Significance
While computational engineering of therapeutic proteins is a desirable goal, in practice the optimization of protein–protein interactions requires substantial experimental intervention. We present here a computational approach that focuses on stabilizing core protein structures rather than engineering the protein–protein interface. Using this approach, we designed thermostabilized interleukin-2 (IL-2) variants that bind tightly to their receptor without experimental optimization, mimicking the properties of the yeast-display engineered IL-2 variant “super-2.” Our results suggest that structure-guided stabilization may be a general method for in silico affinity maturation of protein–protein interactions.
Keywords: protein engineering, in silico design, structural biology, immunology
Abstract
Affinity maturation of protein–protein interactions is an important approach in the development of therapeutic proteins such as cytokines. Typical experimental strategies involve targeting the cytokine-receptor interface with combinatorial libraries and then selecting for higher-affinity variants. Mutations to the binding scaffold are usually not considered main drivers for improved affinity. Here we demonstrate that computational design can provide affinity-enhanced variants of interleukin-2 (IL-2) “out of the box” without any requirement for interface engineering. Using a strategy of global IL-2 structural stabilization targeting metastable regions of the three-dimensional structure, rather than the receptor binding interfaces, we computationally designed thermostable IL-2 variants with up to 40-fold higher affinity for IL-2Rβ without any library-based optimization. These IL-2 analogs exhibited CD25-independent activities on T and natural killer (NK) cells both in vitro and in vivo, mimicking the properties of the IL-2 superkine “super-2” that was engineered through yeast surface display [A. M. Levin et al., Nature, 484, 529–533 (2012)]. Structure-guided stabilization of cytokines is a powerful approach to affinity maturation with applications to many cytokine and protein–protein interactions.
Optimizing binding affinity for a novel protein target is a recurrent challenge in therapeutic development. Binders with improved affinity are typically engineered by constructing combinatorial or random libraries for screening by expression and display on phage, yeast, or other platforms (1–3). Computation offers a faster, more efficient approach to protein engineering. However, approaches that rely solely on computational approaches for interface engineering have proven less fruitful. Instead, significant progress has been made in the structure-based computational design of stable, idealized proteins, whether based on native proteins or generated de novo. This work has led to major advances in de novo protein design (4–6), functional protein design (7, 8), and protein stabilization (9).
An approach to apply these computational advances to affinity maturation would be general and powerful. In this vein, some proteins undergo an increase in rigidity upon binding to target ligands (10–13). This behavior suggests a potential strategy for affinity maturation: by stabilizing the folded binding state of a protein, the “reorganization energy” penalty paid upon binding can be reduced and bound states can be favored. Through global structure optimization, local structural improvements to the binding site can be achieved. Such an approach, while simple in concept, would apply the strengths of computational protein design to affinity maturation.
An important target which can be used to test this strategy for affinity maturation is the cytokine interleukin-2 (IL-2). IL-2 is a key lymphocyte growth factor that is required for the maintenance and expansion of effector T cells (Teffs), regulatory T cells (Tregs), and natural killer (NK) cells. IL-2 signals through a heterodimeric receptor complex consisting of the IL-2Rβ (CD122) and γc (CD132) subunits, which form the “low-affinity” IL-2 receptor complex. Activated lymphocytes and Treg cells also express an IL-2Rα (CD25) subunit, which enables formation of a “high-affinity” receptor complex. By contrast, NK cells and resting CD8 + Teffs express low levels of IL-2Rα and respond to IL-2 primarily through the lower-affinity IL-2Rβ and γc chains (14–18).
The potency of IL-2 as an effector across several immune cell types has led to significant interest in and development of IL-2 as an immunotherapeutic agent in cancer (19, 20). One current view is that IL-2 analogs that are more selective for the “β/γ” receptors would bias IL-2 toward Teffs versus CD25+ Tregs. Thus, affinity-maturing IL-2 for IL-2Rβ could result in such a bias (21–23). In one early study, an unbiased error-prone library was screened to yield variants of IL-2 with increased affinity for IL-2Rβ, termed “super-2” (24). These super-2 cytokines were able to signal directly through binding to IL-2Rβ in the absence of CD25 and showed a Teff bias in vivo as well as superior antitumor properties over IL-2. Of particular interest was the observation that some of the affinity-conferring mutations occurred in the hydrophobic core of IL-2 rather than the receptor binding interface. Molecular dynamics and NMR studies indicated that these mutations worked by stabilizing and simplifying the conformational ensemble of IL-2, preorganizing these super-2 molecules to bind to IL-2Rβ specifically (24, 25). In a subsequent study, a de novo IL-2 analog, neo-2, was designed to eliminate structural nonidealities and reroute the convoluted helical topology of IL-2 by building a de novo helical bundle around the receptor binding helices of native IL-2. After optimization by combinatorial libraries with yeast display, neo-2 was engineered to produce similar effects on cell signaling as super-2 (26).
The comparable success of these two disparate approaches suggests that the plasticity-based mechanism of affinity enhancement in IL-2 can dovetail with the strength of computation in designing idealized, stable structures. We hypothesized that targeted computational remodeling of nonideal, metastable regions on the IL-2 structure would produce better superagonists by allowing us to jointly optimize both sequence and structure while maintaining similarity to the native sequence. Here we report the computational design and validation of “out of the box” thermostable IL-2 variants, stabIL-2 proteins, which display CD25-independent signaling and activity in vitro and in vivo. These in silico variants were tested without any intervening engineering or optimization and shown to exhibit similar properties as IL-2 superkines, validating the approach of cytokine affinity enhancement through computational stabilization.
Results
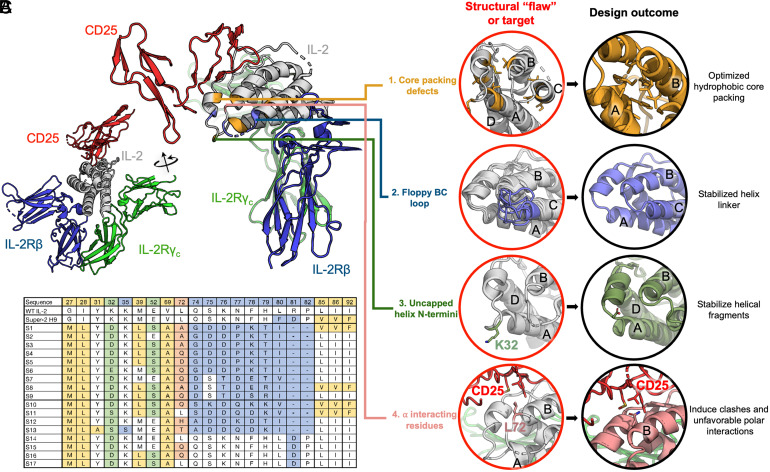
We first sought to find avenues for redesign that would result in improved stability and reduced conformational entropy, which could potentially lead to higher affinity for IL-2Rβ. Examining the structural organization of IL-2 led to four targets for design: 1) packing defects between helices A, B, and C, as well as the BC loop; 2) a long, poorly resolved loop connection between helices B and C; 3) uncapped helices; and 4) an exposed position, Leu72, known to modulate the interaction with CD25 (27) (Fig. 1 A and B). Each of these structural “flaws” or design targets was individually targeted with a different design strategy in Rosetta (28).
Fig. 1.
Design strategies for stabilizing IL-2. (A) Structural context of the IL-2 receptor complex (PDB: 2B5I), with targeted regions of IL-2 (gray) color coded by design strategy. (B) Design strategies for stabilizing IL-2 and improving affinity to IL-2Rβ. Four main strategies were used: thermostabilizing by sequence redesign using the iterative enrichment algorithm (yellow); idealizing the loop structure between helices B and C (blue); N-terminal helix capping (green); and redesigning position 72, which is influential for CD25 interaction (pink). Helices A to D of IL-2 are labeled. (C) Sequence comparisons of stabIL-2 designs with WT IL-2 and super-2 H9. Colors correspond to the strategies described in B.
We implemented strategy 1 using an iterative enrichment design procedure, where the residues on the IL-2 structure (Protein Data Bank [PDB]: 1M47) were first completely redesigned before assessing the mutational behavior of each position one by one. The amino acids highly enriched during sampling at each design round were then fixed to a single or a small subset of amino acids in a subsequent design round. This was done to allow better sampling of the packing environment around frequently observed residues by using them to “seed” future design calculations. At each iteration, backbone torsions were also allowed to vary during energy minimization steps to accommodate these changes. Since design and repacking can be sensitive to small variations in backbone architecture, this allows for design trajectories to be initiated from more varied, energetically favorable conformations. We started this procedure throughout the entire core of the IL-2 structure and we observed locally organized pockets of residues where the mutations were coordinated. In these regions, the packing patterns depended on the identity of the seeding residues. Positions 27, 28, 31, 39, 69, 85, 86, 92, and 115 accumulated as the final set of positions to characterize experimentally.
The BC loop is highly relevant to the placement of the helices on IL-2; several key super-2 mutations played the role of stabilizing this loop. For strategy 2, we completely rebuilt the entire structure of the loop connecting helices B and C, between residues 74 and 82, using a fragment-based design strategy implemented in RosettaRemodel. A loop junction between two helices usually follows conventional α-α turn motifs (29), and we sought to use idealized connections to bridge the helices. Due to the small length deficits in the helix C placement in the PDB structure, to satisfy a canonical α-α turn, we explored extensions of helix C together with varying loop lengths for the BC loop and found optimal sampling convergence with a three-residue extension for helix C and a three-residue loop connection (SI Appendix, Fig. S1). We used RosettaRemodel to specify the loop motifs and helical extensions in one step but sampled extensively at each length combination to derive the most idealized connections for the helices. The final loop motif is an A-G-B-A-A loop (according to the definition in ref. 29) on a length adjusted helix C. We forced a glycine at the “G” position and allowed Rosetta to design the helical capping residue (at the “B” position) and the sequences for the surrounding environment (including the rest of helix C extension and residues that would come in contact with this region). The redesign of the BC loop yielded the connecting sequences reported on S14 to S17 but for the final set of sequences tested experimentally, we further incorporated mutations that would complete helical capping and one that we hypothesized to disrupt CD25 binding.
In strategies 3 and 4, we allowed Rosetta to sample rotamers for allowed amino acids [e.g., D/N/S/T for helix caps at positions 32 and 52 (30) or small/polar residues for position 72]. We did not explicitly model the interface to CD25 in the context of the bound complex. During the sequence exploration steps in strategy 1, we explored polar residues on position 72 because an exposed leucine residue is generally considered unfavorable. We followed the same sampling strategies as described in strategy 1, with fast energy minimization under the Rosetta score function to allow backbone movement and manually inspected the outputs. The resulting structural ensemble was superimposed on the super-2 in the quaternary complex structure (PDB: 2B5I), and it showed that a simple mutation on residue 72 on the redesigned IL-2 may be sufficient to disrupt CD25 interaction. We, therefore, did not further model this site and proceeded with a mutation on residue 72 for experimental validation.
In order to more efficiently explore sequence space, we pursued different strategies in combination with each other. This also allows for interdependent strategies to be designed jointly; for example, the BC loop is also involved in core hydrophobic packing, necessitating both to be remodeled with the other in mind. From low-energy design solutions, we selected 17 designs that include different sequence variations based on a combination of our 4 guiding hypotheses, as well as control sequences (such as those incorporating mutations from super-2) (Fig. 1C). These allowed us to explore the factors that contribute to IL-2 stability and function: for example, 13 (designs S1 to S13) include BC loop remodeling, while 4 (S14 to S17) include the wild-type (WT) BC loop; 4 designs (S1, S8, S10, S11) also include additionally the Val85, Val86, and Phe92 mutations from super-2 as controls. Computational calculations did not suggest these specific mutations, so we included them in a subset in the context of other design features to assess the impact of these mutations. In general, all designs included hydrophobic core redesign and repacking (strategy 1), while some added helical cap optimization (strategy 3) and Rosetta sampling of position 72 (strategy 4); we can compare the results from this panel to assess the relative contribution of the various designed elements.
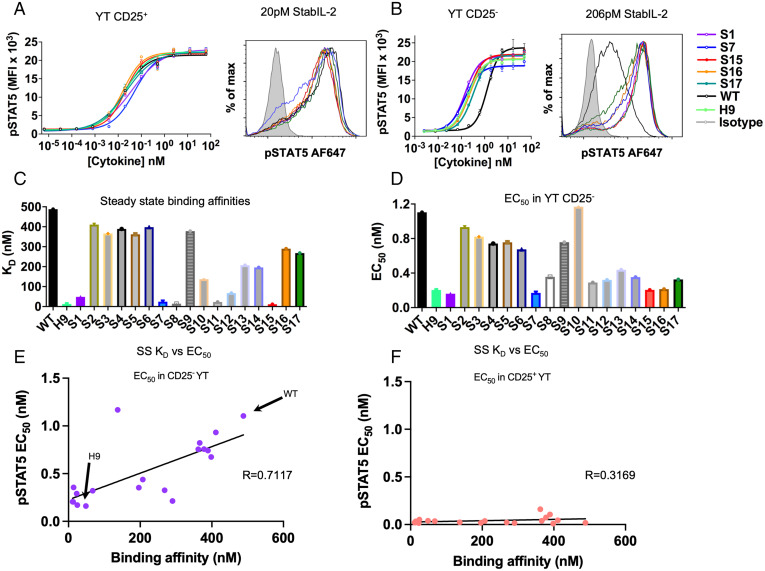
To assess the properties of these designs, we expressed them in Expi293 cells and measured their effects on STAT5 signaling in IL-2 responsive human NK cell lines, unmodified (CD25−) YT and CD25+ YT cells, by phospho-flow cytometry detection of phospho-STAT5 (Tyr694) (SI Appendix, Figs. S2–S4). While most designs exhibited similar signaling potency as WT IL-2 on CD25+ YT cells, nearly all designs showed equal or stronger signaling on CD25− YT cells, suggesting the improved CD25-independent affinity of the designs for IL-2Rβ (Fig. 2 A and B). To confirm this, we measured the designs’ affinity for IL-2Rβ by surface plasmon resonance and found that almost half of the designed sequences showed affinity for IL-2Rβ at a level comparable with super-2 (H9) (Fig. 2C, Table 1, and SI Appendix, Fig. S5 and Table S1). We observed a strong correlation between EC50 in CD25− YT cells and steady-state binding affinity to IL-2Rβ (Fig. 2 D–F). We also noted that the mutations L85V, I86V, and I92F from super-2 H9 seemed to be associated with slow dissociation, though other mutations also conferred the same effect. Across three of the four designs which contained the super-2 mutations (S1, S8, and S11), the off-rate seemed to decrease, while remaining fairly fast for most other designs. S15, however, does not require these mutations to achieve a similar off-rate. Contrasting the S15 mutations with those from the WT and S14 to S16, it is clear that the L72Q mutation in S15, in the context of native M39 and E52, plays an important role in mediating slow dissociation (SI Appendix, Fig. S4). Positions 72 and 39 are proximal in the IL-2 structure, indicating a possible interaction between the bulky Leu39 sidechain of S16 and the helical structure at position 72.
Fig. 2.
Functional properties and steady-state binding of stabIL-2 designs. (A and B) Dose-response curve (Left) and histogram at indicated concentration (Right) showing STAT5 phosphorylation in (A) CD25+ and (B) CD25− YT cells following a 15-min cytokine stimulation. Data are shown as the mean ± SD of triplicate wells and are representative of two independent experiments. (C) Steady-state binding affinities measured by surface plasmon resonance. (D) EC50 values for stat5 phosphorylation, measured on CD25− YT cells. (E) Correlation of improved IL-2Rβ binding affinity with improved signaling in CD25- YT cells. The Pearson correlation coefficient (R) is calculated. (F) Correlation of IL-2Rβ binding affinity with signaling in CD25+ YT cells.
Table 1.
Functional and biophysical parameters of S1 and S15 compared to WT and super-2 H9
| Seq | Tm (°C) | EC50, CD25− YT (nM) | EC50, CD25+ YT (nM) | IL-2Rβ Kd, steady state (nM) | IL-2Rβ ΔH (kcal/mol) | IL-2Rβ ΔS (kcal/mol) | IL-2Rβ Kd (ITC) (nM) |
|---|---|---|---|---|---|---|---|
| WT | 43.2 | 1.104 | 0.01837 | 374 | −7.64 (1) | 4.79 (1) | 144 (1) |
| H9 | 52.0 | 0.204 | 0.02088 | 16 | −15.0 (1) | −13.1 (1) | 2.92 (1) |
| S1 | 79.0 | 0.1606 | 0.03897 | 61 | −14.13 ± 0.46 | −12.3 | 9.4 ± 5.7 |
| S15 | 75.4 | 0.2051 | 0.02011 | 11.5 | 9.4 ± 0.3 | 1.6 | 30 ± 10 |
ITC measurements taken at 288 K.
We sought to examine whether the super-2–like properties of these computational designs were indeed due to improved thermal stability as originally intended. Using the same construct as for the pStat5 assays, we measured the secondary structure formation and irreversible thermal denaturation of these designs by far-UV circular dichroism (CD). Indeed, nearly all designs were significantly more stable than WT IL-2 or H9, as designed (SI Appendix, Fig. S6), suggesting that the structural flaws selected for redesign were real and that WT IL-2 is suboptimally fit in terms of thermal stability.
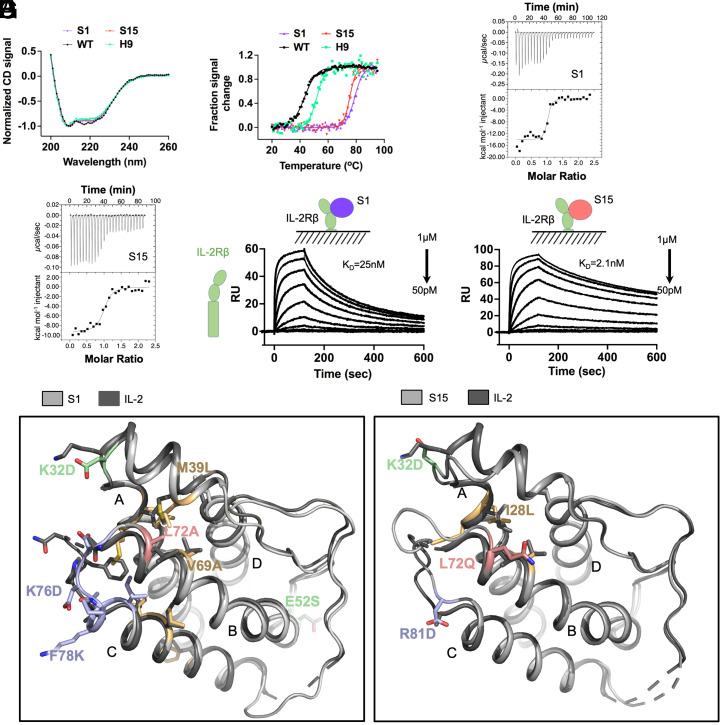
Two of the strongest agonists were selected for further biophysical characterization to determine the origins of the improved affinity. We recombinantly expressed S1 and S15 in Escherichia coli without tags and refolded the protein from inclusion bodies (SI Appendix, Fig. S7). Formation of secondary structure was consistent with super-2 secondary structure, supporting that the variants fold as designed (Fig. 3A). Quantifying the change in melting temperature for these designs indicated a remarkable ∼30 °C increase in melting temperature relative to WT IL-2 and super-2 (Fig. 3B and Table 1). We next measured the thermodynamic binding affinity of S1 and S15 for IL-2Rβ by isothermal titration calorimetry. Interestingly, we observed differential mechanisms for the improved affinity to IL-2Rβ (Fig. 3 C–F and Table 1). Whereas both designs bind IL-2Rβ with a dissociation constant in the tens of nanomoles, S1 (which includes the idealized BC loop) exhibits a significant enthalpic contribution to binding with an unfavorable binding entropy, while S15 (which has the native BC loop) has a slightly enhanced binding enthalpy while retaining a favorable binding entropy (Fig. 3 E and F). This suggests a role for preorganization of the binding conformation.
Fig. 3.
Increased stability and binding affinity, and structures, of stabIL-2 S1 and S15. (A) S1 and S15 designs adopt secondary structure profiles similar to WT and H9 as measured by far-UV CD. (B) Thermal stability of S1, S15, WT, and H9 assessed by heat denaturation monitored by CD at 222 nm. (C and D) Thermodynamic binding affinity of S1 (C) and S15 (D) against IL-2Rβ compared to WT and H9, measured by isothermal titration calorimetry. (E and F) Steady-state binding affinity of S1 (E) and S15 (F) against IL-2Rβ ECD compared to WT and H9, measured by surface plasmon resonance. (G) Crystal structures of S1 (Left, PDB: 7RA9) and S15 (Right, PDB: 7RAA) overlaid against WT (PDB: 1M47). Helices A to D are labeled on each structure.
To investigate the structural basis of these properties, we solved crystal structures of both S1 and S15 (Fig. 3G and SI Appendix, Figs. S8 and S9). The crystal structures were in agreement with the design models, with a Cα rmsd of 0.952 Å (100 atoms) and 0.738 Å (107 atoms), respectively, with the greatest deviations at the CD loop for S1 and the AB and BC loops for S15 (SI Appendix, Fig. S9 A and B). The junction of the BC loop and helix C is a plastic region of IL-2 that can adopt either an “in” state or an “out” state, which is preorganized for binding to IL-2Rβ. In both structures, the structures adopt the out state, with an outward movement at the N terminus of helix C that prearranges the IL-2Rβ binding site. In both S1 and S15, the bulky side chain presented by the nearby G27M mutation occludes the in state, while in seq1 helix C is extended by an additional half turn and held in position by the shortened BC loop (SI Appendix, Fig. S9 D–F). As part of our design process (strategy 3), we allowed the introduction of helical capping residues at the N-termini of helices B and C and the short helix within the AB loop. Small, hydrogen bond accepting side chains at these positions are predicted to stabilize helical conformations. In the case of S1, Ser52 and Asp76 make these stabilizing interactions at helices B and C, while Asp-32 in the AB loop helix points outward and does not make a stabilizing contact (SI Appendix, Fig. S9F). Similarly, in S15 Asp81 stabilizes helix C while Asp32 points outward (SI Appendix, Fig. S9G). The unsuccessful helical caps are both obstructed by the presence of a proline at the i+2 position, suggesting that this motif should be considered when introducing helical caps.
The goal of design strategy 1 was to optimize the core packing of IL-2 by removing packing defects between helices A, B, and C. These mutations were located in the end near the CD25 binding site (Fig. 1A). Comparison of the crystal structures to WT IL-2 (PDB: 1M47) reveals a similar quality of packing for S1 and slightly reduced packing interactions for S15 (SI Appendix, Fig. S9 H–J). These are reflected in mean occluded surface values of 0.396 for wt, 0.397 for S1, and 0.384 for S15 (31, 32). The BC loop optimization that was performed more extensively in S1 may contribute make additional contributions to the packing optimization, and the suboptimal packing of S15 suggests that stabilizing the IL-2Rβ binding conformation of helix C is the key driver of affinity.
Two of the positions targeted during the design process, Val69 and Val115, have high solvent accessibility and we hypothesized that mutating these to polar or smaller, less hydrophobic residues as recommended by Rosetta would improve the solubility and stability of designs, thereby improving the IL-2Rβ affinity. To test this, we reverted these two positions back to the WT sequences (both valine) and tested their ability to induce Stat5 phosphorylation in vitro as before. We found that these two designs in fact maintained the same activity as the parent S1 and S15 designs, while retaining fewer mutations away from the WT IL-2 sequence (Fig. 4 and SI Appendix, Figs. S10 and S11).
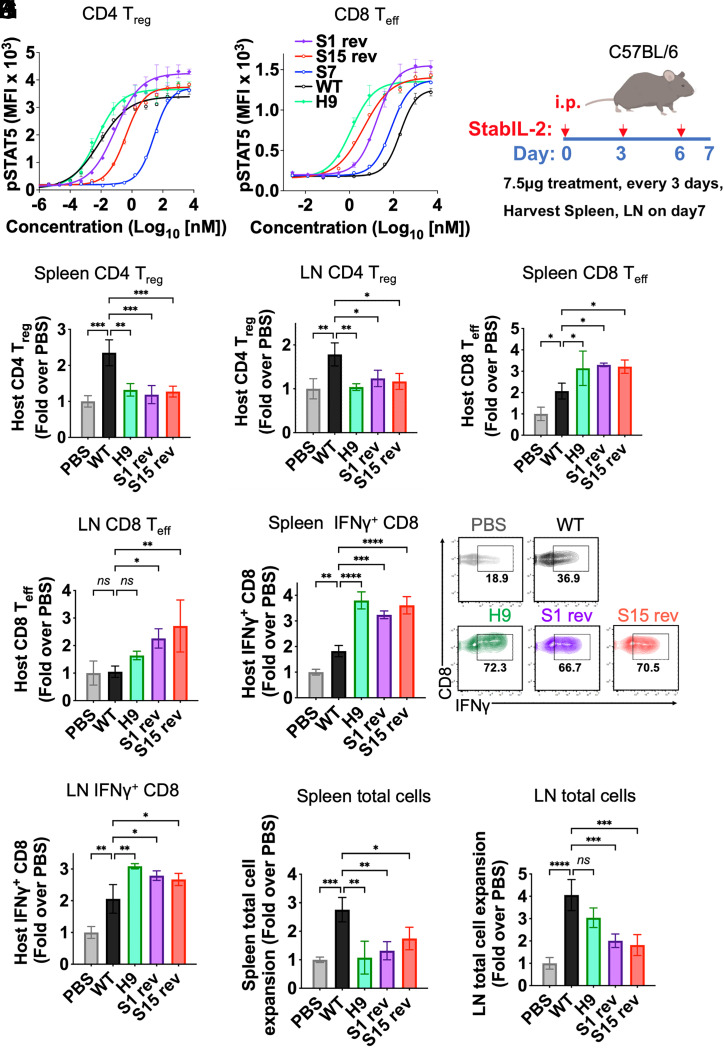
Fig. 4.
In vivo activity of stabIL-2 designs. (A and B) CD4+ Treg and CD8+ Teff were isolated from Foxp3-GFP reporter mice (C57BL6/J). A total of 250,000 cells were incubated with stabIL-2s for 15 min at 37 °C. Dose-response curve showing STAT5 phosphorylation in (A) CD4+ Tregs and (B) CD8+ cells following a 15-min cytokine stimulation. (C) Schematic of the in vivo immune cell profiling of stabIL-2 designs in mice. (D–G) Quantification of stabIL-2 designs administration on BL6 mice (B and C) Treg (CD4+CD25+Foxp3+)and (D and E) CD8+ Teffs (CD44+CD62L−). (H and I), Quantification of cytokine administration on C57BL/6 mice CD8+ production of IFN-γ upon ex vivo restimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin in (F) spleen and (G) lymph nodes (LNs). (J and K) Total cell count of immune cells in the (H) spleen and (I) LNs after stabIL-2 administration. Bar graphs show mean ± SD and were analyzed by one-way ANOVA relative to WT IL-2. Multiple comparisons were corrected using Dunnett’s test. Data are representative of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
IL-2 is a proinflammatory cytokine, which potently induces the activation of multiple immune cells (CD4+ and CD8+ T cells and NK cells) and plays a role in T cell differentiation and exhaustion. Tuning IL-2 signaling toward immune activation is an appealing strategy for immunotherapy. One major limitation of IL-2 in immunotherapy is that IL-2 also strongly activates Tregs, which suppress the effector function of CD8+ Teffs. Tregs express much higher CD25 than CD8+ Teff. Since the top winners of our designs are CD25 independent, we hypothesized that these stabIL-2 designs are CD8+ Teff-biased inducers. To investigate whether the stabIL-2 designs are more proinflammatory than IL-2, we determined the in vivo activity of stabIL-2s together with IL-2 and H9 in C57BL/6 mice. We first isolated Tregs and CD8+ T cells from Foxp3GFP+ transgenic mice and tested stabIL-2s in these cells by measuring Stat5 phosphorylation (pSTAT5) using flow cytometry. Notably, S1 rev and S15 rev induce similar pStat5 signaling potency as WT IL-2 in Tregs, but have superior signaling to IL-2 in CD8+ T cells ex vivo (Fig. 4 A and B), indicating CD8+ T cell biased activity of these stabIL-2 designs. Similarly, when administered to mice systemically, both S1 and S15 induced significantly fewer Tregs and more CD8+ Teff cells than WT IL-2 in both the spleen and lymph nodes (Fig. 4 C–G and SI Appendix, Fig. S11 A and B). Interferon (IFN)-γ production is a key downstream event in IL-2 induced T cells and is a key marker of Teff function. Importantly, S1 rev and S15 rev treatment increased host CD8+ T cell IFN-γ production upon ex vivo restimulation (Fig. 4 H and I). Moreover, S1 rev and S15 rev treatment had no detectable effect on innate immune cell populations (NK cells, monocytes, or granulocytes) (SI Appendix, Fig S11 C–F). However, although stabIL-2s did not show an effect on the spleen weight index, fewer total immune cells were observed compared to mice treated with WT IL-2 (Fig. 4 J and K and SI Appendix, Fig S11G).
Discussion
The ability to redesign proteins for optimized binding by improving stability, which we have demonstrated here, is an approach to computational affinity maturation. By applying principles derived from de novo protein design to native protein templates, we achieved IL-2Rβ affinity and potency at similar or better levels than that achieved in earlier studies. Remarkably, these results were attained directly by screening only 17 designs, compared with other studies which required screening of large libraries and extensive experimental optimization.
This work uncovers modalities for improved IL-2Rβ affinity. Though super-2 affinity was primarily driven by stabilizing the core of the protein, some surface mutations were required to enhance binding to the receptor, resulting in an enthalpically driven binding event. Here, in the case of S15, super-2–like affinity was achieved without making any packing or interface mutations on helix C, demonstrating that a stable conformation of the BC loop is sufficient to prearrange IL-2 for binding to IL-2Rβ.
The application of this design approach is particularly pertinent to IL-2 given current intense interest in engineering IL-2 as a cancer immunotherapeutic. Simultaneous activation of Teffs and Tregs is a common feature of IL-2 therapy (33). However, Tregs suppress the activity of Teffs and limit immune activation (19), suggesting a need for IL-2R “β/γ” biased IL-2 analogs which we have designed here. Many current clinical approaches are focused on IL-2R β/γ receptor biased variants that are intended to show preference for the CD25-negative Teffs versus CD25+ Tregs. Several of these IL-2 variants achieve bias through blocking access to the CD25 binding site of IL-2 but rely on the low affinity of WT IL-2 for IL-2Rβ, which would require dosing at high levels to achieve receptor occupancy (21–23). Super-2 and Neo-2/15 were both engineered for high affinity to IL-2Rβ but required extensive experimental intervention. Here, we find that stabIL-2 S1 and S15 show CD8 Teff bias by virtue of higher affinity for IL-2Rβ, and this was achieved without any experimental engineering intervention. StabIL-2 proteins are a new member of the IL-2 toolkit for cancer immunotherapy.
The significant improvement in IL-2 function from a relatively simple structural idealization and stabilization process suggests that the strategy could potentially be applicable to other proteins as well. While the gamma chain cytokines IL-2 and IL-4 can uniquely be tuned to different cell types by modulating affinity to their different receptors, cytokine affinity and dwell time can also affect signaling outcomes (34, 35). The ability to identify a minimal set of mutations that increase both binding affinity and cell type specificity is a valuable outcome of this research. The ability to obviate the need for interface engineering will have a significant effect on the cost and rate of developing new protein binders. More broadly, we expect the ability to computationally stabilize proteins for improved binding to have broad applications in the development of new research and therapeutic tools across many classes of proteins, including transcription factors, kinases, and others.
Materials and Methods
Cells.
Cell lines were kept in a humidified incubator at 37 °C with 5% CO2. YT-1 cells lacking or expressing CD25 were cultured in RPMI supplemented with 2 mM GlutaMAX (Invitrogen), 10% fetal bovine serum (Sigma), 10 mM Hepes, pH 8.0 (Thermo Fisher), 1 mM sodium pyruvate (Gibco), and 50 U ml−1 penicillin and streptomycin (Thermo Fisher). Expi293 cells (female-derived kidney cell line) were grown in Expi293 Expression Medium (Thermo Fisher) at 37 °C, 125 rpm, and 8% CO2 atmosphere with 80% humidity in plastic flasks with ventilated caps (Corning Erlenmeyer sterile polycarbonate with 0.2-mm ventilated caps, plain bottom).
Iterative Enrichment Design.
Design strategies outlined in Fig. 1 were implemented with backbone fragment sampling and sequence redesign methods in RosettaRemodel (36). Monte Carlo trajectories were initiated from the WT IL-2 crystal structure (PDB: 1M47) which was relaxed and idealized under the Rosetta energy function. Using design blueprints based on the four main design strategies, 1,000 Monte Carlo trajectories were launched, and the results were collected and ranked by Rosetta energy. Sequence design for all strategies was conducted with three iterative rounds of amino acid sampling and rotamer packing using the backbone-dependent rotamer library in Rosetta, followed by energy minimization with ramping repulsion constraints. For the iterative enrichment procedure, this three-round sequence design step was conducted at each round of iteration, followed by sequence distribution analysis. Four successive rounds of iterative enrichment were performed, with the sequence distribution of previous rounds used to guide blueprints for the next round, and low-energy designs were selected as starting points. Example Rosetta scripts and design blueprints are shown in SI Appendix, Fig. S12.
BC Loop Redesign.
Using the design with the lowest free energy, we computationally removed the native BC loop and extended helix C by a range of 1 to 10 residues (SI Appendix, Fig. S1A). We determined extensions by three, six, or seven residues to be the best candidates for loop backbone redesign based on the idealized geometry of helix C facing helix B. Following this, we sampled BC loop backbones ranging in length from one to five residues for each helix C extension length. Based on backbone convergence and apparent loop strain, we narrowed our search space down to a helix C extension of three residues and a redesigned BC loop length of two or three residues (SI Appendix, Fig. S1B). To establish the ideal BC loop length, we then tested α-α turn motifs A-B-B-A, A-G-B-A, and A-G-B-B-A from ref. 29 and settled on the A-G-B-B-A motif due to an overwhelming backbone convergence when compared with the other loop motifs. To finish, we conducted a final iterative enrichment process to optimize the sequence and rotamer packing of the protein.
Mammalian Protein Expression and Purification.
The stabIL-2s were cloned in a pD649 mouse serum alubumin (pD649-MSA) vector. The plasmids were validated by Sanger sequencing. Expi293 cells were cultured at 2.5 × 106/mL. Expi293 cells were transfected with stabIL-2 plasmids by following the ExpiFectamine transfection kit. Enhancers were added in the cells 18 h posttransfection. The supernatants of Expi293 cells were harvested 4 d posttransfection. The proteins were enriched by nickel beads and purified by fast protein liquid chromatography (FPLC) gel filtration. For in vivo study, the proteins will be further cleaned on an endotoxin removal column.
In Vitro Cell Signaling and Screening.
YT-1 CD25+ cells and YT-1 cells, which are CD25 negative, were cultured in complete RPMI 1640 medium. T-1 CD25+ cells and YT-1 cells were mixed 1:1 and stimulated with IL-2 or the stabIL-2s ligands for 15 min at 37 °C, stained with anti-CD25 antibody for 20 min at 4 °C, followed by fixation with 1.2% paraformaldehyde for 10 min at room temperature. After permeabilization with 100% methanol for 20 min or overnight at −20 °C, the cells were then incubated with anti-Stat5 (pY694) at a 1:200 dilution for 1 h at room temperature on a shaker. The cells were washed and analyzed on a CytoFlex flow cytometer instrument (Beckman Coulter). FCS files were exported. The levels of the phospho-Stat5 in CD25+ and CD25− YT cells correspond to the measured values of mean fluorescence intensity (MFI). MFI values were analyzed in FlowJo (Tree Star). The dose response of p-stat5 MFI was plotted in Prism8 (GraphPad).
Ex Vivo Signaling in CD8 T Cells Versus Tregs.
CD8 T cells and CD4+ Foxp3+ Tregs were harvested from Foxp3GFP+ transgenic mice (JAX 023800). Briefly, spleen and lymph nodes from Foxp3GFP+ transgenic mice were collected and minced. Red blood cells were removed by ammonium-chloride–potassium lysis (Biolegend) for 5 min at room temperature. T cells were enriched by the Pan T Cell Isolation Kit (Miltenyi Biotec). The cells were washed and resuspended in complete RPMI 1640 medium. The cells were stimulated with IL-2, H9, or the stabIL-2s ligands for 15 min at 37 °C; stained with anti-CD4, CD8, or CD25 antibodies for 20 min at 4 °C; followed by fixation with 1.2% paraformaldehyde for 10 min at room temperature. After permeabilization with 100% methanol for 20 min or overnight at −20 °C, the cells were then incubated with anti-Stat5 (pY694) at a 1:50 dilution for 1 h at room temperature. The cells were washed and analyzed on CytoFlex and FlowJo. MFI values of phospho-Stat5 in CD8 and Treg populations were analyzed in FlowJo. The dose response of phospho-Stat5 MFI was plotted in Prism8 (GraphPad).
Binding Affinity by SPR.
For affinity measurements, IL-2Rβ was cloned into pAcGP67 vector with a C-terminal AviTag (GLNDIFEAQKIEWHE) and 6× His. Human MSA-stabIL-2s were cloned into pD649 with an N-terminal hemagglutinin signal peptide. P0 and P1 baculoviruses of IL-2Rβ were generated by transfecting SF9 cells. The proteins were expressed in T. ni cells, purified by Ni-nitriloacetic acid chromatography followed by FPLC and incubated with soluble BirA ligase in buffer containing 0.5 mM Bicine (pH 8.3), 100 mM adenosine triphosphate, 100 mM magnesium acetate, and 500 mM biotin overnight at 4 °C. Biotinylated proteins were then purified by FPLC. Affinity measurements were made using a BIAcore T100 instrument. Binding measurements were performed at 25 °C with a flow rate of 50 mL/min. Binding curves were exported for analysis in Prism (GraphPad Software) and steady-state affinity was calculated using BIAevaluation software (Cytiva).
Binding Affinity by ITC.
Calorimetric measurements of MSA-tagged seq1 and seq15 binding to IL-2Rβ were performed as previously described (37). Briefly, titrations were performed on a VP-ITC calorimeter (MicroCal) at 15 °C. Prior to titration, protein and reference water was degassed for 10 min. All samples were extensively dialyzed in 10 mM Hepes, pH 7.3, and 150 mM NaCl (HBS) before titration to minimize heat of dilution effects. Protein concentrations were determined by the Edelhoch method. IL-2Rβ was diluted to 2 to 3 µM as the titrand, and the IL-2 mimetics were injected at 10-fold higher concentration. Data were processed and analyzed using MicroCal Origin 5.0 software.
Bacterial Protein Expression and Purification.
Super-2 was expressed in and purified from E. coli as described previously (24). To express protein, sequences for stabIL-2 S1 and S15 were cloned into pET-24a without tags and transformed into E. coli BL21 (DE3) cells. For each gene, a single colony was picked and used to inoculate 5 mL of 2× YT broth (Research Products International) and grown overnight to saturation. This was used to inoculate 1 L of 2× YT broth and incubated at 37 °C with shaking at 200 rpm, until the culture optical density at 600 nm reached 0.8, about 4 h. The culture was chilled on ice for 10 min, then induced with isopropyl β-d-1-thiogalactopyranoside (MP Biomedicals) and incubated at 16 °C for 4.5 h with shaking at 200 rpm. Cells were harvested by centrifugation and resuspended in 50 mM Tris, pH 7.5, and frozen at −80 °C until extraction and purification.
To extract and refold protein, frozen pellets were thawed and lysed by resuspension in 12.5 mL BugBuster (Millipore), 600 µL of 50 mM phenylmethylsulfonyl fluoride (Roche), 200 µL of 50 mg/mL lysozyme (MP Biomedicals), and 10 µL benzonase nuclease (EMD Millipore), followed by sonication for 30 s, resting for 30 s (total pulse time 3 min). The sample was then washed first in 100 mM Tris, pH 8, 2 mM ethylenediaminetetraacetic acid (EDTA), and 0.1% deoxycholate (Sigma), then twice in 100 mM Tris, pH 8, and 2 mM EDTA by centrifugation for 20 min at 10,000 rpm followed by the same sonication step as before. Guanidinium chloride (GdmCl; EMD Millipore Ultrol Grade) and dithiothreitol (DTT; Thermo Fisher) were added to 6 mL of sample for a final concentration of ∼6 M GdmCl and 1 mM DTT. The sample was incubated for 1 h at room temperature with agitation, then centrifuged as before, chilled at 4 °C, and added dropwise to 100 mL refolding buffer at 4 °C (1.1 M GdmCl, 110 mM Tris, 6.5 mM cysteamine [Santa Cruz Biotechnology], and 0.65 mM cystamine [Santa Cruz Biotechnology], pH 8.0). After 1 d of refolding, the sample was dialyzed twice overnight into 5 L of 20 mM Tris and 50 mM NaCl, pH 8.
Refolded protein was injected onto a MonoQ 5/50 column (Cytiva) and eluted with a gradient to 500 mM NaCl. Protein was then concentrated and injected onto a Superdex S75 10/300 column (Cytiva), then eluted in HBS. The proteins eluted as a single peak and were concentrated on Amicon Ultra centrifugal filtration devices (Millipore) (SI Appendix, Fig. S10).
Far-UV CD.
For WT IL-2, human IL-2 recombinant protein from Proleukin was used. CD spectra were measured on a JASCO CD spectrophotometer in a 1-mm pathlength cuvette (Hellma). Protein sample was at ∼0.22 mg/mL in phosphate-buffered saline solution (Santa Cruz Biotechnology). Heat denaturation curves were obtained by monitoring the CD signal at 222 nm in 1 °C increments every 30 s from 20 to 95 °C, with 10 s of equilibration time and 1 s digital integration time.
X-Ray Crystallography.
Refolded seq1 was concentrated to 7.5 mg/mL and crystallized using the MCSG2 screen in 0.2 M Li2SO4, 0.1 M N-cyclohexyl-3-aminopropanesulfonic acid, pH 10.5, 1.2 M NaH2PO4, and 0.8 M K2HPO4. Crystals were cryoprotected by addition of 10 volumes of 3.4 M sodium malonate, pH 7, and flash cooled in liquid nitrogen. Diffraction data were collected at APS beamline 23ID-B. Data were processed with autoproc (38) and the structure was solved by molecular replacement using Phaser (39) with wt IL-2 as a search model (PDB: 4NEJ). The structure was initially rebuilt using phenix.autobuild (40) with subsequent cycles of interactive rebuilding in coot (41) and reciprocal space refinement in Phenix (42, 43). The final refinement strategy in Phenix included individual B-factor refinement, TLS refinement using a single group, and occupancy refinement of alternate conformations.
Refolded seq15 was concentrated to 19 mg/mL. Initial crystals were obtained using the Index screen (Hampton) in 0.2 M MgCl2, 0.1 M bis-Tris, pH 6.5, and 25% polyethylene glycol (PEG) 3350. These crystals were crushed for seeding. Final crystals were formed by mixing 150 nL seq15, 50 nL microseeds, and 100 nL well solution (0.2 M MgCl2, 0.1 M Tris pH 8.3, 24% PEG 3350) and equilibrating as sitting drops over well solution. Crystals were cryoprotected by addition of glycerol to 30% and flash cooled in liquid nitrogen. Diffraction data were collected at SSRL beamline 12-2. Data were processed with XDS (44), and an anisotropic resolution cutoff was applied using STARANISO (45). The structure was solved by molecular replacement using Phaser, with four copies of the protein found in the asymmetric unit using the predicted structure of seq15 as a search model. The structure was initially rebuilt using noncrystallographic symmetry (NCS)-averaged prime-and-switch maps using phenix.autobuild, with subsequent cycles of manual rebuilding in coot and reciprocal space refinement in Phenix. The final refinement in Phenix included individual B-factor refinement, TLS refinement using groups identified with TLSMD (46), and NCS restraints. Data collection and refinement statistics for both structures are reported in SI Appendix, Table S2. Mean occluded surface measurements were calculated using OS v7.6 (31, 32). Crystallographic software for this project was installed and configured using SBGrid (47).
Mice.
All mice were housed at Stanford University and were maintained according to protocols approved by the Stanford University Institutional Animal Care and Use Committee on Animal Care. C57BL/6J mice (catalog no. 000664) and Foxp3GFP+ transgenic mice (JAX 023800) mice were purchased from Jackson Labs.
Mouse Immune Cell Profiling and Flow Cytometry.
C57BL/6J mice were treated intraperitoneally with 7.5 mg stabIL-2s three times every 3 d. The mice were killed humanely at day 7 post-first treatment, and the spleen and lymph nodes were harvested. Immune cells prepared from mouse spleens and lymph nodes were stained with a mixture of fluorescence-activated cell sorting (FACS) antibodies in FACS buffer (2% fetal bovine serum in phosphate-buffered saline) for 30 min on ice. Immune cell subsets were gated using the following markers: CD4 T cells (CD3+CD4+), CD8 T cells (CD3+CD8+), NK cells (CD3−NK1.1+Nkp46+), CD4 Treg (CD3+CD4+Foxp3+CD25+), CD4 naive (CD3+CD4+CD44−CD62L+), CD4 memory (CD3+CD4+CD44+CD62L+), CD4 effector (CD3+CD4+CD44+CD62L−), CD8 naive (CD3+CD8+CD44−CD62L+), CD8 memory (CD3+CD8+CD44+CD62L+), and CD8 effector (CD3+CD8+CD44+CD62L−). NK activation was quantified via surface staining of CD25. For CD4 Treg cells, immune cells were stained with CD3, CD4, CD25, and fixable live/dead dye (catalog no. 65-0865-14, Invitrogen), intracellular Foxp3 staining was performed by following the protocol of FOXP3 Fix/Perm buffer set (catalog no. 421403, Biolegend). Cells were analyzed using a CytoFlex (Beckman Coulter). For IFN-γ recall, immune cells were stimulated with phorbol 12-myristate 13-acetate, ionomycin, and GolgiStop for 4 h. The cells were stained with CD3, CD4, CD8, CD44, and fixable live/dead dye, and intracellular IFN-γ staining was performed according to the protocol of eBioscience Intracellular Fixation & Permeabilization Buffer Set (catalog no. 88-8824-00, Thermo Fisher).
Statistical Analysis.
Statistical analyses were performed using Prism v9.1.0 (GraphPad Software). Data are expressed as the mean ± SD unless otherwise indicated. We used one-way ANOVA followed by Dunnett’s multiple comparisons tests to analyze experiments with more than two groups. Significance is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank Robert A. Saxton, Caleb R Glassman, Xiang Zhao, and Michelle Yen for helpful advice and discussions; Jingjia Liu for assistance with CD experiments; and Viviana Silva de Paula for providing experimental protocols. This work was supported by NIH Grant R01-AI51321, Howard Hughes Medical Institute, Ludwig Institute, and Mathers Foundation to K.C.G. P.-S.H. is partially supported by the US Department of Energy (DOE), Office of Science, Office of Advanced Scientific Computing Research, Scientific Discovery through Advanced Computing program and a Frederick E. Terman Fellowship from Stanford’s School of Engineering. A.E.C. is supported by a NSF Graduate Research Fellowship. This research used resources of the Advanced Photon Source, a DOE Office of Science User Facility, operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the DOE, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the NIH National Institute of General Medical Sciences (P30GM133894).
Footnotes
Reviewers: T.K., University of California, San Francisco; and K.W., Massachusetts Institute of Technology.
Competing interest statement: The molecules described herein are included in patent filing PCT/US2021/021137 and have been licensed by Dragonfly Therapeutics. K.C.G. is a consultant of Dragonfly Therapeutics. P.-S.H. and Tanja Kortemme are coauthors on a 2020 review article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117401119/-/DCSupplemental.
Data Availability
WT protein sequences were obtained from Uniprot (https://www.uniprot.org/). Structure factors and coordinates have been deposited in PDB (identification nos. 7RA9 and 7RAA), and the corresponding diffraction images have been deposited in the SBGrid Databank: https://doi.org/10.15785/SBGRID/882 (seq1, pdb ID 7RA9) and https://doi.org/10.15785/SBGRID/881 (seq15, pdb ID 7RAA). There are no restrictions on data availability. All other data are included in the article and/or SI Appendix.
References
- 1.Sidhu S. S., Koide S., Phage display for engineering and analyzing protein interaction interfaces. Curr. Opin. Struct. Biol. 17, 481–487 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Gera N., Hussain M., Rao B. M., Protein selection using yeast surface display. Methods 60, 15–26 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Cherf G. M., Cochran J. R., Yeast surface display, methods, protocols, and applications. Methods Mol. Biol. 1319, 155–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang P.-S., Boyken S. E., Baker D., The coming of age of de novo protein design. Nature 537, 320–327 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Korendovych I. V., DeGrado W. F., De novo protein design, a retrospective. Q. Rev. Biophys. 53, e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan X., Kortemme T., Recent advances in de novo protein design: Principles, methods, and applications. J. Biol. Chem. 296, 100558 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C., et al. , Bottom-up de novo design of functional proteins with complex structural features. Nat. Chem. Biol. 17, 492–500 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Sesterhenn F., et al. , De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, eaay5051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldenzweig A., et al. , Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell 63, 337–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger C., et al. , Antigen recognition by conformational selection. FEBS Lett. 450, 149–153 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Lee A. L., Kinnear S. A., Wand A. J., Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat. Struct. Biol. 7, 72–77 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Wang C., Pawley N. H., Nicholson L. K., The role of backbone motions in ligand binding to the c-Src SH3 domain. J. Mol. Biol. 313, 873–887 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Grünberg R., Nilges M., Leckner J., Flexibility and conformational entropy in protein-protein binding. Structure 14, 683–693 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama M., et al. , Interleukin-2 receptor beta chain gene: Generation of three receptor forms by cloned human alpha and beta chain cDNA’s. Science 244, 551–556 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Takeshita T., et al. , Cloning of the gamma chain of the human IL-2 receptor. Science 257, 379–382 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Obar J. J., et al. , CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8+ T cells in primary and secondary responses. Proc. Natl. Acad. Sci. U.S.A. 107, 193–198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Létourneau S., Krieg C., Pantaleo G., Boyman O., IL-2- and CD25-dependent immunoregulatory mechanisms in the homeostasis of T-cell subsets. J. Allergy Clin. Immunol. 123, 758–762 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Handa K., Suzuki R., Matsui H., Shimizu Y., Kumagai K., Natural killer (NK) cells as a responder to interleukin 2 (IL 2). II. IL 2-induced interferon gamma production. J. Immunol. 130, 988–992 (1983). [PubMed] [Google Scholar]
- 19.Jiang T., Zhou C., Ren S., Role of IL-2 in cancer immunotherapy. OncoImmunology 5, e1163462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldmann T. A., The IL-2/IL-15 receptor systems: Targets for immunotherapy. J. Clin. Immunol. 22, 51–56 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Overwijk W. W., Tagliaferri M. A., Zalevsky J., Engineering IL-2 to give new life to T cell immunotherapy. Annu. Rev. Med. 72, 281–311 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Charych D. H., et al. , NKTR-214, an engineered cytokine with biased IL2 receptor binding, increased tumor exposure, and marked efficacy in mouse tumor models. Clin. Cancer Res. 22, 680–690 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Milla M. E., et al. , 1225P THOR-707, a novel not-alpha IL-2, promotes all key immune system anti-tumoral actions of IL-2 without eliciting vascular leak syndrome (VLS). Ann. Oncol. 30, v501 (2019). [Google Scholar]
- 24.Levin A. M., et al. , Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 484, 529–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Paula V. S., et al. , Interleukin-2 druggability is modulated by global conformational transitions controlled by a helical capping switch. Proc. Natl. Acad. Sci. U.S.A. 117, 7183–7192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva D.-A., et al. , De novo design of potent and selective mimics of IL-2 and IL-15. Nature 565, 186–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stauber D. J., Debler E. W., Horton P. A., Smith K. A., Wilson I. A., Crystal structure of the IL-2 signaling complex: Paradigm for a heterotrimeric cytokine receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 2788–2793 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leman J. K., et al. , Macromolecular modeling and design in Rosetta: Recent methods and frameworks. Nat. Methods 17, 665–680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wintjens R. T., Rooman M. J., Wodak S. J., Automatic classification and analysis of α α-turn motifs in proteins. J. Mol. Biol. 255, 235–253 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Doig A. J., Sternberg M. J. E., Side-chain conformational entropy in protein folding. Protein Sci. 4, 2247–2251 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleming P. J., Richards F. M., Protein packing: Dependence on protein size, secondary structure and amino acid composition. J. Mol. Biol. 299, 487–498 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Pattabiraman N., Ward K. B., Fleming P. J., Occluded molecular surface: Analysis of protein packing. J. Mol. Recognit. 8, 334–344 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Khailaie S., Montaseri G., Meyer-Hermann M., An adaptive control scheme for interleukin-2 therapy. iScience 23, 101663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Fabregas J., et al. , Kinetics of cytokine receptor trafficking determine signaling and functional selectivity. eLife 8, e49314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitra S., et al. , Interleukin-2 activity can be fine tuned with engineered receptor signaling clamps. Immunity 42, 826–838 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang P.-S., et al. , RosettaRemodel: A generalized framework for flexible backbone protein design. PLoS One 6, e24109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rickert M., Boulanger M. J., Goriatcheva N., Garcia K. C., Compensatory energetic mechanisms mediating the assembly of signaling complexes between interleukin-2 and its α, β, and γ(c) receptors. J. Mol. Biol. 339, 1115–1128 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Vonrhein C., et al. , Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger T. C., et al. , Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tickle I., et al. , STARANISO. https://staraniso.globalphasing.org/cgi-bin/staraniso.cgi. (2018).
- 46.Painter J., Merritt E. A., Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Morin A., et al. , Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WT protein sequences were obtained from Uniprot (https://www.uniprot.org/). Structure factors and coordinates have been deposited in PDB (identification nos. 7RA9 and 7RAA), and the corresponding diffraction images have been deposited in the SBGrid Databank: https://doi.org/10.15785/SBGRID/882 (seq1, pdb ID 7RA9) and https://doi.org/10.15785/SBGRID/881 (seq15, pdb ID 7RAA). There are no restrictions on data availability. All other data are included in the article and/or SI Appendix.