Abstract
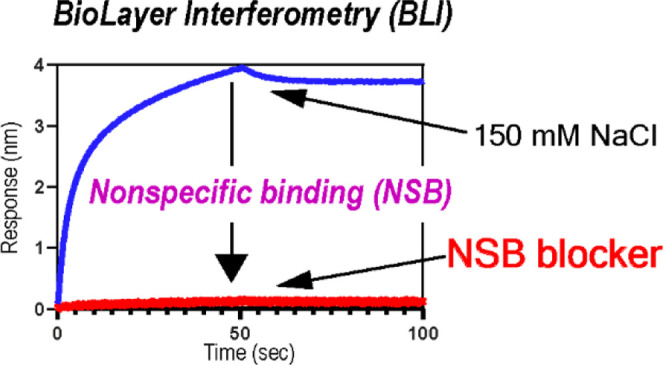
Quantitative analysis of protein–protein interactions (PPIs) using biolayer interferometry (BLI) requires effective suppression of nonspecific binding (NSB) between analytes and biosensors. In particular, the study of weak interactions (i.e., KD > 1 μM) requires high concentrations of analytes, which substantially increases NSB. However, there are only a few so-called NSB blockers compatible with biomolecules, which limits the use of BLI in the accurate analysis of weak interactions. The present study aims to identify a new NSB blocker for the quantitative analysis of weak PPIs using BLI. We find that saccharides, especially sucrose, are potent NSB blockers and demonstrate their compatibility with other blocking additives. We also demonstrate the effects of the new NSB blocker by characterizing the binding between nonstructural protein 1 of the influenza A virus and human phosphoinositide 3-kinase. We anticipate that the new NSB-blocking admixture will find broad applications in studying weak interactions using BLI.
Introduction
Protein–protein (or ligand) interactions (PPIs) play a central role in numerous biological processes, such as cellular signal transduction and proliferation.1 The accurate estimation of binding affinity and kinetics is essential for understanding the molecular mechanism of PPIs and the development of PPI inhibitors.2,3
Biolayer interferometry (BLI) is widely used for the study of diverse biomolecular interactions because the binding affinity and kinetics can be measured in one experiment between biosensor-immobilized ligands and analytes in a well.4−9 One well-established application of BLI is the characterization of tight binding interactions, such as antibody–antigen interactions.10−12 Notable examples include the identification of therapeutic antibodies against SARS-CoV-211 and characterization of the binding between the receptor binding domain (RBD) of SARS-CoV-2 and human ACE2.7,13,14
However, quantitative characterization of weak PPIs (KD > 1 μM) using BLI requires an additional effort to suppress nonspecific binding (NSB). For example, an accurate estimation of the binding affinity (KD) in micromolar ranges requires >10 μM analytes; for reliable estimation of KD values, the analyte concentration should span 0.1–10 times the estimated KD value.15,16 The magnitude of NSB is proportional to the analyte concentration; thus, a significant NSB signal, particularly at high analyte concentrations, can complicate data analysis. Although the double-referencing method can mitigate the influence of NSB, it is still necessary to minimize the NSB signal for a quantitative study because a large NSB signal might cause significant subtraction errors and result in a small net signal change upon ligand–analyte binding.
A standard protocol for minimizing NSB is adding so-called NSB blockers such as bovine serum albumin (BSA) or Tween-20. However, a surprisingly small number of NSB blockers are available. Moreover, their effect of suppressing NSB at high analyte concentrations has not been well-characterized. Currently, available NSB blockers might be ineffective and/or incompatible with analytes of interest. Considering the increasing popularity of BLI, it is essential to identify diverse NSB blockers.
Here, we found that commonly used NSB blockers have limited effects in suppressing the NSB of various protein analytes at high concentrations (>10 μM), preventing the accurate assessment of the binding affinity and kinetics of weak PPIs. Moreover, we found that saccharides, in particular sucrose, are excellent NSB blockers. Their NSB-blocking efficiency is additive to the effects of other blockers, enabling a combinatorial approach to suppress the NSB of analytes of interest. To demonstrate the impact of our new NSB blocker, we characterized the binding between NS1 (nonstructural protein 1) of the 1918 influenza A virus and human PI3K (phosphoinositide 3 kinase).6,17 We show that the new NSB blocker significantly improves the quality of the BLI data, compared to the previous results acquired using a different NSB blocker.18 This comparison highlights the importance of reducing NSB in BLI studies of weak PPIs.
Results and Discussion
Magnitude of NSB is Substantial
To estimate the extent of the NSB signal in BLI experiments, we selected four protein analytes with variable molecular weights and pI values: CRK-II (33.8 kDa; pI = 5.38),19,20 TRIM25 (22.7 kDa; pI = 8.75),21 p85β (19.9 kDa; pI = 8.98),22 and Riplet (10.4 kDa; pI = 4.53).23,24 These proteins are human signaling proteins with binding affinities in the micromolar range to their cognate binding proteins; thus, they represent a collection of common weak PPIs.
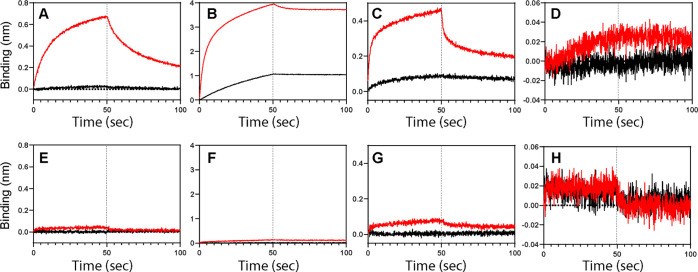
For an accurate estimation of binding affinity (KD) in a micromolar range, concentrations of analytes might need to greatly surpass 10 μM. Thus, we first monitored the intrinsic NSB of the selected proteins at two analyte concentrations, 1 and 40 μM, in a buffer without any additives (Figure 1A–D). Except for Riplet (Figure 1D), all tested proteins showed substantially large NSB signals, especially at 40 μM. Overall, the NSB signal during the association and dissociation phases is concentration-dependent and looks very similar to the regular signal of ligand-analyte binding. It should be noted that these results were acquired without a loaded ligand; thus, they represent the interaction between analytes and Ni-NTA biosensors. These results showed that the NSB is common, and its extent is highly variable; therefore, an explicit test is necessary for individual proteins prior to any quantitative assay.
Figure 1.
BLI sensorgrams of NSB between the Ni-NTA biosensor and various protein analytes in the absence (A–D) and presence (E–H) of the NSB blocker admixture. The analytes are (A,E) CRK II, (B,F) TRIM25, (C,G) p85β, and (D,H) Riplet. Black and red lines correspond to 1 and 40 μM of each analyte, respectively. Vertical dotted lines represent the initiation of the dissociation phase.
It should also be noted that all measurements included in the present study were conducted in the presence of 150 mM NaCl, a commonly suggested concentration to prevent NSB.4,25,26 This result indicates that weak and long-range electrostatic interactions are not the only driving force for the NSB of the tested proteins. We did not try a higher salt concentration because it might perturb the electrostatically driven ligand–analyte interactions.
Common Additives Marginally Suppress NSB
BSA, Tween-20, and casein are common additives used to suppress the NSB during a BLI experiment. To test the effects of the additives on NSB, we selected p85β as a model analyte. Moreover, we have recently studied the interaction of p85β with NS1 of the 1918 influenza A virus;18 thus, a quantitative comparison with the previous result was possible (vide infra).
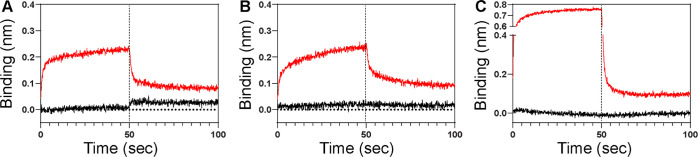
Figure 2 shows the effect of the additives on the NSB signal. Although the additives suppressed the NSB to a certain degree, the effects were rather marginal. Most noticeably, the amplitude of the NSB signal was comparable with that of a typical ligand-analyte binding (0.1–0.6 nm) (vide infra). Moreover, the magnitude of the NSB was even larger in the presence of casein (0.2%) (Figure 2C) than that in the absence of casein (Figure 1C). These results indicated that the double-referencing might be highly error-prone even in the presence of common additives. Overall, our results indicate that commonly used additives are not effective at high analyte concentrations in general and highlight the need for new NSB blockers.
Figure 2.
Effects of common NSB blockers. (A) 0.05% Tween-20. (B) 1% BSA. (C) 0.2% casein. Black and red lines correspond to 1 μM and 40 μM of p85β, respectively. Vertical dotted lines represent the initiation of the dissociation phase.
Saccharides are Effective NSB Blockers
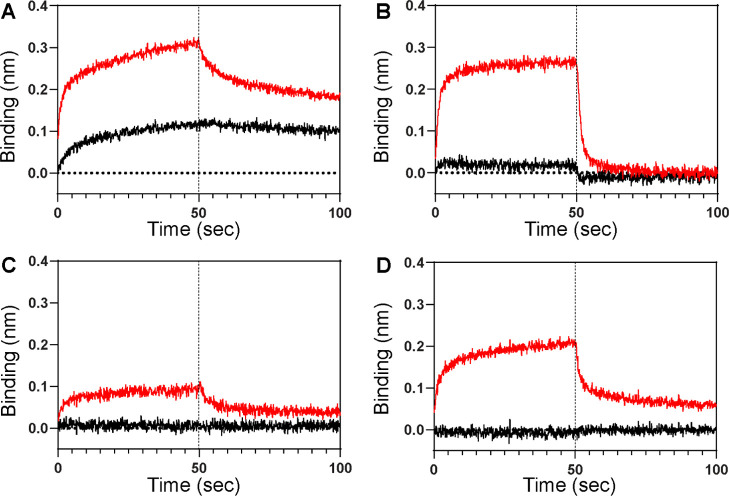
To test whether the NSB is due to the interaction of analytes with the Ni-NTA moiety of the sensor tips, we monitored the NSB in the presence of 50 mM imidazole as a blocking additive (Figure 3A). Although imidazole attenuated the NSB (see Figure 1C for comparison), it can also weaken the binding of a His6-tagged ligand to the sensor tip, resulting in a small BLI signal from a ligand–analyte interaction. Moreover, the reduced interaction between the ligand and the sensor tip can induce baseline drift. As a result, imidazole alone might not be an effective NSB blocker.
Figure 3.
BLI sensorgrams of NSB between the Ni-NTA biosensor and p85β (black; 1 μM, red; 40 μM) in the presence of NSB blocking admixtures containing (A) 50 mM imidazole, (B) 0.6 M trehalose, (C) 0.6 M sucrose, and (D) 0.6 M glucose. See Figure S1 for superimposed sensorgrams. Additionally, all buffers contained 1% BSA. Vertical dotted lines represent the initiation of the dissociation phase.
The marginal effect of the tested NSB blockers might be due to heterogeneous chemical interactions between analytes and biosensor tips. Thus, we tested admixtures of known and new NSB blockers. BSA showed the most desirable effect among the tested additives (Figure 2B); therefore, we selected 1% BSA as our base additive and tested additional compounds that might substantially reduce NSB relative to BSA alone.
Osmolytes enhance the solvation of proteins, resulting in the attenuation of protein aggregation and precipitation.27−29 Among osmolytes, we investigated the effects of three saccharide molecules: glucose, trehalose, and sucrose. These molecules are highly soluble, nonionic, and compatible with BLI sensor tips. Interestingly, we found that these saccharides in combination with BSA attenuated NSB more effectively than BSA alone (Figure 3B–D). Among the three saccharides, sucrose was the most effective in suppressing the NSB (Figure 3C).
Moreover, we observed that the NSB was further reduced by including 20 mM imidazole in buffer containing sucrose (0.6 M) and BSA (1%) (Figures 1E–H and S2). A further comparison also showed that the tri-component admixture (1% BSA, 20 mM imidazole, and 0.6 M sucrose) suppressed NSB more effectively than two-component admixtures (Figure S2). The addition of 20 mM imidazole did not noticeably reduce the affinity of a His-tagged ligand and a Ni-NTA biosensor (Figure S3). The new NSB-blocking admixture was remarkably effective in reducing the NSB of all tested protein analytes (Figure 1E–H), compared to the results without the NSB-blocking admixture (Figure 1A–D). We also found that the new admixture provides the better suppression of NSB than the buffer containing 1% BSA and 0.005% Tween (Figure S4). These results showed that the new NSB-blocking admixture is generally applicable to a broad range of analytes.
New NSB Blocker Enables the Quantitative Study of Weak PPIs Using BLI
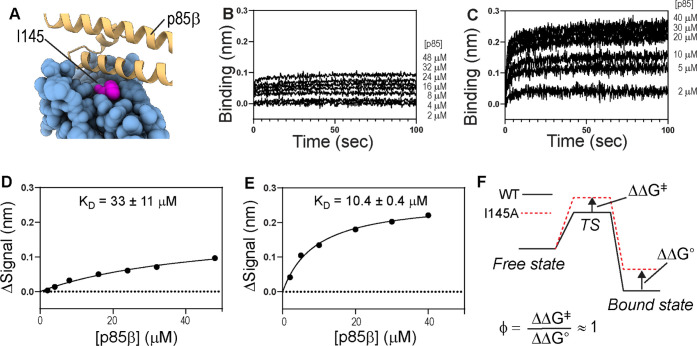
To demonstrate the effectiveness of the new NSB-blocking admixture (i.e., 20 mM imidazole, 0.6 M sucrose, and 1% BSA) in the study of weak PPIs, we characterized the binding between NS1 (ligand) of the 1918 influenza A virus and p85β (analyte) of human PI3K. Recently, we determined the thermodynamic and kinetic contributions of p85β-binding surface residues on NS1 and revealed that I145 (Figure 4A) plays an important role in stabilizing both binding transition and complex states.18
Figure 4.
(A) Interaction between I145 in NS1 and p85β (PDB ID: 6U28). BLI sensorgrams of the binding between immobilized NS1-I145A and p85β in the presence of the buffers containing (B) 50 mM imidazole and 1% BSA as blocking additives and (C) new NSB blocking admixture. The fitted binding kinetic constants are shown on top of each panel. All sensorgrams are the net signal change after double-referencing. Binding isotherms for NS1-I145A and p85β acquired using buffers containing (D) 50 mM imidazole and (E) NSB-blocking admixture. KD values are represented by average ± standard deviation of three repeats. The signal change (ΔSignal) corresponds to the net signal change after subtracting the NSB signal. (F) Schematic showing ϕ-value analysis.
However, the study manifested the difficulty of studying weak PPIs using BLI because of substantial NSB. For example, Ala-scanning mutagenesis of critical residues, including I145A, reduced the binding affinity considerably. As a result, high concentrations of p85β were required for accurate estimation of the binding properties. To suppress the NSB, we used a buffer containing 50 mM imidazole and 1% BSA in the previous study. Although including a high concentration of imidazole reduced the NSB signal, it also weakened the binding of the ligand to the sensor tip (Figure S5). As a result, the binding between NS1-I145A and p85β yielded a small net signal after subtracting the NSB; thus, the KD value was rather poorly defined (KD = 33 ± 11 μM) (Figure 4B,D).
In contrast, the present study using the new NSB-blocking admixture showed a significantly larger net BLI signal change upon the NS1–p85 interaction (Figure 4C,E). This improvement subsequently led to a smaller uncertainty (standard deviation of three repeats) of the KD value (10.4 ± 0.4 μM), compared to the previous result (33 ± 11 μM). This result also indicates that the new NSB-blocking admixture does not interfere with the ligand–analyte interaction; the KD value would be higher in the new admixture if it interferes with the binding process. Moreover, we tested whether sucrose affects the binding affinity by comparing the KD value of wild-type NS1 and p85β using the new admixture. Indeed, the KD value was virtually identical to the previously reported value that was acquired without sucrose (KD = 0.5 ± 0.1 μM) (Figure S6).18 This result indicates that sucrose does not affect the binding.
Based on the new binding parameters (Figure 4), we calculated the ϕ-value30,31 of I145. The ϕ-value reports the degree of an intermolecular interaction at the binding transition state relative to that in the bound state (Figure 4F). Briefly, a ϕ-value close to 1 suggests that the mutated residue develops a bound-like interaction at the transition state, while a ϕ-value close to 0 indicates that the mutated residue does not form intermolecular interactions at the binding transition state. Thus, ϕ-value analysis is a critical tool for the mechanistic understanding of binding kinetics and thermodynamics.18,32−34
The present experiment yielded a ϕ-value close to 1.0 ± 0.3, which indicates that I145 forms a bound-like interaction at the transition state. The new ϕ-value is higher than the previous value (0.8 ± 0.1) acquired in the presence of 50 mM imidazole.18 The present study demonstrates the impact of the new NSB-blocking admixture for accurate, quantitative analysis of weak PPIs using BLI. However, it is worth mentioning that the new result is consistent with our previous interpretation of the role of I145 in the binding to p85β.
This study aimed to identify a new, effective NSB blocker that is compatible with BLI biosensors and diverse protein analytes. We found that commonly used additives are not sufficiently effective for reducing NSB when tested with various protein analytes, especially at high concentrations. Although NSB is often attributed to electrostatic interactions, our results suggest that the chemical interactions underlying NSB are heterogeneous. Thus, employing a multi-component admixture might be a rational approach to reduce NSB.
We demonstrated that saccharides, especially sucrose, are promising NSB blockers. Notably, their effects are additive when combined with other NSB blockers, enabling further optimization for proteins of interest. Moreover, saccharides are nonproteinaceous, inexpensive, homogeneous, and inert with buffer components. These features enable more consistent sample preparation relative to proteinaceous blockers, such as BSA and casein. The mechanistic basis of how osmolytes reduce NSB between protein analytes and a Ni-NTA sensor tip remains unknown. Further research is warranted in future studies to understand the mechanism. We anticipate that our findings here will help broaden the application of BLI toward the quantitative analysis of weak PPIs that underlies numerous cellular signaling processes.
Materials and Methods
Protein Production and Purification
CT10-regulator of kinase II (CRK-II; residues 1–304),19,20 tripartite motif-containing 25 (TRIM25; residues 181–380),21 p85β of human PI3K (residues 431–596),22 and E3 ubiquitin protein ligase RNF135 (Riplet; residues 126–220)23,24 were expressed with a His6 and Sumo tags at the N-terminus of individual proteins in BL21 (DE3) E. coli cells, induced at 0.6 OD600 with 0.5 mM isopropyl-β-thiogalactoside at 37 °C for 4 h. Proteins were purified by immobilized metal affinity chromatography (IMAC), followed by cleavage of the His6-Sumo tag by a sumo protease. The proteins without the tag were further purified by IMAC and size-exclusion chromatography (SEC). NS1 (residues 80–205) of the 1918 influenza A virus was expressed with His6 and Sumo tags at the N-terminus. NS1 was purified by IMAC, followed by SEC. The purity of purified proteins was confirmed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. The quality of proteins was previously tested for structural and biophysical studies.6,19,21,35
Monitoring NSB Using BLI
The NSB between analytes and biosensors was monitored using Ni-NTA biosensors (Sartorius Corp.). The biosensors were pre-incubated for 15 min before each experiment in the binding buffer; 20 mM sodium phosphate (pH 7) and 150 mM sodium chloride. Then, the biosensors were incubated with buffers containing various NSB blockers until the signal was stabilized. All BLI data were obtained at 25 °C using an Octet RED biolayer interferometer (Pall ForteBio). The limit of detection (LOD) and limit of quantification (LOQ) were tested using p85β (Figure S7).
Binding of NS1-I145A and p85β
His6-Sumo tagged NS1 (His-Sumo-NS1) was immobilized on a Ni-NTA biosensor (Sartorius Corp.) using the buffer consisting of 20 mM sodium phosphate (pH 7) and 150 mM sodium chloride. Then, the ligand-loaded biosensors were incubated with the buffer containing NSB blockers until the BLI signal was stabilized before the analyte-association step.
Data Fitting
All BLI data were analyzed using GraphPad Prism 9 (GraphPad Software). Data analysis for binding between NS1-I145A and p85β was conducted after subtracting the NSB using double-referencing. The observed rate constant (kobs) and dissociation rate constant (koff) were calculated by fitting data with single exponential growth and decay functions, respectively. The kon value was calculated using linear regression of the p85β-dependent association rate (kobs vs [p85β]). The plateau value of the single exponential growth function was used for calculating KD values. The reported values are the average and standard deviation of three repeated measurements.
ϕ-Value Analysis
ϕ-values were calculated by dividing ΔΔG⧧ by ΔΔG° which were calculated using following equations
| 1 |
| 2 |
where WT and MT
represent wild-type and I145A
mutant NS1 proteins, respectively. 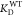 and
and 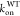 were taken
from the previous report.18
were taken
from the previous report.18
Acknowledgments
B.Z. was supported by the National Science Foundation Research Experiences for Undergraduates (NSF-REU #DBI-1949893).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c05659.
Multiple BLI sensorgrams; binding isotherm plot of NS1 and p85; and estimation of LOD and LOQ (PDF)
Accession Codes
p85β (UniProtKB O00459); CRK II (UniProtKB P46108); Riplet (UniProtKB Q8IUD6); NS1 (UniProtKB Q99AU3); and TRIM25 (UniProtKB Q14258).
Author Present Address
† Department of Microbiology and Immunology, University of Rochester, Rochester, NY 14642, United States; Email: bzuniga@u.rochester.edu
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under grant R01GM127723, the Welch Foundation (A-2028-20200401), and the USDA National Institute of Food and Agriculture (Hatch project 1020344).
The authors declare no competing financial interest.
Supplementary Material
References
- Pawson T.; Scott J. D. Signaling through scaffold, anchoring, and adaptor proteins. Science 1997, 278, 2075–2080. 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Scott D. E.; Bayly A. R.; Abell C.; Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discovery 2016, 15, 533–550. 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- Arkin M. R.; Tang Y.; Wells J. A. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem. Biol. 2014, 21, 1102–1114. 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdiche Y.; Malashock D.; Pinkerton A.; Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008, 377, 209–217. 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Sultana A.; Lee J. E. Measuring Protein-Protein and Protein-Nucleic Acid Interactions by Biolayer Interferometry. Curr. Protoc. Protein Sci. 2015, 79, 19.25.1–19.25.26. 10.1002/0471140864.ps1925s79. [DOI] [PubMed] [Google Scholar]
- Cho J.-H.; Zhao B.; Shi J.; Savage N.; Shen Q.; Byrnes J.; Yang L.; Hwang W.; Li P. Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 6550–6558. 10.1073/pnas.1920582117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forssén P.; Samuelsson J.; Lacki K.; Fornsted T. Advanced Analysis of Biosensor Data for SARS-CoV-2 RBD and ACE2 Interactions. Anal. Chem. 2020, 92, 11520–11524. 10.1021/acs.analchem.0c02475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeramange C. J.; Fairlamb M. S.; Singh D.; Fenton A. W.; Swint-Kruse L. The strengths and limitations of using biolayer interferometry to monitor equilibrium titrations of biomolecules. Protein Sci. 2020, 29, 1018–1034. 10.1002/pro.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y.; Woods R. J. Quantifying Weak Glycan-Protein Interactions Using a Biolayer Interferometry Competition Assay: Applications to ECL Lectin and X-31 Influenza Hemagglutinin. Adv. Exp. Med. Biol. 2018, 1104, 259–273. 10.1007/978-981-13-2158-0_13. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy S.; Tobias R. Label-free kinetic analysis of an antibody-antigen interaction using biolayer interferometry. Methods Mol. Biol. 2015, 1278, 165–182. 10.1007/978-1-4939-2425-7_10. [DOI] [PubMed] [Google Scholar]
- Dzimianski J. V.; Lorig-Roach N.; O’Rourke S. M.; Alexander D. L.; Kimmey J. M.; DuBois R. M. Rapid and sensitive detection of SARS-CoV-2 antibodies by biolayer interferometry. Sci. Rep. 2020, 10, 21738. 10.1038/s41598-020-78895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. M.; Yao X.; Vanam R. P.; Marlow M. S. Measuring the effects of macromolecular crowding on antibody function with biolayer interferometry. MAbs 2019, 11, 1319–1330. 10.1080/19420862.2019.1647744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. K.; Dorosky D.; Sharma P.; Abbasi S. A.; Dye J. M.; Kranz D. M.; Herbert A. S.; Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 2020, 369, 1261–1265. 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I.; AlSadhan I.; Vaidyanathan P. P.; Herschlag D. How to measure and evaluate binding affinities. Elife 2020, 9, e57264 10.7554/eLife.57264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe P. N.; Vaughan C. K.; Daviter T. Measurement of protein-ligand complex formation. Methods Mol. Biol. 2013, 1008, 63–99. 10.1007/978-1-62703-398-5_3. [DOI] [PubMed] [Google Scholar]
- Hale B. G.; Jackson D.; Chen Y.-H.; Lamb R. A.; Randall R. E. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 14194–14199. 10.1073/pnas.0606109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrow A.; Kim I.; Topo E.; Cho J.-H. Understanding the Binding Transition State After the Conformational Selection Step: The Second Half of the Molecular Recognition Process Between NS1 of the 1918 Influenza Virus and Host p85β. Front. Mol. Biosci. 2021, 8, 716477. 10.3389/fmolb.2021.716477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.-H.; Muralidharan V.; Vila-Perello M.; Raleigh D. P.; Muir T. W.; Palmer A. G. Tuning protein autoinhibition by domain destabilization. Nat. Struct. Mol. Biol. 2011, 18, 550–555. 10.1038/nsmb.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge R. B.; Fajardo J. E.; Mayer B. J.; Hanafusa H. Tyrosine-phosphorylated epidermal growth factor receptor and cellular p130 provide high affinity binding substrates to analyze Crk-phosphotyrosine-dependent interactions in vitro. J. Biol. Chem. 1992, 267, 10588–10595. 10.1016/s0021-9258(19)50057-6. [DOI] [PubMed] [Google Scholar]
- Koliopoulos M. G.; Lethier M.; van der Veen A. G.; Haubrich K.; Hennig J.; Kowalinski E.; Stevens R. V.; Martin S. R.; Reis E Sousa C.; Cusack S.; Rittinger K. Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition. Nat. Commun. 2018, 9, 1820. 10.1038/s41467-018-04214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B. G.; Kerry P. S.; Jackson D.; Precious B. L.; Gray A.; Killip M. J.; Randall R. E.; Russell R. J. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 1954–1959. 10.1073/pnas.0910715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H.; Matsumoto M.; Hatakeyama S.; Seya T. Riplet/RNF135, a RING Finger Protein, Ubiquitinates RIG-I to Promote Interferon-β Induction during the Early Phase of Viral Infection. J. Biol. Chem. 2009, 284, 807–817. 10.1074/jbc.m804259200. [DOI] [PubMed] [Google Scholar]
- Gao D.; Yang Y.-K.; Wang R.-P.; Zhou X.; Diao F.-C.; Li M.-D.; Zhai Z.-H.; Jiang Z.-F.; Chen D.-Y. REUL is a novel E3 ubiquitin ligase and stimulator of retinoic-acid-inducible gene-I. PLoS One 2009, 4, e5760 10.1371/journal.pone.0005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat V.; Rafique A. Designing binding kinetic assay on the bio-layer interferometry (BLI) biosensor to characterize antibody-antigen interactions. Anal. Biochem. 2017, 536, 16–31. 10.1016/j.ab.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Tobias R.; Yao D.; Kumaraswamy S.. Analysis of Fc-gamma receptor-IgG interactions on the Octet platform. https://www.sartorius.com/en/pr/octet/analysis-of-fc-gamma-receptor-igg-interactions-on-octet-platform?utm_source=google&medium=cpc&utm_campaign=octet-brand&utm_term=octet&utm_content=product-page&gclid=CjwKCAiA4veMBhAMEiwAU4XRr6M5iLOkg6ASTZhbpXCqTStQfWAICZZ6rff4OY7mF_wTdv94I62PtxoC2hEQAvD_BwE (accessed Nov 20, 2021).
- Auton M.; Rösgen J.; Sinev M.; Holthauzen L. M. F.; Bolen D. W. Osmolyte effects on protein stability and solubility: a balancing act between backbone and side-chains. Biophys. Chem. 2011, 159, 90–99. 10.1016/j.bpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwah N.; Jain V.; Yadav D. Osmolytes: A Possible Therapeutic Molecule for Ameliorating the Neurodegeneration Caused by Protein Misfolding and Aggregation. Biomolecules 2020, 10, 132. 10.3390/biom10010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel C. N.; Noble S. M.; Weatherly G. T.; Tripathy A.; Winzor D. J.; Pielak G. J. Effects of molecular crowding by saccharides on α-chymotrypsin dimerization. Protein Sci. 2002, 11, 997–1003. 10.1110/ps.4450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R.; Sato S. Φ-value analysis and the nature of protein-folding transition states. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 7976–7981. 10.1073/pnas.0402684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.-H.; Meng W.; Sato S.; Kim E. Y.; Schindelin H.; Raleigh D. P. Energetically significant networks of coupled interactions within an unfolded protein. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 12079–12084. 10.1073/pnas.1402054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G.; Haran G.; Zhou H.-X. Fundamental Aspects of Protein–Protein Association Kinetics. Chem. Rev. 2009, 109, 839–860. 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokhovchuk F.; Mesrouze Y.; Meyerhofer M.; Zimmermann C.; Fontana P.; Erdmann D.; Jemth P.; Chène P. An Early Association between the α-Helix of the TEAD Binding Domain of YAP and TEAD Drives the Formation of the YAP:TEAD Complex. Biochemistry 2020, 59, 1804–1812. 10.1021/acs.biochem.0c00217. [DOI] [PubMed] [Google Scholar]
- Horn J. R.; Sosnick T. R.; Kossiakoff A. A. Principal determinants leading to transition state formation of a protein-protein complex, orientation trumps side-chain interactions. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 2559–2564. 10.1073/pnas.0809800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder C.; Ma L.-C.; Krug R. M.; Montelione G. T.; Guan R. Structure of the iSH2 domain of human phosphatidylinositol 3-kinase p85β subunit reveals conformational plasticity in the interhelical turn region. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2010, 66, 1567–1571. 10.1107/s1744309110041333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.