Abstract
Flavonoids are the most widespread polyphenolic compounds and are important dietary constituents present in horticultural crops such as fruits, vegetables, and tea. Natural flavonoids are responsible for important quality traits, such as food colors and beneficial dietary antioxidants, and numerous investigations have shown that intake of flavonoids can reduce the incidence of various non-communicable diseases. Analysis of the thousands of flavonoids reported so far has shown that different hydroxylation modifications affect their chemical properties and nutritional values. These diverse flavonoids can be classified based on different hydroxylation patterns in the B, C, and A rings and multiple structure–activity analyses have shown that hydroxylation decoration at specific positions markedly enhances their bioactivities. This review focuses on current knowledge concerning hydroxylation of flavonoids catalyzed by several different types of hydroxylase enzymes. Flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′5′-hydroxylase (F3′5′H) are important enzymes for the hydroxylation of the B ring of flavonoids. Flavanone 3-hydroxylase (F3H) is key for the hydroxylation of the C ring, while flavone 6-hydroxylase (F6H) and flavone 8-hydroxylase (F8H) are key enzymes for hydroxylation of the A ring. These key hydroxylases in the flavonoid biosynthesis pathway are promising targets for the future bioengineering of plants and mass production of flavonoids with designated hydroxylation patterns of high nutritional importance. In addition, hydroxylation in key places on the ring may help render flavonoids ready for degradation, and the catabolic turnover of flavonoids may open the door for new lines of inquiry.
Introduction
Flavonoids are the most widespread polyphenolic compounds, characterized by possession of at least one aromatic ring with one or more hydroxyl groups attached. Current dietary guidance from the World Health Organization recommends people should consume at least 400 g, i.e. five portions, of fruits and vegetables per day for their optimum health. Besides vitamins, minerals, and dietary fiber, such horticultural crops have the advantage of accumulating high amounts of polyphenols, especially flavonoids [1]. These substances in planta are generated as the products of plant antioxidative defense systems activated in rapid response to both biotic and abiotic stresses and are responsible for important quality traits, such as color, in horticultural crops [2]. Moreover, over recent years, numerous epidemiological and clinical studies have shown that such flavonoids also exhibit strong antioxidant properties in vitro and in vivo, as well as antidiabetic, antiobesity, anti-inflammatory, anticancer, and antibacterial activities [1, 3, 4] (Fig. 1). Hence, flavonoid-abundant foods might be nature’s bountiful gifts to humankind for their excellent health-promoting benefits.
Figure 1.
Health-promoting bioactivities of flavonoids in horticultural crops.
There are >8000 different flavonoids identified from plants [5]. Different modifications, such as hydroxylation, glycosylation, methylation, and acylation, play important roles in generating such diversity of flavonoids, and hydroxylation is the most frequently occurring modification in natural flavonoids (Fig. 2). Hydroxylation of flavonoids improves the chemical solubility and stability of an array of flavonoids and is also correlated with their bioactive properties. For instance, a hydroxylated B ring is crucial for the antioxidant properties of flavonoids [6]. An increased degree of hydroxylation of flavonoids is associated with stronger inhibitory effects on α-glucosidase or α-amylase activities [7, 8]. Inhibition of aldose reductase activity was also found to be remarkably enhanced by the hydroxylation of specific positions, such as the C3′ and C4′ of the B ring of flavonoids [9]. Taken together, the data show that different hydroxylation decoration patterns of flavonoids may affect their chemical properties as well as their bioactivities. So far, there have been several review papers describing the structures and biosynthesis of flavonoids and the introduction of various decorations, such as glycosylation [10–12], methylation [13, 14], and acylation [15]. However, to date, there is no systematic review on the hydroxylation modification of flavonoids, particularly in horticultural crops.
Figure 2.
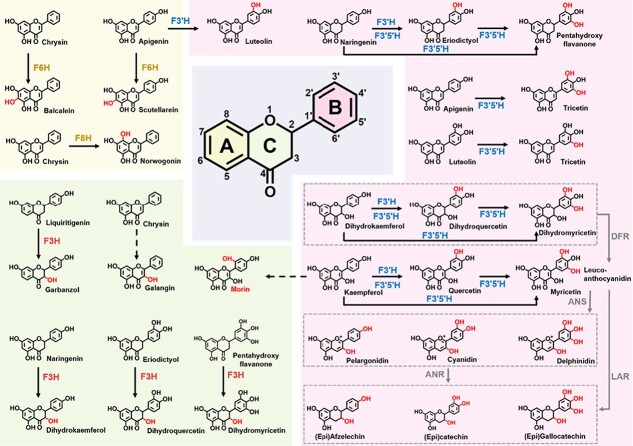
Key hydroxylases that participate in hydroxylation decoration during flavonoid biosynthesis. F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; F3′5′H, flavonoid 3′,5′-hydroxylase; F6H, flavone 6-hydroxylase; F8H, flavone 8-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; LAR, leucoanthocyanidin reductase.
This review focuses on current knowledge concerning hydroxylation decoration of flavonoids and its effects on their chemical and nutritional value. Diverse dietary flavonoids are further classified based on different hydroxylation patterns in the B, C, and A rings of flavonoids and a comparison is made of bioactivities of flavonoids that differ only by hydroxylation patterns. Particular attention is paid to key hydroxylases in the flavonoid biosynthesis pathway, with the objective of future bioengineering-targeted bioactivities in plant resources and mass production of flavonoids with desirable hydroxylation patterns. We also discuss the degradation of natural flavonoids and the relationship between hydroxylation modification and flavonoid catabolism in planta, which requires further investigation in the future.
Divergent hydroxylation patterns and distribution of flavonoids
Flavonoids consist of two aromatic rings (A and B rings) connected by a three-carbon bridge (C ring) containing an embedded oxygen atom, abbreviated as C6-C3-C6 (Fig. 2). According to the oxidative status and hydroxylation degree of the C ring and the connection position of the B ring, flavonoids can be grouped into different subclasses, including flavonols, anthocyanidins, flavones, flavanones, flavanols, and isoflavones (Table 1). Each subclass of flavonoids can be analyzed based on its hydroxylation patterns.
Table 1.
Divergent hydroxylation patterns of flavonoids and their distribution in horticultural crops.
|
Different flavonoids are listed within each subclass according to the position of -OH in the order of A ring, followed by B ring with increasing number of hydroxyl groups. Compounds are listed based on the relative amount of each compound, from high to relatively low, in different horticultural crops.
Hydroxylation and distribution of flavonols
Flavonols are the most widespread subclass of flavonoids and possess the 3-OH and 4-oxo groups on the C6-C3-C6 backbone. Their diversity originates from different substitution of the phenolic -OH groups in both the A and the B ring (Table 1).
The flavonol fisetin is hydroxylated at C7, C3, C3′, and C4′ and is found mainly in strawberry, apple, persimmon, onion, and grape [16]. Galangin, kaempferol, morin, quercetin, and myricetin are common flavonols having the same structures on the A and C rings with hydroxyl groups attached at C5, C7, and C3, but with different hydroxylation modes on their B ring (Table 1). Specifically, galangin, which is a major active component in the root of galangal [17] and is also found in propolis produced from plants by bees [18], has no -OH on the B ring. Kaempferol, with an -OH group substituted at C4′, is abundant in green leafy vegetables such as spinach and kale, and berries, including mulberry and strawberry [19, 20]. Morin also has two -OH groups on the B ring at the C2′ and C4′ positions and is found mainly in mulberry, guava, grape, and fig [21]. Quercetin, which is hydroxylated at both C3′ and C4′, is present in a high concentration in onion, chili pepper, apple, Chinese bayberry, mulberry, and apricot [19, 20, 22]. Notably, myricetin has three hydroxyl groups at C3′, C4′, and C5′. It was originally isolated from the bark of Chinese bayberry [23] and is also present at a relatively high level in its fruit [22]. Myricetin is also widely distributed in many other berries, such as strawberry, blackberry, and blueberry, as well as vegetables like spinach and cauliflower [20, 24]. Quercetagetin which has three -OH groups on the A ring (C5, C6, and C7) is found mainly in Tagetes [25].
Hydroxylation and distribution of anthocyanidins
Anthocyanins are the glycoside conjugates of anthocyanidins (Table 1) and constitute a group of natural pigments that confer a wide spectrum of colors, varying from orange, salmon pink, red, magenta, and violet to dark blue in many fruits, colored leafy vegetables, and tubers. The majority of anthocyanins reported are based on three common anthocyanidins, i.e. pelargonidin, cyanidin, and delphinidin [26]. These three compounds share the same chemical structures at the A and C rings with a characteristic flavylium cation and three -OH groups attached at C5, C7, and C3. They have different degrees of hydroxylation on the B ring (Table 1) and, interestingly, their colors seem to gradually deepen with increasing hydroxylations, i.e. orange/red (pelargonidin), red/magenta (cyanidin), and violet/blue (delphinidin), respectively [27].
Pelargonidin, hydroxylated at C4′, is abundant in strawberry [28], and distributed in red radish, potato, and banana [29]. Cyanidin, with two hydroxyl groups at C3′ and C4′ of the B ring, is the most common anthocyanidin. It is especially rich in berries such as Chinese bayberry [22], elderberry, chokeberry, blackberry, black mulberry, and cherry [28, 29]. Cyanidin is also ubiquitously found in other fruits, like apple, peach, pear, and fig, as well as colored vegetables like red onion and red cabbage [29]. Delphinidin has three hydroxyl groups at C3′, C4′, and C5′ of the B ring, the major sources of which were dark-colored foods such as bilberry, highbush blueberry, blackcurrant, eggplant, etc. [28, 29]. It is also found in passion fruit, green bean, and pomegranate [29].
Hydroxylation and distribution of flavones
Flavones lack the hydroxyl group at C3 compared with the skeleton of flavonols, but have a wide range of hydroxylation patterns on the A and B rings. Typically, chrysin, apigenin, luteolin, and tricetin are common flavones that all have hydroxylation at C5 and C7 but differ from each other by the substitution of -OH groups on their B rings (Table 1). Chrysin has no hydroxyl on the B ring and is found in honey, propolis, and passion fruit [30]. Apigenin (4′-OH) is particularly abundant in parsley and dried flowers of chamomile, and exists in some other vegetables, such as celery, broccoli, spices like thyme, and fruits such as cherry, olive, and legumes [31]. Major food sources of luteolin (3′,4′-OH) include bird chili [32], celery, broccoli, parsley, chrysanthemum flowers, onion leaves, sweet bell pepper, and carrot [33]. Tricetin (3′,4′,5′-OH) is found mainly in Myrtaceae pollen and Eucalyptus honey [34]. In addition, other flavones, such as baicalein or norwogonin, show additional hydroxylations on carbons 6 or 8, based on the structure of chrysin; and scutellarein has an additional -OH group substituted at the C4′ of baicalein or C6 of apigenin (Table 1). These three flavones specifically exist in Scutellaria baicalensis and Scutellaria barbata [35].
Hydroxylation and distribution of flavanones
Flavanones that have a chiral carbon at C2 are the hydrogenated derivatives of flavones. The flavanone liquiritigenin has a C7-hydroxyl on the A ring and C4′-hydroxyl on its B ring and is particularly prevalent in licorice [36] (Table 1). Flavanones including pinocembrin, naringenin, eriodictyol, and 5,7,3′,4′,5′-pentahydroxy flavanone all have dihydroxyl groups at C5 and C7, with the structural difference involving sequential increases in B-ring hydroxylation (Table 1). Pinocembrin has no -OH substitution on the B ring and in the (2S)-form is widely distributed in propolis and Glycyrrhiza glabra [37]. Naringenin (4′-OH) occurs extensively in Citrus fruits like grapefruit, orange, and lemon [38]. Additionally, eriodictyol (3′,4′-OH) is also widely distributed in Citrus fruits such as bitter orange, mandarin, tangerine, and lemon, as well as peanuts and loquats [39]. Pentahydroxy flavanones also exist in some horticultural crops. For instance, 5,7,3′,4′,5′-pentahydroxy flavanone has been detected in Helichrysum bracteatum [40].
Hydroxylation and distribution of flavanols
Compared with other subclasses of flavonoids, flavanols are characterized by the absence of a double bond between C2 and C3 and have no C4 carbonyl in the C ring (Table 1). Therefore, two chiral carbons (C2 and C3) exist in flavanols and the fixed hydroxylation at C3 means each of them has four possible diastereoisomers. For instance, catechin, the most typical monomeric flavanol, exists in four forms, i.e. (−)-catechin (2S,3R), (+)-catechin (2R,3S), (−)-epicatechin (2S,3S), and (+)-epicatechin (2R,3R). Of these, (+)-catechin and (−)-epicatechin are most commonly found in horticultural crops, especially in tea leaves [41, 42]. Catechin has four hydroxyl groups in the basic skeleton of flavan-3-ol at C5, C7, C3′, and C4′, respectively. Afzelechin, which lacks the 3′-OH of catechin, is present in sour jujube [43], cowpea [44], and peanut seed skin [45]. Gallocatechin has one more -OH substitution at C5′ of catechin, and is present at significant levels in tea [41, 42]. The 3-OH on the C ring of catechins is usually esterified with gallic acid, thereby forming the gallated catechins such as epigallocatechin gallate, epicatechin gallate, and catechin gallate.
Hydroxylation and distribution of isoflavones
Unlike other flavonoids, the isoflavone B ring is attached at C3 rather than C2 (Table 1). Isoflavones such as 4',7-dihydroxyisoflavone (daidzein) and 4′,5,7-trihydroxyisoflavone (genistein), are specifically accumulated in legumes, especially soybeans [46]. Thus, soybean-based foods such as tofu, soy milk, and soy yoghurt are excellent dietary sources of isoflavones.
Bioactivities of flavonoids influenced by divergent hydroxylation patterns
For the B ring
With the increasing attention paid to the healthcare benefits of natural products, there are more and more studies focusing on the structure–activity relationships between the different chemical modifications of flavonoids and their bioactivities [47, 48]. Plenty of studies have shown that hydroxylation decoration at specific positions of flavonoids markedly enhances efficacy of their bioactivity (Table 2).
Table 2.
Different bioactivities of flavonoids based on their divergent hydroxylation patterns.
| Compounds | Bioactivities | References | |
|---|---|---|---|
| Kaempferol → quercetin → myricetin | Scavenging hydroxyl radicals ↑ | 49 | |
| 4′-OH → 3′,4′-OH → 3′,4′,5′-OH | Trapping ability of ACR ↑ | 50 | |
| Protection activity against light-mediated photoreceptor damage ↑ | 51 | ||
| Inhibition of α-glucosidase ↑ | 52, 53 | ||
| Inhibits EGF-induced cell transformation ↑ | 54 | ||
| Constituent of active packaging films containing flavonols ↑ | 55 | ||
| Induction of apoptosis in tumor cells ↑ | 56 | ||
| Inhibition of tyrosinase ↑ | 57 | ||
| B ring | Kaempferol → morin | Scavenging hydroxyl radicals ↑ | 49 |
| 4′-OH → 2′,4′-OH | Inhibition of formation of fAβ ↑ | 58 | |
| ACR trapping efficiency ↑ | 50 | ||
|
Galangin → kaempferol
No hydroxyl → 4′-OH |
ACR trapping efficiency ↑ | 50 | |
| Pelargonidin → cyanidin → delphinidin | Superoxide radical- and peroxynitrite-scavenging activity ↑ | 59, 60 | |
| 4′-OH → 3′,4′-OH → 3′,4′,5′-OH | Inhibition of lipid peroxidation ↑ | 61 | |
| Antioxidant activities ↑ | 60 | ||
| C3G → D3G | Stimulation of insulin secretion ↑ | 64 | |
| 3′,4′-OH → 3′,4′,5′-OH | Inhibition of viability of cancer cells ↑ | 65 | |
| Compounds | Bioactivities | References | |
|
Chrysin → apigenin → luteolin
No hydroxyl → 4′-OH → 3′,4′-OH |
Induction of apoptosis in tumor cells ↑ | 66 | |
| Inhibition of proteasome activity ↑ | 66, 67 | ||
| MMP inhibitory effect ↑ | 68 | ||
| B ring | Reactive oxygen scavenging activities ↑ | 68 | |
| Inhibition of pro-inflammatory cytokine production ↑ | 69 | ||
| Pinocembrin → naringenin → eriodictyol | Antioxidant activity ↑ | 70, 71 | |
| No hydroxyl → 4′-OH → 3′,4′-OH | Inhibition of the phosphorylation of PKCδ ↑ | 70 | |
| Inhibition of the formation of AGEs ↑ | 72 | ||
| Tricetin → myricetin 3H → OH | Pro- and antioxidant activity ↑ | 73 | |
| C ring | Apigenin→ kaempferol 3H → OH | Inhibition of α-glucosidase or α-amylase ↑ | 74 |
| Anti-adipogenic action ↑ | 75 | ||
| Fisetin → quercetin 5H → OH | Membrane binding ↑ | 76 | |
| A ring | Daidzein → genistein 5H → OH | Antioxidant activity ↑ | 77 |
| Improves glucose tolerance↑ | 78 |
fAβ, β-amyloid fibrils; C3G, cyanidin 3-O-glucoside; D3G, delphinidin 3-O-glucoside; ACR, acrolein; EGF, epidermal growth factor; MMP, matrix metalloproteinease; PKCδ, protein kinase Cδ; AGEs, advanced glycation end products.
The flavonols kaempferol (4′-OH), quercetin (3′,4′-OH), and myricetin (3′,4′,5′-OH) are a typical group of flavonoids with the same structures for the A and C rings but have different hydroxylation modes on the B ring. Results have shown that quercetin has stronger •OH scavenging properties than kaempferol, and myricetin is the most powerful hydroxyl radical scavenger, indicating that radical scavenging activity increases with the growing number of -OH groups on the aromatic B ring [49] (Table 2). Furthermore, their acrolein-trapping efficiency decreases in the order of myricetin > quercetin > kaempferol [50]. Exposure to blue light for 20 hours induced the death of ~75% of the photoreceptors in bovine retinal cell cultures, while myricetin conferred ~100% protection against light-mediated damage to photoreceptors, whereas quercetin showed relatively poor protective activity, and kaempferol was inactive [51]. Among these three flavonols, myricetin showed the strongest inhibitory effects on either α-glucosidase [52, 53] or epidermal growth factor-induced cell transformation in JB6 P+ cells [54], followed by quercetin and kaempferol. Such flavonols can also be individually mixed with a chitosan-based matrix to develop active packaging films, and myricetin showed the strongest intermolecular interactions with the film matrix, due to the greater number of hydroxyl groups on the B ring [55]. Hence, films containing myricetin have the most satisfactory mechanical properties and the highest ability to provide a barrier to water vapor and oxygen [55]. In addition, studies have reported that myricetin is more effective than quercetin at inhibiting the induction of apoptosis in prostate cancer cell line PC-3 [56]. Also, quercetin has a stronger tyrosinase inhibitory effect than kaempferol [57].
Morin (2′,4′-OH), with an additional hydroxyl at the C2′ position of kaempferol, shows enhanced hydroxyl radical scavenging activity [49]. It has a greater ability to inhibit β-amyloid fibril formation from amyloid β-peptide and destabilizes preformed β-amyloid fibrils, demonstrating its greater potential for the prevention and control of Alzheimer’s disease [58]. Galangin, another flavonol with no hydroxylation on the B ring, as well as morin and kaempferol, all show significant scavenging ability for acrolein, and their trapping efficiency increases in the order galangin < morin < kaempferol [50].
The activities of three anthocyanidins in O2•− and ONOO− scavenging and inhibition of lipid peroxidation have been ranked in the order pelargonidin (4′-OH) < cyanidin (3′,4′-OH) < delphinidin (3′,4′,5′-OH), indicating that the increasing number of -OH groups present on the B ring enhances the antioxidant activities of anthocyanidins [59–61] (Table 2). As mentioned previously, the degree of hydroxylation is related to the gradually deepening colors of these three anthocyanidins and, therefore, dark-colored fruits and vegetables may show greater health benefits [62, 63]. Similarly, compared with cyanidin 3-O-glucoside, delphinidin 3-O-glucoside showed enhanced ability to stimulate insulin secretion [64], and greater ability to inhibit the viability of cancer cells such as HCT 116 cells [65] (Table 2).
Chrysin (no hydroxyl at the B ring), apigenin (4′-OH), and luteolin (3′,4′-OH) belong to group of flavones with increasing numbers of hydroxyl substituents on the B ring. Their apoptosis-inducing potencies in tumor cells [66], proteasome inhibitory activities [66, 67], matrix metalloproteinase (MMP) inhibition effects, and scavenging reactive oxygen capacities [68] are in the order of luteolin > apigenin > chrysin (Table 2). Also, compared with chrysin, apigenin showed a stronger ability to inhibit the production of pro-inflammatory cytokines stimulated by lipopolysaccharide in human peripheral blood mononuclear cells [69].
There are three kinds of flavanones sharing similar chemical structures that differ only in the number of hydroxyl groups on their B ring, namely, pinocembrin (no hydroxyl), naringenin (4′-OH), and eriodictyol (3′,4′-OH). Studies have demonstrated that eriodictyol showed markedly higher antioxidant capacity than naringenin and pinocembrin, as reflected in its stronger inhibition of the phosphorylation of protein kinase Cδ (PKCδ) and p47 in murine macrophage RAW264.7 cells [70, 71] (Table 2). Docking analysis showed that eriodictyol has the most favorable binding with the phorbol ester binding site of PKCδ, which may be due to its greater number of hydroxyl groups on the B ring conferring the ability to form hydrogen bonds with the kinase, and thus strengthened the interaction [70]. In addition, eriodictyol showed stronger inhibitory effects on the formation of advanced glycation end products than naringenin, which may be due partly to its additional -OH in the 3′ position of the B ring [72].
Other ring modifications
Apart from the hydroxylations on the B ring, hydroxyl substituents present on either the C or A ring may also affect the biological activity of flavonoids (Table 2). For instance, myricetin shows enhanced pro- and antioxidant effects compared with tricetin, which may be due to the positive mesomeric effect of the enolic 3-hydroxy group on the C-ring [73]. Kaempferol has a stronger inhibitory effect on α-glucoside and α-amylase with the substitution of an additional hydroxyl group at the C3 position of apigenin [74]. Moreover, treating preadipocytes with kaempferol reduces triacylglycerol content, whereas apigenin treatment has no such anti-adipogenic action [75]. Quercetin (5-OH) has a significantly higher membrane binding constant than fisetin (5-H), indicating that the hydroxylation at the C5 position of the A ring plays an important role in membrane-dependent processes associated with their biological activities [76]. The isoflavone genistein (5-OH) shows stronger antioxidant activity than daidzein (5-H) [77] and supplementing with genistein significantly improved glucose tolerance in diabetic db/db mice, which was not observed in the daidzein-supplemented group [78].
Key hydroxylases in the biosynthesis pathways of flavonoids
Flavonoids are synthesized through the phenylpropanoid metabolic pathway, initiated by transformation of phenylalanine into 4-coumaroyl-CoA. This is combined with malonyl CoA, and naringenin is then synthesized by the decarboxylation and cyclization reactions catalyzed by chalcone synthase (CHS) and chalcone isomerase. Further, naringenin is converted by flavanone 3-hydroxylase (F3H) and other hydroxylases, including flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H), to produce different dihydroflavonols, which can form leucoanthocyanidins catalyzed by dihydroflavonol 4-reductase (Fig. 2). Subsequently, leucoanthocyanidins are converted into anthocyanidins or flavanols by the action of anthocyanidin synthase or leucoanthocyanidin reductase, respectively. Anthocyanidins can also be transformed to flavanols by the catalysis of anthocyanidin reductase.
The basic hydroxylation at C5, C7, and C4' of common flavonoids is shown for naringenin, while additional hydroxyl groups can also occur at C3, C3', C5', C6, and C8 positions (in Table 1). Hydroxylases play a vital role in the biosynthesis of hydroxylated flavonoids and F3′H and F3′5′H are important enzymes for the hydroxylation of the B ring of flavonoids (Fig. 2). F3H is key for the hydroxylation of the C ring and flavone 6-hydroxylase (F6H) and flavone 8-hydroxylase (F8H) are key enzymes for the hydroxylation of the A ring (Fig. 2).
Flavonoid 3′-hydroxylase
F3′H (EC 1.14.13.21) is a cytochrome P450 (CYP450)-dependent monooxygenase requiring NADPH as a cofactor. It catalyzes the hydroxylation of flavanones, flavones, dihydroflavonols, and flavonols at the C3′ position of the B ring to their 3′,4′-hyroxylated states (Fig. 2). F3′H was first cloned from petunia (Petunia hybrida) [79], and then isolated from various resources such as soybean (Glycine max) [80], grapevine (Vitis vinifera) [81], sorghum (Sorghum bicolor) [82], apple (Malus × domestica) [83], strawberry (Fragaria × ananassa) [84], rice (Oryza sativa) [85], and tea (Camellia sinensis) [86] (Table 3).
Table 3.
Transgenic analysis of the function of plant hydroxylase genes in the formation of flavonoids.
| Gene | Gene source | Target plant | Methods | Impact | References |
|---|---|---|---|---|---|
| F3 ′H | Grapevine | Petunia | Overexpression (OE) | Quercetin ↑; peonidin ↑ | 81 |
| Grapevine | Gene silencing | Peonidin ↓; seed tannin ↓ | 89 | ||
| Sorghum | Arabidopsis tt7 mutant | OE | Quercetin ↑; cyanidin ↑; condensed tannin ↑ | 82 | |
| Apple | Arabidopsis tt7 mutant | OE | Cyanidin ↑; pelargonidin ↑; quercetin ↑ | 83 | |
| Tobacco | OE | Cyanidin ↑; quercetin ↑ | |||
| Ginkgo biloba L. | Populus | OE | Epigallocatechin ↑; gallocatechin ↑; catechin ↑ | 90 | |
| F3 ′5′H | Potato | Potato | OE | Petunidin ↑ | 93 |
| Gene silencing | Anthocyanins↓; kaempferol ↑ | 98 | |||
| Pea | Pea | Gene silencing | Delphinidin↓; petunidin↓ | 95 | |
| Grapevine | Petunia | OE | Malvidin ↑; quercetin ↑ | 81 | |
| Tea | Tobacco | OE | Delphinin ↑; cyanidin ↑ | 96 | |
| F3H | Apple | Apple | Gene silencing | Flavanones ↑ | 109 |
| Lycium chinense | Tobacco | OE | Flavanols ↑ | 104 | |
| Tea | Arabidopsis | OE | Flavonols ↑; oligomeric proanthocyanins ↑ | 105 | |
| Tobacco | OE | Flavanols ↑ | 110 | ||
| Tomato | are Tomato mutant | OE | Anthocyanins ↑; flavonols ↑ | 111 | |
| Soya bean | Soya bean | CRISPR/Cas9 | Isoflavone ↑ | 113 | |
| F6H | Scutellaria baicalensis | S. baicalensis | Gene silencing | Baicalein↓; baicalin↓ | 118 |
| Arabidopsis | OE | Baicalein ↑ | |||
| F8H | S. baicalensis | S. baicalensis | Gene silencing | Wogonin↓; wogonoside↓ | 118 |
| Arabidopsis | OE | Norwogonin ↑ |
Heterologous expression in yeast and analysis of F3′Hs from divergent plant species showed significant differences in substrate specificity and catalytic properties (Fig. 2). For instance, F3′H isolated from either tea [86, 87] or montbretia (Crocosmia × crocosmiiflora) [88] catalyzed the introduction of a hydroxyl group at the C3′ position of naringenin, dihydrokaempferol, and kaempferol to produce eriodictyol, dihydroquercetin, and quercetin, respectively (Fig. 3). Similarly, F3′H from strawberry (Fragaria sp.) showed high specificity for naringenin, dihydrokaempferol, and kaempferol, while apigenin was just a minor substrate [84]. However, F3′H encoded by CYP75B4 from rice grain preferred apigenin to other substrates, leading to the formation of luteolin [85] (Fig. 3). Interestingly, CYP75B3, another rice F3′H gene, encodes an enzyme with a higher preference for kaempferol, and CYP75B3 from black rice showed around 2-fold increased catalytic efficiency with naringenin and dihydrokaempferol compared with the enzyme from either white or red rice [85]. It is also possible that different F3′H family members isolated from the same species might also have different catalytic functions.
Figure 3.
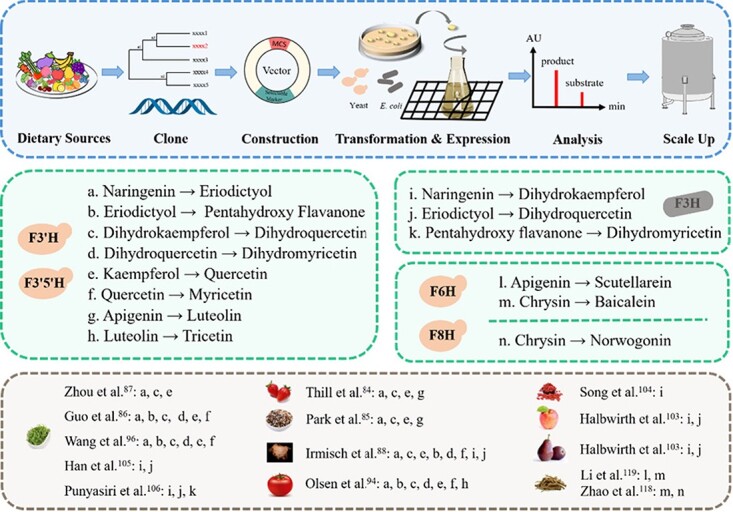
Biofactory scale-up production of desirable hydroxylated flavonoids. Pentahydroxy flavanone in this figure specifically refers to 5,7,3′,4′,5′-pentahydroxy flavanone.
Transgenic analysis has also been used to verify the functionality of F3′H from different plant sources (Table 3). Heterologous overexpression of grapevine VvF3′H in the petunia ht1 mutant showed a significant accumulation of 3′,4′-hydroxylated flavonoids, including peonidin, an O-methylated anthocyanidin derived from cyanidin, and quercetin in the transgenic flowers [81]. In contrast, transgenic grapevines with the F3′H gene silenced had obviously decreased contents of peonidin as well as seed tannins [89]. Transgenic arabidopsis (Arabidopsis thaliana) tt7 seedlings overexpressing F3′H genes isolated from either apple or sorghum showed a restored ability to produce 3′,4′-hydroxylated flavonoids such as cyanidin and quercetin [82, 83]. The increases of these two flavonoids were also shown in transgenic tobacco (Nicotiana tabacum) overexpressing apple F3′H [83]. Overexpression of the ginkgo (Ginkgo biloba) gene GbF3′H1 enhanced flavonoid production in transgenic poplar (Populus davidiana), as shown by the significantly increased red pigmentation and higher concentrations of epigallocatechin, gallocatechin, and catechin compared with that in the wild-type plants [90].
Flavonoid 3′,5′-hydroxylase
F3′5′H (EC 1.14.13.88) is another key hydroxylase responsible for the B-ring hydroxylation of flavonoids and plays a crucial role in the formation of 3′,4′,5′-trihydroxylated derivatives (Fig. 2). Initially, F3′5′H was discovered in delphinidin-rich plants, responsible for the production of blue flower colors, and was therefore known as the blue gene [91]. Later, such genes were also identified widely in other plants that accumulate proanthocyanidins or flavonols possessing trihydroxyls on the B-ring. However, not all plant species have the ability to accumulate such B-ring trihydroxyflavonoids, indicating that the presence or expression of an appropriate F3′5′H gene is not ubiquitous in plants [92]. For example, there is no F3′5′H gene in the genome of the model plantarabidopsis.
To date, a number of F3′5′Hs have been isolated from various horticultural crops, such as potato (Solanum tuberosum) [93], grapevine [81], tomato (Solanum lycopersicum) [94], pea (Pisum sativum) [95], and tea [96, 97]. Through heterologous expression in yeast, F3′5′Hs from tomato [94], tea [86, 96], grapevine [96] or montbretia [88] have been shown to hydroxylate the C3′ and/or C5′ positions of a broad range of flavonoid substrates, including flavanones, dihydroflavonols, flavonols, and/or flavones, with naringenin generally being the optimum substrate (Fig. 2). Thus, F3′5′H generally has a similar function to F3′H, and also possesses catalytic activity capable of converting naringenin or eriodictyol to 5,7,3′,4′,5′-pentahydroxy flavanone, dihydrokaempferol or dihydroquercetin to dihydromyricetin, and kaempferol or quercetin to myricetin, and also transforming luteolin into tricetin (Fig. 3).
In addition, expression levels of genes encoding F3′5′Hs greatly affect the accumulation of flavonoids with diverse B-ring hydroxylation patterns (Table 3). The red-skinned potato cultivar ‘Désirée’ transformed with a potato F3′5′H became purple-skinned, and the major anthocyanins were changed from pelargonidin derivatives to petunidin derivatives [93]. Silencing the F3′5′H in transgenic potato tuber impaired anthocyanin biosynthesis and caused a 100-fold increased level of kaempferol [98]. Similarly, F3′5′H mutants of pea have lost the ability to synthesize delphinidin and petunidin, which are the main pigments in wild-type pea flowers [95]. Additionally, transgenic petunia lines carrying VvF3′5′H1 accumulate the 3′,4′,5′-hydroxylated-based anthocyanin malvidin, and show a shift from kaempferol to quercetin, compared with the non-transgenic control [81]. Overexpression of tea F3′5′H in tobacco plants caused the majority of transgenic flowers to become magenta, compared with the pale pink of the untransformed host, due to the accumulation of a novel 3′,4′,5′-hydroxylated-based anthocyanin, delphinin, and the cyanidin content was also increased [96]. So far, transgenic studies related to F3′5′H genes have focused preferentially on the biosynthesis of anthocyanins that confer the blue or purple colors of plant tissues.
Flavanone 3-hydroxylase
F3H (EC 1.14.11.9) belongs to the FeII/2-ketoglutarate-dependent dioxygenase family. It catalyzes the 3β-hydroxylation of 2S-flavanones at the C3 position to 2R,3R-dihydroflavonols in the presence of O2, 2-oxoglutarate, Fe2+, and ascorbate as cofactors [99, 100] (Fig. 2). The first F3H was cloned from Antirrhinum majus [101], and subsequently additional F3Hs were isolated from other plant-based foods, such as soybean (G. max), apple (Malus domestica), pear (Pyrus communis), Lycium chinense, and tea [102–105]. Heterologous expression of Malus or Pyrus F3H in yeast, and montbretia or tea F3H in Escherichia coli, showed these proteins could all catalyze the conversion of naringenin and eriodictyol into dihydrokaempferol and dihydroquercetin, respectively [88, 103, 105] (Fig. 3). Similarly, when LcF3H was expressed in E. coli, the recombinant enzyme showed the ability to convert naringenin to dihydrokaempferol [104]. Further, recombinant tea F3H was able to accept 5,7,3′,4′,5′-pentahydroxy flavanone as substrate to form dihydromyricetin [106]. Interestingly, in a heterologous assembly of the fisetin biosynthetic pathway in E. coli, the production of fisetin was achieved by the conversion of liquiritigenin into garbanzol as an intermediate under the catalytic action of F3H from arabidopsis [107] (Fig. 2).
F3H shares the flavanones as substrates with other enzymes involved in the synthesis of 3-deoxyflavonoids, and the dihydroflavonols it produces are the key biosynthetic precursors of flavonols, anthocyanins, and proanthocyanins [108]. Hence, altered expression levels of F3H might redirect the flavonoid accumulation patterns in planta (Table 3) and silencing of F3H in apple led to an accumulation of flavanones [109]. Transgenic tobacco overexpressing F3H from either tea or L. chinense showed significantly increased contents of flavanols such as catechin, epicatechin, and epigallocatechin [104, 110]. The anthocyanin reduced (are) tomato mutant has a mutation in the gene encoding F3H and accumulates higher concentrations of naringenin and lower levels of flavonols and anthocyanins, compared with those in the wild type. This phenotype was reversed after the complementation of are with the p35S:F3H transgene [111]. The seeds of transgenic arabidopsis carrying tea F3H showed markedly increased contents of kaempferol glycosides and oligomeric proanthocyanidins [105]. Additionally, as expected, transgenic expression of F3H in sorghum resulted in an enrichment in its flavonoid profile, and CRISPER/Cas9-mediated gene editing of F3H as well as flavone synthase (FNS) in soya bean significantly altered the accumulation of isoflavones, as shown by the doubling in leaf isoflavone content in the T3 generation of homozygous triple mutants compared with that of the wild type [112, 113].
Flavone 6-hydroxylase
During the synthesis of the basic flavonoid skeleton catalyzed by CHS, the hydroxyl groups in the C5 and/or C7 positions are added to the A ring. It has also been found that additional hydroxylations can occur at other carbon sites on the A ring, such as C6 and C8, catalyzed by F6H and F8H, respectively (Fig. 2).
F6H was first identified from soybean and was shown to be a CYP450-dependent hydroxylase [114]. It efficiently catalyzes the hydroxylation at the C6 position of various flavanones but is hardly active with isoflavones. However, in soybean, there are several kinds of isoflavonoid constituents that possess a 6-hydroxyl group. Further investigation indicated that such 6-hydroxylation might occur before the 1,2-aryl migration of the flavonoid B ring in the process of isoflavanone formation [114]. Subsequently, a flavonol 6-hydroxylase was isolated and characterized from Chrysosplenium americanum, which exhibited 2-oxoglutarate-dependent dioxygenase (ODD) activity [115]. Meanwhile, Halbwirth et al. [116] identified another novel flavonol 6-hydroxylase, a microsomal CYP450-dependent monooxygenase in the petals of Tagetes patula and Tagetes erecta that could introduce a hydroxyl at the C6 of quercetin leading to the formation of quercetagetin. An F6H was also found in sweet basil (Ocimum basilicum L.) that could catalyze the 6-hydroxylation of flavones possessing 5-hydroxyl and 7-methoxyl residues [117]. Furthermore, another F6H was isolated from S. baicalensis, which was encoded by a CYP450 enzyme, namely SbCYP82D1.1 [118]. This can convert flavones without 7-O-methyl groups, such as apigenin and chrysin, into scutellarein and baicalein, respectively (Fig. 2). To prove the function of such SbF6H enzymes in planta, hairy root transformants in which SbCYP82D1.1 was knocked down showed significantly reduced levels of baicalein and its glycosides baicalin but higher contents of chrysin glycosides. Also, arabidopsis overexpressing SbCYP82D1.1 accumulated baicalein [118] (Table 3). Moreover, an engineered E. coli that expressed diverse plant flavonoid biosynthetic pathway genes, including SbF6H, produced baicalein and scutellarein successfully, which further verified its 6-hydroxylation function [119] (Fig. 3). In addition, EbF6H, an F6H isolated from Erigeron breviscapus, also converted apigenin to scutellarein, which was demonstrated in a yeast cell factory [120] (Fig. 3).
Flavone 8-hydroxylase
F8H was first isolated from the petals of Chrysanthemum segetum and shown to add a hydroxyl group at position 8 of flavonols and flavones, for instance, converting quercetin to 8-hydroxylquercetin (gossypetin) and luteolin to 8-hydroxyluteolin [103]. An F8H was subsequently identified from sweet basil trichomes and shown to be a specialized Rieske-type oxygenase that could efficiently catalyze the 8-hydroxylation of salvigenin [121]. Another CYP450 enzyme from S. baicalensis, SbCYP82D2, functions as an F8H with high substrate specificity that accepts only chrysin as substrate to produce norwogonin (Fig. 2), the precursor of wogonin, as shown by in vivo yeast assays [118] (Fig. 3). Further investigation in plants found that SbCYP82D2-silenced hairy roots of S. baicalensis showed decreased levels of wogonin and wogonoside, and transgenic arabidopsis overexpressing SbCYP82D2 successfully accumulated norwogonin, which confirmed its 8-hydroxylation function [118] (Table 3). Interestingly, a novel F8H isolated from S. barbata, named SbarCYP82D-6, which has 62% amino acid identity with the above-mentioned F8H encoded by SbCYP82D2, was reported to show increased activity and produce substantial amounts of norwogonin from chrysin [122].
Other hydroxylases
In addition to the common hydroxylases mentioned above, a few flavonoid substances also require the participation of other specialized hydroxylases, such as flavanone 2-hydroxylase (F2H). Flavone synthase II from Medicago truncatula, abbreviated as MtFNSII, converts flavanones to 2-hydroxyflavanone [123]. CYP93G2, a functional F2H, converts naringenin and eriodictyol to the corresponding 2-hydroxyflavanones required for C-glycosylflavone biosynthesis in rice [124]. However, according to the hydroxylation patterns of diverse flavonoid compounds (summarized in Fig. 2), there are some flavonoids whose hydroxylation mechanism has not yet been established, such as morin, which possesses an unusual C2′-OH, and galangin, a flavonol with no hydroxyl group on its B ring. Whether there are corresponding hydroxylases responsible for their formation needs to be investigated.
Towards a biofactory scale-up for production of desirable flavonoids
The excellent bioactivities and biosafety of diverse natural flavonoids are attracting a great deal of attention for their potential applications in food products or as supplements with health-promoting properties. At present, commercially available flavonoids are mainly derived from plant sources. However, the low efficiency and high cost of the preparation of such flavonoids limits their exploitation. The biosynthesis of flavonoids in microorganisms with their mild reaction conditions and high yield is attracting considerable attention (Fig. 3). For instance, vectors constructed with either F3H or FLS from Citrus have both been introduced into E. coli and used to produce 15.1 mg l−1 kaempferol from tyrosine and 1.1 mg l−1 galangin from phenylalanine [125]. By overexpressing F3′H in E. coli, fisetin was synthesized at a concentration of 1.2 mg l−1 by supplementing with 0.05 mmol l−1 resokaempferol [107]. F3H and FLS introduced successfully into Corynebacterium glutamicum produced 23 mg l−1 kaempferol and 10 mg l−1 quercetin [126]. F6H and its partner P450 reductase were very effective for the production of both baicalein (8.5 mg l−1) and scutellarein (47.1 mg l−1) upon supplementation with 0.5 g l−1 phenylalanine and tyrosine in E. coli [119]. Furthermore, Appelhagen et al. [127] succeeded in developing and scaling up production of anthocyanins (90 mg l−1 cyanidin 3-O-rutinoside) in plant cell cultures that could be used as natural food colorants. It seems highly likely that additional applications will be developed as knowledge of flavonoid biosynthesis increases.
Hydroxylation patterns and the degradation of natural flavonoids
Plant secondary metabolites such as flavonoids are not inert end products but they are subject to degradation in the plant [128]. The gradual decrease or complete disappearance of flavonoids observed in specific plant tissues demonstrates that catabolic turnover of flavonoids occurs generally. Myricetin, for example, was recently reported to accumulate to higher levels in younger fruits than in mature fruits of Morella rubra [129], although how it is degraded remains unknown. Thus, biosynthesis and further catabolism of flavonoids occur for both secondary constituents as well as primary metabolites.
There is, however, a quite limited amount of information reported describing the catabolism of flavonoids in plants. Decades ago, Barz and Koster [128] proposed that peroxidative chemistry might be involved in their catabolism. This hypothesis recently received support from investigations in arabidopsis and tomato, which showed that peroxidative cleavage of kaempferol contributed to the biosynthesis of the 4-hydroxybenzenoid moiety of the vital respiratory cofactor ubiquinone [130]. In contrast, there is a lot known about the degradation of flavonoids by microbes, which involves reactions such as dehydroxylation, deglycosylation, demethylation, decarboxylation, or isomerization carried out by rhizosphere microorganisms [131, 132] or gut microbiota [133].
Notably, the free hydroxyl group on C3 of kaempferol was proposed to be crucial for peroxidative cleavage of kaempferol [130]. Similarly, for degradation of plant flavonols by pirin proteins from E. coli or human, the 3-hydroxyl group of the ‘flavonol backbone’ was found to be important for the specific enzyme–substrate interaction [134]. For plant–pathogen interaction, Sclerotinia sclerotiorum was found to be able to catabolize flavonols through enzymes such as quercetin 2,3-dioxygenase [135]. Therefore, hydroxylation in key places on the ring may help render flavonoids ready to participate in degradation through the action of dioxygenases.
Conclusions and perspective
Flavonoids with diverse hydroxylation patterns are important secondary metabolites and food constituents produced by plants during their development or as a defense against various environmental stresses. With the continued development of metabolomics and functional genomics, additional flavonoids are likely to be discovered in food plants with more diverse hydroxylation patterns generated by additional hydroxylases. It is anticipated that further structure–activity investigations, combined with a better understanding of regulatory mechanisms, will identify new bioactive compounds of great nutritional value. The rapid development of synthetic biology in the food and biomedicine industries, combined with new scale-up strategies, opens the prospect for mass production of desirable bioactive flavonoids. This is expected to play an important role in the amelioration and prevention of global high risk of non-communicable diseases. In addition, the recent discovery of ubiquinone biosynthesis from catabolic turnover of flavonoids in planta may open the door for new lines of inquiry.
Acknowledgements
This work was supported by the Key Research and Development Program of Zhejiang Province (2021C02001), the National Natural Science Foundation of China (31872067), the Key Project for New Variety Breeding in Agriculture of Zhejiang Province (2021C02066-3), the 111 project (B17039), and the Fundamental Research Funds for the Central Universities.
Author contributions
Y.L. and X.L. designed the review. Y.L., J.Q., J.L., and M.X. conducted the literature review and wrote the manuscript. D.G. and X.L. carefully compiled and revised the paper. C.S., C.X., and K.C. provided discussion and comments on the paper. All authors approved the final submission.
Conflict of interest
The authors declare no conflict of interests.
References
- 1. Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat Prod Rep. 2009;26:1001–43. [DOI] [PubMed] [Google Scholar]
- 2. Saijo Y, Loo EP. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020;225:87–104. [DOI] [PubMed] [Google Scholar]
- 3. Sun CD, Liua Y, Zhan Let al. Anti-diabetic effects of natural antioxidants from fruits. Trends Food Sci Technol. 2021;117:3–14. [Google Scholar]
- 4. Del Río-Celestino M, Font R. The health benefits of fruits and vegetables. Foods. 2020;9:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alseekh S, Perez de Souza L, Benina Met al. The style and substance of plant flavonoid decoration; towards defining both structure and function. Phytochemistry. 2020;174:112347. [DOI] [PubMed] [Google Scholar]
- 6. Sekher Pannala A, Chan TS, O'Brien PJet al. Flavonoid B-ring chemistry and antioxidant activity: fast reaction kinetics. Biochem Biophys Res Commun. 2001;282:1161–8. [DOI] [PubMed] [Google Scholar]
- 7. Xiao J, Kai G, Yamamoto Ket al. Advance in dietary polyphenols as α-glucosidases inhibitors: a review on structure-activity relationship aspect. Crit Rev Food Sci. 2013;53:818–36. [DOI] [PubMed] [Google Scholar]
- 8. Xiao J, Ni X, Kai Get al. A review on structure-activity relationship of dietary polyphenols inhibiting α-amylase. Crit Rev Food Sci. 2013;53:497–506. [DOI] [PubMed] [Google Scholar]
- 9. Xiao J, Ni X, Kai Get al. Advance in dietary polyphenols as aldose reductases inhibitors: structure-activity relationship aspect. Crit Rev Food Sci. 2015;55:16–31. [DOI] [PubMed] [Google Scholar]
- 10. Khodzhaieva RS, Gladkov ES, Kyrychenko Aet al. Progress and achievements in glycosylation of flavonoids. Front Chem. 2021;9:637994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofer B. Recent developments in the enzymatic O-glycosylation of flavonoids. Appl Microbiol Biotechnol. 2016;100:4269–81. [DOI] [PubMed] [Google Scholar]
- 12. Xiao J, Muzashvili TS, Georgiev MI. Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol Adv. 2014;32:1145–56. [DOI] [PubMed] [Google Scholar]
- 13. Wen LR, Jiang Y, Yang Jet al. Structure, bioactivity, and synthesis of methylated flavonoids. Ann NY Acad Sci. 2017;1398:120–9. [DOI] [PubMed] [Google Scholar]
- 14. Koirala N, Thuan NH, Ghimire GPet al. Methylation of flavonoids: chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzyme Microb Technol. 2016;86:103–16. [DOI] [PubMed] [Google Scholar]
- 15. Chebil L, Humeau C, Falcimaigne Aet al. Enzymatic acylation of flavonoids. Process Biochem. 2006;41:2237–51. [Google Scholar]
- 16. Arai Y, Watanabe S, Kimira Met al. Dietary intakes of flavonols, flavones and isoflavones by Japanese women and the inverse correlation between quercetin intake and plasma LDL cholesterol concentration. J Nutr. 2000;130:2243–50. [DOI] [PubMed] [Google Scholar]
- 17. Zou WW, Xu SP. Galangin inhibits the cell progression and induces cell apoptosis through activating PTEN and caspase-3 pathways in retinoblastoma. Biomed Pharmacother. 2018;97:851–63. [DOI] [PubMed] [Google Scholar]
- 18. Noureddine H, Hage-Sleiman R, Wehbi Bet al. Chemical characterization and cytotoxic activity evaluation of Lebanese propolis. Biomed Pharmacother. 2017;95:298–307. [DOI] [PubMed] [Google Scholar]
- 19. Dabeek WM, Marra MV. Dietary quercetin and kaempferol: bioavailability and potential cardiovascular-related bioactivity in humans. Nutrients. 2019;11:2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sultana B, Anwar F. Flavonols (kaempeferol, quercetin, myricetin) contents of selected fruits, vegetables and medicinal plants. Food Chem. 2008;108:879–84. [DOI] [PubMed] [Google Scholar]
- 21. Akshaya KB, Varghese A, Sudhakar YNet al. Electrocatalytic oxidation of morin on electrodeposited ir-PEDOT nanograins. Food Chem. 2019;270:78–85. [DOI] [PubMed] [Google Scholar]
- 22. Zhang X, Huang H, Zhang Qet al. Phytochemical characterization of Chinese bayberry (Myrica rubra Sieb. et Zucc.) of 17 cultivars and their antioxidant properties. Int J Mol Sci. 2015;16:12467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perkin AG, Hummel JJ. LXXVI.—the colouring principle contained in the bark of Myrica nagi. Part I. J Chem Soc. 1896;69:1287–94. [Google Scholar]
- 24. Skrovankova S, Sumczynski D, Mlcek Jet al. Bioactive compounds and antioxidant activity in different types of berries. Int J Mol Sci. 2015;16:24673–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun C, Dai L, Gao Y. Binary complex based on zein and propylene glycol alginate for delivery of quercetagetin. Biomacromolecules. 2016;17:3973–85. [DOI] [PubMed] [Google Scholar]
- 26. Andersen OM, Markham KR, eds. Flavonoids: Chemistry, Biochemistry and Applications. CRC Press/Taylor & Francis, Boca Raton; 2006. [Google Scholar]
- 27. Tanaka Y, Ohmiya A. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotechnol. 2008;19:190–7. [DOI] [PubMed] [Google Scholar]
- 28. Veberic R, Slatnar A, Bizjak Jet al. Anthocyanin composition of different wild and cultivated berry species. LWT – Food Sci Technol. 2015;60:509–17. [Google Scholar]
- 29. Bueno JM, Sáez-Plaza P, Ramos-Escudero Fet al. Analysis and antioxidant capacity of anthocyanin pigments. Part II: chemical structure, color, and intake of anthocyanins. Crit Rev Anal Chem. 2012;42:126–51. [Google Scholar]
- 30. Manzolli ES, Serpeloni JM, Grotto Det al. Protective effects of the flavonoid chrysin against methylmercury-induced genotoxicity and alterations of antioxidant status, in vivo. Oxid Med Cell Longev. 2015;2015:602360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lefort ÉC, Blay J. Apigenin and its impact on gastrointestinal cancers. Mol Nutr Food Res. 2013;57:126–44. [DOI] [PubMed] [Google Scholar]
- 32. Miean KH, Mohamed S. Flavonoid (myricetin, quercetin, kaempferol, luteolin, and apigenin) content of edible tropical plants. J Agric Food Chem. 2001;49:3106–12. [DOI] [PubMed] [Google Scholar]
- 33. Imran M, Rauf A, Abu-Izneid Tet al. Luteolin, a flavonoid, as an anticancer agent: a review. Biomed Pharmacother. 2019;112:108612. [DOI] [PubMed] [Google Scholar]
- 34. Sun FF, Hu P-F, Xiong Yet al. Tricetin protects rat chondrocytes against IL-1β-induced inflammation and apoptosis. Oxid Med Cell Longev. 2019;2019:4695381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Z, Gao R, Pu Xet al. Comparative genome analysis of Scutellaria baicalensis and Scutellaria barbata reveals the evolution of active flavonoid biosynthesis. Genomics Proteomics Bioinformatics. 2020;18:230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, Sun R, Liu R. Natural products in licorice for the therapy of liver diseases: progress and future opportunities. Pharmacol Res. 2019;144:210–26. [DOI] [PubMed] [Google Scholar]
- 37. Guo L, Chen X, Li L-Net al. Transcriptome-enabled discovery and functional characterization of enzymes related to (2S)-pinocembrin biosynthesis from Ornithogalum caudatum and their application for metabolic engineering. Microb Cell Fact. 2016;15:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Den Hartogh DJ, Tsiani E. Antidiabetic properties of naringenin: a citrus fruit polyphenol. Biomolecules. 2019;9:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He P, Yan S, Zheng Jet al. Eriodictyol attenuates LPS-induced neuroinflammation, amyloidogenesis, and cognitive impairments via the inhibition of NF-κB in male C57BL/6J mice and BV2 microglial cells. J Agric Food Chem. 2018;66:10205–14. [DOI] [PubMed] [Google Scholar]
- 40. Soliman FM, Shehata AH, Khaleel AEet al. Caffeoyl derivatives and flavonoids from three Compositae species. Pharmacogn Mag. 2008;4:1–11. [Google Scholar]
- 41. Del Rio D, Stewart AJ, Mullen Wet al. HPLC-MSn analysis of phenolic compounds and purine alkaloids in green and black tea. J Agric Food Chem. 2004;52:2807–15. [DOI] [PubMed] [Google Scholar]
- 42. Ye JH, Augustin MA. Nano- and micro-particles for delivery of catechins: physical and biological performance. Crit Rev Food Sci. 2019;59:1563–79. [DOI] [PubMed] [Google Scholar]
- 43. Song W, Liu LL, Ren YJet al. Inhibitory effects and molecular mechanism on mushroom tyrosinase by condensed tannins isolation from the fruit of Ziziphus jujuba Mill. var. spinosa (Bunge) Hu ex H. F. Chow. Int J Biol Macromol. 2020;165:1813–21. [DOI] [PubMed] [Google Scholar]
- 44. Ojwang LO, Yang L, Dykes Let al. Proanthocyanidin profile of cowpea (Vigna unguiculata) reveals catechin-O-glucoside as the dominant compound. Food Chem. 2013;139:35–43. [DOI] [PubMed] [Google Scholar]
- 45. Tsujita T, Shintani T, Sato H. Preparation and characterisation of peanut seed skin polyphenols. Food Chem2014;151:15–20. [DOI] [PubMed] [Google Scholar]
- 46. Zaheer K, Humayoun Akhtar M. An updated review of dietary isoflavones: nutrition, processing, bioavailability and impacts on human health. Crit Rev Food Sci. 2017;57:1280–93. [DOI] [PubMed] [Google Scholar]
- 47. Chen L, Teng H, Xie Zet al. Modifications of dietary flavonoids towards improved bioactivity: an update on structure-activity relationship. Crit Rev Food Sci Nutr. 2018;58:513–27. [DOI] [PubMed] [Google Scholar]
- 48. Wang S, Alseekh S, Fernie ARet al. The structure and function of major plant metabolite modifications. Mol Plant. 2019;12:899–919. [DOI] [PubMed] [Google Scholar]
- 49. Husain SR, Cillard J, Cillard P. Hydroxyl radical scavenging activity of flavonoids. Phytochemistry. 1987;26:2489–91. [Google Scholar]
- 50. Zhang D, Jiang X, Xiao Let al. Mechanistic studies of inhibition on acrolein by myricetin. Food Chem. 2020;323:126788. [DOI] [PubMed] [Google Scholar]
- 51. Laabich A, Manmoto CC, Kuksa Vet al. Protective effects of myricetin and related flavonols against A2E and light mediated-cell death in bovine retinal primary cell culture. Exp Eye Res. 2007;85:154–65. [DOI] [PubMed] [Google Scholar]
- 52. Jia Y, Ma Y, Cheng Get al. Comparative study of dietary flavonoids with different structures as α-glucosidase inhibitors and insulin sensitizers. J Agric Food Chem. 2019;67:10521–33. [DOI] [PubMed] [Google Scholar]
- 53. Liu Y, Zhan L, Xu Cet al. α-Glucosidase inhibitors from Chinese bayberry (Morella rubra Sieb. et Zucc.) fruit: molecular docking and interaction mechanism of flavonols with different B-ring hydroxylations. RSC Adv. 2020;10:29347–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumamoto T, Fujii M, Hou DX. Myricetin directly targets JAK1 to inhibit cell transformation. Cancer Lett. 2009;275:17–26. [DOI] [PubMed] [Google Scholar]
- 55. Zhang N, Bi F, Xu Fet al. Structure and functional properties of active packaging films prepared by incorporating different flavonols into chitosan based matrix. Int J Biol Macromol. 2020;165:625–34. [DOI] [PubMed] [Google Scholar]
- 56. Xu R, Zhang Y, Ye Xet al. Inhibition effects and induction of apoptosis of flavonoids on the prostate cancer cell line PC-3 in vitro. Food Chem. 2013;138:48–53. [DOI] [PubMed] [Google Scholar]
- 57. Şöhretoğlu D, Sari S, Barut Bet al. Tyrosinase inhibition by some flavonoids: inhibitory activity, mechanism by in vitro and in silico studies. Bioorg Chem. 2018;81:168–74. [DOI] [PubMed] [Google Scholar]
- 58. Ono K, Yoshiike Y, Takashima Aet al. Potent anti-amyloidogenic and fibril-destabilizing effects of polyphenols in vitro: implications for the prevention and therapeutics of Alzheimer's disease. J Neurochem. 2003;87:172–81. [DOI] [PubMed] [Google Scholar]
- 59. Noda Y, Kaneyuki T, Mori Aet al. Antioxidant activities of pomegranate fruit extract and its anthocyanidins: delphinidin, cyanidin, and pelargonidin. J Agric Food Chem. 2002;50:166–71. [DOI] [PubMed] [Google Scholar]
- 60. Seeram NP, Nair MG. Inhibition of lipid peroxidation and structure-activity-related studies of the dietary constituents anthocyanins, anthocyanidins, and catechins. J Agric Food Chem. 2002;50:5308–12. [DOI] [PubMed] [Google Scholar]
- 61. Rahman MM, Ichiyanagi T, Komiyama Tet al. Superoxide radical- and peroxynitrite-scavenging activity of anthocyanins; structure-activity relationship and their synergism. Free Radic Res. 2006;40:993–1002. [DOI] [PubMed] [Google Scholar]
- 62. Wallace TC, Bailey RL, Blumberg JBet al. Fruits, vegetables, and health: a comprehensive narrative, umbrella review of the science and recommendations for enhanced public policy to improve intake. Crit Rev Food Sci. 2020;60:2174–211. [DOI] [PubMed] [Google Scholar]
- 63. Cömert ED, Mogol BA, Gökmen V. Relationship between color and antioxidant capacity of fruits and vegetables. Curr Res Food Sci. 2019;2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jayaprakasam B, Vareed SK, Olson LKet al. Insulin secretion by bioactive anthocyanins and anthocyanidins present in fruits. J Agric Food Chem. 2005;53:28–31. [DOI] [PubMed] [Google Scholar]
- 65. Mazewski C, Kim MS, Gonzalez de Mejia E. Anthocyanins, delphinidin-3-O-glucoside and cyanidin-3-O-glucoside, inhibit immune checkpoints in human colorectal cancer cells in vitro and in silico. Sci Rep. 2019;9:11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen D, Chen MS, Cui QCet al. Structure-proteasome-inhibitory activity relationships of dietary flavonoids in human cancer cells. Front Biosci. 2007;12:1935–45. [DOI] [PubMed] [Google Scholar]
- 67. Wu YX, Fang X. Apigenin, chrysin, and luteolin selectively inhibit chymotrypsin-like and trypsin-like proteasome catalytic activities in tumor cells. Planta Med. 2010;76:128–32. [DOI] [PubMed] [Google Scholar]
- 68. Sim GS, Lee B-C, Cho HSet al. Structure activity relationship of antioxidative property of flavonoids and inhibitory effect on matrix metalloproteinase activity in UVA-irradiated human dermal fibroblast. Arch Pharm Res. 2007;30:290–8. [DOI] [PubMed] [Google Scholar]
- 69. Hougee S, Sanders A, Faber Jet al. Decreased pro-inflammatory cytokine production by LPS-stimulated PBMC upon in vitro incubation with the flavonoids apigenin, luteolin or chrysin, due to selective elimination of monocytes/macrophages. Biochem Pharmacol. 2005;69:241–8. [DOI] [PubMed] [Google Scholar]
- 70. Kongpichitchoke T, Hsu JL, Huang TC. Number of hydroxyl groups on the B-ring of flavonoids affects their antioxidant activity and interaction with phorbol ester binding site of PKCδ C1B domain: in vitro and in silico studies. J Agric Food Chem. 2015;63:4580–6. [DOI] [PubMed] [Google Scholar]
- 71. Habtemariam S. The Nrf2/HO-1 axis as targets for flavanones: neuroprotection by pinocembrin, naringenin, and eriodictyol. Oxid Med Cell Longev. 2019;2019:4724920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu J, Yang Z, Cheng Yet al. Eriodictyol and naringenin inhibit the formation of AGEs: an in vitro and molecular interaction study. J Mol Recognit. 2020;33:e2814. [DOI] [PubMed] [Google Scholar]
- 73. Chobot V, Hadacek F, Bachmann Get al. In vitro evaluation of pro- and antioxidant effects of flavonoid tricetin in comparison to myricetin. Molecules. 2020;25:5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang H, Du YJ, Song HC. α-Glucosidase and α-amylase inhibitory activities of guava leaves. Food Chem. 2010;123:6–13. [Google Scholar]
- 75. Gómez-Zorita S, Lasa A, Abendaño Net al. Phenolic compounds apigenin, hesperidin and kaempferol reduce in vitro lipid accumulation in human adipocytes. J Transl Med. 2017;15:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sinha R, Srivastava S, Joshi Aet al. In-vitro anti-proliferative and anti-oxidant activity of galangin, fisetin and quercetin: role of localization and intermolecular interaction in model membrane. Eur J Med Chem. 2014;79:102–9. [DOI] [PubMed] [Google Scholar]
- 77. Zhang J, Du F, Peng Bet al. Structure, electronic properties, and radical scavenging mechanisms of daidzein, genistein, formononetin, and biochanin A: a density functional study. J Mol Struc-Theochem. 2010;955:1–6. [Google Scholar]
- 78. Ae Park S, Choi M-S, Cho S-Yet al. Genistein and daidzein modulate hepatic glucose and lipid regulating enzyme activities in C57BL/KsJ-db/db mice. Life Sci. 2006;79:1207–13. [DOI] [PubMed] [Google Scholar]
- 79. Brugliera F, Barri-Rewell G, Holton TAet al. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 1999;19:441–51. [DOI] [PubMed] [Google Scholar]
- 80. Toda K, Akasaka M, Dubouzet EGet al. Structure of flavonoid 3′-hydroxylase gene for pubescence color in soybean. Crop Sci. 2005;45:2212–7. [Google Scholar]
- 81. Bogs J, Ebadi A, McDavid Det al. Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 2006;140:279–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shih CH, Chu IK, Yip WKet al. Differential expression of two flavonoid 3′-hydroxylase cDNAs involved in biosynthesis of anthocyanin pigments and 3-deoxyanthocyanidin phytoalexins in sorghum. Plant Cell Physiol. 2006;47:1412–9. [DOI] [PubMed] [Google Scholar]
- 83. Han Y, Vimolmangkang S, Soria-Guerra REet al. Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010;153:806–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Thill J, Miosic S, Gotame TPet al. Differential expression of flavonoid 3′-hydroxylase during fruit development establishes the different B-ring hydroxylation patterns of flavonoids in Fragaria × ananassa and Fragaria vesca. Plant Physiol Biochem. 2013;72:72–8. [DOI] [PubMed] [Google Scholar]
- 85. Park S, Choi MJ, Lee JYet al. Molecular and biochemical analysis of two rice flavonoid 3′-hydroxylase to evaluate their roles in flavonoid biosynthesis in rice grain. Int J Mol Sci. 2016;17:1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Guo L, Gao L, Ma Xet al. Functional analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylases from tea plant (Camellia sinensis), involved in the B-ring hydroxylation of flavonoids. Gene. 2019;717:144046. [DOI] [PubMed] [Google Scholar]
- 87. Zhou TS, Zhou R, Yu Y-Bet al. Cloning and characterization of a flavonoid 3′-hydroxylase gene from tea plant (Camellia sinensis). Int J Mol Sci. 2016;17:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Irmisch S, Ruebsam H, Jancsik Set al. Flavonol biosynthesis genes and their use in engineering the plant antidiabetic metabolite montbretin A. Plant Physiol. 2019;180:1277–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Robinson SP, Pezhmanmehr M, Speirs Jet al. Grape and wine flavonoid composition in transgenic grapevines with altered expression of flavonoid hydroxylase genes. Aust J Grape Wine Res. 2019;25:293–306. [Google Scholar]
- 90. Wu Y, Wang T, Xin Yet al. Overexpression of the GbF3′H1 gene enhanced the epigallocatechin, gallocatechin, and catechin contents in transgenic Populus. J Agr Food Chem. 2020;68:998–1006. [DOI] [PubMed] [Google Scholar]
- 91. Holton TA, Brugliera F, Lester DRet al. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature. 1993;366:276–9. [DOI] [PubMed] [Google Scholar]
- 92. Seitz C, Eder C, Deiml Bet al. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol Biol. 2006;61:365–81. [DOI] [PubMed] [Google Scholar]
- 93. Jung CS, Griffiths HM, De Jong DMet al. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theor Appl Genet. 2005;110:269–75. [DOI] [PubMed] [Google Scholar]
- 94. Olsen KM, Hehn A, Jugdé Het al. Identification and characterisation of CYP75A31, a new flavonoid 3′5′-hydroxylase, isolated from Solanum lycopersicum. BMC Plant Biol. 2010;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moreau C, Ambrose MJ, Turner Let al. The B gene of pea encodes a defective flavonoid 3′,5′-hydroxylase, and confers pink flower color. Plant Physiol. 2012;159:759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang YS, Xu Y-J, Gao L-Pet al. Functional analysis of flavonoid 3′,5′-hydroxylase from tea plant (Camellia sinensis): critical role in the accumulation of catechins. BMC Plant Biol. 2014;14:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jin JQ, Ma JQ, Yao MZet al. Functional natural allelic variants of flavonoid 3′,5′-hydroxylase gene governing catechin traits in tea plant and its relatives. Planta. 2017;245:523–38. [DOI] [PubMed] [Google Scholar]
- 98. Rommens CM, Richael CM, Yan Het al. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol J. 2008;6:870–86. [DOI] [PubMed] [Google Scholar]
- 99. Britsch L, Grisebach H. Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur J Biochem. 1986;156:569–77. [DOI] [PubMed] [Google Scholar]
- 100. Lukačin R, Gröning I, Pieper Uet al. Site-directed mutagenesis of the active site serine290 in flavanone 3β-hydroxylase from Petunia hybrida. Eur J Biochem. 2000;267:853–60. [DOI] [PubMed] [Google Scholar]
- 101. Martin C, Prescott A, Mackay Set al. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991;1:37–49. [DOI] [PubMed] [Google Scholar]
- 102. Zabala G, Vodkin LO. The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell. 2005;17:2619–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Halbwirth H, Stich K. An NADPH and FAD dependent enzyme catalyzes hydroxylation of flavonoids in position 8. Phytochemistry. 2006;67:1080–7. [DOI] [PubMed] [Google Scholar]
- 104. Song X, Diao J, Ji Jet al. Molecular cloning and identification of a flavanone 3-hydroxylase gene from Lycium chinense, and its overexpression enhances drought stress in tobacco. Plant Physiol Biochem. 2016;98:89–100. [DOI] [PubMed] [Google Scholar]
- 105. Han Y, Huang K, Liu Yet al. Functional analysis of two flavanone-3-hydroxylase genes from Camellia sinensis: a critical role in flavonoid accumulation. Genes. 2017;8:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Punyasiri PA, ISB A, Kumar Vet al. Flavonoid biosynthesis in the tea plant Camellia sinensis: properties of enzymes of the prominent epicatechin and catechin pathways. Arch Biochem Biophys. 2004;431:22–30. [DOI] [PubMed] [Google Scholar]
- 107. Stahlhut SG, Siedler S, Malla Set al. Assembly of a novel biosynthetic pathway for production of the plant flavonoid fisetin in Escherichia coli. Metab Eng. 2015;31:84–93. [DOI] [PubMed] [Google Scholar]
- 108. Martens S, Mithöfer A. Flavones and flavone synthases. Phytochemistry. 2005;66:2399–407. [DOI] [PubMed] [Google Scholar]
- 109. Flachowsky H, Halbwirth H, Treutter Det al. Silencing of flavanone-3-hydroxylase in apple (Malus × domestica Borkh.) leads to accumulation of flavanones, but not to reduced fire blight susceptibility. Plant Physiol Biochem. 2012;51:18–25. [DOI] [PubMed] [Google Scholar]
- 110. Mahajan M, Yadav SK. Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco. Plant Mol Biol. 2014;85:551–73. [DOI] [PubMed] [Google Scholar]
- 111. Maloney GS, DiNapoli KT, Muday GK. The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 2014;166:614–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang L, Lui ACW, Lam PYet al. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol J. 2020;18:2170–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang P, Du H, Wang Jet al. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol J. 2020;18:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Latunde-Dada AO, Cabello-Hurtado F, Czittrich Net al. Flavonoid 6-hydroxylase from soybean (Glycine max L.), a novel plant P-450 monooxygenase. J Biol Chem. 2001;276:1688–95. [DOI] [PubMed] [Google Scholar]
- 115. Anzellotti D, Ibrahim RK. Molecular characterization and functional expression of flavonol 6-hydroxylase. BMC Plant Biol. 2004;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Halbwirth H, Forkmann G, Stich K. The A-ring specific hydroxylation of flavonols in position 6 in Tagetes sp. is catalyzed by a cytochrome P450 dependent monooxygenase. Plant Sci. 2004;167:129–35. [Google Scholar]
- 117. Berim A, Gang DR. The roles of a flavone-6-hydroxylase and 7-O-demethylation in the flavone biosynthetic network of sweet basil. J Biol Chem. 2013;288:1795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhao Q, Cui M-Y, Levsh Oet al. Two CYP82D enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4′-deoxyflavones in Scutellaria baicalensis. Mol Plant. 2018;11:135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Li J, Tian C, Xia Yet al. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab Eng. 2019;52:124–33. [DOI] [PubMed] [Google Scholar]
- 120. Liu X, Cheng J, Zhang Get al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun. 2018;9:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Berim A, Park JJ, Gang DR. Unexpected roles for ancient proteins: flavone 8-hydroxylase in sweet basil trichomes is a Rieske-type. PAO-family oxygenase. Plant J. 2014;80:385–95. [DOI] [PubMed] [Google Scholar]
- 122. Liu X, Cheng J, Zhu Xet al. De novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae. ACS Synth Biol. 2020;9:3042–51. [DOI] [PubMed] [Google Scholar]
- 123. Zhang J, Subramanian S, Zhang Yet al. Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 2007;144:741–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Du Y, Chu H, Chu IKet al. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 2010;154:324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Miyahisa I, Funa N, Ohnishi Yet al. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechno l2006;71:53–8. [DOI] [PubMed] [Google Scholar]
- 126. Kallscheuer N, Vogt M, Bott Met al. Functional expression of plant-derived O-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin. J Biotechnol. 2017;258:190–6. [DOI] [PubMed] [Google Scholar]
- 127. Appelhagen I, Wulff-Vester AK, Wendell Met al. Colour bio-factories: towards scale-up production of anthocyanins in plant cell cultures. Metab Eng. 2018;48:218–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Barz W, Koster J. Chapter 3: Turnover and Degradation of Secondary (Natural) Products. In: Conn EE, ed. The Biochemistry of Plants, Vol. 7. New York: Academic Press, 1981, 35–84. [Google Scholar]
- 129. Xing MY, Cao Y, Ren Cet al. Elucidation of myricetin biosynthesis in Morella rubra of the Myricaceae. Plant J. 2021;108:411–25. [DOI] [PubMed] [Google Scholar]
- 130. Soubeyrand E, Johnson TS, Latimer Set al. The peroxidative cleavage of kaempferol contributes to the biosynthesis of the benzenoid moiety of ubiquinone in plants. Plant Cell. 2018;30:2910–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Hassan S, Mathesius U. The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot. 2012;6:3429–44. [DOI] [PubMed] [Google Scholar]
- 132. Shaw LJ, Morris P, Hooker JE. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ Microbiol. 2006;8:1867–80. [DOI] [PubMed] [Google Scholar]
- 133. Braune A, Blaut M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes. 2016;7:216–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Guo B, Zhang Y, Hicks Get al. Structure-dependent modulation of substrate binding and biodegradation activity of Pirin proteins toward plant flavonols. ACS Chem Biol. 2019;14:2629–40. [DOI] [PubMed] [Google Scholar]
- 135. Chen J, Ullah C, Reichelt Met al. Sclerotinia sclerotiorum circumvents flavonoid defenses by catabolizing flavonol glycosides and aglycones. Plant Physiol. 2019;180:1975–87. [DOI] [PMC free article] [PubMed] [Google Scholar]