Abstract
Glycosylation is an important post-translational modifier of proteins and lipid conjugates critical for the stability and function of these macromolecules. Particularly important are N-linked glycans attached to asparagine residues in proteins. N-glycans have well defined roles in protein folding, cellular trafficking and signal transduction, and alterations to them are implicated in a variety of diseases. However, the non-template driven biosynthesis of these N-glycans leads to significant structural diversity, making it challenging to identify the most biologically and clinically relevant species using conventional analyses. Advances in mass spectrometry instrumentation and data acquisition, as well as in enzymatic and chemical sample preparation strategies, have positioned mass spectrometry approaches as powerful analytical tools for the characterization of glycosylation in health and disease. Imaging mass spectrometry expands upon these strategies by capturing the spatial component of a glycan’s distribution in-situ, lending additional insight into the organization and function of these molecules. Herein we review the ongoing evolution of glycan imaging mass spectrometry beginning with widely adopted tissue imaging approaches and expanding to other matrices and sample types with potential research and clinical implications. Adaptations of these techniques, along with their applications to various states of disease, are discussed. Collectively, glycan imaging mass spectrometry analyses broaden our understanding of the biological and clinical relevance of N-glycosylation to human disease.
Keywords: Imaging mass spectrometry, N-glycan, MALDI, Glycosylation, Mass spectrometry
I. Introduction
A. Glycobiology of N-glycans and mass spectrometry
This chapter is provided in recognition of the many seminal studies and advances from David M. Lubman to the field of mass spectrometry, glycosylation and biomarker research (Mengmeng Wang et al. 2019; Zhu et al. 2019; Patwa et al. 2010). This review will summarize the advantages, limitations and applications of N-glycan imaging mass spectrometry approaches applied to tissues and biofluids from clinical and animal models of disease (Richard R. Drake et al. 2015; Richard R. Drake 2015; R. R. Drake et al. 2017; Angel et al. 2017; Richard R. Drake, West, et al. 2018; Scott and Drake 2019; C. R. K. Blaschke et al. 2021). Glycosylation is the enzymatic addition of carbohydrate residues (or glycans) to a biomolecule, most commonly to a protein or lipid. These sugars can either be attached as monomers or can be linked together to form long, complex chains in a non-template driven synthetic manner. Individual sugar moieties used to produce glycans are taken up directly by the cell or synthesized via numerous metabolic pathways and include, but are not limited to, glucose (Glu), mannose (Man), galactose (Gal), fucose (Fuc), N-acetylglucosamine (GlcNAc), N-acetylgalactosamine (GalNAc) and N-acetylneuraminic acid (Neu5Ac) (Cummings 2009) (Figure 1A). Together these sugar chains form a dense carbohydrate scaffold on the cell’s outer membrane known as the glycocalyx (Reitsma et al. 2007). This surface is comprised of multiple subclasses of glycosylated entities, namely N- and O- linked glycoproteins, glycosaminoglycans (GAGs), proteoglycans and glycolipids (Martinez-Seara Monne et al. 2013). The glycocalyx governs interactions with the local microenvironment by acting as a barrier, filter, substrate and active chemical or enzymatic agent. Glycosylated proteins and lipids in this surface are implicated in cell-cell communication, signal transduction, secretion, interactions with the immune system, cellular migration and adherence, proliferation and regulation amongst other processes (Alphonsus and Rodseth 2014; H. Liu et al. 2020). Critically, the glycocalyx is sensitive to the temporal physiological state of the cell and reflects internal cellular health (Kuo et al. 2018). Thus, it has been well studied that the structure and composition of the glycocalyx changes in response to a variety of diseases (Yilmaz et al. 2019). N-linked glycans, attached to asparagine residues as part of N-X-S/T motif sites on the glycoprotein backbone, are able to play diverse cellular roles due to their structural heterogeneity. Biosynthesized in the endoplasmic reticulum (ER) as a dolichol-linked oligosaccharide substrate, N-glycans are transferred to nascent protein cotranslationally and subsequently processed by a series of glycosidases and glycosyltransferases in the ER and Golgi as part of the secretory pathway (Stanley P 2017). Three main structural classes of N-glycans result from this processing, and are termed oligo- or high mannose, hybrid and complex. Examples of these three N-glycan classes, as well as common N-glycan structural motifs referred to in this review, are shown in Figure 1B,C. The wide variety of possible N-glycoforms interface with an even wider network of molecular interactors to govern cellular processes such as protein folding, immune regulation, cellular mobility and cell signaling.
Figure 1. N-glycan structural annotations.
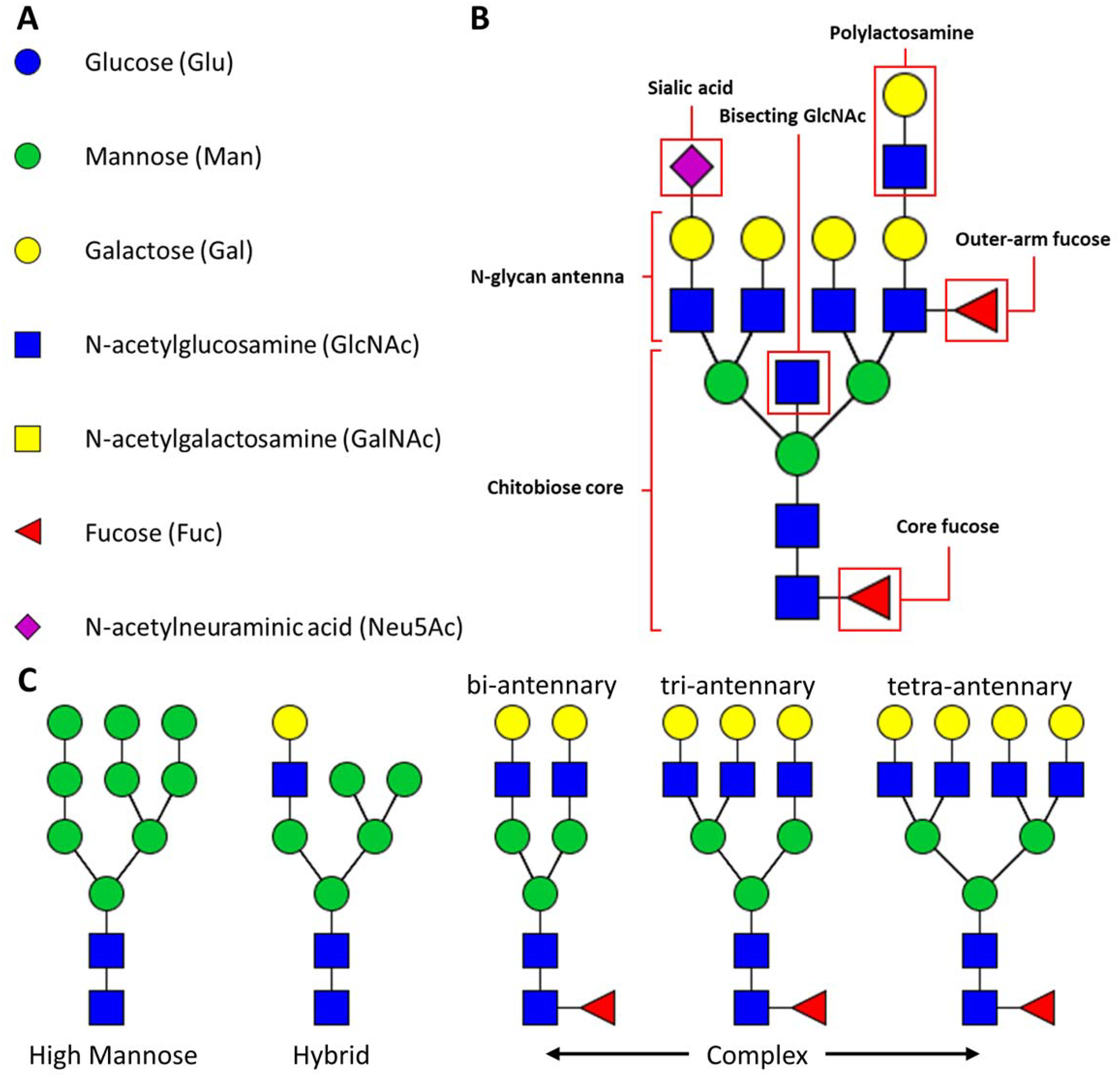
A) individual monosaccharide unit symbols and abbreviations. B) N-glycan structural motifs referenced in this review. C) N-glycan structural classes and examples of N-glycan branching.
N-glycosylation is known to be significantly altered in cancer, both as a response to pathophysiology and as a direct governor of oncogenesis, tumor maintenance, immune evasion and metastatic escape (Mereiter et al. 2019). To this end a variety of methodologies have been developed to analyze N-glycosylation (Mulloy et al. 2017) (Figure 2). Lectins, carbohydrate binding proteins that recognize specific structural epitopes of N-glycans and other glycoconjugates, have been used historically for glycomic analyses, and have since been optimized and adapted into a wide variety of technologies, including microarray and chip formats (Cummings et al. 2017; Patwa et al. 2010). For analysis of tissues and surface-attached targets, lectins are typically used to detect N-glycans through immunohistochemical (IHC), immunofluorescent (IF) or colorimetric assays. Lectins are generally specific for binding glycan structural motifs rather than individual glycan species, and have somewhat variable affinities, limiting their use for precise glycan structural determinations. A wide variety of mass spectrometry approaches have been developed to address the structural characterization of glycans in biological samples (Mengmeng Wang et al. 2019; Ruhaak et al. 2018). Analysis of N-glycans by liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) has emerged as a powerful approach in the last decade. An advantageous feature of LC-MS/MS is the ability to separate N-glycan isomers using online high- or ultra-performance performance liquid chromatography (HPLC/UPLC), facilitated by porous graphitized carbon (PGC) or similar columns (Veillon et al. 2017). When coupled with sequential exoglycosidase digestions or ion mobility-equipped mass spectrometers, more precise structural information can be obtained (Guile et al. 1996; Lane et al. 2019). While effective, in the context of potential diagnostic or clinical applications, these strategies require lengthy purification, chemical derivatization and separation steps which preclude the analysis of large sample cohorts in a feasible timeframe. Analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) of spotted, released N-glycans is another established approach (Ruhaak et al. 2018). While this reduces data acquisition time, most MALDI-TOF MS analyses still require lengthy purification and derivatization steps prior to analysis (Morelle et al. 2009). Imaging mass spectrometry for N-glycans addresses the shortcomings of these previously mentioned technologies as well as introduces advantageous features of its own (Richard R. Drake, West, et al. 2018). Even more complex investigations into precise N-glycan structural linkages can be completed in timeframes acceptable for high-throughput or even clinical applications.
Figure 2. Techniques to analyze N-glycans.
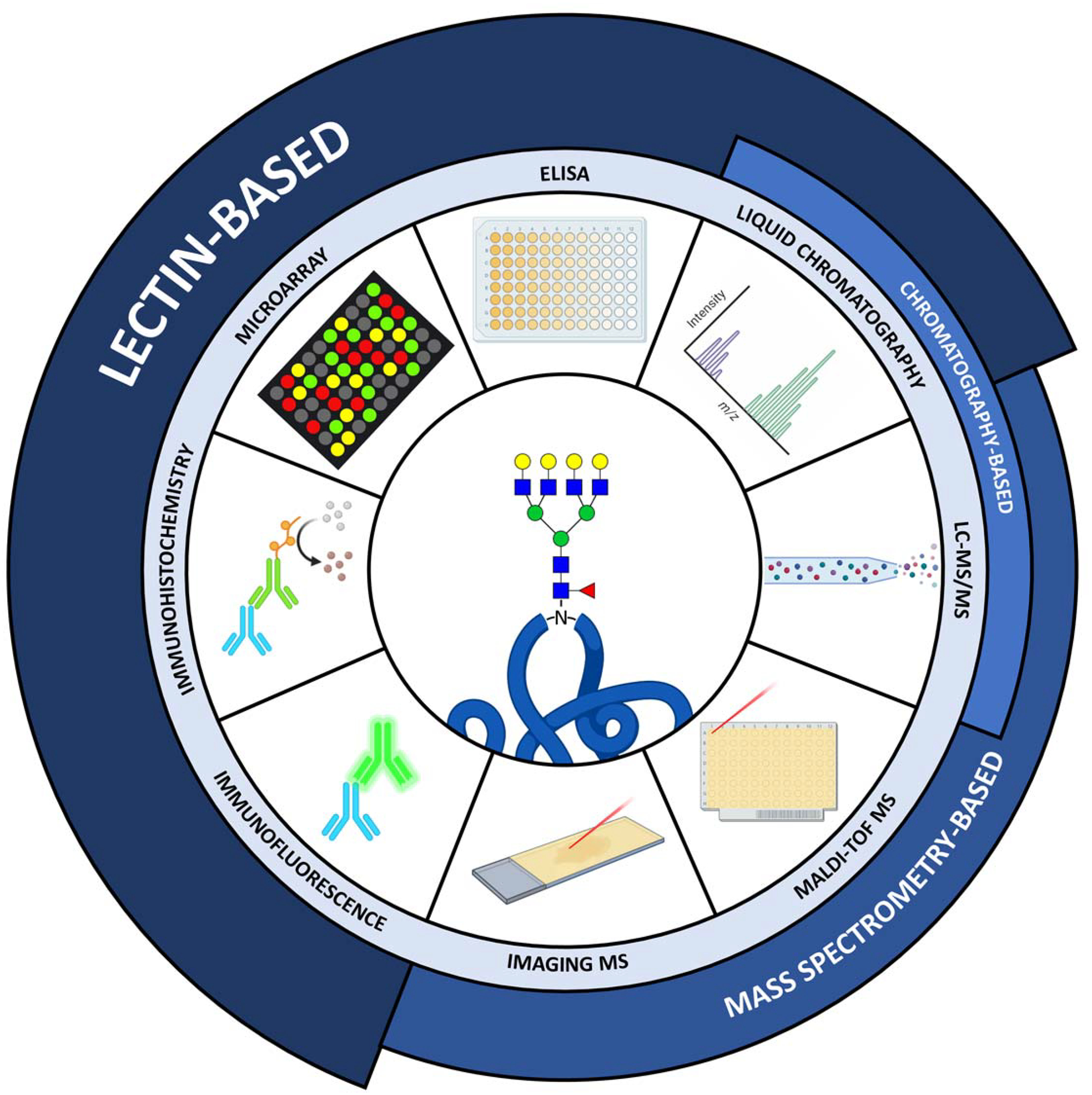
Immunofluorescence, immunohistochemistry, lectin microarray, ELISA, liquid chromatography, LC-MS/MS, MALDI-TOF and Imaging MS techniques for analysis of N-glycosylation. Broadly, these analyses are either lectin-based, chromatography-based or mass spectrometry based, although significant overlap exists between these classes (e.g. liquid chromatography-coupled mass spectrometry or lectin affinity chromatography).
B. Imaging mass spectrometry
Imaging mass spectrometry (IMS) provides the ability to spatially localize analytes detected by MS, and it was first developed for the analysis of metal alloys and semiconductors in industrial applications (Pacholski and Winograd 1999; Galle, Hamburger, and Castaing 1970). Matrix-assisted laser/desorption ionization (MALDI) IMS on biological tissues was first reported in 1997 (Caprioli, Farmer, and Gile 1997). The Caprioli group demonstrated that by rastering the laser beam across the sample surface in a gridded fashion, pseudo-colored heat maps could be generated to show the in situ distributions and abundances of ions of interest where each “pixel” represented a mass spectrum at each tissue location. Imaging experiments utilizing a MALDI source rely on two primary factors: the application of an energy-absorbing organic matrix to the tissue sample and the use of a nitrogen or neodymium-doped yttrium aluminum garnet (Nd:YAG) laser for ion generation from a single coordinate within the tissue, called a data pixel (Robinson, Steven, and Bunch 2018; Holle et al. 2006). These characteristics made MALDI an ideal technology for the initial development of N-glycan IMS, as N-glycan ions have been traditionally difficult to ionize due to their large size and complex structural properties (Powers et al. 2013; Richard R. Drake, Powers, et al. 2018; Powers et al. 2014).
Imaging mass spectrometry has distinct advantages as an imaging modality. Imaging by light microscopy limits the number of identifiable analytes in a single experiment, whereas IMS platforms may detect hundreds of analytes at the same time (Tan et al. 2020). Unlike immunohistochemical or immunofluorescent staining, IMS experiments do not require recognition of an epitope sequence, allowing for in depth and multiplexed profiling of analytes that are not easily recognizable by antibodies, e.g., metabolites, lipids, post-translational modifications. The high-resolution mass analysis afforded by current mass spectrometers allows for reporting of analytes with very similar masses and high confidence in the specificity of molecular assignments, a feature especially useful in metabolomics and lipidomic investigations (Bowman et al. 2020). New advancements in laser, computational and instrumental technologies have facilitated high spatial resolution (≤10 μm) combined with high speed data acquisition (Prentice and Caprioli 2016). A broad range of analytes are detectable by label free imaging of proteins, lipids, nucleic acids, carbohydrates, metabolites and pharmaceuticals ranging from less than 100 to more than 100,000 Da (Mainini et al. 2013). Additionally, the sensitivity of imaging mass spectrometry platforms allows for the detection of molecules of interest at femtomolar levels directly from their biological milieu (Leopold et al. 2018). Most critically, the ability to analyze disease-mediated molecular and morphological features from patient tissue specimens is core to the insights gained by many IMS experiments. Linking histopathologically-defined tissue structures to the expression of specific biomolecules potentiates the discovery of disease markers and therapeutic targets. At its most reductive function, IMS acts as a “molecular microscope” to reveal the biological and chemical mechanisms underpinning pathophysiology, resulting in better understanding of the origins, maintenance and progression of disease.
II. IMS for N-glycans in tissues
The basic N-glycan MALDI IMS schematic is shown in Figure 3. A key component of the N-glycan IMS workflow is the use of a solvent sprayer to apply a thin molecular layer of peptide N-glycosidase F (PNGase F) enzyme onto tissue or cells to release only N-linked glycans from their carrier proteins. Diffusion of the released glycans is minimal allowing spatial co-localization to tissue regions and specific cell types. The first IMS workflows were developed around MALDI platforms, thus, the focus of this review is primarily on MALDI-IMS strategies, although alternative technologies will be briefly covered. Glycan IMS enzymes, chemical matrix, tissue preparation and data analysis strategies are also summarized.
Figure 3. MALDI imaging mass spectrometry for the analysis of N-glycans.
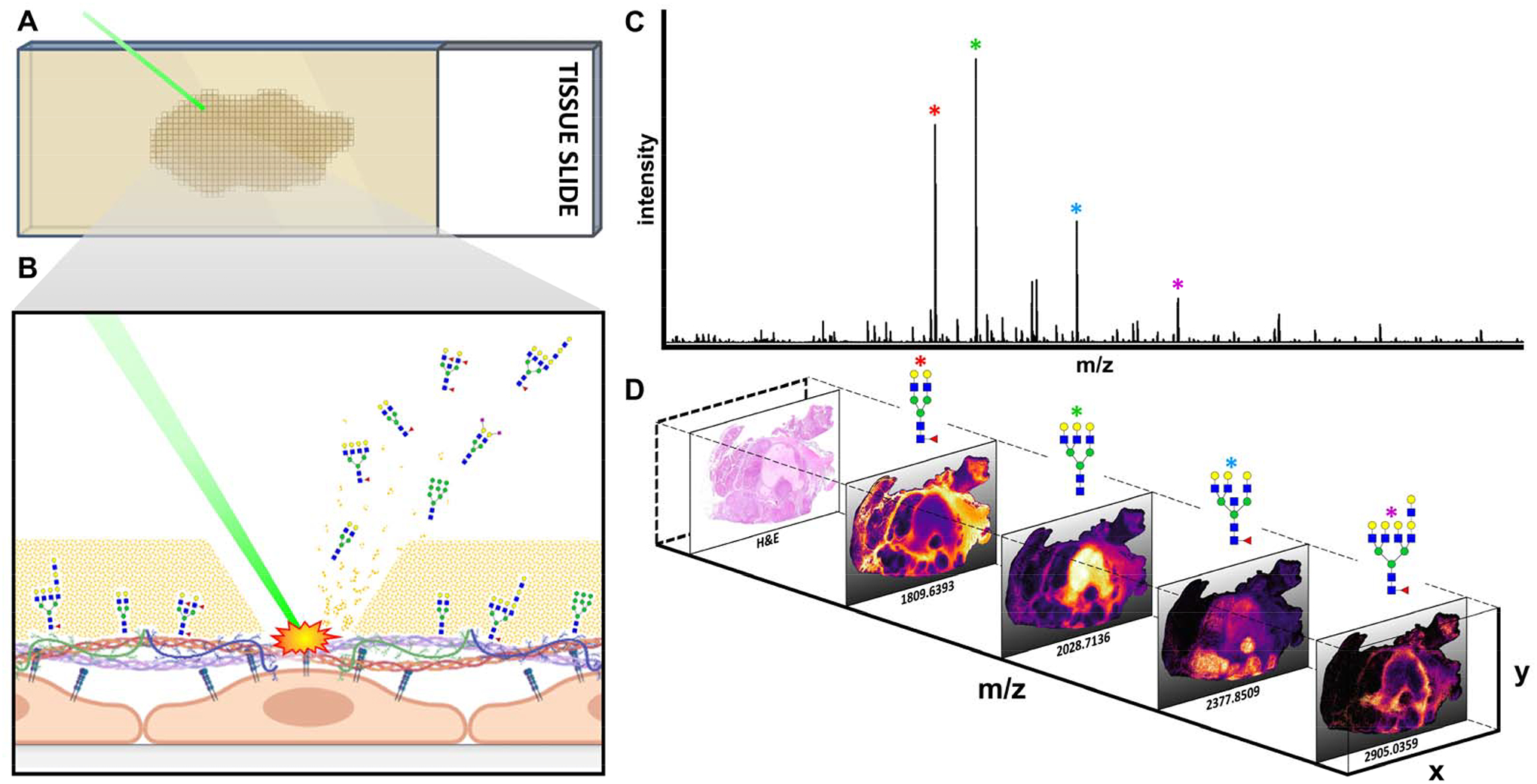
A) MALDI laser striking a rastered FFPE tissue section. B) Close up view of MALDI laser ablating the tissue surface and subsequent desorption/ionization and ejection of N-glycan ions into the mass spectrometer for detection. C) Representative mass spectrum generated from a single MALDI laser strike. D) Spatial mapping of individual pixel spectra back to their original X,Y coordinates generates N-dimensional N-glycan data.
A. Sample Preparation
2.1.1. Tissue sources and sample types
Current MALDI-IMS workflows are able to accommodate both fresh-frozen (FF) and formalin-fixed paraffin-embedded (FFPE) tissue sources from clinical pathology labs, tissue biorepositories, and animal models, providing in essence a nearly unlimited supply of specimens for analysis (Richard R. Drake, Powers, et al. 2018) (Figure 4A). The automated and systematic solvent exchanges required to produce an FFPE block seem to improve and facilitate reproducible detection of N-glycans in FFPE tissue, likely by extensive removal of competing metabolites and lipids. Most N-glycans do not have free amino groups, and thus are not crosslinked in formalin. For optimal results, FF tissue also requires extensive washing to remove metabolites and lipids for detection of N-glycan signal. FFPE tissue slices (3–7 μm; 5 μm standard) are cut from pathology blocks or cores from tissue biopsies and affixed to glass slides (indium tin oxide (ITO)-coated for analyses in TOF instruments). Sections are dewaxed and rehydrated in a graded series of ethanol and water washes prior to heat induced epitope retrieval in a citraconic anhydride-based buffer, required to break bonds created by formalin fixing (Figure 4B,C). For FF tissues, desiccated cryosections (5–12 μm; 10 μm standard) are rinsed in organic solvents to remove lipids, salts and other metabolites, thus increasing downstream N-glycan signal. FF tissues do not require antigen retrieval prior to enzymatic N-glycan release, but heat denaturation improves signal. In addition to whole-tissue IMS, both FFPE and fresh-frozen tissue samples are routinely homogenized for N-glycan extraction and purification (Balog et al. 2012). Doing so enables the analysis of multiple tissues on the same MALDI target plate and can aid in fragmentation and additional structure confirmation tests that support tissue findings. Liquid-phase extraction of N-glycans from tissue homogenate is routinely used orthogonal to whole-tissue MALDI-IMS for precise structural identification via ESI-LC-MS/MS (Holst et al. 2017).
Figure 4. A standardized workflow for in-situ N-glycosylation analysis by MALDI-IMS.
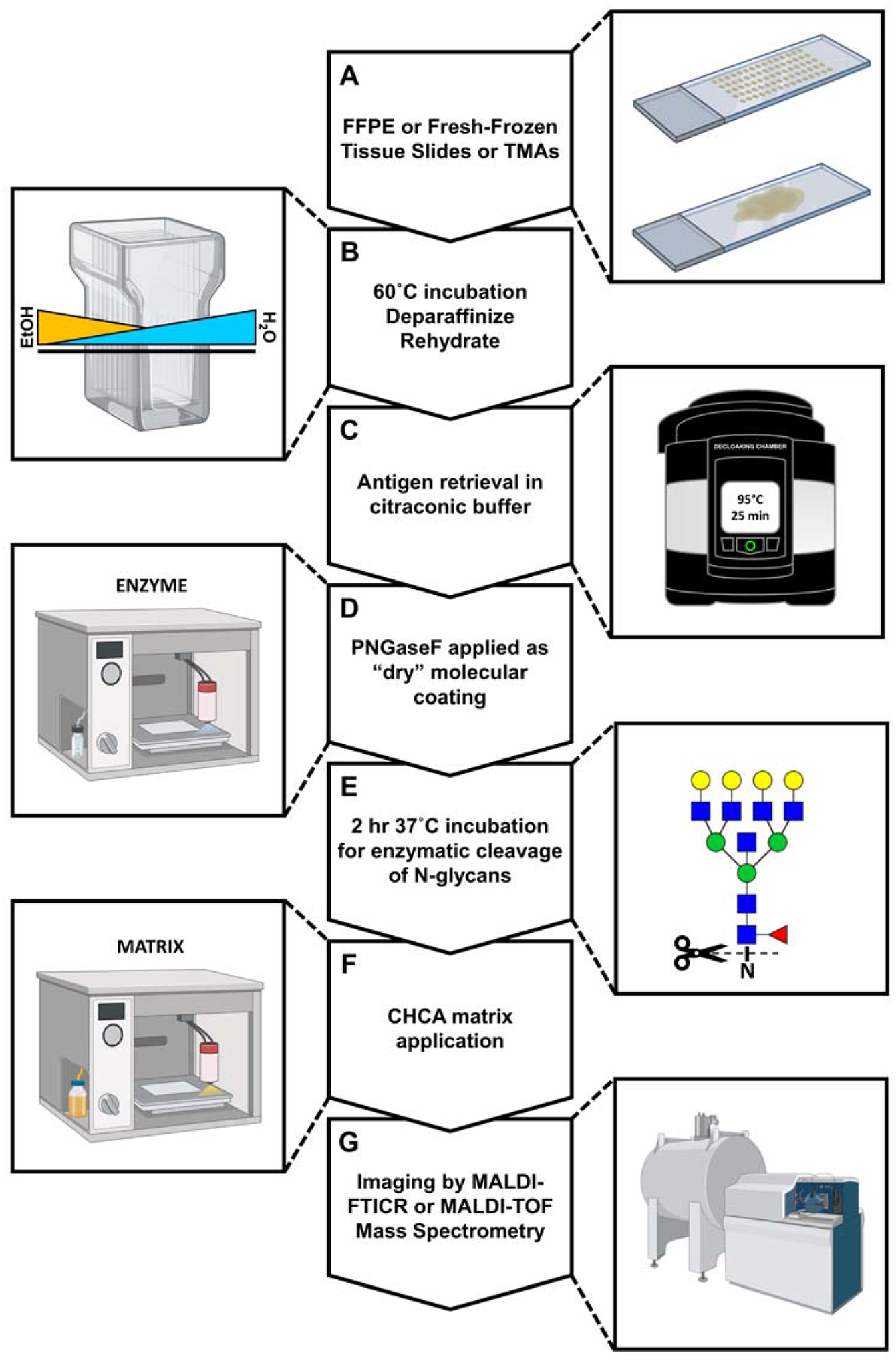
A) FFPE or FF tissue slides. B) Dewaxing and rehydration of tissue specimens prepares them for antigen retrieval and removes signal-suppressing lipids and metabolites. C) Heat-induced epitope exposure in low-pH citraconic buffer. D) Use of an automated solvent sprayer to apply a dry molecular coating of peptide N-glycosidase F. E) Enzymatic cleavage of N-glycans from their glycoprotein carriers. F) Application of a crystalline organic acid matrix by an automated solvent sprayer as in D. G) Spatial analysis of released N-glycans by matrix-assisted laser desorption/ionization and FT-ICR MS detection.
2.1.2. PNGase F de-glycosylation and Matrix application
N-glycans are released from their glycoprotein carriers via on-tissue incubation with peptide N-glycosidase F (PNGase F), which cleaves between asparagine and the first GlcNAc residue on the chitobiose core of the N-glycan (Tarentino and Plummer 1994) (Figure 4D). For N-glycan IMS, the retention of analyte spatial information is accomplished through a “dry” molecular coating of PNGase F that limits diffusion of released N-glycans across the tissue. Current protocols using an automated sprayer system to apply this coating are robust and highly reproducible (Richard R. Drake, Powers, et al. 2018). Tissue slides sprayed with PNGase F are incubated in a pre-warmed (37°C) chamber at ≥ 80% relative humidity for a minimum of two hours to remove the N-glycan structures (Figure 4E). Orthogonal structural analyses can be done using tissue homogenates following deglycosylation with PNGase F followed by N-glycan purification and derivatization steps prior to matrix incorporation and spotting for MALDI-TOF MS (Jensen et al. 2012) or ESI-LC-MS/MS (Holst et al. 2017; Briggs, Condina, Klingler-Hoffmann, et al. 2019).
Desorption and ionization of analytes into a MALDI mass spectrometer for detection is dependent on the application of a crystalline chemical matrix, which absorbs laser energy and transfers it to the substrate below resulting in a transition to the vapor phase and the ionization of analytes (Karas, Bachmann, and Hillenkamp 1985; Karas and Hillenkamp 1988; Jaskolla and Karas 2011) (Figure 4F). The most common organic acids used for N-glycan IMS are α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxybenzoic acid (DHB) (R. R. Drake et al. 2017). Other matrix molecules like 1,5-diaminonaphthalene (DAN) and 2′,4′,6′-trihydroxyacetophenone monohydrate (THAP) have also been used (Hossain and Limbach 2012; Nishikaze 2017). Because matrix crystal size and consistency of matrix application are critical determinants of analyte signal intensity and reproducibility, most current approaches use an automated sprayer for N-glycan imaging experiments. Ideally, the CHCA-coated tissue slides are analyzed within a few days after preparation but can be stored in a desiccator for several weeks (Figure 4G).
B. Instrumentation and ion sources
N-glycan ions span a wide mass range, with the simplest sugar chains detected at ~700 m/z and multiply branched, fucosylated and sialylated structures detected upwards of 4,000 m/z, which approaches the limit of detection for most current MALDI-IMS configurations. There are glycan species of unknown compositions that exceed 4000 m/z, and routine detection of these species represents an area for further study. Further complicating factors for N-glycan MALDI-IMS analysis are variable formation of sodium adducts associated with sialic acids, isotopic overlap between structures, the presence of sulfate groups on some species and interference from the chemical matrix. N-glycans are typically detected in positive mode, however certain special applications are better suited to negative mode analysis (Harvey 2020). To meet these needs, two broad types of MALDI mass spectrometers, each with their own advantages and disadvantages, are typically used to detect released N-glycan ions from tissue, those being Fourier transform ion cyclotron resonance (FT-ICR) and time of flight (TOF) instruments. Recent instrument configurations incorporating a second MALDI laser (termed MALDI-2) and ion mobility separations coupled with MALDI-Q-TOF have recently been used for N-glycan IMS. These are each briefly summarized as follows.
2.2.1. Time of flight MS
Time of flight mass spectrometry, although typically less sensitive, offers access to a broad m/z range of molecules from metabolites to DNA and higher throughput than FT-ICR instruments, and are thus present in many mass spectrometry laboratories. The basic principle of TOF is that ions separate based on the time it takes them to travel through a flight tube of known length and reach the detector (Greaves and Roboz 2013). Although TOF mass spectrometers have generally less resolving power, the rapid time from the laser-ejection of N-glycan ions to detection increases the number of pixels which can be analyzed in a given time frame (up to 50X faster than FTICR-MS for some instruments), resulting in the practical feasibility of much higher spatial resolution analyses (< 5 μm) (Ogrinc Potočnik et al. 2015; Kamata et al. 2020). Also, because of their capacity for higher throughput, TOF mass spectrometers are routinely used for assessment of N-glycans from tissue homogenate via spotting on multi-sample target plates. Another advantage of TOF instruments is that there is no theoretical bound to their upper m/z detection limits. As m/z in these instruments is ultimately a function of flight time, increasing the length of the drift tube allows for detection of analytes with larger masses (J. Lee and Reilly 2011).
2.2.2. FTICR-MS
Ion-Cyclotron Resonance (ICR) detectors excite N-glycan ions using alternating radiofrequency pulses inside a powerful magnetic field (typically ranging from 7T to 21T), resulting in differential cyclotron resonance frequency of the ion packets within the detector proportional to their masses (Karabacak et al. 2010; Smith et al. 2018; Marshall, Hendrickson, and Jackson 1998; Scigelova et al. 2011). This amalgam of resonances is detected by opposed detection plates inside the ICR cell as a single time domain current which is then deconvoluted into its component frequencies by Fourier transformation, for which this class of mass spectrometers bears its name. FT-ICR instruments achieve the highest resolving power (R) in the space (R > 1,600,000) (Bowman et al. 2020) proportional to the strength of the instrument’s magnetic field. However, this resolving power drops off for high-mass analytes (> ~2,000 m/z) without specialized source adjustments (Prentice et al. 2018). Much of the initial N-glycan IMS workflows were developed using a MALDI-FTICR instrument (Drake et. al., 2017).
2.2.3. Ion mobility MS
One of the challenges facing N-glycan MALDI-IMS analysis is the separation of isomeric analytes. The non-linear, non-template driven biosynthesis of N-glycans results in a breadth of structural diversity. Within this heterogeneity are a variety of MS-detected masses that comprise multiple compositionally-identical analytes which differ only with respect to the anomeric linkage of specific residues (Devakumar et al. 2008). Terminal versus bisecting GlcNAc moieties, the specific attachments of sialic acids and core versus outer arm fucosylation, amongst other characteristics, are typically indistinguishable using conventional IMS analyses. Non-imaging mass spectrometry experiments typically resolve glycan isomers using sequential exoglycosidase digestions, potentially coupled with or orthogonal to HPLC/UPLC column-based strategies prior to analysis of electrosprayionized analytes via LC-MS/MS (Mauko et al. 2012; Tao et al. 2014; Yin et al. 2016). Laser ionization of N-glycan ions directly from tissue has not traditionally afforded incorporation of isomeric separation techniques. The recent introduction of MALDI MS instruments with ion mobility capabilities like drift tube ion mobility (DTIMS), traveling wave ion mobility (TWIMS), trapped ion mobility (TIMS) and high-field asymmetric ion mobility (FAIMS), amongst others, helps to deconvolute this complexity (May et al. 2015; Fernandez-Lima 2016; Spraggins et al. 2019). In trapped ion mobility separation instruments, N-glycan isomers within a single m/z ion packet migrate along with an inert drift gas against an oppositely opposed electric field, with their final equilibrium migratory distance dependent on their collisional cross sectional (CCS) areas (Kirk et al. 2019). Gradual reduction of the counter-acting electric field thus elutes these ions in a structural geometry-dependent manner from the mobility cell into the mass analyzer for detection. This technology has already demonstrated utility in parsing out N-glycan isomers (Pu et al. 2016). It should be noted that adding a 4th dimension of data (ion mobility) to IMS experiments exponentially increases the data footprint acquired during a single run. The data handling challenges associated with this new field of analysis are complex and require the development of advanced software solutions before the routine implementation of ion mobility into the imaging space (Sans, Feider, and Eberlin 2018; Rivera et al. 2020).
2.2.4. MALDI-2 MS
One difficulty common to all MALDI platforms is the detection of analytes of very low abundance or very high molecular weight. This complicates the analysis of mammalian N-glycans, which are synthesized to well past 10,000 m/z via the addition of polylactosamine extensions (North et al. 2010). Critical factors contributing to this shortcoming are ionization efficiency and analyte decomposition in the MALDI-source (C. C. Wang et al. 2016). When compared to alternative ionization schema such as ESI, laser-induced ionization of analytes in a MALDI instrument is relatively inefficient (1 out of every ~105 molecules ionized). This is especially true for low-proton affinity substrates like carbohydrates, which ionize at an efficiency of 10−7 – 10−8 in MALDI experiments (Lai and Wang 2017; Page et al. 2007). Recently, novel instrumentation approaches have been developed to overcome this obstacle. Inefficient primary MALDI-ionization is rectified by an additional, subsequent ionization of the analyte plume by a secondary laser oriented perpendicular to the axis of ion ejection in laser positronization-coupled MALDI-IMS (MALDI-2) platforms (Barré et al. 2019). Thus, low abundance structures in the analyte cloud are more likely to be ionized and detected by the mass spectrometer. This new technology has already shown applicability for N-glycan analyses, where a MALDI-2 instrument operating in negative mode demonstrated an order of magnitude more sensitive detection of N-glycans from brain tissue samples than did a positive mode traditional MALDI platform (Heijs et al. 2020). Another possibility afforded by MALDI-2 outfitted mass spectrometers is tissue analysis at subcellular resolution. Coupling a secondary positronization laser with a transition-geometry MALDI mass spectrometer, which positions the primary laser behind the tissue sample, allows pixel sizes on the order of 600 nm and thus the analysis of single cells (Niehaus et al. 2019). Secondary post-ionization is critical for generating acceptable signal at these low rasters, as the volume of sample ejected into the instrument decreases proportionally with reduced pixel size (Zavalin et al. 2012).
2.2.5. Structural analysis of N-glycans by CID fragmentation
Mass spectrometry approaches typically identify N-linked glycans by accurate mass alone, which allows for the precise identification of a particular N-glycan’s carbohydrate composition. Lost in these analyses however is precise information on the linkages which define N-glycan structural features. A traditional approach to deconvoluting N-glycan structural heterogeneity is fragmentation by tandem mass spectrometry (Harvey 2005). The sequential dissolution of glycosidic linkages from the complete N-glycan is typically facilitated by collision induced dissociation (CID) fragmentation. Other tandem mass spectrometry strategies such as higher-energy collisional dissociation (HCD) and electron-transfer dissociation (ETD) are more commonly employed for glycopeptide identification (Ford et al. 2015). Although ESI-based LC-MS/MS traditionally yields more rich fragmentation information, MALDI-MS/MS systems employing on-line CID collision cells now routinely provide fragmentation sufficient for structural identification in imaging experiments (Prentice, Chumbley, and Caprioli 2015; Yang et al. 2007; McDowell et al. 2020) (Figure 5A). Briefly, tandem mass spectrometry by CID involves selective isolation of precursor masses of interest via a multi-pole ion guide followed by introduction of an inert collision gas. Acceleration of precursor ions through application of an electric potential results in gas-phase fragmentation upon collisions of N-glycan ions with the neutral gas molecules (Mitchell Wells and McLuckey 2005) (Figure 5B). Fragment ions are then introduced into the mass analyzer for detection. Sequentially increasing the collision voltage yields a proportional increase in N-glycan fragments. CID fragmentation typically generates both B/Y and C/Z ions and at high energies may also yield A/X cross-ring fragments of individual sugar monomers (Harvey 2000). By observing the sequential loss of sugar monomers from the complete N-glycan, an accurate understanding of the specific linkages composing the structure of interest may be inferred (Figure 5C). These structural inferences are made more concrete by multiplexing CID-based experiments with orthogonal analyses on ion-mobility instruments as previously discussed (Harvey and Struwe 2018).
Figure 5. CID fragmentation for N-glycan structural identification.
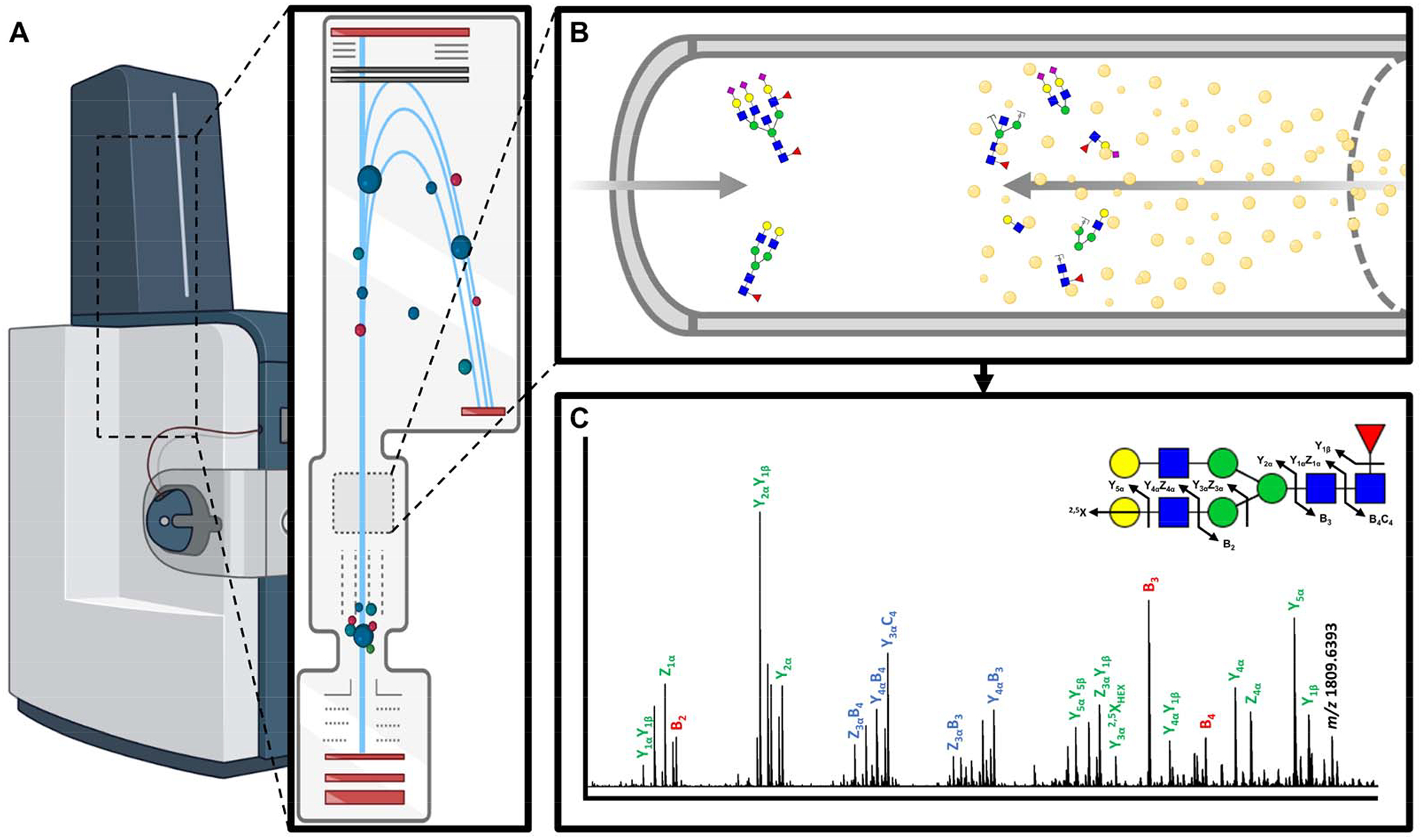
A) MALDI-TOF mass spectrometer with on-line CID collision cell. B) Accelerated precursor N-glycan ions collide with an inert gas (e.g. nitrogen) leading to gas-phase fragmentation. C) Analysis of the N-glycan fragment ions allows for reconstruction of the complete N-glycan structure. CID generates B/Y, C/Z and A/X fragment ions.
2.2.6. Alternative Ionization Strategies
Although MALDI clearly sets itself apart as the ionization strategy of choice for the analysis of N-glycans, additional classes of imaging mass spectrometers, namely desorption electrospray ionization (DESI) and secondary ion mass spectrometry (SIMS) instruments, have potential applicability for N-glycan analyses.
Introduced to the imaging field in 2004, desorption electrospray ionization allows for the analysis of tissue samples under ambient conditions (Takáts et al. 2004). Electrospray-generated charged solvent droplets are directed at the tissue in a rastered fashion similar to that of MALDI experiments (Eberlin 2014). Contact of these droplets with the sample surface extracts molecules and transfers them into the gas phase for introduction into the mass spectrometer (Takáts, Wiseman, and Cooks 2005). Because these sources operate under atmospheric pressure, they can be paired with a wide variety of simpler, less costly mass analyzers making this technology more accessible to both academic and clinical laboratories (Cooks et al. 2011). Another clear advantage of these systems is that desorption by electrospray ionization imparts less energy to the analytes than does a MALDI laser, DESI-IMS may be advantageous for the analysis of oligosaccharides with more labile residues which are typically decomposed in-source during MALDI imaging experiments. Hybrid instruments employing both MALDI and ESI sources (termed MALDESI) are capable of generating multiply charged species, aiding in the detection of low m/z analytes which are not as well resolved in FTICR instruments (Sampson et al. 2006; Nazari et al. 2016). To these ends, multiple groups have demonstrated analyses of carbohydrates, gangliosides and other biomolecules using DESI imaging strategies (Bereman, Williams, and Muddiman 2007; Škrášková et al. 2016; Pace and Muddiman 2020).
Developed prior to MALDI-MS for surface sampling of semiconductors and other inorganic films with industrial applications in mind, secondary ion mass spectrometry (SIMS) employs an ion beam of gaseous clusters which sputters a fixed sample, once again in a rastered manner (Benninghoven 1994). High-energy collisions between these gas molecules and sample molecules results in the ejection of analytes from a sample as secondary ions for analysis by time of flight mass spectrometers. The use of an ion-beam for ionization rather than a laser or microdroplet significantly reduces the footprint of individual pixels and thus allows for sub-micron resolution (< 100 nm) in imaging experiments (Anderton and Gamble 2016; Winograd 2015). Imaging of a wide variety of biomolecules, including glycans from carbohydrate-modified surfaces, by TOF-SIMS has been repeatedly demonstrated (Yoon and Lee 2018; Bolles et al. 2010). Although the complexity of released N-glycans has up-to-now limited their analysis from tissue samples by SIMS, recent chemical derivatization strategies may potentiate N-glycan ionization (Kaya et al. 2018).
C. Data processing for N-glycan IMS
Data processing of the spectral datasets generated from imaging mass spectrometry experiments are challenging to analyze due to their complexity and large data footprint size (Alexandrov 2012). These challenges require significant computational resources to overcome. For example, a typical high spatial resolution MALDI-IMS experiment consists of hundreds of thousands of individual pixels, where each pixel represents a spectrum comprising hundreds of thousands of small m/z bins. These spectra require thresholding and normalization amongst other processing steps and finally spatial mapping to create a composite dataset of pseudo-colored images for every possible m/z of interest. Adding to this complexity, most IMS instruments operate vendor-specific software for initial data processing, making cross-platform comparisons and data sharing traditionally difficult (Chambers et al. 2012).
There is a recent emergence of vendor-neutral IMS processing software options. Open-source packages like MSiReader, Cardinal, and msIQuant utilize MATLAB, R and C++ programming, respectively, to analyze imzML data, the common imaging mass spectrometry file format (Bokhart et al. 2018; Bemis et al. 2015; Källback et al. 2016; Römpp et al. 2011). Even vendor specific software like Bruker Daltonic’s SCiLS Lab and Waters’ High Definition Imaging can accommodate this shared format which allows for the integration of multiple imaging mass spectrometry datasets from different platforms. The complex analyses performed by these software packages are critical for extracting the pathological relevancy of a particular N-glycan’s spatial distribution. Overlaying high-resolution, annotated images of histological staining with N-glycan IMS data reveals co-localization of analytes of interest with distinct tissue morphologies (Aichler and Walch 2015). These programs also allow for statistical considerations such as segmentation, classification and multivariate analyses (Alexandrov et al. 2010; McCombie et al. 2005). New to the space are applications for the multiplexed integration of orthogonal imaging modalities including immunohistochemical and immunofluorescence microscopy, autofluorescence scans and magnetic resonance imaging with N-glycan IMS data to link glycosylation differences to biomarkers of interest or specific pathological features (Porta Siegel et al. 2018; Levenson, Borowsky, and Angelo 2015; Clift, Drake, et al. 2020; Angel et al. 2017).
Unlike peptide, lipid and small molecule imaging which is more established, the relative novelty of N-glycan IMS meant that limited software resources were initially available for peak assignment (Woodin, Maxon, and Desaire 2013). Currently, most laboratories manually assign N-glycan structures to mass spectra via the use of an in-house N-glycan database generated using software such as GlycoWorkbench or GlycoMod or rely on shared online databases like GlyConnect or GlyTouCan (Damerell et al. 2015; Cooper, Gasteiger, and Packer 2001; Alocci et al. 2019; Tiemeyer et al. 2017). Software for automated peak picking of N-glycans from IMS-generated spectra is still in its infancy, but new applications such as Bruker Daltonic’s Metaboscape have shown promise for alignment of mass spectra peaks to an extensive database of known N-glycan masses with matched structures.
III. Applications of N-glycan IMS
The utility of N-glycan IMS has been most thoroughly demonstrated in the field of cancer, as summarized in Table 1, facilitated by access to vast biorepository archives of FFPE tissues. N-glycan IMS has also been applied to non-cancer disease tissues like osteoarthritis and heart valve stenosis (Briggs et al. 2016; Angel et al. 2017; Clift, Drake, et al. 2020). Summaries of N-glycan IMS studies of liver, breast, pancreatic, colorectal, lung, ovarian and renal cancer tissues are discussed in this section. General findings from these studies often compliment serum or plasma N-glycomic studies from the same cancers, although it should be acknowledged that the tumor and circulatory N-glycomes are independently regulated (Kirwan et al. 2015; Gudelj et al. 2018; Dotz et al. 2019). An example panel of N-glycan images for a pancreatic cancer tissue is shown in Figure 6. These investigations have revealed distinct N-glycosylation patterns across tissue subtypes, N-glycan structural motifs associated with particular cancers as well as N-glycan ions whose detection specifically delineates healthy versus diseased tissue or discriminates between cancer stages.
Table 1.
Applications of N-glycan IMS
| Disease | Year | Group | Cohort | Disease-Associated N-glycans |
|---|---|---|---|---|
| Cancer | ||||
| Breast | 2019 | Scott et al. | Whole tissue, TMA | Polylactosamines ↑ (HER2 and TN-associated) |
| 2019 | Herrera et al. | Whole tissue, TMA | Tetra-antennary with LacNAc ↑ (poor survival-associated) | |
| 2019 | Scott et al. | Whole tissue | Sialylation ↑ Fucosylation ↓ (necrosis-associated) | |
| 2019 | Angel et al. | 4T1 cells | Hex9HexNAc2 ↑ | |
| Colorectal | 2016 | Holst et al. | Whole tissue | Sialylation ↑ |
| 2016 | Heijs et al. | Whole tissue | High mannose ↑ | |
| 2020 | Boyaval et al. | Whole tissue | High mannose ↑ | |
| Sialylation ↑ | ||||
| Lung | 2017 | Drake et al. | TMA | High mannose ↑ |
| Hex6HexNAc2 ↑ | ||||
| 2020 | Carter et al. | Whole tissue | High mannose ↑ (hyperplasia-associated) | |
| Fucosylated biantennary ↑ (hyperplasia-associated) | ||||
| Liver | 2014 | Powers et al. | TMA | Hex7HexNAc6 ↑ |
| Hex8HexNAc2 ↑ | ||||
| 2015 | Powers et al. | Whole tissue, TMA | Hex7HexNAc6 ↑ | |
| Core fucosylation ↑ | ||||
| 2018 | West et al. | Whole tissue, TMA | Hex7HexNAc6 ↑ | |
| Branching ↑ | ||||
| Fucosylation ↑ (poor survival-associated) | ||||
| 2019 | Angel et al. | HepG2/C3A cells | Afucosylated bi-/tri-antennary ↑ | |
| Hex9HexNAc2 ↑ | ||||
| 2020 | West et al. | Whole tissue, TMA | Core fucosylation ↑ (poor survival-associated) | |
| 2020 | Black et al. | Serum | Hex3dHex1HexNAc4 ↑ (cirrhosis-associated) | |
| Hex5HexNAc4NeuAc2 ↑ (cirrhosis-associated) | ||||
| Ovarian | 2016 | Everest-Dass et al. | Whole tissue | High mannose ↑ |
| 2019 | Briggs et al. | Whole tissue, TMA | Hex9HexNAc2 ↑ (stage III-associated) | |
| Fucosylation ↑ (stage III-associated) | ||||
| Bisecting GlcNAc ↑ (stage III associated) | ||||
| Sialylation ↑ (stage III-associated) | ||||
| Pancreatic | 2014 | Powers et al. | Whole tissue | Biantennary ± fucose ↑ (fibrosis-associated) |
| Undecorated ↑ (necrosis-associated) | ||||
| 2015 | Powers et al. | Whole tissue | Biantennary ↑ (fibrosis-associated) | |
| 2020 | McDowell et al. | Whole tissue, TMA | Fucosylation ↑ | |
| Branching ↑ | ||||
| Bisecting GlcNAc ↑ | ||||
| Sialylation ↑ | ||||
| Polylactosamines ↑ (tumor margin-associated) | ||||
| Undecorated ↑ (necrosis-associated) | ||||
| High mannose ↓ | ||||
| Sulfated, terminal GalNAc biantennary ↑ (islet-associated) | ||||
| Renal | 2019 | Drake et al. | Whole tissue, TMA | Bisecting GlcNAc ↑ (proximal tubule-associated) |
| Hex7dHex1HexNAc6 ± sialic acid ↑ (glomeruli-associated) | ||||
| Polylactosamine ↑ | ||||
| Branching ↑ | ||||
| Outer-arm fucosylation ↓ | ||||
| Hex5dHex1HexNAc4NeuAc1 ↑ (fibrillar capsule-associated) | ||||
| Fucosylation ↓ (PKD cyst fluid-associated) | ||||
| Non-Cancer | ||||
| Osteoarthritis | 2016 | Briggs et al. | Whole tissue | Hex5HexNAc4NeuAc2 ↑ (bone marrow lesion-associated) |
↑ increased expression relative to normal healthy tissue
↓ decreased expression relative to normal healthy tissue
( ) association with a specific disease or tissue subtype
Figure 6. N-glycan imaging mass spectrometry of a late stage pancreatic ductal adenocarcinoma tissue.
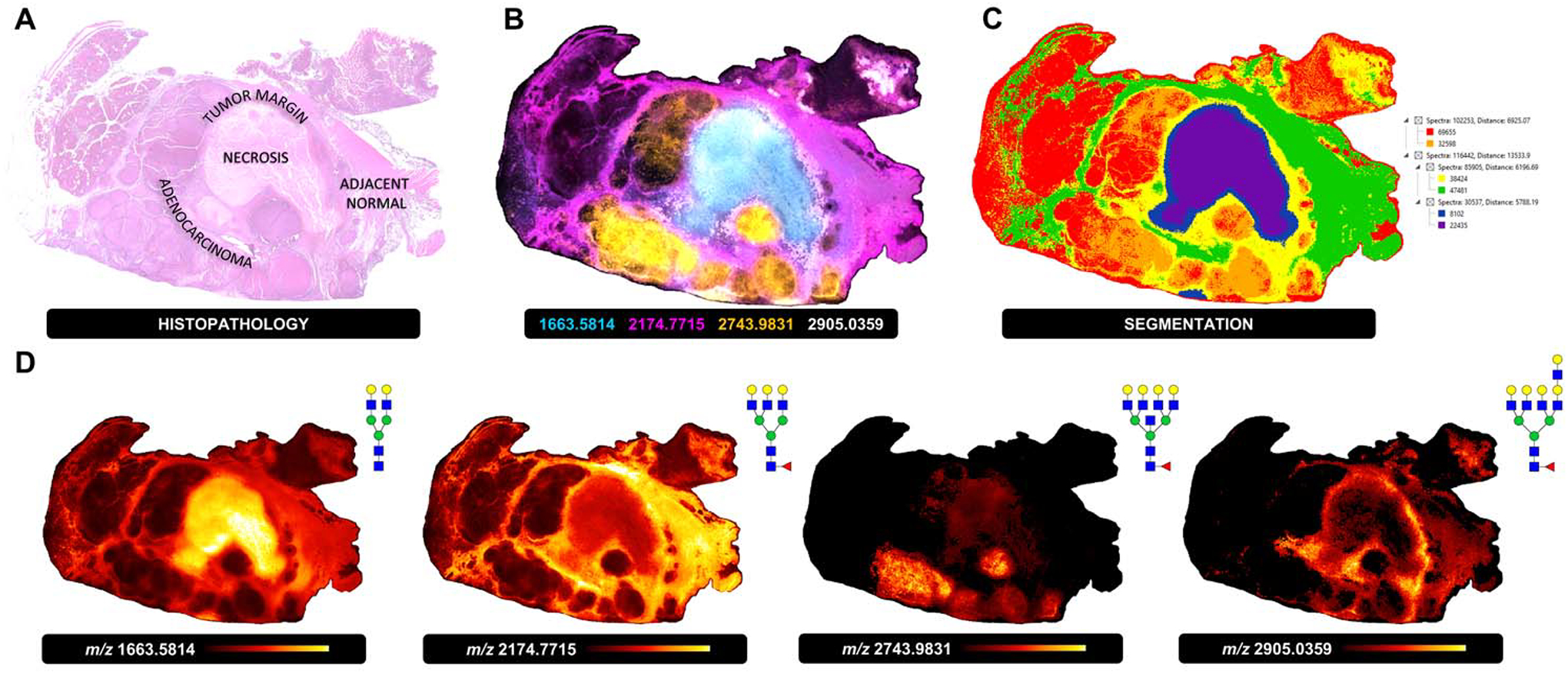
A) H&E staining and histopathological annotation of a late stage PDAC tissue with adenocarcinoma, necrosis, tumor margin and adjacent normal subtypes. B) Overlay image of four representative N-glycan masses specific to each tissue subtype (adenocarcinoma, orange, Hex7dHex1HexNAc7, m/z 2743.9831; necrosis, cyan, Hex5HexNAc4, m/z 1663.5814; tumor margin, white, Hex8dHex1HexNAc7, m/z 2905.0359; adjacent normal, pink, Hex6dHex1HexNAc5, m/z 2174.7715). C) Segmentation analysis based on 85 unique N-glycans detected. D) Individual N-glycan heatmaps for tissue-subtype specific N-glycans m/z 1663.5814, m/z 2174.7715, m/z 2743.9831 and m/z 2905.0359.
A. Liver cancer
Perhaps no other disease has been more frequently associated with N-glycosylation changes than cancers of the liver, specifically hepatocellular carcinoma (HCC) (Mehta, Herrera, and Block 2015; Blomme et al. 2009; Comunale et al. 2006). Interest in liver cancer glycosylation stems from initial studies of alpha-fetoprotein (AFP), which determined core fucosylation as a critical indicator of hepatic dysfunction in HCC (Aoyagi et al. 1988; Zhao et al. 2020). Core fucosylated N-glycans, amongst other structural motifs, have since been associated with a number of circulating glycoproteins from patients with liver cancer (Norton et al. 2008; Comunale et al. 2009; 2010; Mengjun Wang et al. 2009; Comunale et al. 2011; DelaCourt et al. 2021). To this end multiple N-glycan IMS analyses have been directed towards HCC tissue cohorts in hopes of linking in situ expression of N-glycan species directly with those found in circulation. In 2018 West et al. surveyed the N-glycome of a large HCC cohort along with cirrhotic and non-transformed tissue samples (West et al. 2018). Highly branched N-glycans with one or more fucose modification were expressed abundantly in HCC samples while in cirrhotic and adjacent normal tissues their expression was detected minimally or not at all. Critically, this analysis was able to associate expression of the fucosylated tetra-antennary N-glycans Hex7dHex1HexNAc6 and Hex7dHex2HexNAc6 with significantly decreased survival times, demonstrating the functional relevance of N-glycan IMS findings to patient outcomes. A later orthogonal analysis confirmed that the majority of the detected N-glycans with at least one fucose residue in the HCC TMA, especially those associated with poor survival, are core-fucosylated (West et al. 2020). N-glycan imaging of hepatoblastoma (HepG2) cells in 2019 detected afucosylated, bi- and tri-antennary structures as well as the high mannose species Hex9HexNAc2, suggesting disparate glycoprofiles between liver cancers with different origins (Angel et al. 2019). Antibody panel-based N-glycan imaging, described in a later section, was initially developed with screening for liver malignancies in mind (Black, Liang, et al. 2019). The introductory demonstration of the technology featured α-1-antitrypsin (A1AT) and immunoglobulin G (IgG), but expanded panels have since included common serum proteins with well documented N-glycosylation changes in hepatocellular carcinoma. N-glycan signatures between healthy and cirrhotic human serum samples assessed via this platform were distinct, suggesting that future panel designs may facilitate the early detection of HCC transformation in the clinic by N-glycan IMS.
B. Breast cancer
The role of N-glycosylation in the development, maintenance and progression of breast cancer has been extensively studied, but there has been limited translation of results to clinical or diagnostic applications (Scott and Drake 2019). The emergence of imaging mass spectrometry for the investigation of N-glycosylation in breast cancer directly from clinical tissues, however, has shifted this dynamic. In 2018, in situ analyses of breast cancer tissue microarrays (TMAs) and whole tissue sections by MALDI-IMS revealed increases in polylactosamine containing N-glycans specific to the aggressive human epidermal growth factor receptor 2 (HER2) and triple negative (TN) subtypes (Scott, Casadonte, et al. 2019). Polylactosamine extensions are implicated in cellular adhesion, potentially giving mechanistic insight into the metastatic capacity of these subtypes (Srinivasan et al. 2009). This finding has been corroborated by an orthogonal study utilizing MALDI-IMS which implicated tetra-antennary N-glycans containing just a single N-acetyllactosamine branch with poor survival outcomes in breast cancer patients (Herrera et al. 2019). Interestingly, imaging mass spectrometry has revealed patterns of N-glycosylation which are specific to regions of necrosis in breast cancer tissues (Scott, Norris-Caneda, et al. 2019). N-glycans localizing to this area of cellular remnants were uniquely sialylated and lacked fucose residues when compared to the high mannose and multiply branched structures of the tumor tissue. Similar patterns have been observed in other cancer types, suggesting a possible role for N-glycosylation in the pathways leading to cellular death (Seyrek, Richter, and Lavrik 2019).
In 2019, N-glycan IMS of a mouse stage IV breast cancer cell line (4T1) detected increased expression of the high mannose Hex9HexNAc2 N-glycan (Angel et al. 2019). The same high mannose N-glycans seen in breast tumors and the aforementioned cultured cells have also been detected in the sera of breast cancer patients using MALDI-MS (de Leoz et al. 2011). In this context too, the Hex9HexNAc2 structure is especially elevated, which posits the incomplete trimming of pre-golgi N-glycan intermediates as a feature of breast cancer. Other MALDI-based studies of N-glycosylation in breast cancer serum have detected N-glycan signatures which delineates recurrent versus non-recurrent cancer, stage 1 breast cancer from healthy controls and signatures specific to the lymph node invasive subtype (J. W. Lee et al. 2020; S. B. Lee et al. 2020). Taken together, these studies on the complex glycosylation landscape of breast cancer by MALDI imaging mass spectrometry are potentially exploitable for clinical surveillance, early detection or delineation of subtypes.
C. Pancreatic cancer
The role of N-glycosylation in pancreatic ductal adenocarcinoma (PDAC) and associated neoplasia has been previously described (Munkley 2019). Early experiments using N-glycan MALDI-IMS demonstrated biantennary N-glycan structures with and without core fucose residues specific to areas of fibrosis in pancreatic cancer (Powers et al. 2014). Additionally, this study identified undecorated structures (those without fucose, sialic acid or bisecting GlcNAc residues) as characteristic of areas of tumor necrosis. A further exploration of PDAC N-glycosylation by the same group in 2015 corroborated the finding of biantennary N-glycans as features of fibrotic tissue regions in pancreatic cancer (Powers et al. 2015).
Building off these prior studies, an extensive imaging mass spectrometry analysis of the N-glycome of healthy and PDAC pancreatic tissue was reported in 2020 (McDowell et al. 2020). Normal human pancreas samples in this study were defined by an abundance of high mannose N-glycans localized to exocrine acinar tissue while inter- and intralobular ductal tissue regions featured core-fucosylated bi- and triantennary structures. A unique finding of this study was the detection of sulfated, bi-antennary N-glycans with terminal N-acetylgalactosamine residues which localized specifically to pancreatic islets of Langerhans. PDAC tissues from this cohort exhibited a variety of structural themes including increases in fucosylation, sialylation, branching, bisecting N-acetylglucosamines and polylactosamine extensions, amongst others. In addition to these findings, this study demonstrated the utility of a number of the N-glycan IMS adaptations, discussed further later in this review. Sialic acid stabilization by amidation revealed the distinct localizations of α2,3- versus α2,6-sialylated N-glycans in PDAC tissues, suggesting expression of disparate sialyltransferases across different tissue subtypes. Chemoenzymatic cleavage of N-glycans from these tissues using Endo F3 revealed that most structures in this PDAC cohort were predominantly, if not exclusively, core-fucosylated. N-glycan IMS is particularly relevant to pancreatic cancer, as the sole FDA-approved biomarker for pancreatic cancer is carbohydrate-antigen 19–9 (CA19–9), a four-sugar carbohydrate motif attached to both O- and N-linked glycans (E. Poruk et al. 2013). To this end, this analysis demonstrated the potential clinical relevancy for studies which multiplex N-glycan IMS with orthogonal imaging modalities. By combining expression data of select IMS-derived N-glycan masses from PDAC TMAs with immunofluorescence-assessed expression data of current and prospective PDAC biomarkers from the same TMAs, models for classification of pancreas tissue samples as normal or cancerous were built which outperformed modeling based on either dataset alone (Staal et al. 2019; Goonetilleke and Siriwardena 2007). These results suggest the future utility of cross-disciplinary, multi-marker panels which exploit N-glycan IMS in the early detection and diagnosis of pancreatic and other cancers.
D. Colorectal cancer
In colorectal cancer (CRC), N-glycosylation plays a well-defined oncological role (De Freitas-Junior and Morgado-Díaz 2016). Studies utilizing MALDI-TOF MS have analyzed N-glycans from colorectal cancer tissue homogenate, various CRC cell lines and patient serum (Balog et al. 2012; Holst, Deuss, et al. 2016; de Vroome et al. 2018). These studies detected a wide variety of N-glycan structural features specific to CRC tumor tissue, including high mannose, core fucosylation, sialylation, increased branching and even sulfation. These distinctions were born out between healthy and diseased samples but also across molecular subtypes and disease stage. Although the imaging functionality of MALDI-MS has only recently been applied to colorectal cancer, distinct spatial localizations of N-glycan species have been elucidated and the detected structural themes agree with the MALDI-TOF-derived findings from orthogonal sample sources above. In validation of the sialic acid stabilization by amidation protocol developed by the Wuhrer group, elevated levels of sialylated N-glycans were shown in colorectal cancer tissue samples (Holst, Heijs, et al. 2016). An additional proof-of-concept study, which demonstrated both N-glycan and peptide IMS from the same tissue sections, detected high mannose N-glycans localized to tumor regions in a representative colorectal cancer sample (Heijs et al. 2016). These initial findings were solidified in 2020 by a comprehensive survey of the N-glycome of stage II CRC by MALDI-IMS, which confirmed expression of sialylated and high mannose structures in cancer cells (Boyaval et al. 2020). This increased expression was shown to be propagated into the surrounding stroma, suggesting the modification N-glycosylation in the tumor microenvironment ahead of the tumor’s invasive front in advanced stage colorectal cancer.
E. Lung cancer
The role of glycosylation changes in lung cancer are well known and initial studies suggest the existence of disease-specific N-glycosylation (Lemjabbar-Alaoui et al. 2015). One of the proof-of-concept studies which demonstrated the applicability of MALDI-IMS to N-glycan analyses featured TMAs with lung adenocarcinoma cores matched to normal lung tissue. In these cores, high mannose N-glycan structures, especially Hex6HexNAc2, were found to be elevated (R. R. Drake et al. 2017). Although not a direct analysis of lung cancer, a recent study employed the use of radiation to recapitulate pre-neoplastic transformation which may lead to lung cancer in rhesus macaques (Carter et al. 2020). Areas of irradiation-induced hyperplasia in these lung samples analyzed by N-glycan IMS exhibited elevations in high mannose structures consistent with the human lung TMA cores from the aforementioned study, as well as fucosylated bi-antennary structures with and without bisecting GlcNAc residues. This elevation in high mannose N-glycans has also been observed in a MALDI-TOF MS analysis of lung adenocarcinoma patient sera across stages, positing that a further IMS evaluation of the lung cancer N-glycome may discover additional N-glycan features which map to serum expression for possible use as biomarkers (Lattová et al. 2020).
F. Ovarian cancer
Glycosylation changes in ovarian cancer were first observed in the mid 1960’s (Garcia-Bunuel and Monis 1964). Of recent interest is the translation of N-glycosylation analytical applications to clinical strategies for ovarian cancer, as N-glycans are known to be heavily involved in the oncogenesis and maintenance of this disease (Guo and Abbott 2015; Briggs, Condina, Klingler-Hoffmann, et al. 2019). Consistent with other types of cancers, a 2016 study detected increased expression of high mannose N-glycans in ovarian cancer tumor tissue as compared to normal tissue from the same samples (Everest-Dass et al. 2016). This same analysis also detected increases in hybrid-type structures specific to areas of intervening stroma in this cohort. All N-glycan structures in these experiments were orthogonally verified by LC-MS/MS. N-glycans extracted and purified from FFPE ovarian cancer tissue samples for MALDI-TOF analysis by the Li group supported the specificity of high mannose N-glycan expression in ovarian tumor tissue (Chen et al. 2017). This analysis also found that a seven-glycan panel consisting of four high mannose and three fucosylated complex structures was able to delineate epithelial ovarian cancer from healthy controls, while a similar yet distinct N-glycan panel could distinguish specific ovarian cancer grades. N-glycan IMS analyses have also been able to differentiate early- versus late-stage ovarian cancer using specific N-glycosylation signatures (Briggs, Condina, Ho, et al. 2019). The high mannose N-glycan Hex9HexNAc2 was detected abundantly in stage III ovarian cancer samples as compared to stage I tissues. Additionally specific to late-stage ovarian cancer were fucosylated, bisecting and sialylated complex species. These glycosylation changes were not only detected in the whole tissue cohort analyzed in this report, but in corresponding TMAs which lends additional statistical power to these findings. Taken together these early N-glycan IMS studies suggest glycosylation changes as a feature of ovarian cancer potentially exploitable for clinical detection and surveillance of this disease.
G. Renal cancer
The first publication of a standardized N-glycan MALDI-IMS workflow was demonstrated on a normal kidney tissue section which contained both medulla and cortex tissue regions (Powers et al. 2013). Although the technology was in its infancy, the few N-glycan analytes detected were localized to specific tissue subtypes. A subsequent analysis of FFPE murine kidneys demonstrated similar results (Gustafsson et al. 2015). This initial work was furthered by a more comprehensive 2020 study of clear cell renal cell carcinoma (ccRCC), which spatially profiled N-glycosylation in a cohort of normal kidney and renal cell carcinoma tissues (Richard R. Drake, McDowell, et al. 2020). A defining feature of healthy kidney tissue from this study was broad structural diversity between tissue subtypes, with bisecting, high mannose and fucosylated structures localized to the cortex while bi- and tri-antennary N-glycans with core fucose residues localized to medullary tissue. Interestingly, the interface between cortex and medulla also had a distinct glycosylation signature, defined by multiply-fucosylated structures which lacked bisecting GlcNAc residues. A high spatial resolution analysis from this study detected tetra-antennary N-glycans with and without sialic acid modifications specific to kidney glomeruli. These normal-associated structural features were conspicuously absent from ccRCC tumor tissue, which exhibited increased expression of tetra-antennary N-glycans with polylactosamine extensions of varying length. The fibrillar capsule surrounding a Fuhrman grade 2 ccRCC tumor was characterized by a bi-antennary, monosialylated N-glycan. For comparison, a non-ccRCC polycystic kidney disease (PKD) tissue was also analyzed, which again showed a disparate N-glycome as well as distinct glycosylation pattern differences between cyst fluid and tissue regions. N-glycan IMS findings were mapped directly to transcriptomic profiling by RNA sequencing from a prior study, which revealed altered expression of fucosyltransferases FUT3 and FUT6 which attach fucose residues via an a1,3 linkage to N-glycan termini (von Roemeling et al. 2014; Neely et al. 2016; Schneider, Al-Shareffi, and Haltiwanger 2017). The decreased expression of these glycosyltransferases in ccRCC tissue across all stages matches less abundant outer-arm fucosylation of tumor N-glycan structures observed by IMS. Taken together the findings from this multi-omic study comprise the most comprehensive analysis to-date of the healthy and diseased renal N-glycome.
IV. Alternate enzymes and chemical modifications
A. Amidation-amidation to stabilize sialic acids
As previously mentioned, characterization of N-glycan isomers can be challenging using traditional N-glycan IMS workflows. Particularly difficult is the separation of sialylated N-glycan isomers (Nishikaze 2019). Sialic acid monomers are attached to terminal galactose residues on N-glycan branched arms by the β-galactoside sialyltransferases (STGAL) family of enzymes, which catalyze the formation of either α2,3 (ST3GAL1) or α2,6 (ST6GAL1) anomeric linkages (Harduin-Lepers et al. 2001). Adding to this complexity is the labile nature of sialic acid residues, which often decompose in-source during MALDI ionization due to a combination of high vacuum and heat (Harvey 1999). Improved MALDI source configurations have improved overall retention of sialic acids, but not completely (Richard R. Drake, West, et al. 2018). To facilitate analysis by mass spectrometry, multiple chemical derivatization approaches have been developed to stabilize these residues (Xin Liu et al. 2010; Sekiya, Wada, and Tanaka 2005; Powell and Harvey 1996; Reiding et al. 2014; de Haan et al. 2020). Relevant to N-glycan IMS are the strategies that take advantage of the differential chemical properties between α2,3- and α2,6-linked anomers to stabilize each as a distinct moiety. In amidation syntheses developed by the Wuhrer group, α2,3- and α2,6-linked sialic acid residues are modified with amide and dimethyl amine functional groups, respectively, by sequential incubations with 1-ethyl-3(3-dimethylaminopropyl)carbodiimide (EDC)/1-hydroxybenzotriazol (HOBt) and ammonia (Holst, Heijs, et al. 2016; Pongracz et al. 2021) (Figure 7A). These reactions are adapted to use directly on tissue in situ prior to traditional MALDI-IMS N-glycan workflows. The differential incorporation of these moieties introduces well-defined mass shifts for each linkage which aid in their identification by mass spectrometry. This chemical amidation strategy has been successfully used in situ to characterize sialylation differences between healthy and diseased tissue (McDowell et al. 2020; Boyaval et al. 2020). Of interesting note from the aforementioned studies is the finding that differential α2,3 versus α2,6 sialylation of the same base N-glycan structure drove disparate spatial localization within the same tissue, suggesting the compartmentalization of specific sialyltransferases to distinct tissue subtypes or disease morphologies (Figure 7B–D).
Figure 7. Sialic acid stabilization and linkage differentiation by on-tissue amidation chemistry.
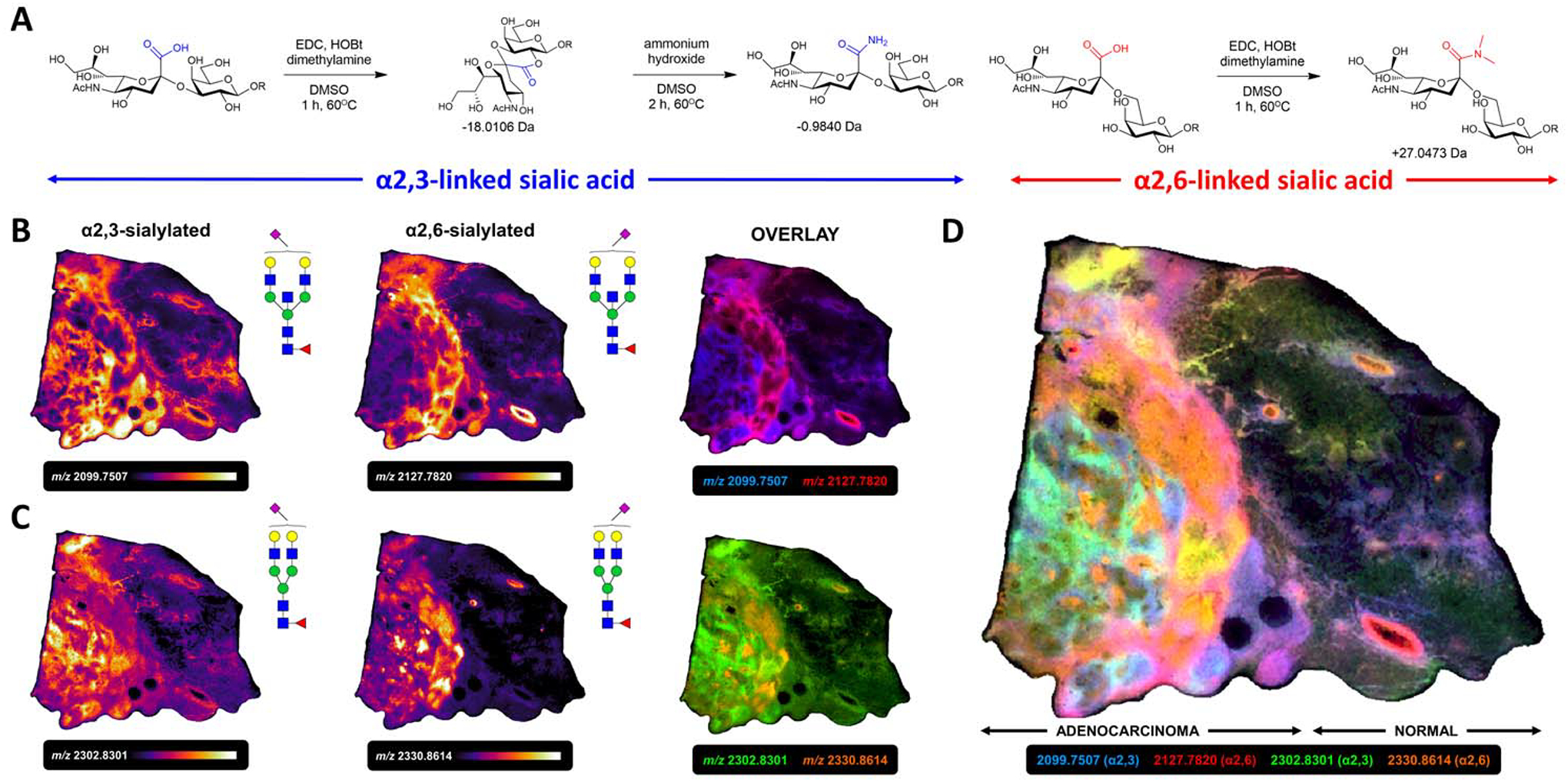
A) Derivatization of α2,3- and α2,6-linked sialic acid anomers with amide (−0.9840 m.u.) and dimethyl amine (+27.0473 m.u.) functional groups. B) α2,3 (m/z 2099.7507) and α2,6 (m/z 2127.7820) linkage isomers of Hex5dHex1HexNAc5Neu5Ac1 drove distinct spatial localization. C) α2,3 (m/z 2302.8301) and α2,6 (m/z 2330.8614) linkage isomers of Hex5dHex1HexNAc4Neu5Ac1. D) Overlay of 4 α2,3- and α2,6-sialylated N-glycans suggests compartmentalization of specific sialyltransferases.
B. Alternate glycosidases
Although PNGase F is the gold standard for enzymatic deglycosylation of N-linked glycoproteins and N-glycan IMS, there are a many different glycosidases reported for N-glycan analysis applications (M. S. Kim and Leahy 2013). Most of these glycosidases have substrate specificities for other parts of N-linked glycan structure, or specific to individual glycoprotein classes in the immunoglobulin family, Incorporation of these enzymes requires minimal modification of existing IMS workflows and allows for the targeted analysis of specific N-glycosylation features. One note, a key consideration when evaluating new glycosidases for spraying on tissue is to identify any salts or stabilizers that may be present in the formulation that could interfere with laser ionization efficiency.
One non-PNGase F example is endoglycosidase F3, which has already been incorporated into N-glycan MALDI IMS workflows alone or combined with PNGase F (West et al. 2020). Similar to sialylation isomers of N-glycans, fucosylation isomers are especially difficult to delineate. Fucose monomers are linked to the growing N-glycan by the fucosyltransferase (FUT) family of enzymes. Core fucose residues, those bonded to the terminal N-acetylglucosamine linked to the glycoprotein’s asparagine, are attached exclusively via an α1,6 linkage by FUT8. Outer arm fucoses are attached to branched N-glycan antennae via α1,2 linkages by FUT1,2 or by α1,3 linkages via FUT3,4,6,7,9–11 (Schneider, Al-Shareffi, and Haltiwanger 2017). Endoglycosidase F3 (Endo F3) hydrolyzes the glycosidic linkage between the N-acetylglucosamine residues of the N-glycan chitobiose core preferentially in the presence of an α1,6-linked core fucose, thus freeing for detection only N-glycan structures that are core fucosylated (Plummer, Phelan, and Tarentino 1996; West et al. 2020). Those N-glycan isomers which are exclusively outer-arm fucosylated are not cleaved and therefore not detected. The process introduces a distinct Δ349 m.u. mass shift loss from the parent N-glycan structure, aiding in the specific detection of these Endo F3-cleaved glycans. A fucose and GlcNAc residue remain on the glycoprotein, providing a searchable peptide modification for glycoproteomic identification of core-fucosylated glycoproteins (Ma et al. 2018). Recent analyses incorporating Endo F3 into standard N-glycan IMS workflows have demonstrated the specific detection of fucosylation isomers in situ (West et al. 2020). The distinction between core and outer-arm fucosylation is an important one, as the location of fucose residues can be a critical determinant of glycoprotein function (Tu, Lin, and Lin 2013; H. I. Kim et al. 2013). Suggestive of their disparate biological functions, core versus outer arm fucosylation isomers often display distinct spatial localization in both healthy and diseased tissues.
C. O-glycans
Although this review is targeted towards N-glycan imaging mass spectrometry, it is worth briefly discussing O-glycan strategies as these carbohydrates make up a large portion of the glycocalyx and are equally relevant to biological and pathological processes in the cell (Varki 2017). Unlike N-glycan analyses, which enjoy a variety of enzymatic and chemical cleavage reagents, O-linked glycan analysis suffers from a lack of reliable enzymes to free these species for detection. This is due in part to the heterogeneity of the modifying sugar directly linked to the glycoprotein’s serine/threonine residue, which could be an N-acetylgalactosamine on mucin glycoproteins or an N-acetylglucosamine, galactose, mannose or fucose on non-mucin glycoproteins (Darula and Medzihradszky 2018). The lack of a consensus motif excludes the broad cleavage of O-glycans by just a single enzyme, making the design of streamlined analysis workflows, like those for N-glycan IMS, more challenging.
Strategies to focus on a subset of O-glycans, namely O-GalNAc-based structures, have been more successful (Saldova and Wilkinson 2020). Endo-α-N-acetylgalactosaminidase (O-glycanase) releases core 1 (Gal[β1,3]GalNAc[α]Ser/Thr) and core 3 (GlcNAc[β1,3]GalNAc[α]Ser/Thr) O-glycans from their glycoprotein carriers, but its efficiency is reduced when the O-glycan contains additional saccharide units beyond the core structure. Both reductive and non-reductive hydrolyses of the O-glycosidic linkage by β-elimination represent alternative chemical approaches to liberate O-glycans, although side reactions with these reagents may generate additional ions that obfuscate analytes of interest (Goso, Tsubokawa, and Ishihara 2009). O-glycans released via these strategies are typically detected after chemical derivatization using LC-MS/MS approaches, although detection by MALDI-TOF of purified O-glycans spotted on target plates has been described (Morelle et al. 2009). An alternative approach for O-glycan analysis is use of a novel, bacteria-derived mucin protease, typified by StcE (secreted protease of C1 esterase inhibitor) (Malaker 2019). These mucin proteases, reported in the literature and also commercially available, cleave at O-glycosylated sites. Detection of the resulting mucin glycopeptides by MALDI IMS could be amenable a workflow similar to those for on-tissue tryptic digestion for imaging mass spectrometry of peptides (Malaker et al. 2019). In situ analyses of released O-glycans and mucin O-glycopeptides by IMS is likely possible, but still requires extensive further optimizations of the release methods and glycosidase conditions specific for tissue IMS approaches.
V. Expanded approaches
A. Tissue microarrays
Tissue microarrays are single pathology slides to which multiple, small tissue punches, collected from biopsies or surgical resection, are affixed in a manner similar to the preparation of clinical FFPE tissue sections (Jawhar 2009). Histological staining of resected tissues allows clinical pathologists to sample healthy or diseased tissue subtypes of interest and compile samples from large numbers of patients into the same TMA block for the creation of individual FFPE TMA sections. Often, samples will be taken of both healthy and malignant tissue from the same patient, resulting in matched pairs for analysis. While the spatial information aspect of imaging mass spectrometry is less relevant to these analyses, TMAs provide the opportunity to assay N-glycosylation information from hundreds of patient samples in a high throughput manner (Powers et al. 2014) (Figure 8A). For example, one tissue microarray representing over 100 patient biopsies can be analyzed via N-glycan MALDI-IMS in the same (or less) time as it takes to run a single whole tissue section. Critically, microarray formats enable the cost effective and rapid high throughput screening required for biomarker discovery (Hewitt 2012). An additional advantage of this tissue specimen layout is that the large sample sizes afforded by TMAs provides adequate statistical power for complex analyses of differences in N-glycosylation between healthy and diseased tissue types (Xueli Liu et al. 2004). In multiple studies, N-glycan imaging of tissue microarrays has demonstrated utility in the screening of large patient cohorts for N-glycan expression differences in a variety of cancers (Powers et al. 2015; West et al. 2020; 2018; Richard R. Drake, McDowell, et al. 2020; Conroy et al. 2020; Herrera et al. 2019; Scott, Casadonte, et al. 2019). Like whole-tissue sections, N-glycan IMS data from TMAs may be multiplexed with immunohistochemical, immunofluorescence or autofluorescence microscopy for comparison with biomarkers or other molecules of interest (McDowell et al. 2020).
Figure 8. Expanded N-glycan IMS approaches for alternative clinical and laboratory sample types.
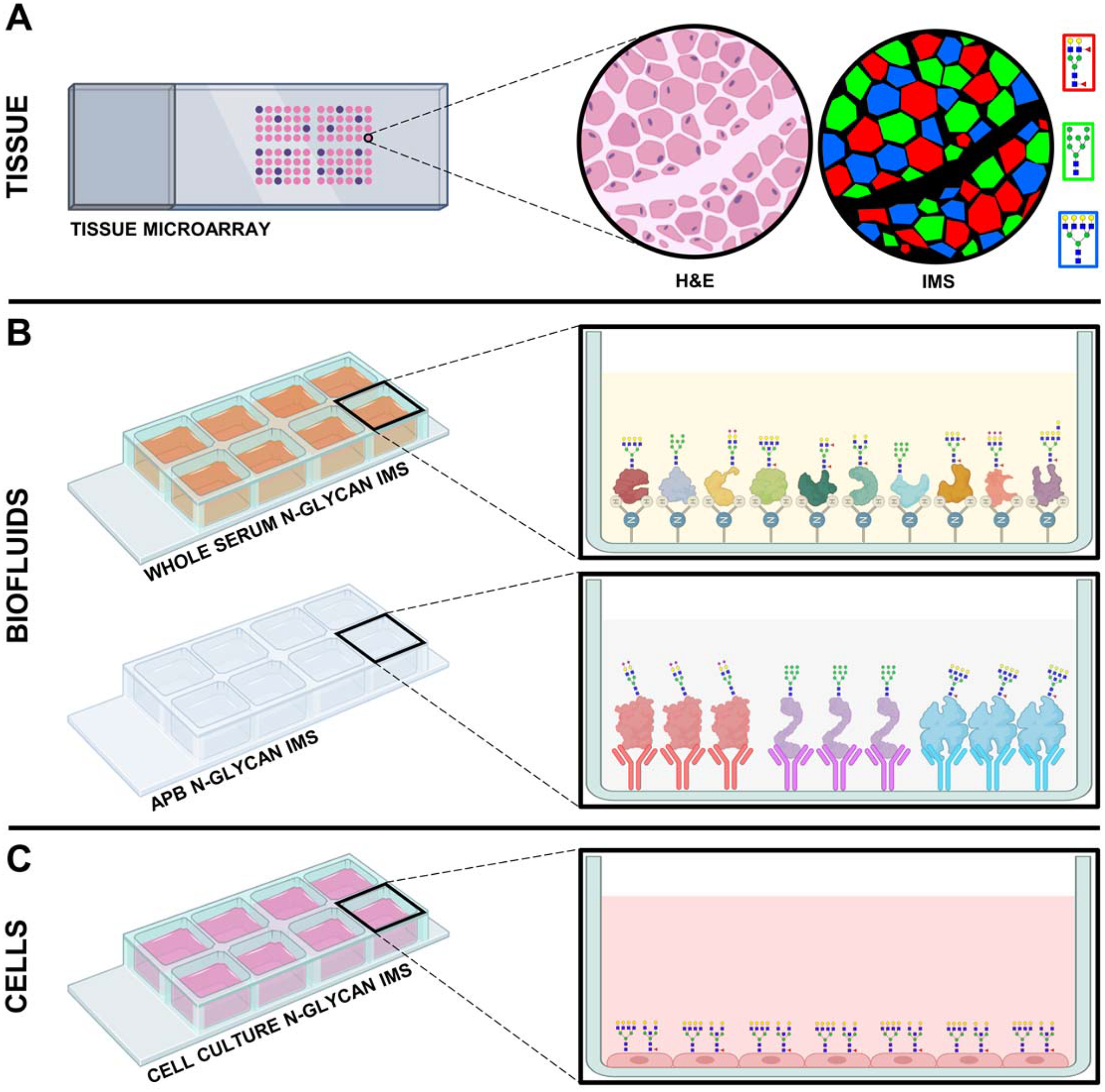
A) N-glycan imaging mass spectrometry of tissue microarray cores correlates histopathologically-annotated tissue features with N-glycan analytes in a high-throughput fashion. B) N-glycan imaging of biofluids. Whole-serum N-glycan analysis examines amine reactive slide-captured serum total protein, while APB N-glycan imaging employs targeted N-glycan analysis of specific glycoproteins through high-affinity capture by slide-bound antibodies. C) N-glycan imaging mass spectrometry of cultured cells fixed to a glass microscope slide.
B. Biofluids
An emerging role for the field of imaging mass spectrometry is that of clinical diagnostics for the specific detection of disease. The utilization of IMS for carbohydrates to assess healthy versus pathological states is potentiated by the fact that all current FDA-approved cancer biomarkers are glycoproteins or are themselves glycan motifs (Adamczyk, Tharmalingam, and Rudd 2012; Kailemia, Park, and Lebrilla 2017). The adaptation of N-glycan imaging strategies to other clinical sample types is an attractive alternative to FFPE or FF tissues. By far the most clinically accessible medium for analysis are patient biofluids, namely sera and plasma, which can be collected via minimally invasive blood draws as part of routine diagnostic screenings. Discussed below are two novel adaptations of N-glycan IMS towards clinical biofluids (Figure 8B).
5.2.1. Biofluid N-glycan profiling
Human serum and plasma contains thousands of proteins and metabolites that are reflective of the disease state and overall health of the donor patient. (Anderson and Anderson 2002). These complex mixtures are comprised of a relatively high-abundance (10X order of magnitude) of a small number of glycoproteins which have the potential to obfuscate detection of low-abundance yet potentially clinically relevant analytes, thus making these matrices sometimes difficult to analyze (Qian et al. 2006). N-glycosylation of human serum is traditionally assessed by high- or ultra-performance liquid chromatography (HPLC/UPLC), capillary gel electrophoresis (CGE) or MALDI-TOF of purified N-glycans (Stöckmann et al. 2015; Ruhaak et al. 2010; Vreeker et al. 2018). While these technologies have helped elucidate N-glycan functions in disease, lengthy derivatization, labeling and cleanup steps in these workflows make them incompatible with the rapidity required for clinical diagnoses.
To address the necessity for rapid, specific diagnostics of serum glycosylation, a novel imaging mass spectrometry approach has been developed (C. Blaschke et al. 2020). A single microliter of patient serum is diluted in sodium bicarbonate and spotted on amine-reactive hydrogel-coated glass slides to immobilize serum glycoproteins, followed by desalting and delipidating through a series of organic washes and drying under desiccation. The dried, serum-spotted slides are then processed with the standard N-glycan MALDI-IMS workflow described in section 2.1.1, starting with enzymatic release of N-glycans from serum proteins by PNGase F. Analysis by MALDI-FTICR or MALDI-QTOF instruments has demonstrated the specific detection of over 75 N-glycan structures from human serum using this method. This new imaging platform is also compatible with Endo F3 and sialic acid amidation approaches traditionally applied to tissue samples as described above. MALDI-MS is already routinely used in the clinical laboratory for identification of microbial samples via analyte “barcodes” corresponding to particular pathological signatures (Kostrzewa 2018; Richard R. Drake, Boggs, and Drake 2011; Mellmann et al. 2009). It is foreseeable that this biofluid IMS method or some further iteration of the technology could be used to generate N-glycan barcodes for the identification of various diseases in the clinic.
5.2.2. Antibody panel-based N-glycan IMS
In 2019 a more targeted imaging mass spectrometry strategy for the identification of N-glycosylation signatures on common serum glycoproteins was developed (Black, Angel, et al. 2019; Black, Liang, et al. 2019). Antibody Panel-Based N-glycan Imaging (APB) takes advantage of established slide-based antibody array formats (Jones et al. 2020) repurposed for mass spectrometry analysis of N-glycans.
APB N-glycan imaging capitalizes on decades of antibody engineering, using high binding affinity anti-human antibodies to specifically capture individual serum glycoproteins (Black, Angel, et al. 2019; Black, Liang, et al. 2019). Briefly, antibodies targeting specific serum glycoproteins are manually spotted on nitrocellulose slides then washed and blocked with bovine serum albumin (BSA). Incubation with human serum samples results in the capture from the total protein pool of only the glycoprotein antigens specific to the antibody spots on the array. Initial demonstrations of this technology focused on six abundant serum proteins, alpha-1 antitrypsin (A1AT), haptoglobin, hemopexin, immunoglobulin G (IgG), low molecular weight kininogen and transferrin, although any combination of antigens of interest may be used dependent on the availability of an appropriate antibody. Similar to the biofluid imaging strategy described above, slides with captured glycoproteins can subsequently be incorporated into MALDI-IMS workflows at the point of enzymatic deglycosylation (Black, Liang, et al. 2019). This method has demonstrated the specific detection of over 20 individual glycan species with high sensitivity and the capability to detect ~3 fmol of glycan from just 10 ng of captured protein. Initial results from analyses of healthy versus cirrhotic human serum revealed differential glycosylation signatures between healthy and diseased samples. Currently under development is a neural network framework for the analysis of N-glycan IMS data from these panels which seeks to assign specific glycosylation barcodes to disparate disease states. Like whole-serum N-glycan IMS, further development could see this technology integrated into the clinical laboratory.
C. Cell culture
Cultured cells are one of the most frequently-used tools for the study of human disease, as they model the basic biological processes underlying both healthy and diseased states and are inherently manipulatable for experimental design (Gillet, Varma, and Gottesman 2013). Cultured cells have long been used as model systems to probe the biosynthesis, function and regulation of N-glycans and glycosylated proteins (Montreuil 1995). Comprehensive studies of N-glycans in cell culture conducted today typically utilize LC-MS or MALDI-TOF MS approaches (Aich, Lakbub, and Liu 2016; Shajahan et al. 2017; Parry et al. 2006).
In 2019, a novel approach for the analysis of N-glycans directly from cultured cells was developed utilizing imaging mass spectrometry (Angel et al. 2019; Angel, Mehta, and Drake 2021) (Figure 8C). Cells cultured on standard chambered glass slides were fixed and delipidated in organic solvents prior to microscopy imaging to confirm appropriate cellular density. Using conditions optimized specifically for cultured cells, the standard N-glycan IMS processing workflow was applied to the cells on the slide. Protocol modifications include the adjustment of both PNGase F and CHCA spraying parameters and the incorporation of a post-matrix ammonium phosphate spray to limit the formation of signal-suppressing matrix clusters (Chubatyi and McEwen 2015; Ucal and Ozpinar 2018). Analyses of a panel of human and animal cell lines cultured and prepared using this method by MALDI-FTICR MS detected over 70 N-glycoforms. This assay is innovative in a number of ways. Cells for this method can be cultured using standard approaches without requiring further optimization. The inclusion of media blanks for background signal subtraction excludes obfuscation by glycans derived from fetal bovine serum (FBS) (Carr, Huddleston, and Bean 1993; Lin, Franc, and Heck 2018). Incorporation of Coomassie blue staining after IMS data acquisition to infer total protein expression allows for both intra-and inter-run normalization, making this method inherently quantitative (Bradford 1976; Butt and Coorssen 2013). Expanded use of this method allows for the monitoring of N-glycan turnover through stable isotopic labeling of amino acids in cell culture (SILAC) using an isotopic detection of amino sugars with glutamine (IDAWG) approach (Orlando et al. 2009). N15 labeling in this way could also be used to measure cellular N-glycosylation response to stress or therapeutics interventions induced with a variety of chemical or pharmaceutical antagonists (Mechref et al. 2013; Xu et al. 2019; Delafield and Li 2020). Consequently, revealing the N-glycan profiles of cultured cell lines allows assessment of how faithfully these models recapitulate glycosylation seen in clinically-derived tissue specimens and thus in human biology. Linking these glycosylation states will help better understand which cell lines are most representative of a given disease state for studying N-glycosylation in both basic science and therapeutic contexts.
VI. Conclusions
The specific adaptation of imaging mass spectrometry as a tool for N-glycan analysis has exponentially expanded glycomics research efforts into organ, tissue and cell-type specific glycosylation studies. Capturing the in situ spatial component of a particular N-glycan’s expression has led to insights into the multifaceted role these molecules play in a variety of diseases, with potential for multiple clinical applications. Discussed in this review are the most common neoplasias studied by N-glycan IMS, summarized in Table 1. The broad applicability of the approach suggests that any type of cancer may be investigated, as well as non-cancer diseases, assuming availability of appropriate clinical specimens. Emphasis on the MALDI IMS application to human disease was discussed, but tissue and biofluids from any biological source could be used with these workflows. For many organisms, the N-glycomes of most organisms, as well as individual organs in humans, remain largely uncharacterized. While the basic structural features of high mannose and bi-antennary glycans are fairly conserved across all species, there are many known species-specific glycans synthesized by a broader class of glycosyltransferases and glycosidases than that found in humans. For example, different sialic acid structural variants are prevalent in non-human mammals (Varki 2008; Irie et al. 1998). Analysis of non-mammalian N-linked glycosylation by N-glycan IMS is a wide-open space for application of this method.
No single MS method is sufficient to fully characterize the structural complexities of N-glycans, However, in regard to providing N-glycan co-localization information with tissue histopathology, N-glycan IMS approaches have proven to be highly effective and informative. The utility of N-glycan IMS to distinguish isomers, novel glycans and larger structures will continue to evolve with improvements in instrumentation, data analysis and incorporation of other approaches. This includes use of other ionization approaches, in particular atmospheric pressure methods like DESI and IR-MALDESI, hybrid instruments and increased use of integrated ion mobility separation (Bereman, Williams, and Muddiman 2007; Pu et al. 2016; Škrášková et al. 2016; Lane et al. 2019; Pace and Muddiman 2020). N-glycan IMS is amenable to multiplexing with immunohistochemical and immunofluorescence imaging, generating multi-modal data for both glycan and protein markers of interest which has demonstrated utility in the characterization of disease states (Levenson, Borowsky, and Angelo 2015; Tan et al. 2020; McDowell et al. 2020; Clift, Mehta, et al. 2020; Yagnik et al. 2021). Linkage of N-glycan IMS with more advanced molecular imaging modalities like mass cytometry/mibiTOF (Keren et al. 2019), and in the other resolution direction, clinical MRI combinations are being pursued (Abdelmoula et al. 2019; Richard R. Drake, Angel, et al. 2020; Basu and Agar 2021).
While initially developed for N-glycans, glycan IMS is adaptable to a wide range of enzymatic and chemical release agents, which expands the repertoire of detectable analytes beyond N-linked structures to O-linked glycans, glycosaminoglycans and glycolipids amongst other species (Morelle et al. 2009; Goto-Inoue, Hayasaka, and Setou 2010; Malaker et al. 2019; Saldova and Wilkinson 2020; Clift, Drake, et al. 2020). Alternative endoglycosidases like Endo F3, Endo H and Endo S recognize distinct epitopes on glycans, cleaving them in can be advantageous for the delineation of specific glycan anomeric linkages, specific glycan classes and glycans specific to certain proteins or cell populations (Plummer, Phelan, and Tarentino 1996; Collin et al. 2001; Freeze and Kranz 2010; West et al. 2020). A recent study incorporating isoamylase, to free glycogen polymers for detection, into the N-glycan IMS workflow has demonstrated a novel use of imaging mass spectrometry to link glucosamine metabolism and storage to the biosynthesis of N-linked glycans (Sun et al. 2021). As more specific enzymes and chemistries are discovered or engineered, an even wider array of carbohydrate analytes will become available for detection by IMS.
Together, glycan imaging mass spectrometry approaches have deepened our understanding of the glycobiome and the complex biology and chemistry which governs the synthesis, function and physiological effects of these carbohydrates. The continued evolution and expanded adaptations of this technology promises further insight into the biochemical underpinnings of health and disease in the context of glycosylation. Ultimately, in the near future glycan imaging mass spectrometry platforms may be integrated into clinical laboratories for disease surveillance and early discovery by the detection of specific glycan molecular signatures.
Acknowledgements
This research was supported in part National Institutes of Health grants U01CA242096 (R.R. Drake, A.S. Mehta, P.M. Angel), R41DK124058 (A.S. Mehta), U01CA226052 (A.S. Mehta, R.R. Drake), T32GM132055 (C.T. McDowell), U01CA168896 (B.B. Haab), P30DK123704 (R.R. Drake, P.M. Angel), the South Carolina Smart State Centers of Economic Excellence (R.R. Drake, A.S. Mehta) and the Biorepository and Tissue Analysis Shared Resource, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313).
VII Abbreviations
- A1AT
α-1-antitrypsin
- AFP
α-fetoprotein
- APB
Antibody panel-based
- BSA
Bovine serum albumin
- CA19–9
Carbohydrate-antigen 19–9
- ccRCC
Clear cell renal cell carcinoma
- CCS
Collisional cross-sectional area
- CGE
Capillary gel electrophoresis
- CHCA
α-cyano-4-hydroxycinnamic
- CID
Collision-induced dissociation
- CRC
Colorectal cancer
- DAN
1,5-diaminonaphthalene
- DESI
Desorption electrospray ionization
- DHB
2,5-dihydroxybenzoic acid
- dHex
Deoxy hexose
- DTIMS
Drift tube ion mobility separation
- ECM
Extracellular matrix
- EDC
1-ethyl-3(3-dimethylaminopropyl)carbodiimide
- Endo F3
Endoglycosidase F3
- ER
Endoplasmic reticulum
- ESI
Electrospray ionization
- ETD
Electron-transfer dissociation
- FAIMS
High-field asymmetric ion mobility separation
- FBS
Fetal bovine serum
- FDA
Food and Drug Administration
- FF
Fresh-frozen
- FFPE
Formalin-fixed paraffin-embedded
- FT-ICR
Fourier transform ion cyclotron resonance
- Fuc
Fucose
- FUT
Fucosyltransferase
- GAG
Glycosaminoglycan
- Gal
Galactose
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- Glu
Glucose
- HCC
Hepatocellular carcinoma
- HCD
Higher-energy collisional dissociation
- HER2
Human epidermal growth factor receptor 2
- Hex
Hexose
- HexNAc
N-acetylhexosamine
- HOBt
1-hydroxybenzotriazole
- HPLC
High-performance liquid chromatography
- IDAWG
Isotopic detection of amino sugars with glutamine
- IF
Immunofluorescence
- IgG
Immunoglobulin G
- IHC
Immunohistochemistry
- IR
Infrared
- ITO
Indium tin oxide
- IMS
Imaging mass spectrometry
- LC-MS
Liquid chromatography-coupled mass spectrometry
- MALDESI
Matrix-assisted laser desorption electrospray ionization
- MALDI
Matrix assisted laser desorption/ionization
- Man
Mannose
- MIBI-TOF
Multiplexed ion beam imaging by time of flight
- MRI
Magnetic resonance imaging
- MS
Mass spectrometry
- Nd:YAG
Neodymium-doped yttrium aluminum garnet
- Neu5Ac
N-acetylneuraminic acid
- PGC
Porous graphitized carbon
- PDAC
Pancreatic ductal adenocarcinoma
- PKD
Polycystic kidney disease
- PNGase F
Peptide N-glycosidase F
- QTOF
Quadrupole time-of-flight
- SILAC
Stable isotopic labeling of amino acids in cell culture
- SIMS
Secondary ion mass spectrometry
- StcE
Secreted protease of C1 esterase inhibitor
- STGAL
B-galactoside sialyltransferase
- THAP
2′,4′,6′-trihydroxyacetophenone monohydrate
- TIMS
Trapped ion mobility separation
- TMA
Tissue microarray
- TN
Triple negative
- TOF
Time-of-flight
- TWIMS
Traveling wave ion mobility separation
- UPLC
Ultrahigh-performance liquid chromatography
References
- Abdelmoula Walid M., Regan Michael S., Lopez Begona G.C., Randall Elizabeth C., Lawler Sean, Mladek Ann C., Nowicki Michal O., et al. 2019. “Automatic 3D Nonlinear Registration of Mass Spectrometry Imaging and Magnetic Resonance Imaging Data.” Analytical Chemistry 91 (9): 6206–16. 10.1021/acs.analchem.9b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk Barbara, Tharmalingam Tharmala, and Rudd Pauline M.. 2012. “Glycans as Cancer Biomarkers.” Biochimica et Biophysica Acta - General Subjects. 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Aich Udayanath, Lakbub Jude, and Liu Aston. 2016. “State-of-the-Art Technologies for Rapid and High-Throughput Sample Preparation and Analysis of N-Glycans from Antibodies.” ELECTROPHORESIS 37 (11): 1468–88. 10.1002/elps.201500551. [DOI] [PubMed] [Google Scholar]
- Aichler Michaela, and Walch Axel. 2015. “MALDI Imaging Mass Spectrometry: Current Frontiers and Perspectives in Pathology Research and Practice.” Laboratory Investigation 95 (4): 422–31. 10.1038/labinvest.2014.156. [DOI] [PubMed] [Google Scholar]
- Alexandrov Theodore. 2012. “MALDI Imaging Mass Spectrometry: Statistical Data Analysis and Current Computational Challenges.” BMC Bioinformatics. BioMed Central. 10.1186/1471-2105-13-s16-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov Theodore, Becker Michael, Deininger Sören Oliver, Ernst Günther, Wehder Liane, Grasmair Markus, Von Eggeling Ferdinand, Thiele Herbert, and Maass Peter. 2010. “Spatial Segmentation of Imaging Mass Spectrometry Data with Edge-Preserving Image Denoising and Clustering.” Journal of Proteome Research 9 (12): 6535–46. 10.1021/pr100734z. [DOI] [PubMed] [Google Scholar]
- Alocci Davide, Mariethoz Julien, Gastaldello Alessandra, Gasteiger Elisabeth, Karlsson Niclas G., Kolarich Daniel, Packer Nicolle H., and Lisacek Frédérique. 2019. “GlyConnect: Glycoproteomics Goes Visual, Interactive, and Analytical.” Journal of Proteome Research 18 (2): 664–77. 10.1021/acs.jproteome.8b00766. [DOI] [PubMed] [Google Scholar]
- Alphonsus CS, and Rodseth RN. 2014. “The Endothelial Glycocalyx: A Review of the Vascular Barrier.” Anaesthesia. Blackwell Publishing Ltd. 10.1111/anae.12661. [DOI] [PubMed] [Google Scholar]
- Anderson N. Leigh, and Anderson Norman G.. 2002. “The Human Plasma Proteome: History, Character, and Diagnostic Prospects.” Molecular & Cellular Proteomics : MCP. American Society for Biochemistry and Molecular Biology. 10.1074/mcp.R200007-MCP200. [DOI] [PubMed] [Google Scholar]
- Anderton Christopher R., and Gamble Lara J.. 2016. “Secondary Ion Mass Spectrometry Imaging of Tissues, Cells, and Microbial Systems.” Microscopy Today 24 (2): 24–31. 10.1017/s1551929516000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel Peggi M., Baldwin H. Scott, Sen Danielle Gottlieb, Su Yan Ru, Mayer John E., Bichell David, and Drake Richard R.. 2017. “Advances in MALDI Imaging Mass Spectrometry of Proteins in Cardiac Tissue, Including the Heart Valve.” Biochimica et Biophysica Acta - Proteins and Proteomics. Elsevier B.V. 10.1016/j.bbapap.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel Peggi M., Mehta Anand S., and Drake Richard R.. 2021. “Array-Based N-Glycan Profiling of Cells in Culture.” In Methods in Molecular Biology, 2271:331–42. Humana Press Inc. 10.1007/978-1-0716-1241-5_23. [DOI] [PubMed] [Google Scholar]
- Angel Peggi M., Saunders Janet, Clift Cassandra L., White-Gilbertson Shai, Voelkel-Johnson Christina, Yeh Elizabeth, Mehta Anand, and Drake Richard R.. 2019. “A Rapid Array-Based Approach to N-Glycan Profiling of Cultured Cells.” Journal of Proteome Research 18 (10): 3630–39. 10.1021/acs.jproteome.9b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi Yutaka, Suzuki Yasufumi, Isemura Mamoru, Nomoto Minoru, Sekine Chuichi, Igarashi Kentarou, and Ichida Fumihiro. 1988. “The Fucosylation Index of Alpha-fetoprotein and Its Usefulness in the Early Diagnosis of Hepatocellular Carcinoma.” Cancer 61 (4): 769–74. . [DOI] [PubMed] [Google Scholar]
- Balog Crina I.A., Stavenhagen Kathrin, Fung Wesley L.J., Koeleman Carolien A., McDonnell Liam A., Verhoeven Aswin, Mesker Wilma E., Tollenaar Rob A.E.M., Deelder André M., and Wuhrer Manfred. 2012. “N-Glycosylation of Colorectal Cancer Tissues: A Liquid Chromatography and Mass Spectrometry-Based Investigation.” Molecular and Cellular Proteomics 11 (9): 571–85. 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré Florian P.Y., Paine Martin R.L., Flinders Bryn, Trevitt Adam J., Kelly Patrick D., Ait-Belkacem Rima, Garcia João P., et al. 2019. “Enhanced Sensitivity Using Maldi Imaging Coupled with Laser Postionization (Maldi-2) for Pharmaceutical Research.” Analytical Chemistry 91 (16): 10840–48. 10.1021/acs.analchem.9b02495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu Sankha S., and Agar Nathalie Y.R.. 2021. “Bringing Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging to the Clinics.” Clinics in Laboratory Medicine 41 (2): 309–24. 10.1016/j.cll.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis Kyle D., Harry April, Eberlin Livia S., Ferreira Christina, van de Ven Stephanie M., Mallick Parag, Stolowitz Mark, and Vitek Olga. 2015. “Cardinal: An R Package for Statistical Analysis of Mass Spectrometry-Based Imaging Experiments.” Bioinformatics 31 (14): 2418–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benninghoven Alfred. 1994. “Chemical Analysis of Inorganic and Organic Surfaces and Thin Films by Static Time-of-Flight Secondary Ion Mass Spectrometry (TOF-SIMS).” Angewandte Chemie International Edition in English. John Wiley & Sons, Ltd. 10.1002/anie.199410231. [DOI] [Google Scholar]
- Bereman Michael S., Williams Taufika Islam, and Muddiman David C.. 2007. “Carbohydrate Analysis by Desorption Electrospray Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry.” Analytical Chemistry 79 (22): 8812–15. 10.1021/ac0713858. [DOI] [PubMed] [Google Scholar]
- Black Alyson P., Angel Peggi M., Drake Richard R., and Mehta Anand S.. 2019. “Antibody Panel Based N -Glycan Imaging for N -Glycoprotein Biomarker Discovery.” Current Protocols in Protein Science 98 (1). 10.1002/cpps.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black Alyson P., Liang Hongyan, West Connor A., Wang Mengjun, Herrera Harmin P., Haab Brian B., Angel Peggi M., Drake Richard R., and Mehta Anand S.. 2019. “A Novel Mass Spectrometry Platform for Multiplexed N-Glycoprotein Biomarker Discovery from Patient Biofluids by Antibody Panel Based N-Glycan Imaging.” Analytical Chemistry 91 (13): acs.analchem.9b01445. 10.1021/acs.analchem.9b01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke Calvin, Black Alyson, Mehta Anand S, Angel Peggi M, and Drake Richard R. 2020. “Rapid N-Glycan Profiling of Serum and Plasma by a Novel Slide Based Imaging Mass Spectrometry Workflow.” Journal of the American Society for Mass Spectrometry. 10.1021/jasms.0c00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke Calvin R.K., McDowell Colin T., Black Alyson P., Mehta Anand S., Angel Peggi M., and Drake Richard R.. 2021. “Glycan Imaging Mass Spectrometry: Progress in Developing Clinical Diagnostic Assays for Tissues, Biofluids, and Cells.” Clinics in Laboratory Medicine 41 (2): 247–266. 10.1016/j.cll.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme Bram, Van Steenkiste Christophe, Callewaert Nico, and Van Vlierberghe Hans. 2009. “Alteration of Protein Glycosylation in Liver Diseases.” Journal of Hepatology 50 (3): 592–603. 10.1016/j.jhep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Bokhart Mark T., Nazari Milad, Garrard Kenneth P., and Muddiman David C.. 2018. “MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses.” Journal of the American Society for Mass Spectrometry 29 (1): 8–16. 10.1007/s13361-017-1809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles Kathryn M., Cheng Fang, Burk-Rafel Jesse, Dubey Manish, and Ratner Daniel M.. 2010. “Imaging Analysis of Carbohydrate-Modified Surfaces Using Tof-SIMS and Spri.” Materials 3 (7): 3948–64. 10.3390/ma3073948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman Andrew P., Blakney Greg T., Hendrickson Christopher L., Ellis Shane R., Heeren Ron M.A., and Smith Donald F.. 2020. “Ultra-High Mass Resolving Power, Mass Accuracy, and Dynamic Range MALDI Mass Spectrometry Imaging by 21-T FT-ICR MS.” Analytical Chemistry 92 (4): 3133–42. 10.1021/acs.analchem.9b04768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyaval Fanny, Van Zeijl René, Dalebout Hans, Holst Stephanie, van Pelt Gabi W., Fariña-Sarasqueta Arantza, Mesker Wilma E., et al. 2020. “N-Glycomic Signature of Stage II Colorectal Cancer and Its Association with the Tumor Microenvironment.” Molecular & Cellular Proteomics, October, mcp.RA120.002215. 10.1074/mcp.ra120.002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Marion M. 1976. “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding.” Analytical Biochemistry 72 (1–2): 248–54. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briggs Matthew T., Condina Mark R., Ho Yin Ying, Everest-Dass Arun V., Mittal Parul, Kaur Gurjeet, Oehler Martin K., Packer Nicolle H., and Hoffmann Peter. 2019. “MALDI Mass Spectrometry Imaging of Early and Late-Stage Serous Ovarian Cancer Tissue Reveals Stage-Specific N-Glycans.” PROTEOMICS 19 (21–22): 1800482. 10.1002/pmic.201800482. [DOI] [PubMed] [Google Scholar]
- Briggs Matthew T., Condina Mark R., Klingler Hoffmann Manuela, Arentz Georgia, Everest Dass Arun V., Kaur Gurjeet, Oehler Martin K., Packer Nicolle H., and Hoffmann Peter. 2019. “Translating N-Glycan Analytical Applications into Clinical Strategies for Ovarian Cancer.” PROTEOMICS – Clinical Applications 13 (3): 1800099. 10.1002/prca.201800099. [DOI] [PubMed] [Google Scholar]
- Briggs Matthew T., Kuliwaba Julia S., Muratovic Dzenita, Everest-Dass Arun V., Packer Nicolle H., Findlay David M., and Hoffmann Peter. 2016. “MALDI Mass Spectrometry Imaging of N-Glycans on Tibial Cartilage and Subchondral Bone Proteins in Knee Osteoarthritis.” PROTEOMICS 16 (11–12): 1736–41. 10.1002/pmic.201500461. [DOI] [PubMed] [Google Scholar]
- Butt R. Hussain, and Coorssen Jens R.. 2013. “Coomassie Blue as a Near-Infrared Fluorescent Stain: A Systematic Comparison with Sypro Ruby for in-Gel Protein Detection.” Molecular and Cellular Proteomics 12 (12): 3834–50. 10.1074/mcp.M112.021881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli Richard M., Farmer Terry B., and Gile Jocelyn. 1997. “Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS.” Analytical Chemistry 69 (23): 4751–60. 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Carr Steven A., Huddleston Michael J., and Bean Mark F.. 1993. “Selective Identification and Differentiation of N- and O-linked Oligosaccharides in Glycoproteins by Liquid Chromatography-mass Spectrometry.”Protein Science 2 (2): 183–96. 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter Claire L., Parker George A., Hankey Kim G., Farese Ann M., MacVittie Thomas J., and Kane Maureen A.. 2020. “MALDI-MSI Spatially Maps N-Glycan Alterations to Histologically Distinct Pulmonary Pathologies Following Irradiation.” Scientific Reports 10 (1): 11559. 10.1038/s41598-020-68508-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers Matthew C., MacLean Brendan, Burke Robert, Amodei Dario, Ruderman Daniel L., Neumann Steffen, Gatto Laurent, et al. 2012. “A Cross-Platform Toolkit for Mass Spectrometry and Proteomics.” Nature Biotechnology. NIH Public Access. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Huanhuan, Deng Zaian, Huang Chuncui, Wu Hongmei, Zhao Xia, and Li Yan. 2017. “Mass Spectrometric Profiling Reveals Association of N-Glycan Patterns with Epithelial Ovarian Cancer Progression.” Tumor Biology 39 (7): 101042831771624. 10.1177/1010428317716249. [DOI] [PubMed] [Google Scholar]
- Chubatyi Nicholas D., and McEwen Charles N.. 2015. “Improving the Sensitivity of Matrix-Assisted Ionization (MAI) Mass Spectrometry Using Ammonium Salts.” Journal of the American Society for Mass Spectrometry 26 (10): 1649–56. 10.1007/s13361-015-1205-z. [DOI] [PubMed] [Google Scholar]
- Clift Cassandra L., Drake Richard R., Mehta Anand, and Angel Peggi M.. 2020. “Multiplexed Imaging Mass Spectrometry of the Extracellular Matrix Using Serial Enzyme Digests from Formalin-Fixed Paraffin-Embedded Tissue Sections.” Analytical and Bioanalytical Chemistry, November, 1–11. 10.1007/s00216-020-03047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift Cassandra L., Mehta Anand S., Drake Richard R., and Angel Peggi M.. 2020. “Multiplexed Imaging Mass Spectrometry of Histological Staining, N-Glycan and Extracellular Matrix from One Tissue Section: A Tool for Fibrosis Research.” In Multiplexed Imaging: Methods and Protocols, In Press. [DOI] [PubMed] [Google Scholar]
- Collin M, Olsén A, Mattias Collin, and Arne Olsen. 2001. “EndoS, a Novel Secreted Protein from Streptococcus Pyogenes with Endoglycosidase Activity on Human IgG.” The EMBO Journal 20 (12): 3046–55. 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale Mary Ann, Lowman Melissa, Long Ronald E., Krakover Jonathan, Philip Ramila, Seeholzer Steven, Evans Alison A., Hann Hie Won L., Block Timothy M., and Mehta Anand S.. 2006. “Proteomic Analysis of Serum Associated Fucosylated Glycoproteins in the Development of Primary Hepatocellular Carcinoma.” Journal of Proteome Research 5 (2): 308–15. 10.1021/pr050328x. [DOI] [PubMed] [Google Scholar]
- Comunale Mary Ann, Rodemich-Betesh Lucy, Hafner Julie, Wang Mengjun, Norton Pamela, Di Bisceglie Adrian M., Block Timothy, and Mehta Anand. 2010. “Linkage Specific Fucosylation of Alpha-1-Antitrypsin in Liver Cirrhosis and Cancer Patients: Implications for a Biomarker of Hepatocellular Carcinoma.” Edited by Ryu Wang-Shick. PLoS ONE 5 (8): e12419. 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale Mary Ann, Wang Mengjun, Hafner Julie, Krakover Jonathan, Rodemich Lucy, Kopenhaver Brent, Long Ronald E., et al. 2009. “Identification and Development of Fucosylated Glycoproteins as Biomarkers of Primary Hepatocellular Carcinoma.” Journal of Proteome Research 8 (2): 595–602. 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale Mary Ann, Wang Mengjun, Rodemich-Betesh Lucy, Hafner Julie, Lamontagne Anne, Klein Andrew, Marrero Jorge, et al. 2011. “Novel Changes in Glycosylation of Serum Apo-J in Patients with Hepatocellular Carcinoma.” Cancer Epidemiology Biomarkers and Prevention 20 (6): 1222–29. 10.1158/1055-9965.EPI-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy Lindsey R, Young Lyndsay E A, Stanback Alexandra E, Austin Grant L, Liu Jinpeng, Liu Jinze, Allison Derek B, and Sun Ramon C. 2020. “Mass Spectrometry Imaging of N-Glycans Reveals Racial Discrepancies in Low Grade Prostate.” BioRxiv, August, 2020.08.20.260026. 10.1101/2020.08.20.260026. [DOI] [Google Scholar]
- Cooks R. Graham, Manicke Nicholas E., Dill Allison L., Ifa Demian R., Eberlin Livia S., Costa Anthony B., Wang He, Huang Guangming, and Ouyang Zheng. 2011. “New Ionization Methods and Miniature Mass Spectrometers for Biomedicine: DESI Imaging for Cancer Diagnostics and Paper Spray Ionization for Therapeutic Drug Monitoring.” Faraday Discussions 149: 247–67. 10.1039/c005327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper Catherine A., Gasteiger Elisabeth, and Packer Nicolle H.. 2001. “GlycoMod - A Software Tool for Determining Glycosylation Compositions from Mass Spectrometric Data.” Proteomics 1 (2): 340–49. . [DOI] [PubMed] [Google Scholar]
- Cummings Richard D. 2009. “The Repertoire of Glycan Determinants in the Human Glycome.” Molecular BioSystems 5 (10): 1087–1104. 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- Cummings Richard D., Darvill Alan G., Etzler Marilynn E., and Hahn Michael G.. 2017. “Glycan-Recognizing Probes as Tools.” 10.1101/GLYCOBIOLOGY.3E.048. [DOI] [Google Scholar]
- Damerell David, Ceroni Alessio, Maass Kai, Ranzinger René, Dell Anne, and Haslam Stuart M.. 2015. “Annotation of Glycomics MS and MS/MS Spectra Using the Glycoworkbench Software Tool.” Methods in Molecular Biology 1273: 3–15. 10.1007/978-1-4939-2343-4_1. [DOI] [PubMed] [Google Scholar]
- Darula Zsuzsanna, and Medzihradszky Katalin F.. 2018. “Analysis of Mammalian O-Glycopeptides - We Have Made a Good Start, but There Is a Long Way to Go.” Molecular and Cellular Proteomics. American Society for Biochemistry and Molecular Biology Inc. 10.1074/mcp.MR117.000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelaCourt Andrew T. and Mehta Anand S. 2021. “Liver Cancer (Current Therapies).” Reference Module in Biomedical Sciences 2021. 10.1016/B978-0-12-820472-6.00007-4. [DOI] [Google Scholar]
- Delafield Daniel G, and Li Lingjun. 2020. “Recent Advances in Analytical Approaches for Glycan and Glycopeptide Quantitation.” Molecular & Cellular Proteomics, June, mcp.R120.002095. 10.1074/mcp.r120.002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devakumar Arugadoss, Mechref Yehia, Kang Pilsoo, Novotny Milos V., and Reilly James P.. 2008. “Identification of Isomeric N-Glycan Structures by Mass Spectrometry with 157 Nm Laser-Induced Photofragmentation.” Journal of the American Society for Mass Spectrometry 19 (7): 1027–40. 10.1016/j.jasms.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotz Victoria and Wuhrer Manfred. 2019. “N-Glycome Signatures in Human Plasma: Associations with Physiology and Major Diseases.” FEBS Letters 593 (21): 2966–2976. 10.1002/1873-3468.13598. [DOI] [PubMed] [Google Scholar]
- Drake RR, Powers TW, Jones EE, Bruner E, Mehta AS, and Angel PM. 2017. “MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues.” In Advances in Cancer Research, 134:85–116. Academic Press Inc. 10.1016/bs.acr.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake Richard R. 2015. “Glycosylation and Cancer: Moving Glycomics to the Forefront.” In Advances in Cancer Research, 126:1–10. Academic Press Inc. 10.1016/bs.acr.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Drake Richard R., Angel Peggi M., Wu Jennifer, Pachynski Russell K., and Ippolito Joseph E.. 2020. “How Else Can We Approach Prostate Cancer Biomarker Discovery?” Expert Review of Molecular Diagnostics 20 (2): 123–25. 10.1080/14737159.2019.1665507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake Richard R., Boggs Sarah R., and Drake Steven K.. 2011. “Pathogen Identification Using Mass Spectrometry in the Clinical Microbiology Laboratory.” Journal of Mass Spectrometry 46 (12): 1223–32. 10.1002/jms.2008. [DOI] [PubMed] [Google Scholar]
- Drake Richard R., Jones E. Ellen, Powers Thomas W., and Nyalwidhe Julius O.. 2015. “Altered Glycosylation in Prostate Cancer.” In Advances in Cancer Research, 126:345–82. Academic Press Inc. 10.1016/bs.acr.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Drake Richard R., McDowell Colin, West Connor, David Fred, Powers Thomas W., Nowling Tamara, Bruner Evelyn, et al. 2020. “Defining the Human Kidney N-Glycome in Normal and Cancer Tissues Using MALDI Imaging Mass Spectrometry.” Journal of Mass Spectrometry 55 (4): e4490. 10.1002/jms.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake Richard R., Powers Thomas W., Norris-Caneda Kim, Mehta Anand S., and Angel Peggi M.. 2018. “In Situ Imaging of N-Glycans by MALDI Imaging Mass Spectrometry of Fresh or Formalin-Fixed Paraffin-Embedded Tissue.” Current Protocols in Protein Science 94 (1): e68. 10.1002/cpps.68. [DOI] [PubMed] [Google Scholar]
- Drake Richard R., West Connor A., Mehta Anand S., and Angel Peggi M.. 2018. “MALDI Mass Spectrometry Imaging of N-Linked Glycans in Tissues.” In Advances in Experimental Medicine and Biology, 1104:59–76. Springer New York LLC. 10.1007/978-981-13-2158-0_4. [DOI] [PubMed] [Google Scholar]
- Eberlin Livia S. 2014. “DESI-MS Imaging of Lipids and Metabolites from Biological Samples.” Methods in Molecular Biology 1198: 299–311. 10.1007/978-1-4939-1258-2_20. [DOI] [PubMed] [Google Scholar]
- Poruk KE, Gay DZ, Brown K, Mulvihill JD, Boucher KM, Scaife CL, Firpo MA, and Mulvihill SJ. 2013. “The Clinical Utility of CA 19–9 in Pancreatic Adenocarcinoma: Diagnostic and Prognostic Updates.” Current Molecular Medicine 13 (3): 340–51. 10.2174/156652413805076876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everest-Dass Arun V., Briggs Matthew T., Kaur Gurjeet, Oehler Martin K., Hoffmann Peter, and Packer Nicolle H.. 2016. “N-Glycan MALDI Imaging Mass Spectrometry on Formalin-Fixed Paraffin-Embedded Tissue Enables the Delineation of Ovarian Cancer Tissues.” Molecular and Cellular Proteomics 15 (9): 3003–16. 10.1074/mcp.M116.059816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lima Francisco. 2016. “Trapped Ion Mobility Spectrometry: Past, Present and Future Trends.” International Journal for Ion Mobility Spectrometry. Springer Verlag. 10.1007/s12127-016-0206-3. [DOI] [Google Scholar]
- Ford Kristina L., Zeng Wei, Heazlewood Joshua L., and Bacic Antony. 2015. “Characterization of Protein N-Glycosylation by Tandem Mass Spectrometry Using Complementary Fragmentation Techniques.” Frontiers in Plant Science 6 (AUG): 674. 10.3389/fpls.2015.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze Hudson H., and Kranz Christian. 2010. “Endoglycosidase and Glycoamidase Release of N-Linked Glycans.” Current Protocols in Molecular Biology 2010 (January). 10.1002/0471142727.mb1713as89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Freitas-Junior, Madureira Julio Cesar, and Morgado-Díaz José Andrés. 2016. “The Role of N-Glycans in Colorectal Cancer Progression: Potential Biomarkers and Therapeutic Applications.” Oncotarget. Impact Journals LLC. 10.18632/oncotarget.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle P, Hamburger J, and Castaing R. 1970. “Sur Une Nouvelle Méthode d’analyse Cellulaire Utilisant Le Phénomène d’émission Ionique Secondaire.” Annales de Physique Biologique et Médicale 42: 83–94. [Google Scholar]
- Garcia-Bunuel Rafael, and Monis Benito. 1964. “Histochemical Observations on Mucins in Human Ovarian Neoplasms.” Cancer 17 (9): 1108–18. . [DOI] [PubMed] [Google Scholar]
- Gillet Jean Pierre, Varma Sudhir, and Gottesman Michael M.. 2013. “The Clinical Relevance of Cancer Cell Lines.” Journal of the National Cancer Institute. Oxford University Press. 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke KS, and Siriwardena AK. 2007. “Systematic Review of Carbohydrate Antigen (CA 19–9) as a Biochemical Marker in the Diagnosis of Pancreatic Cancer.” European Journal of Surgical Oncology. W.B. Saunders. 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Goso Y, Tsubokawa D, and Ishihara K. 2009. “Evaluation of Conditions for Release of Mucin-Type Oligosaccharides from Glycoproteins by Hydrazine Gas Treatment.” Journal of Biochemistry 145 (6): 739–49. 10.1093/jb/mvp031. [DOI] [PubMed] [Google Scholar]
- Goto-Inoue Naoko, Hayasaka Takahiro, and Setou Mitsutoshi. 2010. “Imaging Mass Spectrometry of Glycolipids.” In Methods in Enzymology, 478:287–301. Academic Press Inc. 10.1016/S0076-6879(10)78014-9. [DOI] [PubMed] [Google Scholar]
- Greaves John, and Roboz John. 2013. Mass Spectrometry for the Novice. 1st ed. CRC Press. [Google Scholar]
- Gudelj Ivan, Lauc Gordan and Pezer Marija. 2018. “. Immunoglobulin G Glycosylation in Aging and Diseases.” Cellular Immunology 333: 65–79. 10.1016/J.CELLIMM.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Guile Geoffrey R., Rudd Pauline M., Wing David R., Prime Sally B., and Dwek Raymond A.. 1996. “A Rapid High-Resolution High-Performance Liquid Chromatographic Method for Separating Glycan Mixtures and Analyzing Oligosaccharide Profiles.” Analytical Biochemistry 240 (2): 210–26. 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- Guo Huabei, and Abbott Karen L.. 2015. “Functional Impact of Tumor-Specific N-Linked Glycan Changes in Breast and Ovarian Cancers.” In Advances in Cancer Research, 126:281–303. Academic Press Inc. 10.1016/bs.acr.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Gustafsson Ove J. R., Briggs Matthew T., Condina Mark R., Winderbaum Lyron J., Pelzing Matthias, McColl Shaun R., Everest-Dass Arun V., Packer Nicolle H., and Hoffmann Peter. 2015. “MALDI Imaging Mass Spectrometry of N-Linked Glycans on Formalin-Fixed Paraffin-Embedded Murine Kidney.” Analytical and Bioanalytical Chemistry 407 (8): 2127–39. 10.1007/s00216-014-8293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan Noortje, Yang Shuang, Cipollo John, and Wuhrer Manfred. 2020. “Glycomics Studies Using Sialic Acid Derivatization and Mass Spectrometry.” Nature Reviews Chemistry. Nature Research. 10.1038/s41570-020-0174-3. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, and Delannoy P. 2001. “The Human Sialyltransferase Family.” Biochimie 83 (8): 727–37. 10.1016/S0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- Harvey David J. 1999. “Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Carbohydrates.” Mass Spectrometry Reviews 18 (6): 349–450. . [DOI] [PubMed] [Google Scholar]
- Harvey David J. 2000. “Electrospray Mass Spectrometry and Fragmentation of N-Linked Carbohydrates Derivatized at the Reducing Terminus.” Journal of the American Society for Mass Spectrometry 11 (10): 900–915. 10.1016/S1044-0305(00)00156-2. [DOI] [PubMed] [Google Scholar]
- Harvey David J. 2005. “Fragmentation of Negative Ions from Carbohydrates: Part 1. Use of Nitrate and Other Anionic Adducts for the Production of Negative Ion Electrospray Spectra from N-Linked Carbohydrates.” Journal of the American Society for Mass Spectrometry 16 (5): 622–30. 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Harvey David J. 2020. “Negative Ion Mass Spectrometry for the Analysis of N-Linked Glycans.” Mass Spectrometry Reviews 39 (5–6): 586–679. 10.1002/mas.21622. [DOI] [PubMed] [Google Scholar]
- Harvey David J., and Struwe Weston B.. 2018. “Structural Studies of Fucosylated N-Glycans by Ion Mobility Mass Spectrometry and Collision-Induced Fragmentation of Negative Ions.” Journal of the American Society for Mass Spectrometry 29 (6): 1179–93. 10.1007/s13361-018-1950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijs Bram, Holst Stephanie, Briaire-De Bruijn Inge H., Van Pelt Gabi W., De Ru Arnoud H., Van Veelen Peter A., Drake Richard R., et al. 2016. “Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section.” Analytical Chemistry 88 (15): 7745–53. 10.1021/acs.analchem.6b01739. [DOI] [PubMed] [Google Scholar]
- Heijs Bram, Potthoff Alexander, Soltwisch Jens, and Dreisewerd Klaus. 2020. “MALDI-2 for the Enhanced Analysis of N-Linked Glycans by Mass Spectrometry Imaging.” Analytical Chemistry 92 (20): 13904–11. 10.1021/acs.analchem.0c02732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera Harmin, Dilday Tinslee, Uber Allison, Scott Danielle, Zambrano Joelle N., Wang Mengjun, Angel Peggi M., et al. 2019. “Core-Fucosylated Tetra-Antennary n-Glycan Containing a Single n-Acetyllactosamine Branch Is Associated with Poor Survival Outcome in Breast Cancer.” International Journal of Molecular Sciences 20 (10): 2528. 10.3390/ijms20102528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt Stephen M. 2012. “Tissue Microarrays as a Tool in the Discovery and Validation of Predictive Biomarkers.” Methods in Molecular Biology 823: 201–14. 10.1007/978-1-60327-216-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle Armin, Haase Andreas, Kayser Markus, and Höhndorf Jens. 2006. “Optimizing UV Laser Focus Profiles for Improved MALDI Performance.” Journal of Mass Spectrometry. John Wiley & Sons, Ltd. 10.1002/jms.1041. [DOI] [PubMed] [Google Scholar]
- Holst Stephanie, Belo Ana I., Giovannetti Elisa, Van Die Irma, and Wuhrer Manfred. 2017. “Profiling of Different Pancreatic Cancer Cells Used as Models for Metastatic Behaviour Shows Large Variation in Their N-Glycosylation.” Scientific Reports 7 (1): 1–15. 10.1038/s41598-017-16811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst Stephanie, Deuss Anna J.M., Van Pelt Gabi W., Van Vliet Sandra J., Garcia-Vallejo Juan J., Koeleman Carolien A.M., Deelder André M., et al. 2016. “N-Glycosylation Profiling of Colorectal Cancer Cell Lines Reveals Association of Fucosylation with Differentiation and Caudal Type Homebox 1 (CDX1)/Villin MRNA Expression.” Molecular and Cellular Proteomics 15 (1): 124–40. 10.1074/mcp.M115.051235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst Stephanie, Heijs Bram, De Haan Noortje, Van Zeijl René J.M., Briaire-De Bruijn Inge H., Van Pelt Gabi W., Mehta Anand S., et al. 2016. “Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues.” Analytical Chemistry 88 (11): 5904–13. 10.1021/acs.analchem.6b00819. [DOI] [PubMed] [Google Scholar]
- Hossain Mahmud, and Limbach Patrick A.. 2012. “A Comparison of MALDI Matrices.” In Electrospray and MALDI Mass Spectrometry, 215–44. Hoboken, NJ, USA: John Wiley & Sons, Inc. 10.1002/9780470588901.ch7. [DOI] [Google Scholar]
- Irie Atsushi, Koyamat Susumu, Kozutsumi Yasunori, Kawasaki Toshisuke, and Suzuki Akemi. 1998. “The Molecular Basis for the Absence of N-Glycolylneuraminic Acid in Humans.” Journal of Biological Chemistry 273 (25): 15866–71. 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- Jaskolla Thorsten W., and Karas Michael. 2011. “Compelling Evidence for Lucky Survivor and Gas Phase Protonation: The Unified MALDI Analyte Protonation Mechanism.” Journal of the American Society for Mass Spectrometry 22 (6): 976–88. 10.1007/s13361-011-0093-0. [DOI] [PubMed] [Google Scholar]
- Jawhar Nazar M.T. 2009. “Tissue Microarray: A Rapidly Evolving Diagnostic and Research Tool.” Annals of Saudi Medicine. King Faisal Specialist Hospital and Research Centre. 10.4103/0256-4947.51806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen Pia H., Karlsson Niclas G., Kolarich Daniel, and Packer Nicolle H.. 2012. “Structural Analysis of N- and O-Glycans Released from Glycoproteins.” Nature Protocols 7 (7): 1299–1310. 10.1038/nprot.2012.063. [DOI] [PubMed] [Google Scholar]
- Jones Abby, Dhanapala Lasangi, Kankanamage Rumasha N.T., Kumar Challa V., and Rusling James F.. 2020. “Multiplexed Immunosensors and Immunoarrays.” Analytical Chemistry. American Chemical Society. 10.1021/acs.analchem.9b05080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kailemia Muchena J., Park Dayoung, and Lebrilla Carlito B.. 2017. “Glycans and Glycoproteins as Specific Biomarkers for Cancer.” Analytical and Bioanalytical Chemistry. Springer Verlag. 10.1007/s00216-016-9880-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källback Patrik, Nilsson Anna, Shariatgorji Mohammadreza, and Andrén Per E.. 2016. “MsIQuant - Quantitation Software for Mass Spectrometry Imaging Enabling Fast Access, Visualization, and Analysis of Large Data Sets.” Analytical Chemistry 88 (8): 4346–53. 10.1021/acs.analchem.5b04603. [DOI] [PubMed] [Google Scholar]
- Kamata Tooru, Shima Noriaki, Miki Akihiro, Matsuo Eiichi, Yamamoto Takushi, Tsuchihashi Hitoshi, Sato Takako, Shimma Shuichi, and Katagi Munehiro. 2020. “High Spatial-Resolution Matrix-Assisted Laser Desorption/Ionization-Ion Trap-Time-of-Flight Tandem Mass Spectrometry Imaging for Depicting Longitudinal and Transverse Distribution of Drugs Incorporated into Hair.” Analytical Chemistry 92 (8): 5821–29. 10.1021/acs.analchem.9b05401. [DOI] [PubMed] [Google Scholar]
- Karabacak N. Murat, Easterling Michael L., Agar Nathalie Y.R., and Agar Jeffrey N.. 2010. “Transformative Effects of Higher Magnetic Field in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry.” Journal of the American Society for Mass Spectrometry 21 (7): 1218–22. 10.1016/j.jasms.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas Michael, Bachmann Doris, and Hillenkamp Franz. 1985. “Influence of the Wavelength in High-Irradiance Ultraviolet Laser Desorption Mass Spectrometry of Organic Molecules.” Analytical Chemistry 57 (14): 2935–39. 10.1021/ac00291a042. [DOI] [Google Scholar]
- Karas Michael, and Hillenkamp Franz. 1988. “Laser Desorption Ionization of Proteins with Molecular Masses Exceeding 10 000 Daltons.” Analytical Chemistry. American Chemical Society. 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- Kaya Ibrahim, Brülls Steffen M., Dunevall Johan, Jennische Eva, Lange Stefan, Martensson Jerker, Ewing Andrew G., Malmberg Per, and Fletcher John S.. 2018. “On-Tissue Chemical Derivatization of Catecholamines Using 4-(N-Methyl)Pyridinium Boronic Acid for ToF-SIMS and LDI-ToF Mass Spectrometry Imaging.” Analytical Chemistry 90 (22): 13580–90. 10.1021/acs.analchem.8b03746. [DOI] [PubMed] [Google Scholar]
- Keren Leeat, Bosse Marc, Thompson Steve, Risom Tyler, Vijayaragavan Kausalia, McCaffrey Erin, Marquez Diana, et al. 2019. “MIBI-TOF: A Multiplexed Imaging Platform Relates Cellular Phenotypes and Tissue Structure.” Science Advances 5 (10): eaax5851. 10.1126/sciadv.aax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Han Ie, Saldova Radka, Park Jun Hyoung, Lee Young Hun, Harvey David J., Wormald Mark R., Wynne Kieran, et al. 2013. “The Presence of Outer Arm Fucose Residues on the N -Glycans of Tissue Inhibitor of Metalloproteinases-1 Reduces Its Activity.” Journal of Proteome Research 12 (8): 3547–60. 10.1021/pr400276r. [DOI] [PubMed] [Google Scholar]
- Kim Min Sung, and Leahy Dan. 2013. “Enzymatic Deglycosylation of Glycoproteins.” In Methods in Enzymology, 533:259–63. Academic Press Inc. 10.1016/B978-0-12-420067-8.00019-2. [DOI] [PubMed] [Google Scholar]
- Kirk Ansgar T., Bohnhorst Alexander, Raddatz Christian Robert, Allers Maria, and Zimmermann Stefan. 2019. “Ultra-High-Resolution Ion Mobility Spectrometry—Current Instrumentation, Limitations, and Future Developments.” Analytical and Bioanalytical Chemistry. Springer Verlag. 10.1007/s00216-019-01807-0. [DOI] [PubMed] [Google Scholar]
- Kirwan Alan, Utratna Marta, O’Dwyer Michael E., Joshi Lokesh and Kilcoyne Michelle. 2015. “Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics.” Biomed Research International 2015. 10.1155/2015/490531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrzewa Markus. 2018. “Application of the MALDI Biotyper to Clinical Microbiology: Progress and Potential.” Expert Review of Proteomics 15 (3): 193–202. 10.1080/14789450.2018.1438193. [DOI] [PubMed] [Google Scholar]
- Kuo Joe Chin Hun, Gandhi Jay G., Zia Roseanna N., and Paszek Matthew J.. 2018. “Physical Biology of the Cancer Cell Glycocalyx.” Nature Physics 14 (7): 658–69. 10.1038/s41567-018-0186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Yin-Hung, and Wang Yi-Sheng. 2017. “Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry: Mechanistic Studies and Methods for Improving the Structural Identification of Carbohydrates.” Mass Spectrometry 6 (3): S0072–S0072. 10.5702/massspectrometry.s0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane Catherine S., McManus Kirsty, Widdowson Philip, Flowers Sarah A., Powell Gerard, Anderson Ian, and Campbell J. Larry. 2019. “Separation of Sialylated Glycan Isomers by Differential Mobility Spectrometry.” Analytical Chemistry 91 (15): 9916–24. 10.1021/acs.analchem.9b01595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattová Erika, Skřičková Jana, Hausnerová Jitka, Frola Lukáš, Křen Leoš, Ihnatová Ivana, Zdráhal Zbyněk, Bryant Joseph, and Popovič Mikuláš. 2020. “N-Glycan Profiling of Lung Adenocarcinoma in Patients at Different Stages of Disease.” Modern Pathology 33 (6): 1146–56. 10.1038/s41379-019-0441-3. [DOI] [PubMed] [Google Scholar]
- Lee Jeonghoon, and Reilly Peter T.A.. 2011. “Limitation of Time-of-Flight Resolution in the Ultra High Mass Range.” Analytical Chemistry 83 (15): 5831–33. 10.1021/ac201537b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Jong Won, Lee Kyungsoo, Ahn Sei Hyun, Son Byung Ho, Ko Beom Seok, Kim Hee Jeong, Chung Il Yong, et al. 2020. “Potential of MALDI-TOF-Based Serum N-Glycan Analysis for the Diagnosis and Surveillance of Breast Cancer.” Scientific Reports 10 (1): 1–8. 10.1038/s41598-020-76195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Sae Byul, Bose Shambhunath, Ahn Sei Hyun, Son Byung Ho, Ko Beom Seok, Kim Hee Jeong, Chung Il Yong, et al. 2020. “Breast Cancer Diagnosis by Analysis of Serum N-Glycans Using MALDI-TOF Mass Spectroscopy.” Edited by Halama Anna. PLOS ONE 15 (4): e0231004. 10.1371/journal.pone.0231004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemjabbar-Alaoui Hassan, McKinney Andrew, Yang Yi Wei, Tran Vy M., and Phillips Joanna J.. 2015. “Glycosylation Alterations in Lung and Brain Cancer.” In Advances in Cancer Research, 126:305–44. Academic Press Inc. 10.1016/bs.acr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold Jenny, Popkova Yulia, Engel Kathrin M., and Schiller Jürgen. 2018. “Recent Developments of Useful MALDI Matrices for the Mass Spectrometric Characterization of Lipids.” Biomolecules. MDPI AG. 10.3390/biom8040173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leoz Maria Lorna A., Young Lawrence J. T., An Hyun Joo, Kronewitter Scott R., Kim Jaehan, Miyamoto Suzanne, Borowsky Alexander D., Chew Helen K., and Lebrilla Carlito B.. 2011. “High-Mannose Glycans Are Elevated during Breast Cancer Progression.” Molecular & Cellular Proteomics 10 (1): M110.002717. 10.1074/mcp.m110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson Richard M., Borowsky Alexander D., and Angelo Michael. 2015. “Immunohistochemistry and Mass Spectrometry for Highly Multiplexed Cellular Molecular Imaging.” Laboratory Investigation 95 (4): 397–405. 10.1038/labinvest.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Yu Hsien, Franc Vojtech, and Heck Albert J.R.. 2018. “Similar Albeit Not the Same: In-Depth Analysis of Proteoforms of Human Serum, Bovine Serum, and Recombinant Human Fetuin.” Journal of Proteome Research 17 (8): 2861–69. 10.1021/acs.jproteome.8b00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Huan-qiu, Li Ji, Xuan Cheng-luan, and Ma Hai-chun. 2020. “A Review on the Physiological and Pathophysiological Role of Endothelial Glycocalyx.” Journal of Biochemical and Molecular Toxicology 34 (11). 10.1002/jbt.22571. [DOI] [PubMed] [Google Scholar]
- Liu Xin, Qiu Hongyu, Lee Rhonda Kuo, Chen Wangxue, and Li Jianjun. 2010. “Methylamidation for Sialoglycomics by MALDI-MS: A Facile Derivatization Strategy for Both A2,3- and A2,6-Linked Sialic Acids.” Analytical Chemistry 82 (19): 8300–8306. 10.1021/ac101831t. [DOI] [PubMed] [Google Scholar]
- Liu Xueli, Minin Vladimir, Huang Yunda, Seligson David B., and Horvath Steve. 2004. “Statistical Methods for Analyzing Tissue Microarray Data.” Journal of Biopharmaceutical Statistics. J Biopharm Stat. 10.1081/BIP-200025657. [DOI] [PubMed] [Google Scholar]
- Ma Junfeng, Sanda Miloslav, Wei Renhuizi, Zhang Lihua, and Goldman Radoslav. 2018. “Quantitative Analysis of Core Fucosylation of Serum Proteins in Liver Diseases by LC-MS-MRM.” Journal of Proteomics 189 (October): 67–74. 10.1016/j.jprot.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainini Veronica, Bovo Giorgio, Chinello Clizia, Gianazza Erica, Grasso Marco, Cattoretti Giorgio, and Magni Fulvio. 2013. “Detection of High Molecular Weight Proteins by MALDI Imaging Mass Spectrometry.” Molecular BioSystems 9 (6): 1101–7. 10.1039/c2mb25296a. [DOI] [PubMed] [Google Scholar]
- Malaker Stacy A., Pedram Kayvon, Ferracane Michael J., Bensing Barbara A., Krishnan Venkatesh, Pett Christian, Yu Jin, et al. 2019. “The Mucin-Selective Protease StcE Enables Molecular and Functional Analysis of Human Cancer-Associated Mucins.” Proceedings of the National Academy of Sciences of the United States of America 116 (15): 7278–87. 10.1073/pnas.1813020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Alan G., Hendrickson Christopher L., and Jackson George S.. 1998. “Fourier Transform Ion Cyclotron Resonance Mass Spectrometry: A Primer.” Mass Spectrometry Reviews 17 (1): 1–35. . [DOI] [PubMed] [Google Scholar]
- Monne Martinez-Seara, Hector Reinis Danne, Róg Tomasz, Ilpo Vattulainen, and Gurtovenko Andrey. 2013. “Structure of Glycocalyx.” Biophysical Journal 104 (2): 251a. 10.1016/j.bpj.2012.11.1412. [DOI] [Google Scholar]
- Mauko Lea, Lacher Nathan A., Pelzing Matthias, Nordborg Anna, Haddad Paul R., and Hilder Emily F.. 2012. “Comparison of ZIC-HILIC and Graphitized Carbon-Based Analytical Approaches Combined with Exoglycosidase Digestions for Analysis of Glycans from Monoclonal Antibodies.” Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences 911 (December): 93–104. 10.1016/j.jchromb.2012.10.043. [DOI] [PubMed] [Google Scholar]
- May Jody C., and McLean John A. 2015. “Ion Mobility-Mass Spectrometry: Time-Dispersive Instrumentation” Analytical Chemistry 87 (3): 1422–1436. 10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCombie Gregor, Staab Dieter, Stoeckli Markus, and Knochenmuss Richard. 2005. “Spatial and Spectral Correlations in MALDI Mass Spectrometry Images by Clustering and Multivariate Analysis.” Analytical Chemistry 77 (19): 6118–24. 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- McDowell Colin T., Klamer Zachary, Hall Johnathan, West Connor A., Wisniewski Luke, Powers Thomas W., Angel Peggi M., et al. 2020. “Imaging Mass Spectrometry and Lectin Analysis of N-Linked Glycans in Carbohydrate Antigen Defined Pancreatic Cancer Tissues.” Molecular & Cellular Proteomics 20 (January): Manuscript submitted for review. 10.1074/mcp.ra120.002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechref Yehia, Hu Yunli, Desantos-Garcia Janie L., Hussein Ahmed, and Tang Haixu. 2013. “Quantitative Glycomics Strategies.” Molecular and Cellular Proteomics. American Society for Biochemistry and Molecular Biology. 10.1074/mcp.R112.026310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta Anand, Herrera Harmin, and Block Timothy. 2015. “Glycosylation and Liver Cancer.” In Advances in Cancer Research, 126:257–79. Academic Press Inc. 10.1016/bs.acr.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A, Bimet F, Bizet C, Borovskaya AD, Drake RR, Eigner U, Fahr AM, et al. 2009. “High Interlaboratory Reproducibility of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry-Based Species Identification of Nonfermenting Bacteria.” Journal of Clinical Microbiology 47 (11): 3732–34. 10.1128/JCM.00921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereiter Stefan, Meritxell Balmaña Diana Campos, Gomes Joana, and Reis Celso A.. 2019. “Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading?” Cancer Cell. Cell Press. 10.1016/j.ccell.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Mitchell Wells J, and McLuckey Scott A.. 2005. “Collision-Induced Dissociation (CID) of Peptides and Proteins.” Methods in Enzymology. Academic Press Inc. 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- Montreuil Jean. 1995. “The History of Glycoprotein Research, a Personal View.” New Comprehensive Biochemistry 29 (PA): 1–12. 10.1016/S0167-7306(08)60584-0. [DOI] [Google Scholar]
- Morelle Willy, Faid Valegh, Chirat Frédéric, and Michalski Jean Claude. 2009. “Analysis of N-and O-Linked Glycans from Glycoproteins Using MALDI-TOF Mass Spectrometry.” Methods in Molecular Biology. Humana Press, Totowa, NJ. 10.1007/978-1-59745-022-5_1. [DOI] [PubMed] [Google Scholar]
- Mulloy Barbara, Dell Anne, Stanley Pamela, and Prestegard James H.. 2017. “Structural Analysis of Glycans.” 10.1101/GLYCOBIOLOGY.3E.050. [DOI] [Google Scholar]
- Munkley Jennifer. 2019. “The Glycosylation Landscape of Pancreatic Cancer.” Oncology Letters 17 (3): 2569–75. 10.3892/ol.2019.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari Milad, Muddiman David C. 2016. MALDESI: Fundamentals, Direct Analysis, and MS Imaging. Advances in MALDI and Laser-Induced Soft Ionization Mass Spectrometry 2016: 169 – 182. 10.1007/978-3-319-04819-2_9. [DOI] [Google Scholar]
- Neely Benjamin A., Wilkins Christopher E., Marlow Laura A., Malyarenko Dariya, Kim Yunee, Ignatchenko Alexandr, Sasinowska Heather, et al. 2016. “Proteotranscriptomic Analysis Reveals Stage Specific Changes in the Molecular Landscape of Clear-Cell Renal Cell Carcinoma.” PloS One 11 (4): e0154074. 10.1371/journal.pone.0154074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus M, Soltwisch J, Belov ME, and Dreisewerd K. 2019. “Transmission-Mode MALDI-2 Mass Spectrometry Imaging of Cells and Tissues at Subcellular Resolution.” Nature Methods 16 (9): 925–31. 10.1038/s41592-019-0536-2. [DOI] [PubMed] [Google Scholar]
- Nishikaze Takashi. 2017. “Sensitive and Structure-Informative N-Glycosylation Analysis by MALDI-MS; Ionization, Fragmentation, and Derivatization.” Mass Spectrometry 6 (1): A0060–A0060. 10.5702/massspectrometry.a0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikaze Takashi. 2019. “Sialic Acid Derivatization for Glycan Analysis by Mass Spectrometry.” Proceedings of the Japan Academy Series B: Physical and Biological Sciences. Japan Academy. 10.2183/pjab.95.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North Simon J., Huang Hung Hsiang, Sundaram Subha, Jang-Lee Jihye, Etienne A. Tony, Trollope Alana, Chalabi Sara, Dell Anne, Stanley Pamela, and Haslam Stuart M.. 2010. “Glycomics Profiling of Chinese Hamster Ovary Cell Glycosylation Mutants Reveals N-Glycans of a Novel Size and Complexity.” Journal of Biological Chemistry 285 (8): 5759–75. 10.1074/jbc.M109.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton Pamela A., Comunale Mary Ann, Krakover Jonathan, Rodemich Lucy, Pirog Natalie, D’Amelio Anthony, Philip Ramila, Mehta Anand S., and Block Timothy M.. 2008. “N-Linked Glycosylation of the Liver Cancer Biomarker GP73.” Journal of Cellular Biochemistry 104 (1): 136–49. 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrinc Potočnik Nina, Porta Tiffany, Becker Michael, Heeren Ron M. A., and Ellis Shane R.. 2015. “Use of Advantageous, Volatile Matrices Enabled by next-Generation High-Speed Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Imaging Employing a Scanning Laser Beam.” Rapid Communications in Mass Spectrometry 29 (23): 2195–2203. 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- Orlando Ron, Lim Jae Min, Atwood James A., Angel Peggi M., Fang Meng, Aoki Kazuhiro, Alvarez-Manilla Gerardo, et al. 2009. “IDAWG: Metabolic Incorporation of Stable Isotope Labels for Quantitative Glycomics of Cultured Cells.” Journal of Proteome Research 8 (8): 3816–23. 10.1021/pr8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace Crystal L., and Muddiman David C.. 2020. “Direct Analysis of Native N -Linked Glycans by IR-MALDESI.” Journal of the American Society for Mass Spectrometry 31 (8): 1759–62. 10.1021/jasms.0c00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholski ML, and Winograd N. 1999. “Imaging with Mass Spectrometry.” Chemical Reviews 99 (10): 2977–3005. 10.1021/cr980137w. [DOI] [PubMed] [Google Scholar]
- Page Jason S., Kelly Ryan T., Tang Keqi, and Smith Richard D.. 2007. “Ionization and Transmission Efficiency in an Electrospray Ionization-Mass Spectrometry Interface.” Journal of the American Society for Mass Spectrometry 18 (9): 1582–90. 10.1016/j.jasms.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Parry Simon, Hanisch Franz Georg, Leir Shih-Hsing, Sutton-Smith Mark, Morris Howard R., Dell Anne, and Harris Ann. 2006. “N-Glycosylation of the MUC1 Mucin in Epithelial Cells and Secretions.” Glycobiology 16 (7): 623–34. 10.1093/glycob/cwj110. [DOI] [PubMed] [Google Scholar]
- Patwa Tasneem, Li Chen, Simeone Diane M., and Lubman David M.. 2010. “Glycoprotein Analysis Using Protein Microarrays and Mass Spectrometry.” Mass Spectrometry Reviews 29 (5): 830–44. 10.1002/mas.20269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer Thomas H., Phelan Arthur W., and Tarentino Anthony L.. 1996. “Porcine Fibrinogen Glycopeptides: Substrates for Detecting Endo-β-N-Acetylglucosaminidases F2 and F3 1.” Analytical Biochemistry 235 (1): 98–101. 10.1006/abio.1996.0096. [DOI] [PubMed] [Google Scholar]
- Pongracz Tamas, Verhoeven Aswin, Wuhrer Manfred, and de Haan Noortje. 2021. “The Structure and Role of Lactone Intermediates in Linkage-Specific Sialic Acid Derivatization Reactions.” Glycoconjugate Journal 38 (2): 157–66. 10.1007/s10719-020-09971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel Porta, Tiffany Gregory Hamm, Bunch Josephine, Cappell Jo, Fletcher John S., and Schwamborn Kristina. 2018. “Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues.” Molecular Imaging and Biology. Springer New York LLC. 10.1007/s11307-018-1267-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell Andrew K., and Harvey David J.. 1996. “Stabilization of Sialic Acids in N-Linked Oligosaccharides and Gangliosides for Analysis by Positive Ion Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry.” Rapid Communications in Mass Spectrometry 10 (9): 1027–32. . [DOI] [PubMed] [Google Scholar]
- Powers Thomas W., Holst Stephanie, Wuhrer Manfred, Mehta Anand S., Drake Richard R., Powers Thomas W., Holst Stephanie, Wuhrer Manfred, Mehta Anand S., and Drake Richard R.. 2015. “Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry.” Biomolecules 5 (4): 2554–72. 10.3390/biom5042554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers Thomas W., Jones E. Ellen, Betesh Lucy R., Romano Patrick R., Gao Peng, Copland John A., Mehta Anand S., and Drake Richard R.. 2013. “Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry Workflow for Spatial Profiling Analysis of N-Linked Glycan Expression in Tissues.” Analytical Chemistry 85 (20): 9799–9806. 10.1021/ac402108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers Thomas W., Neely Benjamin A., Shao Yuan, Tang Huiyuan, Troyer Dean A., Mehta Anand S., Haab Brian B., and Drake Richard R.. 2014. “MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays.” Edited by Batra Surinder K.. PLoS ONE 9 (9): e106255. 10.1371/journal.pone.0106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice Boone M., and Caprioli Richard M.. 2016. “The Need for Speed in Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry.” Postdoc Journal 4 (3): 3. 10.14304/surya.jpr.v4n3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice Boone M., Chumbley Chad W., and Caprioli Richard M.. 2015. “High-Speed MALDI MS/MS Imaging Mass Spectrometry Using Continuous Raster Sampling.” Journal of Mass Spectrometry 50 (4): 703–10. 10.1002/jms.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice Boone M., Ryan Daniel J., Van De Plas Raf, Caprioli Richard M., and Spraggins Jeffrey M.. 2018. “Enhanced Ion Transmission Efficiency up to m / z 24 000 for MALDI Protein Imaging Mass Spectrometry.” Analytical Chemistry 90 (8): 5090–99. 10.1021/acs.analchem.7b05105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Yi, Ridgeway Mark E., Glaskin Rebecca S., Park Melvin A., Costello Catherine E., and Lin Cheng. 2016. “Separation and Identification of Isomeric Glycans by Selected Accumulation-Trapped Ion Mobility Spectrometry-Electron Activated Dissociation Tandem Mass Spectrometry.” Analytical Chemistry 88 (7): 3440–43. 10.1021/acs.analchem.6b00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Wei Jun, Jacobs Jon M., Liu Tao, Camp David G., and Smith Richard D.. 2006. “Advances and Challenges in Liquid Chromatography-Mass Spectrometry-Based Proteomics Profiling for Clinical Applications.” Molecular and Cellular Proteomics. American Society for Biochemistry and Molecular Biology. 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiding Karli R., Blank Dennis, Kuijper Dennis M., Deelder André M., and Wuhrer Manfred. 2014. “High-Throughput Profiling of Protein N-Glycosylation by MALDI-TOF-MS Employing Linkage-Specific Sialic Acid Esterification.” Analytical Chemistry 86 (12): 5784–93. 10.1021/ac500335t. [DOI] [PubMed] [Google Scholar]
- Reitsma Sietze, Slaaf Dick W., Vink Hans, Van Zandvoort Marc A.M.J., and Oude Egbrink Mirjam G.A.. 2007. “The Endothelial Glycocalyx: Composition, Functions, and Visualization.” Pflugers Archiv European Journal of Physiology. Springer. 10.1007/s00424-007-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera Emilio S., Djambazova Katerina V., Neumann Elizabeth K., Caprioli Richard M., and Spraggins Jeffrey M.. 2020. “Integrating Ion Mobility and Imaging Mass Spectrometry for Comprehensive Analysis of Biological Tissues: A Brief Review and Perspective.” Journal of Mass Spectrometry 55 (12). 10.1002/jms.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson Kenneth N., Steven Rory T., and Bunch Josephine. 2018. “Matrix Optical Absorption in UV-MALDI MS.” Journal of the American Society for Mass Spectrometry 29 (3): 501–11. 10.1007/s13361-017-1843-4. [DOI] [PubMed] [Google Scholar]
- von Roemeling Christina A., Marlow Laura A., Radisky Derek C., Rohl Austin, Larsen Hege Ekeberg, Wei Johnny, Sasinowska Heather, et al. 2014. “Functional Genomics Identifies Novel Genes Essential for Clear Cell Renal Cell Carcinoma Tumor Cell Proliferation and Migration.” Oncotarget 5 (14): 5320–34. 10.18632/oncotarget.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römpp Andreas, Schramm Thorsten, Hester Alfons, Klinkert Ivo, Both Jean Pierre, Heeren Ron M.A., Stöckli Markus, and Spengler Bernhard. 2011. “ImzML: Imaging Mass Spectrometry Markup Language: A Common Data Format for Mass Spectrometry Imaging.” Methods in Molecular Biology (Clifton, N.J.) 696: 205–24. 10.1007/978-1-60761-987-1_12. [DOI] [PubMed] [Google Scholar]
- Ruhaak L. Renee, Hennig René, Huhn Carolin, Borowiak Matthias, Dolhain Radboud J.E.M., Deelder André M., Rapp Erdmann, and Wuhrer Manfred. 2010. “Optimized Workflow for Preparation of APTS-Labeled N-Glycans Allowing High-Throughput Analysis of Human Plasma Glycomes Using 48-Channel Multiplexed CGE-LIF.” Journal of Proteome Research 9 (12): 6655–64. 10.1021/pr100802f. [DOI] [PubMed] [Google Scholar]
- Ruhaak L.Renee, Xu Gege, Li Qiongyu, Goonatilleke Elisha, and Lebrilla Carlito B.. 2018. “Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses.” Chemical Reviews 118 (17): 7886–7930. 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldova Radka, and Wilkinson Hayden. 2020. “Current Methods for the Characterization of O-Glycans.” Journal of Proteome Research. American Chemical Society. 10.1021/acs.jproteome.0c00435. [DOI] [PubMed] [Google Scholar]
- Sampson Jason S., Hawkridge Adam M. and Muddiman David C. 2006. Generation and Detection of Multiply-Charged Peptides and Proteins by Matrix-Assisted Laser Desorption Electrospray Ionization (MALDESI) Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Journal of the American Society for Mass Spectrometry 17 (12): 1712 – 1716. 10.1016/J.JASMS.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Sans Marta, Feider Clara L., and Eberlin Livia S.. 2018. “Advances in Mass Spectrometry Imaging Coupled to Ion Mobility Spectrometry for Enhanced Imaging of Biological Tissues.” Current Opinion in Chemical Biology. Elsevier Ltd. 10.1016/j.cbpa.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider Micheal Michael, Al-Shareffi Esam, and Haltiwanger Robert S.. 2017. “Biological Functions of Fucose in Mammals.” Glycobiology 7 (27): 601–18. 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scigelova Michaela, Hornshaw Martin, Giannakopulos Anastassios, and Makarov Alexander. 2011. “Fourier Transform Mass Spectrometry.” Molecular and Cellular Proteomics. American Society for Biochemistry and Molecular Biology. 10.1074/mcp.M111.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott Danielle A., Casadonte Rita, Cardinali Barbara, Spruill Laura, Mehta Anand S., Carli Franca, Simone Nicole, et al. 2019. “Increases in Tumor N-Glycan Polylactosamines Associated with Advanced HER2-Positive and Triple-Negative Breast Cancer Tissues.” PROTEOMICS – Clinical Applications 13 (1): 1800014. 10.1002/prca.201800014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott Danielle A., and Drake Richard R.. 2019. “Glycosylation and Its Implications in Breast Cancer.” Expert Review of Proteomics. Taylor and Francis Ltd. 10.1080/14789450.2019.1645604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott Danielle A., Norris-Caneda Kim, Spruill Laura, Bruner Evelyn, Kono Yuko, Angel Peggi M., Mehta Anand S., and Drake Richard R.. 2019. “Specific N-Linked Glycosylation Patterns in Areas of Necrosis in Tumor Tissues.” International Journal of Mass Spectrometry 437 (March): 69–76. 10.1016/j.ijms.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya Sadanori, Wada Yoshinao, and Tanaka Koichi. 2005. “Derivatization for Stabilizing Sialic Acids in MALDI-MS.” Analytical Chemistry 77 (15): 4962–68. 10.1021/ac050287o. [DOI] [PubMed] [Google Scholar]
- Seyrek Kamil, Richter Max, and Lavrik Inna N.. 2019. “Decoding the Sweet Regulation of Apoptosis: The Role of Glycosylation and Galectins in Apoptotic Signaling Pathways.” Cell Death and Differentiation. Nature Publishing Group. 10.1038/s41418-019-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan Asif, Heiss Christian, Ishihara Mayumi, and Azadi Parastoo. 2017. “Glycomic and Glycoproteomic Analysis of Glycoproteins—a Tutorial.” Analytical and Bioanalytical Chemistry 409 (19): 4483–4505. 10.1007/s00216-017-0406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Škrášková Karolina, Claude Emmanuelle, Jones Emrys A., Towers Mark, Ellis Shane R., and Heeren Ron M.A.. 2016. “Enhanced Capabilities for Imaging Gangliosides in Murine Brain with Matrix-Assisted Laser Desorption/Ionization and Desorption Electrospray Ionization Mass Spectrometry Coupled to Ion Mobility Separation.” Methods 104 (July): 69–78. 10.1016/j.ymeth.2016.02.014. [DOI] [PubMed] [Google Scholar]
- Smith Donald F., Podgorski David C., Rodgers Ryan P., Blakney Greg T., and Hendrickson Christopher L.. 2018. “21 Tesla FT-ICR Mass Spectrometer for Ultrahigh-Resolution Analysis of Complex Organic Mixtures.” Analytical Chemistry 90 (3): 2041–47. 10.1021/acs.analchem.7b04159. [DOI] [PubMed] [Google Scholar]
- Spraggins Jeffrey M., Djambazova Katerina V., Rivera Emilio S., Migas Lukasz G., Neumann Elizabeth K., Fuetterer Arne, Suetering Juergen, et al. 2019. “High-Performance Molecular Imaging with MALDI Trapped Ion-Mobility Time-of-Flight (TimsTOF) Mass Spectrometry.” Analytical Chemistry 91 (22): 14552–60. 10.1021/acs.analchem.9b03612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan Nithya, Bane Sanjay M., Ahire Shashikant D., Ingle Arvind D., and Kalraiya Rajiv D.. 2009. “Poly NAcetyllactosamine Substitutions on N- and Not O-Oligosaccharides or Thomsen-Friedenreich Antigen Facilitate Lung Specific Metastasis of Melanoma Cells via Galectin-3.” Glycoconjugate Journal 26 (4): 445–56. 10.1007/s10719-008-9194-9. [DOI] [PubMed] [Google Scholar]
- Staal Ben, Liu Ying, Barnett Daniel, Hsueh Peter, He Zonglin, Chong Feng Gao Katie Partyka, et al. 2019. “The Stra Plasma Biomarker: Blinded Validation of Improved Accuracy over CA19–9 in Pancreatic Cancer Diagnosis.” Clinical Cancer Research 25 (9): 2745–54. 10.1158/1078-0432.CCR-18-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Cummings RD. 2017. Stanley P, Cummings RD. Structures Common to Different Glycans. Essentials of Glycobiology [Internet]. Cold Spring Harbor Laboratory Press. 10.1101/glycobiology.3e.014. [DOI] [Google Scholar]
- Stöckmann H, O’Flaherty R, Adamczyk B, Saldova R, and Rudd PM. 2015. “Automated, High-Throughput Serum Glycoprofiling Platform.” Integrative Biology (United Kingdom) 7 (9): 1026–32. 10.1039/c5ib00130g. [DOI] [PubMed] [Google Scholar]
- Sun Ramon C., Young Lyndsay E.A., Bruntz Ronald C., Markussen Kia H., Zhou Zhengqui, Conroy Lindsey R., Hawkinson Tara R., Clarke Harrison A., Stanback Alexandra E., Macedo Jessica K.A., Emanuelle Shane, Brewer M. Kathryn, Rondon Alberto L., Mestas Annette, Sanders William C., Mahalingan Krishna K., Tang Buyun, Chikwana Vimbai M., Segvich Dyann M., Contreras Cristopher J., Allenger Elizabeth J., Brainson Christine F., Johnson Lance A., Taylor Richard E., Armstrong Dustin D., Shaffer Robert, Waechter Charles J., Vander Kooi Craig W., DePaoli-Roach Anna A., Roach Peter J., Hurley Thomas D., Drake Richard R. and Gentry Mathew S. 2021. “Brain glycogen serves as a critical glucosamine cache required for protein glycosylation.” Cell Metabolism 33 (7): 1404–1417. 10.1016/j.cmet.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts Zoltán, Wiseman Justin M., and Cooks Graham. 2005. “Ambient Mass Spectrometry Using Desorption Electrospray Ionization (DESI): Instrumentation, Mechanisms and Applications in Forensics, Chemistry, and Biology.” Journal of Mass Spectrometry. John Wiley & Sons, Ltd. 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- Takáts Zoltán, Wiseman Justin M., Gologan Bogdan, and Cooks R. Graham. 2004. “Mass Spectrometry Sampling under Ambient Conditions with Desorption Electrospray Ionization.” Science 306 (5695): 471–73. 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Tan Wei Chang Colin, Nerurkar Sanjna Nilesh, Cai Hai Yun, Ng Harry Ho Man, Wu Duoduo, Wee Yu Ting Felicia, Lim Jeffrey Chun Tatt, Yeong Joe, and Lim Tony Kiat Hon. 2020. “Overview of Multiplex Immunohistochemistry/Immunofluorescence Techniques in the Era of Cancer Immunotherapy.” Cancer Communications. John Wiley and Sons Inc. 10.1002/cac2.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Shujuan, Huang Yining, Boyes Barry E., and Orlando Ron. 2014. “Liquid Chromatography-Selected Reaction Monitoring (LC-SRM) Approach for the Separation and Quantitation of Sialylated N-Glycans Linkage Isomers.” Analytical Chemistry 86 (21): 10584–90. 10.1021/ac5020996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarentino Anthony L., and Plummer Thomas H.. 1994. “Enzymatic Deglycosylation of Asparagine-Linked Glycans: Purification, Properties, and Specificity of Oligosaccharide-Cleaving Enzymes from Flavobacterium Meningosepticum.” Methods in Enzymology 230 (C): 44–57. 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- Tiemeyer Michael, Aoki Kazuhiro, Paulson James, Cummings Richard D, York William S, Karlsson Niclas G, Lisacek Frederique, et al. 2017. “GlyTouCan: An Accessible Glycan Structure Repository.” Glycobiology 27 (10): 915–19. 10.1093/glycob/cwx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Zhijay, Lin Yu Nong, and Lin Chun Hung. 2013. “Development of Fucosyltransferase and Fucosidase Inhibitors.” Chemical Society Reviews 42 (10): 4459–75. 10.1039/c3cs60056d. [DOI] [PubMed] [Google Scholar]
- Ucal Yasemin, and Ozpinar Aysel. 2018. “Improved Spectra for MALDI MSI of Peptides Using Ammonium Phosphate Monobasic in MALDI Matrix.” Journal of Mass Spectrometry 53 (8): 635–48. 10.1002/jms.4198. [DOI] [PubMed] [Google Scholar]
- Varki Ajit. 2008. “Sialic Acids in Human Health and Disease.” Trends in Molecular Medicine. NIH Public Access. 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki Ajit. 2017. “Biological Roles of Glycans.” Glycobiology 27 (1): 3–49. 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillon Lucas, Huang Yifan, Peng Wenjing, Dong Xue, Cho Byeong Gwan, and Mechref Yehia. 2017. “Characterization of Isomeric Glycan Structures by LC-MS/MS.” Electrophoresis. Wiley-VCH Verlag. 10.1002/elps.201700042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeker Gerda C.M., Nicolardi Simone, Bladergroen Marco R., Van Der Plas Corné J., Mesker Wilma E., Tollenaar Rob A.E.M., Van Der Burgt Yuri E.M., et al. 2018. “Automated Plasma Glycomics with Linkage-Specific Sialic Acid Esterification and Ultrahigh Resolution MS.” Analytical Chemistry 90 (20): 11955–61. 10.1021/acs.analchem.8b02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vroome Stefan W., Holst Stephanie, Girondo Mar Rodriguez, van der Burgt Yuri E.M., Mesker Wilma E., Tollenaar Rob A.E.M., and Wuhrer Manfred. 2018. “Serum N-Glycome Alterations in Colorectal Cancer Associate with Survival.” Oncotarget 9 (55): 30610–23. 10.18632/oncotarget.25753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Chia Chen, Lai Yin Hung, Ou Yu Meng, Chang Huan Tsung, and Wang Yi Sheng. 2016. “Critical Factors Determining the Quantification Capability of Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry.” Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. Royal Society of London. 10.1098/rsta.2015.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Mengjun, Long Ronald E., Comunale Mary Ann, Junaidi Omer, Marrero Jorge, Di Bisceglie Adrian M., Block Timothy M., and Mehta Anand S.. 2009. “Novel Fucosylated Biomarkers for the Early Detection of Hepatocellular Carcinoma.” Cancer Epidemiology Biomarkers and Prevention 18 (6): 1914–21. 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Mengmeng, Zhu Jianhui, Lubman David M., and Gao Chunfang. 2019. “Aberrant Glycosylation and Cancer Biomarker Discovery: A Promising and Thorny Journey.” Clinical Chemistry and Laboratory Medicine. De Gruyter. 10.1515/cclm-2018-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West Connor A., Liang Hongyan, Drake Richard R., and Mehta Anand S.. 2020. “New Enzymatic Approach to Distinguish Fucosylation Isomers of N-Linked Glycans in Tissues Using MALDI Imaging Mass Spectrometry.” Journal of Proteome Research, June. 10.1021/acs.jproteome.0c00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West Connor A., Wang Mengjun, Herrera Harmin, Liang Hongyan, Black Alyson, Angel Peggi M., Drake Richard R., and Mehta Anand S.. 2018. “N-Linked Glycan Branching and Fucosylation Are Increased Directly in Hcc Tissue As Determined through in Situ Glycan Imaging.” Journal of Proteome Research 17 (10): 3454–62. 10.1021/acs.jproteome.8b00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd Nicholas. 2015. “Imaging Mass Spectrometry on the Nanoscale with Cluster Ion Beams.” Analytical Chemistry 87 (1): 328–333. 10.1021/ac503650p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodin Carrie L., Maxon Morgan, and Desaire Heather. 2013. “Software for Automated Interpretation of Mass Spectrometry Data from Glycans and Glycopeptides.” Analyst. Royal Society of Chemistry. 10.1039/c2an36042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Gege, Wong Maurice, Li Qiongyu, Park Dayoung, Cheng Zhi, and Lebrilla Carlito B.. 2019. “Unveiling the Metabolic Fate of Monosaccharides in Cell Membranes with Glycomic and Glycoproteomic Analyses.” Chemical Science 10 (29): 6992–7002. 10.1039/c9sc01653h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagnik Gargey, Liu Ziying, Rothschild Kenneth J., and Lim Mark J.. 2021. “Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues.” Journal of the American Society for Mass Spectrometry 32 (4): 977–88. 10.1021/jasms.0c00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yong, Zhang Sheng, Howe Kevin, Wilson David B., Moser Felix, Irwin Diana, and Thannhauser Theodore W.. 2007. “A Comparison of NLC-ESI-MS/MS and NLC-MALDI-MS/MS for GeLC-Based Protein Identification and ITRAQ-Based Shotgun Quantitative Proteomics.” Journal of Biomolecular Techniques 18 (4): 226–37. http://docs.appliedbiosystems.com/search. [PMC free article] [PubMed] [Google Scholar]
- Yilmaz Onur, Afsar Baris, Ortiz Alberto, and Kanbay Mehmet. 2019. “The Role of Endothelial Glycocalyx in Health and Disease.” Clinical Kidney Journal. Oxford University Press. 10.1093/ckj/sfz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Haidi, Zhu Jianhui, Wu Jing, Tan Zhijing, An Mingrui, Zhou Shiyue, Mechref Yehia, and Lubman David M.. 2016. “A Procedure for the Analysis of Site-Specific and Structure-Specific Fucosylation in Alpha-1-Antitrypsin.” Electrophoresis 37 (20): 2624–32. 10.1002/elps.201600176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Sohee, and Lee Tae Geol. 2018. “Biological Tissue Sample Preparation for Time-of-Flight Secondary Ion Mass Spectrometry (ToF–SIMS) Imaging.” Nano Convergence. 5 (1):24. 10.1186/s40580-018-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavalin Andre, Todd Erik M., Rawhouser Patrick D., Yang Junhai, Norris Jeremy L., and Caprioli Richard M.. 2012. “Direct Imaging of Single Cells and Tissue at Sub-Cellular Spatial Resolution Using Transmission Geometry MALDI MS.” Journal of Mass Spectrometry 47 (11): i–i. 10.1002/jms.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Ting, Jia Li, Li Jun, Ma Chen, Wu Jingyu, Shen Jiechen, Dang Liuyi, et al. 2020. “Heterogeneities of Site-Specific N-Glycosylation in HCC Tumors With Low and High AFP Concentrations.” Frontiers in Oncology 10 (April): 496. 10.3389/fonc.2020.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Jianhui, Warner Elisa, Parikh Neehar D., and Lubman David M.. 2019. “Glycoproteomic Markers of Hepatocellular Carcinoma-Mass Spectrometry Based Approaches.” Mass Spectrometry Reviews. John Wiley and Sons Inc. 10.1002/mas.21583. [DOI] [PMC free article] [PubMed] [Google Scholar]


