycerol as cryoprotectant and flash-frozen in liquid nitrogen. Diffraction data sets were collected at the beamline BL17U1 or BL19U1 of the Shanghai Synchrotron Radiation Facility (SSRF) (54). Data sets were processed with XDS and autoPROC software suite (55, 56). The apo form structure of IpaH1.4 LRR was solved by molecular replacement using the IpaH1880 structure (PDB: 5KH1) as the searching template. The refined IpaH1.4 LRR, HOIP RING1 (PDB: 5EDV), and HOIL-1L UBL (PDB: 4DBG) structures were used as searching templates to solve the IpaH1.4 LRR/HOIP RING complex and the IpaH1.4 LRR/HOIL-1L UBL complex structures. Similarly, the UBE2L3/HOIP RING1 complex structure was solved by molecular replacement using the structure of UBE2L3 from PDB entry 4Q5H. All structure models were manually adjusted in Coot (57) and refined with Phenix suite (58). The final refinement statistics of solved structures in this study are listed in SI Appendix, Tables S1 and S2. All figures of protein structures were prepared with PyMol software (https://www.pymol.org).
In Vitro Ubiquitination Assay.
Recombinant murine UBA1 (E1), human UBE2L3 (E2), and Ub expressed in E. coli cells were used for the assay. The typical reaction system contained 1 μL of E1, 2 μM E2, 100 μM Ub, and 1 μM HOIP or IpaH1.4 in a buffer with 20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM DTT, 10 mM ATP, and 10 mM MgCl2. Reaction mixtures were incubated at 37 °C for the desired time, and aliquots of sample were immediately denatured by mixing with 2× SDS-PAGE sample buffer.
Luciferase Assay.
HEK293T cells were cultured in 12-well plates to a confluency of ∼60% in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum before transfection. Each well was transfected with 200 ng of pGL-SV40, 20 ng of pRL-NF-κB, 200 ng of FLAG-SHARPIN, 200 ng of FLAG-HOIL-1L, 800 ng of FLAG-HOIP, and 800 ng of FLAG-cherry-IpaH1.4/2.5 plasmids by Lipofectamine 2000 (Thermo Scientific) according to the manufacturer’s protocol. After 24 h, cells were washed once with phosphate-buffered saline (PBS) and collected for dual-luciferase assay using Promega’s DLR kit (Promega) according to manufacturer’s protocol.
S. flexneri Mutation.
Double knockout of the ipaH1.4/ipaH2.5 gene in the S. flexneri M90T strain was constructed by λ red recombinase–mediated replacement by homologous recombination with a kanamycin resistance cassette, followed by flippase recombinase–catalyzed removal of the cassette. To recomplement the ipaH1.4/ipaH2.5-knockout strain, the wild-type or mutant ipaH1.4 gene, which is C-terminally fused with a His tag gene sequence, was introduced back into the ipaH1.4 locus on the chromosome in the knockout strain by a CRISPR–Cas12a-assisted recombination method. Successful recomplements of different ipaH1.4 genes were verified by PCR amplification of the ipaH1.4 locus with a pair of primers (up: gaacgtaacaacctgaccctcc; down: gttagtgacgtatctgacgtgg) targeting the upstream and downstream regions of the ipaH1.4 gene.
Western blotting.
To facilitate the detection of protein expression levels of ipaH1.4 variants, a His tag gene sequence was fused to the C terminus of each recomplemented ipaH1.4 variant gene. Protein expression levels of these ipaH1.4 variants in mutant bacterial strains were detected by Western blotting with a specific anti-His antibody (Sangon Biotech, AB102-02). Meanwhile, the DnaK protein expression level was also detected as a control using a specific anti-DnaK antibody (Abcam, ab69617).
HeLa Cell Infection by S. flexneri.
HeLa cells were preseeded into 15-mm dishes at a density of 2 × 105 cells per dish and infected with S. flexneri strains at a multiplicity of infection of 100. After incubating at 37 °C for 3 h, the infected cells were washed three times with PBS. To kill extracellular bacteria, the medium was then replaced with DMEM supplemented with 100 μg/mL gentamycin and incubated for an additional 2 h.
Immunofluorescence Microscopy.
HeLa cells on coverslips were washed twice in PBS and fixed with 4% (wt/vol) formaldehyde for 10 min at 25 °C; after washing three time in PBS, cells were permeabilized (in PBS with 0.2% [vol/vol] Triton X-100) for 10 min and incubated with rabbit anti-p65 poly-antibody (Cell Signaling Technology, 8242) in PBS buffer with 10% (wt/vol) bovine serum albumin (BSA) for 45 min. After washing three times in PBS, cells were incubated with AlexaFluor 568–conjugated goat anti-rabbit secondary antibody in PBS with 10% (wt/vol) BSA and washed three times in PBS. The DNA was stained with 0.1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) in PBS and washed three times in PBS. Images were collected with a fluorescent confocal microscope (DMi8, Leica Microsystems Inc.). The number of cells with p65 translocation into the nucleus was counted manually. Only infected cells (with presence of DAPI-stained bacteria in the cytoplasm of cells) were included in the calculation of percentage of p65 nuclear enrichment. A p65 signal higher in the nucleus than in the cytoplasm was scored as nuclear translocation.
Supplementary Material
Acknowledgments
We thank SSRF BL19U1, BL02U1, and BL10U2 for X-ray beam time, Professor Jiahuai Han for the HOIP, SHARPIN, HOIL-1L, and UBE2D1 cDNA, Professor Yufeng Yao for the IpaH1.4 and IpaH2.5 cDNA, and Professor Ronggui Hu for the E1 (mouse UBA1) plasmid used in this study. This work was supported by grants from the National Natural Science Foundation of China (21822705, 91753113, 21621002, and 32071219), the National Key R&D Program of China (2016YFA0501903), the Science and Technology Commission of Shanghai Municipality (20XD1425200), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB20000000), the start-up fund from State Key Laboratory of Bioorganic and Natural Products Chemistry and Chinese Academy of Sciences (to L.P.), and grants from the National Natural Science Foundation of China (32071297) (to J.L.), National Natural Science Foundation of China (31900111) (to D.W.), and National Natural Science Foundation of China (31800646) (to Yingli Wang).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116776119/-/DCSupplemental.
Data Availability
The atomic coordinates and structure factors of the crystal structures of the apo form IpaH1.4 LRR, the IpaH1.4 LRR/HOIP RING1 complex, the UBE2L3/HOIP RING1 complex, and the IpaH1.4 LRR/HOIL-1L UBL complex have been deposited in the PDB under the accession codes 7V8H, 7V8G, 7V8F, and 7V8E, respectively. All other study data are included in the article and/or SI Appendix.
References
- 1.Weissman A. M., Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2, 169–178 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Pickart C. M., Eddins M. J., Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Kerscher O., Felberbaum R., Hochstrasser M., Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hicke L., Schubert H. L., Hill C. P., Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Popovic D., Vucic D., Dikic I., Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Hershko A., Ciechanover A., The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Iha H., et al. , Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 27, 629–641 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z. J., Sun L. J., Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Komander D., Rape M., The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Kirisako T., et al. , A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieser E., Cordier S. M., Walczak H., Linear ubiquitination: A newly discovered regulator of cell signalling. Trends Biochem. Sci. 38, 94–102 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Iwai K., Fujita H., Sasaki Y., Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Ikeda F., Linear ubiquitination signals in adaptive immune responses. Immunol. Rev. 266, 222–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu Y., Taraborrelli L., Walczak H., Linear ubiquitination in immunity. Immunol. Rev. 266, 190–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokunaga F., et al. , SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Ikeda F., et al. , SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlach B., et al. , Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Hrdinka M., Gyrd-Hansen M., The Met1-linked ubiquitin machinery: Emerging themes of (de)regulation. Mol. Cell 68, 265–280 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Schreiber A., Peter M., Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 163–181 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Keusekotten K., et al. , OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiil B. K., et al. , OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol. Cell 50, 818–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hrdinka M., et al. , CYLD limits Lys63- and Met1-linked ubiquitin at receptor complexes to regulate innate immune signaling. Cell Rep. 14, 2846–2858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smit J. J., et al. , The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 31, 3833–3844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stieglitz B., et al. , Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature 503, 422–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lechtenberg B. C., et al. , Structure of a HOIP/E2∼ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529, 546–550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis M. J., et al. , UBE2L3 polymorphism amplifies NF-κB activation and promotes plasma cell development, linking linear ubiquitination to multiple autoimmune diseases. Am. J. Hum. Genet. 96, 221–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuseya Y., et al. , The HOIL-1L ligase modulates immune signalling and cell death via monoubiquitination of LUBAC. Nat. Cell Biol. 22, 663–673 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Sato Y., et al. , Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc. Natl. Acad. Sci. U.S.A. 108, 20520–20525 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stieglitz B., Haire L. F., Dikic I., Rittinger K., Structural analysis of SHARPIN, a subunit of a large multi-protein E3 ubiquitin ligase, reveals a novel dimerization function for the pleckstrin homology superfold. J. Biol. Chem. 287, 20823–20829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., et al. , Structural insights into SHARPIN-mediated activation of HOIP for the linear ubiquitin chain assembly. Cell Rep. 21, 27–36 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Fujita H., et al. , Cooperative domain formation by homologous motifs in HOIL-1L and SHARPIN plays a crucial role in LUBAC stabilization. Cell Rep. 23, 1192–1204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noad J., et al. , LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat. Microbiol. 2, 17063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Wijk S. J. L., et al. , Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nat. Microbiol. 2, 17066 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Fiil B. K., Gyrd-Hansen M., Met1-linked ubiquitination in immune signalling. FEBS J. 281, 4337–4350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokunaga F., et al. , Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Sanada T., et al. , The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626 (2012). [DOI] [PubMed] [Google Scholar]
- 37.de Jong M. F., Liu Z., Chen D., Alto N. M., Shigella flexneri suppresses NF-κB activation by inhibiting linear ubiquitin chain ligation. Nat. Microbiol. 1, 16084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiskin E., Bionda T., Dikic I., Behrends C., Global analysis of host and bacterial ubiquitinome in response to Salmonella Typhimurium infection. Mol. Cell 62, 967–981 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Khan M., Syed G. H., Kim S. J., Siddiqui A., Hepatitis B virus-induced parkin-dependent recruitment of linear ubiquitin assembly complex (LUBAC) to mitochondria and attenuation of innate immunity. PLoS Pathog. 12, e1005693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., et al. , The hepatitis C virus protein NS3 suppresses TNF-α-stimulated activation of NF-κB by targeting LUBAC. Sci. Signal. 8, ra118 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Jing H., et al. , Porcine reproductive and respiratory syndrome virus nsp1α inhibits NF-κB activation by targeting the linear ubiquitin chain assembly complex. J. Virol. 91, e01911-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., et al. , The linear ubiquitin assembly complex modulates latent membrane protein 1 activation of NF-κB and interferon regulatory factor 7. J. Virol. 91, e01138-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jennison A. V., Verma N. K., Shigella flexneri infection: Pathogenesis and vaccine development. FEMS Microbiol. Rev. 28, 43–58 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Mattock E., Blocker A. J., How do the virulence factors of Shigella work together to cause disease? Front. Cell. Infect. Microbiol. 7, 64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newton H. J., et al. , The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-κB p65. PLoS Pathog. 6, e1000898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y., et al. , Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat. Struct. Mol. Biol. 15, 1302–1308 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Rohde J. R., Breitkreutz A., Chenal A., Sansonetti P. J., Parsot C., Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1, 77–83 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Duda D. M., et al. , Structure of a glomulin-RBX1-CUL1 complex: Inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol. Cell 47, 371–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang L., Zhang Z., Yang J., McLaughlin S. H., Barford D., Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522, 450–454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiskin E., et al. , Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA. Nat. Commun. 8, 14004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elton L., Carpentier I., Verhelst K., Staal J., Beyaert R., The multifaceted role of the E3 ubiquitin ligase HOIL-1: Beyond linear ubiquitination. Immunol. Rev. 266, 208–221 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Norkowski S., Schmidt M. A., Rüter C., The species-spanning family of LPX-motif harbouring effector proteins. Cell. Microbiol. 20, e12945 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q. S., et al. , The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 26, 12–17 (2015). [Google Scholar]
- 55.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of the crystal structures of the apo form IpaH1.4 LRR, the IpaH1.4 LRR/HOIP RING1 complex, the UBE2L3/HOIP RING1 complex, and the IpaH1.4 LRR/HOIL-1L UBL complex have been deposited in the PDB under the accession codes 7V8H, 7V8G, 7V8F, and 7V8E, respectively. All other study data are included in the article and/or SI Appendix.