Significance
Increasing evidence demonstrates that small RNAs can serve as trafficking effectors to mediate bidirectional transkingdom RNA interference (RNAi) in interacting organisms, including plant–pathogenic fungi systems. Previous findings demonstrated that plants can send microRNAs (miRNAs) to fungal pathogen Verticillium dahliae to trigger antifungal RNAi. Here we report that V. dahliae is able to secret an effector to the plant nucleus to interfere with the nuclear export of AGO1–miRNA complexes, leading to an inhibition in antifungal RNAi and increased virulence in plants. Thus, we reveal an antagonistic mechanism that can be exploited by fungal pathogens to counteract antifungal RNAi immunity via manipulation of plant small RNA function.
Keywords: transkingdom RNAi, miRNA, suppressor, V. dahliae, AGO1
Abstract
Communication between interacting organisms via bioactive molecules is widespread in nature and plays key roles in diverse biological processes. Small RNAs (sRNAs) can travel between host plants and filamentous pathogens to trigger transkingdom RNA interference (RNAi) in recipient cells and modulate plant defense and pathogen virulence. However, how fungal pathogens counteract transkingdom antifungal RNAi has rarely been reported. Here we show that a secretory protein VdSSR1 (secretory silencing repressor 1) from Verticillium dahliae, a soil-borne phytopathogenic fungus that causes wilt diseases in a wide range of plant hosts, is required for fungal virulence in plants. VdSSR1 can translocate to plant nucleus and serve as a general suppressor of sRNA nucleocytoplasmic shuttling. We further reveal that VdSSR1 sequesters ALY family proteins, adaptors of the TREX complex, to interfere with nuclear export of the AGO1–microRNA (AGO1–miRNA) complex, leading to a great attenuation in cytoplasmic AGO1 protein and sRNA levels. With this mechanism, V. dahliae can suppress the accumulation of mobile plant miRNAs in fungal cells and succedent transkingdom silencing of virulence genes, thereby increasing its virulence in plants. Our findings reveal a mechanism by which phytopathogenic fungi antagonize antifungal RNAi-dependent plant immunity and expand the understanding on the complex interaction between host and filamentous pathogens.
Communication between different organisms via bioactive molecules is widespread in nature. Recently, growing evidence has demonstrated that regulatory small RNAs (sRNAs) can serve as trafficking effectors to travel between interacting organisms (1–4). Bidirectional transmission of sRNAs has been reported between host plants and their interacting organisms, including parasitic plants (5) and bacterial and fungal pathogens (6–10). These mobile sRNAs are biologically active and able to silence targets in trans in recipient organisms. Transkingdom RNAi plays a significant role in the modulation of plant–pathogen interactions. Fungal pathogens can send sRNAs into plants to silence immune genes and promote infection (6, 10, 11). Moreover, plant hosts can export mobile sRNAs into fungal cells to compromise the expression of virulence genes (8). For example, in the plant– Botrytis cinerea system, the necrotrophic fungus B. cinerea can deliver sRNAs to Arabidopsis and tomato plants to trigger the silencing of immune genes (6). Similarly, a recent report has demonstrated that a trafficking microRNA (miRNA) from the pathogenic fungus Fusarium oxysporum can be exported into tomato cells to silence disease resistance genes and promote infection (10). In the interaction between plants and the pathogenic fungus Verticillium dahliae, plants can export miR159 and miR166 to fungal cells to trigger transkingdom RNAi of virulence genes (8). Regarding the transport mechanism, current evidence supports a model in which host plants secrete exosome-like extracellular vesicles (EVs) to deliver sRNAs into fungal cells (12). A very recent report has indicated that several RNA-binding proteins, including Argonaute 1 (AGO1), selectively bind to EV-enriched sRNAs and are secreted by Arabidopsis (13). Mutating these RNA-binding proteins results in reduced secretion of sRNAs in EVs (13), demonstrating that AGO1 and these RNA-binding proteins are required for sRNA loading and/or stabilization in EVs.
During the arms race between plant hosts and viruses, viruses can encode suppressor proteins to counteract host antiviral RNA silencing (14). Recently, emerging evidence indicates that similar strategies can also be exploited by filamentous pathogens to antagonize plant immunity and promote infection (14–18). For example, the effector PSR2 from the oomycete plant pathogen Phytophthora sojae and the effector PgtSR1 from wheat stem rust fungus Puccinia graminis f. sp. tritici affect the production of secondary small interfering RNAs (siRNAs) (16–18). These secondary siRNAs are found in EVs and are believed to silence Phytophthora genes during natural infection (18). However, whether such a suppressive phenomenon is widespread in the transkingdom RNAi between plant and filamentous pathogens and whether there are alternative mechanisms are still unclear.
Here we demonstrated that the secretary protein VdSSR1 from V. dahilae, a causal agent of plant wilt diseases, is a general suppressor of RNA silencing in plant cells. VdSSR1 sequesters ALY family proteins, the adaptors of the TREX (Transcription-Export) mRNA transport complex, to interfere with TREX/TREX-2 complexes–mediated nuclear export of AGO1–miRNA complex and mRNAs, leading to a reduced accumulation of transkingdom miR159 and miR166 in fungal cells and increased expression of fungal virulence genes. Our findings identify a mechanism by which phytopathogenic fungi antagonize RNA silencing-dependent plant immunity and advance our understanding of the arms race between plants and phytopathogenic fungi.
Results
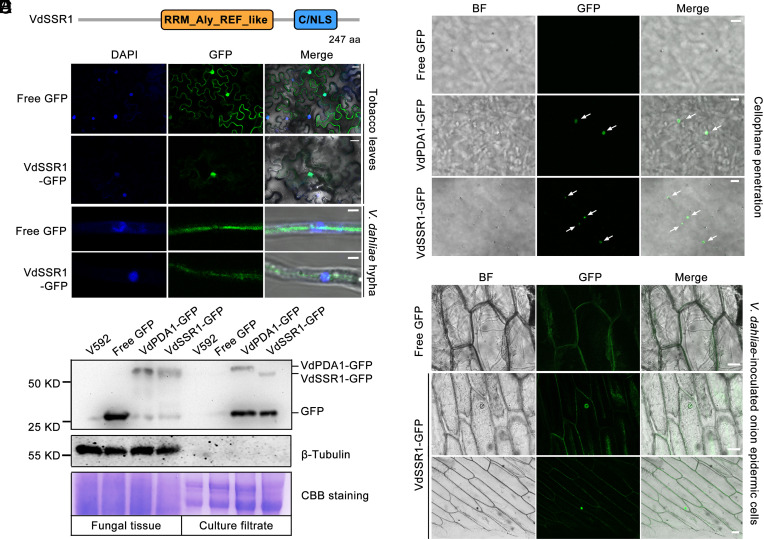
The V. dahliae Secretory Protein VdSSR1 Can Translocate to the Plant Nucleus.
Recent studies reported that plants can send small RNAs to interacting organisms, including phytopathogenic fungi, to trigger transkingdom RNAi (8, 9). In the interacting system between plant and V. dahliae, a causal agent of plant wilt diseases that cause very serious harm to numerous crops, miR159 and miR166 can be sent to fungal cells to trigger transkingdom RNAi and antagonize fungal virulence. We are interested in the question of whether fungi can send effector to plant cells to antagonize this process. It is well known that miRNAs are processed mainly in nucleus. To answer this question, we first performed mass spectrometry (MS) assay in the culture filtrates of a virulent defoliating V. dahliae isolate V592 from cotton and identified potential effectors from the MS data that can be located in plant nuclei and possess silencing suppression activity in plants. In this way, we identified that a 247–amino acid protein VdSSR1, which contains a conserved RRM_Aly_REF_like domain (cd12418) (19) and a C/NLS motif (Fig. 1A and Dataset S1), is a potential candidate. VdSSR1 is expressed in both mycelia and spores and displays a higher expression in spores (SI Appendix, Fig. S1). VdSSR1 is present in both the nucleus and cytoplasm of fungal cells but localized mainly in the nucleus of plant cells (Fig. 1B). VdSSR1 mainly localizes to the cytoplasm in fungal cells (Fig. 1B). VdSSR1 is encoded by gene VDAG_04215. According to the annotation of the Fungal Secretome Database, VDAG_04215 in VdLs. 17 strain is a class NS secreted protein that lacks a typical signal peptide (http://fsd.snu.ac.kr/contig.php?a=protein_dv&bid=533&id=4215&sv=VDAG_04215&sf=SEQUENCE_NAME). To test whether VdSSR1 can be secreted into plant cells during infection, we first examined the accumulation of VdSSR1 protein in the culture filtrates of VdSSR1-GFP–expressing V592 strain via Western blotting as previously described (20). Culture filtrates from the strains expressing free GFP and VdPDA1-GFP, a characterized secretory protein of V. dahliae (21), were also used for nonsecretory and secretory controls. As shown in Fig. 1C, similar to VdPDA1-GFP, VdSSR1-GFP protein was detected in the culture filtrates of VdSSR1-GFP strain but not in that of V592, suggesting that VdSSR1-GFP protein can be secreted into the extracellular space from fungal cells. To confirm the secretion, a cellophane penetration assay was performed as previously reported (22). As shown in Fig. 1D, both VdSSR1-GFP and VdPDA1-GFP can penetrate into the cellophane and were observed in the infectious structure, which also functions as a secretory structure (22). Considering that VdSSR1 lacks a typical signal peptide, we asked whether it is secreted via EVs. To answer this question, EVs were isolated from the VdSSR1-GFP–expressing strain. Unfortunately, we cannot detect the accumulation of VdSSR1 (SI Appendix, Fig. S2). We speculate that VdSSR1 may be secreted through an unconventional method as reported previously (23). To further verify that VdSSR1 could be translocated from V. dahliae to plant cells, conidial spores of VdSSR1-GFP and free GFP strain were inoculated on onion epidermal cells. At 7 d postinoculation (dpi), a nuclear fluorescence signal was observed in the nucleus of onion epidermal cells inoculated with VdSSR1-GFP conidial spores (Fig. 1E). In contrast, in the free GFP strain–inoculated cells, fluorescence was only observed in conidial spores (Fig. 1E). Combined with these data, we concluded that VdSSR1 could translocate from V. dahliae to the plant nucleus.
Fig. 1.
The secretory V. dahliae protein VdSSR1 can translocate to the plant nucleus. (A) Domain structure of VdSSR1 protein. aa, amino acid. (B) The subcellular localization of VdSSR1 protein in plant (top two rows) and fungal cells (bottom two rows). In the top two rows, free GFP and VdSSR1-GFP were expressed in N. benthamiana leaves. (Scale bar, 20 µm.) In the bottom two rows, the hyphae of V. dahliae strains expressing free GFP and VdSSR1-GFP were subjected to GFP fluorescence microscopy. DAPI staining indicates the nuclei. (Scale bar, 2 µm.) (C) Immunoblotting result showing the accumulation of VdSSR1-GFP protein in the total proteins of fungal tissue and culture filtrates. V592 strain serves as a negative control. The strains expressing free GFP and VdPDA1-GFP serve as controls of nonsecretory and secretory proteins. CBB, Coomassie brilliant blue. β-Tubulin levels serve as internal control of total fungal proteins. (D) VdSSR1 is a secretory protein. The V. dahliae strains expressing VdSSR1-GFP, free GFP, and VdPDA1-GFP were incubated on solid minimal medium overlaid with cellophane for induction of infectious structure and hyphal neck. The secretory protein VdPDA1-GFP-expressing strain serves as a positive control. VdSSR1-GFP was observed at the hyphal septa (white arrows). Images were taken at 3 dpi. (Scale bar, 5 μm.) (E) Nuclear targeting of VdSSR1-GFP in onion epidermal cells. Conidial suspensions of the V. dahliae strains expressing free GFP and VdSSR1-GFP were inoculated on the inner layer of onion epidermal cells for fungal growth. After 3 to 7 d growth, the onion epidermal cells were subjected to fluorescence observation. Photographs were taken at 7 dpi. (Scale bar, 50 µm.)
VdSSR1 Is Required for V. dahliae Virulence in Host Plants.
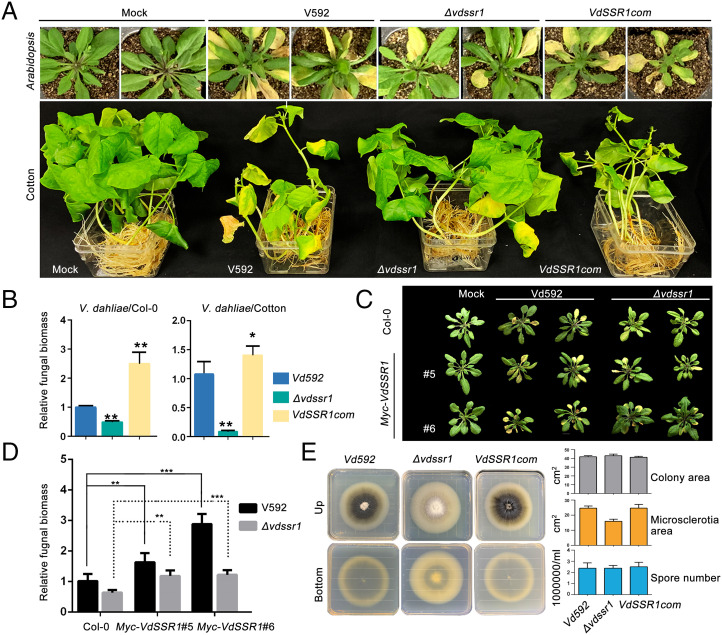
We next investigated whether VdSSR1 is required for V. dahliae virulence in host plants. To this end, the VdSSR1 knockout mutant Δvdssr1 strain and the complementation strain VdSSR1com (constitutive Tef promoter) in the Δvdssr1 background were generated (SI Appendix, Fig. S3) according to a previously reported method (24). The wild-type V592, Δvdssr1, and VdSSR1com strains were subjected to inoculation assays in Arabidopsis (Col-0 ecotype) and cotton seedlings through an unimpaired root dip inoculation approach (24). At 18 to 21 dpi, typical wilt disease symptoms, including leaf wilt and defoliation, developed in most V592-inoculated plants (Fig. 2A). In contrast, the disease symptoms were much milder in the Δvdssr1 mutant strain–inoculated plants, whereas the VdSSR1com strain–inoculated plants displayed significantly more severe disease symptoms than the V592-inoculated plants (Fig. 2A). Consistently, fungal biomass analysis demonstrated a significantly reduced and increased accumulation of V. dahliae in the Δvdssr1 mutant and VdSSR1com strain–inoculated plants compared with V592 (Fig. 2B). To further investigate the contribution of VdSSR1 to fungal virulence in plants, VdSSR1 was constitutively expressed in Arabidopsis (Col-0) driven by the cauliflower mosaic virus 35S promoter, and the generated Myc-VdSSR1 transgenic plants were inoculated with wild-type V592 and the Δvdssr1 mutant strains. The results indicated that constitutive expression of VdSSR1 significantly enhanced the infection of both wild-type V592 and the Δvdssr1 mutant strains (Fig. 2 C and D). These data suggested that VdSSR1 was required for V. dahliae virulence in plants.
Fig. 2.
VdSSR1 is required for V. dahliae virulence in host plants. (A) Disease symptoms of (Upper) Arabidopsis (Col-0 ecotype) and (Bottom) cotton plants infected with wild-type V592, Δvdssr1 mutant, and VdSSR1com strains at 18 to 21 dpi. (B) Relative fungal biomass analysis of (Left) Col-0 and (Right) cotton plants infected with the V592, ΔVdssr1 mutant, and VdSSR1com strains. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. *P < 0.5. **P < 0.05. (C) Disease symptoms of Myc-VdSSR1 transgenic Arabidopsis and wild-type Col-0 plants infected with V592 and the ΔVdssr1 mutant strains at 18 to 21 dpi. Two random transgenic lines were selected for analysis. (D) Relative fungal biomass analysis of Col-0 and Myc-VdSSR1 transgenic plants infected with V592 and the ΔVdssr1 mutant at 18 dpi. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. **P < 0.05. ***P < 0.01. Solid and dashed lines represent the significance analysis of the difference between V592- and Δvdssr1-inoculated plants, respectively. (E) (Left) Colony morphology of V592, the Δvdssr1 mutant, and the complementation VdSSR1com strains on PDA medium. Photographs were taken at 2 wk postinoculation. (Right) Column diagrams showing the comparison of the colony area, microsclerotia area, and spore number of the V592, Δvdssr1, and VdSSR1com strains.
We next investigated that whether VdSSR1 knockout has an effect on fungal growth and development. Compared with V592, the Δvdssr1 strain exhibited a similar morphology in hyphal growth and spore number (Fig. 2E). Intriguingly, the production of melanin was not affected in the early growth stage but was reduced in the later stage of Δvdssr1 strain (Fig. 2E and SI Appendix, Fig. S4). No obvious difference was observed between the VdSSR1com and wild-type strains (Fig. 2E and SI Appendix, Fig. S4). Although a previous study has shown that V. dahliae virulence in plants is less affected by the production of melanin (25), we cannot rule out the possibility that VdSSR1 dysfunction may have certain effect on V. dahliae virulence.
VdSSR1 Suppresses RNA Silencing in Plants.
To investigate whether VdSSR1 has RNA silencing suppressor activity, VdSSR1 was transiently coexpressed with the reporter gene GFP in Nicotiana benthamiana leaves. The reported silencing suppressors 2b (26) from cucumber mosaic virus (CMV-2b) and P19 from tomato bushy stunt virus (TBSV) served as positive controls, and β-glucuronidase (GUS) protein served as a parallel control. As shown in Fig. 3A, compared with the GUS control, dramatically enhanced GFP fluorescence signals were observed in VdSSR1-, CMV-2b-, and P19-infiltrated leaf regions. Consistently, greatly increased levels of GFP mRNA and protein were detected (Fig. 3B). These results suggested that VdSSR1 could suppress sense exogenous gene-triggered RNA silencing. To further characterize the suppressor activity of VdSSR1, the Myc-VdSSR1 transgene was crossed into a SUC:SUL silencing reporter system (Suli) (27). In this system, an inverted repeat of the endogenous SULFUR (SUL) gene is expressed in phloem companion cells and leads to the production of siRNAs (siSULs) and the ensuing RNA silencing, resulting in a leaf bleaching phenotype in the silenced cells (28). The results demonstrated that the expression of VdSSR1 greatly inhibited the spread of SUL silencing (Fig. 3C) and fully rescued the reduced levels of SUL mRNA in Suli plants (Fig. 3D). Moreover, the siSUL levels were obviously reduced in VdSSR1-expressing plants compared with Suli plants (Fig. 3E). Taken together, these data demonstrated that VdSSR1 not only suppressed sense exogenous gene-triggered silencing but also interfered with inverted repeat dsRNA-mediated RNA silencing. Consistent with the RNA silencing suppressor activity, VdSSR1 plants exhibited higher sensitivity to CMV infection (SI Appendix, Fig. S5).
Fig. 3.
VdSSR1 represses RNA silencing in plants and transkingdom RNAi in the interacting V. dahliae. (A) VdSSR1 suppresses sense exogenous gene-triggered RNA silencing. GFP was transiently coexpressed with VdSSR1, GUS, CMV-2b, and TBSV-P19 in tobacco leaves. Photographs were taken at 2 d postinfiltration. (B) RNA and protein blotting showing the accumulation of GFP mRNA and protein, respectively, in coexpressed tobacco leaves. Methyl blue staining of ribosomal RNA (rRNA) and CBB staining served as loading controls of total RNAs and proteins, respectively. (C) VdSSR1 suppresses phloem-specific silencing of the endogenous SUL gene triggered by the Suli reporter system. (D) Relative accumulation levels of endogenous SUL mRNA in Col-0, Suli, and two lines of VdSSR1/Suli transgenes. The mRNA levels were first normalized to ACT2 and then to Col-0. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. **P < 0.05. (E) RNA blotting results showing the accumulation levels of siSULs in different genotypes. Results from three biological replicates were shown. The U6 level serves as RNA loading control. (F) RNA blotting results showing the accumulation of cotton miR159 and miR166 in cultured V. dahliae strains recovered from the infected cotton or in the uninfected V. dahliae strains. RNAs from Col-0 and cotton serve as plant controls. Plant U6 and Fungal 5S RNA serve as loading control. (G) Relative transcript levels of fungal genes VdHic-15 and VdClp-1 targeted by plant miR159 and miR166, respectively. (Right) The fold change of VdHic-15 and VdClp-1 levels. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. **P < 0.05. ***P < 0.01. ns, no significance.
We next asked whether nucleus targeting is indispensable for VdSSR1-dependent silencing suppression and fungal virulence in plants. To this end, NLS deletion protein VdSSR1Δnls and VdSSR1Δnls-expressing strain in Δvdssr1 background were generated, and silencing suppression and fungal inoculation assays were performed. As shown in SI Appendix, Fig. S6A, NLS deletion greatly reduced the nuclear accumulation of VdSSR1 but dramatically increased its cytoplasmic accumulation. The RNA silencing suppression activity of VdSSR1Δnls was almost lost (SI Appendix, Fig. S6 B and C), and expression of VdSSR1Δnls cannot rescue the reduced virulence of Δvdssr1 strain (SI Appendix, Fig. S6D). These results indicated that nucleus targeting is indispensable for VdSSR1-dependent silencing suppressor activity and fungal virulence in plants.
VdSSR1 Suppresses Transkingdom RNAi in the Plant–V. dahliae System.
In some plant–pathogenic fungus systems, including the plant–V. dahliae system, small RNAs can be trafficked bidirectionally to guide transkingdom RNA silencing, resulting in the modulation of plant immunity and fungal virulence (2, 6, 8, 9). Cotton and Arabidopsis plants have been shown to export miR159 and miR166 to V. dahliae cells to trigger silencing of the fungal genes VdHic-15 and VdClp-1, respectively, thereby antagonizing the virulence of V. dahliae (8). To test whether VdSSR1 plays a role in the transkingdom silencing mediated by plant trafficking miRNAs, we examined the accumulation of plant miR159 and miR166 in V. dahliae cultured from hyphae recovered from infected cotton plants (V. dahliae-Cotton) as previously reported (8). Consistent with previous findings, RNA blotting results demonstrated that trafficking miR159 and miR166 were detected in the cultured V. dahliae recovered from the infected cotton plant (V592-Cotton) but not in the uninfected V592 (Fig. 3F). Intriguingly, much higher levels of trafficking miR159 and miR166 were detected in Δvdssr1-Cotton compared with V592-Cotton (Fig. 3F). In line with the increased accumulation of trafficking miRNAs, the levels of VdHic-15 and VdClp-1 mRNAs, the targets of plant miR159 and miR166, respectively, were significantly reduced in Δvdssr1-Cotton (Fig. 3G), demonstrating that transkingdom RNA silencing was enhanced by the knockout of VdSSR1. Importantly, no significant difference in VdHic-15 and VdClp-1 mRNA levels was detected between the uninfected V592 and Δvdssr1 strains (Fig. 3G), implying that the down-regulation of VdHic-15 and VdClp-1 mRNA levels observed in Δvdssr1-Cotton was attributed to the increased accumulation of trafficking miRNAs but not a direct effect of VdSSR1 knockout on these two genes. Altogether, these data demonstrated that VdSSR1 could suppress the transkingdom RNAi of fungal virulence genes by inhibiting the accumulation of trafficking plant miRNAs in recipient cells.
VdSSR1 Affects the Nucleocytoplasmic Partitioning of sRNAs in Plants.
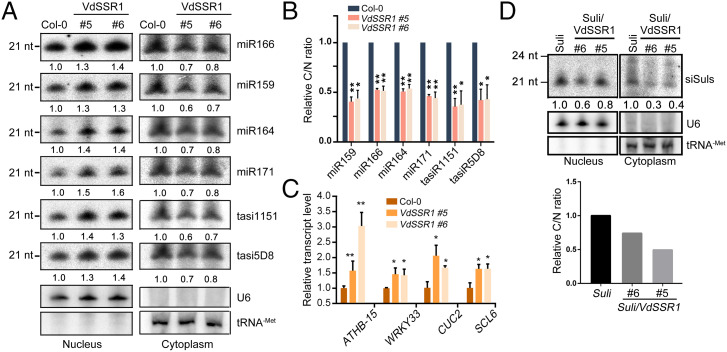
To decipher the mechanism underlying VdSSR1-mediated silencing suppression, we first examined small RNA accumulation in transgenic Myc-VdSSR1 plants by RNA blotting. To our surprise, in contrast to the strong suppression activity of VdSSR1, the transgenic expression of VdSSR1 only had a weak repression effect on the accumulation of four representative miRNAs, miR159, miR166, miR171, and miR164, and two trans-acting siRNAs (tasiRNAs), tasi5D8 and tasi1151 (29) (SI Appendix, Fig. S7). We next measured primary miRNA (pri-miRNA) levels and found that the selected pri-miRNAs exhibited no obvious change or slightly reduced levels in the Myc-VdSSR1 plants compared with Col-0 (SI Appendix, Fig. S8). To gain further insight into the effect of VdSSR1 on plant sRNA biogenesis, small RNA sequencing was performed in Col-0 and Myc-VdSSR1#6 plants. The results indicated that VdSSR1 exerted a weak effect on global small RNA patterns, including the composition of sRNAs in different genomic elements and sRNA length distribution (SI Appendix, Fig. S9). Consistent with the RNA blotting results, most of the annotated miRNAs had a similar accumulation in Col-0 and VdSSR1 plants (SI Appendix, Fig. S10).
Nuclear export is a necessary step in miRNA maturation. We next examined whether the nucleocytoplasmic partitioning of the tested sRNAs was affected by VdSSR1. RNA was extracted from the nucleus and cytoplasm of Col-0 and Myc-VdSSR1 plants to measure small RNA levels. Small RNA blotting results demonstrated that the cytoplasmic accumulation of all the tested miRNAs as well as tasiRNAs was significantly reduced in Myc-VdSSR1 plants compared with Col-0 plants. In contrast, the nuclear levels were increased (Fig. 4A and SI Appendix, Fig. S11), leading to a great reduction in the ratio of cytoplasmic/nuclear (C/N) sRNA levels (Fig. 4B). In line with the reduction in cytoplasmic miRNAs, an obvious increase in the mRNA levels of representative target genes was observed in Myc-VdSSR1 plants (Fig. 4C). In addition to miRNAs and tasiRNAs, we also measured the nucleocytoplasmic distribution of siSuls from Suli and Myc-VdSSR1/Suli plants and found that the C/N siSul ratio was also reduced in Myc-VdSSR1/Suli compared with Suli plants (Fig. 4D). Intriguingly, unlike tested miRNAs and tasiRNAs, nuclear siSuls were reduced in Myc-VdSSR1/Suli plants. This result is consistent with the obvious reduction of total siSuls in Myc-VdSSR1/Suli plants (Fig. 3E). We speculate that this discrepancy may be due to a different biogenesis of miRNA and artificial siRNAs, as reported previously (28). In brief, these data support the notion that VdSSR1 suppresses RNA silencing by modulating the nucleocytoplasmic partitioning of sRNAs.
Fig. 4.
VdSSR1 affects the nucleocytoplasmic partitioning of sRNAs in plant cells. (A) RNA blotting results showing the nuclear and cytoplasmic accumulation of representative sRNAs in Col-0 and Myc-VdSSR1 plants. U6 and tRNA-Met served as nuclear and cytoplasmic RNA loading controls, respectively. The nuclear and cytoplasmic sRNA signals were quantified and normalized to those of U6 and tRNA-Met, respectively. One representative result of three biological replicates (SI Appendix, Fig. S11) was shown. (B) The relative C/N ratios of sRNAs as determined in three RNA blotting results (Fig. 4A and SI Appendix, Fig. S11). Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. *P < 0.1. **P < 0.01. (C) The relative transcript levels of representative miRNA target genes in Col-0 and two Myc-VdSSR1 lines. The mRNA levels were first normalized to ACT2 and then to Col-0. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. *P < 0.01. **P < 0.01. (D) Small RNA blotting results showing the nuclear and cytoplasmic accumulation of siSuls from Suli and VdSSR1/Suli plants. (Lower) The relative C/N ratios of sRNAs as determined in Upper.
VdSSR1 Associates with TREX/TREX-2 Complexes In Vivo by Interacting with ALY Family Adaptors.
To decipher how VdSSR1 affects the nucleocytoplasmic partitioning of sRNAs, a VdSSR1 immunoprecipitation assay followed by MS (IP–MS) was performed in Myc-VdSSR1 transgenic Arabidopsis. Intriguingly, three RNA export complex-related proteins, ALY4, DSS1-V, and SAC3B, were copurified with Myc-VdSSR1 protein (Fig. 5A and Dataset S2). Arabidopsis encodes four ALY family proteins (ALY1-4), which have been shown to function as adaptor proteins in the TREX complex–mediated mRNA export pathway (30). DSS1-V and SAC3B are conserved components of the TREX-2 mRNA export receptor complex (30–32). In yeast and metazoans, the TREX complex associates with nascent transcripts, leading to the recruitment of export adaptors that bind to the mRNA and deliver it to the TREX-2 export receptor complex to facilitate translocation of the mRNP through the nuclear pore complex (NPC) (30). Intriguingly, phylogenetic analysis indicated that VdSSR1 is evolutionarily related to Arabidopsis ALYs but displays closer evolutionary relationship to its fungal homologous proteins (SI Appendix, Fig. S12). Supporting this notion, the RRM domains of VdSSR1 and ALY family proteins displayed sequence similarity (SI Appendix, Fig. S13). The copurification of VdSSR1 with RNA export-related proteins suggested that VdSSR1 might interfere with the TREX/TREX-2–related pathway to regulate miRNA nucleocytoplasmic shuttling. To test our hypothesis, protein interactions were examined between VdSSR1 and copurified RNA export proteins. The yeast two-hybrid (Y2H) data indicated that VdSSR1 directly interacted with all four ALY proteins but not DSS1-V and SAC3B (Fig. 5B). The interaction between VdSSR1 and ALYs was further supported by evidence from the split luciferase assay in N. benthamiana leaves (Fig. 5C). In addition, both ALYs and VdSSR1 could self-interact (SI Appendix, Fig. S14).
Fig. 5.
VdSSR1 interacts with ALY family proteins in plants. (A) VdSSR1 IP–MS analysis in transgenic Myc-VdSSR1 Arabidopsis. The numbers of recovered peptides of bait and prey proteins and potential functions are shown. (B) VdSSR1 interacts with ALY family proteins in the Y2H assay. The interaction of VdSSR1 with ALY1/2/3/4, DSS1-V, and SAC3B was tested. (C) Split luciferase assays in N. benthamiana leaves showing the interaction of VdSSR1 and ALY family proteins. Photographs were taken at 3 d postinfiltration. (D) Subcellular assay showing the colocalization of ALY1/2/3/4 with VdSSR1 in N. benthamiana leaves. VdSSR1-RFP was coexpressed with ALY1/2/3/4-GFP in tobacco leaves. Photographs were taken at 2 d postinfiltration. DAPI staining indicates the nucleus. (Scale bar, 10 µm.)
In a previous report, the silencing suppressor P19 encoded by TBSV could interact with ALY family adaptors and cause the relocalization of Arabidopsis ALY2 and ALY4 from the nucleus to the cytoplasm (33). Intriguingly, the expression of Arabidopsis ALY1 and ALY3, which are not relocalized by P19 (33), could cause some P19 proteins to translocate from the cytoplasm to the nucleus to compromise their silencing suppressor activity (34). To clarify whether the suppressor activity of VdSSR1 was related to a possible relocalization of ALY proteins, as observed for P19, fluorescence protein-fused ALY proteins and VdSSR1 were coexpressed in tobacco leaves, and their subcellular localization patterns were determined. As shown in Fig. 5D, all four ALY proteins colocalized with VdSSR1 in the nucleus, suggesting that VdSSR1 suppressed RNA silencing through a different mechanism from the P19.
VdSSR1 Sequesters ALY Adaptors to Inhibit Its Association with UAP56 and Disrupt mRNA Export.
In Arabidopsis, metazoans, and yeast, ALY export adaptors are recruited by the DEAD-box RNA helicase UAP56 (Sub2 in yeast) (30, 35, 36). We found that V. dahliae encoded one UAP56-like protein (VdUAP56L) and three ALY-like proteins (SI Appendix, Fig. S15), including VdSSR1. Y2H data demonstrated that all four Arabidopsis ALY proteins could interact with UAP56 (Fig. 6A). However, VdSSR1 could interact neither with V. dahliae VdUAP56L nor with Arabidopsis UAP56 (Fig. 6A and SI Appendix, Fig. S15). In addition, RNA binding activity was indispensable for the function of ALY adaptors in RNA export (37–39). We examined the RNA binding activity of VdSSR1 in an electrophoretic mobility shift assay (EMSA) with Arabidopsis ALY4 as a positive control. Compared with ALY4, VdSSR1 lost its RNA binding activity (Fig. 6B). The above evidence supports the conclusion that VdSSR1 may be an unfunctional analog of ALY adaptors, or act as a different mechanism from Arabidopsis adaptors. Based on these data, another hypothesis was proposed: VdSSR1 may mimic and sequester ALY adaptors to interfere with the functioning of TREX/TREX-2 complexes. To test this hypothesis, an immunoprecipitation assay was performed to determine the effect of VdSSR1 on the ALY-UAP56 interaction in vivo. In this assay, myc-tagged ALY4 was coexpressed with GFP-tagged UAP56 in tobacco leaves. A recombinant VdSSR1 protein purified from Escherichia coli was added to the lysate of tobacco leaves. α-GFP immunoprecipitation results demonstrated that ALY4 could be pulled down by UAP56 (Fig. 6C). Importantly, the pulled-down ALY4 level was greatly reduced by the addition of recombinant VdSSR1 protein to the lysate. In contrast, addition of the MBP control protein had no obvious effect on the pulled-down ALY4 level. These results support our hypothesis that VdSSR1 sequesters ALY adaptors to inhibit its interaction with UAP56.
Fig. 6.
VdSSR1 sequesters ALY adaptors to hijack the nucleocytoplasmic shuttling of AGO1-miRNA complex and mRNA export. (A) Y2H results showing that Arabidopsis ALY adaptors, but not fungal VdSSR1, can interact with UAP56. (B) EMSA results showing a comparison of MBP, MBP-ALY4, and MBP-VdSSR1 in RNA binding activity. (C) Immunoblotting results of the α-GFP coimmunoprecipitation assay showing that the addition of purified MBP-VdSSR1 protein could inhibit the pulled-down ALY-Myc level by UAP56-GFP. MBP and GUS proteins served as negative controls. (D) (Upper) Fluorescence in situ hybridization assay showing the fluorescence signals of poly(A) RNA in Col-0 and Myc-VdSSR1 plants. The nua mutant serves as a positive control. (Lower) Relative nuclear/cytoplasmic poly(A) RNA ratio. Data are the mean, and error bars represent ± SD from the number of cells counted for each genotype. n = 10 to 16. An unpaired one-tailed t test was performed. ***P < 0.01. (E) Western blotting results showing the nucleocytoplasmic partitioning of AGO1 proteins in different genotypes. H3 and cFBPase served as nuclear and cytoplasmic markers, respectively. AGO1 signals were first normalized to H3 and cFBPase and then to Col-0. The experiment was repeated three times with similar results. (F) RNA gel blotting results showing the nucleocytoplasmic partitioning of sRNAs in Col-0, aly2/3, and aly4 mutants. Nuclear and cytoplasmic sRNA signals were first normalized to U6 and tRNA-Met, respectively, and then to Col-0. (G) The relative C/N sRNA ratios as determined in F. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. *P < 0.1. **P < 0.01.
It has been reported that knockout of ALY adaptors leads to a nuclear retention of mRNA (37). We next asked whether VdSSR1 influences the nucleocytosolic mRNA transport. To this end, a fluorescence in situ hybridization assay was performed in Col-0, Myc-VdSSR1 plants, and a mutant of Nuclear Pore Anchor (NUA), in which mRNA retention has been reported (40). Consistent with previous finding (40), a great increase of nuclear mRNA signal was observed in the nua mutant (Fig. 6D). As expected, increased fluorescence signal in the nucleus was also observed in VdSSR1 plants, suggesting that VdSSR1-ALY interaction has a negative impact on the mRNA export process.
VdSSR1 Interferes with the Nucleocytoplasmic Shuttling of the AGO1–miRNA Complex.
In plants, miRNA biogenesis undergoes stepwise nuclear maturation before engaging cytosolic target transcripts. During nuclear maturation, the pri-miRNAs are first processed by a dicing complex containing DCL1, HYL1, and SE, and the resulting miRNA–miRNA* duplexes are 2′-O–methylated by the small RNA methyltransferase HUA ENHANCER1 (HEN1) (41, 42). Mature miRNA duplexes were originally believed to translocate to the cytosol in an EXPORTIN5 homolog HASTY (HST)-dependent manner and to be incorporated into AGO1 (43–46). However, recent studies have revealed that HST participates in the miRNA pathway independent of its cargo-exporting activity in Arabidopsis because hst mutants with impaired shuttling exhibit a normal subcellular distribution of miRNAs (47). The current literature supports a model of AGO1–miRNA complex-dependent nucleocytosolic shuttling (47, 48). In this model, miRNA is incorporated into AGO1 in the nucleus, and the resulting AGO1–miRNA complexes are then exported to the cytosol through an interaction of the AGO1 nuclear export signal (NES) with EXPORTIN 1 (EXP1/XPO1) in the nuclear pore (48). Consistently, TREX is associated with the NPC, and a very recent study has reported that the Arabidopsis TREX-2 RNA export complex is essential for multiple steps in miRNA biogenesis, including transcription and nuclear export, through its interactions with RNA polymerase II and the nucleoporin protein NUP1, respectively (49). Dysfunction of the TREX-2 complex leads to increased retention of AGO1 and compromised nuclear export of miRNAs (49).
Here the compromised nucleocytoplasmic shuttling of miRNAs, together with the interaction of VdSSR1 with TREX complex adaptor, inspired us to propose that VdSSR1 may influence the nuclear export of the AGO1–miRNA complex through the connection between TREX and TREX-2 complexes. To test this hypothesis, the nucleocytoplasmic distributions of the AGO1 proteins were measured by Western blotting in Col-0 and Myc-VdSSR1 plants as well as the aly2/3, aly4, and thp1 mutants, which is a component of TREX-2 complex (SI Appendix, Fig. S16). The data revealed a similar accumulation of total AGO1 proteins in all tested genotypes (Fig. 6E). Importantly, AGO1 protein showed increased accumulation in the nuclear fraction in the tested mutants and Myc-VdSSR1 plants compared with Col-0. In contrast, cytoplasmic AGO1 protein levels were greatly reduced. In line with the increased retention of nuclear AGO1, compromised miRNA export (Fig. 6F and SI Appendix, Fig. S17) and increased accumulation of representative miRNA target genes (SI Appendix, Fig. S18) were also detected in the aly2/3 and aly4 mutants. We noted that the reduction of C/N sRNA levels in aly4 mutant is weaker than in aly2/3 mutant (Fig. 6G), which may be attributed to the functional redundancy between ALY family adaptors (37). In addition, like Myc-VdSSR1 plants, the levels of tested pri-miRNAs were slightly reduced in aly2/3 and aly4 mutants (SI Appendix, Fig. S8).
The similar inhibition of AGO1–miRNA nuclear export inspires us to investigate the global effects of VdSSR1 and ALY dysfunction on miRNA target genes. To this end, RNA sequencing was performed in Col-0, VdSSR1#6, and aly2/3 mutant plants. The results indicate that most of the miRNA target genes were commonly regulated in VdSSR1#6 and aly2/3 mutant plants compared with Col-0 (SI Appendix, Fig. S19), among which 370 genes were commonly up-regulated and 270 genes were commonly down-regulated. The expression pattern of miRNA target genes may attribute to a comprehensive effect of VdSSR1–ALY interaction on the inhibition of nuclear export of both AGO1–miRNA and mRNA.
VdSSR1-Mediated Interference of AGO1–miRNA Nuclear Export Contributes to V. dahliae Virulence in Plants.
Next, we asked whether VdSSR1–ALY interaction-mediated interference of plant AGO1–miRNA nuclear export contributes to V. dahliae virulence in plants. To this end, Myc-VdSSR1 plants were crossed into aly2/3 mutant to generate Myc-VdSSR1/aly2/3 plants. The sensitivities of Myc-VdSSR1, aly2/3, and Myc-VdSSR1/aly2/3 plants to V. dahliae were measured. As shown in Fig. 7 A and B, all tested genotypes exhibited similar sensitivity to V. dahliae compared with Col-0. Compared with Myc-VdSSR1 and aly2/3 plants, no obvious additive effect was observed in Myc-VdSSR1/aly2/3 plants, suggesting that VdSSR1 promote V. dahliae infection in host plants mainly through hijacking ALY functions.
Fig. 7.
VdSSR1-mediated suppression of transkingdom RNA silencing in plant–V. dahliae system. (A) Wilt disease symptoms of Col-0, Myc-VdSSR1, Myc-VdSSR1/aly2/3, and aly2/3 plants infected with the wild-type V592 strain at 30 dpi. (B) Relative fungal biomass analysis of Arabidopsis plants infected with V592 at 30 dpi. Data are the means ± SD of three biological repeats. An unpaired one-tailed t test was performed. **P < 0.05. (C) A working model of VdSSR1-mediated suppression of the nuclear export of AGO1–miRNA complex and mRNA and its roles in counteracting antifungal RNAi.
To further verify the contribution of inhibiting AGO1–miRNA nuclear export in VdSSR1-dependent virulence regulation, mutants of XPO1A, a nuclear export receptor in Arabidopsis (50), and NUP96, a component of NPC, as well as ago1-27, were subjected to fungal inoculation assay. Consistent with previous reports that XPO1A and NUP1, another component of NPC, were indispensable for the nuclear export of AGO1–miRNA complex (48, 49), xpo1a and nup96 were more sensitive to both V592 and Δvdssr1 strains compared with Col-0 (SI Appendix, Fig. S20). Intriguingly, we found that ago1-27 mutant was more resistant to both V592 and Δssr1 strains compared with Col-0 (SI Appendix, Fig. S20). This result is in line with the previous findings of ago1 in V. dahliae (51) and B. cinerea (6) inoculation assays. The enhanced resistance of ago1 mutant to Δssr1 strain conflicts with the notion that VdSSR1 promotes V. dahliae virulence by interfering with AGO1–miRNA nuclear export. We speculate that this may be due to the multifaceted function of AGO1 in plant–fungus interaction. AGO1 is indispensable not only for sRNA-mediated posttranscriptional silencing of target genes, including defense genes, but also for transkingdom RNAi triggered by fungal sRNAs (6). In the latter case, AGO1 dysfunction will lead to a disability of fungal sRNA-mediated transkingdom silencing of plant defense genes, thereby enhancing plant resistance against fungal pathogens (6). Therefore, we think that the increased resistance of ago1 to Δssr1 strain is a comprehensive result of the interaction of multiple AGO1-dependent processes.
Based on the above evidence, we proposed an AGO1–miRNA nuclear export-dependent mechanism of VdSSR1-mediated virulence regulation in the plant–V. dahliae system (Fig. 7C). In this mechanism, VdSSR1 is translocated from fungal cells to the plant nucleus, possibly through a noncanonical secretion pathway. Nucleus-localized VdSSR1 sequesters ALY adaptors to inhibit their association with the UAP56–TREX complex, which directly/indirectly interferes with TREX-2 complex-dependent nuclear export of the AGO1–miRNA complex and mRNAs. Therefore, VdSSR1 is a general suppressor of plant miRNAs, including the transkingdom miR159 and miR166 in the plant–V. dahliae system. The compromised accumulation of cytoplasmic miR166 and miR159 results in attenuated transport to V. dahliae cells, finally suppressing the transkingdom silencing of fungal virulence genes and promoting infection. In addition, VdSSR1-mediated inhibition of the mRNA export of certain defense genes may also contribute to the enhanced virulence in plants.
Discussion
In this study, we show that the fungal effector VdSSR1 from the phytopathogenic fungus V. dahliae sequesters host ALY adaptors to interfere with the nuclear export of the AGO1–miRNA complex and mRNAs, leading to a reduction in the levels of cytoplasmic miRNAs, including mobile miRNAs, and succedent attenuation of transkingdom RNAi. With this mechanism, V. dahliae antagonizes transkingdom sRNA-dependent plant immunity and establishes an efficient infection in hosts.
Some biomolecules, such as proteins, antimicrobial peptides, and metabolites, can move from pathogens and pests to hosts or vice versa to modulate cellular processes in recipient organisms (52–55). Recently, increasing evidence has shown that regulatory sRNAs can also move between hosts and pathogens to modulate plant immunity and pathogenic virulence in recipient organisms (3, 15). In this mechanism, plant regulatory sRNAs are assembled into EVs and are transported into the recipient cells of filamentous pathogens to confer transkingdom RNAi of virulent genes (3). Thus, efficient transkingdom RNAi mediated by host sRNAs is beneficial for plants in establishing a successful defense system. It is well known that encoding silencing suppressors is a general strategy for viruses to counteract plant antiviral RNA silencing, and diverse mechanisms are adopted by viruses to hijack host RNA silencing (14). Recently, a few studies have shown that such a strategy can also be adopted by filamentous pathogens to counteract plant transkingdom RNAi (15). Qiao et al. reported that two effectors from the oomycete phytopathogen P. sojae suppress RNA silencing by inhibiting sRNA biogenesis (17). Among the two suppressors, PSR1 is reported to regulate the accumulation of both miRNAs and endogenous siRNAs in Arabidopsis by binding to nuclear PSR1-interacting protein 1 (PINP1), which may facilitate sRNA processing by affecting the assembly of dicing complexes (29, 56). A recent report from the same group further showed that PSR2 specifically inhibits the biogenesis of secondary siRNAs, some of which are found in EVs and likely silence target genes in Phytophthora during natural infection (18).
In this study, we report a mechanism of fungal effector-mediated suppression of transkingdom RNAi-dependent plant immunity: interfering with the nuclear export of the AGO1–miRNA complex. In this case, our evidence supports the notion that VdSSR1 serves as a general regulator of plant sRNA biogenesis. VdSSR1 expression in plants inhibits the accumulation of mature sRNAs in cytoplasm, leading to an attenuation in the levels of mobile miRNAs and succedent transkingdom silencing of virulent genes in fungal cells. As a result, the sensitivity of VdSSR1-expressing plants to V. dahliae is enhanced. Nuclear export is essential for the maturation of regulatory sRNAs and ensuing targeting. A recent report has shown that miRNAs are mature, methylated, loaded into AGO1 in the nucleus, and exported to the cytoplasm as AGO1–miRNA complexes in a CRM1(EXPO1)/NES-dependent manner (48). Moreover, a recent study from another group further revealed that the TREX-2 complex coordinates with the NPC to link the key steps of miRNA biogenesis, including transcription, processing, and nuclear export, in an AGO1-dependent manner (49). In this study, VdSSR1 interacted with ALY family proteins, adaptors of the TREX complex, to interfere with nucleocytoplasmic shuttling of both the AGO1–miRNA complex and mRNAs. In line with our findings, previous reports have shown that a number of TREX complex mutants display defects in miRNA and siRNA biosynthesis, including tasiRNAs (57, 58). In the tex1 mutant, inverted repeat siRNAs and tasiRNAs are down-regulated, but miR173/miR173* levels are not obviously affected (58). In the case of THO2, miRNAs, tasiRNAs, and 24-nt siR1003 are down-regulated in its mutant (59). This evidence suggests that different mutations of the TREX complex have specificity in the regulation of sRNA biogenesis. In this study, we showed that the total levels of miRNAs and tasiRNAs were slightly down-regulated in VdSSR1 plants (SI Appendix, Fig. S7A), and compromised nucleocytoplasmic shuttling of miRNAs, tasiRNAs, and AGO1 proteins was observed in both VdSSR1 and aly mutants (Fig. 6F). These findings suggest that the TREX complex is also involved in the nuclear transport of the AGO1–miRNA complex, although the detailed mechanism remains elusive. In the TREX-2 complex, THP1, one of the core subunits, has been shown to interact and colocalize with the nucleoporin protein NUP1 at the nuclear envelope (49). Thus, we speculate that the TREX complex may coordinate with the TREX-2 complex to facilitate the nuclear export of the AGO1–miRNA complex. This phenomenon may explain the increase in nuclear sRNAs in VdSSR1 plants. Notably, the transcript levels of tested pri-miRNAs are only slightly reduced in Myc-VdSSR1 plants and aly mutants (SI Appendix, Fig. S8) compared with an obvious reduction in TREX-2 mutant (49). We propose that the TREX complex or ALY adaptors have a different effect on transcription with TREX-2 complex.
Here we also observed increased nuclear retention of mRNAs in Myc-VdSSR1 plants, which is consistent with a previous report showing that ALY family proteins are required for nucleocytoplasmic mRNA transport (37). Thus, it is reasonable to propose that a reduced cytoplasmic accumulation of certain defense-related mRNAs may also contribute to the enhanced sensitivity of VdSSR1 plants to V. dahliae infection. This issue is worth addressing in future research.
Although our evidence supports that VdSSR1 is a secretory protein, the pathway through which VdSSR1 is secreted remains not completely understood. We speculate that VdSSR1 is secreted through an uncanonical method, as reported previously in a study of two effectors from P. sojae and V. dahliae (23). In addition, considering the reduced melanin production in Δvdssr1 strain (Fig. 2E) and the increased accumulation of V592 in VdSSR1-expressing plants compared with Δvdssr1 (Fig. 2D), the possibility that VdSSR1 may possess dual functions in both plants and fungi cannot be ruled out, although a previous study has shown that V. dahliae virulence in plants is less affected by the production of melanin (25). Indeed, VdSSR1 and Arabidopsis ALYs belong to similar protein families but display closer evolutionary relationship with fungal homologs (SI Appendix, Fig. S12). Importantly, VdSSR1 lose the ability of RNA-binding activity and interaction with UAP56 (Fig. 6 A and B). There are two possibilities: one is that VdSSR1 may function in a different way from Arabidopsis ALYs, and the other is that VdSSR1 has evolved into an effector without ALY-like function. All of these questions deserve further clarity in future studies. Altogether, we revealed an antagonizable mechanism adopted by filamentous pathogens in counteracting transkingdom sRNA-dependent plant immunity. Considering that nuclear export is an essential process for sRNA biogenesis, this mechanism may also be used by other pathogens.
Materials and Methods
Materials.
Arabidopsis (Col-0 ecotype) and N. benthamiana plants were grown in a growth chamber at 22 ± 2 °C with a 16-h light/8-h dark photoperiod. Gossypium hirsutum plants were grown in a growth chamber at 25 ± 2 °C with a 16-h light/8-h dark photoperiod. The virulent defoliating V. dahliae strain V592 (from diseased cotton in Xinjiang Province, China) was grown on PDA plates at 25 °C in the dark.
Plasmid Constructs and Transformation.
To generate Myc-VdSSR1 lines, the VdSSR1 coding sequence was cloned into pCAMBIA1300 driven and fused to a Myc tag at the N terminus. The 35S-Myc-VdSSR1 was transformed into Col-0 through the floral dip method (60) to generate Myc-VdSSR1 plants. For the VdSSR1 knockout strain, a 1-kb genomic sequence upstream and downstream of the VdSSR1 gene was amplified using high-fidelity Pfu polymerase (Vazyme). The PCR products were ligated to the pGKO-HYG plasmid simultaneously with the ClonExpress MultiS One Step Cloning Kit (Vazyme), producing a construct for homologous recombination. For the VdSSR1com strain, the coding sequence was ligated into the Tef-GFP vector with a ClonExpress II One Step Cloning Kit (Vazyme). All constructs were transformed into Agrobacterium tumefaciens strain EHA105 individually. The Agrobacterium-mediated transformation method (24) was used for knockout and complementation transformation in the V592 and Δvdssr1 strains, respectively. All the primers used in this study were listed in Dataset S3.
Fungal Inoculation Assay.
Plants were inoculated by the root-dipping inoculation method as described previously (51, 61). At 18 to 21 dpi, Arabidopsis rosette leaves and cotton stem tissues were harvested per plant and flash-frozen in liquid nitrogen. The DNA from each sample (100 mg) was isolated for fungal biomass analysis through RT-qPCR calculation of V. dahliae–specific gene expression (51). AtRuBisCo gene was used for normalization.
Suppressor Activity Assay.
Suppressor activity assays were carried out according to a previous report (26). In brief, 4-wk-old N. benthamiana leaves were coinfiltrated with cultures of A. tumefaciens GV3101 carrying different constructs. The final concentration of each culture in the mixture (per mL) was adjusted to an OD600 of 0.5.
Purification of Recombinant Proteins and EMSA Assay.
The recombinant plasmids were transformed into BL21 cells and induced with 0.3 mM isopropyl b-D-1-thiogalactopyranoside (Sigma-Aldrich) in Luria-Bertani (LB) medium at 16 °C for 12 h. Fusion proteins were purified using Glutathione Sepharose 4B (GE Healthcare) and amylose-conjugated agarose beads (NEB), according to the manufacturer’s instructions. For the EMSA, synthesized RNA oligos were radiolabeled in 50-pmol quantities with 0.3 mM [γ-32P] ATP and 20 units of T4 PNK (NEB) and annealed. Binding reactions were performed according to a previous method (26). After 1 h of electrophoresis at 4 °C, the gel was dried and exposed to a storage phosphor screen (GE Healthcare). The image was scanned using an Amersham Typhoon (GE Healthcare).
RNA Isolation, Gene Quantification, and RNA Blotting.
Total RNA was isolated using the RNAprep Pure Plant Kit (TIANGEN). The cDNA was synthesized using HiScript II Q Select RT SuperMix (Vazyme). Quantitative PCR was performed with AceQ qPCR SYBR Green Master Mix (Vazyme). The relative gene expression level was normalized to AtACT2 or VdElf1. RNA blotting assay was performed as reported previously (62). For mRNA blotting, 10 µg of total RNA was separated by electrophoresis on 1.2% agarose gels with formaldehyde and transferred to nylon N+ membranes through capillary transfer. GFP DNA probe was labeled with [α-32P] dCTP using the Rediprime II system (GE Healthcare). For sRNA blotting, 60 µg of total RNA was separated by electrophoresis on 17% PAGE gels and electrically transferred to nylon N+ membranes. [γ-32P] ATP was used to label gene-specific oligo sequences by polynucleotide kinase (NEB) for hybridization (8). After washing and drying, the membrane was exposed to a storage phosphor screen (GE Healthcare). Images were scanned using an Amersham Typhoon (GE Healthcare).
Secretion Assay and Confocal Observation.
For the effector secretion assay (20), the VdSSR1-GFP strain was inoculated on the inner layer of onion epidermal cells and incubated on a 1% agar plate at room temperature for 5 to 7 d, followed by confocal imaging (TCS SP8, Leica). To collect fungal culture filtrate and fungal tissue for protein isolation and Western blot, 1 mL fungal spore (106 spore mL−1) was cultured in 50 mL liquid Czapek–Dox medium for 7 d. After centrifugation at 1,614 × g for 10 min. The pellets of fungal tissues were frozen and stored at −80 °C for protein isolation. The supernatants were filtered through 0.22-μm-diameter filters to obtain culture filtrates and ultrafiltrated by Amicon Ultra-15 (3 kDa NMWCO) (Millipore) at 4 °C. Approximately 50 mL supernatant of each sample was ultrafiltrated to 100 μL and stored at −80 °C for immunoblotting.
Fungal Recovery Assay.
The fungus was recovered from infected cotton stems or Arabidopsis leaves as previously described (8). V. dahliae hyphae that grew from cotton stems or Arabidopsis leaves were transferred to PDA medium for further growth for another 2 to 3 d. Then, the hyphae were cut from the agar and placed in CM liquid medium for another 3 d of culture. The final large number of hyphae were collected for RNA blotting and gene expression examination.
IP–MS Assay.
Twelve-day-old Arabidopsis seedlings were ground to a fine powder in liquid nitrogen, and the powder was resuspended in IP buffer (40 mM Tris⋅HCl, pH 8.0, 300 mM NaCl, 4 mM MgCl2, 2% glycerol, 0.1% Triton-X100, 1 mM PMSF, and 1X mixture) and incubated for 30 min with gentle rotation at 4 °C. The protein suspensions were centrifuged at 14,000 rpm for 20 min to remove debris. IP–MS was performed as previously reported (63). In brief, the supernatant was incubated with Myc-conjugated beads (Invitrogen) for 2 h at 4 °C and then was washed with IP buffer two times following a fivefold wash with PBS buffer. The pellet was subjected to MS analysis.
Y2H and Split Luciferase Assay.
The Y2H assays were performed according to the Matchmaker Gold Y2H system user manual (Clontech). The pGBKT7 and pGADT7 plasmids containing tested genes were transformed into the AH109 yeast strain. Each combination of single colonies from the SD-LW medium culture was further transferred to synthetically defined (SD) medium lacking leucine, tryptophan, histidine (SD-LWH) or lacking leucine, tryptophan, histidine, alanine (SD-LWHA) culture media. For the split luciferase assay (64), the proteins fused to split luciferase were coinfiltrated into 4-wk-old N. benthamiana leaves. The luciferase activity was determined using a charge-coupled device (CCD) camera equipped with Winview software (Princeton Instruments).
Fluorescence In Situ Hybridization Assay.
To determine the relative distribution of mRNA in nuclei and cytosol, 6-d-old seedlings grown on solid MS medium were used as previously described (65). Hybridization was performed in PerfectHyb plus solution (Sigma) with an Alexa Fluor 488-labeled 48-nucleotide oligo (dT) probe. Fluorescent signals of seedling root tip cells were analyzed using a Leica SP8 microscope.
Co-IP Assay.
Four-week-old N. benthamiana leaves were infiltrated with a culture of A. tumefaciens GV3101 carrying 35S-ALY4-Myc and 35S-UAP56-GFP or 35S-UAP56-GFP and 35S-GUS. After 2 d, the protein extraction and Co-IP procedures were performed as previously described (66), excluding the addition of MBP or MBP-SSR1 before the GFP-conjugated bead Co-IP.
Nuclear–Cytoplasmic Fractionation.
The nucleocytoplasmic fractionation assay was conducted as previously described (49). Five grams of 12-d-old seedlings were used for each sample. The 5-mL cytoplasmic fraction and final nuclear pellet were used for RNA isolation. For the protein immunoblot in nucleocytoplasmic fractionation, 12-d-old seedlings were cross-linked in formaldehyde as previously described (49). The frozen tissue was then subjected to the same procedure as described above.
Supplementary Material
Acknowledgments
We sincerely thank Prof. Yingfang Zhu of Henan University for kindly providing the seeds of nup96 mutant. C.-G.D. was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences ( Grant XDB27040203) and the National Natural Science Foundation of China (Grants 31770155 and 31800131). H.-S.G. was supported by the National Natural Science Foundation of China (Grant 32020103003). This work was partially supported by the National Natural Science Foundation of China (Grant 31771349 to Z.Z.) and Guangdong Provincial Pearl River Talent Plan (Grant 2019QN01N108 to Z.Z.)
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.I. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114583119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. The small RNA and mRNA sequencing data have been deposited in the GEO with the accession code GSE190467.
References
- 1.Zhao J. H., Zhang T., Liu Q. Y., Guo H. S., Trans-kingdom RNAs and their fates in recipient cells: Advances, utilization, and perspectives. Plant Commun. 2, 100167 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu C., Liu T., Chang Y. N., Duan C. G., Small RNA functions as a trafficking effector in plant immunity. Int. J. Mol. Sci. 20, E2816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Q., He B., Jin H., A safe ride in extracellular vesicles—Small RNA trafficking between plant hosts and pathogens. Curr. Opin. Plant Biol. 52, 140–148 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Huang C. Y., Wang H., Hu P., Hamby R., Jin H., Small RNAs—Big players in plant-microbe interactions. Cell Host Microbe 26, 173–182 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Shahid S., et al. , MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Weiberg A., et al. , Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren B., Wang X., Duan J., Ma J., Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Zhang T., et al. , Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 16153 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Wang M., et al. , Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 16151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji H. M., et al. , Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol. 232, 705–718 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner B. T., et al. , Fusarium graminearum DICER-like-dependent sRNAs are required for the suppression of host immune genes and full virulence. PLoS One 16, e0252365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Q., et al. , Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He B., et al. , RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 7, 342–352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z., Li Y., Ding S. W., Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 19, 31–44 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Qiao Y., et al. , Small RNAs in plant immunity and virulence of filamentous pathogens. Annu. Rev. Phytopathol. 59, 265–288 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Yin C., et al. , A novel fungal effector from Puccinia graminis suppressing RNA silencing and plant defense responses. New Phytol. 222, 1561–1572 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Qiao Y., et al. , Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou Y., et al. , A phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25, 153–165.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S., et al. , CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., et al. , The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 215, 368–381 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Gao F., et al. , Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 5, 1167–1176 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Zhou T. T., Zhao Y. L., Guo H. S., Secretory proteins are delivered to the septin-organized penetration interface during root infection by Verticillium dahliae. PLoS Pathog. 13, e1006275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu T., et al. , Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5, 4686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao F., et al. , A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS One 5, e15319 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klimes A., Dobinson K. F., A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet. Biol. 43, 283–294 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Duan C. G., et al. , Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24, 259–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Himber C., Dunoyer P., Moissiard G., Ritzenthaler C., Voinnet O., Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22, 4523–4533 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunoyer P., Himber C., Ruiz-Ferrer V., Alioua A., Voinnet O., Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 39, 848–856 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Qiao Y., Shi J., Zhai Y., Hou Y., Ma W., Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. U.S.A. 112, 5850–5855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrnsberger H. F., Grasser M., Grasser K. D., Nucleocytosolic mRNA transport in plants: Export factors and their influence on growth and development. J. Exp. Bot. 70, 3757–3763 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Sørensen B. B., et al. , The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol. Biol. 93, 283–298 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Lu Q., et al. , Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J. 61, 259–270 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Uhrig J. F., Canto T., Marshall D., MacFarlane S. A., Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol. 135, 2411–2423 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canto T., Uhrig J. F., Swanson M., Wright K. M., MacFarlane S. A., Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J. Virol. 80, 9064–9072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfab A., Bruckmann A., Nazet J., Merkl R., Grasser K. D., The adaptor protein ENY2 Is a component of the deubiquitination module of the arabidopsis SAGA transcriptional co-activator complex but not of the TREX-2 complex. J. Mol. Biol. 430, 1479–1494 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Heath C. G., Viphakone N., Wilson S. A., The role of TREX in gene expression and disease. Biochem. J. 473, 2911–2935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaff C., et al. , ALY RNA-binding proteins are required for nucleocytosolic mRNA transport and modulate plant growth and development. Plant Physiol. 177, 226–240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo M. L., et al. , Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413, 644–647 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z., et al. , The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407, 401–405 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Xu X. M., et al. , NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19, 1537–1548 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu Y., Zhang Y., Chen X., Chen Y., Plant noncoding RNAs: Hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 35, 407–431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji L., Chen X., Regulation of small RNA stability: Methylation and beyond. Cell Res. 22, 624–636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter C. A., Aukerman M. J., Sun H., Fokina M., Poethig R. S., PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol. 132, 2135–2143 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park M. Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R. S., Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 3691–3696 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi R., Qin Y., Macara I. G., Cullen B. R., Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui J., You C., Chen X., The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 35, 61–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cambiagno D. A., et al. , HASTY modulates miRNA biogenesis by linking pri-miRNA transcription and processing. Mol. Plant 14, 426–439 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Bologna N. G., et al. , Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell 69, 709–719.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Zhang B., et al. , Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis. Nat. Plants 6, 957–969 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu G., et al. , EXPORTIN 1A prevents transgene silencing in Arabidopsis by modulating nucleo-cytoplasmic partitioning of HDA6. J. Integr. Plant Biol. 61, 1243–1254 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Ellendorff U., Fradin E. F., de Jonge R., Thomma B. P., RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui H., Tsuda K., Parker J. E., Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Stuart J., Insect effectors and gene-for-gene interactions with host plants. Curr. Opin. Insect Sci. 9, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Lehrer R. I., Ganz T., Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 11, 23–27 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Hegedüs N., Marx F., Antifungal proteins: More than antimicrobials? Fungal Biol. Rev. 26, 132–145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang P., et al. , The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytol. 223, 839–852 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Francisco-Mangilet A. G., et al. , THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 82, 1018–1029 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Yelina N. E., et al. , Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 13948–13953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choudury S. G., et al. , The RNA export factor ALY1 enables genome-wide RNA-directed DNA methylation. Plant Cell 31, 759–774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Zhou B. J., Jia P. S., Gao F., Guo H. S., Molecular characterization and functional analysis of a necrosis- and ethylene-inducing, protein-encoding gene family from Verticillium dahliae. Mol. Plant Microbe Interact. 25, 964–975 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Liu X., Zhao J., Guo H. S., IBM1-dependent H3K9 demethylation enables self-silencing of an exogenous silencer for the non-cell autonomous silencing of an endogenous target gene. J. Genet. Genomics 46, 149–153 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y. Z., et al. , Coupling of H3K27me3 recognition with transcriptional repression through the BAH-PHD-CPL2 complex in Arabidopsis. Nat. Commun. 11, 6212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duan C. G., et al. , A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 27, 226–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong Z., et al. , A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17, 256–267 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu F., et al. , NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog. 10, e1004312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. The small RNA and mRNA sequencing data have been deposited in the GEO with the accession code GSE190467.