Abstract
Genetic engineering of economically important traits in plants is an effective way to improve global welfare. However, introducing foreign DNA molecules into plant genomes to create genetically engineered plants not only requires a lengthy testing period and high developmental costs but also is not well-accepted by the public due to safety concerns about its effects on human and animal health and the environment. Here, we present a high-throughput nucleic acids delivery platform for plants using peptide nanocarriers applied to the leaf surface by spraying. The translocation of sub-micrometer-scale nucleic acid/peptide complexes upon spraying varied depending on the physicochemical characteristics of the peptides and was controlled by a stomata-dependent-uptake mechanism in plant cells. We observed efficient delivery of DNA molecules into plants using cell-penetrating peptide (CPP)-based foliar spraying. Moreover, using foliar spraying, we successfully performed gene silencing by introducing small interfering RNA molecules in plant nuclei via siRNA-CPP complexes and, more importantly, in chloroplasts via our CPP/chloroplast-targeting peptide-mediated delivery system. This technology enables effective nontransgenic engineering of economically important plant traits in agricultural systems.
Keywords: DNA, siRNA, peptide, spraying, plant cell, chloroplast, nanocarrier
Introduction
The use of conventional transgenic approaches for industrial-scale quality trait improvement in crops is notoriously uneconomical due to high upstream production costs, including the laborious processes of plant regeneration and the subsequent propagation of an elite line.1 Transgenic crops also prompt public concerns regarding biosafety issues for humans, animals, and the environment.2 Therefore, the high-throughput application of bioactive molecules via foliar spraying could represent a superior technique for crop improvement. This technique enables the rapid, simple introduction of biomolecules of interest into plant cells without the need for costly, laborious, biomolecule transfer techniques.3 For example, the foliar application of naked double-stranded DNA fragments and small interfering RNAs (siRNAs) enable the engineering of metabolic traits and economically important characteristics of plants without permanently changing the plant genome.4,5
Nanocarrier-based molecule delivery is a promising technique for plant improvement. Various metallic, nonmetallic, and polymer-based nanocarriers have been conjugated to bioactive molecules, and these biomolecule/nanocarrier composites have been translocated through the plant cell wall network to the cytoplasm.6 Recent studies have demonstrated that biomolecule/nanocarrier conjugates could be translocated into plant cells through leaf cuticular or stomatal pathways following foliar spraying.7−10 Nanocarriers have also been used to protect these biomolecules from various degradation processes in plant cells.11 These studies highlight the potential use of nanocarriers for the systematic improvement of quality traits in commercial crops.
Among nanocarriers, polypeptide-based carriers can not only translocate biomolecules of interest across the rigid plant cell boundaries but also target the transport of the biomolecules to specific organelles in the cells. Cell-penetrating peptides (CPPs) are short chains of amino acids that can spontaneously infiltrate the plant cell wall and plasma membrane.12,13 Cationic CPPs such as KH9 and R9 passively transfer biomolecules into plant cells.14 The amphipathic CPP BP100 and its cationic derivatives have greater abilities to bind to and deliver various types of biomolecules into different plant cells.15−18 Organelle-targeting peptides (OTPs) are functional peptides that target biomolecule transport to specific organelles. OTPs such as mitochondria-targeting peptide (MTP) and chloroplast-targeting peptide (CTP) are generally derived from organelle-targeting domains of nucleus-encoded organellar proteins.19,20 MTP and CTP recognize specific organellar membrane moieties or chosen transporters and transfer the associated biomolecules into target organelles.19,20
Engineered CPPs and OTPs bind different biomolecules via their corresponding amino acid residues. DNA and RNA molecules spontaneously bind to positively charged side chains of amino acids in polypeptides via noncovalent, positive-to-negative charge interactions and create sub-micrometer-sized peptide/nucleic acid complexes.21−23 Plant cells transfected by gene expression cassette/CPP complexes showed efficient translocation of DNA molecules into the nucleus as well as transient expression of the exogenous gene.16,24 Transgenic GFP-overexpressing plants infiltrated by double-stranded GFP-interfering RNA/CPP complexes exhibited lower GFP transcript levels than the wild type, along with reduced GFP accumulation.17 In addition, functional proteins were successfully internalized into plant cells via functionalized CPPs.18,24
Recent advances in organelle transformation involve the use of nanocarriers to precisely transport biomolecules of interest into the desired plant organelles. OTP-based nanocarriers were successfully used to transport biomolecules into plant mitochondria and chloroplasts.25,26 Moreover, surface modification of plasmid DNA/OTP complexes with CPP significantly enhanced the expression of recombinant proteins in mitochondria and various types of plastids (including chloroplasts).25,27 The use of these functionalized CPPs and OTPs highlights the advanced development of peptide-based biomolecule delivery systems for plants. Various combinations of cationic CPPs and OTPs have been successfully used to deliver plasmid DNA harboring reporter gene expression cassettes to nuclei and targeted organelles in plant cells using syringe infiltration,16−18,24−27 vacuum/compression infiltration,26,28 or injection.27 However, these techniques are impractical for reprogramming plant quality traits under agricultural conditions. Therefore, an efficient high-throughput, cost-effective technique for biomolecule delivery is clearly needed.
In the current study, to develop a large-scale platform for peptide-based biomolecule delivery into plants, we designed a foliar spraying technique to introduce nucleic acid/peptide nanocarriers into the cytosol, nuclei, and chloroplasts of plant cells. We evaluated the ability of naturally derived and artificially synthesized CPPs for cell penetration via spray application. Factors influencing the spray efficiency included the buffer system, guard cell density, and leaf trichome density. Foliar spraying of the CPP-based complexes of plasmid DNA (pDNA) or siRNA significantly increased the nucleic acid delivery efficiency into plant cells. Interestingly, siRNA molecules were successfully targeted to chloroplasts to suppress the functions of chloroplast-expressed proteins via a process mediated by sprayable clustered CTP/CPP nanocarriers. Our peptide-based nucleic acids spraying platform enables highly efficient, comprehensive engineering of commercially important characteristics and metabolic traits of cultivated crops under agricultural conditions.
Results and Discussion
Cell-Penetration Efficiencies of CPPs upon Spraying
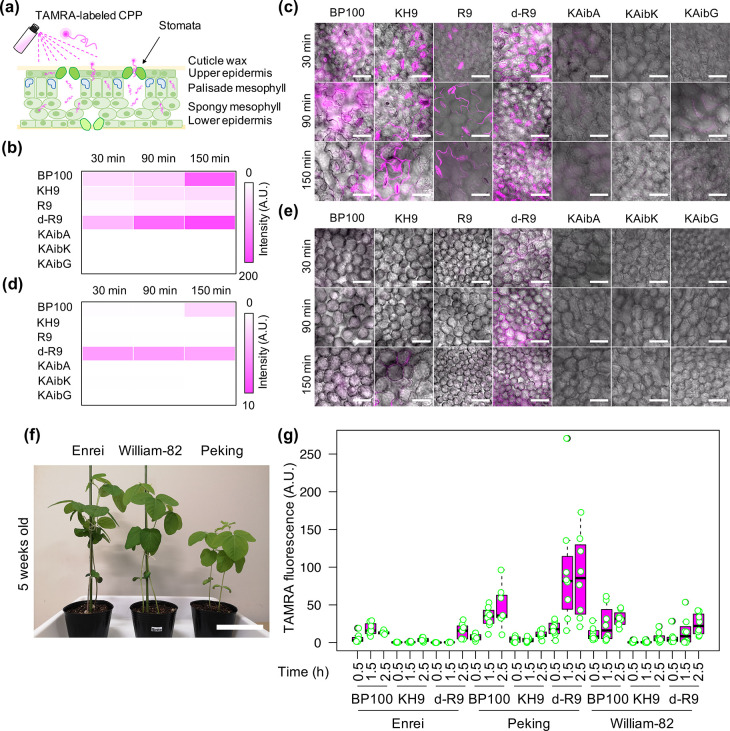
Different CPPs exhibited different levels of translocation into plant cells. Fluorescein tetramethylrhodamine (TAMRA)-labeled CPPs such as amphipathic BP100, cationic nonameric lysine/histidine (KH9), and nonameric arginine (R9) and its d-configurated form (dR9) showed moderate to high penetration efficiencies into various plant cells after syringe infiltration (see the CPP’s amino acid sequences in Supporting Information Table S1 and Figure S1a–c).14 Moreover, artificially designed CPPs containing periodic α-aminoisobutyric acids (Aib) such as KAibA(Ala), KAibG(Gly), and KAibK (Table S1 and Figure S1a–c) showed significantly improved cell-penetrating abilities and stabilities in plant and animal cells.29 To test the cell-penetration efficiencies of these CPPs upon spraying, we sprayed solutions containing 0.1 μg/mL of CPPs that have been chemically labeled with TAMRA14,29 onto the leaves of Arabidopsis thaliana (Arabidopsis ecotype Col-0) and compared TAMRA fluorescence in plant cells at different time points after spraying (Figure 1a) via confocal laser-scanning microscopy (CLSM). The cell-penetration efficiencies of CPPs refer to the different fluorescence intensities of TAMRA-CPPs, that being adsorbed and translocated to leaf cells after spraying. After spray application, 14.8–29.1% of TAMRA-labeled CPPs reached the surface of plant leaves (Figure S2a). The fluorescence in the epidermal cells on the adaxial (upper) sides of leaves gradually increased in plants sprayed with TAMRA-labeled BP100, KH9, R9, and dR9 (Figure 1b,c and Figure S2b). However, TAMRA-labeled artificial CPPs KAibA, KAibG, and KAibK showed lower penetration efficiencies into these cells (Figure 1b,c and Figure S2b). We also observed strong TAMRA fluorescence at the guard cells of leaves sprayed with TAMRA-labeled KH9, R9, and dR9, suggesting that these cationic CPPs passively infiltrated the leaves through stomata and highly accumulated in substomatal cavities (Figure 1a–c). TAMRA-labeled dR9 exhibited higher penetration efficiency into the palisade mesophyll than the other CPPs examined (Figure 1d,e and Figure S2c).
Figure 1.
Cell-penetration and translocation of CPPs in leaves after spraying. (a) Leaf cell architecture and possible uptake mechanisms of CPP into different leaf cell layers after spraying. (b–e) Retention and translocation of different tetramethylrhodamine (TAMRA)-labeled CPPs into the upper epidermal cells (b, c) and palisade mesophyll cells (d, e) of Arabidopsis thaliana leaves at various time points after spraying. Two independent experiments (two leaves per experiment) were performed for each TAMRA-CPPs and two regions of interest (ROIs) in one leaf were observed by CLSM. The average fluorescence intensities of 8 ROIs (n = 8) in leaves at different time points after spraying are shown as heat maps for upper epidermal cells (b) and palisade mesophyll cells (d). The distributions of TAMRA fluorescence signals in plant cells were presented in Figure S2. Colored bars represent the range of fluorescence intensity in the heat map in arbitrary units (A.U.). (c, e) Scale bars = 50 μm. (f) Plant characteristics of three commercially important soybean cultivars (5 weeks old). Scale bar = 15 cm. (g) TAMRA fluorescence intensities in epidermal cells in soybean leaves at different time points after spraying with TAMRA-labeled CPPs. The distribution of TAMRA fluorescence in plant cells is presented as a box plot from 8 different regions of interest (2 ROIs per leaf, 2 leaves per experiment, 2 independent experiments, n = 8). Black bars show the median values.
We expanded our spraying method to commercially important crops, including soybean (Glycine max) and tomato (Solanum lycopersicum). We examined the activities of six TAMRA-labeled CPPs with high cell-penetration efficiency in soybean, including BP100, KH9, and dR9, and three artificial Aib-containing CPPs. First, we sprayed TAMRA-labeled CPP solutions onto the leaves of Japanese soybean cultivar (cv.) Enrei and examined TAMRA fluorescence. Only 26.3–46.8% of TAMRA-labeled CPPs were accumulated on plant leaves after foliar spraying (Figure S3). All TAMRA-labeled CPPs showed progressive translocation into epidermal cells in soybean leaves, especially BP100, dR9, and KAibA (Figure S4a,b). TAMRA-labeled KAibA exhibited higher translocation efficiency into the palisade mesophyll than the other CPPs (Figure S4c,d). The difference between this result and that in Arabidopsis could be due to the distinctive stabilities and penetration efficiencies of these compounds in Arabidopsisvs soybean.29
Due to their high translocation and penetration efficiencies into the leaf epidermis (Figure S4), we chose bio-inspired BP100, dR9, and KH9 for analysis in Enrei and two other commercially important soybean cultivars, Williams-82 and Peking (Figure 1f). These CPPs displayed different translocation efficiencies into the leaf epidermal cells of these soybean cultivars (Figure 1g and Figures S4 and S5). dR9 showed the highest progressive translocation efficiency in all three cultivars, especially Peking (Figure 1g and Figures S4 and S5). Although there were substantial losses (75–83%) of foliarly applied CPPs to the aerosols after spraying (Figure S6a), TAMRA-labeled CPPs BP100, dR9, KH9, and KAibA displayed different translocation efficiencies in the epidermal cells of tomato leaves after spraying, with dR9 exhibiting the greatest translocation efficiency (Figure S6b,c). After spraying, CPPs would likely be degraded by cellular peptidases and proteases, which could abolish their cell-penetration functions,30 but the incorporation of unusual, chirality-reversed d-arginine into dR9 enhances its stability and cellular uptake, as observed in both animal cells and plant cells.14,31 Our study of CPP translocation upon spraying highlights the great potential of dR9 for the development of a high-throughput biomolecule delivery system into plant cells.
Transfection Efficiencies of Nucleic Acid/CPP Complexes after Spraying
We demonstrated that different CPPs showed different translocation efficiencies into plant cells after spraying (Figure 1). However, the CPPs had to be further engineered to enhance their ability to transport biomolecules into plant cells. The highly efficient cell-penetrating BP100 peptide was previously fused with the cationic, biomolecule-binding domains KH9 and R9 to increase the translocation efficiency of biomolecules into plant cells.15−18,32,33 To re-evaluate the abilities of these different functionalized CPPs (Table S1) to deliver nucleic acids into plant cells upon spraying, we performed comparative internalization studies of plasmid DNA/cationic CPP complexes into plant cells. We formed complexes between different BP100-derived cationic CPPs and cyanine 3 (Cy3)-labeled pBI221 (Figure S7) and performed physicochemical and morphological characterizations of these different Cy3-labeled pBI221/CPP complexes. In aqueous solution, Cy3-labeled pBI221 formed positively charged complexes (54–77 nm in diameter) with various BP100-derived cationic CPPs at N/P ratio = 2.0 (a molar ratio of NH3+ groups of polypeptides to PO4– groups of plasmid DNA) (Figure S8a,b and Table S2). These Cy3-labeled pBI221/CPP complexes showed electrostatic mobility shifts in gel-retardation assays (Figure S8c). The Cy3-labeled pBI221/BP100, /BP100-KH9, and/KH9-BP100 complexes appeared as intact, globular complexes when viewed by atomic force microscopy (AFM) (Figure S8d). By contrast, the Cy3-labeled pBI221/BP100-R9 and/R9-BP100 complexes appeared enlarged, with plasmid DNA segregating from the irregularly shaped complexes on the mica surface after drying, suggesting that these complexes are less stable than Cy3-labeled pBI221/BP100-KH9 and/KH9-BP100 complexes (Figure S8d).
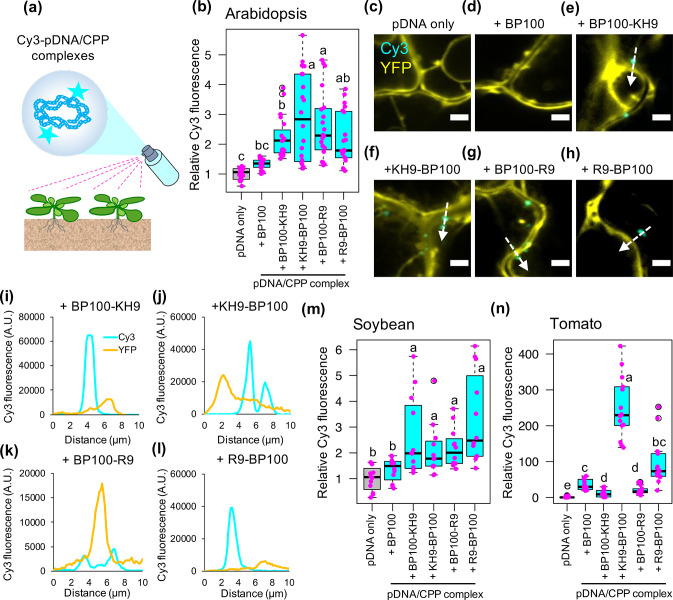
We sprayed solutions of the resulting complexes onto the leaves of transgenic Arabidopsis plants overexpressing yellow fluorescent protein (YFP) and examined the fluorescence of Cy3-labeled pBI221 in leaf cells by CLSM at 2 h post-spraying (Figure 2a). Thus, the difference of Cy3 fluorescence in plant cells suggests differential transfection efficiencies of various Cy3-labeled pBI221/CPPs complexes after spraying. Compared to leaves sprayed with Cy3-labeled pBI221 only and Cy3-labeled pBI221/BP100 complex, notably stronger Cy3 fluorescent signals (2- to 5-fold) were observed in epidermal cells sprayed with all four Cy3-labeled pBI221/BP100-derived cationic CPP complexes (Figure 2b). These Cy3-labeled pBI221/cationic CPP complexes entered the transfected cells and localized to the cytoplasm (Figure 2c–l and Figure S9).
Figure 2.
Transfection of plasmid DNA/CPP complexes into plant cells after spraying. (a) Spray application of different Cy3-labeled pBI221/CPP complexes onto plants for transfection analysis. Cy3-pBI221/CPP complexes were formed using five different BP100-derived CPPs and were sprayed onto leaves. The Cy3 fluorescence signals in transfected cells were observed by CLSM at 2 h post-spraying. (b) Fluorescence intensities of Cy3 in transgenic Arabidopsis leaf cells overexpressing YFP at 2 h after spraying with different Cy3-pBI221/CPP complexes. The distribution of Cy3 signals in 20 CLSM images collected from four experimental independent leaves (5 ROIs per leaf, one leaf per experiment) for each treatment is shown as a box plot. Black bars indicate medians of distribution. Each data point is represented by a magenta dot. (c–h) Internalization of Cy3-pBI221 (cyan) in YFP-overexpressing Arabidopsis leaf cells (yellow) after spraying with different Cy3-pBI221/CPP complexes at 2 h post-spraying. Scale bars = 5 μm. White arrows indicate the detection trajectories of the Cy3 and YFP fluorescence profiles shown in (i–l). (m) Box plot of Cy3 fluorescence in soybean leaf cells after spraying with different Cy3-pBI221/CPP complexes (n = 12 ROIs from three independent experiments, 4 ROIs per leaf per experiment). (n) Box plot of Cy3 fluorescence in transgenic tomato leaf cells overexpressing GFP after spraying with Cy3-pBI221/CPP complexes (n = 15 ROIs collected from three independent leaves, 5 ROIs per leaf per experiment). Different letters in the box plots indicate significant differences of means among the six treatments analyzed by one-way ANOVA with Tukey’s HSD test at p = 0.05.
We then analyzed the transfection efficiencies of different Cy3-labeled pBI221/cationic CPP complexes in soybean and tomato leaf cells. Consistent with the results in Arabidopsis leaf cells, all four Cy3-labeled pBI221/cationic CPP complexes showed significantly higher transfection efficiencies than Cy3-labeled pBI221 only or Cy3-labeled pBI221/BP100 complexes in soybean leaf cells after spraying (Figure 2m and Figure S10a). In tomato leaf cells, only KH9-BP100 demonstrated higher transfection efficiency of plasmid DNA molecules after spraying (Figure 2n and Figure S10b). Adding biomolecule-binding domains to either or both ends of BP100 influenced its cell-penetration ability as well as the reciprocal functions of the conjugated biomolecules.15,33 In a previous study, KH9-BP100 demonstrated higher transfection efficiency of DNA molecules into plant cells than R9-BP100 after syringe infiltration.16 The current results suggest that, among all four cationic domain-fused BP100 CPPs, the N-terminal fused KH9-BP100 provides the most effective tool for the transport of exogenous DNA molecules into plant cells via foliar spraying. Additionally, our Cy3-fluorescence imaging in different tissues of fluorescent protein-overexpressing Arabidopsis and tomatoes suggested auxiliary activities of KH9-BP100 in facilitating long-distance transports of biomolecule cargos and stabilizing the Cy3-plasmid DNA molecules while being translocated from leaf to other plant tissues after spraying (Figure S11). Nevertheless, the considerable delivery ability of KH9-BP100 to effectively transport biomolecules to plants upon spraying can be influenced by the leaf features such as stomata density, appearance of trichome, and difference in leaf architectures.10,34,35 Moreover, the complexity of extracellular cuticle components and the stiffness of cell wall compositions of leaf cells can critically determine the transfection efficiencies of foliarly applied biomolecule/CPP complexes in different plant species.36
Foliar Spraying of Plasmid DNA/CPP Complexes for Transgene Expression in Plant Cells
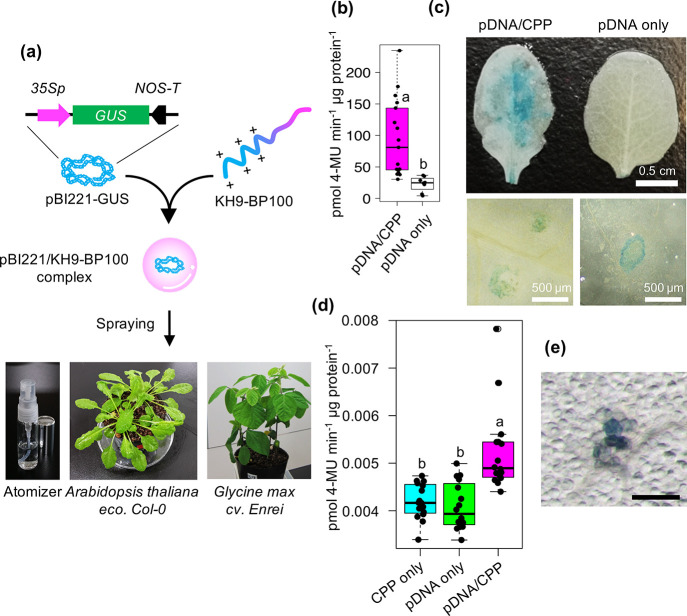
To validate the efficiency of peptide-based foliar spray application for gene delivery into plant cells, we generated plasmid DNA/CPP complexes of the β-glucuronidase (GUS) reporter gene expression vector pBI221 with the highly efficient CPP KH9-BP100 at different N/P ratios (Figure 3a and Figure S12 and Table S3).16,17,24 The hydrodynamic diameters of pBI221/KH9-BP100 complexes in aqueous solutions gradually decreased from 190 to 79 nm, while the surface charges of the resulting complexes progressively increased at increasing N/P ratios (Figure S12a,b and Table S3). Increasing the added amount of KH9-BP100 to formed pBI221/KH9-BP100 complexes at various N/P ratios slightly decreased pH of the complex solutions (Table S3). The mobilities of plasmid DNA molecules in pBI221/KH9-BP100 complex solutions in an electrostatic field gradually decreased with increasing N/P ratio (Figure S12c). AFM imaging of pBI221/KH9-BP100 complexes formed at N/P ratio = 2.0 revealed intact, monodispersed spherical complexes on the mica surface compared to complexes formed at N/P ratio = 0.5 (Figure S12d,e). On the basis of the physicochemical properties and morphological appearance of plasmid DNA/CPP complexes formed at different N/P ratios, we chose pBI221/KH9-BP100 complex solution formed at N/P ratio = 2.0 for spray application.
Figure 3.
Uptake efficiency of cell-penetrating peptide-based DNA cargos into plant cells after foliar application. (a) Foliar application of plasmid DNA (pDNA) into A. thaliana leaf cells mediated by cell-penetrating peptide (CPP; KH9-BP100). The pBI221/KH9-BP100 complexes formed in aqueous solution and were applied onto leaves using a spray atomizer. (b) GUS activity in leaves at 24 h after spraying with a solution containing pDNA/CPP complexes. The distributions of GUS activity in at least 20 sprayed leaves are shown as a box plot. (c) Histochemical staining of GUS reporter in leaves sprayed with pDNA/CPP complex solution. (d, e) GUS activity and histological staining of GUS reporter in Glycine max (soybean) leaves transfected with pDNA/CPP complex via spraying. Scale bar in panel e is 500 μm. The distributions of GUS activity in at least 16 sprayed leaves are shown as a box plot. Dots represent individual data points. Different letters indicate significant differences in GUS activity, as analyzed by one-way ANOVA with Tukey’s HSD test at p = 0.05.
Silwet L-77 and other agricultural surfactants facilitate transport of biomolecules and nanostructures across the multifarious leaf surface by disrupting the complex cuticular wax layer and the first layer of epidermis of plant leaves and by inducing stomatal uptake of foreign molecules.35 Appropriate concentrations of Silwet L-77 and sucrose in transformation solutions improved transformation efficiencies of Agrobacterium to plants.37−39 Moreover, exogenously applied sucrose prominently increased stomatal density on plant leaves.40 The elevated sucrose concentration also enhanced endocytic uptake of extracellular molecules.41 We diluted the pBI221/KH9-BP100 complex solution at N/P ratio = 2.0 with spray solution to a final concentration of 5.0% (w/v) sucrose + 0.05% (v/v) Silwet L-77 to theoretically increase the transfection efficiency of plasmid DNA cargos into plant cells.42 This spray solution did not cause coagulation of the sprayed droplets of pBI221/KH9-BP100 complex solution (Figure S13a,b). Spraying of plasmid DNA/CPP complexes diluted in spray solution containing Silwet L-77 onto Arabidopsis and soybean leaves significantly enhanced GUS activity in transfected leaf cells (Figure 3b–e and Figure S13c). The transfection efficiency of the DNA/CPP complexes was much lower in soybean leaves than in Arabidopsis (Figure 3b,d). Perhaps Arabidopsis and soybean differ in their cell surface compositions,43 cellular responses to foreign molecules,44 nanoparticle-uptake mechanisms,45 and/or transgene expression machineries.46
An advanced development in DNA transfer techniques using Agrobacterium-based spraying to plant tissues allows industrial-scale manufacturing of valuable proteins and biologically active molecules.3 To compare the efficacies between Agrobacterium-based spray application and our CPP-mediated plasmid DNA delivery via foliar spraying, we performed spray experiments of solutions containing Agrobacterium harboring plant binary vector; pBI121 (a T-DNA vector containing GUS reporter gene expression cassette) and pBI121/KH9-BP100 complexes formed at N/P ratio = 2.0 (Figure S14a–c and Table S4) to Arabidopsis plants. Histochemical staining of GUS catalytic activity in Arabidopsis leaves at 24 h post-spraying showed that both Agrobacterium and pBI121/KH9-BP100 complex efficiently transfected and induced GUS expression in leaflets (Figure S14d). However, spraying of pBI121/KH9-BP100 complexes showed significantly (∼0.5 times) lower transfection efficiency in plant leaves than Agrobacterium-mediated foliar spraying (Figure S14e). This is due to lacking of virulence factors and replicative ability of plasmid DNA/CPP complexes as well as lower stability of plasmid DNA in CPP carriers.
The d-configurated form of R9 CPP (dR9) exhibited high translocation efficiency after it was sprayed onto plant leaves (Figure 1 and Figures S2, S4, S5, and S6) and could potentially be superior to BP100 for creating designed cationic CPPs. We chemically synthesized the cationic CPPs dR9-KH9 and KH9-dR9 (Table S1 and Figure S15) and tested their biomolecule translocation efficiencies into plant cells after spraying. Cy3-labeled plasmid DNA/dR9-KH9 and KH9-dR9 complexes formed at N/P ratio = 2.0 displayed comparable physicochemical properties to plasmid DNA/CPP complexes containing other CPPs (Figures S8 and S16). Both KH9-dR9 and dR9-KH9 showed efficiencies similar to that of KH9-BP100 for introducing Cy3-labeled plasmid DNA into plant cells and increasing the number of Cy3 particles inside the transfected cells (Figure S17a–c). Moreover, the spraying of pBI221/CPP complexes containing KH9-dR9 and dR9-KH9 (N/P ratio = 2.0) did not induce greater GUS activity compared to the pBI221/KH9-BP100 complex (Figure S17d). The unusual d-arginine in the dR9 portion may compete with KH9 to interact with plasmid DNA molecules, which would decrease the cell-penetration activity of dR9-derived cationic CPPs.47,48 However, engineering of a chirality d-configured KH9-BP100 would increase protease resistance and enhance the transfection efficiency of CPP-based spray application of biomolecules into plant cells.
We then investigated biological and physiological factors in plants that influence the efficiency of sprayable peptide-mediated biomolecule application. Stomata strongly affect the infiltration of foreign particles into leaves. Nanoparticles ranging in size from several nanometers to 1–2 μm passively penetrating through stomatal pores to substomatal cavities before they disperse into other layers of the leaf cell.7,10,49 Our fluorescent imaging and three-dimentional model reconstruction of plant leaf sprayed with Cy3-labeled pBI221/KH9-BP100 complexes elucidated that the nucleic acid/peptide complexes entered the stomata and accumulated in substomatal cavities (Figure S18). These biomolecule cargos could further vigorously translocate over intercellular space and transfect the adjacent epidermal and mesophyll cells (Figure S18d). Additionally, the number of stomata in the epidermal cell layer of a leaf could significantly affect the transfection efficiency upon spraying of biomolecule/peptide complexes. We therefore analyzed the transfection efficiency of plasmid DNA/CPP complexes sprayed onto the leaves of transgenic Arabidopsis plants with high (STOMAGEN-OX) or low stomatal density (STOMAGEN-amiR).50 The transfection efficiency of the plasmid DNA/CPP complex was significantly reduced in transgenic STOMAGEN-amiR leaves, which have reduced numbers of stomata (Figure S19a). In agreement with a previous study,10 this result suggests that the transfection of plasmid DNA/CPP complexes via spraying is a stomata-dependent mechanism. Besides, complexing double-stranded DNA with cationic CPPs improves the stability of DNA molecules against extracellular DNases in substomatal cavity.17 Moreover, the positively charged amino acids of BP100 CPP domain prominently exposing on the surface of pDNA/CPP complexes stringently interact with the negatively charged extracellular components and feasibly recognize the receptor proteins on guard cells to trigger systemic foreign molecules uptake and internalization mechanisms.15,33 However, higher numbers of stomata on leaves did not increase the transfection efficiency of the plasmid DNA/CPP complex (Figure S19a). Although stomatal aperture and the number of stomata control the loading of plasmid DNA/CPP complexes into leaves, their transfection efficiency still strongly depends on the distribution of the complexes inside the leaf. Additionally, regardless of the stomatal features on a plant leaf, the uptake and translocation of foliarly applied biomolecule/peptide complexes can be partially contributed by cuticular pathways.10,35
The trichome is a specialized epidermal structure that strongly influences nanoparticle distribution in plants.51 Foliarly applied ionic nanoparticles, such as iron and gold nanoparticles 10–43 nm in size, strongly accumulated in trichomes and were partially discharged from plant cells via a cellular detoxification mechanism.52,53 The presence of trichomes on leaves may affect the transfection of relatively large plasmid DNA/peptide complexes as well. To test this hypothesis, we sprayed pBI221/KH9-BP100 complexes formed at N/P ratio = 2.0 (93 nm, +25 mV) onto the leaves of an Arabidopsis mutant lacking trichomes (gl1–2)54 (Figure S19b) and compared the transfection efficiency to that of the wild type (Col-0). Plasmid DNA/CPP complexes exhibited higher transfection efficiency when sprayed onto gl1–2 leaves (∼2.0-fold) vs Col-0 leaves (Figure S19c), implying that trichomes on leaves could lower the active amount reaching the leaf surface and restrain translocation of the sprayed plasmid DNA/CPP complexes into other plant cells.
Gene Silencing in Plant Cells Mediated by the siRNA/CPP Complex after Foliar Spraying
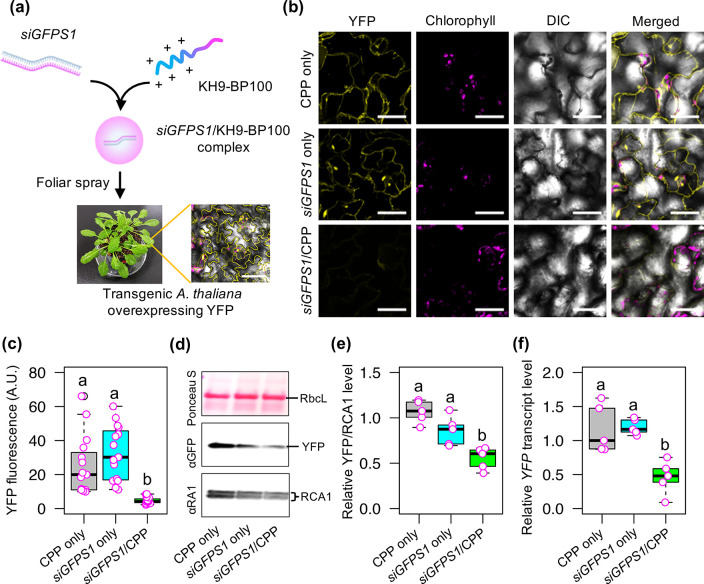
Engineering of important plant traits using RNA interference (RNAi) technology has considerable benefits, as this approach can be accomplished in a nontransgenic manner.5 Free double-stranded small interfering RNA (siRNA) molecules were previously applied to plant cells using high-pressure spraying, which successfully suppressed the levels of target mRNA molecules.55 Additionally, recent studies demonstrated that combining small RNA molecules with nanocarriers such as CPP,17 three-dimentional (3-D) DNA nanostructures,56 and carbon nanotubes57 enhanced gene silencing efficiency and the stability of RNA molecules in plant cells after infiltration. Hence, we developed a high-throughput spray application technique to introduce siRNA/CPP complex into plant cells for efficient gene knockdown (Figure 4a). We synthesized 27-bp siGFPS1 RNA duplexes, which potentially showed superior activity for silencing the expression of GFP variants (GFP, enhanced-GFP [eGFP], and yellow fluorescent protein [YFP]) in transfected plant cells (Figure S20a and Table S5).58 The double-stranded siGFPS1 RNA molecules were complexed with KH9-BP100 to form ∼264 nm negatively charged siRNA/CPP complexes at N/P ratio = 2.0 (Figure S20b,c, Table S6). These globular siRNA/CPP complexes showed the expected gel-retardation pattern of siRNA molecules in an electrostatic field (Figure S20d,e).
Figure 4.
Suppression of gene expression in plant cells mediated by sprayable peptide-based siRNA cargos. (a) Formulation of the siGFPS1/KH9-BP100 complex for YFP transgene suppression in transgenic Arabidopsis leaves. (b) YFP fluorescence in plant cells at 3 days after spraying (3 DAS) with a solution containing siGFPS1/KH9-BP100 complex. Scale bars = 50 μm. (c) Quantitative fluorescence intensity of YFP in plant cells at 3 DAS with siRNA/CPP complex. The distributions of YFP fluorescence from 15 ROIs (n = 15, 5 ROIs were collected from one leaf, 3 biological independent experiments) are shown as a box plot. Dots represent the fluorescence values. Black bars show the median values. (d) Immunoblot analysis of YFP and endogenous RubisCo Activase 1 protein (RCA1; a highly abundant intracellular plant protein) in soluble proteins extracted from Arabidopsis leaves at 3 DAS with siRNA/CPP complex. The membrane was stained with Ponceau S before probing with antibodies. RbcL = RubisCo large subunit. (e) Relative abundance of YFP/RCA1 in total leaf proteins at 3 DAS with siRNA/CPP complex determined by immunoblotting. Amounts of YFP relative to RCA1 are shown as a box plot. Magenta circles represent the distribution of data (n = 5). Black bars show median values. (f) Relative YFP transcript levels in leaves at 3 DAS with siRNA/CPP complex. Magenta dots represent the relative YFP transcript levels in five different experiments (n = 5). Different letters in the box plots indicate significant differences, as analyzed by one-way ANOVA with Tukey’s HSD test at p = 0.05.
To test the gene silencing activity of siGFPS1 in plant cells, we sprayed a solution containing siRNA/CPP complexes onto transgenic Arabidopsis leaves overexpressing YFP. Spraying of siGFPS1/KH9-BP100 complex solution onto YFP-overexpressing plants drastically reduced YFP fluorescence in plant cells (Figure 4b,c) and resulted in a 45.5% decrease in YFP protein level in leaves (Figure 4d,e and Figure S21) at 3 days after spraying (DAS). Moreover, we observed a 54.1% decrease in YFP transcript levels in transgenic Arabidopsis leaves sprayed with siRNA/CPP complexes at 3 DAS (Figure 4f). These results imply that siGFPS1/KH9-BP100 complexes efficiently induce transgene silencing in plant cells after foliar spraying.
To expand the application of spray-induced gene silencing mediated by functional peptides to economically important crop species, we sprayed a solution containing siGFPS1/KH9-BP100 complexes onto the fully expanded leaves of transgenic tomato overexpressing GFP. Compared to the control, tomato leaves sprayed with siGFPS1/KH9-BP100 complex showed significantly lower GFP fluorescence (∼83% decrease) at 3 DAS (Figure 5a,b). Immunoblot analysis confirmed the lower abundance (28% remained) of GFP in leaves sprayed with siRNA/CPP complex at 3 DAS vs the control (Figure 5c,d and Figure S22). Furthermore, GFP transcript levels in tomato leaves sprayed with siRNA/CPP complex decreased by 59% at 3 DAS, respectively (Figure 5e). The silencing efficiency of the sprayable KH9-BP100-mediated siRNA delivery system in both Arabidopsis and tomato leaf cells is comparable to the efficiency previously achieved by infiltration of a double-stranded GFP5 siRNA/CPP complex.17 However, this efficiency is lower than that of GFP silencing mediated by the infiltration of 3-D DNA nanostructures, carbon dots, or carbon nanotubes into plant cells or spraying with other siRNA/nanoparticle complexes.56,57,59 This could be due to the different physicochemical properties, surface coating chemistries, or cellular uptake and distribution of different nanoparticles. Taken together, these results highlight the potential of using CPP-mediated siRNA delivery via foliar spraying to suppress the functions of genes related to commercially important plant characteristics.
Figure 5.
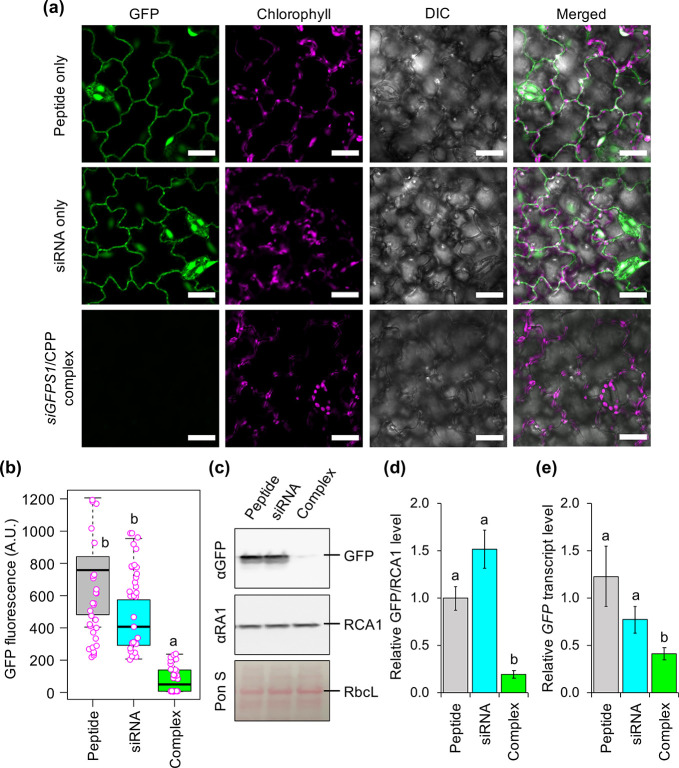
Transgene suppression in tomato leaves mediated by spraying with siRNA/CPP complexes. (a) CLSM images of epidermal cells of transgenic tomato leaves overexpressing GFP after spraying with siGFPS1/KH9-BP100 complexes at 3 days after spraying (DAS). Scale bars = 20 μm. (b) Distribution of GFP fluorescence intensities in leaves sprayed with siRNA/CPP complexes at 3 DAS. Data points (magenta circles) of GFP fluorescence analyzed from CLSM images by ImageJ are shown in a box plot (n = 30 ROIs, 10 ROIs per leaf, three independent experiments), and black bars represent the median values. (c) Immunoblot analysis of GFP and RCA1 (endogenous plant protein control) in tomato leaves sprayed with KH9-BP100 only (P), siGFPS1 only (S), and siGFPS1/KH9-BP100 complex formed at N/P ratio = 2.0 (C). The membrane was stained by Ponceau S (Pon S) to confirm equal protein loading, as indicated by the RbcL bands on the membrane. (d) Quantitative analysis of GFP accumulation in transgenic tomato leaves sprayed with siRNA/CPP complex. Relative GFP/RCA1 protein levels were analyzed from three experimentally independent immunoblot membranes by ImageJ. Error bar = standard deviation (SD). (e) Transcript suppression in leaves after spraying with siRNA/CPP complex. GFP transcript levels in three independent tomato leaves were analyzed by qRT-PCR at 3 DAS. Error bar = SD. Significant differences among treatments in panels b, d, and e analyzed by one-way ANOVA with Tukey’s HSD test at p = 0.05 are indicated by different letters in the graphs.
Spray Application of Biomolecules for Delivery to Chloroplasts
Nanoparticle-based targeted biomolecule delivery to plastids (including chloroplasts) is an evolving biotechnological tool for plastid engineering. Following leaf infiltration, plasmid DNAs are specifically released inside chloroplasts, a process mediated by a clustered CTP/CPP-based DNA delivery system27 and single-walled carbon nanotubes.60 Bioactive compounds could be selectively transferred into chloroplasts using surface-modified cadmium-based nanoparticles to fine-tune the oxidative status of plant cells.61 These techniques provide feasible nanoparticle-based platforms for manipulating organellar functions in plant cells.
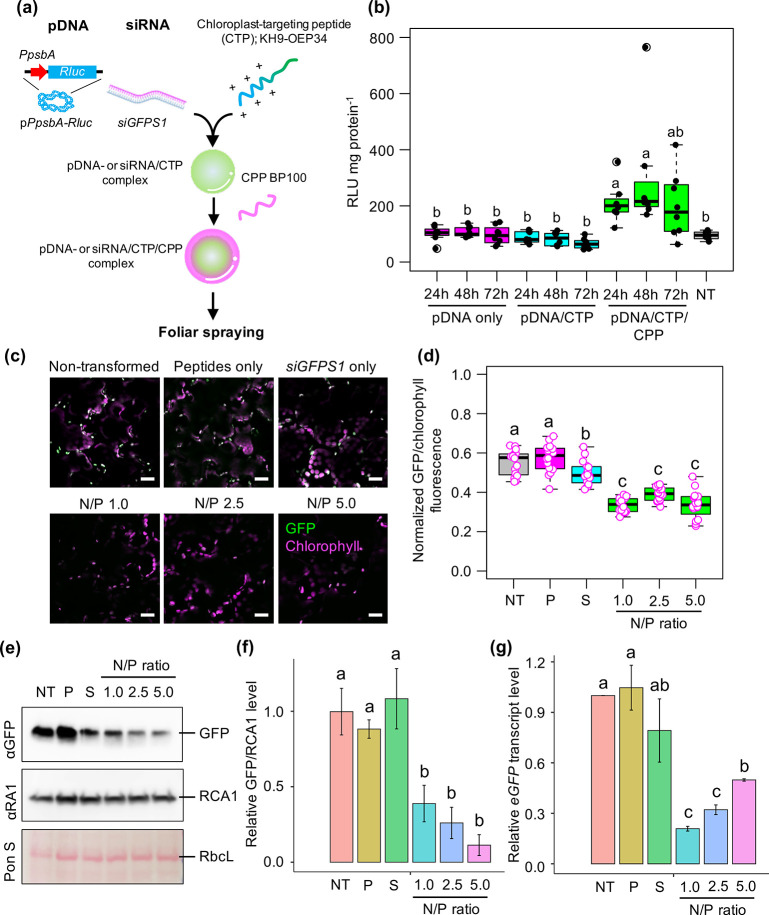
To examine the efficiency of CTP/CPP-based biomolecule targeting into plastids via spray application, we generated clustered complexes of plasmid DNA harboring the chloroplast-specific Renilla luciferase (Rluc) expression cassette pPpsbA::Rluc with CTP KH9-OEP34 (see amino acid sequence in Table S1) and CPP BP100 at N/P ratio = 1.0. The resulting complexes in spray solution (5% sucrose + 0.05% Silwet L-77) were applied onto Arabidopsis leaves (eco. Col-0) by spraying and the Rluc activities in leaves were measured up to 72 h after spraying (Figure 6a). The transfection efficiency of the clustered plasmid DNA/CTP/CPP complexes gradually increased (up to 7.6 times, 2.7 times in average) in Arabidopsis leaves at 24–72 h after spraying (Figure 6b) compared to leaves sprayed with a solution containing only plasmid DNA molecules or plasmid DNA/CTP complexes formed at N/P ratio = 1.0 (Figure 6b). These results suggest that nucleic acid molecules can be efficiently targeted to chloroplasts using peptide-based foliar spraying.
Figure 6.
Targeted delivery of plasmid DNA and siRNA molecules into chloroplasts by peptide-based foliar spraying. (a) Formation of clustered biomolecule/chloroplast-targeting peptide/cell-penetrating peptide complex for targeted delivery of biomacromolecules to plastids in plant cells. (b) Renilla luciferase activities in Arabidopsis leaves sprayed with clustered pPpsbA::Rluc/peptide complexes formed at N/P ratio = 1.0 at different times post-spraying. Distribution of luciferase activities in the plant cells was shown as a box plot. Dots represent the distribution of luciferase activity in 8 different samples per treatment (n = 8). Black bars are medians of the distributed data. NT = nontransformed leaves. (c) GFP fluorescence in chloroplasts of transplastomic eGFP-overexpressing tobacco leaf cells after spraying with siGFPS1/peptide complexes formed at different N/P ratios for 3 days. GFP fluorescence in the chloroplast of epidermal cell is usually stronger than that of mesophyll (see also Figure S25). Scale bars = 20 μm. (d) Normalized GFP/chlorophyll fluorescence in plant cells sprayed with siRNA/peptide complexes at 3 DAS. Distribution of fluorescence values in plant cells was shown as a box plot. Dots represent the distribution of fluorescence values in 16 different samples per treatment (n = 16). NT = nontransformed leaves; P = leaves sprayed by equivalent mole of peptides used to form siRNA/CTP/CPP complex at N/P ratio = 5.0; S = siGFPS1 only sprayed leaves. (e) Immunoblot analysis of total leaf proteins from transplastomic tobacco leaves sprayed with siRNA/peptide complexes. The membrane was stained with Ponceau S staining solution to confirm equal protein loading (indicated by RbcL bands on membrane) prior to probing with anti-RubisCo Activase 1 (αRA1) and anti-GFP antibodies. (f) Quantitative GFP/RCA1 levels in three experimentally independent siRNA/peptide complexes-sprayed leaves. Relative GFP/RCA1 levels were calculated from respective protein bands on immunoblotted membranes by ImageJ. Error bars = SD (n = 3). (g) Decreased expression of eGFP transcripts in chloroplasts of plant leaves sprayed by siRNA/peptide complexes at 3 DAS. Transcriptional changes of eGFP gene in plant leaves were determined by qRT-PCR (n = 3). Error bars = SD. Letters in box plots and bar graphs show statistical significance of difference analyzed by one-way ANOVA with Tukey’s HSD test at p = 0.05.
Suppressing gene expression in genome-containing plant organelles (mitochondria and plastids) is a promising approach for engineering organellar functions, especially metabolic processes related to economically important quality traits. However, there is insufficient evidence to support an RNAi mechanism in plant mitochondria and plastids.5 Numerous small noncoding RNA molecules have been recurrently identified in chloroplasts.62 However, their involvement in post-transcriptional regulation of plastidial mRNA is still underexplored. Perhaps this is due to the lack of an effective carrier to transport a small RNA molecule into the target organelle to study its function in an organellar RNA suppression mechanism.
To develop a peptide carrier-based, high-throughput siRNA method for gene silencing in chloroplasts, we performed foliar application of clustered siGFPS1/KH9-OEP34/BP100 complex solutions onto a transplastomic tobacco line overexpressing eGFP. We first established a homoplasmic tobacco line (PRV-3 line) that exhibited strong eGFP fluorescence inside the chloroplasts of leaf cells for use as a reporter model (Figure S23). Cell-membrane penetration and compartmentalization of nanostructured complexes including metallic nanoparticles and biomolecule/peptide complexes to chloroplasts are prominently influenced by the differences of their physicochemical properties and coating chemistries.10,16,17,27,35,63 Regarding this, we rationally formulated different clustered siGFPS1/KH9-OEP34/BP100 complexes at N/P ratios = 1.0, 2.5, and 5.0 to test their transfection abilities and targeted gene suppression function in GFP-overexpressing chloroplasts (Figure S24). The hydrodynamic diameters of the clustered siRNA/peptide complexes progressively decreased from 560 to 107 nm with increasing N/P ratios (Figure S24a and Table S7). The surface charges of the complexes gradually increased while the pH of the siRNA/peptide complex solutions slightly reduced as the N/P ratio increased from 1.0 to 5.0 (Figure S24b and Table S7). The mobilities of the siRNA molecules in these globular-shaped, clustered siRNA/peptide complexes were retarded in an electrostatic field (Figure S24c–f).
To test the gene silencing efficiencies of the sprayable siRNA/peptide complexes, we sprayed fully expanded leaves of the PRV-3 line with solutions containing clustered siRNA/peptide complexes formed at different N/P ratios. At 3 DAS, eGFP fluorescence was significantly decreased in chloroplasts in leaf cells transfected by clustered siRNA/CTP/CPP complexes (Figure 6c,d and Figure S25). Small chloroplasts in the epidermal cells exhibited stronger GFP fluorescence than that of the palisade mesophylls (Figure 6c,d and Figure S25). No substantial change in eGFP signal was observed in the chloroplasts of leaf cells spray-transfected by solutions containing siGFPS1 only or peptides only (Figure 6c,d and Figure S25). Immunoblot analysis of total leaf proteins revealed significantly lower accumulation of eGFP in leaves sprayed with clustered siRNA/CTP/CPP complex solutions than leaves sprayed with control solutions or nontransformed leaves (Figure 6e,f and Figure S26). Moreover, transcript analysis of transplastomic tobacco leaves sprayed with clustered siRNA/peptide complex solutions revealed significantly reduced eGFP transcript levels at 3 DAS (Figure 6f). These results indicate that siRNA molecules can be delivered into chloroplasts in transfected plant cells via peptide carrier-based spray application to suppress the functions of chloroplast-expressed genes. However, the differences of physicochemical properties of siRNA/peptide complexes formed at different N/P ratios did not influence their cellular uptake and chloroplast-targeting function upon spraying. Our platform provides a feasible tool for engineering organellar functions in plant cells without altering the genetic background of the target organelle.
The mechanisms underlying post-transcriptional regulation of organellar RNAs in plants are still unclear. Our results point to the possible existence of a small-RNA-mediated transcription suppression mechanism in chloroplasts. A recent study reported an active argonaute 2-dependent RNAi mechanism in controlling targeted mRNA levels in animal and human mitochondria (mitoRNAi).64 However, there is no evidence of the co-occurrence of nuclear-encoded RNA-induced silencing complex (RISC) components in plant chloroplasts.62 Interestingly, overexpression of long dsRNA molecules in plastomes partially produced unexpected lower molecular weight transcripts than the desired full-length double-stranded structures, suggesting the existence of an active RNA processing mechanism in plastids.65,66 Molecular genetics and transplastomic expression studies suggested that antisense-strand mRNAs of chloroplast rrn5 gene (AS5) formed RNA duplexes with 5S-rRNA precursors which served as apparent dsRNA substrates of plastidial dsRNA-dependent RNase III enzymes.67−70 The siGFPS1 dsRNA molecule delivered to chloroplasts by peptide carriers may dissociate into single-stranded antisense RNA that subsequently form RNA duplexes with the target eGFP transcript. These antisense siGFPS1/eGFP RNA duplexes are then cleaved by chloroplast RNase III endonucleases, resulting in lower abundance of eGFP transcripts and decreasing in eGFP accumulation in chloroplasts (Figure 6c–g). The disassociated antisense siGFPS1 strands can also bind to complementary DNA sequence of eGFP gene in unwound transplastomes during DNA replication and transcription initiation.71 Finally, the antisense siGFPS1/eGFP DNA heteroduplexes inhibit endonuclease activity of plastidial RNase H1 in resolving the collisions of R-loop replisomes and in initiating transcription of the highly transcribed eGFP expression cassette in plastomes.69,71 Certainly, the exact mechanism underlying siRNA-based gene suppression in our research must be further explored.
An effective protocol for improving a particular economically important metabolic pathway leading to the synthesis of an individual metabolite of interest in plant requires comprehensive combinations of multiple controllable regulators to completely govern the bifurcated reactions that coexist in distinct cellular compartments. For example, biosynthesis of isopentenyl diphosphate, the central intermediate of isoprenoid end products, in plants has two distinct routes, a cytosolic mevalonate (MVA) biosynthesis pathway and plastid-localized methylerythritol phosphate (MEP) pathway.72 Attractively, catalytic activities of rate-limiting enzymes in these two distinctive pathways could be either genetically regulated or biochemically impaired by exogenously applied biomolecules.73−76 Functional peptide-based carriers have abilities to (1) recognize diverse biomolecules and chemical compounds, (2) efficiently transport the conveying cargos into plant cells, and (3) selectively deliver assigned molecules to specific cellular compartments, especially mitochondria and plastids.25−27 Our recent peptide-based spray delivery technology enables a coordinated introduction of siRNA molecules and chemical compounds to simultaneously control the targeted multifaceted metabolic pathway in cytoplasm and plastids such as the astounding IPP biosynthesis pathway. However, sequence engineering and large-scale production of the sophisticated cell-penetrating/organelle-targeting peptides as well as optimizing the physiological appearances of biomolecule/peptide complexes potentially maximize biotechnological features of our peptide-based spraying technology. Furthermore, designing an integrative platform comprising drone technology, Internet of Things (IoT), and this spray technology allows effectual smart farming and precision agriculture.
Conclusion
We demonstrated that bioactive molecules could be successfully applied to plants using peptide-based spraying without the need for costly equipment and tedious preparation procedures. Sub-micrometer-sized nucleic acid/peptide complexes were transferred into plant cells via guard cells after foliar spraying. The sprayable peptide nanocarrier-based DNA delivery platform is effective in different plant species, including crop plants. Moreover, siRNA molecules were successfully transported into plant cells using our targeting peptide-based biomolecule spray application system to specifically induce gene silencing in chloroplasts. Our high-throughput peptide-based nucleic acid spray technology enables the comprehensive engineering of economically important traits and metabolic processes in plants under agricultural conditions without introducing transgenes.
Methods
Peptide Synthesis
The 5-carboxyltetramethylrhodamine (TAMRA)-labeled cell-penetrating peptides were chemically synthesized and purified as previously described.14,29 The integrities and purities of purified TAMRA-labeled CPPs were analyzed by reverse-phase HPLC (RP-HPLC) (COSMOSIL 5C18-MS-II) with fluorescence detection (excitation/emission wavelength of 545/575 nm) eluting with CH3CN/water containing 0.1% TFA (5/95 over 75 min) (Figure S1a). Changes of absorption and fluorescence emission spectra of TAMRA after conjugation to CPPs were analyzed by UV–visible spectrophotometry (V-750 spectrophotometer, JASCO, Tokyo, Japan) and fluorescence spectrometry with excitation wavelength of 550 nm (FP-8500 spectrophotometer, JASCO) (Figure S1b,c). The CPP BP100 and its cationic peptide-conjugated derivatives and chloroplast-targeting peptide (CTP) KH9-OEP34 were prepared as described previously.16,17,26,27 To generate dR9-KH9 and KH9-dR9, the CPPs were synthesized on Fmoc-NH-SAL resin (Watanabe Chemical, Hiroshima, Japan, 0.21 mmol/g) using Fmoc-based coupling reactions (4 equiv of Fmoc amino acids). Solutions of (1-cyano-2-ethoxy-2-oxoethylidenaminooxy)(dimethylamino)morpholinocarbenium hexafluorophosphate (COMU) and diisopropylamine in N-methylpyrrolidone (NMP) were used as coupling reagents. A 20% amount of piperidine in N,N-dimethylformamide (DMF) was used for Fmoc deprotection. Progression of the coupling reaction and Fmoc deprotection were confirmed using a TNBS kit (Tokyo Chemical Industry, Tokyo, Japan). The peptidyl resin was washed with NMP. After loading the amino acid, the peptidyl resin was added to a mixture of 25% acetic anhydride DMF solution to protect the N-terminal amine with the acetyl group. The CPPs were deprotected and cleaved from the resins by treatment with a mixture of trifluoroacetic acid (TFA):water:triisopropylsilane = 9.5:0.25:0.25 at room temperature for 90 min. The reaction mixtures were filtered to remove the resin, and the filtrates were concentrated under a vacuum. The CPPs were precipitated by adding diethyl ether to the residue, and the supernatant was decanted. After repeating the washing step with diethyl ether three times, the precipitated CPPs were dried and crude products were purified by RP-HPLC (COSMOSIL 5C18-MS-II) eluting with a linear gradient of CH3CN/water containing 0.1% TFA (10.5/89.5 to 15/85 over 60 min). The fractions containing CPPs were lyophilized to give 12.7 mg of a flocculent solid (2.6% yield) for dR9-KH9 and 7.2 mg of a flocculent solid (1.5% yield) for KH9-dR9. Both CPPs had MALDI-TOF-MS profiles at (matrix:α-CHCA): m/z = 3853 [M+H]+ (Figure S15a,b). These CPPs were dissolved in water to a concentration of 1.0 mg/mL and stored at −20 °C. The sequences and physicochemical properties of these CPPs are listed in Table S1.
Plasmid DNA Molecules
Plasmid DNA molecules, pBI221 and pBI121, harboring a GUS gene expression cassette for translocation study and transient expression of β-glucuronidase (GUS) enzyme activity in plant cells were purchased from Addgene (Addgene, Watertown, MA, USA). The chloroplast-specific expression vector pPpsbA::Rluc was constructed as previously described.27 Plasmid DNA was extracted from a liquid culture of Escherichia coli using a QIAGEN Plasmid Giga kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany) and stored at −20 °C until use.
Labeling of plasmid DNA pBI221 with Cy3-fluorescent dye was performed using Label IT Nucleic Acid Labeling kit regarding the manufacturer’s protocols (Mirus Bio, Madison, WI, USA). Cy3-labeled pBI221 was purified using QIAGEN PCR purification kit (Qiagen). The exact concentration of purified Cy3-labeled pBI221 and purity were determined by spectrophotometry. Binding of Cy3 to pBI221 was confirmed by nucleic acid mobility shift assays and band quantification using Fiji ImageJ.77 Binding efficiency and integrity of Cy3-labeled pBI221 is shown in Figure S7.
Plant Cultivation
Seeds of Arabidopsis thaliana ecotype Col-0, CS6000, STOMAGEN-OX, STOMAGEN-amiR,50 and the trichrome-less mutant gl1–2(54) were germinated in soil (Promix, Rivièredu-Loup, Canada) supplemented with vermiculite at a ratio of 2:1 at 22 °C under a 16/8 h light/dark photoperiod at 100 μmol photons m–2 s–1 for 7 days. The plants were transferred and cultivated individually at 22 °C under an 8/16 h light/dark photoperiod at 80 μmol photons m–2 s–1 with 50% relative humidity (RH) in a growth chamber. Fully expanded leaves of 4 week old plants were used in the spraying experiments.
Transplastomic Nicotiana tabacum lines overexpressing eGFP specifically in chloroplasts were generated by particle bombardment as previously described.78Nicotiana tabacum, Glycine max (cvs Enrei, Williams-82, and Peking), and Solanum lycopersicum (cv. Micro-Tom) seeds were sown in soil supplemented with vermiculite and cultured at 25 °C under a 16/8 h light/dark period at 80 μmol photons m–2 s–1 with 50% RH for 5–8 weeks. Plants with 5–6 fully expanded leaves were used in the syringe-infiltration and spraying experiments.
Formation of Nucleic Acid/Peptide Complexes
Various amounts of CPP and CTP were added to solutions containing 10 μg of plasmid DNA or 2 μg of siGFPS1 in 200 μL to form nucleic acid/peptide complexes at different N/P ratios. The N/P ratio represents the molar ratio of positively charged NH3+ groups of polypeptides to negatively charged PO4– groups of the corresponding nucleic acid molecules.21 For GUS activity assays in sprayed leaves, pBI221/KH9-BP100 and pBI121/KH9-BP100 complexes were formed at an N/P ratio = 2.0. The complex solutions were vortexed vigorously and incubated at 25 °C for 30 min without shaking. The ternary pPpsbA::Rluc/CTP/CPP complex for chloroplast transfection was formulated via the sequential addition of CTP and CPP to a solution containing plasmid DNA molecules as described previously.27 Briefly, a complex of pPpsbA::Rluc and CTP KH9-OEP34 was formed at an N/P ratio = 1.0. CPP BP100 was added to the plasmid DNA/CTP complex solution to form clustered plasmid DNA/CTP/CPP complexes at an N/P ratio = 1.0. The complex solutions were diluted with 800 μL of water and used for subsequent experiments.
Double-stranded siGFPS1(58) molecules were synthesized on the Eurofin Custom RNA Oligos synthesis platform (Eurofin Genomics, Ebersberg, Germany) (see siRNA sequences in Table S5). A 2 μg amount of siGFPS1 was mixed with KH9-BP100 in 200 μL of DEPC-treated water to form an siRNA/CPP complex at N/P ratio = 2.0. This complex was mixed well and incubated at 25 °C for 30 min without shaking. The clustered siGFPS1/KH9-OEP34/BP100 complex was formed at different N/P ratios in 200 μL of DEPC-treated water using the same procedure used for plasmid DNA/CTP/CPP complex formation. These complex solutions were diluted with 800 μL of DEPC-treated water and used for subsequent experiments.
The hydrodynamic diameter and polydispersity index (PDI) of biomolecule/peptide complexes were characterized with a Zeta Nanosizer using the dynamic light scattering (DLS) mode with a 633 nm He–Ne laser at 25 °C with a backscatter detection angle of 173° (Malvern Instruments, Ltd., Worcestershire, U.K.). Surface charges of the obtained biomolecule/peptide complexes were determined with a Zeta potentiometer. Gel-mobility shift assays were carried out in 1.0% (w/v) agarose gel matrices in 1× TAE buffer for plasmid DNA/peptide complexes and 4.0% (w/v) agarose gel matrices in 0.5× TBE buffer for siRNA/peptide complexes. The morphologies of the nucleic acid/peptide complexes were observed by tapping-mode atomic force microscopy as previously described.27
Foliar Spraying with TAMRA-Labeled CPPs and Biomolecule/Peptide Complex Solutions
Plant materials, functional peptides, and nucleic acid molecules used in different spray experiments were summarized in Tables S8 and S9. For CPP internalization assays, a 1 mL solution of TAMRA-labeled CPPs at a concentration of 0.1 μg/mL was loaded into a 5 mL spray atomizer, allowing approximately 34 μL of solution per spray. The spray nozzle was set 20 cm away from the plants, and the solution was sprayed until the spray mist covered the entire leaf area. The plants were then covered with a plastic lid and returned to standard cultivation conditions. Exposure of TAMRA-labeled CPPs to the leaf surface after spraying was analyzed by fluorospectrometry. Briefly, leaf samples collected at 30 min post-spraying were weighed out and leaf lysates were extracted from ∼100 mg of leaf powder ground in liquid N2 with 100 μL of 1× PBS buffer (pH 7.4) supplemented with 0.1% Triton-X100. TAMRA fluorescence in leaf lysate was determined by fluorescence microplate reader at excitation and emission wavelengths of 545/580 nm. Quantities of different TAMRA-labeled CPPs in sprayed leaves were computed against the linear regression equations generated for each TAMRA-labeled CPP.
Prior to foliar spraying with biomolecule/peptide complexes, the leaf surfaces of plants were washed by sprinkling with water and allowed to dry under standard cultivation conditions at 50% RH for 3–4 h. The solutions containing biomolecule/peptide complexes were diluted with spray solution to a final concentration of 5% (w/v) sucrose and 0.05% (v/v) Silwet L-77 (PhytoTechnology Laboratories, Shawnee Mission, KS, USA). The diluted solutions of biomolecule/peptide complex were loaded into the atomizers and sprayed onto fully expanded leaves until the leaf surfaces were completely covered with the spray mist. The plants were returned to standard culture conditions. Leaves were collected and immediately used in subsequent experiments or stored at −80 °C.
Measuring Droplet Size of Spray Mist
The droplet size of spray mist after spraying through a nozzle was analyzed using a Malvern Spraytec laser diffraction system (Malvern Panalytical, Malvern, U.K.) according to the manufacturer’s protocol.
GUS Activity Assays and Histochemical Staining
For comparison of transfection efficiencies of plasmid DNA/CPP complex with a standard Agrobacterium spraying technique, Agrobacterium tumefaciens strain LBA4404 harboring plant binary vector, pBI121, was prepared by freeze–thaw protocol.79 The overnight cultured cells of Agrobacterium were resuspended and incubated in 10 mM MES, pH 5.7 + 10 mM MgCl2 buffer supplemented with 200 μM acetosyringone in the dark for 2 h. The bacterial culture was then diluted to OD600 nm = 1.0 in 5% sucrose + 0.05% Silwet L-77 prior to spraying to Arabidopsis plants.
Transfection efficiencies in leaves spray-transfected with plasmid DNA harboring GUS expression vector/CPP complexes or Agrobacterium solutions were reported as relative GUS activities. Plant leaves were collected at 24 h after spraying. Total leaf proteins were extracted with a MarkerGene β-glucoronidase (GUS) Reporter Gene Activity Detection kit according to the manufacturer’s protocol (Marker Gene Technology Inc., Eugene, OR, USA). A 50 μL aliquot of total protein solution was incubated with 0.1 mM GUS assay buffer at 37 °C in the dark for 2 h. The reaction was stopped by adding 200 μL of 0.2 M Na2CO3 buffer. The fluorescence activity of the fluorophore 4-methylumbelliferone (4-MU, the catalytic product of GUS from 4-methylumbelliferyl β-d-glucuronide [4-MUG]) was determined using the fluorescence mode in a 96-well plate microplate reader (SpectraMax M3) with emission/excitation wavelengths of 360/465 nm (Molecular Devices, San Jose, CA, USA). The relative fluorescence of a sample was computed from a linear regression equation of a 4-MU standard solution. Total protein concentration was determined by Bradford assay. GUS activity ((pmol of 4-MU/min)/(mg of protein)) was calculated according to the following equation:
For GUS histochemical staining, leaves were collected at 24 h post-spraying. The samples were fixed with ice-cold 0.3% formaldehyde in MES buffer, pH 5.7. The samples were washed twice with ice-cold 50 mM NaH2PO4 solution, pH 7.0, and vacuum-infiltrated with X-gluc substrate solution containing 0.5 mM 5-bromo-4-chloro-3-indolyl-β-glucoronic acid (X-gluc), 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.05% Triton X-100, and 50 mM NaH2PO4, pH 7.0, for 5 min. The leaves were incubated in this solution at 37 °C in the dark for 16–24 h. Chlorophylls were completely removed from the leaves via serial washing with 25%, 50%, 75%, and 95% ethanol. GUS signals were observed under a Leica M165 FC fluorescent stereomicroscope (Leica Microsystems, Tokyo, Japan).
Renilla Luciferase Activity Assay
Renilla luciferase (Rluc) activity in leaves sprayed with pPpsbA::Rluc/peptide complex was assayed as described by the manufacturer (Promega Corp., Madison, WI, USA). Leaves were collected at different time points after spraying with plasmid DNA/peptide complex. Total leaf proteins were extracted from the samples using 200 μL of Rluc lysis buffer. A 50 μL aliquot of protein solution was mixed with 150 μL of Rluc assay buffer by pipetting and the chemiluminescence (RLU) in the mixture determined using a Glomax 20/20 luminometer (Promega). The concentration of total leaf proteins was determined by Bradford assay. Rluc activity was reported as RLU per mg of protein.
Fluorescence Microscopy
Plant leaf surface was rinsed by spraying 1 mL of water for 5 times prior to fluorescence imaging. Confocal laser-scanning microscopy (CLSM; ZeissLSM 700, Carl Zeiss, Oberkochen, Germany) was used to study the internalization of TAMRA-labeled CPPs in plant cells. The ex/em for the detection of TAMRA signals in plant tissue were 555 and 580 nm, respectively. TAMRA fluorescence was observed in both the epidermal and mesophyll cell layers on the adaxial side of the leaf. Green and yellow fluorescent proteins were observed under CLSM with an ex/em of 488/510–535 nm. An ex/em of 488/640–700 nm was used to image chlorophyll autofluorescence in plant cells. Cyanine-3 fluorescence was observed with 555/560–580 nm ex/em wavelengths. The fluorescence filter setups and image acquisition parameters for different fluorophores were provided in Table S10. The fluorescence images were converted to 8-bit type. Background corrections of fluorescence images were performed by setting lower and upper “threshold” levels to 30 and 255,80 respectively, before quantifying the fluorescence signal in a region of interest by Fiji ImageJ.77 Guard cells have different features from epidermal and mesophyll cells, and fluorescence signals in guard cells should be negligible and should not affect the analysis.
Transcript Analysis
Quantitative reverse-transcriptase PCR (qRT-PCR) was used to analyze transcriptional changes in fluorescent reporter genes in leaves after spraying. Total RNA was extracted from leaves with an RNeasy Plant Mini Kit (Qiagen). Complementary DNA molecules were synthesized with a QuantiTect Reverse Transcription kit (Qiagen). qRT-PCR was performed using SYBR Green RealTime Master Mix Plus (Toyobo, Osaka, Japan) with gene-specific primers listed in Table S11. Arabidopsis thaliana ACTIN2 (AtACT2) and Nicotiana tabacum ACTIN4 (NtACT4) were used as constitutive housekeeping gene expression controls in qRT-PCR. The comparative CT (2-ΔΔCT)81 method was employed to compare the differential expression of the reporter gene in plants.
Immunoblotting
Total proteins were extracted from leaves with protein extraction solution containing 6 M urea, 200 mM Tris-HCl, pH 6.8, 10 mM NaCl, 10% (v/v) SDS, 20% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, and cOmplete EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany). A 10 μg amount of total leaf proteins was resolved on a 4–20% Mini PROTEAN TGX Precast gel (Bio-Rad Laboratories, Hercules, CA, USA) and blotted onto a 0.45 μm Hybond-P PVDF membrane (GE Healthcare, Buckinghamshire, U.K.). The GFP and YFP bands on the membrane were detected using a 1:5000 dilution of rabbit anti-GFP polyclonal antibody (NB600-308) as the primary antibody (Novus Biologicals, Littleton, CO, USA). RubisCo Activase 1 (RCA1) protein was detected using a 1:5000 dilution of rabbit anti-RA1 polyclonal antibody (AS10700) as the primary antibody (Agrisera, Vännäs, Sweden). The secondary antibody was a 1:20,000 dilution of horseradish peroxidase (HRP)-conjugated goat antirabbit IgG polyclonal antibody (ab6721) (Abcam, Tokyo, Japan). The HRP signal on the membrane was detected using an LAS3000 imaging system (FujiFilm, Tokyo, Japan) after applying 1 mL of SuperSignal West Pico PLUS chemiluminescence substrate (Thermo Scientific, Waltham, MA, USA) onto the membrane. The band intensities on the membrane were quantified using Fiji ImageJ.77
Statistical Analysis
Graphical figures were created using R studio (R studio, Boston, MA, USA) or Jamovi version 1.6 (The Jamovi Project, Sydney, Australia). The statistical differences among groups in an experiment were analyzed by one-way ANOVA with Tukey’s HSD test at p = 0.05. All statistical analysis in this study was performed using Jamovi.
Acknowledgments
This research was financially supported by Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO; K.N., Grant JPMJER1602), JST COI-NEXT, and BASF SE.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c07723.
Peptide chemical identification experimental data, fluorescence dye-labeled analyses, characteristics of different complexes, translocation and transfection studies, and gene expression and suppression analyses (Figures S1–S26); sequence information and properties, physicochemical properties, qRT-PCR analysis primers, fluorescence imaging parameters, and experimental frameworks (Tables S1–S11) (PDF)
Author Contributions
C.T. and K.N. conceived the research, designed the experiments, and prepared the manuscript draft. C.T., Mi.O, Ma.O., Y.H., and M.M. performed the spray experiments and analyzed the data. S.F. and K.T. designed and synthesized the peptides. C.T., Y.K., Ma.O., and K.N. interpreted the experimental data and finalized the manuscript. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Dunwell J. M. Transgenic Approaches to Crop Improvement. J. Exp. Bot. 2000, 51, 487–496. 10.1093/jexbot/51.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- Goodman R. E.; Vieths S.; Sampson H. A.; Hill D.; Ebisawa M.; Taylor S. L.; van Ree R. Allergenicity Assessment of Genetically Modified Crops—What Makes Sense?. Nat. Biotechnol. 2008, 26, 73–81. 10.1038/nbt1343. [DOI] [PubMed] [Google Scholar]
- Hahn S.; Giritch A.; Bartels D.; Bortesi L.; Gleba Y. A Novel and Fully Scalable Agrobacterium Spray-Based Process for Manufacturing Cellulases and Other Cost-Sensitive Proteins in Plants. Plant Biotechnol. J. 2015, 13, 708–716. 10.1111/pbi.12299. [DOI] [PubMed] [Google Scholar]
- Wang M.; Jin H. Spray-Induced Gene Silencing: A Powerful Innovative Strategy for Crop Protection. Trends Microbiol. 2017, 25, 4–6. 10.1016/j.tim.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakouras A.; Wassenegger M.; Dadami E.; Ganopoulos I.; Pappas M. L.; Papadopoulou K. Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants. Plant Physiol. 2020, 182, 38–50. 10.1104/pp.19.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F. J.; Goh N. S.; Demirer G. S.; Matos J. L.; Landry M. P. Nanoparticle-Mediated Delivery towards Advancing Plant Genetic Engineering. Trends Biotechnol. 2018, 36, 882–897. 10.1016/j.tibtech.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raliya R.; Franke C.; Chavalmane S.; Nair R.; Reed N.; Biswas P. Quantitative Understanding of Nanoparticle Uptake in Watermelon Plants. Front. Plant Sci. 2016, 7, 1288. 10.3389/fpls.2016.01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Wang P.; Lombi E.; Cheng M.; Tang C.; Howard D. L.; Menzies N. W.; Kopittke P. M. Absorption of Foliar-Applied Zn Fertilizers by Trichomes in Soybean and Tomato. J. Exp. Bot. 2018, 69, 2717–2729. 10.1093/jxb/ery085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karny A.; Zinger A.; Kajal A.; Shainsky-Roitman J.; Schroeder A. Therapeutic Nanoparticles Penetrate Leaves and Deliver Nutrients to Agricultural Crops. Sci. Rep. 2018, 8, 7589. 10.1038/s41598-018-25197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avellan A.; Yun J.; Zhang Y.; Spielman-Sun E.; Unrine J. M.; Thieme J.; Li J.; Lombi E.; Bland G.; Lowry G. V. Nanoparticle Size and Coating Chemistry Control Foliar Uptake Pathways, Translocation, and Leaf-to-Rhizosphere Transport in Wheat. ACS Nano 2019, 13, 5291–5305. 10.1021/acsnano.8b09781. [DOI] [PubMed] [Google Scholar]
- Zhao P.; Cao L.; Ma D.; Zhou Z.; Huang Q.; Pan C. Translocation, Distribution and Degradation of Prochloraz-Loaded Mesoporous Silica Nanoparticles in Cucumber Plants. Nanoscale 2018, 10, 1798–1806. 10.1039/C7NR08107C. [DOI] [PubMed] [Google Scholar]
- Eudes F.; Chugh A. Cell-Penetrating Peptides: From Mammalian to Plant Cells. Plant Signal. Behav. 2008, 3, 549–550. 10.4161/psb.3.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh A.; Eudes F.; Shim Y. S. Cell-Penetrating Peptides: Nanocarrier for Macromolecule Delivery in Living Cells. IUBMB Life 2010, 62, 183–193. 10.1002/iub.297. [DOI] [PubMed] [Google Scholar]
- Numata K.; Horii Y.; Oikawa K.; Miyagi Y.; Demura T.; Ohtani M. Library Screening of Cell-Penetrating Peptide for BY-2 Cells, Leaves of Arabidopsis, Tobacco, Tomato, Poplar, and Rice Callus. Sci. Rep. 2018, 8, 10966. 10.1038/s41598-018-29298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger K.; Mink C.; Wadhwani P.; Ulrich A. S.; Nick P. Using the Peptide BP100 As a Cell-Penetrating Tool for the Chemical Engineering of Actin Filaments within Living Plant Cells. Chembiochem 2011, 12, 132–137. 10.1002/cbic.201000402. [DOI] [PubMed] [Google Scholar]
- Lakshmanan M.; Kodama Y.; Yoshizumi T.; Sudesh K.; Numata K. Rapid and Efficient Gene Delivery into Plant Cells Using Designed Peptide Carriers. Biomacromolecules 2013, 14, 10–16. 10.1021/bm301275g. [DOI] [PubMed] [Google Scholar]
- Numata K.; Ohtani M.; Yoshizumi T.; Demura T.; Kodama Y. Local Gene Silencing in Plants via Synthetic DsRNA and Carrier Peptide. Plant Biotechnol. J. 2014, 12, 1027–1034. 10.1111/pbi.12208. [DOI] [PubMed] [Google Scholar]
- Ng K. K.; Motoda Y.; Watanabe S.; Sofiman Othman A.; Kigawa T.; Kodama Y.; Numata K. Intracellular Delivery of Proteins via Fusion Peptides in Intact Plants. PLoS One 2016, 11, e0154081. 10.1371/journal.pone.0154081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienk H. L. J.; Wechselberger R. W.; Czisch M.; De Kruijff B. Structure, Dynamics, and Insertion of a Chloroplast Targeting Peptide in Mixed Micelles. Biochemistry 2000, 39, 8219–8227. 10.1021/bi000110i. [DOI] [PubMed] [Google Scholar]
- Lin R.; Zhang P.; Cheetham A. G.; Walston J.; Abadir P.; Cui H. Dual Peptide Conjugation Strategy for Improved Cellular Uptake and Mitochondria Targeting. Bioconjugate Chem. 2015, 26, 71–77. 10.1021/bc500408p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niidome T.; Ohmori N.; Ichinose A.; Wada A.; Mihara H.; Hirayama T.; Aoyagi H. Binding of Cationic α-Helical Peptides to Plasmid DNA and Their Gene Transfer Abilities into Cells. J. Biol. Chem. 1997, 272, 15307–15312. 10.1074/jbc.272.24.15307. [DOI] [PubMed] [Google Scholar]
- Tecle M.; Preuss M.; Miller A. D. Kinetic Study of DNA Condensation by Cationic Peptides Used in Nonviral Gene Therapy: Analogy of DNA Condensation to Protein Folding. Biochemistry 2003, 42, 10343–10347. 10.1021/bi034325e. [DOI] [PubMed] [Google Scholar]
- Vieregg J. R.; Lueckheide M.; Marciel A. B.; Leon L.; Bologna A. J.; Rivera J. R.; Tirrell M. V. Oligonucleotide-Peptide Complexes: Phase Control by Hybridization. J. Am. Chem. Soc. 2018, 140, 1632–1638. 10.1021/jacs.7b03567. [DOI] [PubMed] [Google Scholar]
- Thagun C.; Motoda Y.; Kigawa T.; Kodama Y.; Numata K. Simultaneous Introduction of Multiple Biomacromolecules into Plant Cells Using a Cell-Penetrating Peptide Nanocarrier. Nanoscale 2020, 12, 18844–18856. 10.1039/D0NR04718J. [DOI] [PubMed] [Google Scholar]
- Chuah J. A.; Yoshizumi T.; Kodama Y.; Numata K. Gene Introduction into the Mitochondria of Arabidopsis Thaliana via Peptide-Based Carriers. Sci. Rep. 2015, 5, 7751. 10.1038/srep07751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi T.; Oikawa K.; Chuah J. A.; Kodama Y.; Numata K. Selective Gene Delivery for Integrating Exogenous DNA into Plastid and Mitochondrial Genomes Using Peptide-DNA Complexes. Biomacromolecules 2018, 19, 1582–1591. 10.1021/acs.biomac.8b00323. [DOI] [PubMed] [Google Scholar]
- Thagun C.; Chuah J. A.; Numata K. Targeted Gene Delivery into Various Plastids Mediated by Clustered Cell-Penetrating and Chloroplast-Targeting Peptides. Adv. Sci. 2019, 6, 1902064. 10.1002/advs.201902064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midorikawa K.; Kodama Y.; Numata K. Vacuum/Compression Infiltration-Mediated Permeation Pathway of a Peptide-PDNA Complex as a Non-Viral Carrier for Gene Delivery In Planta. Sci. Rep. 2019, 9, 271. 10.1038/s41598-018-36466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada K.; Gimenez-Dejoz J.; Miyagi Y.; Oikawa K.; Tsuchiya K.; Numata K. Artificial Cell-Penetrating Peptide Containing Periodic α-Aminoisobutyric Acid with Long-Term Internalization Efficiency in Human and Plant Cells. ACS Biomater. Sci. Eng. 2020, 6, 3287–3298. 10.1021/acsbiomaterials.0c00182. [DOI] [PubMed] [Google Scholar]
- Saric T.; Graef C. I.; Goldberg A. L. Pathway for Degradation of Peptides Generated by Proteasomes: A Key Role for Thimet Oligopeptidase and Other Metallopeptidases. J. Biol. Chem. 2004, 279, 46723–46732. 10.1074/jbc.M406537200. [DOI] [PubMed] [Google Scholar]
- Verdurmen W. P. R.; Bovee-Geurts P. H.; Wadhwani P.; Ulrich A. S.; Hällbrink M.; Van Kuppevelt T. H.; Brock R. Preferential Uptake of L- versus D-Amino Acid Cell-Penetrating Peptides in a Cell Type-Dependent Manner. Chem. Biol. 2011, 18, 1000–1010. 10.1016/j.chembiol.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Ferre R.; Melo M. N.; Correia A. D.; Feliu L.; Bardaji E.; Planas M.; Castanho M. Synergistic Effects of the Membrane Actions of Cecropin-Melittin Antimicrobial Hybrid Peptide BP100. Biophys. J. 2009, 96, 1815–1827. 10.1016/j.bpj.2008.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenberger K.; Sanyal P.; Hundt S.; Wadhwani P.; Ulrich A. S.; Nick P. Challenge Integrity: The Cell-Penetrating Peptide BP100 Interferes with the Auxin-Actin Oscillator. Plant Cell Physiol. 2016, 58, 71–85. 10.1093/pcp/pcw161. [DOI] [PubMed] [Google Scholar]
- Kranjc E.; Mazej D.; Regvar M.; Drobne D.; Remškar M. Foliar Surface Free Energy Affects Platinum Nanoparticle Adhesion, Uptake, and Translocation from Leaves to Roots in Arugula and Escarole. Environ. Sci. Nano 2018, 5, 520–532. 10.1039/C7EN00887B. [DOI] [Google Scholar]
- Hu P.; An J.; Faulkner M. M.; Wu H.; Li Z.; Tian X.; Giraldo J. P. Nanoparticle Charge and Size Control Foliar Delivery Efficiency to Plant Cells and Organelles. ACS Nano 2020, 14, 7970–7986. 10.1021/acsnano.9b09178. [DOI] [PubMed] [Google Scholar]
- Avellan A.; Yun J.; Morais B. P.; Clement E. T.; Rodrigues S. M.; Lowry G. V. Critical Review: Role of Inorganic Nanoparticle Properties on Their Foliar Uptake and In Planta Translocation. Environ. Sci. Technol. 2021, 55, 13417–13431. 10.1021/acs.est.1c00178. [DOI] [PubMed] [Google Scholar]
- Clough S. J.; Bent A. F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis Thaliana. Plant J. 1998, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Chung M.-H.; Chen M.-K.; Pan S.-M. Floral Spray Transformation Can Efficiently Generate Arabidopsis. Transgenic Res. 2000, 9, 471–486. 10.1023/A:1026522104478. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Henriques R.; Lin S.-S.; Niu Q.-W.; Chua N.-H. Agrobacterium-Mediated Transformation of Arabidopsis Thaliana Using the Floral Dip Method. Nat. Protoc. 2006, 1, 641–646. 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Han C.; Qiao Y.; Yao L.; Hao W.; Liu Y.; Shi W.; Fan M.; Bai M.-Y.. TOR and SnRK1 Fine Tune SPEECHLESS Transcription and Protein Stability to Optimize Stomatal Development in Response to Exogenously Supplied Sugar. New Phytol.. 2022, . 10.1111/nph.17984. [DOI] [PubMed] [Google Scholar]
- Etxeberria E.; Baroja-Fernandez E.; Muñoz F. J.; Pozueta-Romero J. Sucrose-Inducible Endocytosis As a Mechanism for Nutrient Uptake in Heterotrophic Plant Cells. Plant Cell Physiol. 2005, 46, 474–481. 10.1093/pcp/pci044. [DOI] [PubMed] [Google Scholar]
- Keates S. E.; Kostman T. A.; Anderson J. D.; Bailey B. A. Altered Gene Expression in Three Plant Species in Response to Treatment with Nep1, a Fungal Protein That Causes Necrosis. Plant Physiol. 2003, 132, 1610–1622. 10.1104/pp.102.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.; Ashworth V.; Kim C.; Adeleye A. S.; Rolshausen P.; Roper C.; White J.; Jassby D. Delivery, Uptake, Fate, and Transport of Engineered Nanoparticles in Plants: A Critical Review and Data Analysis. Environ. Sci. Nano 2019, 6, 2311–2331. 10.1039/C9EN00461K. [DOI] [Google Scholar]
- Ma C.; White J. C.; Zhao J.; Zhao Q.; Xing B. Uptake of Engineered Nanoparticles by Food Crops: Characterization, Mechanisms, and Implications. Annu. Rev. Food Sci. Technol. 2018, 9, 129–153. 10.1146/annurev-food-030117-012657. [DOI] [PubMed] [Google Scholar]
- Lv J.; Christie P.; Zhang S. Uptake, Translocation, and Transformation of Metal-Based Nanoparticles in Plants: Recent Advances and Methodological Challenges. Environ. Sci. Nano 2019, 6, 41–59. 10.1039/C8EN00645H. [DOI] [Google Scholar]
- Ma J. K. C.; Drake P. M. W.; Christou P. The Production of Recombinant Pharmaceutical Proteins in Plants. Nat. Rev. Genet. 2003, 4, 794–805. 10.1038/nrg1177. [DOI] [PubMed] [Google Scholar]
- Ito M.; Sakakura A.; Miyazawa N.; Murata S.; Yoshikawa K. Nonspecificity Induces Chiral Specificity in the Folding Transition of Giant DNA. J. Am. Chem. Soc. 2003, 125, 12714–12715. 10.1021/ja036745x. [DOI] [PubMed] [Google Scholar]
- Akitaya T.; Seno A.; Nakai T.; Hazemoto N.; Murata S.; Yoshikawa K. Weak Interaction Induces an ON/OFF Switch, Whereas Strong Interaction Causes Gradual Change: Folding Transition of a Long Duplex DNA Chain by Poly-L-Lysine. Biomacromolecules 2007, 8, 273–278. 10.1021/bm060634j. [DOI] [PubMed] [Google Scholar]
- Eichert T.; Kurtz A.; Steiner U.; Goldbach H. E. Size Exclusion Limits and Lateral Heterogeneity of the Stomatal Foliar Uptake Pathway for Aqueous Solutes and Water-Suspended Nanoparticles. Physiol. Plant. 2008, 134, 151–160. 10.1111/j.1399-3054.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- Sugano S. S.; Shimada T.; Imai Y.; Okawa K.; Tamai A.; Mori M.; Hara-Nishimura I. Stomagen Positively Regulates Stomatal Density in Arabidopsis. Nature 2010, 463, 241–244. 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- Schreel J. D. M.; Leroux O.; Goossens W.; Brodersen C.; Rubinstein A.; Steppe K. Identifying the Pathways for Foliar Water Uptake in Beech (Fagus Sylvatica L.): A Major Role for Trichomes. Plant J. 2020, 103, 769–780. 10.1111/tpj.14770. [DOI] [PubMed] [Google Scholar]
- Corredor E.; Testillano P. S.; Coronado M. J.; González-Melendi P.; Fernández-Pacheco R.; Marquina C.; Ibarra M. R.; De La Fuente J. M.; Rubiales D.; Pérez-De-Luque A.; Risueño M.-C. Nanoparticle Penetration and Transport in Living Pumpkin Plants: In Situ Subcellular Identification. BMC Plant Biol. 2009, 9, 45. 10.1186/1471-2229-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes Z.; Custardoy L.; de la Fuente J. M.; Marquina C.; Ibarra M. R.; Rubiales D.; Pérez-de-Luque A. Absorption and Translocation to the Aerial Part of Magnetic Carbon-Coated Nanoparticles through the Root of Different Crop Plants. J. Nanobiotechnology 2010, 8, 26. 10.1186/1477-3155-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y.; Sano R.; Wada T.; Takabayashi J.; Okada K. Jasmonic Acid Control of GLABRA3 Links Inducible Defense and Trichome Patterning in Arabidopsis. Development 2009, 136, 1039–1048. 10.1242/dev.030585. [DOI] [PubMed] [Google Scholar]
- Dalakouras A.; Wassenegger M.; McMillan J. N.; Cardoza V.; Maegele I.; Dadami E.; Runne M.; Krczal G.; Wassenegger M. Induction of Silencing in Plants by High-Pressure Spraying of In Vitro-Synthesized Small RNAs. Front. Plant Sci. 2016, 7, 1327. 10.3389/fpls.2016.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Zhang H.; Demirer G. S.; González-Grandío E.; Fan C.; Landry M. P. Engineering DNA Nanostructures for SiRNA Delivery in Plants. Nat. Protoc. 2020, 15, 3064–3087. 10.1038/s41596-020-0370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G. S.; Zhang H.; Goh N. S.; Pinals R. L.; Chang R.; Landry M. P. Carbon Nanocarriers Deliver SiRNA to Intact Plant Cells for Efficient Gene Knockdown. Sci. Adv. 2020, 6, eaaz0495. 10.1126/sciadv.aaz0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H.; Behlke M. A.; Rose S. D.; Chang M. S.; Choi S.; Rossi J. J. Synthetic DsRNA Dicer Substrates Enhance RNAi Potency and Efficacy. Nat. Biotechnol. 2005, 23, 222–226. 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Schwartz S. H.; Hendrix B.; Hoffer P.; Sanders R. A.; Zheng W. Carbon Dots for Efficient Small Interfering RNA Delivery and Gene Silencing in Plants. Plant Physiol. 2020, 184, 647–657. 10.1104/pp.20.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S. Y.; Lew T. T. S.; Sweeney C. J.; Koman V. B.; Wong M. H.; Bohmert-Tatarev K.; Snell K. D.; Seo J. S.; Chua N. H.; Strano M. S. Chloroplast-Selective Gene Delivery and Expression In Planta Using Chitosan-Complexed Single-Walled Carbon Nanotube Carriers. Nat. Nanotechnol. 2019, 14, 447–455. 10.1038/s41565-019-0375-4. [DOI] [PubMed] [Google Scholar]
- Santana I.; Wu H.; Hu P.; Giraldo J. P. Targeted Delivery of Nanomaterials with Chemical Cargoes in Plants Enabled by a Biorecognition Motif. Nat. Commun. 2020, 11, 2045. 10.1038/s41467-020-15731-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A.; Pandi G. Noncoding RNA: An Insight into Chloroplast and Mitochondrial Gene Expressions. Life 2021, 11, 49. 10.3390/life11010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew T. T. S.; Wong M. H.; Kwak S. Y.; Sinclair R.; Koman V. B.; Strano M. S. Rational Design Principles for the Transport and Subcellular Distribution of Nanomaterials into Plant Protoplasts. Small 2018, 14, 1802086. 10.1002/smll.201802086. [DOI] [PubMed] [Google Scholar]
- Gao K.; Cheng M.; Zuo X.; Lin J.; Hoogewijs K.; Murphy M. P.; Fu X. D.; Zhang X. Active RNA Interference in Mitochondria. Cell Res. 2021, 31, 219–228. 10.1038/s41422-020-00394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.; Singh N. D.; Li L.; Zhang X.; Daniell H. Engineered Chloroplast DsRNA Silences Cytochrome P450 Monooxygenase, V-ATPase and Chitin Synthase Genes in the Insect Gut and Disrupts Helicoverpa Armigera Larval Development and Pupation. Plant Biotechnol. J. 2015, 13, 435–446. 10.1111/pbi.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Khan S. A.; Hasse C.; Ruf S.; Heckel D. G.; Bock R. Full Crop Protection from an Insect Pest by Expression of Long Double-Stranded RNAs in Plastids. Science 2015, 347, 991–994. 10.1126/science.1261680. [DOI] [PubMed] [Google Scholar]
- Hotto A. M.; Huston Z. E.; Stern D. B. Overexpression of a Natural Chloroplast-Encoded Antisense RNA in Tobacco Destabilizes 5S RRNA and Retards Plant Growth. BMC Plant Biol. 2010, 10, 213. 10.1186/1471-2229-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood R. E.; Hotto A. M.; Bollenbach T. J.; Stern D. B. Overaccumulation of the Chloroplast Antisense RNA AS5 Is Correlated with Decreased Abundance of 5S RRNA In Vivo and Inefficient 5S RRNA Maturation In Vitro. RNA 2011, 17, 230–243. 10.1261/rna.2336611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R.; Meurer J. The Cutting Crew – Ribonucleases Are Key Players in the Control of Plastid Gene Expression. J. Exp. Bot. 2012, 63, 1663–1673. 10.1093/jxb/err401. [DOI] [PubMed] [Google Scholar]
- Hotto A. M.; Castandet B.; Gilet L.; Higdon A.; Condon C.; Stern D. B. Arabidopsis Chloroplast Mini-Ribonuclease III Participates in RRNA Maturation and Intron Recycling. Plant Cell 2015, 27, 724–740. 10.1105/tpc.114.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Hou Q.; Cheng L.; Xu W.; Hong Y.; Li S.; Sun Q. RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. Plant Cell 2017, 29, 2478–2497. 10.1105/tpc.17.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B. M.; Rujan T.; Martin W.; Croteau R. Isoprenoid Biosynthesis: The Evolution of Two Ancient and Distinct Pathways across Genomes. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 13172–13177. 10.1073/pnas.240454797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stermer B. A.; Bianchini G. M.; Korth K. L. Regulation of HMG-CoA Reductase Activity in Plants. J. Lipid Res. 1994, 35, 1133–1140. 10.1016/S0022-2275(20)39958-2. [DOI] [PubMed] [Google Scholar]
- Estévez J. M.; Cantero A.; Reindl A.; Reichler S.; León P. 1-Deoxy-D-Xylulose-5-Phosphate Synthase, a Limiting Enzyme for Plastidic Isoprenoid Biosynthesis in Plants. J. Biol. Chem. 2001, 276, 22901–22909. 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- Pollier J.; Moses T.; González-Guzmán M.; De Geyter N.; Lippens S.; Bossche R.; Marhavý P.; Kremer A.; Morreel K.; Guérin C. J.; Tava A.; Oleszek W.; Thevelein J. M.; Campos N.; Goormachtig S.; Goossens A. The Protein Quality Control System Manages Plant Defence Compound Synthesis. Nature 2013, 504, 148–152. 10.1038/nature12685. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú.; Sauret-Güeto S.; Gas E.; Jarvis P.; Rodríguez-Concepción M. A Mutant Impaired in the Production of Plastome-Encoded Proteins Uncovers a Mechanism for the Homeostasis of Isoprenoid Biosynthetic Enzymes in Arabidopsis Plastids. Plant Cell 2008, 20, 1303–1315. 10.1105/tpc.108.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J.; Arganda-Carreras I.; Frise E.; Kaynig V.; Longair M.; Pietzsch T.; Preibisch S.; Rueden C.; Saalfeld S.; Schmid B.; Tinevez J. Y.; White D. J.; Hartenstein V.; Eliceiri K.; Tomancak P.; Cardona A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z.; Maliga P. High-Frequency Plastid Transformation in Tobacco by Selection for a Chimeric AadA Gene. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 913–917. 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D.; Glazebrook J. Transformation of Agrobacterium Using the Freeze-Thaw Method. Cold Spring Harb. Protoc. 2006, 2006, pdb.prot4666. 10.1101/pdb.prot4666. [DOI] [PubMed] [Google Scholar]
- Shihan M. H.; Novo S. G.; Le Marchand S. J.; Wang Y.; Duncan M. K. A Simple Method for Quantitating Confocal Fluorescent Images. Biochem. Biophys. Reports 2021, 25, 100916. 10.1016/j.bbrep.2021.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D.; Livak K. J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.