Significance
There are various diseases caused by protein aggregation such as Alzheimer’s, Parkinson’s, and Huntington’s diseases. From the diversity in the fibril structure, aggregation is expected to occur via heterogeneous pathways. However, characterization of this heterogeneity is extremely difficult because it requires following individual fibril formation in a mixture from early oligomerization stages. In this work, we investigated aggregation of the 42-residue isoform of amyloid β (Aβ42) using single-molecule fluorescence imaging and deep learning. We could track the growth of individual fibrils, which allows for a quantitative description of heterogeneous fibril formation and discovery of a new fibril nucleation mechanism. Further characterization of heterogeneity involving Aβ42 will be important for better understanding the disease mechanism.
Keywords: single-molecule spectroscopy, amyloid, protein aggregation, deep neural network, fluorescence lifetime imaging
Abstract
Polymorphism in the structure of amyloid fibrils suggests the existence of many different assembly pathways. Characterization of this heterogeneity is the key to understanding the aggregation mechanism and toxicity, but in practice it is extremely difficult to probe individual aggregation pathways in a mixture. Here, we present development of a method combining single-molecule fluorescence lifetime imaging and deep learning for monitoring individual fibril formation in real time and their high-throughput analysis. A deep neural network (FNet) separates an image of highly overlapping fibrils into single fibril images, which allows for tracking the growth and changes in characteristics of individual fibrils. Using this method, we investigated aggregation of the 42-residue amyloid-β peptide (Aβ42). We demonstrate that highly heterogeneous fibril formation can be quantitatively characterized in terms of the number of cross-β subunits, elongation speed, growth polarity, and conformation of fibrils. Tracking individual fibril formation and growth also leads to the discovery of a general nucleation mechanism (termed heterogeneous secondary nucleation), where a fibril is formed on the surface of an oligomer with a different structure. Our development will be broadly applicable to characterization of heterogeneous aggregation processes of other proteins.
Protein aggregation is implicated as the cause of pathology in various diseases (1–3). During aggregation, proteins form amyloid fibrils, long fibers with parallel (or antiparallel) β-sheet structures (cross-β structure). Interestingly, there are variations in the fibril structures (4–6), depending on various factors such as aggregation conditions. This polymorphism indicates that the entire aggregation process, including oligomerization, should be heterogeneous. Understanding this diversity can be clinically very important, as revealed by recent studies that found correlations between the disease/clinical phenotype and fibril structures (7–10). However, biophysical and biochemical characterizations of the parallel aggregation pathways are very difficult, and a lack of quantitative experimental results on heterogeneity prevents building comprehensive aggregation models. Single-molecule spectroscopy can be an effective tool to probe heterogeneity because molecular species can be observed one at a time without separation (11). This technique has the potential to identify toxic species. In this study, we combine single-molecule fluorescence lifetime imaging (FLIM) and image analysis using a newly developed deep neural network (FNet) to interrogate the aggregation process of proteins.
For a complete and high-throughput individual fibril analysis of the entire dataset it is necessary to identify and separate individual fibrils and follow their changes over time. However, as the aggregation proceeds, existing fibrils grow and new fibrils appear, which leads to overlap of many fibrils. In many cases, it is not possible to split the fluorescence intensity of overlapping regions into different fibrils in a single image. This problem can be solved by using FNet, which is trained to distinguish the growth of existing individual fibrils and appearance of new fibrils by tracking the history of the entire FLIM movies. Starting from the easiest problem, the initial frames where fibrils (i.e., oligomers) do not overlap, the deep neural network builds up information to track, characterize, and separate overlapping fibrils in later frames iteratively.
We emphasize that FNet and the experimental and analysis methods presented in this study are generally applicable to quantitative characterization of any amyloid fibril formation. As a first step, we applied this method to studying aggregation of the 42-residue amyloid-β (Aβ) peptide (Aβ42). Aβ is a peptide fragment consisting of 37 to 43 amino acid residues, which is produced by successive proteolytic cleavages of the amyloid precursor protein (APP) (12). Its aggregation to form fibrils that are found in brain tissue is one of the key characteristics of Alzheimer’s disease. Despite tremendous effort to understand aggregation of Aβ to form oligomers and fibrils, experimental results vary widely and there is no consensus on the model for these processes (13). For example, diverse morphologies and elongation of individual fibrils of Aβ and their fragments have been studied using total internal reflection fluorescence (TIRF) microscopy (14–17) and atomic force microscopy (18–22) at various conditions. However, the fibril growth rate differs by several orders of magnitude. One of the difficulties in studying Aβ may result from its hydrophobicity because it is a part of the transmembrane domain of APP. Aβ also interacts with many proteins either specifically or nonspecifically as a monomer and various oligomeric forms (23). A more fundamental reason for the difficulty may be the heterogeneity of the aggregation process, which can be very sensitive to the environment and experimental conditions (24). Various simulation studies have shown the diversity of aggregation pathways depending on how and when fibril-like conformations are formed (25, 26), which can be as early as small oligomers including monomers (27–32).
Our method provides direct experimental visualization of various heterogeneous characteristics of individual fibril formation and growth and their high-throughput analysis. Previous studies of Aβ fibril formation relied mostly on manual picking of individual fibrils, which led to the analysis of a small fraction of the data (up to 200 to 300 fibrils). Since our method analyzes the entire data and extracts individual fibril images (7,050 fibrils), quantitative information such as length, fluorescence intensity, and lifetime can be determined accurately for further analysis with better statistics and for the detection of rare events. We show that diverse Aβ42 fibrils can be categorized into three groups based on the fluorescence lifetime (i.e., conformation) and the monomer density in fibrils (i.e., number of fibril subunits), each of which consists of rapid-growing and nongrowing fibrils. In addition, most fibrils grow in one direction (polarized growth). We also identify the growth of fibrils from oligomers with different structures, which we term “heterogeneous secondary nucleation,” a previously unknown type of nucleation mechanism in Aβ42 aggregation. Importantly, these heterogeneous characteristics vary over different experiments at the same condition, which explains highly diverse results of previous experimental studies.
Results
FLIM of Aβ42 Aggregation.
To monitor formation of individual fibrils using FLIM, we incubated 500 nM of Aβ42 labeled with Alexa 594 at the N terminus (SI Appendix, Fig. S1) in a polyethylene glycol (PEG)-coated glass surface chamber (Fig. 1A). We verified that the aggregation features of dye-labeled Aβ42 are similar to those of unlabeled Aβ42 (SI Appendix, Figs. S2 and S3). First, the aggregation time is similar between labeled and unlabeled Aβ42 (SI Appendix, Fig. S2 A and B). In addition, 5% addition of the sonicated sample after aggregation of labeled Aβ42 eliminates the lag phase, indicating that dye-labeled Aβ42 can form aggregation seeds similar to unlabeled Aβ42 (SI Appendix, Fig. S2C). Furthermore, electron microscope images of fibrils of Alexa 594-labeled Aβ42 are indistinguishable from those of unlabeled Aβ42 (SI Appendix, Fig. S3). Together, these results strongly suggest that fluorophores interfere minimally with aggregation.
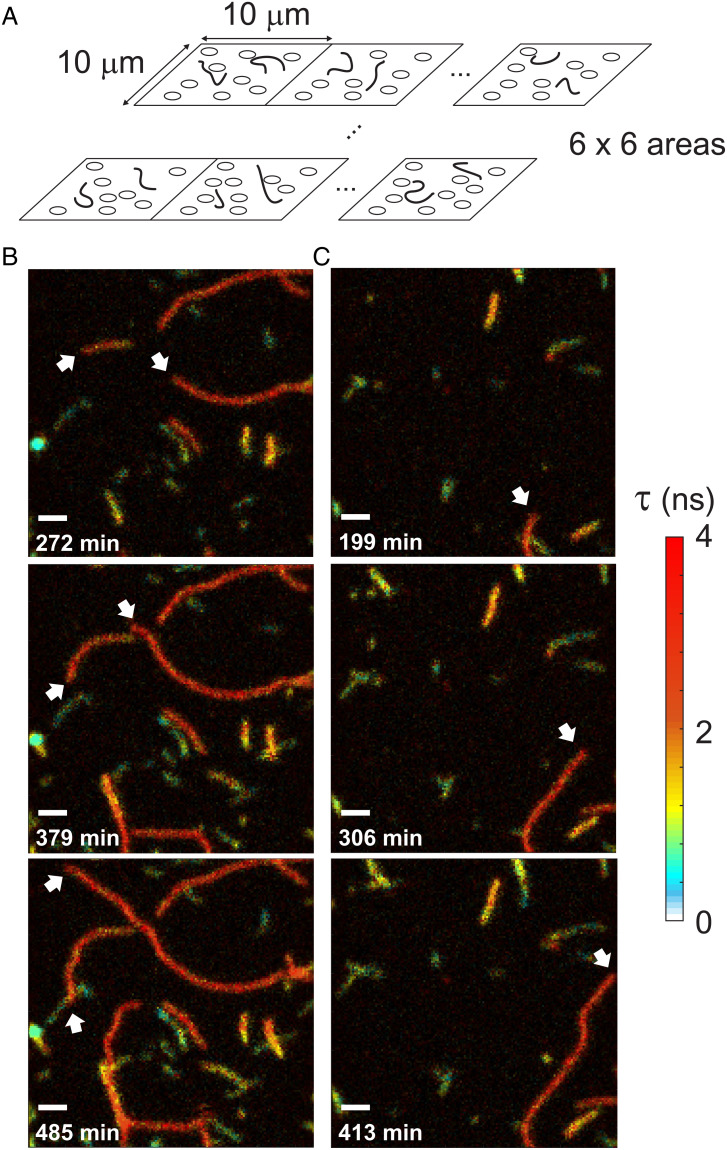
Fig. 1.
Monitoring of Aβ42 aggregation by FLIM. (A) A PEG-coated surface was incubated with 500 nM of Alexa 594-Aβ42. Immediately after starting incubation at room temperature, 16 (4 × 4) – 36 (6 × 6) 10- × 10-μm2 regions were sequentially scanned. After completing one set of scans, the stage was moved back to the first region and the scans were repeated. (B and C) Three snapshots of Movie S1 (B) and Movie S2 (C). Fast growing ends are indicated by white arrows. Incubation times are indicated on the lower left corner of each image. Fluorescence lifetime (τ) images were masked by count rates and smoothed using total variation denoising (36). (Scale bars: 1 μm.)
Immediately after the start of incubation of Alexa 594-labeled Aβ42 at room temperature, 16 (4 × 4) – 36 (6 × 6) 10- × 10-μm2 regions of the sample were sequentially scanned. After completion of one set of scans, the stage was moved back to the first region and scans were repeated. Scanning 36 regions takes about 50 min, for example, 24 repetitions produce 36, 20-h-long movies (∼50 min frame rate) of fluorescence intensity and lifetime of oligomers and fibril elongation (Movies S1–S3; see also Movies S4–S8 for the entire aggregation movies of five different experiments). Fig. 1 B and C show snapshots of Movies S1 and S2 that capture various features of large oligomer formation and fibril elongation. Since Alexa 594-labeled Aβ42 is not immobilized, all detected short and long fibrils are those that sedimented from the solution. The appearance of fibrils on the surface does not result from permanent sticking because some fibrils suddenly disappear (dissociation from the surface, 37% of the fibrils that appeared during the measurement dissociated before the end of the measurement) and small movements of fibrils are observed over time.
The movies and snapshots in Fig. 1 show that fibril elongation is highly heterogeneous. There are relatively short fibrils, which do not grow or grow slowly (green/blue). These molecules could be large nonfibrillar oligomers or protofibrils (33, 34). In addition, there are long fibrils that elongate much faster (orange and red). Overall, the fluorescence lifetime is shorter than that of Alexa 594 of the monomer (3.67 ns), which can be explained by self-quenching of fluorescence when dyes are placed close to one another (35). Interestingly, the fluorescence lifetimes of short, slowly growing fibrils (green and blue) are shorter than those of long, fast-growing fibrils (orange and red). Different lifetimes indicate that the structure (i.e., arrangement of monomers) is different for short, slowly growing and long, fast growing fibrils.
Architecture of FNet.
More detailed and quantitative aggregation features can be obtained from characterization of individual fibrils. For the analysis of individual fibrils, it is necessary to identify and separate fibrils and follow their changes over time. However, as fibrils grow they start to overlap, and in many cases it is unclear how to split the changes of photon counts in overlapping regions into different fibrils in a single image (SI Appendix, Fig. S4B).
A state-of-the-art method for semantic segmentation of touching and overlapping biological objects such as cells is a mixed two-dimensional (2D)–3D deep neural segmentation network using object bounding boxes, which are located at specific reference points (37). In the case of overlapping fibrils, defining such reference points and bounding boxes is impossible due to the simple shape of fibrils (there is no structure like a nucleus in a cell as a reference point). Therefore, instead of segmenting fibrils directly from an image of a single frame, we exploited temporal information by comparing two consecutive image frames. We assume that there exists a correct fibril segmentation in the previous image frame and using this segmentation it is possible to predict the segmentation in the next frame iteratively. The first assumption is always true, because at the beginning of an experiment only a few small oligomers are present and there is no overlap. To predict the next frame segmentation, we first tried a classification-based image segmentation deep neural network, U-Net (38), which shows good performances in segmentation of biological objects. However, U-Net showed very poor performance in segmentation of overlapping regions of fibrils (Discussion). Therefore, we developed and trained a neural network (FNet) for the photon count estimation of highly overlapping transparent biological objects (Fig. 2).
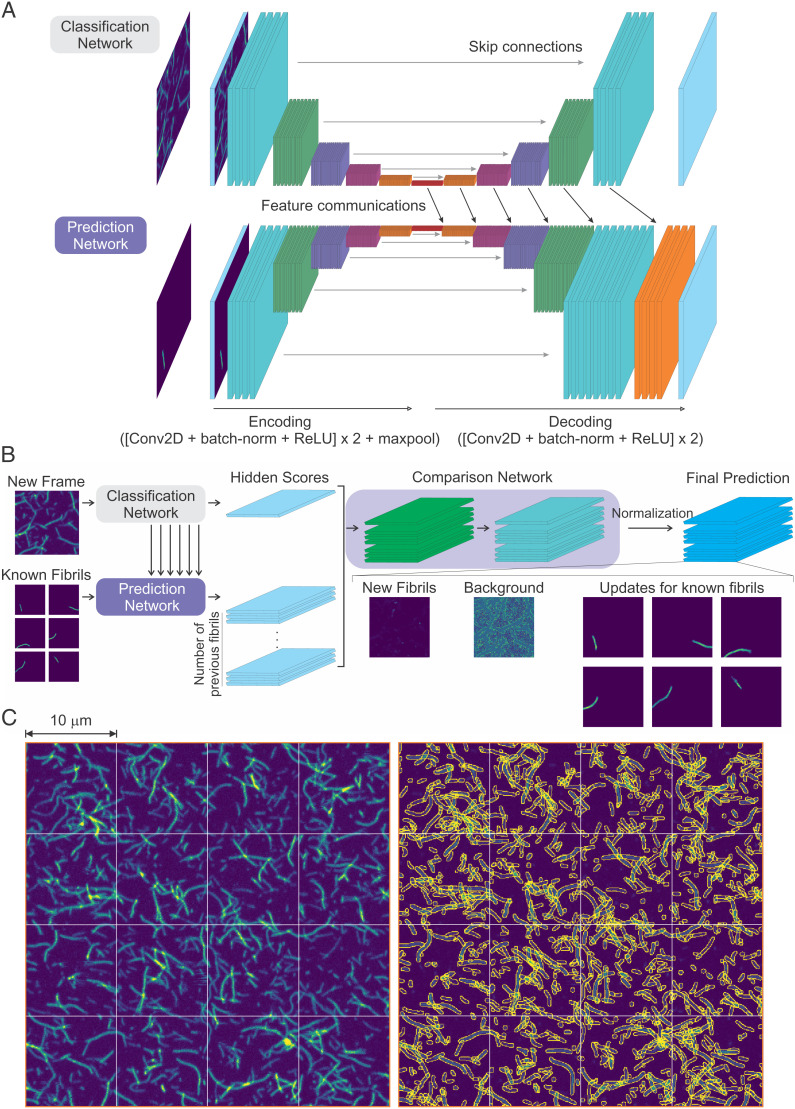
Fig. 2.
Deep neural network (FNet) architecture. (A) Classification network and prediction network. Each network takes an image as an input. The input of the classification network is a new frame image. The background prediction network uses a background prediction image of the previous frame as an input, and the growth prediction network uses prediction images of individual fibrils of the previous frame (known fibrils) as an input. The prediction network connects the hidden features of the classification network to generate predictions (feature communications). (B) The output of the classification network and the prediction networks are compared to generate the final prediction. The comparison network has two applications of a convolution, a batch-normalization and an ReLU activation layer, followed by a convolution and a PReLU activation layer. The output of PReLU activation is normalized pixel by pixel to make the number of photons in each pixel of the summed output image equal to that of the original input image of the new frame. This results in an image of new fibrils, a background image, and the updated images of the known fibrils from the previous frame (see Fig. 3A). (C, Left) Highly overlapping fibril images from Experiment 1. The 4 × 4 (10- × 10-μm2) scanned images are the last frames of the experiment (26-h incubation). (C, Right) Segmentation of fibrils in the same image data on the left side using FNet. Peripheries of individual fibrils are colored in yellow. The fibrils touching the boundaries of 10- × 10-μm2 areas were merged with the continuing fibrils in the adjacent areas.
This neural network consists of four subnetworks: 1) classification network, 2) growth prediction network, 3) background prediction network, and 4) comparison network (Fig. 2). The overall information flow is as follows. First, the classification network encodes features from a new input image and decodes them into feature maps of different resolution. The growth prediction network encodes features of individual known fibrils from the previous image and then decodes them together with the feature maps of the same resolution from the classification network which contains the information of the new image. The background prediction network has the same structure as that of the prediction network, but it takes the previous background image as an input and uses independent weights. The classification, the growth prediction, and the background prediction networks generate single feature image outputs which stand for their prediction power for how many photons of each pixel result from known fibrils, new fibrils, and background, respectively. Then, the comparison network (Fig. 2B) compares relative prediction powers from the prediction networks and generates photon count images of known fibrils, newly appearing fibrils, and background (codes are available at https://github.com/hoisunglab/FNet). See Materials and Methods and SI Appendix, Fig. S4 for more details of the networks, training, validation, and performance of FNet on an image of extremely highly overlapping fibrils, in which it is impossible to separate fibrils manually. The segmentation results of highly overlapping fibrils from the last frames of the experiments are shown in Fig. 2C and SI Appendix, Fig. S5 (Movies S4–S8 for the entire aggregation movies and the segmentation of fibrils of five different experiments).
Individual Fibril Analysis.
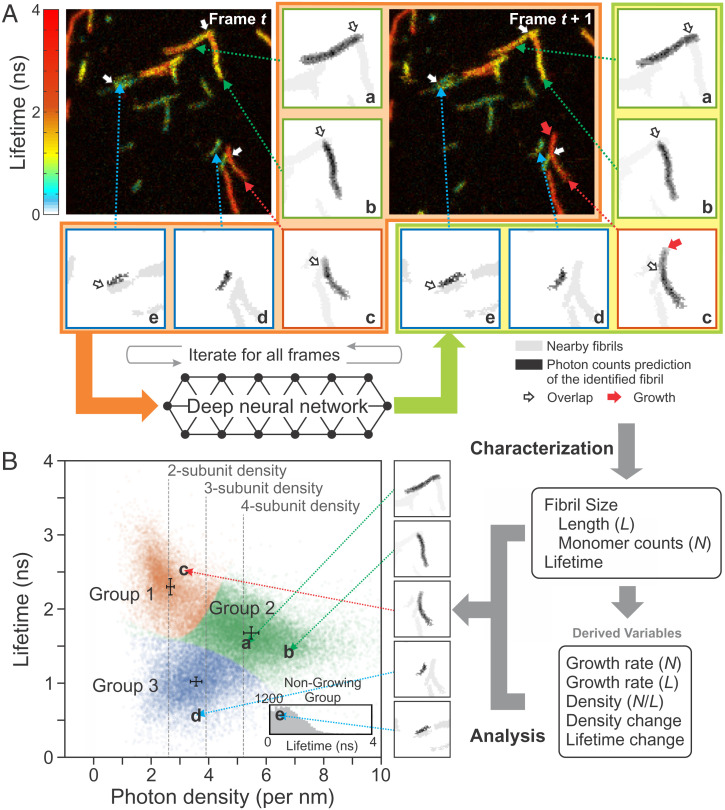
Using FNet, we analyzed 3,893 image frames from five experiments and characterized 116,504 images of 7,050 individual fibrils. Figs. 3 and 4 illustrate separation of fibrils and quantitative characterization of the heterogeneity of aggregation. Five fibrils, a through e, are shown as examples in Fig. 3. After separation, the lengths of the fibrils are measured and the number of photons and fluorescence lifetimes are calculated from the photons and their delay times (i.e., photon arrival times after pulsed laser excitation) collected in those fibril regions. This results in a 2D plot of fluorescence lifetime vs. photon density (i.e., number of photons per unit length), which is proportional to the monomer density in fibrils. As seen in Movies S1–S3 and in Fig. 1, there are large variations in the fluorescence lifetime and intensity. By clustering these 2D data, we classified fibrils longer than 500 nm, which is about twice the size of the point spread function (PSF, 2σ = 247 nm), into three groups using Gaussian mixture models (39) (see Materials and Methods). Based on their lifetimes, fibrils in groups 1, 2, and 3 appear in red (fibril c), yellow (fibril a, b), and green/blue (fibril d, e), respectively, in the image (Fig. 3A). Using the photon density and fluorescence lifetime, it is also possible to estimate the number of subunits in a fibril (i.e., polymorphism) from the length of a fibril and the number of monomers in it (see Materials and Methods for the calculation). Fig. 3B shows that group 1, 2, and 3 fibrils consist of two, four, and three subunits, respectively. In addition, there are nongrowing fibrils (fibril e in Fig. 3A), the majority of which are shorter than the measurable size (i.e., twice the PSF size). Since the density cannot be defined, these fibrils (or oligomers) are classified based on their lifetimes (group 1: τ > 2.04 ns, group 2: 1.35 ns < τ ≤ 2.04 ns, group 3: τ ≤ 1.35 ns) (Fig. 3 B, Inset).
Fig. 3.
Flowchart of individual fibril analysis. (A) Identification and separation of individual fibrils using FNet. Individual fibril images of frame t and the new image of frame t + 1 are processed in FNet (images inside the orange area), which results in the updated individual fibril images (inside the yellowish-green area) and new fibrils. The procedure is repeated for the entire time series of images of the same region. Five individual fibrils a through e with different characteristics are shown in the subregions on the right and bottom sides of the image as examples. Each subregion consists of only one fibril (black), but nearby fibrils (gray) are also shown for comparison. Red, green, and blue squares of the subregion indicate fibril groups 1, 2, and 3 in B, respectively. White arrows indicate the overlapping regions of fibrils before separation. Red arrow in the subregion of fibril c at t + 1 indicates the growth of a fibril. (B, Right) After separation, fibrils in each image are characterized by various properties: length (L) measured on the fibril image, fluorescence lifetime, and size in terms of the number of monomers (N), which is calculated using the fluorescence intensity and lifetime. Using these parameters, other time-dependent variables are extracted: growth rate in terms of the length and size (number of monomers) and changes of the monomer density and lifetime of fibrils. (B, Left) Using the variables from these characterizations, a 2D plot of fluorescence lifetime vs. photon density (number of photons per unit length) is constructed. The distribution of individual fibrils with a measurable length (i.e., twice larger than the PSF size) are clustered into three groups (three different colors). The error bars located at the center of each cluster indicate the shot noise (SD) of the density and lifetime determined from the smallest number of photons of the fibrils in the experiment (412, 436, and 323 photons for groups 1, 2, and 3). The much smaller error compared to the width of the distribution of all three groups may indicate additional fibril heterogeneity. Nongrowing fibrils (SI Appendix, SI Text for the definition) are characterized only by the fluorescence lifetime (Inset). Letters a through e and arrows indicate the locations of the five fibrils in the plot.
Fig. 4.
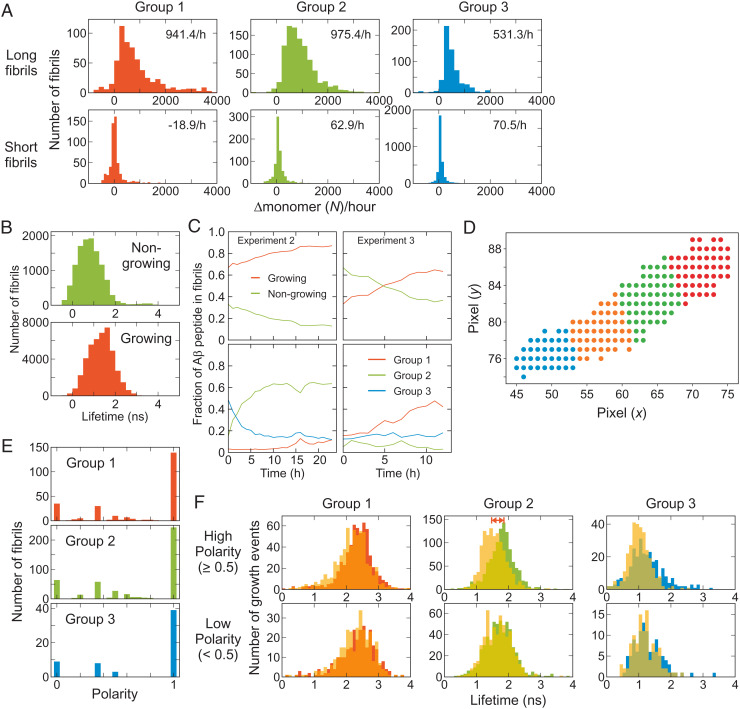
Individual fibril analysis. (A) Growth rate of long and short fibrils of the three fibril groups. (B) Fluorescence lifetime distributions of nongrowing and growing fibrils. (C) Time-dependent changes of the fractions of Aβ peptide incorporated in nongrowing and growing fibrils (Upper) and those incorporated in the three fibril groups of growing fibrils (Lower) from Experiments 2 and 3. SI Appendix, Fig. S9 shows the results from other experiments. (D) Fibril fragmentation (see Materials and Methods for the details). (E) Polarized growth of fibrils. Fibril growth polarity is calculated as the difference of the growing events of the two ends divided by the total growing events (see Materials and Methods for the definition [Eq. 3]). (F) Comparison of the fluorescence lifetimes of the fragments at the growing (solid bars) and nongrowing ends (yellow translucent bars) of the three groups of fibrils with the high (Upper) and low (Lower) polarity growth. The average lifetimes are different for the growing and nongrowing ends of group 2 fibrils with high polarity (double red arrow).
Fig. 4 A–C shows detailed statistics of fibril elongation analysis that reveals highly heterogenous aggregation features even in the same fibril groups. Some of long fibrils touch the boundaries of 10- × 10-μm2 image areas (see Fig. 1). We identified continuing fibrils in the images of the adjacent regions and combined information of those fibrils (see Materials and Methods, Fig. 2C, and SI Appendix, Fig. S5 for stitched fibrils in adjacent image regions). We first compared the average growth rate (increase of the number of monomers per hour) of the long (i.e., measurable) and short (i.e., unmeasurable) fibrils of different groups (Fig. 4A). The growth rate of the short fibrils is very low for all three groups, indicating these fibrils are classified as mostly nongrowing fibrils. In addition, most short fibrils belong to group 3 with short fluorescence lifetimes (compare the height of the histograms in Fig. 4A), which is consistent with the observation from the example movies (Fig. 1). Among the long fibrils, the growth rate of group 3 is also the lowest. The growth rates of group 1 and 2 are similar, but the number of subunits of group 2 is twice as many as that of group 1 (Fig. 3B). Therefore, the apparent growing speed of group 1 in terms of length in movies looks twice as fast as that of group 2. In addition, there is a long tail in the group 1 distribution (Fig. 4A), resulting from extremely fast-growing fibrils as seen in Fig. 1 (fibrils colored in red). The variation of the growth rate distribution of growing fibrils over different experiments is not large for all three groups (SI Appendix, Fig. S6A). The growth trajectories of individual fibrils (SI Appendix, Fig. S6B) show that the growth rate vary largely over time, which is consistent with stop-and-go behavior observed in previous studies (14, 16, 17, 19–21). The maximum size of fibrils that is reached at the end of the experiments ranges widely between 1,000 and 50,000 monomers (SI Appendix, Fig. S7).
We define nongrowing fibrils as the fibrils that do not grow to the measurable size (∼500 nm, twice the PSF size) at the end of experiments (24 h), the growth rate of which corresponds to 132 monomers per h (79.2 photons per h). Fig. 4 B and C compare the characteristics of growing and nongrowing fibrils. First, the average fluorescence lifetime of nongrowing fibrils is shorter than that of the growing fibrils, consistent with the visual characterization of the movies (Fig. 1). The distribution varies depending on the sample batches. SI Appendix, Fig. S8 shows the comparison of the fluorescence lifetimes of nongrowing and growing fibrils from five experiments with two different sample batches (i.e., two different expressions of Aβ42 and labeling) (Experiments 1 and 2 are from batch 1 and 3 to 5 are from batch 2).
In addition to this overall distribution and classification, it is important to characterize how these different fibril groups change over time as aggregation proceeds. Fig. 4C compares the evolution of the population of nongrowing and growing fibrils of Experiments 2 and 3. SI Appendix, Fig. S9 shows the results of all five experiments. In general, the fraction of growing fibrils increases because of their higher growth rates. However, at the beginning of aggregation, the fraction of the growing fibrils varies widely between 20 and 80%. The changes of the fraction of different fibril groups are more diverse. Overall, group 3 is dominant in the nongrowing fibril group (SI Appendix, Fig. S9B). On the other hand, for growing fibrils, group 3 dominates at the beginning of aggregation (except for Experiment 3), but other groups catch up at later times (SI Appendix, Fig. S9C). In batch 1 (Experiments 1 and 2), the fraction of group 1 fibrils is very low over the entire time course, whereas both group 1 and 2 increase with time in batch 2 data.
Polarized Growth and Heterogeneity within Fibrils.
Fig. 1B shows a fibril (indicated by an arrow on the left side) that grows to only one direction. This polarized fibril growth has been observed experimentally (14, 16, 19, 40) and in molecular dynamics simulations (41). To obtain statistics of unidirectional and bidirectional fibril growth, we divided fibrils into short fragments (10 pixels in length) (Fig. 4D) and tracked the growth of individual fibrils. Fig. 4E shows that the fibril growth is predominantly unidirectional in all three fibril groups. This result indicates that fibrils may not be structurally homogeneous. Therefore, we compared 2D fluorescence lifetime-density plots of the entire fibrils of each group and their fragments. As shown in SI Appendix, Fig. S10, most fragments belong to the same group of the fibrils, indicating fibrils are predominantly homogeneous. The penetration of the distributions of fragments into other groups results from larger errors in the determination of the lifetime and density values from smaller number of photons. However, when we compare the lifetimes of the growing and nongrowing end fragments, there is a clear difference for the group 2 fibrils with high polarity (≥0.5) (Fig. 4F, indicated by double red arrow). The lifetime of the nongrowing end is shorter than that of the growing end, implying that the character of a different group (i.e., group 3) is mixed in the nongrowing end fragment. In addition, we observed fibrils with mixed density and fluorescence lifetime characteristics in more complex ways. These will be further discussed in the next section.
Heterogeneous Secondary Nucleation from Oligomers.
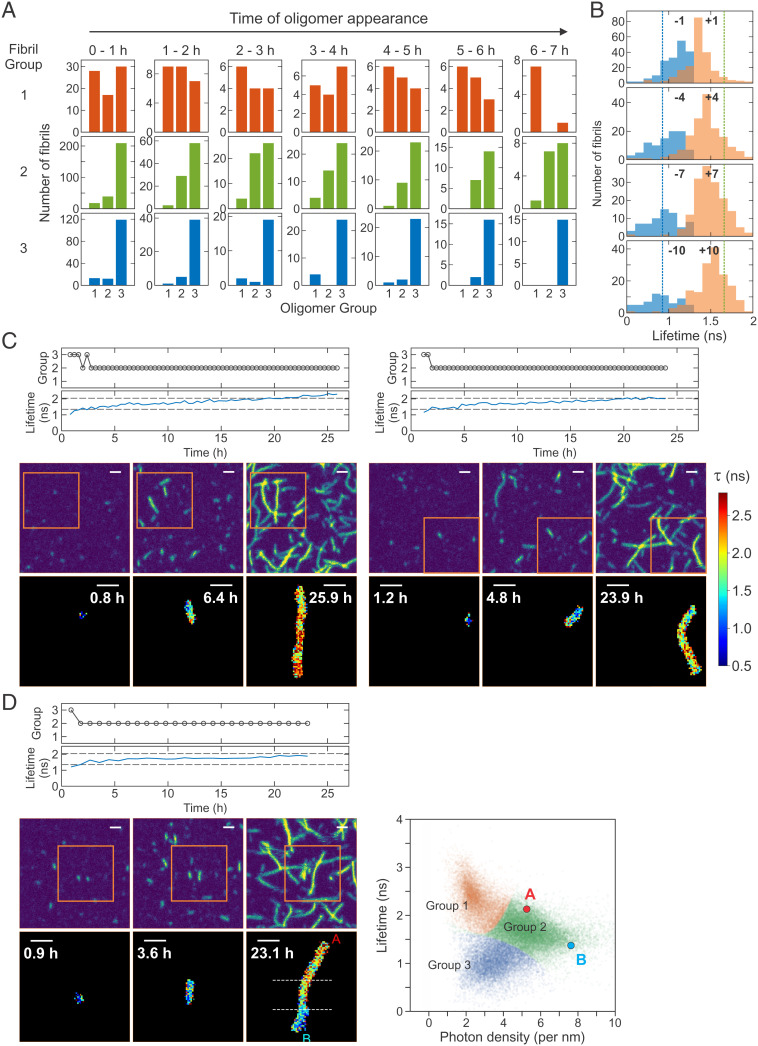
Since it is possible to follow the growth of each individual fibrils, we backtracked the growth of fibrils with a measurable length to identify the fibril group at their first appearance (i.e., origin) as an oligomer (or protofibrils, <500 nm). Fig. 5A shows this distribution. First, group 3 fibrils originate mostly from oligomers of their own group (group 3). However, the majority of group 2 fibrils grow from group 3 oligomers when they appear at the beginning of aggregation (0 to 1 h). The fraction of fibrils grown from the same group (group 2) increases as the oligomers appear at later times. Finally, the origin of group 1 fibrils is more diverse at the early stage of aggregation but gradually converges to group 1 oligomers at later times. This apparent interconversion between different groups may support the mechanism of the aggregation seed formation by conformational conversion (42, 43). However, abrupt structural conversion of large oligomers would be highly improbable because many monomers need to almost simultaneously convert conformations into the same structure. Instead, we interpret this apparent interconversion of groups as the formation of a new fibril (nucleation) on the surface of an oligomer with a different structure. In this case, as a fibril grows, the group identity will change gradually from one group (original oligomer) to another (new fibril) as new monomers with a different structure group are added over time. Indeed, the fluorescence lifetime of individual fibrils in Fig. 5C (lifetime trajectories) gradually increases from group 3 to group 2 as fibrils grow (Movies S9 and S10), supporting this mechanism (SI Appendix, Figs. S11D and S12 and Movies S11–S18). This behavior is summarized in the lifetime distributions of fibrils before and after the transition from group 3 to group 2 in Fig. 5B. The lifetime distributions of the frames immediately before (blue) and after (orange) the identified transition (±1) (see group trajectories in Fig. 5 C and D) are close and overlap, which are located in the middle of the average lifetimes of group 2 and group 3 fibrils (vertical dashed lines). However, the distributions are separated more and become closer to the average values as the frames are away from the transition interval. This type of nucleation is similar to the secondary nucleation mechanism (44, 45) but different because the nucleation occurs on the surface of oligomer/protofibril rather than long fibrils. In addition, the structure of a newly formed fibril is different from the structure of the original oligomer/protofibril. Therefore, we call this “heterogeneous secondary nucleation” (see Fig. 6).
Fig. 5.
Heterogeneous secondary nucleation from oligomers. (A) Oligomer origin of the three fibril groups (three columns). The growth of individual fibrils of the three groups was backtracked to identify the group of oligomers or short fibrils (<500 nm) at their appearance during the time period of 0 to 7 h. The majority of group 3 fibrils originate from the oligomers of their own group, whereas the majority of group 2 fibrils originate from group 3 oligomers when they appear at the beginning of aggregation (0 to 1 h). The origin of group 1 fibrils is more diverse at the early stage of aggregation, which gradually converges to group 1 oligomers at later times. (B) The distributions of fluorescence lifetimes of fibrils before (blue) and after (orange) the transition from group 3 to group 2 at different frame separations from the transition interval: ±1 to ±10 frames. Vertical dashed lines show the average fluorescence lifetimes of group 2 (green) and group 3 (blue) fibrils. (C) Time-dependent changes of the fibril group and average fluorescence lifetime of individual fibrils and snapshot images of two fibrils exhibiting heterogeneous secondary nucleation (group 3 → 2). See Movies S9 and S10 for these fibrils and additional examples in SI Appendix, Fig. S11D and Movies S11 and S12. (D) Example of fibrils that grow both directions with different lifetimes. The fibril grows from a group 3 oligomer. In the last frame (23.1 h) upper (red A, above the upper horizonal dashed line) and lower (cyan B, below the lower dashed line) parts of the fibril show distinct density and fluorescence lifetimes as shown in the 2D plot on the right side (double heterogeneous secondary nucleation). See Movie S13 and more examples in SI Appendix, Fig. S12 and Movies S14–S18. (C and D) Horizontal dashed lines in the lifetime trajectories indicate the lifetime criteria for the separation of the oligomer groups (1.35 ns and 2.04 ns). Note that the fibril density is also used for the fibrils of measurable size at later stages of aggregation. Upper image panels are fluorescence intensity images of a 10- × 10-μm2 area at three different time points. Lower panels are fluorescence lifetime images (5 × 5 μm2) of single fibrils inside the orange square in the corresponding intensity images. (Scale bars: 1 μm.)
Fig. 6.
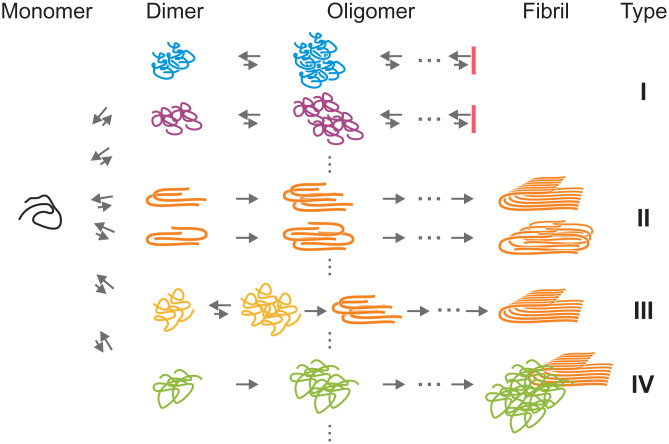
Model of heterogeneous Aβ42 oligomerization and aggregation. Initially, monomers form dimers or small oligomers with different structures, which are followed by diverse oligomerization and aggregation pathways. In type I pathways, the assembly stops in oligomeric stages. Type II and III pathways lead to fibril elongation, but the stage where fibril-like conformations are formed is different for the two types (see Discussion). Type III requires conformational conversion from non-β structures to cross-β structures. There are fibrils growing on the surface of oligomers with different structures (heterogeneous secondary nucleation, type IV pathway). The heterogeneity observed in the experiment suggests that multiple different pathways exist in each type of aggregation pathways indicated by vertical dots.
The oligomers that lead to heterogeneous secondary nucleation may be nongrowing oligomers because growing oligomers will grow into fibrils of the same group (SI Appendix, Fig. S11 A–C for the fibrils that grow homogeneously). In this case, the growth of fibrils should appear polarized, and the lifetime of the nongrowing end can be different from the growing end as shown in Fig. 5C and SI Appendix, Fig. S11D. This is also consistent with the result for the group 2 fibrils with the high growth polarity (Fig. 4F). In addition, there are other variations in this heterogeneous growth of fibrils as shown in Fig. 5D and SI Appendix, Fig. S12. In several cases, fibrils grow bidirectionally from group 3 oligomers and one end shows group 1 characteristics whereas the other end shows group 2 characteristics, which looks like double heterogeneous secondary nucleation. This result indicates that there are much more complex and diverse aggregation pathways that may not even be detected in our experiments.
Discussion
The key technical development in this work is FNet that can separate a highly crowded fibril image into individual fibril images for further quantitative analyses. As shown in Fig. 2C and SI Appendix, Figs. S4 and S5, it is impossible to distinguish individual fibrils in a single frame image. FNet uses temporal information (i.e., history) of fibril growth to overcome this problem. Since fibrils do not overlap at the beginning of an experiment, using earlier frame information fibrils at later frames can be separated iteratively. Another big challenge in the fibril segmentation is proper allocation of the photon counts in overlapping regions to the corresponding individual fibrils. The nature of classification-based image segmentation deep neural networks [e.g., U-Net (38)], which classify a pixel into a certain class, introduces discontinuity in the classification probability (p) in a single fibril image (e.g., p = 1 for the nonoverlapping region and p = 0.5 for the region where two fibrils overlap). This is not appropriate for splitting fluorescence intensity of a pixel into multiple fibrils. FNet solves this issue by introducing a modified classification network which can generate continuous prediction images.
The major problem in characterizing Aβ42 aggregation is that the experimental results vary widely depending on experimental methods and often vary even at the same condition. The size of stable oligomers, for example, which have been reported to show biological toxicity, ranges from dimers to large protofibrils (13). The most quantitative model for the aggregation mechanism that explains the long lag time and its nonlinear concentration dependence is the double (primary and secondary) nucleation model (44, 45) of Ferrone et al. (46) that accounted for the lag phase and large nucleus size for the aggregation of sickle hemoglobin. However, there is no connection between the kinetic model and the heterogeneity of the aggregation process and polymorphs of fibrils, which can be critical for the assessment of the clinical phenotype and toxicity (7, 8).
Our experimental results directly demonstrate the heterogeneity in the aggregation in terms of the number of subunits, length, elongation speed, and structure of fibrils, which varies widely over samples prepared at different times (different batches) as well as experiments performed using the same batch of sample. These results suggest that a wide variation in experimental results using different protocols and techniques is fully expected due to the heterogeneous nature of Aβ42 aggregation. In one experiment, one set of oligomerization and aggregation pathways are preferred, while under even slightly different conditions a different set of pathways is preferred, or the selection of aggregation pathways can be entirely stochastic.
A model of heterogeneous aggregation based on our observations is shown in Fig. 6. Initially, monomers form dimers or small oligomers with various conformations, which can grow into oligomers of larger size. However, many of these oligomers may stop growing into long fibrils (i.e., nonfibrillar oligomers) as indicated by two type I pathways in Fig. 6 as examples. These pathways correspond to the formation of short and slowly growing or nongrowing fibrils in Fig. 4. Since these oligomers are observed in all three fibril groups, there must be much more pathways that belong to type I.
Some of the oligomers would grow into long fibrils, which correspond to fast elongating fibrils in the experiment (two type II pathways in Fig. 6). However, the stage where the conformations of Aβ peptide become similar to those in the fibrils is unknown. Type II pathways in Fig. 6 are drawn such that fibril-like structures are formed at the dimerization stage. Simulation studies by Thirumalai and coworkers (28, 47) have shown that the dimerization in type II pathways can be initiated from transiently populated fibril-like monomer conformations. For example, two fibril-like monomers can form a dimer or a fibril-like monomer can serve as a template for the dimer formation with an unfolded monomer, which can lead to the diversity of the fibril conformation. It has been proposed that Aβ forms oligomers without β-structures first, which is followed by the structural conversion to cross-β structures that promote the fibril elongation (42, 43) (type III pathway in Fig. 6). Simulation studies by Wolynes and coworkers (30, 31) have shown that this conformational conversion can occur at early aggregation stages (i.e., small oligomers) by breaking prefibrillar contacts and forming fibrillar contacts, which is called “backtracking” observed in Aβ40 simulation. If this structural conversion happens, it may appear as transitions between different groups as shown in Fig. 5. However, abrupt structural conversion of large oligomers (larger than 100-mer) would be highly improbable because many monomers need to almost simultaneously convert conformations. We therefore propose another mechanism in which fibril elongation occurs on the surface of (potentially) nongrowing oligomers. The gradual changes in fluorescence lifetime support this mechanism (Fig. 5 B and C and SI Appendix, Fig. S11D). This is similar to the secondary nucleation mechanism, in which the nucleation occurs on the surface of existing fibrils, but in our case, the structure of the new fibril is different from that of the parent oligomer (or short fibril). To distinguish our observation from the original mechanism, we call this process “heterogeneous secondary nucleation” (type IV pathway in Fig. 6). A recent study by Knowles and coworkers (17) using TIRF imaging has reported the detection of the branch-like fibril formation in the middle of existing fibrils as the original (homogeneous) secondary nucleation process. In our data, we could not detect such events that are clearly distinguishable from a simple overlap of two fibrils. However, this does not reject the original mechanism because small nuclei would not be detected in our experiment and oligomers that nucleate on parent fibrils may rapidly detach (48). In Fig. 5A, the origins of group 1 and 2 fibrils change over time from different groups (0 to 1 h) to their own groups (6 to 7 h), which suggests that original (homogeneous) secondary nucleation process becomes more dominant as aggregation proceeds. In addition, our observation does not exclude the possibility of conformational conversions between small oligomers or backtracking (type III pathway in Fig. 6), which cannot be detected in our experiment due to the low photon count rates from small oligomers and their low populations.
The observation of highly heterogeneous pathways that may be sensitive to the environment suggests that oligomerization and aggregation pathways in vivo may be quite different from those observed in vitro. The physiological concentration of Aβ42 is much lower than 1 μM, and, therefore, aggregation by Aβ42 alone would be very improbable. Since Aβ can interact with many other cellular components nonspecifically, these interactions may promote oligomerization, nucleation, and eventually fibril formation similar to the heterogeneous secondary nucleation process that we observed. Difference in the structures between the fibrils grown in vitro without seeding and those grown from brain-derived aggregation seed supports this hypothesis (7, 49). Given that various structural polymorphs of fibrils are found in other neurodegenerative proteins (6), characterization of the heterogeneity in the context of cellular toxicity will be critical to understand the disease mechanism and to discover targets for drug therapy for various diseases involving protein aggregation. As demonstrated in this paper, our fluorescence imaging and detailed single fibril analyses using FNet are best suited for this type of characterizations.
Materials and Methods
Sample Preparation, Plate Reader Experiments, Electron Microscopy, and FLIM.
Details of the sample preparations including protein expression, purification and dye labeling, and experimental procedures for characterization of Aβ aggregation including plate reader experiments, electron microscopy and FLIM are described in SI Appendix, SI Text.
Deep Neural Network.
Classification network and prediction network.
The base structure of the classification network and the prediction networks are U-Net (38)-like deep encoder–decoder networks. The classification network takes a photon count image as an input (160 × 160 × 1, 10 pixels were padded to the original image of 150 × 150 × 1 pixels) and the growth prediction network takes photon count images of individual fibrils from the previous frame as inputs (126 images) (Fig. 2A). The encoding blocks (convolution, batch-normalization, rectified linear unit [ReLU] activation, and max pooling) of the classification network initially encodes four features (160 × 160 × 4). The following encoder blocks reduce the image width and height by half and add four additional features. The decoder blocks are similar to the encoder blocks but double the image width and height and reduce the number of features by four. The decoder blocks also have skip connections from the encoder blocks of the same image size. The prediction network has the same structure as that of the classification network, but decoder blocks of the prediction network have connections from the decoder blocks of the classification network (feature communications; Fig. 2A). The background prediction network has the same structure as that of the growth prediction network but with independent weights.
Comparison network.
Each input from the classification network and the prediction networks generates single-channel image output. The comparison network takes these images as an input and generates final-result images by twice operations of a convolution, a batch-normalization, and an ReLU activation, followed by a convolution and a PReLU activation. The result is photon count prediction images of newly appearing fibrils, background, and updated known fibrils in the new image frame. The result images are then normalized pixel by pixel so that the number of photons of a pixel of the summed result image is equal to that of the corresponding pixel of the original input image. Multiple fibrils in the prediction image of newly appearing fibrils are subsequently separated into single fibril images for the prediction of the next time frame image. Overlapping new fibrils were occasionally observed and these fibrils were excluded from the analysis.
Training data generation.
Using the Experiment 1 data, we identified potential single fibril locations by clustering high-intensity pixels. By visual inspection, we extracted 1,483 non-overlapping single-fibril movies. Training images were generated by randomly rotating, reflecting, and placing fibrils with variations of photon counts by multiplying a random factor ranging from 0.5 to 1.5 to the original fibril movies. Background photons with Poissonian statistics were generated with a mean value of 3.
Adaptive supervised learning.
A training image set that mimics the actual experimental data resulted in many incorrect segmentations for overlapped and fast-growing fibrils, probably due to the relatively small fraction of photons resulting from those rare events. Therefore, we employed an adaptive learning strategy to enhance learning. In this method, new training data were generated based on the previous training result by changing four parameters: 1) number of initial fibrils, 2) number of newly appearing fibrils, 3) length distribution of fibrils, and 4) growth speed (acceleration). For the length distribution of fibrils, we categorized fibrils into three groups, short (number of pixels < 200), mid (200 ≤ number of pixels < 400), and long (number of pixels ≥ 400) fibrils and adjusted their relative population. For the growth speed acceleration, a certain number of frames were omitted when generating a next frame image. For each generation, three to seven training datasets were generated with different parameters. Starting from the initial model (a set of weights of the neural network) that was trained with the data that mimic the real experimental data, we trained models with these modified training datasets. Once training was completed, we tested each model with the real data of Experiment 1. The best model inherits its weights to the next generation model and new training datasets were generated by increasing the occurrences of the poorly characterized events. For examples, if a model predicts a new fibril as growth of a nearby existing fibril, we increased the number of newly appearing fibrils in the new training. If a model predicts a fast-growing event as the appearance of a new fibril, we increased the growth speed acceleration. This process was repeated until the prediction is indistinguishable from the result by human inspection. The neural network was trained using TensorFlow 1.14 with a Tesla P100 GPU of NIH HPC Biowulf cluster.
Hyperparameters.
An Adam optimizer (50) with a learning rate of 0.001 was used with dice loss metric.
Loss function and validation of image segmentation.
For the training, we introduced a dice loss function L:
| [1] |
where and are the pixel intensities at location (i, j) of the prediction image and the label image, respectively.
Eighty percent of simulated images were used for training and 20% of images were used for the validation using the same loss function L in Eq. 1.
Validation Statistics and Prediction Accuracy of FNet.
We performed detailed analyses of the accuracy of the fibril segmentation as shown in SI Appendix, Fig. S4. We simulated 1,000 fibril growth images (10 × 10 μm2) for four cases with different fibril densities mimicking various stages of aggregation: 1) an early phase with 5 to 10 fibrils per image consisting of 80% short (< 150 pixels in area) and 20% intermediate (150 ≤ pixels < 250) fibrils; 2) a midphase with 11 to 20 fibrils consisting of 60% short, 35% intermediate, and 5% long (≥ 250 pixels) fibrils; 3) a late phase with 21 to 30 fibrils consisting of 40% short, 50% mid, and 10% long fibrils; and 4) an extreme case used for the training, with 126 fibrils consisting of 10% short, 30% mid, and 60% long fibrils with a 10 times accelerated growth speed. Example images are shown in SI Appendix, Fig. S4D. SI Appendix, Fig. S4C shows the comparison of image segmentations with the ground-truth images for the extreme case (SI Appendix, Fig. S4B). Examples of mispredicted fibrils are shown inside the orange rectangle. We calculated the fraction of mispredicted fibrils using Dice coefficient (1 – L in Eq. 1) (SI Appendix, Fig. S4D), which is very low even at the late phase of aggregation.
Identification and Stitching of Continuing Fibrils in Adjacent Image Regions.
Long and fast-growing fibrils reach boundaries of image areas at certain time points. In this case, continuing fibrils were identified in the adjacent image regions if they have more than three neighboring pixels and a continuous fibril can be determined uniquely, and the information was combined for the analysis of fluorescence intensity, lifetime, and length (Fig. 2C and SI Appendix, Fig. S5). However, when multiple fibrils are present at the boundary in the adjacent image, a unique image of a continuous fibril cannot be reconstructed. In addition, in some cases a single fibril was fragmented into multiple fibrils. To collect all the available information, we inspected all images and manually stitched the remaining fibrils that can be uniquely stitched (77 fibrils were manually stitched). Finally, all the fibril images were inspected again to exclude artifacts from the stitching process, which resulted in total 7,050 fibrils for further analyses.
Determination of Fluorescence Lifetime of Each Fibril.
The fluorescence lifetime of each fibril was calculated using the mean delay time of the photons contained in the image pixels of a fibril corrected for the mean delay time of background photons and the offset by the instrument response function of the detector (51). We determined the average lifetime of background photons by averaging lifetimes of background pixels, the intensity of which is lower than 90% of the average count rate of the predicted background image.
Estimation of Number of Subunits in a Fibril.
Solid-state NMR structures have revealed the polymorphism of Aβ fibrils, which consist of different numbers of subunits with distinct structures. We estimated the number of subunits of each fibril using the length from the image and the number of monomers comprising the fibril.
To determine the length of a fibril, the individual fibril image was rotated to make the fibril axis approximately parallel to the x axis using a linear fitting. Next, pixels were segmented to have a length along the x axis smaller than 10 pixels for a segment. Individual segments were fitted to a third-order polynomial from the left to the right with restricting the ends of two consecutive segments are continuously connected. Fibrils shorter than 500 nm (approximately twice the size of PSF) were excluded from the further analyses that use length information.
The number of monomers in a fibril (N) was calculated by comparing the number of photons and fluorescence lifetime of the fibril (Np,fibril and τfibril) with those of the monomer (Np,monomer and τmonomer) measured at the same illumination intensity to account for the reduced intensity of Alexa 594 in fibrils due to fluorescence quenching as
| [2] |
Here, a conversion factor A (= 2.7) was used to make the photon densities of different fibril groups integer multiples of a common photon density because the number of subunits should be an integer. However, small oligomer signals with only a few photons often lead to unreasonable lifetime values after background correction (shorter than 0 ns or longer than the unquenched monomer lifetime). Therefore, we set the maximum and the minimum values of the lifetime for the quenching correction in Eq. 2. The minimum lifetime for the quenching correction is 0.15 ns, which is the 10th percentile of an exponential distribution with the shortest lifetime (1.4 ns) of fibrils with more than 5,000 photons which show strong quenching in the late phase of the experiments. The maximum lifetime for the quenching correction is 3.67 ns, the unquenched lifetime of the monomer.
In the calibration experiment, a direct measurement of the number of photons from the monomer after excitation at 485 nm is not possible due to the very low fluorescence intensity from the monomer at low illumination intensity for fibril imaging. Therefore, we used pulse-interleaved excitation using 2-ps pulsed lasers (485 nm, LDH-D-C-485 and 595 nm, LDH-D-TA-595; PicoQuant). Monomers can be easily identified in an image collected using 595-nm excitation. The pixels comprising each monomer image were saved and used for the calculation of photons emitted by 485-nm excitation after subtraction of the background. The average number of photons emitted from the monomer, Np,monomer = 1.60 and the lifetime of Alexa 594 attached to the monomer is τmonomer = 3.67 ns.
Clustering of 2D Plot of Lifetime and Photon Density.
Fibrils from five experiments were clustered into three groups using Gaussian mixture models (39) (Fig. 3B). However, we observed slight fluctuations of the photon density in different experiments. Therefore, we normalized photon counts using group 1 (the longest-lifetime group), which shows the smallest overlap with the other groups in the 2D plot. After normalization, we calculated the average photon density of each group and conversion factors to convert the number of photons into the number of monomers. Since the ratios of the photon densities of groups 1, 2, and 3 are close to 2:4:3, we assumed that the number of subunits of the three groups are 2, 4, and 3, respectively, in the calculation of the conversion factors. The average of the conversion factors of the three groups was used in further analyses (A in Eq. 2). Short fibrils without length information were clustered using their lifetime distances from the average lifetimes of the fibril groups (group 1: τ > 2.04 ns, group 2: 1.35 ns < τ ≤ 2.04 ns, group 3: τ ≤ 1.35 ns).
Fibril Growth Analysis.
In the analysis of individual fibril growth, we removed the frames of a fibril containing less than 10 photons which can result from the background fluctuation. When a fibril grows and reaches the boundary region (three pixels from the edge of a 10- × 10-μm2 image), continuing fibrils are identified in the adjacent area images and analyzed together as described above. We selected the longest continuous frame sequence for the growth analysis of each fibril.
In the growing and nongrowing fibril analysis, to minimize the possibility of including stochastically slowly growing fibrils in the nongrowing group, the fibrils that do not grow to the measurable size (∼500 nm, twice the PSF size) at the end of experiments (24 h), which corresponds to 132 monomers/h (79.2 photons per h), were identified as nongrowing fibrils.
For the lifetime histograms (Fig. 4B and SI Appendix, Fig. S8), lifetimes were calculated from fibrils containing more than 200 photons.
Fragmentation Analysis of Fibrils and Polarized Fibril Growth Analysis.
To characterize the heterogeneity within individual fibrils, we divided fibrils into multiple fragments. A fibril image was fitted to a straight line and then the fibril was divided into fragments of 10 pixels long along the straight line (the last fragment can be shorter than 10 pixels; see Fig. 4D). The 2D lifetime-photon density plots of the three group fibrils and their fragments are compared in SI Appendix, Fig. S10.
To find the locations of growing parts of a fibril (longer than 500 nm) between two consecutive image frames for the polarized growth analysis, newly appeared pixel locations of a fibril were collected by excluding the area of the fibril in the previous frame. The newly appeared pixels were clustered by the mean shift clustering algorithm (52). To select significant growth which can be distinguished from the changes by the stage drift and thermal motion of fibrils, clusters with a size of 15 pixels or less were discarded. For each growing event detected, we compared the proximity of the newly appeared growing part to the previous growing part and to the rest of the fibril of the previous frame (nongrowing part). If the newly growing part is closer to the previous nongrowing part, we recorded the event as a switching event of the growing direction. The polarity (p) of a growing fibril was calculated by counting the growing events at the two ends as
| [3] |
where N1 and N2 are the numbers of growing events of fibril ends 1 and 2, respectively.
Supplementary Material
Acknowledgments
We thank W. A. Eaton, A. Szabo, R. Tycko, I. V. Gopich, and J.-Y. Kim for numerous helpful discussions and comments and J. M. Louis for advice and suggestions on protein expression and purification. This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov/). This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116736119/-/DCSupplemental.
Data Availability
Custom codes for the deep neural network, training with examples, and construction of individual fibril database are available at https://github.com/hoisunglab/FNet. All other data are available in the article and/or supporting information. Materials used in this study are available upon request.
References
- 1.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Knowles T. P. J., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Iadanza M. G., Jackson M. P., Hewitt E. W., Ranson N. A., Radford S. E., A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 19, 755–773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tycko R., Amyloid polymorphism: Structural basis and neurobiological relevance. Neuron 86, 632–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meier B. H., Riek R., Böckmann A., Emerging structural understanding of amyloid fibrils by solid-state NMR. Trends Biochem. Sci. 42, 777–787 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Li D., Liu C., Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat. Chem. Biol. 17, 237–245 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Qiang W., Yau W.-M., Lu J.-X., Collinge J., Tycko R., Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 541, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schweighauser M., et al. , Structures of α-synuclein filaments from multiple system atrophy. Nature 585, 464–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick A. W. P., et al. , Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcon B., et al. , Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metskas L. A., Rhoades E., Single-molecule FRET of intrinsically disordered proteins. Annu. Rev. Phys. Chem. 71, 391–414 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Selkoe D. J., Alzheimer’s disease. Cold Spring Harb. Perspect. Biol. 3, a004457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benilova I., Karran E., De Strooper B., The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 15, 349–357 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Ban T., et al. , Direct observation of Abeta amyloid fibril growth and inhibition. J. Mol. Biol. 344, 757–767 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Ogi H., et al. , Ultrafast propagation of β-amyloid fibrils in oligomeric cloud. Sci. Rep. 4, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young L. J., Kaminski Schierle G. S., Kaminski C. F., Imaging Aβ(1-42) fibril elongation reveals strongly polarised growth and growth incompetent states. Phys. Chem. Chem. Phys. 19, 27987–27996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmermann M. R., et al. , Mechanism of secondary nucleation at the single fibril level from direct observations of Aβ42 aggregation. J. Am. Chem. Soc. 143, 16621–16629 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Goldsbury C., Frey P., Olivieri V., Aebi U., Müller S. A., Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J. Mol. Biol. 352, 282–298 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kellermayer M. S. Z., Karsai A., Benke M., Soós K., Penke B., Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 141–144 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiang W., Kelley K., Tycko R., Polymorph-specific kinetics and thermodynamics of β-amyloid fibril growth. J. Am. Chem. Soc. 135, 6860–6871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe-Nakayama T., et al. , High-speed atomic force microscopy reveals structural dynamics of amyloid β1-42 aggregates. Proc. Natl. Acad. Sci. U.S.A. 113, 5835–5840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nirmalraj P. N., et al. , Complete aggregation pathway of amyloid β (1-40) and (1-42) resolved on an atomically clean interface. Sci. Adv. 6, eaaz6014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masters C. L., Selkoe D. J., Biochemistry of amyloid β-protein and amyloid deposits in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasica-Labouze J., et al. , Amyloid β protein and Alzheimer’s disease: When computer simulations complement experimental studies. Chem. Rev. 115, 3518–3563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellarin R., Schuetz P., Guarnera E., Caflisch A., Amyloid fibril polymorphism is under kinetic control. J. Am. Chem. Soc. 132, 14960–14970 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Bellesia G., Shea J. E., Diversity of kinetic pathways in amyloid fibril formation. J. Chem. Phys. 131, 111102 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Tarus B., Straub J. E., Thirumalai D., Dynamics of Asp23-Lys28 salt-bridge formation in Abeta10-35 monomers. J. Am. Chem. Soc. 128, 16159–16168 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Chakraborty D., Straub J. E., Thirumalai D., Differences in the free energies between the excited states of Aβ40 and Aβ42 monomers encode their aggregation propensities. Proc. Natl. Acad. Sci. U.S.A. 117, 19926–19937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenman D. J., Connors C. R., Chen W., Wang C., García A. E., Aβ monomers transiently sample oligomer and fibril-like configurations: Ensemble characterization using a combined MD/NMR approach. J. Mol. Biol. 425, 3338–3359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng W., Tsai M. Y., Chen M., Wolynes P. G., Exploring the aggregation free energy landscape of the amyloid-β protein (1-40). Proc. Natl. Acad. Sci. U.S.A. 113, 11835–11840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng W., Tsai M. Y., Wolynes P. G., Comparing the aggregation free energy landscapes of amyloid beta(1-42) and amyloid beta(1-40). J. Am. Chem. Soc. 139, 16666–16676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrone F. A., Assembly of Aβ proceeds via monomeric nuclei. J. Mol. Biol. 427, 287–290 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Harper J. D., Wong S. S., Lieber C. M., Lansbury P. T., Observation of metastable Abeta amyloid protofibrils by atomic force microscopy. Chem. Biol. 4, 119–125 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Walsh D. M., Lomakin A., Benedek G. B., Condron M. M., Teplow D. B., Amyloid β-protein fibrillogenesis. Detection of a protofibrillar intermediate. J. Biol. Chem. 272, 22364–22372 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Loyter A., Citovsky V., Blumenthal R., “The use of fluorescence dequenching measurements to follow viral membrane fusion events” in Methods of Biochemical Analysis, Glick D., Ed. (John Wiley & Sons Inc., 1988), pp. 129–164. [DOI] [PubMed] [Google Scholar]
- 36.Beck A., Teboulle M., Fast gradient-based algorithms for constrained total variation image denoising and deblurring problems. IEEE Trans. Image Process. 18, 2419–2434 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Bohm A., Tatarchenko M., Falk T., “ISOO V2 DL - Semantic instance segmentation of touching and overlapping objects” in IEEE 16th International Symposium on Biomedical Imaging (IEEE, 2019), pp. 343–347.
- 38.Falk T., et al. , U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Dempster A. P., Laird N. M., Rubin D. B., Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. B 39, 1–22 (1977). [Google Scholar]
- 40.Amin L., Harris D. A., Aβ receptors specifically recognize molecular features displayed by fibril ends and neurotoxic oligomers. Nat. Commun. 12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han W., Schulten K., Fibril elongation by Aβ(17-42): Kinetic network analysis of hybrid-resolution molecular dynamics simulations. J. Am. Chem. Soc. 136, 12450–12460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chimon S., et al. , Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat. Struct. Mol. Biol. 14, 1157–1164 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lee J., Culyba E. K., Powers E. T., Kelly J. W., Amyloid-β forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 7, 602–609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen S. I. A., et al. , Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 9758–9763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meisl G., et al. , Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl. Acad. Sci. U.S.A. 111, 9384–9389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrone F. A., Hofrichter J., Eaton W. A., Kinetics of sickle hemoglobin polymerization. II. A double nucleation mechanism. J. Mol. Biol. 183, 611–631 (1985). [DOI] [PubMed] [Google Scholar]
- 47.Straub J. E., Thirumalai D., Toward a molecular theory of early and late events in monomer to amyloid fibril formation. Annu. Rev. Phys. Chem. 62, 437–463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Šarić A., et al. , Physical determinants of the self-replication of protein fibrils. Nat. Phys. 12, 874–880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J.-X., et al. , Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154, 1257–1268 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kingma D. P., Ba J. L., (2015) “Adam: A method for stochastic optimization” in Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), pp. 1–15.
- 51.Chung H. S., Louis J. M., Gopich I. V., Analysis of fluorescence lifetime and energy transfer efficiency in single-molecule photon trajectories of fast-folding proteins. J. Phys. Chem. B 120, 680–699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comaniciu D., Meer P., Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 24, 603–619 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom codes for the deep neural network, training with examples, and construction of individual fibril database are available at https://github.com/hoisunglab/FNet. All other data are available in the article and/or supporting information. Materials used in this study are available upon request.