Significance
This study shows that Fragile X mental retardation protein (FMRP) promotes messenger RNA (mRNA)-dependent recombination via facilitating ten-eleven translocation protein 1 (TET1)-mediated mRNA methyl-5-cytosine (m5C) demethylation. Loss of FMRP leads to damage induced mRNA m5C and R-loop accumulation at sites of active transcription, defective recombination repair, and increased radiosensitivity of tumor cells. FMRP-dependent RNA m5C demethylation and R-loop resolving during DNA repair are important for repair completion and the maintenance of genome stability. The removal of m5C by the FMRP–TET1 axis is coupled with R-loop dissolution, which ensures proper completion of DNA repair and survival of cells after DNA damage. These findings significantly advance our understanding of the regulation of RNA modifications in R-loop dynamics during DNA repair.
Keywords: mRNA modification, FMRP, TRDMT1, m5C, DNA damage repair
Abstract
RNA modifications regulate a variety of cellular processes including DNA repair. The RNA methyltransferase TRDMT1 generates methyl-5-cytosine (m5C) on messenger RNA (mRNA) at DNA double-strand breaks (DSBs) in transcribed regions, promoting transcription-coupled homologous recombination (HR). Here, we identified that Fragile X mental retardation protein (FMRP) promotes transcription-coupled HR via its interaction with both the m5C writer TRDMT1 and the m5C eraser ten-eleven translocation protein 1 (TET1). TRDMT1, FMRP, and TET1 function in a temporal order at the transcriptionally active sites of DSBs. FMRP displays a higher affinity for DNA:RNA hybrids containing m5C-modified RNA than for hybrids without modification and facilitates demethylation of m5C by TET1 in vitro. Loss of either the chromatin- or RNA-binding domain of FMRP compromises demethylation of damage-induced m5C in cells. Importantly, FMRP is required for R-loop resolving in cells. Due to unresolved R-loop and m5C preventing completion of DSB repair, FMRP depletion or low expression leads to delayed repair of DSBs at transcriptionally active sites and sensitizes cancer cells to radiation in a BRCA-independent manner. Together, our findings present an m5C reader, FMRP, which acts as a coordinator between the m5C writer and eraser to promote mRNA-dependent repair and cell survival in cancer.
It is known that DNA damage becomes deleterious to cells at transcriptionally active coding regions because RNA polymerase II (RNA Pol II) is generally stalled at DNA strand breaks, and nascent RNA transcript is able to rehybridize with DNA template and displaces nontemplate single-stranded DNA, forming a so-called R-loop structure (1). R-loops are susceptible to breakage and cause genome instability. Meanwhile, R-loops serve as a signal to initiate transcription-coupled homologous recombination (TC-HR) after damage at the transcribed genome (1, 2). Such beneficial impact of transcription on DNA repair was discovered in transcription-coupled nucleotide excision repair (TC-NER) (3, 4) for ultraviolet (UV)-induced damage and TC-HR for DNA nuclease-induced double-strand breaks (DSBs) and oxidative damage-induced DSBs at the transcribed genome (5–8). Although the impact of transcription on DNA repair has been long appreciated, the functions of RNAs in DNA repair have just begun to unfold. The RNAs newly synthesized upon DSB generation, including small RNA and long noncoding RNA, have been demonstrated to activate DNA damage response or promote the DSB repair (9–12). Recent studies indicate that RNA modifications, e.g., N6-methyladenosine (m6A) and methyl-5-cytosine (m5C), contribute to R-loop stabilization and are involved in TC-HR (13, 14). Among RNA modifications, m5C is less studied, and “readers” of m5C are not well identified, while emerging studies have revealed m5C RNA modification as a new layer in tumorigenesis and tumor progression. For example, elevated m5C RNA modification has been reported in the circulating tumor cells from lung cancer patients compared to the whole blood cells (15). More recently, m5C RNA modification was found to be increased in human urothelial carcinoma of the bladder (16) and in human hepatocellular carcinoma (17). Importantly, m5C RNA modification promotes the pathogenesis of bladder cancer through stabilizing messenger RNAs (mRNAs) (16). A growing body of studies has shown that just like phosphorylation and ubiquitination, dephosphorylation (18, 19) and deubiquitination (20, 21) play important roles in the DNA damage repair process. Thus, we are very interested in how m5C RNA modification is dynamically regulated in cells and subsequently contributes to repair and cancer-cell survival. Furthermore, the removal of R-loops is required for the completion of DNA repair, but how it is regulated is also unknown. Thus, elucidating the regulation of m5C and R-loops during DNA repair is critical for understanding how DNA repair is uniquely regulated in the transcribed genome.
In this study, we identified the Fragile X mental retardation protein (FMRP) as an interacting partner of the m5C writer, TRDMT1, using immunoprecipitation (IP) and mass spectrometry (MS). FMRP, encoded by FMR1, is a predominantly cytoplasmic RNA-binding protein that regulates protein translation (22, 23). The deficiency of FMRP causes Fragile X syndrome, which is associated with inherited intellectual disability, premature ovarian failure, autism, Parkinson's disease, developmental delays, and other cognitive deficits (24). Here, we found that the knockdown (KD) or knockout (KO) of FMRP delays the demethylation of m5C RNA modification and resolving of R-loops after DNA damage. FMRP preferentially binds DNA:RNA hybrids containing m5C-modified RNA. Mechanistically, FMRP is recruited to sites of damage by the m5C writer, TRDMT1, and then promotes the removal of m5C by its eraser ten-eleven translocation protein 1 (TET1). Both the chromatin-binding domain and RNA-binding domain are required for FMRP to promote m5C demethylation. Loss of FMRP leads to defective HR repair and increased radiosensitivity of tumor cells. Thus, FMRP-dependent m5C RNA-modification demethylation and R-loop resolving during DNA repair are important for repair completion and the maintenance of genome stability.
Results
FMRP Interacts with m5C RNA-Modification Writer TRDMT1 and Promotes m5C Demethylation after DNA Damage.
Most cancer cells are consistently burdened by oxidative stress due to altered oxidation metabolism. Our recent study revealed that oxidative damage and oxidative damage-derived DSBs trigger the methyltransferase TRDMT1-mediated m5C RNA modification, which contributes to DNA repair and cell survival (14). To understand how m5C RNA modification is dynamically regulated in cells and subsequently contributes to repair and cancer-cell survival, we sought to identify regulators of the m5C RNA-modification writer TRDMT1. TRDMT1 stably expressed Flp-in 293 cells with or without H2O2 damage were subjected to IP, and the pull-down elution fractions were applied to MS analysis (Fig. 1A). Under the unperturbed conditions, most of the pulled-down proteins by green fluorescent protein (GFP)-tagged TRDMT1 are involved in four specific biological processes: transfer RNA (tRNA) or mRNA biogenesis, DNA synthesis, DNA repair, and cell-cycle regulation (SI Appendix, Fig. 1A). Level of exogenously expressed TRDMT1 was identical to endogenous TRDMT1 expression (SI Appendix, Fig. 1B). Specifically, a number of aminoacyl–tRNA synthetases were detected, including phenylalanyl–tRNA synthetase, tyrosyl–tRNA synthetase, and glycyl–tRNA synthetase (SI Appendix, Fig. 1A), which is consistent with the role of TRDMT1 as a tRNA methyltransferase. In addition, the RNA Pol II subunit, POLR2B, was detected as a TRDMT1-interacting protein (Fig. 1B), which is consistent with a previous report that TRDMT1 forms a complex with RNA Pol II (25). We also confirmed that the interaction between TRDMT1 and RNA Pol II increases upon DNA damage (Fig. 1B). As previously reported, we also detected Top2A, MSH2, MSH6, and HDAC2 as TRDMT1-interacting partners (26). For the H2O2-damaged condition, 10 of the top 15 enriched proteins are RNA-binding proteins. Among them, FMRP was identified as one of the top proteins. Interestingly, it has been shown that many FMRP-binding sites in RNA overlap with sites of m5C modification in RNA (27); however, the role of FMRP in the regulation of m5C RNA modification is unknown. The relative abundance of FMRP in different groups of MS was analyzed, and FMRP was found to be mostly enriched in the H2O2 damage group after TRDMT1 pull-down (Fig. 1B). The interaction between FMRP and TRDMT1 was confirmed by co-IP detected with the anti-FMRP antibody. As expected, H2O2 treatment increased their interaction (Fig. 1C). In contrast, EtBr treatment, RNaseH, or RNaseA treatment did not abolish the interaction, indicating that the interaction is not mediated by DNA, RNA, or DNA:RNA hybrid (SI Appendix, Fig. 1C). FMRP and TRDMT1 interacted with each other in the in vitro pull-down with purified FMRP and TRDMT1 proteins, indicating a direct protein–protein interaction between FMRP and TRDMT1 (Fig. 1D).
Fig. 1.
FMRP interacts with the m5C RNA-modification methyltransferase TRDMT1 and contributes to m5C RNA-modification demethylation after damage. (A) Schematic diagram of the MS to identify the interacting partners of TRDMT1. (A, Right) Top 15 proteins enriched in the H2O2 damage group. RNA-binding proteins were marked with orange color. (B) The relative abundance of POLR2B and FMRP in different groups of mass spectrometry. The relative intensity of three identified peptides for FMRP in three groups of MS was calculated (mean ± SEM). (C) Flp-in 293 cells were transfected with GFP-TRDMT1 or GFP empty vector with or without treatment of 1 mM H2O2 for 1 h. Interaction of FMRP and TRDMT1 was detected with Co-IP. Anti-GFP antibody was used for pull-down, and anti-FMRP was used for detection in Western blot (WB). (D) In vitro pull-down of FMRP and TRDMT1. We used 0.3 μg of purified His-FMRP and His-TRDMT1 for pull-down assay with the anti-FMRP antibody. WB of anti-His antibody is shown. (E and F) WT and FMRP KO U2OS–TRE cells were treated with or without 2 mM H2O2 for 1 h and then recovered for 8 h after H2O2 removal. After treatment, mRNA was extracted for m5C dot-blot assay at the indicated time point. Quantification of the level of m5C (mean ± SD) from three independent experiments normalized with WT no H2O2 is shown in F. (G) siFMRP, siTRDMT1, or control siRNA pretreated U2OS–TRE cells were transfected with TA-KR. The cells were exposed to light for KR activation for 25 min and followed with recovery for the indicated time before fixation. m5C foci frequency at TA-KR sites at the indicated time point is shown. (Scale bar: 10 μm.) (n = 3; 50 cells per replicate; mean ± SD.) Statistical analysis was done with the unpaired two-tailed Student t test. ns. not significant. **P < 0.01.
To investigate if m5C RNA-modification kinetics is regulated by FMRP upon DNA damage, we created the FMRP KO U2OS cells (SI Appendix, Fig. 1D) and examined the level of m5C in mRNA using the dot-blot assay. After the cells were treated with H2O2 for 1 h, H2O2 was removed from the medium, and cells were allowed to recover for 8 h. The level of m5C in mRNA isolated from cells was analyzed. In the wild-type (WT) U2OS cells, an increase of m5C in mRNA was detected after 1-h H2O2 treatment (Fig. 1E), as reported (14). To be noticed, the m5C level in mRNA gradually decreased after damage induction. In the FMRP KO cells, the induction of m5C at 1 h after treatment with H2O2 was the same as in WT cells (Fig. 1E). However, at 8 h after H2O2 removal, the m5C RNA-modification level in FMRP KO cells remained significantly higher than in WT cells (Fig. 1 E and F). m5C RNA modification is a signal to promote repair, while the resolving of m5C is also required with repair progression. FMRP depletion affects the resolving of m5C at the late stage of repair, indicating that FMRP may be required for repair completion.
We previously established the DNA damage at RNA-transcribed sites (DART) system to monitor the kinetics of DNA repair at transcribed vs. nontranscribed genome in real time in live cells (8, 28). In the DART system, we used KillerRed (KR), a light-excitable chromophore, to release free radicals upon light activation in a dose-dependent manner. A tandem tetracycline-responsive element array cassette with an adjacent reporter gene was stably integrated into the genome of U2OS cells. When KR is fused with either tetR (tetR-KR) or tetR with transcription activator VP16 (TA-KR) and expressed in cells, the fusion protein tetR-KR or TA-KR binds to the TRE locus. TetR-KR binds to the TRE locus, but does not activate transcription, while TA-KR binds and activates the reporter gene transcription locally (8, 28). After KR activation with visible light exposure, either tetR-KR or TA-KR releases free radicals and causes the equivalent amount of local oxidative damage-derived DSBs at the locus (8, 28). With this system, we have detected robust m5C at the TRE locus specifically at sites of TA-KR, but not at sites of tetR-KR, despite the similar induction of damage at both sites after light activation (14) (SI Appendix, Fig. 1E). Without light activation, KR did not release free radicals, and there were no m5C foci at this locus (SI Appendix, Fig. 1E). We have monitored the kinetics of local m5C with different recovery time points after light activation-induced damage in the DART system with or without small interfering TRDMT1 (siTRDMT1) and/or small interfering FMRP (siFMRP) (Fig. 1G). In the small interfering Control (siCtrl)-treated cells, the percentage of m5C-positive cells increased and reached the maximum at damage sites at 1-h recovery, then decreased gradually. In siFMRP-treated cells, although m5C was similarly induced at 1-h recovery, the disappearance of m5C was delayed with time (Fig. 1G and SI Appendix, Fig. 1F). These results confirmed that FMRP is required for local m5C resolving at the transcribed genome after damage. Moreover, m5C kinetics in both siTRDMT1- and siFRMP-treated cells is identical to that in siTRDMT1 cells with reduced m5C induction 1 h after damage. This result supports the idea that TRDMT1 acts upstream of FMRP in the m5C pathway at a DNA damage site (Fig. 1G).
FMRP Preferentially Binds DNA:RNA Hybrids Containing m5C-Modified RNA and its RNA Binding Is Required for m5C Demethylations.
To understand the molecular basis of the regulation on the m5C RNA-modification kinetics by FMRP, we examined if FMRP could bind m5C-modified RNA:DNA hybrids. We synthesized a set of biotin-labeled RNA oligos of 30 nucleotide long, with or without five m5Cs, and used them to generate DNA:RNA hybrids (Fig. 2 A, Left). The 30-bp DNA:RNA hybrids containing m5Cs captured FMRP in cell lysates more efficiently than the hybrids containing unmodified RNA (Fig. 2 A and B). Thus, FMRP preferentially binds m5C RNA-modified RNA:DNA hybrids.
Fig. 2.
FMRP preferentially binds m5C RNA-modified RNA:DNA hybrids and contributes to m5C demethylation dependently on its RNA binding in cells. (A) Lysates of Flp-in 293 cells with overexpression of GFP–FMRP were incubated with biotin-labeled DNA–RNA hybrids with or without m5C RNA modification coated on streptavidin magnetic beads. Pull-down fractions were detected by FMRP–antibody. The presence of m5C in the RNA oligo was confirmed by m5C dot blot. (B) Relative binding efficiency of FMRP with biotin-labeled DNA–RNA hybrids with or without m5C RNA modification was calculated from three pull-down experiments (mean ± SD). (C and D) Flp-in 293 FMRP KO cells were transfected with Flag, Flag-FMRP, or Flag-FMRP I304N treated with 1 mM H2O2 for 1 h. Cell lysates were pulled down with the anti-FMRP antibody. The m5C RNA:DNA hybrid was incubated with the pulled-down fraction at room temperature for 3 h following the demethylation assay protocol. The m5C level after demethylation assay was checked with dot blot and quantified in D (n = 3, mean ± SD). Statistical analysis was done with the unpaired two-tailed Student t test. ns, not significant. **P < 0.01; ***P < 0.001.
To understand how the m5C RNA-modified hybrids-preferred binding affinity of FMRP affects the demethylation process of m5C RNA modification, we performed the demethylation assay using the FMRP pull-down fraction from FMRP-expressed FMRP KO Flp-in 293 cell lysates. The FMRP pull-down fraction was incubated with m5C-modified RNA:DNA hybrids. The m5C level in the FMRP-expressed cells was lower compared to that in FMRP KO cells (Fig. 2 C and D), indicating that FMRP is involved in the demethylation of the m5C-modified RNA:DNA hybrids. In contrast, the FMRP mutant I304N, which is defective in RNA binding (29), failed to remove the m5C-modified DNA:RNA hybrids (Fig. 2 C and D). These results together suggest that FMRP preferentially binds m5C RNA-modified RNA:DNA hybrids and subsequently contributes to the demethylation of m5C in cells.
FMRP Is Epistasis of TRDMT1 and Is Recruited to DNA Damage Sites by TRDMT1.
Since FMRP is required for damage-induced m5C RNA-modification demethylation, we asked if FMRP itself is recruited to the sites of m5C locally. Cells transfected with Flag-FMRP and TA-KR or TA-Cherry were light-irradiated and allowed to recover for 1 h before fixation and staining with Flag antibody. Cherry was used as an additional nondamage control for KR, and TA-Cherry activates transcription at the TRE array, but does not induce damage upon light exposure. The majority of FMRP was distributed in the cytoplasm, which is consistent with previous literature (22). Importantly, after damage induction by KR in the nucleus, FMRP was preferentially recruited to sites of TA-KR, but not to TA-Cherry (Fig. 3 A and B), indicating that transcription itself does not trigger the recruitment of FMRP. The chromatin IP (ChIP)-qPCR assay confirmed the specific binding of Flag-FMRP to sites of TA-KR compared to the Flag empty vector (Fig. 3C). Besides the Flag-tagged FMRP, the endogenous FMRP was also recruited to the sites of TA-KR after damage induction (SI Appendix, Fig. 2A). Importantly, the recruitment of FMRP at sites of TA-KR was diminished in TRDMT1 KO cells (Fig. 3 D and E), indicating that TRDMT1 may facilitate the recruitment of FMRP to m5C sites via the TRDMT1–FMRP interaction.
Fig. 3.
FMRP is recruited to the transcribed damage sites by TRDMT1 and cooperates with TRDMT1 to promote efficient damage removal. (A) U2OS–TRE cells transfected with TA-KR/TA-Cherry and Flag-FMRP plasmids were light-irradiated and allowed to recover for 1 h before fixation. Cells were stained with Flag antibody, and represented figures are shown. (Scale bar: 10 μm.) (B, Left) FMRP foci frequency at TA-KR or TA-Cherry in U2OS–TRE cells (n = 3; 50 cells per replicate; mean ± SEM). (B, Right) Fold increase of FMRP foci intensity was quantified. Mean intensity of FMRP at TA-KR or TA-Cherry/mean intensity of background is shown (n = 15, mean ± SEM). (C) ChIP PCR detected the DNA at the loci near the TRE region in cells transfected with TA-KR and GFP/GFP-FMRP. (D) WT and TRDMT1 KO U2OS–TRE cells transfected with TA-KR and Flag-FMRP plasmids were light-irradiated and allowed to recover for 1 h before fixation. Cells were stained with Flag antibody, and representative figures are shown. (Scale bar: 10 μm.) (E, Left) FMRP foci frequency at TA-KR in U2OS–TRE and TRDMT1 KO cells (n = 3; 50 cells per replicate; mean ± SEM). (E, Right) Fold increase of FMRP foci intensity = mean intensity of FMRP at TA-KR/mean intensity of background (n = 15; mean ± SEM). (F and G) γH2AX foci frequency at sites of TA-KR (F) or tetR-KR (G) at early (1 h) and late (36 h) time points after damage induction in U2OS–TRE WT and FMRP KO cells. (Scale bar: 10 μm.) (n = 3; 50 cells per replicate; mean ± SEM.) (H) WT and FMRP KO U2OS–TRE cells transfected with siTRDMT1 or control siRNA were exposed to 4 Gy of IR. Survival rate was measured via colony-formation assay (n = 3; mean ± SEM). (I) γH2AX foci frequency at TA-KR at early (1 h) and late (36 h) time points after damage induction in WT or TRDMT1 KO U2OS–TRE cells pretreated with siFMRP or control siRNA (n = 3; 50 cells per replicate; mean ± SEM). Statistical analysis was done with the unpaired two-tailed Student t test. ns, not significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Light-activated KR induces oxidative damage-derived DSBs, and the repair kinetics can be monitored by γH2AX kinetics at local damage sites (14). We characterized the γH2AX kinetics in FMRP KO cells to understand if the FMRP-dependent m5C demethylation is important for DNA repair. We observed the equivalent induction of TA-KR–induced γH2AX foci in the WT and FMRP KO cells 1 h after damage induction. In the WT cells, the frequency of TA-KR–induced γH2AX foci was significantly reduced 36 h after damage induction; however, the decline of γH2AX foci at the TRE array was compromised in FMRP KO cells (Fig. 3F), suggesting that FMRP is required for efficient repair of DSBs at the transcribed genome. We also examined the effect of FMRP KO on the repair of DSBs at the sites of tetR-KR, in which DSBs were induced within heterochromatin structure without the presence of mRNA and active transcription. At sites of tetR-KR, the rate of γH2AX clearance did not show any differences between the U2OS WT and FMRP KO cells (Fig. 3G). To rule out the possibility that the γH2AX clearance defect of the FMRP KO cell is due to the checkpoint defect caused by FMRP deficiency, we checked the activation of the ATM signaling pathway upon ionizing radiation (IR) damage. Under the condition of no IR damage, no activation of ATM or CHK2 was observed in the FMRP KO cells compared to U2OS WT cells. After cells were exposed to IR damage, similar activation of ATM and CHK2 was observed for FMRP KO cells and U2OS WT cells (SI Appendix, Fig. 2B), indicating that the ATM signaling pathway was not affected by knocking out FMRP. Thus, the FMRP-dependent m5C demethylation specifically contributes to the repair of DSBs within actively transcribed chromatin. We also used reporter assays to examine the HR-mediated repair of DSBs. Using the direct-repeat GFP (DR-GFP) assay, we found that FMRP KD decreased the repair of I-SceI–generated DSBs (SI Appendix, Fig. 2C). Furthermore, the HR-mediated integration of mClover in a site of CRISPR/Cas9-generated DSB in the endogenous LAMIN A gene was reduced in FMRP KD cells compared to control cells (SI Appendix, Fig. 2D). The genome of U2OS–TRE cells contains 96 copies of I-SceI sites within the TRE array. Thus, coexpression of the I-SceI endonuclease with TA-Cherry induced I-SceI–generated DSBs at the TRE array. FMRP was found to be recruited to the I-SceI–generated DSBs (SI Appendix, Fig. 2E), which has also been reported for TRDMT1 (14). Thus, FMRP is required for efficient HR for DSBs in an exogenously or endogenously transcribed gene.
TRDMT1 is likely to be upstream of FMRP to recruit FMRP to sites of damage, as double depletion of TRDMT1 and FRMP didn’t alter the methylation status of damage-induced m5C RNA modification at the TRE array compared to the single depletion of TRDMT1 (Fig. 1G). Next, we examined how the depletion of TRDMT1 and/or FMRP affects cell survival after IR with the colony-forming assay. FMRP KO cells displayed increased IR sensitivity compared to WT U2OS cells. Importantly, TRDMT1 KD in FMRP KO cells did not further increase the cell sensitivity of FMRP KO cells (Fig. 3H). In addition, FMRP KD in TRDMT1 KO cells didn’t further delay the γH2AX clearance compared to single KD or KO (Fig. 3I). FMRP is largely distributed in the cytoplasm and can be shuttled to the nucleus. Both IR and H2O2 treatments increased the nuclear distribution of FMRP (SI Appendix, Fig. 2F), suggesting that the nuclear function of FMRP might contribute to cell survival and damage repair. These results also suggest that FMRP and TRDMT1 are in the same epistasis group for damage removal at the transcriptionally active sites of damage.
The Chromatin- and RNA-Binding Domains of FMRP Are Required for m5C RNA-Modification Demethylation.
Having shown that TRDMT1 is required the recruitment of FMRP to the damage sites, we next determined the interacting domain of FMRP that interacts with TRDMT1 for their cooperation. The N-terminal domain and K Homology (NKH) domain and the Nuclear Export Sequence and the C-terminal (NESC) domain were constructed for this study (30, 31) (Fig. 4A). The N-terminal NKH fragment of FMRP contains a nuclear localization signal, and the C-terminal NESC fragment of FMRP contains a nuclear export signal (NES). Therefore, the NKH fragment predominantly localizes in the nucleus, whereas the NESC fragment is exclusively cytoplasmic. Furthermore, the NKH fragment is recruited to sites of damage induced by TA-KR, but the NESC fragment is not (Fig. 4A). The NKH fragment, but not NESC, interacted with TRDMT1 in co-IP (Fig. 4B). To understand if the NKH domain could rescue m5C resolving, we first used the small interfering RNA (siRNA)-targeted 3′ untranslated region (UTR) of FMRP to suppress endogenous FMRP. Treatment with siRNA targeting 3′ UTR of FMRP led to m5C accumulation 8 h after light activation at TA-KR damage sites as well as the other FMRP KD did in Fig. 1G (Fig. 4C and SI Appendix, Fig. 1F). Expression of full-length FMRP in 3′ UTR of FMRP pretreated cells facilitated m5C clearance at 8 h after light activation. The expression of the NKH fragment rescued m5C clearance as the full-length FMRP, while the NESC did not (Fig. 4C). Similarly, comparable to the expression of full-length FMRP in FMRP KO cells, expression of the NKH fragment, but not the NESC, restored the decline of global m5C level at 8 h after H2O2 damage from mRNA extracts (Fig. 4D). Thus, the NKH domain of FMRP, which interacts with TRDMT1, is important for FMRP-dependent m5C RNA-modification resolving.
Fig. 4.
Both chromatin-binding and RNA-binding domains of FMRP are required for m5C RNA-modification resolving. (A, Upper) Schematic diagram of the FMRP fragments. (A, Lower) U2OS–TRE cells transfected with TA-KR and GFP-FMRP fragment plasmids were light-irradiated and allowed to recover for 1 h before fixation. Representative figures are shown. (Scale bar: 10 μm.) (B) Interaction of FMRP fragments and TRDMT1 detected with Co-IP. Flp-in 293 FMRP KO cells were transfected with FMRP fragments and GFP-TRDMT1, treated with 1 mM H2O2 for 1 h before co-IP analysis with the anti-GFP antibody. (C) U2OS–TRE cells pretreated with control siRNA or siFMRP targeting in the 3′ UTR region of FMRP were transfected with TA-KR and FMRP fragment plasmids. The cells were exposed to light activation for 25 mins and then left to recover for 8 h before fixation. m5C foci frequency at TA-KR sites is shown (n = 3; 50 cells per replicate; mean ± SEM). (D) FMRP KO U2OS–TRE cells transfected with full-length FMRP or FMRP fragment plasmids were treated with or without 2 mM H2O2 for 1 h and then recovered for 8 h after H2O2 removal. After treatment, mRNA was extracted for m5C dot-blot assay at the indicated time point. (E, Upper) Schematic diagram of the FMRP mutants. (E, Lower) Interaction of FMRP point mutants and TRDMT1 detected with Co-IP. Flp-in 293 FMRP KO cells were transfected with FMRP point mutants and GFP-TRDMT1 or GFP empty vector, treated with 1 mM H2O2 for 1 h before co-IP analysis with the anti-GFP antibody. (F) U2OS–TRE cells pretreated with control siRNA or siFMRP targeting in the 3′ UTR region of FMRP were transfected with TA-KR and FMRP mutant plasmids. The cells were exposed to light activation for 25 mins and then left to recover for 8 h before fixation. m5C foci frequency at TA-KR sites is shown (n = 3; 50 cells per replicate; mean ± SEM). (G) FMRP KO U2OS–TRE cells transfected with full-length FMRP or FMRP fragments plasmids were exposed to 2 Gy of IR. Survival rate was measured via colony-formation assay (n = 3, mean ± SEM). Statistical analysis was done with the unpaired two-tailed Student t test. ns, not significant. *P < 0.05; **P < 0.01.
The NKH domain also contains chromatin-binding activity or RNA-binding ability, which may be important for FMRP to regulate m5C (29, 32). Next, we made chromatin-binding defective mutant Y103L and RNA-binding defective mutant I304N of FMRP to test if chromatin-binding ability or RNA-binding ability is important for FMRP to regulate m5C (29, 32). Both Y103L and I304N FMRP still interacted with TRDMT1 (Fig. 4E); however, none of them rescued the function of FMRP to resolve the m5C RNA modification at the late stage of damage repair (Fig. 4F). These results indicate that both intrinsic chromatin-binding and RNA-binding domains of FMRP are required for m5C RNA modification resolving after FRMP is recruited to sites of damage by TRDMT1. The colony-forming assay with IR demonstrated that the NKH fragment, but not the NESC fragment, rescues survival of FMRP KO cells after IR as full-length FMRP does (Fig. 4G). Together, these results indicate that the nucleus-localized NKH domain of FMRP with TRDMT1 interaction, chromatin, and RNA-binding activity is crucial for its functions at DNA damage sites.
Loss of FMRP Prevents the Resolving of R-Loops at the Late Stage of DNA Repair.
To understand the connection between m5C regulation and contribution to downstream damage repair by FMRP upon DNA damage, we monitored the key structure, R-loops, in the TC-HR pathway (Fig. 5A). Considering that the m5C modification occurs within the R-loop structure, we followed the R-loop kinetics with S9.6 antibody at the TRE locus upon light activation of TA-KR. The R-loop was greatly induced within 4 h after light activation (Fig. 5A). The R-loop level at the damage sites induced by TA-KR was abolished by the expression of RNase H1, but not by the catalytically inactive mutant D210N (SI Appendix, Fig. 3A), confirming the specificity of S9.6 staining-dependent R-loop detection. In the siCtrl-treated cells, the R-loop level largely decreased 8 h and 24 h after light activation. However, in siFMRP-treated cells, the clearance of R-loops was significantly delayed (Fig. 5A). To test whether the delay of m5C resolution or R-loop disappearance was caused by the FMRP depletion-mediated interruption of RNA degradation, the RNA transcripts at the TRE array were monitored by using MS2 foci as a read-out. At the TRE array, an RNA transcript containing MS2-binding sites is generated, and YFP-MS2 binds to the RNA and acts as an RNA and transcription reader (2, 33). MS2 foci decreased similarly in both WT and FMRP KO cells 1 h after damage induction, which is consistent with the transcription repression after DNA damage (2). At 24 h after damage induction, MS2 foci recovered similarly in WT and FMRP KO cells (SI Appendix, Fig. 3B). These results suggest that loss of FMRP does not affect the levels of RNA during the DNA damage response. Although temporary R-loops at damage sites facilitate TC-HR, persistent R-loop structure is a cause of DNA damage and is a threat to genome stability due to its unique structure. Therefore, the delay of m5C erasing in FMRP-deficient cells could affect the timely resolving of R-loops, thus further impairing the repair progression and completion.
Fig. 5.
FMRP prevents the resolving of R-loops at the late stage of repair. (A) U2OS–TRE cells pretreated with control siRNA or siFMRP were transfected with TA-KR plasmid. The cells were exposed to light activation for 25 mins and then left to recover for different time points before fixation and S9.6 staining. R-loop foci frequency at TA-KR sites at different time points after light activation is shown. (Scale bar: 10 μm.) (n = 3; 50 cells per replicate; mean ± SEM.) (B) U2OS–TRE cells pretreated with control siRNA or siFMRP were transfected with TA-KR. The relative foci intensity of RPA, CSB, TRDMT1, RAD52, and RAD51 at TA-KR (recover 1 h after light activation) are shown (n ≥ 10; mean ± SD). (C) WT and FMRP KO U2OS–TRE cells were transfected with TA-KR. The cells were exposed to light activation for 25 mins and then left to recover for different time points before fixation and RAD51 staining. (Scale bar: 10 μm.) RAD51 foci intensity at TA-KR sites at different time points after light activation is shown (n > 15; mean ± SD). (D) WT and FMRP KO U2OS–TRE cells were cotransfected with TA-KR and GFP-RAD52. The cells were exposed to light activation for 25 mins and then left to recover for different time before fixation. RAD52 foci frequency at TA-KR sites at different time points after light activation is shown. (Scale bar: 10 μm.) (n = 3; 50 cells per replicate; mean ± SEM.) Statistical analysis was done with the unpaired two-tailed Student t test. ns, not significant. *P < 0.05; **P < 0.01.
Next, we examined the effect of FMRP knocking down on the damage response of a set of R-loop sensors, recognizers, and downstream repair factors, which are involved in TC-HR. RPA is known to be an R-loop sensor (34). CSB and RAD52 recognize R-loops (7). RAD51 is required for R-loop–initiated TC-HR (7). Above R-loop regulators either are involved in R-loop formation or play a role in the context of R-loops (Fig. 5B). FMRP KD only had a mild effect on the damage response of RAD51, while it didn’t affect the recruitment of RPA, CSB, TRDMT1, and RAD52 to the R-loop structure at the early stage of damage repair (Fig. 5B and SI Appendix, Fig. 3C). Meanwhile, the analysis of the RNA-sequencing (RNA-seq) data from the breast cancer patients showed that high expression of FMRP was correlated with slightly increased expression of RPA1, CSB, and RAD51, with no correlation for RAD52 (SI Appendix, Fig. 4). These results suggest that FMRP is not required for R-loop formation and may be the downstream regulator of R-loop players. To comprehensively investigate the influence of FMRP on the RAD51, a downstream factor after R-loop formation, we assayed the RAD51 foci at the damage sites at different time points after light activation of KR. At the late stage of damage repair, the disassociation of RAD51 from damage sites was delayed (Fig. 5C). Consistently, we observed that the R-loop and m5C recognizer RAD52, which regulates RAD51 foci in TC-HR, was retained at damage sites in FMRP KO cells (Fig. 5D). These results indicate that the TC-HR process could be less efficient or prolonged in the FMRP KO cells.
FMRP Stimulates TET1-Mediated Demethylation of m5C RNA Modification both in Cells and In Vitro.
Although we have shown that FMRP pull-down cell fraction could demethylate the m5C RNA modification in DNA:RNA hybrids (Fig. 2 C and D), it is still unknown if FMRP is a cofactor of demethylase or a direct demethylase by itself for m5C RNA modification. FMRP does not contain any conserved motif with other known demethylases on RNA m6A or m5C, e.g., FMRP and FTO or ALKBH5 or TET family (35, 36). Furthermore, the demethylation activity of purified FMRP was not detected with the substrate of m5C-modified single-stranded RNA (ssRNA) or DNA:RNA hybrid under the in vitro demethylation assay (SI Appendix, Fig. 5 A and B). These results indicate that FMRP itself is not sufficient for demethylation reaction in vitro, although FMRP facilitates demethylation in cells. Since TET1 is the founding member of the TET family, which are the only established demethylases for m5C RNA modification (37), we tested if FMRP is a cofactor of TET1 activity (36, 38). In FMRP stably expressed cells identical to endogenous FMRP level (SI Appendix, Fig. 5C), we found that FMRP could pull down TET1 in cells (Fig. 6A), and benzonase treatment didn’t abolish their interaction (SI Appendix, Fig. 5D), indicating that the interaction between FMRP and TET1 is also not mediated by nucleotides. Moreover, in the ChIP-qPCR assay, TET1 was recruited to the TA-KR damage sites, as well as TRDMT1 and FMRP. Importantly, the recruitment of TET1 to damage sites was decreased in FMRP KO cells (Fig. 6B). As well as FMRP KD, TET1 KD delayed the m5C disappearance (Fig. 6C). Furthermore, the kinetics of m5C disappearance after TET1 KD in WT cells was similar with TET1 KD in FMRP KO cells (Fig. 6D). While TET1 KD decreased the HR repair efficiency as FMRP KD, double depletion of FMRP and TET1 didn’t further decrease the HR repair efficiency (Fig. 6E). These data collectively support that FMRP and TET1 are also epistasis in m5C demethylation. Together, the TRDMT1–FMRP–TET1 axis plays an important role to regulate the m5C dynamics for repair progression in cells. Unlike FMRP, the demethylation activity of TET1 on m5C RNA modification in the context of DNA:RNA hybrids was detected in in vitro demethylation assay analyzed by liquid chromatography (LC)/MS. Furthermore, in the in vitro reaction, the presence of FMRP largely enhanced the demethylation activity of TET1 on m5C RNA modification within DNA:RNA hybrids (Fig. 6F). Thus, FMRP is not only critical to recruiting TET1 to sites of damage in cells, but also is an enhancer for TET1 to demethylate m5C RNA modification within DNA:RNA hybrids.
Fig. 6.
FMRP stimulates TET1-mediated demethylation of m5C. (A) Interaction of FMRP and TET1 detected with Co-IP. Flp-in 293 cells were cotransfected with GFP-FMRP and Flag-TET1, treated with 1 mM H2O2 for 1 h before co-IP analysis with anti-Flag antibody. (B) ChIP-qPCR detected the DNA at the loci near the TRE region in cells transfected with TA-KR and Flag/Flag-TET1. (C and D) siTET1 or control siRNA pretreated U2OS–TRE or FMRP KO cells were transfected with TA-KR. The cells were exposed to light for KR activation for 25 min and followed with recovery for the indicated time before fixation. m5C foci frequency at TA-KR sites at the indicated time point is shown. (Scale bar: 10 μm.) (n = 3; 50 cells per replicate; mean ± SD.) (E) U2OS–TRE cells pretreated with siFMRP, siTET1, or control siRNA were transfected with Cas9-sgLMNA-mCherry and LMNA-mClover. The fraction of mClover-positive cells in mCherry-positive population was analyzed by flow cytometry (n = 3; mean ± SEM). (F) On the LC/MS system, the amount of each ribonucleoside was quantified by its integration area (in a rectangle) in the corresponding chromatogram (blue for m5C, red for guanosine). The relative abundance of m5C was calculated based on the amount of m5C relative to guanosine. Statistical analysis was done with the unpaired two-tailed Student t test. ns, not significant. **P < 0.01; ***P < 0.001; ****P < 0.0001.
FMRP Promotes Radioresistance and TC-HR in Tumor Cells Independently of BRCA.
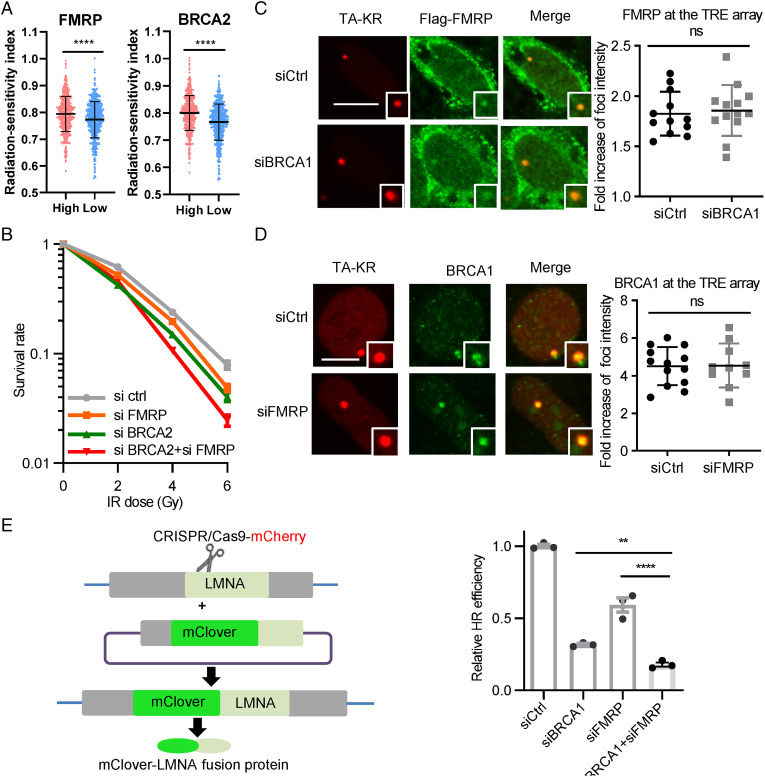
Given that HR is an essential repair mechanism for IR-induced DNA damage and FMRP deficiency leads to increased IR sensitivity, we analyzed the clinical relevance of FMRP expression and radio response in patients. Based on clinical information and RNA-seq gene expression in breast cancer downloaded from The Cancer Genome Atlas (TCGA), the radiosensitivity index (RSI) showed a significant difference in FMRP low and high groups. Low-FMRP-group patients correlated significantly with higher sensitivity and vice versa (Fig. 7A). The same correlation was observed in BRCA2 expression. However, double KD of FMRP and BRCA2 further increased the IR sensitivity compared to single KD of FMRP or BRCA2 (Fig. 7B), indicating that FMRP is not in the same epistasis group with BRCA2 in protecting cell survival after damage.
Fig. 7.
FMRP promotes radioresistance and TC-HR in tumor cells independently of BRCA. (A) Clinical and RNA-seq gene-expression data were downloaded from TCGA. RSI was calculated by rank-based linear regression algorithm. The median value of FMRP TPM was defined as the cutoff value to divide patients into high-expression and low-expression groups. (B) U2OS–TRE cells transfected with siFMRP, siBRCA2, or control siRNA were exposed to a gradient dose of IR. Survival rate was measured via colony-formation assay (n = 3; mean ± SEM). (C) U2OS–TRE cells pretreated with control siRNA or siBRCA1 were transfected with TA-KR and Flag-FMRP. The foci intensity of Flag-FMRP at TA-KR (recovered 1 h after light activation) is shown. (Scale bar: 10 μm.) (n ≥ 10; mean ± SD.) (D) U2OS–TRE cells pretreated with control siRNA or siFMRP were transfected with TA-KR and GFP-BRCA1. The foci intensity of GFP-BRCA1 at TA-KR (recovered 1 h after light activation) is shown. (Scale bar: 10 μm.) (n ≥ 10, mean ± SD.) (E) U2OS–TRE cells pretreated with siFMRP, siBRCA1, or control siRNA were transfected with Cas9-sgLMNA-mCherry and LMNA-mClover. The fraction of mClover-positive cells in the mCherry-positive population was analyzed by flow cytometry (n = 3, mean ± SEM). Statistical analysis was done with the unpaired two-tailed Student t test. **P < 0.01; ****P < 0.0001.
TC-HR was shown to be the predominant repair mechanisms for DSBs at transcriptionally active sites, in which the recruitment of RAD51 is essential for repair (5–8). Although BRCA1 and BRCA2 facilitate RAD51 loading in canonical HR, they are not essential for RAD51 recruitment at R-loops (7). Moreover, TRDMT1, which is required for the TC-HR to repair reactive oxygen species-induced DSBs, is not epistatic to BRCA1 and BRCA2 in the DSB repair (14). Here, the recruitment of FMRP was not affected by the loss of BRCA1 and vice visa (Fig. 7 C and D). Since FMRP affects RAD51 kinetics in the context of TC-HR (Fig. 5), we further explored if the role of FMRP is in the same epistasis with BRCA genes. The decrease of HR efficiency mediated by FMRP KD was further decreased in BRCA1 KD cells, reinforcing that FMRP promotes DSB repair in a BRCA1-independent manner (Fig. 7E). These results indicate that in the absence of BRCA functions, FMRP-dependent TC-HR could serve as a backup repair for cell survival.
Discussion
Various RNA modifications have emerged as important posttranscriptional regulators of a wide range of cellular processes (39), including the DNA damage response (40). m5C RNA modification was first identified in tRNA and rRNA (41). Recently, the presence of m5C in mRNA has been reported in mammals and plants (27, 42, 43). Our recent studies suggest that the RNA methyltransferase TRDMT1 generates m5C in DNA-damage-induced R-loops at sites of DSBs to promote TC-HR (14). m5C RNA modification has been shown to be elevated in many types of cancer (15–17), which is consistent with the high oxidative damage level in cancer cells (44), as oxidative damage and oxidative damage-derived DSBs at transcribed sites generate m5C RNA modification in R-loop (14). Moreover, the m5C at DNA damage sites is gradually resolved during DNA repair. Here, we identified FMRP as the interacting partner of TRDMT1. Depletion of FMRP delays the demethylation of m5C in R-loops. The persistence of m5C delays R-loop removal, which eventually impairs the completion of DNA repair (Fig. 8). While m5C RNA modification and R-loop promote the damage repair at an early stage, their persistence at the damage sites impairs the repair completion at a late stage of repair. FMRP, which contributes to m5C RNA-modification demethylation and following R-loop resolution at damage sites, is highly expressed in human breast cancer (45), hepatocellular carcinoma (46), and melanoma (47), reinforcing the importance of FMRP to facilitate m5C demethylation and to maintain genome stability of cancer cells.
Fig. 8.
Working model of TRDMT1–FMRP–TET1-mediated m5C regulation in TC-HR.
FMRP is largely a cytoplasmic RNA-binding protein that represses translation of the proteins in the metabotropic glutamate receptor pathway, contributing to FXS pathogenesis. We found that FMRP is recruited to DNA damage sites in transcribed regions via association with TRDMT1, which is consistent with the previous report that FMRP was recruited onto chromatin in response to replication-stress-induced DNA damage (32, 48). We show that FMRP is an m5C reader in the context of RNA:DNA hybrids and facilitates the m5C demethylation at DNA damage sites, thereby contributing to DSB repair through HR and to the maintenance of genome stability. Similarly, FMRP has been recently reported to prevent R-loop accumulation and decrease DSB formation during replication stress (49). Although we compared the binding of purified FMRP to m5C-modified RNA:DNA hybrid and unmodified hybrid using an electrophoretic mobility shift assay experiment, we didn’t see preferential binding of FMRP to m5C-modified hybrid. This is not consistent with the result that m5C-modified RNA:DNA hybrids captured FMRP in cell lysates more efficiently. We anticipate that posttranslational modification may be required for FMRP to perform the m5C reader role. The previous reports of increased DNA damage and apoptosis in Fragile X patients also support the role of FMRP in protecting genome stability (50, 51). Apart from regulating m5C demethylation, FMRP was also found to preferentially bind m6A-modified RNA and modulate its stability or nuclear export (52, 53). RNA m6A was primarily induced by UVC-generated DNA damage via the METTL3–METTL14 complex (40). Interestingly, the METTL3–m6A-YTHDC1 axis was just reported to promote accumulation of DNA–RNA hybrids at DSB sites, which then recruit RAD51 and BRCA1 for HR repair (54). The involvement of FMRP in the regulation of m5C and m6A induced by various DNA damage suggests that the writer and readers of these RNA modifications must be intricately regulated in different contexts. Protein complexes that contain both ubiquitin ligases and deubiquitinases have been shown to localize to DNA damage sites (55). FMRP is recruited by the m5C RNA-modification writer, TRDMT1, which also suggests that the formation and removal of m5C during DNA repair are coordinated.
The removal of m5C involves the oxidation of m5C, which is catalyzed by the demethylase. TET proteins belong to the Fe(II)-dependent and 2-oxoglutarate–dependent dioxygenases family and could induce oxidation of m5C in DNA (56). Recently, the function of TET proteins to oxidize m5C RNA modification has been reported (36, 57). Our result confirmed that TET1 responds to DNA damage and catalyzes the demethylation of m5C RNA modification at damage sites. Importantly, our data suggest that TRDMT1–FMRP–TET1 factors function in a temporal order to coordinate the dynamic m5C in cells. The fact that FMRP interacts with TET1 and stimulates its demethylase activity of TET1 further suggests that these proteins function as a dynamic complex to regulate m5C at damage sites. We also showed that although TRDMT1 is required for the damage response of FMRP to sites of m5C, chromatin- and RNA-binding domains of FMRP are also required for m5C demethylation. The binding of FMRP to chromatin and DNA:RNA hybrids may cooperate with TET for demethylation of m5C (Fig. 8). Therefore, the m5C in DNA damage-induced R-loops is dynamically regulated by the TRDMT1–FMRP–TET1 axis.
While R-loops are a source of genomic instability, the R-loops induced by DNA damage have positive roles in DNA repair (7, 58, 59). R-loop-initiated TC-HR plays an important role in maintaining cancer-cell survival. TRDMT1–FMRP–TET1-mediated m5C regulation promotes TC-HR, which also serves as a backup repair for cancer-cell survival. Thus, targeting enzymes in TC-HR in cancer cells provide a promising approach to kill cancer cells of high oxidative stress level and/or sensitize tumors to radiation and other DNA-damage drugs. In these DNA-damage-induced R-loops, m5C RNA modification serves as a signal to promote the early phase of DNA repair. However, as m5C RNA modification increases the stability of RNA (60), the timely resolution of R-loops in the late phase of DNA repair requires the removal of m5C RNA modification. Depletion of FMRP doesn’t affect the recruitment of the DNA-repair proteins that we examined, but it delays the clearance of m5C and R-loops at the DNA-damage sites, which impairs the completion of DNA repair. As the demethylation of m5C is a multistep biochemical process (36) and many proteins may go on and off the DNA-damage-induced R-loops during the process, future studies are needed to fully understand how m5C RNA-modification demethylation occurs during HR and how R-loop stability is regulated by this dynamic RNA modification.
Materials and Methods
U2OS and Flp-in 293 cells were cultured in Dulbecco’s modified Eagle medium (catalog no. 12-604F, Lonza) with 10% (volume [vol]/vol) fetal bovine serum at 37 °C with 5% CO2. The U2OS–TRE cells used for the DART system have been described in previous articles (28). For plasmid and siRNA transfection, Lipofectamine 2000 and Lipofectamine RNAiMax (Invitrogen) were used following the manufacturer’s standard protocol, respectively. The siRNA for TRDMT1 was purchased from Invitrogen (siRNA ID s4219; catalog no. 4392420). The siRNA for FMRP was purchased from Dharmacon (catalog no. L-019631-00-0005); the sequences of siRNA targeting FMRP 3′ UTR are 5′- GUACUGAGCAGUGAUAUUCdTdT-3′ and 5′- GAAUAUCACUGCUCAGUACdTdT-3′. Other siRNAs include siTET1 (catalog no. AM16708, assay ID:147892, Thermo), siBRCA1 (catalog no. L-003461-00, Dharmacon), and siBRCA2 (catalog no. GS675, Qiagen). Antibodies used in this study are summarized in SI Appendix, Table 1. Other methods used in the study are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grants GM118833 (to L.L.) and GM076388 and CA197779 (to L.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116251119/-/DCSupplemental.
Data Availability
The LC/MS raw files and search results were deposited into the MassIVE data repository with accession code MSV000085313.
References
- 1.Crossley M. P., Bocek M., Cimprich K. A., R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A., ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Marteijn J. A., Lans H., Vermeulen W., Hoeijmakers J. H., Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15, 465–481 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Aymard F., et al. , Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keskin H., et al. , Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teng Y., et al. , ROS-induced R loops trigger a transcription-coupled but BRCA1/2-independent homologous recombination pathway through CSB. Nat. Commun. 9, 4115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei L., et al. , DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 112, E3495–E3504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Francia S., et al. , Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W., et al. , A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Michelini F., et al. , Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S., et al. , RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination. Cell 184, 1314–1329 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Zhang C., et al. , METTL3 and N6- methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol. Cell 79, 425–442 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Chen H., et al. , m5C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat. Commun. 11, 2834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang W., et al. , Determination of DNA and RNA methylation in circulating tumor cells by mass spectrometry. Anal. Chem. 88, 1378–1384 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Chen X., et al. , 5-Methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q., Zheng Q., Yu X., He Y., Guo W., Overview of distinct 5-methylcytosine profiles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues. J. Transl. Med. 18, 245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman A. K., Monteiro A. N., Phosphatases in the cellular response to DNA damage. Cell Commun. Signal. 8, 27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos F., Villoria M. T., Alonso-Rodríguez E., Clemente-Blanco A., Role of protein phosphatases PP1, PP2A, PP4 and Cdc14 in the DNA damage response. Cell Stress 3, 70–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler L. R., et al. , The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 31, 3918–3934 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson S. P., Durocher D., Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Feng Y., et al. , Fragile X mental retardation protein: Nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 17, 1539–1547 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown V., et al. , Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Penagarikano O., Mulle J. G., Warren S. T., The pathophysiology of fragile X syndrome. Annu. Rev. Genomics Hum. Genet. 8, 109–129 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Cheng J. X., et al. , RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat. Commun. 9, 1163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amort T., et al. , Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 18, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan L., et al. , Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 42, 2330–2345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siomi H., Choi M., Siomi M. C., Nussbaum R. L., Dreyfuss G., Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77, 33–39 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Chen E., Joseph S., Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie 114, 147–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dockendorff T. C., Labrador M., The Fragile X protein and genome function. Mol. Neurobiol. 56, 711–721 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Alpatov R., et al. , A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell 157, 869–881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janicki S. M., et al. , From silencing to gene expression: Real-time analysis in single cells. Cell 116, 683–698 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen H. D., et al. , Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65, 832–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue Y., Liu J., He C., RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu L., et al. , Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 136, 11582–11585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H., et al. , TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc. Natl. Acad. Sci. U.S.A. 110, 11994–11999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delatte B., et al. , RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Gilbert W. V., Bell T. A., Schaening C., Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang Y., et al. , RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trixl L., Lusser A., The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 10, e1510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X., et al. , 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27, 606–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David R., et al. , Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29, 445–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liou G. Y., Storz P., Reactive oxygen species in cancer. Free Radic. Res. 44, 479–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucá R., et al. , The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol. Med. 5, 1523–1536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y., et al. , Identification of differential expression of genes in hepatocellular carcinoma by suppression subtractive hybridization combined cDNA microarray. Oncol. Rep. 18, 943–951 (2007). [PubMed] [Google Scholar]
- 47.Zalfa F., et al. , The fragile X mental retardation protein regulates tumor invasiveness-related pathways in melanoma cells. Cell Death Dis. 8, e3169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W., et al. , A feed-forward mechanism involving Drosophila fragile X mental retardation protein triggers a replication stress-induced DNA damage response. Hum. Mol. Genet. 23, 5188–5196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty A., et al. , Replication stress induces global chromosome breakage in the fragile X genome. Cell Rep. 32, 108179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon S. J., et al. , Cellular stress-induced up-regulation of FMRP promotes cell survival by modulating PI3K-Akt phosphorylation cascades. J. Biomed. Sci. 18, 17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakraborty A., et al. , Fragile X mental retardation protein regulates R-loop formation and prevents global chromosome fragility. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/601906v2.article-info (Accessed 24 October 2020).
- 52.Edens B. M., et al. , FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang F., et al. , Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C., et al. , METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol. Cell 79, 425–442 (2020). [DOI] [PubMed] [Google Scholar]
- 55.He M., et al. , The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 6, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo J. U., Su Y., Zhong C., Ming G. L., Song H., Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen Q., et al. , Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 554, 123–127 (2018). [DOI] [PubMed] [Google Scholar]
- 58.García-Muse T., Aguilera A., R loops: From physiological to pathological roles. Cell 179, 604–618 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Ouyang J., Lan L., Zou L., Regulation of DNA break repair by transcription and RNA. Sci. China Life Sci. 60, 1081–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y., et al. , RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell 75, 1188–1202 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The LC/MS raw files and search results were deposited into the MassIVE data repository with accession code MSV000085313.