Abstract
Simple Summary
Osimertinib has become the standard of care for the first-line treatment of EGFR-mutant NSCLC patients. The aim of this current translational research study was to assess the clinical relevance of liquid biopsy in 47 patients receiving osimertinib. Effects on circulating tumor cells (CTCs) and plasma-DNA (ctDNA) were investigated before, after one treatment cycle, and at the end of treatment. ctDNA and CTCs decreased after one treatment cycle, but increased at the end of treatment. The detection of ctDNA before and after one treatment cycle was associated with shorter progression-free and overall survivals (PFS and OS), whereas ctDNA clearance after one treatment cycle resulted in a significantly longer PFS and OS. ctDNA at baseline emerged as an independent predictor of shorter PFS. Thus, changes in liquid biopsy status (CTCs, ctDNA) during osimertinib treatment can be used as a tool for treatment efficacy.
Abstract
Introduction: Liquid biopsy is a useful tool for monitoring treatment outcome in solid tumors, including lung cancer. The relevance of monitoring CTCs and plasma ctDNA as predictors of clinical outcome was assessed in EGFR-mutant NSCLC patients treated with osimertinib. Methods: Forty-seven EGFR-mutant NSCLC patients who had progressed on prior first- or second-generation EGFR inhibitors were enrolled in the study and treated with osimertinib, irrespective of the presence of the T790M mutation in the primary tumor or the plasma. Peripheral blood was collected at baseline (n = 47), post-Cycle 1 (n = 47), and at the end of treatment (EOT; n = 39). CTCs were evaluated in 32 patients at the same time points (n = 32, n = 27, and n = 21, respectively) and phenotypic characterization was performed using triple immunofluorescence staining (CK/VIM/CD45). Results: Osimertinib resulted in an ORR of 34% (2 CR) and a DCR of 76.6%. The median PFS and OS values were 7.5 (range, 0.8–52.8) and 15.1 (range, 2.1–52.8) months, respectively. ctDNA was detected in 61.7%, 27.7%, and 61.5% of patients at baseline, post-Cycle 1, and EOT, respectively. CTCs (CK+/CD45-) were detected in 68.8%, 48.1%, and 61.9% of patients at the three time points, respectively. CTCs expressing both epithelial and mesenchymal markers (CK+/VIM+/CD45-) were detected in 56.3% and 29.6% of patients at baseline and post-Cycle 1, respectively. The detection of ctDNA at baseline and post-Cycle 1 was associated with shorter PFS and OS, whereas the ctDNA clearance post-Cycle 1 resulted in a significantly longer PFS and OS. Multivariate analysis revealed that male sex and the detection of ctDNA at baseline were independent predictors of shorter PFS (HR: 2.6, 95% C.I.: 1.2–5.5, p = 0.015 and HR: 3.0, 95% C.I.: 1.3–6.9; p = 0.009, respectively). Conclusions: The decrease in both CTCs and ctDNA occurring early during osimertinib treatment is predictive of better outcome, implying that liquid biopsy monitoring may be a valuable tool for the assessment of treatment efficacy.
Keywords: osimertinib, CTCs, ctDNA, EGFR mutant, NSCLC
1. Introduction
Non-small cell lung cancer (NSCLC) harboring somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) represents a molecular subgroup of NSCLC requiring treatment with EGFR tyrosine kinase inhibitors (TKIs) [1,2,3,4,5,6,7,8]. In almost 60% of EGFR-mutant NSCLC patients, acquired resistance to first- and second-generation EGFR TKIs has been attributed to the emergence of the exon 20 EGFR T790M mutation [9,10,11]. The third-generation EGFR TKI osimertinib was initially approved for the treatment of patients with EGFR T790M-positive NSCLC who had progressed on prior EGFR TKIs [12,13]. However, osimertinib has now become the standard of care for the first-line treatment of EGFR-mutant NSCLC [8,14] and it was recently approved for use in the adjuvant setting following tumor resection in patients with EGFR-mutant NSCLC [15].
EGFR-mutant NSCLC is characterized by tumor heterogeneity that, under treatment pressure, may lead to the emergence of tumor clones with additional genetic alterations including MET or HER2 amplification, PIK3CA mutations, or with histological transformation to small cell lung cancer (SCLC) and the epithelial-to-mesenchymal transition (EMT). All these genetic changes have been associated with resistance to EGFR TKIs [15,16,17,18]. Considering that tumor tissue is not always available for the detection of resistance mechanisms, liquid biopsy (LB) including circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), is an alternative approach [19,20,21]. CTCs represent a heterogeneous cell population [19]. CTCs often undergo EMT during their migration and harbor a mesenchymal phenotype [18,19,20,21] that cannot be recognized by epithelial marker-based detection assays such as the CellSearch platform [21,22,23,24,25,26,27,28]. Our group has previously reported that a substantial proportion of CTCs in cancer patients may express stem cell and EMT markers, such as aldehyde dehydrogenase 1 (ALDH1), twist, and vimentin (VIM) [29,30]. Therefore, the high prevalence of CTCs with a mesenchymal phenotype must be taken into consideration for the detection of CTCs in NSCLC patients.
In NSCLC patients, the detection of CTC before treatment is associated with shorter progression-free survival (PFS) and overall survival (OS) [31,32]. In EGFR-mutant NSCLC patients, the genotyping of CTCs and tumor biopsies gave comparable results [31]. The detection of EGFR mutations in ctDNA from NSCLC patients has proved to be complementary to tissue biopsy in clinical practice and is correlated with both the tumor baseline lesion size and response to EGFR TKIs [33,34].
The aim of this translational research study was to investigate the effect of osimertinib on the changes in LB (both CTCs and ctDNA) status in patients with EGFR-mutant NSCLC and to assess the clinical relevance of those changes.
2. Materials and Methods
2.1. Study Design
Patients with histologically documented EGFR-mutant NSCLC and disease progression on first- or second-generation EGFR TKIs were treated with osimertinib and the changes in CTCs and ctDNA (LB status) were assessed during treatment. Patients had to fulfill the standard clinical study inclusion and exclusion eligibility criteria (Materials and Methods). Patients were treated in the Hellenic Oncology Research Group’s (HORG) collaborative centers and the study was approved by the Ethics Committees and the Institutional Review Boards of the participating hospitals, the National Ethic Committee (no: 35/00-03/16), the National Drug Organization (no: IS 28/16), and registered in the clinicaltrials.gov platform (number: NCT02771314, registration date: 13 May 2016) and EudraCT (number: 2016-001335-12, registration date: 13 April 2016). All patients provided written informed consent for participation in the study.
2.2. Blood Samples
Peripheral blood (20 mL in EDTA) was obtained before the administration of osimertinib (baseline sample; Pre), after one month of treatment (Post-1 sample), and at the end of treatment (EOT sample), which corresponded to the time of disease progression. All blood samples were obtained from the middle of vein puncture, after the first 5 mL of blood was discarded to avoid contamination with epithelial cells from the skin. A 10 mL sample of blood was used for the capture of CTCs using the ISET-based filtration system; the sample was centrifuged at 2500 rpm for 10 min, the plasma was removed, and aliquots of 2 mL were stored at −80 °C until use.
2.3. ISET Isolation Platform and Immunofluorescence Staining
CTCs were isolated using the ISET system according to the manufacturer’s instructions. Briefly, 10 mL of peripheral blood were diluted 1:10 in ISET buffer (Rarecells, Paris, France) for 10 min at RT. Diluted samples were filtered using depression tab adjusted at 10 KPa. Membranes were dried for 2 h at RT and stored at −20 °C. Each spot on the membrane was used for identification of CTCs after IF staining.
CTCs from patients and cells from lung cancer cell lines, used as controls, were evaluated for cytokeratin (CK)/CD45 expression and for CK/Vimentin (VIM)/CD45 expression by double and triple IF staining, respectively, using the appropriate antibodies as previously described [31]; samples were evaluated using confocal laser scanning microscopy (details of the antibodies used and the immunofluorescence (IF) staining are presented in Materials and Methods. The cytomorphological criteria described by Meng et al. [35] were used for the characterization of a cell as a CTC.
2.4. ctDNA Isolation and PNA-Q-PCR Assay for Mutation Testing
ctDNA was extracted from two aliquots of 2 mL of plasma using the QIAsymphony® DSP Virus/Pathogen Midi Kit and a QIAsymphony robot (Qiagen, Amtsgericht Düsseldorf, HRB 45822, Hilden, Germany), following the manufacturer’s instructions. The final elution volume was 30 µL per 2 mL aliquot. EGFR sensitizing (exons 19, 21) and resistance mutations (T790M and C797S) were detected using a quantitative real-time PCR (Taqman®, Rue Bois Saint-Jean 5, Rue du Bois Saint-Jean 14, 4102, Seraing, Belgium) assay in the presence of a PNA clamp (Eurogentec, Rue Bois Saint-Jean 5, Rue du Bois Saint-Jean 14, 4102, Seraing, Belgium), which inhibits the amplification of the wild-type alleles, as previously described [36]; the details for the assay are presented in Materials and Methods.
2.5. Statistical Analysis
This was a prospective, non-randomized, multi-center, translational research study. No formal statistical sample size calculation was performed. Progression-free survival (PFS) was defined as the time elapsed between the start date of treatment and the date of clinical or radiological progression or death from any reason. Overall survival (OS) was defined as the time elapsed between the start date of treatment until the date of death from any reason or the date of last follow-up. Qualitative factors were compared by Pearson’s Chi-square test or Fisher’s exact test, whenever appropriate. Differences in positivity rates were assessed using the McNemar test and differences in terms of continuous variables were assessed by the non-parametric Wilcoxon test. PFS and OS for all patients were estimated using the Kaplan–Meier analysis and the comparisons were computed with the log-rank test. Associations between prognostic factors and PFS or OS were examined using Cox proportional hazards regression models. All statistical tests were two-sided and p-values < 0.05 were considered statistically significant. Data were analyzed using the SPSS statistical software, version 22.0 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Study Population
In total, 50 EGFR-mutant NSCLC patients who had progressed on prior first- or second-generation EGFR TKIs, were registered and 48 of them were treated since two of them withdrew their consent. LB status was assessed in 47 patients since one patient had no blood sampling at baseline for technical reasons (CONSORT Diagram; Figure S1). The baseline characteristics of the patients are summarized in Table 1. The median age was 66 years (range: 43–87), 34 (72.3%) were women, all had adenocarcinoma, and 57.4% had a PS (ECOG) of 0. Molecular analysis of the primary tumor, which was performed in the initial tissue sample used for diagnosis, is presented in Table S1. The tumoral detection of the EGFR exon 20 T790M mutation was detected in 14 (29.8%) patients; moreover, three additional patients harbored the T790M mutation in the baseline blood sample but not in the primary tumor. Twenty-three (48.9%) patients had received more than two prior lines of treatment with either first- or second-generation EGFR TKIs and/or chemotherapy. Thirteen (27.7%) and twelve (25.5%) patients had liver and CNS metastases, respectively, and the median number of involved sites was three (range, 1–5).
Table 1.
Clinical characteristics and clinical outcome.
| N = 47 | N (%) |
|---|---|
| Age->Median (Min–Max) | 66.0 (43–87) |
| Sex | |
| Male | 13 (27.7) |
| Female | 34 (72.3) |
| PS | |
| 0 | 27 (57.4) |
| 1 | 20 (42.6) |
| T790M (tissue and/or plasma) mutations | |
| Detected | 17 (36.2) |
| Not detected | 30 (63.8) |
| Line osimertinib admin | |
| Second Line | 24 (51.1) |
| >Second Line | 23 (48.9) |
| Previous Treatment | |
| Chemo and TKI | 23 (48.9) |
| TKI only | 24 (51.1) |
| Site of Disease | |
| Lung | 39 (83.0) |
| LNs | 24 (51.1) |
| Pleura | 11 (23.4) |
| Liver | 13 (27.7) |
| Bones | 21 (44.7) |
| CNS | 12 (25.5) |
| Other | 9 (19.1) |
| Median sites involved | 3 (1–5) |
| Response to osimertinib | |
| Complete response (CR) | 2 (4.3) |
| Partial response (PR) | 14 (29.8) |
| Stable disease (SD) | 20 (42.6) |
| Progressive disease (PD) | 11 (23.4) |
| ORR, 95% C.I | 16 (34.0%; 20.5–47.6%) |
| DCR, 95% C.I | 36 (76.6%; 64.5–88.7% |
| Relapses | 41 (87.2) |
| PFS | |
| Median (mo; min–max) | 7.5 (0.8–52.8) |
| 95% C.I | 6.0–9.0 |
| Deaths | 35 (74.5) |
| OS | |
| Median (mo; min–max) | 15.1 (2.1–52.8) |
| 95% C.I | 10.8–19.4 |
| 1-year OS | 69.8% |
| Follow up | |
| Median (mo; min–max) | 41.9 (2.1–52.8) |
ORR: overall response rate; DCR: disease control rate; OS: overall survival.
3.2. Efficacy
A summary of the efficacy results is presented in Table 1. Briefly, there were 2 patients with complete response (CR) and 14 patients with partial response (PR) for an overall objective response rate (ORR) of 34.0% (95% C.I., 20.5–47.6%) and a disease control rate (DCR) of 76.6% (95% C.I., 64.5–88.7%). For a median follow-up period of 41.9 months, the median PFS was 7.5 months (95% C.I., 6.0–9.0; range, 0.8–52.8), the median OS was 15.1 months (95% C.I., 10.8–19.4; range, 2.1–52.8 months), and the 1-year survival rate was 69.8%. Four patients were alive and without disease progression for 53, 45, 42, and 36 months, whereas 35 (74.5%) had died by the time of analysis. Adverse events were as expected and are presented in Table S2.
3.3. CTC and ctDNA Status before Study Treatment (Pre Sample)
Adequate biological material for the assessment of CTC status at baseline was available in 32 (68.1%) patients; the assessment of 15 patients failed for technical reasons. CTC could be detected in 22 (68.8%) patients by double IF staining [total number of CTCs = 56 with a median of 2.5/mL of blood (range, 1–5/mL)]. Triple IF staining in these patients revealed a heterogeneous population of CTCs based on the expression of CK (CK+) and VIM (VIM+). In 12 (37.5%) patients, CTCs had exclusively an EMT (CK+/VIM+/CD45-; n = 8) or epithelial (CK+/VIM-/CD45-; n = 4) phenotype, whereas in 10 (31.2%) patients, both subpopulations could be detected (Table 2). Representative images of triple IF staining are presented in Figure S2.
Table 2.
CTCs and phenotype status at baseline, after Cycle 1, and at the EOT.
| Time Points | CTCs | Phenotype | ||
|---|---|---|---|---|
| CTCs Detected | CK+VIM+CD45- | CK+VIM-CD45- | CK+VIM-CD45-/CK+VIM+/CD45- | |
| Baseline (n = 32) | 22 (68.8%) | 8 | 4 | 10 |
| Post 1st cycle (n = 27) | 13 (48.1%) | 5 | 5 | 3 |
| EOT (n = 21) | 13 (61.9%) | 4 | 5 | 4 |
ctDNA molecular profiling of baseline samples was available for all patients and EGFR mutations were detected in 29 (61.7%) patients (Table 3). There was no correlation between the detection of CTCs and ctDNA in the baseline samples, since seven (21.9%) patients had detectable ctDNA in their plasma, but not CTCs in the same blood drawn (p = 0.703).
Table 3.
Detection of ctDNA at each time point.
| Detectable ctDNA | Baseline (n = 47) | Post-1 (n = 47) | EOT (n = 39) |
|---|---|---|---|
| ctDNA (at least one) | 29 (61.7) | 13 (27.7) | 24 (61.5) |
| * T790M | 10 (21.3) | 1 (2.1) | 1 (2.6) |
| Del19 | 18 (38.3) | 8 (17.0) | 11 (28.2) |
| L858R | 8 (17.0) | 4 (8.5) | 9 (23.1) |
| S768I & G719X | 3 (6.4) | 1 (2.1) | 3 (7.7) |
| H773_V774insNPH | - | - | 1 (2.6) |
| C797S | - | - | 1 (2.6) |
* T790M mutation co-existed with other exon 19 and 21 EGFR mutations.
3.4. CTC and ctDNA Status after One Month of Study Treatment (Post-1 Sample)
CTCs were also assessed by double IF staining after the first cycle in 39 (83%) patients, and 20 (51.3%) patients had detectable CTCs [total number of CTCs = 35 (median number: 1/mL of blood; range, 1–13)]. There was a statistically significant decrease in the total number of CTCs at the Post-1 time point compared to the baseline sample (p = 0.037; Wilcoxon test). ISET filters from 27 (69.2%) patients were also available for triple IF staining and in 23 (85.2%) patients, CTCs detection was available in both time points (Pre and Post-1). Eight (34.8%) patients with CTCs detectable in the baseline sample had CTCs non-detectable in the Post-1 sample, whereas 3 (13.0%) patients with CTCs non-detectable in the baseline sample had CTCs detectable in the Post-1 sample (Table S3). Ten (37.0%) patients had exclusively CK+/VIM+/CD45- (n = 5) or CK+/VIM-/CD45- (n = 5) CTCs, whereas three additional patients harbored both CTC subpopulations (Table 2). There were no significant differences in the frequency of the various CTC subpopulations between the baseline and the Post-1 samples (p = 0.065; Wilcoxon test).
ctDNA in the Post-1 was significantly decreased compared to baseline, as EGFR mutations were detected in the Post-1 sample in only 13 (27.7%) of the 47 patients who had LB status assessed in the study (p < 0.001). The EGFR T790M mutation was cleared in the Post-1 sample in all but one patient (Table 3). In 17 (36.2%) patients, there was a molecular ctDNA response with the disappearance of the EGFR mutation in the Post-1 sample, whereas for 1 (2.1%) patient the EGFR mutation was non-detected in the baseline sample but appeared in the Post-1 sample (Table S3).
There was no correlation between the detection of CTCs and ctDNA in the post-Cycle 1 samples, since 6 (22.2%) patients had detectable ctDNA in their plasma, but not CTCs in the same blood drawn (p = 0.420).
3.5. CTC and ctDNA Status at the End of Study Treatment (EOT Sample)
In 34 (72.3%) patients, CTCs were measured at the EOT sample and 18 (52.9%) patients had detectable CTCs (total number of CTCs = 37 with a median number of 3/mL of blood; range, 1–6). Fifteen of these patients had available matched Post-1 and EOT samples: for one patient CTCs, were detectable Post-1, but were non-detectable at the EOT, whereas five (33.3%) patients with no detectable CTCs Post-1 had CTCs detectable at the EOT (Table S3). Triple IF staining in 13 of these patients with available ISET filters revealed that CK+/VIM+/CD45- and CK+/VIM-/CD45- CTCs were exclusively detected in four and five patients, respectively, whereas in four additional patients, both subpopulations of CTCs were present (Table 2).
The EOT results of ctDNA molecular profiling was available for 39 (83.0%) patients and EGFR mutations were detected in 24 (61.5%) patients; the detected EGFR mutations in the EOT samples are presented in Table 3; 12 (30.8%) patients with undetectable ctDNA in the Post-1 sample turned positive in the EOT sample (Table S3). There was no correlation between the detection of CTCs and ctDNA in the end of treatment samples, since 7 (33.3%) patients had detectable ctDNA in their plasma but not CTCs in the same blood collection (p = 0.336).
3.6. Changes in LB Status during Treatment
Considering that the detection of CTCs or/and ctDNA represent a positive LB status, the changes in LB status during treatment with osimertinib were further assessed. One treatment cycle turned negative the LB status in 18 (38.3%) patients, whereas 3 (6.4%) patients with negative LB status at baseline turned positive in Post-1 sample; similarly, 3 (7.7%) patients with positive LB status post-Cycle 1 turned negative at the EOT (Table S4).
3.7. Clinical Outcome according to CTC, ctDNA and LB Status
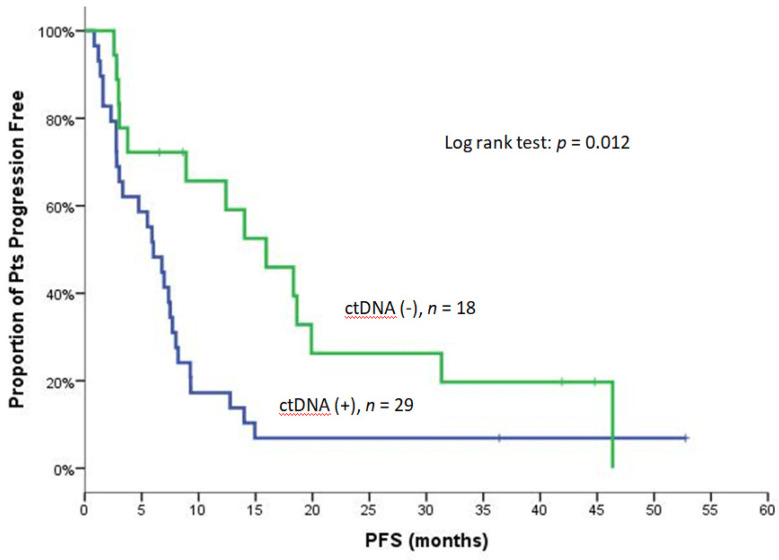
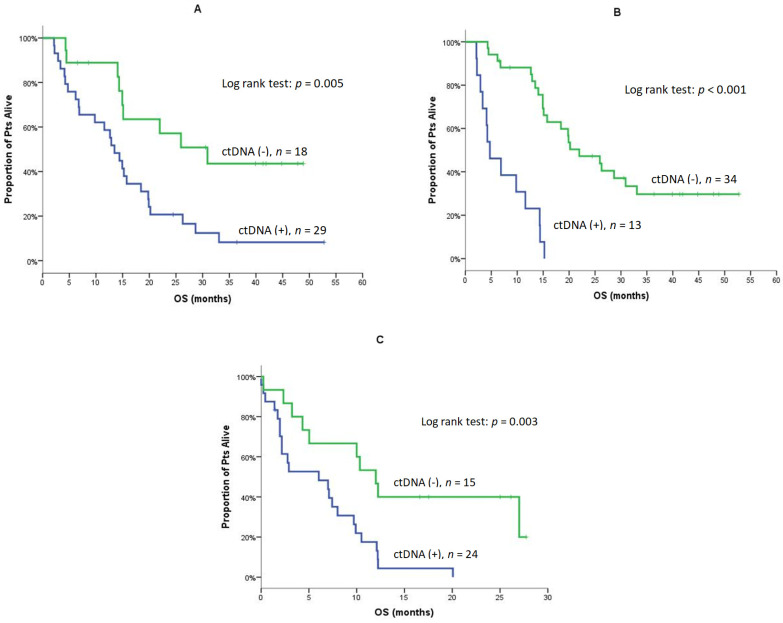
The clinical response in terms of ORR and DCR was not associated with the detection of CTCs, ctDNA, or LB status at baseline or Post-1. Similarly, the detection of CTCs at any time point was not associated with PFS and OS (data not shown). Conversely, the median PFS was significantly shorter in patients with detectable ctDNA at baseline [6.0 vs. 15.9 months; 95% C.I., 3.8–8.3 vs. 8.6–23.3; p = 0.012) (Figure 1) and Post-1 (2.8 vs. 9.3 months; 95% C.I., 1.4–4.1 vs. 4.4–14.2, p < 0.001) (Table 4). The positive LB status at baseline was associated with shorter PFS (6.0 vs. 18.6 months; 95% C.I., 3.8–8.2 vs. 15.0–22.2, p = 0.020), but this was not the case for the positive LB status at Post-1 (p = 0.056) (Table 4 and Figure S3). The median OS was significantly shorter in patients with detectable ctDNA at baseline (13.5 vs. 30.9 months; 95% C.I., 10.4–16.6 vs. 14.6–47.2, p = 0.005), Post-1 (4.7 vs. 22.0 months; 95% C.I., 1.5–8.0 vs. 13.7–30.2, p < 0.001), and the EOT (6.0 vs. 12.0 months; 95% C.I., 0–12.6 vs. 9.2–14.8, p = 0.003) (Table 4; Figure 2A–C). The positive LB status was not associated with OS at any time point (Table 4).
Figure 1.
PFS for patients with detectable and non-detectable ctDNA at baseline.
Table 4.
PFS and OS based on ctDNA and LB status.
| PFS | Baseline (n = 47) | |||||
| ctDNA (+) | ctDNA (−) | p | Liquid (+) | Liquid (−) | p | |
| N/Events | 29/27 | 18/14 | 38/35 | 9/6 | ||
| Median | 6.0 | 15.9 |
0.012 (Figure 1) |
6.0 | 18.6 |
0.020 (Figure S3) |
| Min–Max | 0.8–52.8 | 2.6–46.4 | 0.8–52.8 | 3.0–44.8 | ||
| 95% C.I. | 3.8–8.3 | 8.6–23.3 | 3.8–8.2 | 15.0–22.2 | ||
| Post-1 (n = 47) | ||||||
| ctDNA (+) | ctDNA (−) | p | Liquid (+) | Liquid (−) | p | |
| N/Events | 13/13 | 34/28 | 23/22 | 24/19 | ||
| Median | 2.8 | 9.3 | <0.001 | 3.3 | 8.2 | 0.056 |
| Min–Max | 0.8–5.9 | 2.6–52.8 | 0.8–52.8 | 2.6–44.8 | ||
| 95% C.I. | 1.4–4.1 | 4.4–14.2 | 0.6–6.1 | 5.8–10.6 | ||
| OS | Baseline (n = 47) | |||||
| ctDNA (+) | ctDNA (−) | p | Liquid (+) | Liquid (−) | p | |
| N/Events | 29/26 | 18/9 | 38/30 | 9/5 | ||
| Median | 13.5 | 30.9 |
0.005 (Figure 2A) |
14.9 | 25.9 | 0.093 |
| Min–Max | 2.1–52.8 | 4.3–48.9 | 2.1–52.8 | 6.5–44.8 | ||
| 95% C.I. | 10.4–16.6 | 14.6–47.2 | 12.3–17.5 | 13.5–38.3 | ||
| 1-year OS | 58.6% | 88.9% | 62.9% | 100% | ||
| Post-1 (n = 47) | ||||||
| ctDNA (+) | ctDNA (−) | p | Liquid (+) | Liquid (−) | p | |
| N/Events | 13/13 | 34/22 | 23/18 | 24/17 | ||
| Median | 4.7 | 22.0 |
<0.001 (Figure 2B) |
14.3 | 20.2 | 0.269 |
| Min–Max | 2.1–15.2 | 4.3–52.8 | 2.1–52.8 | 4.3–48.9 | ||
| 95% C.I. | 1.5–8.0 | 13.7–30.2 | 9.9–18.8 | 14.9–25.5 | ||
| 1-year OS | 23.1% | 88.1% | 56.5% | 83.1% | ||
| EOT (n = 39) | ||||||
| ctDNA (+) | ctDNA (−) | p | Liquid (+) | Liquid (−) | p | |
| N/Events | 24/23 | 15/10 | 29/25 | 10/8 | ||
| Median | 6.0 | 12.0 |
0.003 (Figure 2C) |
7.1 | 10.0 | 0.250 |
| Min–Max | 0–20.1 | 0.2–27.7 | 0–27.7 | 0.2–27.0 | ||
| 95% C.I. | 0–12.6 | 9.2–14.8 | 1.3–12.9 | 0–20.8 | ||
| 1-year OS | 17.5% | 46.7% | 25.1% | 40.0% | ||
p-values marked with bold indicate statistically significant.
Figure 2.
OS for Patients with detectable and non-detectable ctDNA at baseline (A), Post-1 (B) and, at the EOT (C).
3.8. CTCs and ctDNA Changes during Treatment and Clinical Outcome
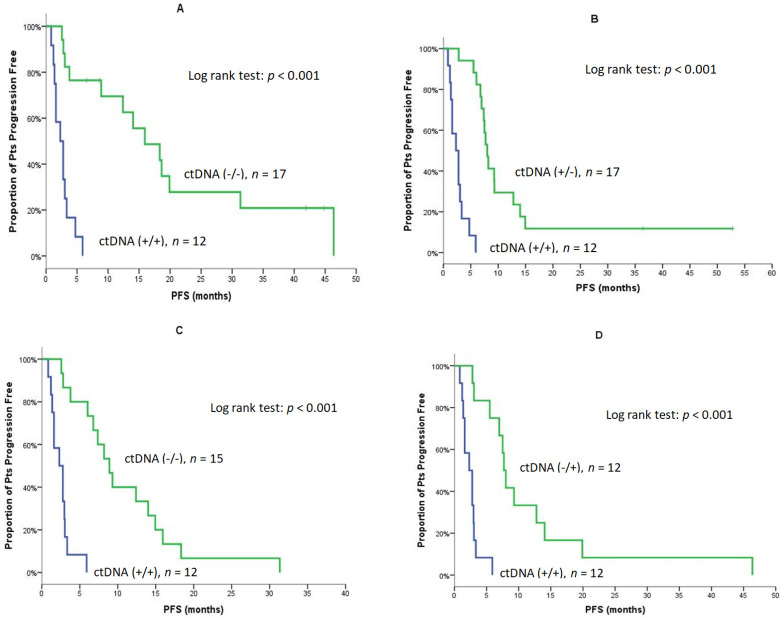
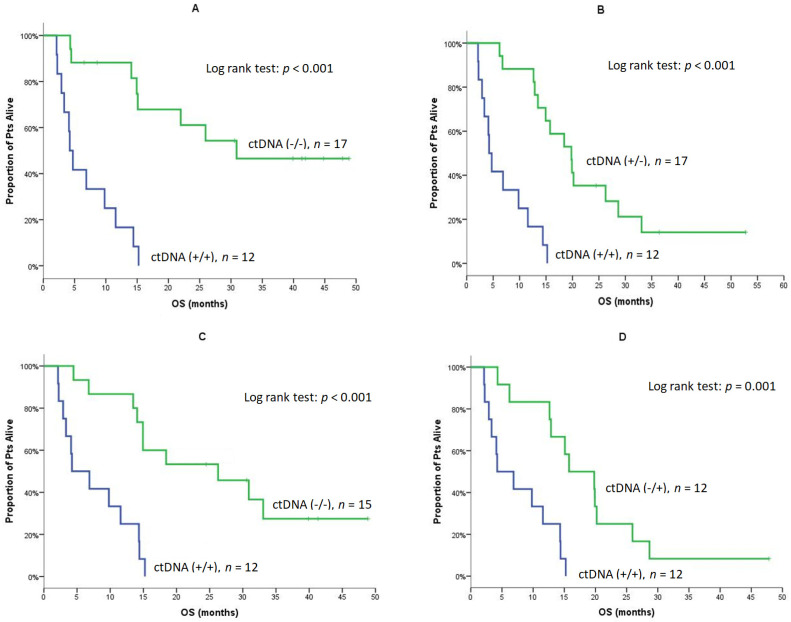
The monitoring of ctDNA during treatment revealed that patients with detectable ctDNA at baseline and Post-1 [Pre (+)/Post-1 (+)] experienced a significantly shorter PFS compared to patients without ctDNA at both time points [Pre (−)/Post-1 (−)] (2.3 vs. 15.9 months, p < 0.001; Table 5 and Figure 3A); in addition, patients with detectable ctDNA at baseline and Post-1 [Pre (+)/Post-1 (+)] had significantly shorter PFS compared to patients who turned ctDNA negative post Cycle 1 [Pre (+)/Post-1 (−)] (2.3 vs. 8.0 months, p < 0.001; Table 5 and Figure 3B). Furthermore, the PFS of patients with detectable ctDNA Post-1 and at the EOT [Post-1 (+)/EOT (+)] was significantly shorter compared to patients without detectable ctDNA at both time points [Post-1 (−)/EOT (−)] (2.3 vs. 8.9 months, p < 0.001; Table 5 and Figure 3C) as well as compared to those with non-detectable ctDNA Post-1 but with detectable ctDNA at the EOT [Post-1 (−)/EOT (+)]: (2.3 vs. 7.7 months, p < 0.001; Table 5 and Figure 3D). For OS, similar associations between ctDNA status at baseline and Post-1 were observed [Pre (+)/Post-1 (+) vs. Pre (−)/Post-1 (−): 4.2 vs. not reached; p < 0.001] and [Pre (+)/Post-1 (+) vs. Pre (+)/Post-1 (−): (4.2 vs. 19.8 months, p < 0.001; Table 5 and Figure 4A,B). Moreover, the median OS of patients with a ctDNA Post-1 (+)/EOT (+) was significantly shorter compared to patients with ctDNA Post-1 (−)/EOT (−): (4.2 vs. 26.3 months, p < 0.001; Table 5 and Figure 4C). In addition, patients with ctDNA Post-1 (+)/EOT (+) had significantly shorter OS compared to patients with non-detectable ctDNA Post-1 but with detectable ctDNA at the EOT [Post-1 (−)/EOT (+)]: (4.2 vs. 15.8 months, p < 0.001; Table 5 and Figure 4D).
Table 5.
PFS and OS according to changes in ctDNA at Baseline, Post-1, and the EOT.
| PFS | Baseline/Post-1 | |||||
| +/+ | −/− | p | +/+ | +/− | p | |
| N/Events | 12/12 | 17/13 | 12/12 | 17/15 | ||
| Median | 2.3 | 15.9 |
<0.001 (Figure 3A) |
2.3 | 8.0 |
<0.001 (Figure 3B) |
| Min–Max | 0.8–5.9 | 2.6–46.4 | 0.8–5.9 | 2.8–52.8 | ||
| 95% C.I. | 1.0–3.6 | 8.1–23.7 | 1.0–3.6 | 7.0–8.9 | ||
| PFS | Post-1/EOT | |||||
| +/+ | −/− | p | +/+ | −/+ | p | |
| N/Events | 12/12 | 15/15 | 12/12 | 12/12 | ||
| Median | 2.3 | 8.9 |
<0.001 (Figure 3C) |
2.3 | 7.7 |
<0.001 (Figure 3D) |
| Min–Max | 0.8–5.9 | 2.6–31.3 | 0.8–5.9 | 2.8–46.4 | ||
| 95% C.I. | 1.0–3.6 | 6.5–11.3 | 1.0–3.6 | 6.9–8.5 | ||
| OS | Baseline/Post-1 | |||||
| +/+ | −/− | p | +/+ | +/− | p | |
| N/Events | 12/12 | 17/8 | 12/12 | 17/14 | ||
| Median | 4.2 | NE |
<0.001 (Figure 4A) |
4.2 | 19.8 |
<0.001 (Figure 4B) |
| Min–Max | 2.1–15.2 | 4.3–48.9 | 2.1–15.2 | 6.2–52.8 | ||
| 95% C.I. | 3.1–5.3 | - | 3.1–5.3 | 14.2–25.3 | ||
| 1-year OS | 16.7% | 88.2% | 16.7% | 88.2% | ||
| OS | Post-1/EOT | |||||
| +/+ | −/− | p | +/+ | −/+ | p | |
| N/Events | 12/12 | 15/10 | 12/12 | 12/11 | ||
| Median | 4.2 | 26.3 |
<0.001 (Figure 4C) |
4.2 | 15.8 |
0.001 (Figure 4D) |
| Min-Max | 2.1–15.2 | 4.5–48.9 | 2.1–15.2 | 4.3–47.8 | ||
| 95% C.I. | 0–8.9 | 8.8–43.8 | 0–8.9 | 7.8–23.7 | ||
| 1-year OS | 25.0% | 86.7% | 25.0% | 83.3% | ||
p-values marked with bold indicate statistically significant.
Figure 3.
PFS of patients according to changes of ctDNA at Baseline, Post-1 and, at the EOT (A). Median PFS of patients with detectable ctDNA at Baseline and Post-1 [Pre (+)/Post-1 (+)] compered to those without detectable ctDNA at both time points [Pre (−)/Post-1 (−)]. (B) Median PFS of patients with detectable ctDNA at Baseline and Post-1 [Pre (+)/Post-1 (+)] compared to those with detectable ctDNA at baseline and non-detactable ctDNA Post-1 [Pre (+)/Post-1 (−)]. (C) Median PFS of patients with detectable ctDNA Post-1 and at the EOT [Post-1 (+)/EOT (+)] compared to patients without detectable ctDNA at both time points [Post-1 (−)/EOT (−)]. (D) Median PFS of patients with detectable ctDNA Post-1 and at the EOT [Post-1 (+)/EOT (+)] compared to patients without detectable ctDNA at Post-1 and detectable ctDNA at the EOT [Post-1 (−)/EOT (+)].
Figure 4.
OS of patients according to changes of ctDNA at Baseline, Post-1 and at the EOT (A). Median OS of patients with detectable ctDNA at Baseline and Post-1 [Pre (+)/Post-1 (+)] compered to those without detectable ctDNA at both time points [Pre (−)/Post-1 (−)]. (B) Median OS of patients with detectable ctDNA at Baseline and Post-1 [Pre (+)/Post-1 (+)] compared to those with detectable ctDNA at baseline and non-detactable ctDNA Post-1 [Pre (+)/Post-1 (−)]. (C) Median OS of patients with detectable ctDNA Post-1 and at the EOT [Post-1 (+)/EOT (+)] compared to patients without detectable ctDNA at both time points [Post-1 (−)/EOT (−)]. (D) Median OS of patients with detectable ctDNA Post-1 and at the EOT [Post-1 (+)/EOT (+)] compared to patients without detectable ctDNA at Post-1 and detectable ctDNA at the EOT [Post-1 (−)/EOT (+)].
3.9. Univariate and Multivariate Analyses
The univariate analysis revealed that male sex and detection of ctDNA at baseline were significantly correlated with shorter median PFS (p = 0.002 and p = 0.015, respectively) and OS (p = 0.007 for both) (Table 6).
Table 6.
Univariate for PFS and OS.
| Independent Factors | PFS | OS | ||
|---|---|---|---|---|
| Hazard Ratio (95% Confidence Interval) |
p | Hazard Ratio (95% Confidence Interval) |
p | |
| Performance status | ||||
| PS 0 | 1 (reference) | 1 (reference) | ||
| PS 1 | 1.2 (0.7–2.3) | 0.534 | 1.8 (0.9–3.5) | 0.094 |
| Gender | ||||
| Male | 3.2 (1.5–6.6) | 0.002 | 2.6 (1.3–5.2) | 0.007 |
| Female | 1 (reference) | 1 (reference) | ||
| Line of osimertinib | ||||
| Second line | 1.0 (0.5–1.7) | 0.835 | 1.1 (0.6–2.2) | 0.751 |
| >Second line | 1 (reference) | 1 (reference) | ||
| T790M (plasma and/or tissue) | ||||
| Detected | 1.0 (0.5–2.0) | 0.886 | 1.0 (0.5–2.0) | 0.955 |
| Not detected | 1 (reference) | 1 (reference) | ||
| Baseline ctDNA | ||||
| Detected | 2.3 (1.2–4.5) | 0.015 | 2.9 (1.3–6.2) | 0.007 |
| Not detected | 1 (reference) | 1 (reference) | ||
p-values marked with bold indicate statistically significant.
The multivariate analysis revealed gender (HR: 2.6; 95% C.I.: 1.2–5.5, p = 0.015) and ctDNA at baseline (HR: 3.0, 95% C.I.: 1.3–6.9, p = 0.009) as independent factors associated with PFS. For overall survival, only the detection of ctDNA at baseline emerged as independent factor associated with shorter OS (HR: 2.3; 95% C.I.: 1.1–5.1, p = 0.041), while only gender had a marked trend (HR: 2.0, 95% C.I.: 1.0–4.1, p = 0.05).
4. Discussion
We observed a significant decrease in ctDNA and CTCs after osimertinib treatment in EGFR-mutant NSCLC patients who had progressed on first-generation EGFR TKIs. Changes in ctDNA status between baseline and post-Cycle 1 emerged as an important and independent factor associated with clinical outcome. Although the heterogeneity of CTCs has been previously described in breast cancer patients [29,30], our study is the first to describe the heterogeneous phenotype of CTCs in EGFR-mutant NSCLC.
In a previous study focused on a subgroup of the patients enrolled in the current study, we investigated the mRNA expression of epithelial, mesenchymal/EMT, and stem cell markers of CTCs and found a high prevalence of VIM-positive CTCs, indicating their EMT status during osimertinib treatment [37]. In addition, in the current study, we demonstrated that osimertinib resulted in a significant decrease of the CTCs after one month of treatment compared to pre-treatment values (p = 0.037). This is consistent with previous studies indicating that the number of CTCs can be used for monitoring drug efficacy [38,39]. Although the number of patients harboring CK+/VIM+/CD45- decreased post-Cycle 1 and increased at the end of treatment, the observed differences were not statistically significant, probably due to the low number of patients in each group. Nevertheless, this seems to support our previous observation that osimertinib has no effect on the expression of mesenchymal markers [37].
The molecular ctDNA response has been associated with longer PFS to first-line EGFR TKIs [40]. In the present study, the ctDNA status during treatment with osimertinib was significantly correlated with the clinical outcome. ctDNA detection at baseline emerged as an independent factor associated with significantly shorter PFS and OS. The disappearance of ctDNA post-Cycle 1 was associated with longer PFS and OS, while the number of patients with detectable ctDNA increased at the end of treatment. Furthermore, among the 10 patients with detectable T790M in ctDNA at baseline, only 1 continued harboring T790M in ctDNA post-Cycle 1 and at the end of treatment, underlying the known effect of osimertinib on T790M-positive tumor cells [34]. Intriguingly, at the time of data analysis, four patients had not progressed on osimertinib for more than 3 years and all four had undetectable ctDNA at baseline and post-Cycle 1. These findings reinforce the clinical utility of the molecular ctDNA response [41]. In addition, the monitoring of ctDNA during osimertinib treatment could reveal patients with refractory or resistant oligometastatic disease who may benefit from radiation therapy, as has been shown by Wang et al. [42].
Our study did not confirm the previously reported predictive role of more than 5 CTCs per 7.5 mL of blood for osimertinib treatment [43]. We were also not able to find any correlation between the detection of CTCs and ctDNA, which implies that they represent distinct biologic phenomena. Whether CTC count and ctDNA could have a complementary prognostic value requires a larger cohort of patients, or even closer monitoring with liquid biopsies than in our study.
5. Conclusions
In conclusion, the CTCs and ctDNA detected were affected by osimertinib treatment in EGFR-mutant NSCLC patients, indicating a role of these blood biosources in addressing and monitoring drug efficacy.
Acknowledgments
We thank Vasso Athanasaki for the secretarial support with the preparation of the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14061574/s1, Supplement Materials and Methods, Figure S1: Consort Diagram, Table S1: Somatic EGFR mutations, Table S2: Adverse Events possibly or probably related to study treatment, Figure S2: Expression of Cytokeratin (CK) and Vimentin (VIM) in patients’ CTCs, Table S3: Changing status in CTCs and ctDNA, Table S4: Liquid-Changing status [ctDNA and/or CTCs (ISET) positive], Figure S3: PFS for Patients with detectable and non-detectable LB at baseline.
Author Contributions
Conceptualization: V.G. and A.K. (Athanasios Kotsakis); methodology: G.K., E.K., V.G. and A.K. (Athanasios Kotsakis); software: V.G., E.L., A.K. (Athanasios Kotsakis) and R.R.; validation: V.G. and A.K. (Athanasios Kotsakis); formal analysis: A.N., D.H. and E.L.; investigation: V.G. and A.K. (Athanasios Kotsakis); data curation: G.K., E.K., N.J.-A., N.K., E.P., H.A.C., A.P., E.T., I.B., A.K. (Anna Koumarianou), V.G., R.R. and A.K. (Athanasios Kotsakis); writing—original draft preparation: G.K., E.K., V.G. and A.K. (Athanasios Kotsakis); writing—review and editing: all authors; visualization: G.K. and E.K.; supervision: V.G. and A.K. (Athanasios Kotsakis); project administration: D.H.; funding acquisition: V.G. and A.K. (Athanasios Kotsakis). All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially funded by an unrestricted grant from AstraZeneca (ESR-15-10926/D5160C00030) to the Hellenic Oncology Research group. This research has also been financed by the European Regional Development Fund of the European Union and Greek funds through the Operational Program Competitiveness Entrepreneurship and Innovation, under the call RESEARCH–CREATE–INNOVATE (T2ΕΔΚ-01562).
Institutional Review Board Statement
Patients were treated in the Hellenic Oncology Research Group’s (HORG) collaborative centers and the study was approved by the Ethics Committees and the Institutional Review Boards of the participating hospitals, the National Ethic Committee (no: 35/00-03/16), the National Drug Organization (no: IS 28/16) and registered in the clinicaltrials.gov platform (number: NCT02771314, registration date: 22 February 2019) and EudraCT (number: 2016-001335-12, registration date: 12 May 2016). All patients provided written informed consent for participation in the study.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data generated in this study are available upon reasonable request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R., Moran T., Queralt C., Porta R., Cardenal F., Camps C., Majem M., Lopez-Vivanco G., Isla D., Provencio M., et al. Screening for epidermal growth factor receptor mutations in lung cancer. N. Engl. J. Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 4.Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., Seto T., Satouchi M., Tada H., Hirashima T., et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R., Carcereny E., Gervais R., Vergnenegre A., Massuti B., Felip E., Palmero R., Garcia-Gomez R., Pallares C., Sanchez J.M., et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y.L., Zhou C., Hu C.P., Feng J., Lu S., Huang Y., Li W., Hou M., Shi J.H., Lee K.Y., et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 8.Soria J.C., Ohe Y., Vansteenkiste J., Reungwetwattana T., Chewaskulyong B., Lee K.H., Dechaphunkul A., Imamura F., Nogami N., Kurata T., et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi S., Boggon T.J., Dayaram T., Janne P.A., Kocher O., Meyerson M., Johnson B.E., Eck M.J., Tenen D.G., Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 10.Balak M.N., Gong Y., Riely G.J., Somwar R., Li A.R., Zakowski M.F., Chiang A., Yang G., Ouerfelli O., Kris M.G., et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 11.Yun C.H., Mengwasser K.E., Toms A.V., Woo M.S., Greulich H., Wong K.K., Meyerson M., Eck M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross D.A., Ashton S.E., Ghiorghiu S., Eberlein C., Nebhan C.A., Spitzler P.J., Orme J.P., Finlay M.R., Ward R.A., Mellor M.J., et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janne P.A., Yang J.C., Kim D.W., Planchard D., Ohe Y., Ramalingam S.S., Ahn M.J., Kim S.W., Su W.C., Horn L., et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N. Engl. J. Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 14.Ramalingam S.S., Vansteenkiste J., Planchard D., Cho B.C., Gray J.E., Ohe Y., Zhou C., Reungwetwattana T., Cheng Y., Chewaskulyong B., et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N. Engl. J. Med. 2020;382:41–50. doi: 10.1056/NEJMoa1913662. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y.L., Tsuboi M., He J., John T., Grohe C., Majem M., Goldman J.W., Laktionov K., Kim S.W., Kato T., et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N. Engl. J. Med. 2020;383:1711–1723. doi: 10.1056/NEJMoa2027071. [DOI] [PubMed] [Google Scholar]
- 16.Engelman J.A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J.O., Lindeman N., Gale C.M., Zhao X., Christensen J., et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 17.Takezawa K., Pirazzoli V., Arcila M.E., Nebhan C.A., Song X., de Stanchina E., Ohashi K., Janjigian Y.Y., Spitzler P.J., Melnick M.A., et al. HER2 amplification: A potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–933. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashi K., Sequist L.V., Arcila M.E., Moran T., Chmielecki J., Lin Y.L., Pan Y., Wang L., de Stanchina E., Shien K., et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc. Natl. Acad. Sci. USA. 2012;109:E2127–E2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeinali M., Lee M., Nadhan A., Mathur A., Hedman C., Lin E., Harouaka R., Wicha M.S., Zhao L., Palanisamy N., et al. High-throughput label-free isolation of heterogeneous circulating tumor cells and CTC clusters from non-small-cell lung cancer patients. Cancers. 2020;12:127. doi: 10.3390/cancers12010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofman V., Ilie M., Long E., Guibert N., Selva E., Washetine K., Mograbi B., Mouroux J., Venissac N., Reverso-Meinietti J., et al. Detection of circulating tumor cells from lung cancer patients in the era of targeted therapy: Promises, drawbacks and pitfalls. Curr. Mol. Med. 2014;14:440–456. doi: 10.2174/1566524014666140414205455. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Wei L., Li J., Zheng J., Zhang S., Zhou J. Epithelialmesenchymal transition phenotype of circulating tumor cells is associated with distant metastasis in patients with NSCLC. Mol. Med. Rep. 2019;19:601–608. doi: 10.3892/mmr.2018.9684. [DOI] [PubMed] [Google Scholar]
- 22.Punnoose E.A., Atwal S., Liu W., Raja R., Fine B.M., Hughes B.G., Hicks R.J., Hampton G.M., Amler L.C., Pirzkall A., et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: Association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:2391–2401. doi: 10.1158/1078-0432.CCR-11-3148. [DOI] [PubMed] [Google Scholar]
- 23.Krebs M.G., Sloane R., Priest L., Lancashire L., Hou J.M., Greystoke A., Ward T.H., Ferraldeschi R., Hughes A., Clack G., et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 24.Krebs M.G., Hou J.M., Sloane R., Lancashire L., Priest L., Nonaka D., Ward T.H., Backen A., Clack G., Hughes A., et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer. 2012;7:306–315. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 25.Papadaki M.A., Messaritakis I., Fiste O., Souglakos J., Politaki E., Kotsakis A., Georgoulias V., Mavroudis D., Agelaki S. Assessment of the efficacy and clinical utility of different circulating tumor cell (CTC) detection assays in patients with chemotherapy-naive advanced or metastatic non-small cell lung cancer (NSCLC) Int. J. Mol. Sci. 2021;22:925. doi: 10.3390/ijms22020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallo M., De Luca A., Maiello M.R., D’Alessio A., Esposito C., Chicchinelli N., Forgione L., Piccirillo M.C., Rocco G., Morabito A., et al. Clinical utility of circulating tumor cells in patients with non-small-cell lung cancer. Transl. Lung Cancer Res. 2017;6:486–498. doi: 10.21037/tlcr.2017.05.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamminga M., de Wit S., Schuuring E., Timens W., Terstappen L., Hiltermann T.J.N., Groen H.J.M. Circulating tumor cells in lung cancer are prognostic and predictive for worse tumor response in both targeted- and chemotherapy. Transl. Lung Cancer Res. 2019;8:854–861. doi: 10.21037/tlcr.2019.11.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milaki G., Messaritakis I., Koinis F., Kotsakis A., Apostolaki S., Dermitzaki E.K., Perraki M., Hatzidaki D., Georgoulias V. Prognostic value of chemotherapy-resistant CK19mRNA-positive circulating tumor cells in patients with advanced/metastatic non-small cell lung cancer. Cancer Chemother. Pharmacol. 2017;80:101–108. doi: 10.1007/s00280-017-3339-0. [DOI] [PubMed] [Google Scholar]
- 29.Kallergi G., Papadaki M.A., Politaki E., Mavroudis D., Georgoulias V., Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. BCR. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papadaki M.A., Kallergi G., Zafeiriou Z., Manouras L., Theodoropoulos P.A., Mavroudis D., Georgoulias V., Agelaki S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maheswaran S., Sequist L.V., Nagrath S., Ulkus L., Brannigan B., Collura C.V., Inserra E., Diederichs S., Iafrate A.J., Bell D.W., et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundaresan T.K., Sequist L.V., Heymach J.V., Riely G.J., Janne P.A., Koch W.H., Sullivan J.P., Fox D.B., Maher R., Muzikansky A., et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016;22:1103–1110. doi: 10.1158/1078-0432.CCR-15-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray J.E., Okamoto I., Sriuranpong V., Vansteenkiste J., Imamura F., Lee J.S., Pang Y.K., Cobo M., Kasahara K., Cheng Y., et al. Tissue and plasma EGFR mutation analysis in the FLAURA trial: Osimertinib versus comparator EGFR tyrosine kinase inhibitor as first-line treatment in patients with egfr-mutated advanced non-small cell lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:6644–6652. doi: 10.1158/1078-0432.CCR-19-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadimitrakopoulou V.A., Han J.Y., Ahn M.J., Ramalingam S.S., Delmonte A., Hsia T.C., Laskin J., Kim S.W., He Y., Tsai C.M., et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer. 2020;126:373–380. doi: 10.1002/cncr.32503. [DOI] [PubMed] [Google Scholar]
- 35.Meng S., Tripathy D., Frenkel E.P., Shete S., Naftalis E.Z., Huth J.F., Beitsch P.D., Leitch M., Hoover S., Euhus D., et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2004;10:8152–8162. doi: 10.1158/1078-0432.CCR-04-1110. [DOI] [PubMed] [Google Scholar]
- 36.Karachaliou N., Mayo-de las Casas C., Queralt C., de Aguirre I., Melloni B., Cardenal F., Garcia-Gomez R., Massuti B., Sanchez J.M., Porta R., et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol. 2015;1:149–157. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 37.Ntzifa A., Strati A., Kallergi G., Kotsakis A., Georgoulias V., Lianidou E. Gene expression in circulating tumor cells reveals a dynamic role of EMT and PD-L1 during osimertinib treatment in NSCLC patients. Sci. Rep. 2021;11:2313. doi: 10.1038/s41598-021-82068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Z.J., Guo Y.H., Zhao Z., Yao J.T., Xu R., Nan K.J. Gemcitabine inhibits the micrometastasis of non-small cell lung cancer by targeting the EpCAM-positive circulating tumor cells via the HGF/cMET pathway. Int. J. Oncol. 2014;45:651–658. doi: 10.3892/ijo.2014.2464. [DOI] [PubMed] [Google Scholar]
- 39.Park C.K., Cho H.J., Choi Y.D., Oh I.J., Kim Y.C. A phase II trial of osimertinib as the first-line treatment of non-small cell lung cancer harboring activating EGFR mutations in circulating tumor DNA: LiquidLung-o-cohort 1. Cancer Res. Treat. 2021;53:93–103. doi: 10.4143/crt.2020.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou C., Imamura F., Cheng Y., Okamoto I., Cho B., Lin M.-C., Majem M., Gautschi O., Gray J., Boyer M., et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J. Clin. Oncol. 2019;37:9020. doi: 10.1200/JCO.2019.37.15_suppl.9020. [DOI] [Google Scholar]
- 41.Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of american pathologists joint review. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 42.Wang X.S., Bai Y.F., Verma V., Yu R.L., Tian W., Ao R., Deng Y., Xia J.L., Zhu X.Q., Liu H., et al. Randomized trial of first-line tyrosine kinase inhibitor with or without radiotherapy for synchronous oligometastatic EGFR-mutated NSCLC. J. Natl. Cancer Inst. 2022;114:djac015. doi: 10.1093/jnci/djac015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang B., Zheng D., Zeng U., Qin A., Gao J., Yu G. Circulating tumor cells predict prognosis following secondline AZD 9291 treatment in EGFR-T790M mutant non-small cell lung cancer patients. J. BUON Off. J. Balk. Union Oncol. 2018;23:1077–1081. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available upon reasonable request from the corresponding author.