Abstract
Background
Macrophage migration inhibitory factor (MIF) is a multifunctional cytokine that promotes neurogenesis and neuroprotection. MIF is predominantly expressed in astrocytes in the brain. The serum MIF level and microsatellites/single nucleotide polymorphisms (SNPs) in the MIF gene promoter region are known to be associated with schizophrenia (SCZ). Interestingly, previous studies reported that hypoxia, an environmental risk factor for SCZ, induced MIF expression through binding of the hypoxia inducible factor (HIF)-1 to the hypoxia response element (HRE) in the MIF promoter.
Methods
We investigated the involvement of MIF in SCZ while focusing on the HIF pathway. First, we conducted an association study of the SNP rs17004038 (C>A) in the HRE of the MIF promoter between 1758 patients with SCZ and 1507 controls. Next, we investigated the effect of hypoxia on MIF expression in primary cultured astrocytes derived from neonatal mice forebrain.
Results
SNP rs17004038 was significantly associated with SCZ (p = 0.0424, odds ratio = 1.445), indicating that this SNP in the HRE of the MIF promoter was a genetic risk factor for SCZ. Hypoxia induced MIF mRNA expression and MIF protein production and increased HIF-1 binding to the MIF promoter, while the activity of the MIF promoter was suppressed by mutations in the HRE and by deletion of the HRE in astrocytes.
Conclusion
These results suggest that SNP rs17004038 in the HRE of the MIF promoter was significantly associated with SCZ and may be involved in the pathophysiology of SCZ via suppression of hypoxia and HIF pathway-induced MIF expression.
Introduction
Schizophrenia (SCZ) is a chronic and disabling psychiatric disorder [1] that affects approximately 1% of the general population worldwide [2]. Although the onset of SCZ is commonly observed in adolescence, premorbid features, including cognitive impairments, often precede the diagnosis by years [3]. Recent genetic studies, including genome-wide association studies (GWASs), have contributed to the common theory that SCZ is a neurodevelopmental disorder [4–7]. According to the multiple-hit model, SCZ is attributable to the cumulative effects of genetic susceptibility and environmental insults during brain development [8]. Emerging evidence shows that gene–environment interactions underlie critical environmental contributions in the perinatal period [9–11]. In human research, there is consistent evidence linking perinatal hypoxia to SCZ in later life [12–16]. In addition, perinatal hypoxia has been reported to be a crucial environmental risk factor in the neurodevelopmental model of SCZ [17–20]. Indeed, oxygen restriction of perinatal murine has been established as an animal model of SCZ [21–23]. A recent review concluded that perinatal hypoxia, as well as the maternal–offspring immune activation and maternal hypothalamic–pituitary–adrenal axis, can contribute to the pathogenesis of SCZ based on early environmental upheavals, including prenatal maternal stress, obstetric complications, infections, and maternal lifestyle-related factors [24]. However, the detailed molecular mechanisms underlying the involvement of hypoxia in the pathophysiology of SCZ remain unclear.
Macrophage migration inhibitory factor (MIF) is a multifunctional cytokine that serves as a regulator in both innate and adaptive immunity. MIF is also expressed in nervous cells such as neurons [25, 26] and (predominantly) astrocytes [27–29]. In addition, MIF performs crucial functions in neurogenesis and neuroprotection [30], which are suggested to be involved in the pathophysiology of SCZ [31]. Several comprehensive biomarker studies have shown that MIF is a potential biomarker for SCZ [32, 33]. We also reported that the serum MIF level is higher in patients with SCZ and positively correlated with antipsychotic doses, and that a higher-expression allele of the MIF−794CATT5−8 microsatellite (rs5844572) in the MIF promoter is significantly lower frequent in female patients with adolescent-onset SCZ [34]. Furthermore, we have shown that clozapine, an atypical antipsychotic, increased MIF expression via histone acetylation in neonatal mice-derived primary cultured astrocytes (PCAs) [35]. Taken together, these findings suggest that MIF may have an important role in the pathophysiology of SCZ and the functional mechanisms of antipsychotics.
The adaptive responses to hypoxia that restore oxygen homeostasis in tissues are predominantly regulated by hypoxia inducible factor (HIF)-1, a transcription factor activated by hypoxia [36, 37]. Recent studies have shown that hypoxia is a potent and rapid inducer of MIF expression via the HIF pathway, and that hypoxia-induced MIF expression is dependent on the hypoxia response element (HRE) in the 5′ UTR of the MIF gene [38, 39]. In addition, the single nucleotide polymorphism (SNP) rs17004038, which maps to the functional HRE in the MIF gene, prevents hypoxia-induced MIF expression [39].
Based on past studies including ours described above, we hypothesized that MIF may be involved in the pathophysiology of SCZ via the HIF pathway. To test this hypothesis, we first conducted an association study of SNP rs17004038 located in the HRE of the MIF promoter between patients with SCZ and controls. Subsequently, we investigated the molecular mechanisms underlying the effect of hypoxia on MIF expression in PCAs.
Materials and methods
Ethics statements
This study was conducted in accordance with the Declaration of Helsinki. The clinical part of this study was approved by the Ethical Committee for Genetic Studies of Kobe University Graduate School of Medicine and the Ethics Committee of Genetics at Niigata University. Written informed consent from all participants was obtained after the details of the procedures of the clinical part of this study had been fully explained. The animal study protocol was approved by the Institutional Animal Care and Use Committee and carried out according to the Kobe University Animal Experimentation Regulations. All efforts were made to minimize suffering of mice.
Participants
In the association study, all participants were of Japanese descent and recruited from the suburbs of Kobe city (first set, 915 patients and 836 controls) and Niigata city (second set, 843 patients and 671 controls) in Japan. A diagnosis of SCZ was made by at least two psychiatrists according to the DSM-5 criteria for SCZ. Control participants were healthy volunteers who were screened for psychiatric disorders by a psychiatrist. None of the control participants had any present, past, or family (first-degree relatives) history of psychiatric disorders, use of neuroleptic medication, and substance abuse outside of nicotine. The demographic and clinical characteristics are shown in S1 Table.
Genotyping of SNP rs17004038 in the HRE of the MIF gene
Genotyping was performed as described elsewhere [34, 40]. The MIF gene is located on chromosome 22q11.23 (GenBank accession No. NM_002415). We analyzed SNP rs17004038 (C>A), which is located in the HRE of the MIF promoter and is involved in hypoxia-induced MIF expression [39] (Fig 1).
Fig 1. Human and mouse macrophage migration inhibitory factor (MIF) gene promoter region.
Peripheral blood samples were obtained from the participants, and DNA was extracted with the QIAamp DNA Blood Midi Kit (Qiagen Inc., Valencia, CA, USA). The quantity and purity of the DNA were determined with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). DNA samples were kept frozen at −80°C until analysis. We obtained the pre-designed TaqMan SNP genotyping assay for SNP rs17004038 from the Applied Biosystems database and performed genotyping with a 7500 Real-Time PCR System (Applied Biosystems) in accordance with the manufacturer’s protocol.
Cell culture
PCAs were prepared from neonatal C57BL/6J mice as described elsewhere [35, 41]. Briefly, the isolated forebrain was minced and incubated with DNase I (FUJIFILM Wako Pure Chemical) and trypsin (Thermo Fisher Scientific) for dissociation. The dissociated tissues were suspended in Dulbecco’s modified Eagle medium/Nutrient Mixture F-12 (DMEM/F12; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin/streptomycin (PS; Thermo Fisher). The suspended cells were seeded in poly-l-lysine (PLL; ScienCell, Carlsbad, CA, USA)-coated 75-mm2 flasks (10–15 × 106 cells/flask). The plated cells were cultured in a 5% CO2 incubator at 37°C. Every 8–12 days, the confluent cells were shaken for 10 min to separate them from microglial cells, released from the flasks with trypsin, and seeded onto non-coated flasks. These cells were seeded onto non-coated 8-well chamber slides (2.5 × 104 cells/well) and evaluated with immunocytochemistry for anti-glial fibrillary acidic protein (GFAP) antibody (Abcam).
Drugs
We used ML228 (Tocris Bioscience, Bristol, UK) as a HIF pathway activator and YC-1 (Abcam, Cambridge, UK) as a HIF inhibitor.
Hypoxic treatments
For hypoxic treatments, we applied the BIONIX-1 hypoxic culture kit (Sugiyamagen, Tokyo, Japan) as previously described [42, 43]. This system consists of an AnaeroPack-Anaero 5% (oxygen absorber; Mitsubishi Gas Chemical, Tokyo, Japan), an OXY-1 oxygen monitor (JIKCO, Tokyo, Japan), an AnaeroPouch (Mitsubishi Gas Chemical), and plastic clips for sealing the pouch. Briefly, PCAs, the oxygen absorber, and the oxygen monitor were arranged in the pouch, and the left open side was sealed with a clip. The O2 concentration in the pouch rapidly decreased after sealing; once a low O2 concentration (approximately 0.1%) was reached, the pouch was sealed with another clip between the culture dish and the oxygen absorber to stop further oxygen absorption. Then, the pouch was maintained in an incubator at 37°C.
Total RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay
PCAs were seeded onto non-coated 6-well plates (2 × 105 cells/well) in DMEM/F12 supplemented with 10% FBS and PS. After 3 days, the medium was changed to FBS-free medium. After overnight incubation, drug administration or hypoxic treatments were performed and followed by incubation. Total RNA was extracted from PCAs with RNeasy Mini Kit (Qiagen, Hilden, Germany), followed by conversion to cDNA with Quantitect Reverse Transcription Kit (Qiagen). Quantitative PCR was performed with TB Green Advantage qPCR Premix (Takara Bio, Tokyo, Japan) and the QuantiStudio 3 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. Beta-actin (ACTB) was used as an endogenous control. The sequences of the forward and reverse primers are described in S2 Table. The PCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was performed as described elsewhere [35]. Briefly, PCAs were seeded onto non-coated 6-well plates (2 × 105 cells/well) in DMEM/F12 supplemented with 10% FBS and PS. After 3 days, the culture medium was changed to an FBS-free medium. After overnight incubation, hypoxic treatment was performed and followed by incubation. The culture medium and cell lysate were collected from PCAs and stored at −80°C. Total protein concentration of the samples was quantified using Qubit 3.0 Fluorometer (Thermo Fisher Scientific). HIF-1α protein concentration was measured with the Mouse HIF-1-alpha SimpleStep ELISA Kit (Abcam, Cambridge, UK) according to the manufacturer’s protocol. MIF protein concentration was measured with the Mouse MIF SimpleStep ELISA Kit (Abcam) according to the manufacturer’s protocol. The absorbance at 450 nm was detected with Multiskan FC (Thermo Fisher Scientific). All samples and standards were measured in duplicate. The protein level per 1 μg of total protein was calculated.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed as described elsewhere [35]. Briefly, PCAs were seeded onto a non-coated 90-mm dish (2 × 106 cells/dish) in DMEM/F12 supplemented with 10% FBS and PS. After 3 days, the culture medium was changed to FBS-free medium. After overnight incubation, hypoxic treatment was performed and followed by incubation. For the ChIP assay, we used the ChIP-IT express kit (Active Motif, La Hulpe, Belgium) according to the manufacturer’s protocol. Briefly, PCAs were fixed with 37% formaldehyde. For shearing the chromatin of the fixed PCAs, sonication was performed with Bioruptor II (BM Equipment, Tokyo, Japan) over 10 homogenization cycles. Each homogenization cycle consisted of homogenization at the maximum setting for 30 s followed by a 30 s interval. The sheared chromatin was utilized for immunoprecipitation with the anti-HIF-1α antibody (ab1, Abcam). Real-time PCR for the immunoprecipitated DNA was performed with TB Green Advantage qPCR Premix (Takara Bio) and the QuantiStudio 3 Real-Time PCR System (Applied Biosystems). The forward and reverse primers were designed to cover 151 base pairs of the MIF promoter (Fig 1 and S2 Table). The PCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 20 s and 72°C for 34 s.
Plasmid construction
We subcloned a part of the MIF promoter region (−335 to −1), which included the HRE, as the insert region for the luciferase reporter assay (Fig 1). The forward and reverse primers ware designed with the online Primer Design tool for In-Fusion (S2 Table). Mouse genomic DNA extracted from PCAs with the AllPrep DNA/RNA Mini Kit (Qiagen) was used as the template for PCR amplification of the insert region. The reporter plasmid was generated by incorporating the PCR-amplified insert region upstream of the Nanoluc luciferase reporter gene of the pNL1.2 [NlucP] vector (Promega, Madison, WI, USA) with the In-Fusion HD Cloning Kit (Takara Bio) and Escherichia coli Competent High DH5α (Toyobo, Osaka, Japan), according to the manufacturer’s protocol. All of the mutated HRE plasmids, such as the SNP HRE, the mutant HRE, and the deletion HRE, were generated by site-directed mutagenesis with the KOD Plus Mutagenesis Kit (Toyobo). The primers harboring the desired mutations are shown in S2 Table.
Luciferase reporter assay
PCAs were seeded onto non-coated 6-well plates (2 × 105 cells/well) in DMEM/F12 supplemented with 10% FBS and PS. After 3 days, the medium was changed to FBS-free medium. After overnight incubation, each well was transfected with a DNA mixture containing 1.25 μg of the indicated reporter plasmid and 1.25 μg of pGL4.13[luc2/SV40] Vector (Promega) encoding the firefly luciferase under the control of a SV40 promoter using Lipofectamine LTX Reagent with PLUS Reagent (Thermo Fisher Scientific). At 24 h after transfection, PCAs were transferred to hypoxic conditions (0.1% O2) or left under normoxic conditions for 24 h. Cell lysate was collected from PCAs with Passive Lysis Buffer (Promega) and stored at −80°C. NanoLuc and firefly luciferase activities were determined with Nano-Glo Dual-Luciferase Reporter Assay (Promega) and GloMax Navigator Microplate Luminometer (Promega) according to the manufacturer’s protocol. In order to correct for transfection efficiency, the NanoLuc luciferase activity was normalized to the firefly luciferase activity.
Statistical analyses
Statistical analyses were performed with R version 3.6.1 (The R Foundation for Statistical Computing, Vienna, Austria) and EZR version 1.41 (Jichi Medical University, Saitama, Japan) [44]. For the SNP rs17004038 association study, we used Haploview version 4.2 (Daly Lab at the Broad Institute Cambridge, MA, USA) [45] to determine the Hardy–Weinberg equilibrium (HWE), allele frequencies, and genetic associations. Genotypic association was examined with the Cochran-Armitage trend test. Allelic association was examined with the χ2 test. For the PCA study, the group differences were analyzed using one-way ANOVA with Dunnett’s or Tukey’s multiple comparison tests, as appropriate. Statistical significance was defined as two-tailed p-value < 0.05.
Results
Association study of SNP rs17004038 in the HRE of the human MIF gene
The distribution of SNP rs17004038 (C>A) in patients with schizophrenia and controls was investigated. We found a non-significant trend for the association of SNP rs17004038 with SCZ in the first set of subjects (p = 0.0509) (S3 Table), but not in the second set (p = 0.364) (S4 Table). Next, we performed an analysis using both sets combined and found a significant association of SNP rs17004038 with SCZ (p = 0.0424). The odds ratio (1.445) suggests that the minor A allele of SNP rs17004038 may be involved in the pathophysiology of SCZ (Table 1).
Table 1. Distribution of rs17004038 in patients with schizophrenia and controls in this study.
| Genotype | Allele | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | HWE | C/C | C/A | A/A | P-valuea | C | A | MAF | P-valueb | Odds ratio (95% CI) | Power | |
| Overall | ||||||||||||
| SCZ | 1758 | 1.00 | 1677 | 80 | 1 | 0.0467 | 3434 | 82 | 0.0233 | 0.0424 | 1.445 (1.011–2.065) | 0.520 |
| CTL | 1507 | 0.012 | 1461 | 43 | 3 | 2965 | 49 | 0.0163 | ||||
| Male | ||||||||||||
| SCZ | 946 | 0.77 | 904 | 41 | 1 | 0.389 | 1849 | 43 | 0.0227 | 0.368 | 1.250 (0.769–2.032) | 0.141 |
| CTL | 739 | 0.0025 | 715 | 21 | 3 | 1451 | 27 | 0.0183 | ||||
| Female | ||||||||||||
| SCZ | 812 | 1.00 | 773 | 39 | 0 | 0.0456 | 1585 | 39 | 0.0240 | 0.0478 | 1.693 (0.999–2.869) | 0.509 |
| CTL | 768 | 1.00 | 746 | 22 | 0 | 1514 | 22 | 0.0143 | ||||
CI, Confidence interval; CTL, control; HWE, Hardy-Weinberg equilibrium; MAF, minor allele frequency; SCZ, schizophrenia.
Boldface type indicates statistical significance.
a Genotypic p-values were calculated with Cochran-Armitage trend test.
b Allelic p-values were calculated with the χ2 test.
We additionally performed subgroup analyses divided by sex. Each set of subjects showed no significant difference in the frequency of the allele between SCZ and control (S3 and S4 Tables). Combining the sets revealed a significant difference in the samples from female subjects (p = 0.0478) but not male subjects (p = 0.368), consistent with the overall analysis (Table 1).
Effects of hypoxia on mouse HIF mRNA expression and HIF protein level
First, the effect of hypoxia on HIF-1α/β mRNA expression was examined. The HIF-1α/β mRNA expression levels were slightly changed by hypoxia, although significant changes were observed in HIF-1α mRNA level at 48 h and HIF-1β mRNA level at 24 h (S1A and S1B Fig). Next, the effect of hypoxia on HIF-1α protein level was examined with ELISA. Hypoxic treatments for 24 and 48 h increased HIF-1α protein level in the cell lysates (S1C Fig).
Effects of hypoxia on mouse MIF mRNA expression
First, the effect of ML228, a HIF pathway activator, on MIF mRNA expression was examined. ML228 increased MIF mRNA expression in a dose-dependent manner 48 h after its administration (Fig 2A). Next, the effect of hypoxia on MIF mRNA expression was examined. Hypoxia increased MIF mRNA expression in a time-dependent manner, which reached significance at 6 h and increased until 48 h after the start of hypoxic treatment (Fig 2B). On the other hand, YC-1, a HIF inhibitor, significantly suppressed MIF mRNA expression 6 h after the start of hypoxic treatments (Fig 2C).
Fig 2. Hypoxia induced macrophage migration inhibitory factor (MIF) mRNA expression and protein production via the hypoxia inducible factor (HIF) pathway.
A. The effects of ML228, a HIF activator, on MIF mRNA expression in primary cultured astrocytes (PCAs). PCAs were incubated for 48 h in 0, 0.25, 0.5, or 1.0 μM ML228. MIF mRNA expression was analyzed by qRT-PCR. The values are shown as the ratio of MIF mRNA to beta-actin (ACTB) mRNA (Dunnett’s test vs. vehicle; n = 3). B. The time-dependent effects of hypoxia on MIF mRNA expression in PCAs. PCAs were incubated for 3, 6, 12, 24, or 48 h in 21.0% or 0.1% O2. MIF mRNA expression was analyzed by qRT-PCR. The values are shown as the ratio of MIF mRNA to ACTB mRNA (Student’s t-test; n = 5–6). C. The effects of the HIF inhibitor YC-1 on hypoxia-induced MIF mRNA expression in PCAs. PCAs were incubated for 6 h in 21.0% or 0.1% O2 and treated with 0 or 10 μM of YC-1. MIF mRNA expression was analyzed by qRT-PCR. The values are shown as the ratio of MIF mRNA to ACTB mRNA (Tukey’s test; n = 6). D. The effects of hypoxia on MIF protein production in cell lysate of PCAs. PCAs were incubated for 24 or 48 h in 21.0% or 0.1% O2. The MIF protein level per 1 μg of total protein was analyzed using a MIF enzyme-linked immunosorbent assay (ELISA) (Student’s t-test; n = 6). The data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, or ***p < 0.001.
Effects of hypoxia on mouse MIF protein production
To examine whether hypoxia increases MIF protein production as in the case of mRNA, the effect of hypoxia on MIF protein production was examined with ELISA. Hypoxic treatments for 24 and 48 h increased MIF protein production in the cell lysates (Fig 2D) but not in the cell culture medium (S2 Fig).
Effects of hypoxia on HIF-1α binding to the HRE of mouse MIF promoter
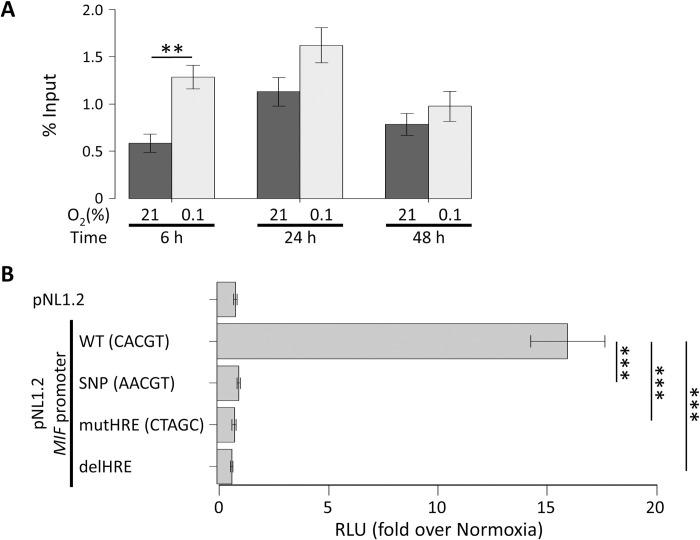
Under hypoxic conditions, the HIF-1α subunit translocates to the cell nucleus and associates with the HIF-1β subunit, and then this HIF-1α/1β complex binds to the HRE of the promoter region of its target genes. Therefore, we investigated the effects of hypoxia on the binding of HIF-1α to the MIF promoter with the ChIP assay. We found a significant increase in HIF-1α binding to the MIF promoter 6 h after the start of hypoxic treatments but not after 24 or 48 h (Fig 3A).
Fig 3. Hypoxia-inducible factor (HIF) binding to the hypoxia response element (HRE) induced activation of the macrophage migration inhibitory factor (MIF) gene promoter under hypoxia, that was abrogated by the allelic variant of the HRE.
A. The effects of hypoxia on HIF binding at the MIF gene promoter in primary cultured astrocytes (PCAs). PCAs were incubated for 6, 24, or 48 h in 21.0% or 0.1% O2. HIF-1α binding to the MIF gene promoter was analyzed by chromatin immunoprecipitation (ChIP) assay. Real-time PCR was performed on DNA purified from each of the ChIP reactions by using a primer set specific for the MIF gene promoter. The % input values indicate the ratio of immunoprecipitated DNA fragments to the input DNA fragments (Student’s t-test after logarithmic transformation; n = 6). B. The allelic variant of the HRE abrogates MIF induction by hypoxia in PCAs. PCAs were transfected with a reporter plasmid containing the MIF genomic region (−335 to −1) upstream of the Nanoluc luciferase reporter gene and a control plasmid containing the firefly luciferase reporter gene. Where indicated, the wild type (WT) HRE sequence (CACGT) was mutated to AACGT (SNP), CTAGC (mutHRE), or deletion (delHRE). At 24 h after transfection, PCAs were incubated for 24 h in 21.0% or 0.1% O2. The graph represents the corrected luciferase activity values of each construct in cells exposed to hypoxia over the luciferase activity obtained in normoxic cells (Dunnett’s test; n = 9–14). The data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, or ***p < 0.001.
Effects of the SCZ-associated SNP in the HRE on hypoxia-induced activation of the MIF promoter
The adaptation to hypoxia is mainly dependent on HIF-mediated gene expression, which means that the effect of the SCZ-associated SNP on the HRE of the MIF promoter might alter the response to hypoxia in individuals with this SNP. The HRE sequence (CACGT) of the MIF promoter identified in the human genome also exists in the mouse genome (Fig 1). In order to investigate the biological effect of the SCZ-associated SNP rs17004038, we constructed plasmids for a luciferase reporter assay including the wild-type mouse MIF promoter, the C→A variant corresponding to the SNP rs17004038, and mutation/deletion of the HRE placed upstream of the Nanoluc luciferase gene. Then, we performed luciferase reporter assays with these plasmids. Like previous studies using human cell lines [38, 39], the activity of mouse MIF promoter (WT) was robustly promoted by hypoxia in PCAs (Fig 3B). In contrast, mutation (mutHRE) and deletion (delHRE) versions of the HRE completely suppressed hypoxia-induced activation of MIF promoter (Fig 3B). In addition, the variant allele C→A also completely suppressed hypoxia-induced activation of MIF promoter as effectively as mutHRE and delHRE (Fig 3B).
Discussion
First, we investigated the SNP rs17004038 (C>A) in the HRE of the MIF promoter. We found that rs17004038 is significantly associated with SCZ. The odds ratio indicates the minor A allele of rs17004038 is a risk allele for SCZ. Sex distribution was significantly unequal between control and SCZ groups in the first set of participants. Thus, we additionally performed subgroup analyses divided by sex. The samples from female subjects showed a significant difference, consistent with overall results, but the samples from male subjects did not. One reason is that the sample size of male subjects was relatively small, considering that the same minor allele frequency was present in both samples of male (SCZ: 0.0227 vs. CTL: 0.0183) and female (SCZ: 0.0240 vs. CTL: 0.0143) subjects, and that the samples from male subjects had smaller sample power (0.141) than that of female subjects (0.509). Another reason may be the influence of sex difference in the development of SCZ. Further studies with larger samples are required.
Next, we investigated the functional role of the SNP rs17004038 on the HRE of the MIF promoter by performing biological experiments with neonatal mice-derived PCAs. First, we confirmed that hypoxia increased HIF-1α protein level in the cell lysates of PCAs, although HIF-1α/β mRNA levels were little changed. Under normoxic conditions, the intracellular concentration of HIF-1α protein is low, not due to lower protein expression but because it is negatively regulated by proteolysis by the ubiquitin-proteasome system via the von Hippel–Lindau tumor suppressor; under hypoxic conditions, HIF-1α levels are stable and it translocates to the cell nucleus and associates with HIF-1β, and the HIF-1α/1β complex binds to the HRE of the DNA resulting in the transcription of various genes [46–48]. Our experimental system using PCAs under hypoxia seems to be in agreement with the results of previous studies.
Both hypoxia and ML228, a HIF activator, increased MIF mRNA expression. Hypoxia also induced MIF protein production in cell lysates. Conversely, YC-1, a HIF inhibitor, disturbed hypoxia-induced MIF mRNA expression. In addition, a ChIP assay showed that hypoxia increased the binding of the HIF-1α subunit to the MIF promoter region including the HRE. These findings strongly indicate that the HRE plays a role in the regulation of MIF expression. Finally, the luciferase reporter assay showed that hypoxia-induced activation of the MIF promoter was disrupted by variants of the HRE including that corresponding to the SNP rs17004038, which suggests that SNP rs17004038 may be involved in the pathophysiology of SCZ through disruption of hypoxia-induced MIF expression. Our present study is the first study to show the involvement of MIF in hypoxia-associated mechanisms of SCZ.
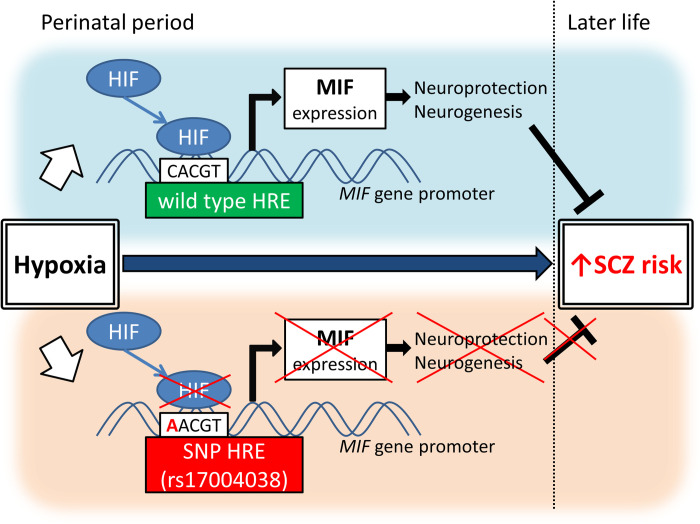
Previous studies have shown that blood MIF is increased in patients with SCZ, including initial-onset, drug-naive patients [32, 33]. Our previous studies showed that antipsychotic dosage positively correlated with MIF blood levels, that a higher-expression polymorphism of the MIF promoter is significantly lower frequent in female patients with adolescent-onset SCZ [34], and that clozapine and other antipsychotics increased MIF expression in PCAs [35]. These findings suggest that MIF may function as a physiological protective factor against SCZ risk [35]. The present study showed that SNP rs17004038 in the HRE of the MIF gene is significantly more common in patients with SCZ, and that this SNP inhibits hypoxia-induced MIF expression in PCAs from mice. Taken together, these studies suggest that elevated levels of hypoxia-induced MIF may have a protective function against perinatal hypoxia-associated SCZ development, and that SNP rs17004038 may inhibit the protective function of MIF by blocking MIF expression in response to hypoxia. MIF may play a role in the pathophysiology of SCZ through both genetic factors (i.e., the SNP) and environmental factors (i.e., hypoxia), indicating that MIF may mediate the gene-environment interaction underlying the pathophysiology of SCZ. Putative roles of SNP rs17004038 in the HRE of the MIF gene promoter in perinatal hypoxia-associated SCZ risk are shown in Fig 4.
Fig 4. Putative roles of single nucleotide polymorphism (SNP) rs17004038 in the hypoxia response element (HRE) of the macrophage migration inhibitory factor (MIF) gene promoter in perinatal hypoxia-associated schizophrenia (SCZ) risk.
Abbreviation: HIF, Hypoxia-inducible factor.
MIF is widely expressed during development in the nervous system [49, 50], which suggests that it plays a role in neural development. In fact, MIF is predominantly expressed in astrocytes, a major regulator of hippocampal neurogenesis [51], in the brain; regulates the proliferation, differentiation, and survival of neural stem/progenitor cells [30, 52–54]; and plays a role in hippocampal development [55]. Interestingly, the hippocampus is one of the most hypoxia-sensitive regions in the brain [56] and a major SCZ-associated brain region [31]. In addition, perinatal hypoxia is related to the risk of SCZ [12–16]. Taken together, these studies easily lead to the hypothesis that perinatal hypoxia increases the risk of SCZ by affecting MIF-mediated hippocampal development. Our present results showing that a SCZ-associated SNP in the HRE of the MIF promoter disrupted hypoxia-induced MIF expression in neonatal mice-derived astrocytes can be considered to validate this hypothesis.
Although we showed the association of SNP rs17004038 (C>A) with SCZ, recent large GWASs of SCZ did not identify this SNP [5–7]. We showed that the A allele frequency of rs17004038 is around 2%. This result is consistent with the Japanese data from the 1000 Genomes Project database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/) [57]. However, the A allele is found only in Asian populations, and the global A allele frequency is 0.0044 (S5 Table). Thus, rs17004038 may not have been adopted in SNP arrays used in previous GWASs because its minor allele frequency is relatively low. Therefore, our findings for SNP rs17004038 indicate the need for further studies that also consider such relatively rare variants.
Hypoxia is a common phenomenon in malignant tumors, and its roles in cancer progression, angiogenesis, and metastasis have been well studied [58]. In addition, numerous studies have shown that MIF plays an important role in advocating such tumorigenic processes [59, 60], and that HIF-induced MIF expression is associated with tumorigenesis [38]. On the other hand, hypoxia may be involved in neurological diseases such as ischemic stroke, and a few studies reported that MIF plays a beneficial role in neurological recovery after ischemic stroke [61–63]. Moreover, recombinant MIF administration is effective for inducing the expression of brain-derived neurotrophic factor and promotes neuroprotection of neuronal cells in human neuroblastoma cell lines under oxygen-glucose deprivation [64]. A recent review concluded that the role of MIF in neurological disorders is controversial and depends on the pathophysiological context and/or cellular microenvironment [65]. Considering our results and the findings of previous studies, further elucidation is required for not only the pathological but also neuroprotective role of MIF in neurological and psychiatric diseases via HIF pathway under hypoxic conditions.
Since hypoxia increased MIF mRNA expression and MIF is a secreted protein, we predicted that hypoxia may increase the MIF protein level in cell culture medium. However, contrary to our expectation, hypoxia increased the MIF protein level in the cell lysate of PCAs but not in the cell culture medium. MIF is a type of cytokine, and produced cytokines are usually stored in secretory vesicles or granules via the Golgi and then secreted by exocytosis in response to various stimuli [66]. Merk et al. showed that p115, a Golgi-associated protein, binds to MIF and facilitates its secretion [67]. These findings suggest that hypoxia may directly increase MIF protein production in astrocytes but does not affect its secretion, and that some indirect pathways may be involved in MIF protein secretion in actual brain tissues.
Our study had several limitations. First, our association study included only a Japanese population. Therefore, the results of our association study might not be generalizable to other populations. Second, we used mice-derived cells. Thus, the findings should be validated in analyses focusing on human astrocytes performed using cell lines derived from human brain tissues or induced-pluripotent stem cells. Third, the present study is an in vitro study. Therefore, the role of MIF and the HIF pathway in the pathophysiology of SCZ under hypoxia should be further investigated in in vivo models of SCZ.
Conclusions
We have shown that a SNP of the HRE in the MIF promoter is associated with SCZ, and that this SNP disrupts hypoxia-induced MIF expression via the HIF pathway in astrocytes. These findings indicate the potential role of astrocyte-derived MIF in the hypoxia-related pathophysiology of SCZ.
Supporting information
A. The time-dependent effects of hypoxia on HIF-1α mRNA expression in primary cultured astrocytes (PCAs). PCAs were incubated for 3, 6, 12, 24, or 48 h in 21.0% or 0.1% O2. HIF-1α mRNA expression was analyzed by qRT-PCR. The values are shown as the ratio of HIF-1α mRNA to beta-actin (ACTB) mRNA (Student’s t-test; n = 6). B. The time-dependent effects of hypoxia on HIF-1β mRNA expression in PCAs. PCAs were incubated for 3, 6, 12, 24, or 48 h in 21.0% or 0.1% O2. HIF-1β mRNA expression was analyzed by qRT-PCR. The values are shown as the ratio of HIF-1β mRNA to ACTB mRNA (Student’s t-test; n = 6). C. The effects of hypoxia on HIF-1α protein level in cell lysate of PCAs. PCAs were incubated for 24 or 48 h in 21.0% or 0.1% O2. The HIF-1α protein level per 1 μg of total protein was analyzed using a HIF-1α enzyme-linked immunosorbent assay (ELISA) (Student’s t-test; n = 6). The data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, or ***p < 0.001.
(TIF)
PCAs were incubated for 24 or 48 h in 21.0% or 0.1% O2. The MIF protein level per 1 μg of total protein was analyzed using a MIF enzyme-linked immunosorbent assay (ELISA). The data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, or ***p < 0.001 (Student’s t-test; n = 6).
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
We thank Y. Nagashima and H. Maeda for their expert technical assistance, as well as M. Takebayashi, K. Hisaoka-Nakashima, and N. Kajitani for their helpful advice about primary cultured astrocytes.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partly supported by grants from Japan Society of the Promotion of Science (JSPS) KAKENHI grant numbers 15K19727, 18K15483, and 21K07520 (SO), 15K09805 and 18K07556 (SB), as well as 17H04249 (AH), https://www.jsps.go.jp/j-grantsinaid/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marder SR, Cannon TD. Schizophrenia. N Engl J Med. 2019;381(18):1753–61. doi: 10.1056/NEJMra1808803 . [DOI] [PubMed] [Google Scholar]
- 2.Mueser KT, McGurk SR. Schizophrenia. Lancet. 2004;363(9426):2063–72. doi: 10.1016/S0140-6736(04)16458-1 . [DOI] [PubMed] [Google Scholar]
- 3.Dickinson D, Zaidman SR, Giangrande EJ, Eisenberg DP, Gregory MD, Berman KF. Distinct Polygenic Score Profiles in Schizophrenia Subgroups With Different Trajectories of Cognitive Development. Am J Psychiatry. 2020;177(4):298–307. doi: 10.1176/appi.ajp.2019.19050527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattane N, Richetto J, Cattaneo A. Prenatal exposure to environmental insults and enhanced risk of developing Schizophrenia and Autism Spectrum Disorder: focus on biological pathways and epigenetic mechanisms. Neurosci Biobehav Rev. 2020;117:253–78. doi: 10.1016/j.neubiorev.2018.07.001 . [DOI] [PubMed] [Google Scholar]
- 5.Ikeda M, Takahashi A, Kamatani Y, Momozawa Y, Saito T, Kondo K, et al. Genome-Wide Association Study Detected Novel Susceptibility Genes for Schizophrenia and Shared Trans-Populations/Diseases Genetic Effect. Schizophr Bull. 2019;45(4):824–34. doi: 10.1093/schbul/sby140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–8. doi: 10.1038/s41588-019-0512-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–7. doi: 10.1038/nature13595 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis J, Eyre H, Jacka FN, Dodd S, Dean O, McEwen S, et al. A review of vulnerability and risks for schizophrenia: Beyond the two hit hypothesis. Neurosci Biobehav Rev. 2016;65:185–94. doi: 10.1016/j.neubiorev.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clifton NE, Hannon E, Harwood JC, Di Florio A, Thomas KL, Holmans PA, et al. Dynamic expression of genes associated with schizophrenia and bipolar disorder across development. Translational psychiatry. 2019;9(1):74. doi: 10.1038/s41398-019-0405-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Brien HE, Hannon E, Hill MJ, Toste CC, Robertson MJ, Morgan JE, et al. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19(1):194. doi: 10.1186/s13059-018-1567-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ursini G, Punzi G, Chen Q, Marenco S, Robinson JF, Porcelli A, et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat Med. 2018;24(6):792–801. doi: 10.1038/s41591-018-0021-y . [DOI] [PubMed] [Google Scholar]
- 12.Cannon TD, Bearden CE, Hollister JM, Rosso IM, Sanchez LE, Hadley T. Childhood cognitive functioning in schizophrenia patients and their unaffected siblings: a prospective cohort study. Schizophr Bull. 2000;26(2):379–93. doi: 10.1093/oxfordjournals.schbul.a033460 . [DOI] [PubMed] [Google Scholar]
- 13.Cannon TD, Yolken R, Buka S, Torrey EF, Collaborative Study Group on the Perinatal Origins of Severe Psychiatric D. Decreased neurotrophic response to birth hypoxia in the etiology of schizophrenia. Biol Psychiatry. 2008;64(9):797–802. doi: 10.1016/j.biopsych.2008.04.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies C, Segre G, Estrade A, Radua J, De Micheli A, Provenzani U, et al. Prenatal and perinatal risk and protective factors for psychosis: a systematic review and meta-analysis. The lancet Psychiatry. 2020;7(5):399–410. doi: 10.1016/S2215-0366(20)30057-2 . [DOI] [PubMed] [Google Scholar]
- 15.Fineberg AM, Ellman LM, Buka S, Yolken R, Cannon TD. Decreased birth weight in psychosis: influence of prenatal exposure to serologically determined influenza and hypoxia. Schizophr Bull. 2013;39(5):1037–44. doi: 10.1093/schbul/sbs084 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zornberg GL, Buka SL, Tsuang MT. Hypoxic-ischemia-related fetal/neonatal complications and risk of schizophrenia and other nonaffective psychoses: a 19-year longitudinal study. Am J Psychiatry. 2000;157(2):196–202. doi: 10.1176/appi.ajp.157.2.196 . [DOI] [PubMed] [Google Scholar]
- 17.Giannopoulou I, Pagida MA, Briana DD, Panayotacopoulou MT. Perinatal hypoxia as a risk factor for psychopathology later in life: the role of dopamine and neurotrophins. Hormones (Athens). 2018;17(1):25–32. doi: 10.1007/s42000-018-0007-7 . [DOI] [PubMed] [Google Scholar]
- 18.Howell KR, Pillai A. Long-Term Effects of Prenatal Hypoxia on Schizophrenia-Like Phenotype in Heterozygous Reeler Mice. Mol Neurobiol. 2016;53(5):3267–76. doi: 10.1007/s12035-015-9265-4 . [DOI] [PubMed] [Google Scholar]
- 19.Schmidt-Kastner R, van Os J, Esquivel G, Steinbusch HW, Rutten BP. An environmental analysis of genes associated with schizophrenia: hypoxia and vascular factors as interacting elements in the neurodevelopmental model. Mol Psychiatry. 2012;17(12):1194–205. doi: 10.1038/mp.2011.183 . [DOI] [PubMed] [Google Scholar]
- 20.Schmidt-Kastner R, van Os J, H WMS, Schmitz C. Gene regulation by hypoxia and the neurodevelopmental origin of schizophrenia. Schizophr Res. 2006;84(2–3):253–71. doi: 10.1016/j.schres.2006.02.022 . [DOI] [PubMed] [Google Scholar]
- 21.El-Khodor BF, Boksa P. Long-term reciprocal changes in dopamine levels in prefrontal cortex versus nucleus accumbens in rats born by Caesarean section compared to vaginal birth. Exp Neurol. 1997;145(1):118–29. doi: 10.1006/exnr.1997.6437 . [DOI] [PubMed] [Google Scholar]
- 22.Hefter D, Marti HH, Gass P, Inta D. Perinatal Hypoxia and Ischemia in Animal Models of Schizophrenia. Frontiers in psychiatry. 2018;9:106. doi: 10.3389/fpsyt.2018.00106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadri C, Belmaker RH, Agam G. Oxygen restriction of neonate rats elevates neuregulin-1alpha isoform levels: possible relationship to schizophrenia. Neurochem Int. 2007;51(6–7):447–50. doi: 10.1016/j.neuint.2007.03.013 . [DOI] [PubMed] [Google Scholar]
- 24.Paquin V, Lapierre M, Veru F, King S. Early Environmental Upheaval and the Risk for Schizophrenia. Annu Rev Clin Psychol. 2021;17:285–311. doi: 10.1146/annurev-clinpsy-081219-103805 . [DOI] [PubMed] [Google Scholar]
- 25.Bacher M, Meinhardt A, Lan HY, Dhabhar FS, Mu W, Metz CN, et al. MIF expression in the rat brain: implications for neuronal function. Mol Med. 1998;4(4):217–30. . [PMC free article] [PubMed] [Google Scholar]
- 26.Savaskan NE, Fingerle-Rowson G, Buchfelder M, Eyupoglu IY. Brain miffed by macrophage migration inhibitory factor. Int J Cell Biol. 2012;2012:139573. doi: 10.1155/2012/139573 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conboy L, Varea E, Castro JE, Sakouhi-Ouertatani H, Calandra T, Lashuel HA, et al. Macrophage migration inhibitory factor is critically involved in basal and fluoxetine-stimulated adult hippocampal cell proliferation and in anxiety, depression, and memory-related behaviors. Mol Psychiatry. 2011;16(5):533–47. doi: 10.1038/mp.2010.15 . [DOI] [PubMed] [Google Scholar]
- 28.Gellen B, Volgyi K, Gyorffy BA, Darula Z, Hunyadi-Gulyas E, Baracskay P, et al. Proteomic investigation of the prefrontal cortex in the rat clomipramine model of depression. J Proteomics. 2017;153:53–64. doi: 10.1016/j.jprot.2016.06.027 . [DOI] [PubMed] [Google Scholar]
- 29.Su Y, Wang Y, Zhou Y, Zhu Z, Zhang Q, Zhang X, et al. Macrophage migration inhibitory factor activates inflammatory responses of astrocytes through interaction with CD74 receptor. Oncotarget. 2017;8(2):2719–30. doi: 10.18632/oncotarget.13739 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta S, Misawa A, Fukaya R, Inoue S, Kanemura Y, Okano H, et al. Macrophage migration inhibitory factor (MIF) promotes cell survival and proliferation of neural stem/progenitor cells. J Cell Sci. 2012;125(Pt 13):3210–20. doi: 10.1242/jcs.102210 . [DOI] [PubMed] [Google Scholar]
- 31.Kusumi I, Boku S, Takahashi Y. Psychopharmacology of atypical antipsychotic drugs: From the receptor binding profile to neuroprotection and neurogenesis. Psychiatry Clin Neurosci. 2014. doi: 10.1111/pcn.12242 . [DOI] [PubMed] [Google Scholar]
- 32.Chan MK, Krebs MO, Cox D, Guest PC, Yolken RH, Rahmoune H, et al. Development of a blood-based molecular biomarker test for identification of schizophrenia before disease onset. Translational psychiatry. 2015;5:e601. doi: 10.1038/tp.2015.91 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz E, Guest PC, Rahmoune H, Harris LW, Wang L, Leweke FM, et al. Identification of a biological signature for schizophrenia in serum. Mol Psychiatry. 2012;17(5):494–502. doi: 10.1038/mp.2011.42 . [DOI] [PubMed] [Google Scholar]
- 34.Okazaki S, Hishimoto A, Otsuka I, Watanabe Y, Numata S, Boku S, et al. Increased serum levels and promoter polymorphisms of macrophage migration inhibitory factor in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2018;83:33–41. doi: 10.1016/j.pnpbp.2018.01.001 . [DOI] [PubMed] [Google Scholar]
- 35.Okazaki S, Boku S, Otsuka I, Horai T, Kimura A, Shimmyo N, et al. Clozapine increases macrophage migration inhibitory factor (MIF) expression via increasing histone acetylation of MIF promoter in astrocytes. J Psychiatr Res. 2021;135:237–42. doi: 10.1016/j.jpsychires.2021.01.033 . [DOI] [PubMed] [Google Scholar]
- 36.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12(12):5447–54. doi: 10.1128/mcb.12.12.5447-5454.1992 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–4. doi: 10.1073/pnas.92.12.5510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baugh JA, Gantier M, Li L, Byrne A, Buckley A, Donnelly SC. Dual regulation of macrophage migration inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1. Biochem Biophys Res Commun. 2006;347(4):895–903. doi: 10.1016/j.bbrc.2006.06.148 . [DOI] [PubMed] [Google Scholar]
- 39.Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38(7):2332–45. doi: 10.1093/nar/gkp1205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okazaki S, Watanabe Y, Hishimoto A, Sasada T, Mouri K, Shiroiwa K, et al. Association analysis of putative cis-acting polymorphisms of interleukin-19 gene with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014;50:151–6. doi: 10.1016/j.pnpbp.2013.12.006 . [DOI] [PubMed] [Google Scholar]
- 41.Maruyama S, Boku S, Okazaki S, Kikuyama H, Mizoguchi Y, Monji A, et al. ATP and repetitive electric stimulation increases leukemia inhibitory factor expression in astrocytes: A potential role for astrocytes in the action mechanism of electroconvulsive therapy. Psychiatry Clin Neurosci. 2020;74(5):311–7. doi: 10.1111/pcn.12986 . [DOI] [PubMed] [Google Scholar]
- 42.Itoi F, Tokoro M, Terashita Y, Yamagata K, Fukunaga N, Asada Y, et al. Offspring from mouse embryos developed using a simple incubator-free culture system with a deoxidizing agent. PLoS One. 2012;7(10):e47512. doi: 10.1371/journal.pone.0047512 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaida A, Miura M. Differential dependence on oxygen tension during the maturation process between monomeric Kusabira Orange 2 and monomeric Azami Green expressed in HeLa cells. Biochem Biophys Res Commun. 2012;421(4):855–9. doi: 10.1016/j.bbrc.2012.04.102 . [DOI] [PubMed] [Google Scholar]
- 44.Kanda Y. Investigation of the freely available easy-to-use software ’EZR’ for medical statistics. Bone Marrow Transplant. 2013;48(3):452–8. doi: 10.1038/bmt.2012.244 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457 . [DOI] [PubMed] [Google Scholar]
- 46.Kaelin WG Jr., The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8(11):865–73. doi: 10.1038/nrc2502 . [DOI] [PubMed] [Google Scholar]
- 47.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399(6733):271–5. doi: 10.1038/20459 . [DOI] [PubMed] [Google Scholar]
- 48.Semenza GL. Hydroxylation of HIF-1: Oxygen Sensing at the Molecular Level. Physiology. 2004;19(4):176–82. doi: 10.1152/physiol.00001.2004 [DOI] [PubMed] [Google Scholar]
- 49.Ito K, Yoshiura Y, Ototake M, Nakanishi T. Macrophage migration inhibitory factor (MIF) is essential for development of zebrafish, Danio rerio. Dev Comp Immunol. 2008;32(6):664–72. doi: 10.1016/j.dci.2007.10.007 . [DOI] [PubMed] [Google Scholar]
- 50.Suzuki M, Takamura Y, Maeno M, Tochinai S, Iyaguchi D, Tanaka I, et al. Xenopus laevis macrophage migration inhibitory factor is essential for axis formation and neural development. J Biol Chem. 2004;279(20):21406–14. doi: 10.1074/jbc.M311416200 . [DOI] [PubMed] [Google Scholar]
- 51.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417(6884):39–44. doi: 10.1038/417039a . [DOI] [PubMed] [Google Scholar]
- 52.Ohta S, Misawa A, Lefebvre V, Okano H, Kawakami Y, Toda M. Sox6 up-regulation by macrophage migration inhibitory factor promotes survival and maintenance of mouse neural stem/progenitor cells. PLoS One. 2013;8(9):e74315. doi: 10.1371/journal.pone.0074315 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohta S, Yaguchi T, Okuno H, Chneiweiss H, Kawakami Y, Okano H. CHD7 promotes proliferation of neural stem cells mediated by MIF. Molecular brain. 2016;9(1):96. doi: 10.1186/s13041-016-0275-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Chen L, Wang Y, Ding Y, Peng Z, Duan L, et al. Macrophage migration inhibitory factor promotes proliferation and neuronal differentiation of neural stem/precursor cells through Wnt/beta-catenin signal pathway. Int J Biol Sci. 2013;9(10):1108–20. doi: 10.7150/ijbs.7232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chai X, Zhang W, Li L, Wu Y, Zhu X, Zhao S. Profile of MIF in Developing Hippocampus: Association With Cell Proliferation and Neurite Outgrowth. Front Mol Neurosci. 2020;13:147. doi: 10.3389/fnmol.2020.00147 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hossmann KA. The hypoxic brain. Insights from ischemia research. Adv Exp Med Biol. 1999;474:155–69. . [PubMed] [Google Scholar]
- 57.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia. 2015:83. doi: 10.2147/hp.s93413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bach JP, Deuster O, Balzer-Geldsetzer M, Meyer B, Dodel R, Bacher M. The role of macrophage inhibitory factor in tumorigenesis and central nervous system tumors. Cancer. 2009;115(10):2031–40. doi: 10.1002/cncr.24245 . [DOI] [PubMed] [Google Scholar]
- 60.O’Reilly C, Doroudian M, Mawhinney L, Donnelly SC. Targeting MIF in Cancer: Therapeutic Strategies, Current Developments, and Future Opportunities. Med Res Rev. 2016;36(3):440–60. doi: 10.1002/med.21385 . [DOI] [PubMed] [Google Scholar]
- 61.Chang MC, Park CR, Rhie SH, Shim WH, Kim DY. Early treadmill exercise increases macrophage migration inhibitory factor expression after cerebral ischemia/reperfusion. Neural regeneration research. 2019;14(7):1230–6. doi: 10.4103/1673-5374.251330 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Zis O, Ma G, Shan Z, Zhang X, Wang S, et al. Upregulation of macrophage migration inhibitory factor gene expression in stroke. Stroke. 2009;40(3):973–6. doi: 10.1161/STROKEAHA.108.530535 . [DOI] [PubMed] [Google Scholar]
- 63.Zis O, Zhang S, Dorovini-Zis K, Wang L, Song W. Hypoxia signaling regulates macrophage migration inhibitory factor (MIF) expression in stroke. Mol Neurobiol. 2015;51(1):155–67. doi: 10.1007/s12035-014-8727-4 . [DOI] [PubMed] [Google Scholar]
- 64.Bae SH, Yoo MR, Kim YY, Hong IK, Kim MH, Lee SH, et al. Brain-derived neurotrophic factor mediates macrophage migration inhibitory factor to protect neurons against oxygen-glucose deprivation. Neural regeneration research. 2020;15(8):1483–9. doi: 10.4103/1673-5374.274340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leyton-Jaimes MF, Kahn J, Israelson A. Macrophage migration inhibitory factor: A multifaceted cytokine implicated in multiple neurological diseases. Exp Neurol. 2018;301(Pt B):83–91. doi: 10.1016/j.expneurol.2017.06.021 . [DOI] [PubMed] [Google Scholar]
- 66.Stanley AC, Lacy P. Pathways for cytokine secretion. Physiology. 2010;25(4):218–29. doi: 10.1152/physiol.00017.2010 . [DOI] [PubMed] [Google Scholar]
- 67.Merk M, Baugh J, Zierow S, Leng L, Pal U, Lee SJ, et al. The Golgi-associated protein p115 mediates the secretion of macrophage migration inhibitory factor. J Immunol. 2009;182(11):6896–906. doi: 10.4049/jimmunol.0803710 . [DOI] [PMC free article] [PubMed] [Google Scholar]