Abstract
A study of eight sulfonamide-resistant clinical isolates of Streptococcus pneumoniae revealed chromosomal mutations within the gene encoding dihydropteroate synthase that play a role in conferring resistance to sulfamethoxazole. The presence of the suld mutation, found previously only in a laboratory mutant, was shown to occur in three of the wild-type clinical isolates. The duplication of Ser61, the other previously defined mutation in the dihydropteroate synthase gene of S. pneumoniae, was observed in only one of the isolates characterized. We report two previously unidentified amino acid alterations, namely, a duplication of Arg58 and Pro59 and an insertion of an arginine residue between Gly60 and Ser61 in trimethoprim-sulfamethoxazole-resistant strains. The significance of these mutations was confirmed by site-directed mutagenesis and by the transformation of a susceptible strain of S. pneumoniae to sulfamethoxazole resistance. Two resistant isolates did not contain any mutations within the gene encoding dihydropteroate synthase. The results presented suggest the independent generation of resistant mutations among South African clinical isolates. It is also proposed that the mechanism of sulfonamide resistance in S. pneumoniae involves the expansion of a specific region within dihydropteroate synthase, which probably forms part of the sulfonamide binding site.
The sulfonamide class of drugs has played an important role in the treatment of pneumococcal diseases. On the recommendation of the World Health Organization, trimethoprim in combination with sulfamethoxazole (co-trimoxazole) has been widely administered for the treatment of respiratory tract infections in children (29). In recent years, high rates of resistance to co-trimoxazole have been reported worldwide, especially in Spain, Portugal, Hungary, and South Africa (8, 10, 11, 18, 32). In South Africa, co-trimoxazole resistance among systemic isolates increased from 32% in 1985 to 44% in 1991, while co-trimoxazole resistance in association with multiple antibiotic resistance increased from 3.8% in 1985 to 14.8% in 1991 (10).
Despite the fact that trimethoprim-sulfamethoxazole resistance is widespread in pneumococci, there is little information on the molecular basis of resistance to this agent. Our group (1) has recently identified the mutations in the dihydrofolate reductase gene that confer resistance to trimethoprim. We have now investigated the mechanism of resistance to sulfamethoxazole in Streptococcus pneumoniae.
The target for sulfonamide action is dihydropteroate synthase (DHPS), which catalyzes the condensation of para-aminobenzoic acid with 7,8-dihydro-6-hydroxymethylopterine-pyrophosphate to form 7,8-dihydropteroate (28). The dihydropteroate is subsequently converted to tetrahydrofolate, an essential metabolite for the synthesis of purines, thymidylate, glycine, methionine, pantothenic acid, and N-formylmethionyl tRNA. Sulfonamides are structural analogues of para-aminobenzoic acid and therefore competitively inhibit DHPS by acting as alternative substrates (4, 25, 28).
In most gram-negative bacteria, sulfonamide resistance is largely plasmid borne and due to the acquisition of alternative drug-resistant variants of DHPS. Two such plasmid genes, sulI and sulII have been characterized and sequenced (24, 30). Chromosomal mutations in the dhps gene that confer resistance to sulfonamides have been identified in a number of bacteria. In Escherichia coli, a single change of Phe28 to Leu in DHPS has been demonstrated to be responsible for sulfonamide resistance (5). Horizontal transfer has been implicated in the acquisition of a 6-bp insert in the gene encoding DHPS in Neisseria meningitidis (7), while in Staphylococcus aureus as many as 14 mutations are thought to be involved in conferring resistance to sulfonamides (9). Studies by Lopez and coworkers (14) on the dhps gene of a sulfonamide-resistant S. pneumoniae strain initially revealed a 6-bp repeat that duplicated amino acids 66 and 67, in an area distinct from that observed in N. meningitidis. Recent studies by Maskell and coworkers (19) have demonstrated that sulfonamide resistance in S. pneumoniae may be caused by the presence of 3- or 6-bp duplications within the gene in the area encoding Arg58 to Tyr63, close to, but distinct from, the mutations observed by Lopez et al. (14).
In this study, we attempted to better understand the mechanisms involved in conferring sulfonamide resistance in S. pneumoniae by characterization of the mutations with the gene encoding DHPS.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. pneumoniae strains used in this study are listed in Table 1. The cloning vector pGEM-7Zf(+) (Promega, Madison, Wis.), allowing blue-white screening of transformants, was used. The S. pneumoniae recipient strain CP1015, derived from Rx, a strain that is only partly related to R6, was used in transformation experiments. S. pneumoniae R6, ATCC 49619, and CP1015 were used as susceptible controls when determining MICs.
TABLE 1.
Clinical isolates of S. pneumoniae used in this study
| Isolatea | Hospitalb | Sourcec | Serotype | Antibiotic resistance patternd | MIC (μg/ml)e
|
||
|---|---|---|---|---|---|---|---|
| SMX | TMP | CoT (SMX/TMP) | |||||
| 111 | GS | CSF | 14 | E | 4,096 | 16 | 8/152 |
| 13 | JHB | NS | 6B | TCd | 2,048 | 512 | 2/38 |
| 8 | JHB | NS | 6B | PTCdE | 1,024 | 128 | 8/152 |
| 11 | CHB | BC | 14 | TE | 1,024 | 16 | 2/38 |
| 45 | CHB | NS | 6A | PTCdE | 512 | 256 | 8/152 |
| 104 | HB | BC | 23F | CT | 512 | 16 | 2/38 |
| 85 | RC | BC | 1 | Sens | 256 | 32 | 0.5/9.5 |
| 42 | RC | TA | 23F | PCT | 256 | 128 | 8/152 |
| 405 | HB | BC | 15 | Sens | 16 | 1 | 0.125/2.375 |
| 540 | RC | BC | 23F | Sens | 8 | 1 | 0.125/2.375 |
| 544 | CHB | BC | 8 | Sens | 16 | 1 | 0.125/2.375 |
| 688 | JHB | NS | 8 | Sens | 4 | 1 | 0.125/2.375 |
| 49619 | Sens | 16 | 4 | 0.25/4.75 | |||
| CP1015 | Sens | 64 | 2 | 0.25/4.75 | |||
| R6 | Sens | 16 | 2 | 0.25/4.75 | |||
ATCC 49619 is a control strain, S. pneumoniae R6 is an unencapsulated laboratory strain, and CP1015 is a recipient strain used in transformation experiments.
Isolates were collected from the following hospitals: JHB, Johannesburg; CHB, Chris Hani Baragwanath; GS, Groote Schuur; RC, Red Cross; and HB, Hillbrow.
CSF, cerebral spinal fluid; NS, nasal swab; BC, blood culture; TA, tracheal aspirate.
Antibiotic resistance patterns were obtained for penicillin (P), chloramphenicol (C), tetracycline (T), clindamycin (Cd), erythromycin (E). Sens, sensitivity.
SMX, sulfamethoxazole; TMP, trimethoprim; CoT, sulfamethoxazole-trimethoprim (co-trimoxazole). The breakpoints for resistance to the antibiotics were as follows: SMX, ≥128 μg/ml; TMP, ≥8 μg/ml; CoT, ≥1/19 μg/ml.
Susceptibility testing.
MICs were determined by using the agar dilution method described by the National Committee for Clinical Laboratory Standards (20). For the susceptibility testing of sulfamethoxazole (Sigma Chemical Company, St. Louis, Mo.), Mueller-Hinton agar (Difco, Detroit, Mich.) supplemented with 5% lysed defibrinated horse blood was used. Plates were incubated at 37°C in 5% CO2 for 48 h.
DNA extraction.
Chromosomal DNA was extracted based on the method described by Paton et al. (23). Cultures were harvested from blood agar plates and resuspended in 90 μl of a suspension buffer (10 mM Tris-HCl, 0.14 M NaCl, 0.1 M sodium citrate, 1 mM EDTA). Thereafter, sodium deoxycholate (1%) was added to a final concentration of 0.1%, and the suspension was left to stand at room temperature for 10 min. The aqueous phase was extracted twice with TE (10 mM Tris-HCl, 1 mM EDTA)-saturated phenol and once with chloroform. The DNA was recovered by adding 2.5 volumes of ice-cold ethanol and incubating at −70°C for 30 min. After centrifugation, the pellet obtained was washed with 70% ethanol, resuspended in 50 to 100 μl of distilled water containing 20 μg of RNase A per ml and stored at −20°C.
PCR.
The PCR primer sequences were based on the published sequence of the DHPS gene of S. pneumoniae R6 (14). The following sets of primers were used to amplify the DHPS gene: F1, 5′-ATGTCAAGTAAAGCCAATCAT-3′ (position 1 to 21); F2, 5′-GACTCCTTTTCGGACGGT-3′ (position 61 to 78); F3, 5′-GATATCGGCGGAGAATCG-3′ (position 150 to 171); F4, 5′-CTGGTATTGCACCAGAAAATA-3′ (position 572 to 592); R1, 5′-TGGAACAACACGCTGGATTTC-3′ (position 208 to 225); R2, 5′-AGCAGCCAAAGCCTCTGC-3′ (position 291 to 312); R3, 5′-TGAGGTCGCGCCATAACTGGAT-3′ (position 410 to 431); R4, 5′-GCCAATTCCTGGATCCAA-3′ (position 598 to 615); and R5, 5′-CCGGTAGTTAGCAATCCATTG-3′ (position 967 to 988).
DNA amplification was performed in 50-μl volumes containing a 1 μM concentration of each primer, 1.5 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, 1 U of Taq polymerase, and 2 μl of DNA in a buffer provided by the manufacturer. Amplification reactions were performed with an Omnigene thermocycler (Hybaid, Middlesex, United Kingdom) using the following program: denaturation at 95°C for 5 min, followed by 32 cycles at 95°C for 1 min, primer-specific annealing temperatures for 1 min, 72°C for 2 min, and a final extension at 72°C for 7 min. An annealing temperature of 58°C was used for PCRs with primer pairs F1-R3 and F5-R6, 54°C reactions were used for F2-R5, and F3-R2 were annealed at 62°C.
Sequence analysis.
Sequence analysis was carried out with both manual sequencing according to the chain termination sequencing method of Sanger and colleagues (27) and with an automated sequencer. Single-stranded PCR products were prepared for manual sequencing by using streptavidin magnetic particles (Boehringer, Mannheim, Germany). Briefly, a standard amplification reaction (100-μl mixture) was performed with a 5′ biotinylated forward primer and an unlabelled reverse primer. Nonincorporated deoxynucleoside triphosphates and primers were removed by precipitation with polyethylene glycol as described previously (2), and the pellet was resuspended in 40 μl of TE buffer. Single-stranded DNA bound to the magnetic beads was prepared according to the manufacturer’s recommendations. The unlabelled DNA was salt precipitated from the supernatant and resuspended in 12 μl of water. Sequence analysis was carried out by using the Sequenase, version 2.0, sequencing kit (United States Biochemicals, Cleveland, Ohio) according to the manufacturer’s recommendations, with 3 μl of single-stranded DNA per dideoxy chain termination sequencing reaction.
PCR products for automated sequence analysis were treated with shrimp alkaline phosphatase and exonuclease I, from a presequencing kit (Amersham, Little Chalfont, Buckinghamshire, England). Fluorescent cycle sequencing was then performed according to the manufacturer’s recommendations by using the ABI Prism dRhodamine terminator sequencing kit containing AmpliTaq DNA polymerase FS (Perkin-Elmer) and 3.2 pmol of primer under the following cycling parameters: 96°C for 30 s, 50°C for 15 s, and 60°C for 4 min for 25 cycles. Sequencing products were purified with Centricep purification columns (Princeton Separations, Adelphia, N.J.) to remove excess terminators before sequence analysis on an ABI Prism 377 DNA sequencer (Applied Biosystems Inc., Foster City, Calif.).
Cloning PCR products.
The amplified PCR products of the DHPS gene were prepared according to the method of Sambrook et al. (26). Blunt end ligations were then performed into a SmaI-digested expression vector pGEM-7Zf(+). The ligations were carried out in 12-μl reaction volumes with 0.1 μg of SmaI-restricted vector, 0.3 μg of PCR product, 8% polyethylene glycol 6000, 4.34 mM ATP, 5 U of SmaI, and 2 U of T4 DNA ligase (United States Biochemicals) in a ligation buffer provided by the manufacturer. The mixture was then subjected to cycling overnight between 10 and 30°C for 30 s according to the method described by Lund et al. (17). Competent E. coli JM109 cells were electrotransformed by using the purified ligated DNA as described previously (6). Transformants were screened for the lack of α-complementation on Luria-Bertani agar containing 50 μg of ampicillin per ml, 20 μl of 50-mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) stock solution and 100 μl of a 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside) stock solution.
Transformations.
The sulfonamide-susceptible S. pneumoniae recipient strain CP1015 was grown at 37°C to an A400 of 0.4 in competence and transformation medium containing 10 g of Casitone (Difco), 5 g of tryptone (Difco), 4 g of yeast extract, and 5 g of NaCl per liter supplemented with 0.2% glucose and 17 mM K2HPO4. Glycerol was added to a final concentration of 15% before cells were stored at −70°C. Competent cells were produced by diluting thawed cells to a density of 1:100 in a complete transformation medium, pH 7 competence and transformation medium supplemented with final concentrations of 1 mM CaCl2 and 0.4% bovine serum albumin). After incubation at 37°C for 2 h (A400 ≅ 0.05) the pH of the suspension was raised to 7.8 with 1 M NaOH and then incubated for a further 20 min.
For transformations, 1 μg of the cloned PCR product per ml of cells was incubated for 30 min at 30°C, followed by 2 h of incubation at 37°C. Cells were then diluted 10-fold before being plated onto a selection medium containing 128 or 256 μg of sulfamethoxazole per ml.
The plates were incubated for 48 h at 37°C in 5% CO2. DNA-free controls were also tested in order to confirm that any growth observed was the result of transformation and not mutation. Cells were also plated onto an antibiotic-free medium for the determination of viable counts. The MICs of transformants were determined as described above.
Site-directed mutagenesis.
Mutagenesis was performed using the Muta-Gene M13 in vitro mutagenesis kit, version 2 (Bio-Rad, Hercules, Calif.) according to the manufacturer’s instructions. Mutagenic primers were produced to contain mutations at Glu45-Gly, Leu115-Phe, His123-Asp, Phe156-Val, Gly157-Asn, Thr170-Lys, Glu175-Asp, Leu185-Val, Lys210-Asn, Asn247-Ser, Arg282-Cys, Val283-Leu, and Leu301-Gln. These primers were designed to be 28-mers and showed 100% identity to the S. pneumoniae R6 DNA sequence on either side of the mutagenic amino acid. The presence of the mutagenic base in the synthesized cDNA was confirmed by sequence analysis before transformation was attempted. Transformants were selected on antibiotic plates, containing sulfamethoxazole concentrations of 8, 16, 32, 128, and 256 μg/ml, as described above. The insertion mutations from the resistant isolates, shown in Table 1, were confirmed to play a role in sulfamethoxazole resistance. These insertion mutants were used as the controls to test the efficacy of the transformation system.
DHFR analysis.
The dihydrofolate reductase (DHFR) fragments encompassing the published S. pneumoniae (1) mutation was generated with primers F1 (5′-GGAAGCATGACTAAGAAAATCG-3′) and R1 (5′-TTAGACTTCCTTTCTCTTG-3′), based on the S. pneumoniae DHFR gene from the EMBL database (accession no. Z74777). Standard PCRs were performed as described above. The cycling parameters were denaturation at 93°C for 3 min, followed by 32 cycles of 93°C for 1 min, 53°C for 1 min, and 72°C for 1 min, and a final extension of 72°C for 2 min. Sequence analysis of the DHFR gene was performed by using the 5′ biotinylated primer F1 as described above.
Nucleotide sequence accession numbers.
The DNA sequences containing the novel mutations identified in this study have been assigned accession no. AJ132956 (DNA sequence of isolate 13) and AJ132957 (DNA sequence of isolate 42) in the EMBL database.
RESULTS
Sequence analysis of pneumococcal DHPS genes in sulfamethoxazole-resistant and -susceptible strains.
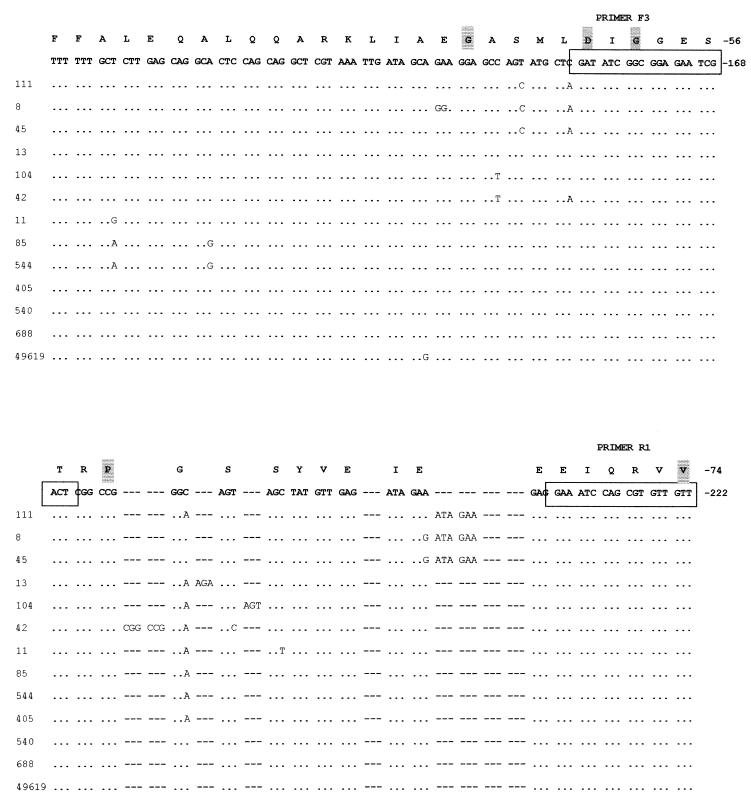
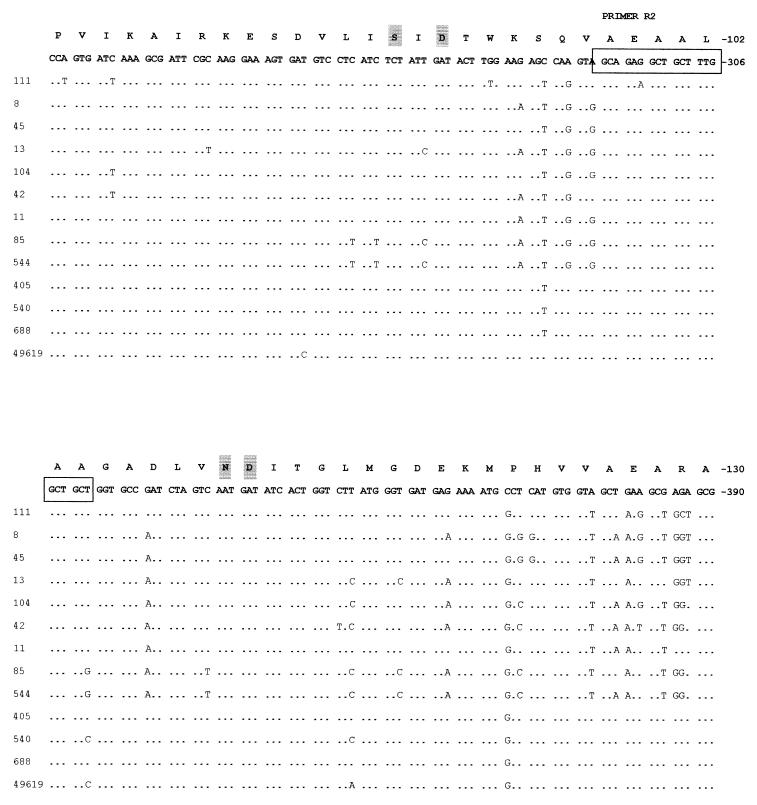
Eight sulfamethoxazole-resistant and five sulfamethoxazole-susceptible isolates, including S. pneumoniae ATCC 49619, for which there is a broad range of MICs, were randomly chosen from a collection of clinical isolates from hospitals around South Africa (Table 1). Sequence analysis of sulA revealed a number of polymorphic changes in both susceptible and resistant isolates when compared to sulA of R6. A section of the deduced DNA sequence containing the mutations observed in the resistant strains is presented in Fig. 1. In isolates 8, 45, and 111, we observed the presence of the suld mutation (13), the duplication of Ile66 and Glu67. Strain 104 contained a duplication of an existing serine residue at position 61 (19). Sequence analysis of isolate 13 showed that it contained an insertion of an arginine residue between Glu60 and Ser61, while isolate 42 was found to contain a duplication of Arg58 and Pro59. These mutations are being reported for the first time. The above-mentioned changes were absent in all the susceptible isolates studied.
FIG. 1.
A comparison of the nucleotide sequences of sulfonamide-resistant and -susceptible isolates of S. pneumoniae to the R6 sequence described by Lopez et al. (14). Only a section of the deduced sequence, containing the mutations observed, is shown above. The complete sulA gene sequence is available as EMBL accession no. U16156. Identical residues are indicated by dots, while asterisks have been inserted where insertions or duplications have been detected to allow sequence alignment. The amino acid sequence has been translated and appears above the nucleotide sequence. Amino acids that are conserved in all DHPS sequences are highlighted.
We noted with interest that the sulfamethoxazole-resistant isolates 11 and 85 showed no changes in this area between Arg58 and Glu68. There was considerable sequence diversity elsewhere in the gene, resulting in amino acid changes among the resistant strains not seen in susceptible isolates. Comparison of the nucleotide sequence of the sulfonamide-resistant isolates revealed other amino acid changes at Glu45-Gly, Leu115-Phe, His123-Asp, Phe156-Val, Gly157-Asn, Glu163-Lys, Thr170-Lys, Glu175-Asp, Leu185-Val, Ala189-Glu, Lys210-Asn, Asn247-Ser, Arg282-Cys, Val283-Leu, and Leu301-Gln. Of these changes only Lys210-Asn and Asn247-Ser occur exclusively in isolates 11 and 85, respectively.
Transformation to sulfonamide resistance.
Whole DNA and PCR products from the sulfonamide-resistant isolates were used for transformation of recipient S. pneumoniae CP1015. Whole DNA from each isolate was capable of transforming CP1015 to sulfamethoxazole resistance. PCR products of primer pairs F2-R5 and F3-R2 from isolates 111, 8, 45, 13, 104, and 42 transformed CP1015 to sulfonamide resistance (Table 2). The products with DNA from strains 11 and 85 did not transform CP1015 to resistance. The MICs for the transformants obtained by using whole DNA or PCR products were within 1 dilution of donor DNA (Table 2), except for the strain for which the MIC was the highest (MIC for strain 111, 4,096 μg/ml). In this instance, whole DNA transformed CP1015 to full resistance while the PCR product transformed CP1015 to a level of resistance at which the MIC was 1,024 μg/ml. The higher MICs observed for the recipient strain than for the donor strains may be due to the higher background MIC for CP1015, which was determined to be 64 mg/liter (19). In the DNA-free controls no spontaneous resistant mutations were detected.
TABLE 2.
MICs determined for sulfamethoxazole-resistant transformantsa
| Donor strain | Sulfamethoxazole MIC (μg/ml)
|
||
|---|---|---|---|
| Donor | Transformants (whole DNA) | Transformants (PCR product) | |
| 111 | 4,096 | 4,096 | 1,024 |
| 8 | 1,024 | 1,024 | 1,024 |
| 45 | 512 | 512 | 1,024 |
| 13 | 2,048 | 2,048 | 1,024 |
| 104 | 512 | 512 | 1,024 |
| 42 | 256 | 256 | 512 |
| 11 | 1,024 | 1,024 | |
| 85 | 512 | 512 | |
Transformants were selected on antibiotic plates containing 16 and 32 μg of sulfamethoxazole per ml. MICs were determined for two to four transformants, according to the method outlined in the experimental section. The frequencies of the transformations achieved were dependent on the size of the fragment used and the degree of homology between the PCR product and the recipient strain. The average frequencies of transformation obtained for the PCR products obtained with primer pairs F3-R2 and F3-R5 and with whole DNA were 5.5 × 10−6, 4.9 × 10−3, and 2.5 × 10−2, respectively.
Site-directed mutagenesis.
By using site-directed mutagenesis, it was determined that the amino acid substitutions observed, Glu45-Gly, Leu115-Phe, His123-Asp, Phe156-Val, Gly157-Asn, Glu163-Lys, Thr170-Lys, Glu175-Asp, Leu185-Val, Ala189-Glu, Lys210-Asn, Asn247-Ser, Arg282-Cys, Val283-Leu, and Leu301-Gln, did not confer resistance to sulfamethoxazole. Of these changes only Lys210-Asn and Asn247-Ser occur exclusively in isolates 11 and 85, respectively. The amino acid substitutions that occur at, or close to, conserved areas may, however, be particularly important, not so much in conferring resistance but rather in that they may contribute to the resistant phenotype observed by affecting the tertiary conformation of the SulA protein.
DHFR characterization.
Sequence analysis of the DHFR gene of the resistant isolates revealed the presence of the Iso100-Leu substitution previously reported to be responsible for trimethoprim resistance in S. pneumoniae (1). Isolates 11 and 85 did not, however, contain this mutation, and neither did any of the susceptible isolates.
DISCUSSION
Two separate duplications within sulA, the gene encoding DHPS, have previously been shown to confer sulfonamide resistance on S. pneumoniae. The 6-bp repeat described by Lopez et al. (14), which results in the duplication of Ile66 and Glu67, and the duplication of Ser61 observed by Maskell et al. (19) were found among the isolates in this study. An insertion of arginine between Gly60 and Ser61 and the duplication of amino acids Arg58 and Pro59 are novel changes and have never been reported among any of the bacteria investigated for sulfonamide resistance. The observation that two independent additions to the protein in the area of Arg58 and Gly68 result in sulfonamide resistance suggests that an expansion of the length of the protein in that area is the critical determinant of resistance. Our data show that wild-type strains have developed at least four strategies to effect this change.
Studies performed in the 1960s (33) showed that pneumococcal resistance to sulfanilamide could be explained by the production of enzymes with altered affinities for the drug. These resistant strains expressed an enzyme containing a mutation, referred to as Fd, which had a lowered affinity for ρ-aminobenzoic acid as well as an altered capability of binding the drug. It is suggested that the mutations identified in this study produced resistant isolates that also exhibited such altered DHPS enzymes.
Site-directed mutagenesis showed that none of the other single amino acid changes observed in the genes of resistant strains were capable of conferring resistance to sulfamethoxazole.
As whole DNA conferred higher resistance on CP1015 than did the PCR products obtained from the amplification of dhps, this suggests at least one other mechanism of sulfonamide resistance in S. pneumoniae. The failure to find the molecular basis of resistance in strains 11 and 85 further supports this idea, as does the fact that the MICs observed could not always be correlated with the type of mutation identified. It appears therefore that there must be some other contributing factor(s) involved in sulfonamide resistance in S. pneumoniae.
The existence of more than one mechanism of resistance in a single bacterium has been alluded to by a number of researchers. At least two different types of mutations can contribute to rendering E. coli resistant to sulfonamides (22). Some mutations produce altered enzymes that differ structurally from the wild-type strain in that they do not combine as readily with sulfonamide. Other mutants have enzymes that resemble those of the wild-type strains, but it is thought that these mutants have different permeability characteristics that reduce sulfonamide uptake into the cells. The permeability of sulfonamides or its analogues have to date not been investigated in S. pneumoniae, so this mechanism of resistance remains a possible contributing factor to sulfonamide resistance in this species.
Other possible mechanisms that may confer resistance to sulfonamides had been alluded to after observations made in other bacteria. In S. aureus some resistant strains were observed to display an overproduction of para-aminobenzoic acid (13).
The presence of an efflux mechanism for the active transport of sulfonamides has been alluded to in E. coli. The sur gene sequence identified in sulfathiazole-resistant E. coli was found to match the E. coli bcr gene, which is responsible for conferring bicyclomycin resistance when it is overproduced (3). The product of sur (bcr) is similar to that of the family of proton motive force-dependent drug-H+ antiporters, and it may be that sur (bcr) functions in an analogous manner effluxing para-aminobenzoic acid or another intermediate in folate biosynthesis, in addition to sulfathiazole from the cell (21). These observations have yet to be investigated in pneumococci and hence cannot be ruled out as having some role in conferring resistance to sulfonamides.
The operon dedicated to folate synthesis in S. pneumoniae consists of four genes, namely, sulA, sulB, sulC, and sulD (15). The functions of SulB, SulC, and SulD have been determined. It appears that SulC has cyclohydrolase activity and catalyzes the first step in the folate biosynthetic pathway (12). SulD is a bifunctional protein, which catalyzes two successive steps in folate biosynthesis (16), and sulB encodes dihydrofolate synthetase, which catalyzes the last step of the folate pathway (12). Mutations in these genes could play a role in conferring resistance although no evidence has as yet been discovered. Efforts are currently being made to sequence sulB, sulC, and sulD. The absence of mutations in sulA in sulfonamide-resistant isolates of S. pneumoniae could, however, imply the presence of mutations in any of the other genes making up the operon. Mutations in these genes may allow the pneumococcus to synthesize folate by a different pathway in the presence of sulfonamides.
The isolates studied are all co-trimoxazole resistant, implying that resistance to both components occurs and may be attributable to any combination of the resistance mechanisms. Besides a permeability change, which may affect the action of both trimethoprim and sulfamethoxazole, a single mutation has been suggested to confer resistance to trimethoprim, sulfamethoxazole, and their combination, namely, the mutation to thymine auxotrophy (31). It is possible therefore that since isolates 11 and 85 in our study do not contain mutations in either of their enzymes (although they are resistant to both trimethoprim and sulfamethoxazole), they might have reverted to auxotrophy. The growth of these isolates was, however, obtained on minimal media and supplemented media, suggesting that these strains remain prototrophs.
The mechanism for sulfonamide resistance in S. pneumoniae appears to involve the expansion of the region that probably forms the sulfonamide binding site, therefore leading to alterations in the structural conformation of the site. The presence of distinct resistance-mediating alterations identified in this study lends support for the independent generation of resistant alleles that contribute to the dissemination of sulfonamide resistance among clinical isolates of S. pneumoniae. Sulfonamide resistance does, however, appear to be more complex, and the results presented in this work further substantiate the possibility that additional mechanisms are involved in conferring sulfonamide resistance on S. pneumoniae.
ACKNOWLEDGMENTS
We are grateful for the recipient strain CP1015 received from M. C. Trombe and J. P. Maskell.
This work was supported by the Medical Research Council, SAIMR, and the University of the Witwatersrand.
REFERENCES
- 1.Adrian P V, Klugman K P. Mutations in the dihydrofolate reductase gene of trimethoprim-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2406–2413. doi: 10.1128/aac.41.11.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J, Smith A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 3.Bentley J, Hyatt L S, Ainley K, Parish J H, Herbert R B, White G R. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene. 1993;127:117–120. doi: 10.1016/0378-1119(93)90625-d. [DOI] [PubMed] [Google Scholar]
- 4.Brown G M. The biosynthesis of folic acid: inhibition by sulfonamides. J Biol Chem. 1962;237:536–540. [PubMed] [Google Scholar]
- 5.Dallas W S, Gowen J E, Ray P H, Cox M J, Dev I K. Cloning, sequencing, and enhanced expression of the dihydropteroate synthase gene of Escherichia coli MC4100. J Bacteriol. 1992;174:5961–5970. doi: 10.1128/jb.174.18.5961-5970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fermér C, Kristiansen B-E, Sköld O, Swedberg G. Sulfonamide resistance in Neisseria meningitidis as defined by site-directed mutagenesis could have its origin in other species. J Bacteriol. 1995;177:4669–4675. doi: 10.1128/jb.177.16.4669-4675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-de-Lomas J. Antimicrobial susceptibility of Streptococcus pneumoniae isolated from pediatric carriers in Spain. Eur J Microbiol Infect Dis. 1997;16:11–13. doi: 10.1007/BF01575112. [DOI] [PubMed] [Google Scholar]
- 9.Hampele I C, D’Arcy A, Dale G E, Kostrewa D, Nielsen J, Oefner C, Page M G P, Schönfeld H-J, Strüber D, Then R L. Structure and function of the dihydropteroate synthase from Staphylococcus aureus. J Mol Biol. 1997;268:21–30. doi: 10.1006/jmbi.1997.0944. [DOI] [PubMed] [Google Scholar]
- 10.Klugman K P. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koornhof H J, Wasas A, Klugman K P. Antimicrobial resistance in Streptococcus pneumoniae: a South African perspective. Clin Infect Dis. 1992;15:84–94. doi: 10.1093/clinids/15.1.84. [DOI] [PubMed] [Google Scholar]
- 12.Lacks S A, Greenberg B, Lopez P. A cluster of four genes encoding enzymes for five steps in the folate biosynthetic pathway of Streptococcus pneumoniae. J Bacteriol. 1995;177:66–74. doi: 10.1128/jb.177.1.66-74.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landy M, Larkum N W, Oswald E J, Streightoff F. Increased synthesis of p-aminobenzoic acid associated with the development of sulfonamide resistance in Staphylococcus aureus. Science. 1943;97:265–267. doi: 10.1126/science.97.2516.265. [DOI] [PubMed] [Google Scholar]
- 14.Lopez P, Espinosa M, Greenberg B, Lacks S. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987;169:4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez P, Greenberg B, Lacks S A. DNA sequence of folate biosynthesis gene sulD, encoding hydroxymethyldihydropterin pyrophosphokinase in Streptococcus pneumoniae, and characterization of the enzyme. J Bacteriol. 1990;172:4766–4774. doi: 10.1128/jb.172.9.4766-4774.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez P, Lacks S A. A bifunctional protein in the folate biosynthetic pathway of Streptococcus pneumoniae with dihydroneopterin aldolase and hydroxymethlydihydropterin pyrophosphokinase activities. J Bacteriol. 1993;175:2214–2220. doi: 10.1128/jb.175.8.2214-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lund A H, Duch M, Pedersen F S. Increased cloning efficiency by temperature-cycle ligation. Nucleic Acids Res. 1996;24:800–801. doi: 10.1093/nar/24.4.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marton A. Epidemiology of resistant pneumococci in Hungary. Microb Drug Resist. 1995;1:127–130. doi: 10.1089/mdr.1995.1.127. [DOI] [PubMed] [Google Scholar]
- 19.Maskell J P, Sefton A M, Hall L M C. Mechanism of sulfonamide resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2121–2126. doi: 10.1128/aac.41.10.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 21.Nichols B P, Guay G G. Gene amplification contributes to sulfonamide resistance in Escherichia coli. Antimicrob Agents Chemother. 1989;33:2242–2248. doi: 10.1128/aac.33.12.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pato M L, Brown G M. Mechanisms of resistance of Escherichia coli to sulfonamides. Arch Biochem Biophys. 1963;103:443–448. doi: 10.1016/0003-9861(63)90435-1. [DOI] [PubMed] [Google Scholar]
- 23.Paton J C, Berry A M, Lock R A, Hansman D, Manning P A. Cloning and expression in Escherichia coli of the Streptococcus pneumoniae gene encoding pneumolysin. Infect Immun. 1986;54:50–55. doi: 10.1128/iai.54.1.50-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rådström P, Swedberg G. RSF1010 and a conjugative plasmid contain sulII, one of two known genes for plasmid-borne sulfonamide resistance dihydropteroate synthase. Antimicrob Agents Chemother. 1988;32:1684–1692. doi: 10.1128/aac.32.11.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roland S, Ferone F, Harvey R J, Styles V L, Morrison R W. The characteristics and significance of sulfonamides as substrates for Escherichia coli dihydropteroate synthase. J Biol Chem. 1979;254:10337–10345. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiota T, Baugh M, Jackson R, Dillard R. The enzymatic synthesis of hydroxymethyldihydropteridine pyrophosphate and dihydrofolate. Biochemistry. 1969;8:5022–5028. doi: 10.1021/bi00840a052. [DOI] [PubMed] [Google Scholar]
- 29.Straus W L, Qasi S A, Kundi Z, Nomani N K, Schwartz B. Antimicrobial resistance and clinical effectiveness of co-trimoxazole versus amoxycillin for pneumonia among children in Pakistan: randomised controlled trial. Pakistan Co-trimoxazole Study Group. Lancet. 1998;352:270–274. doi: 10.1016/s0140-6736(97)10294-x. [DOI] [PubMed] [Google Scholar]
- 30.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 31.Then R L. Mechanisms of resistance to trimethoprim, the sulfonamides, and trimethoprim-sulfamethoxazole. Rev Infect Dis. 1982;4:261–269. doi: 10.1093/clinids/4.2.261. [DOI] [PubMed] [Google Scholar]
- 32.Vaz Pato M V, Belo de Carvalho C, Tomasz A the Multicenter Study Group. Antibiotic susceptibility of Streptococcus pneumoniae isolates in Portugal. A multicenter study between 1989 and 1993. Microb Drug Resist. 1995;1:59–69. doi: 10.1089/mdr.1995.1.59. [DOI] [PubMed] [Google Scholar]
- 33.Wolf B, Hotchkiss R D. Genetically modified folic acid synthesizing enzymes of pneumococcus. Biochemistry. 1962;2:145–150. doi: 10.1021/bi00901a026. [DOI] [PubMed] [Google Scholar]