Abstract
Early detection of biomarkers in lung cancer is one of the best preventive strategies. Although many attempts have been made to understand the early events of lung carcinogenesis including cigarette smoking (CS) induced lung carcinogenesis, the integrative metabolomics and next-generation sequencing approaches are lacking. In this study, we treated the female A/J mice with CS carcinogen 4-[methyl(nitroso)amino]-1-(3-pyridinyl)-1-butanone (NNK) and naturally occurring organosulphur compound, diallyl sulphide (DAS) for 2 and 4 weeks after NNK injection and examined the metabolomic and DNA CpG methylomic and RNA transcriptomic profiles in the lung tissues. NNK drives metabolic changes including mitochondrial tricarboxylic acid (TCA) metabolites and pathways including Nicotine and its derivatives like nicotinamide and nicotinic acid. RNA-seq analysis and Reactome pathway analysis demonstrated metabolism pathways including Phase I and II drug metabolizing enzymes, mitochondrial oxidation and signaling kinase activation pathways modulated in a sequential manner. DNA CpG methyl-seq analyses showed differential global methylation patterns of lung tissues from week 2 versus week 4 in A/J mice including Adenylate Cyclase 6 (ADCY6), Ras-related C3 botulinum toxin substrate 3 (Rac3). Oral DAS treatment partially reversed some of the mitochondrial metabolic pathways, global methylation and transcriptomic changes during this early lung carcinogenesis stage. In summary, our result provides insights into CS carcinogen NNK’s effects on driving alterations of metabolomics, epigenomics and transcriptomics and the chemopreventive effect of DAS in early stages of sequential lung carcinogenesis in A/J mouse model.
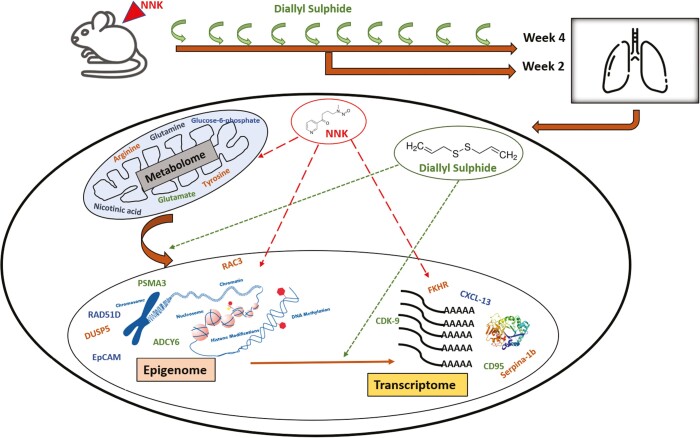
Graphical Abstract
Graphical Abstract.
NNK induced metabolic rewiring and epigenetic reprogramming and prevention with diallyl sulphide (DAS).
Introduction
Lung cancer is the leading cause of cancer death in the past few decades worldwide. Early monitoring of lung cancer development is critical and essential for improving prognosis and prolonging life/survival. Various methods including liquid biopsy, complete blood cell count-derived inflammatory biomarkers, screening of circulating tumor cells are some of the emerging approaches for early biomarkers discovery (1–3). Multiple early diagnosis and monitoring approaches of lung cancer at different biological levels have all been validated to be effective and applicable for clinical practice; such methods include evaluation of DNA variations (4), CpG island methylations (5), and miRNA (6) distributions in the circulatory system. Although early detection is a promising approach, the technological limitation, not only limited to low dose computed tomography (LD-CT), but sensitivity of circulating biomarker detection remains challenging.
Cancer chemoprevention by administration of dietary phytochemicals with relatively low toxicity is one of the oldest and proven approach to delay the progression of carcinogenesis including lung carcinogenesis. Dietary polyphenols including phenethyl isothiocyanate (PEITC), curcumin, tea polyphenols have been shown to be effective against various lung cancer types including adenocarcinoma, small cell lung carcinoma (7–11). Dietary cancer chemopreventive agents including polyphenols generally are known to act by affecting metabolism of carcinogens by Phase I and II enzymes (12), modulation of signaling kinases (13), regulating the epigenetics including miRNA, histone modifications and regulating DNA methyl transferases (DNMTs) and histone deacetylases (HDACs) (14). Gradually advancing technology and most recently, the advent of next-generation sequencing (NGS), combined with bioinformatics analytic tools, have revolutionized our ability to interrogate cancer cells. The ultimate goal is to apply these high-throughput technologies to the various aspects of cancer studies including target identification for treatment and prevention. For instance, the application of NGS in studying biomarkers like the transcriptomic and epigenomic alterations in UVB induced skin carcinogenesis (15), gene expression profiling signatures for the diagnosis and prevention of oral cavity carcinogenesis induced by 4-nitroquinoline 1-oxide (4-NQO) (16).
Recently efforts have been made to understand the connectivity between metabolic rewiring and epigenetic reprogramming in the various cancers (17). Methylation of DNA is mediated by DNA methyltransferases (DNMT) that use S-adenosylmethionine (SAM) as methyl donor. Methylation of histones also requires SAM where the methylation occurs at the lysine or arginine residue and the reaction is catalyzed by histone methyltransferases (HMT). Therefore, the abundance or availability of SAM can directly affect the methylation status of DNA and histone proteins (18). Frequently activated pathways including PI3K pathway, AKT has been known as the major regulator of glucose uptake and enhances glucose metabolism via glycolysis and PPP (19). In non-small cell lung cancer (NSCLC) cells with oncogenic K-Ras or B-Raf fueling the tumors, autophagy has been found to sustain mitochondrial function by providing intracellular supply of glutamine (20). Qu et al. had demonstrated cigarette smoke carcinogens influences metabolic homeostasis in the benzo[a]pyrene (B(a)P) and 4-[methyl(nitroso)amino]-1-(3-pyridinyl)-1-butanone (NNK) treated A/J mouse model (21). However, the attempts to understand the interlink of metabolic rewiring and its influence on epigenetic reprogramming influencing transcriptional regulation was limited.
The objective of the present study is to identify the early events in the tobacco-specific carcinogen (TSCs) induced lung carcinogenesis. We have used carcinogen, 4-[methyl(nitroso)amino]-1-(3-pyridinyl)-1-butanone, from the class nicotine-derived nitrosamine ketone NNK in A/J mice model system to understand the early transcriptomic, epigenomic and metabolomic changes in the Week 2 and 4 postcarcinogen treatment using LCMS metabolomics and next-generation sequencing. Our study identified the signature molecular markers in the sequential lung carcinogenesis process, which acts as an early driver in the process of carcinogenesis.
Material and methods
Glyceryl trioctonoate and corn oil are purchased from Sigma-Aldrich (St. Louis, MO, USA) while 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK, 98% purity) and diallyl sulfide (DAS) was obtained from Cayman Chemicals (Ann Arbor, MI, USA). NNK was dissolved in glyceryl trioctonoate to the final concentration of 24 μM in 0.1 ml. DAS was dissolved in corn oil to the concentration 100 μM/0.1 ml. DNA/RNA isolation was carried out by AllPrep DNA/RNA Mini kit by Qiagen (Valencia, CA). Mouse SureSelectXT RNA Direct Library Preparation kit and mouse SureSelect Methyl-seq Target Enrichment kit was obtained from Agilent (Santa Clara, CA).
Animal treatment
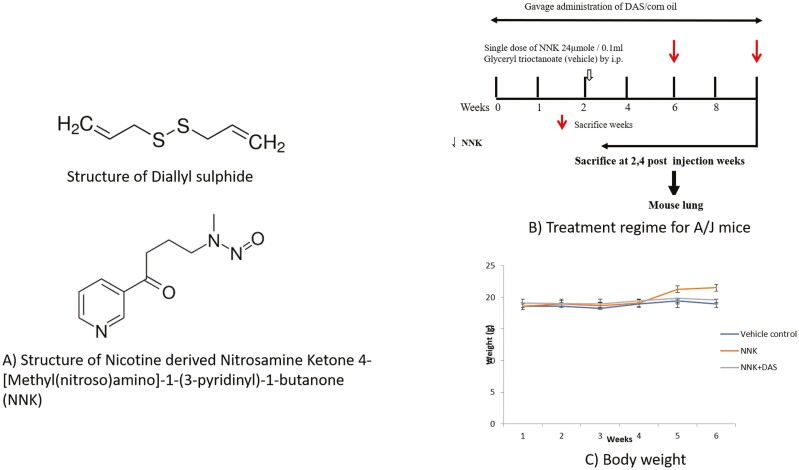
All mice were housed in our animal facility in accordance with the protocol approved (PROTO201800155) by the Rutgers University Institutional Animal Care and Use Committee (IACUC). All mice were maintained under standard 12-h light/12-h dark cycles with water and diet provided ad libitum. Female A/J mice, 6–8 weeks old were received from Jackson lab (Maine, USA) and randomized into three different groups each group containing three animals. Vehicle Control (VC) and NNK group received corn oil with oral gavage (0.1 ml) while NNK + DAS received DAS dissolved in corn oil (100 μM/0.1 ml) (Figure 1A). After 2 weeks of priming, mice from NNK and NNK + DAS group were received single dose of NNK (24 μM/0.1 ml) while VC group received glyceryl trioctonoate by intraperitoneal injection (Figure 1B). Body weights were monitored every week. Mice were sacrificed 2- and 4-weeks post injection. Lung tissue was excised and harvested and snap-frozen in liquid nitrogen followed by storage at −80°C.
Figure 1.
(A) Structure of diallyl sulphide (DAS) and nicotine-derived nitrosamine ketone (NNK). (B) Experimental design for studying the early molecular changes induced by DAS on NNK-treated A/J mice. 7 weeks old female A/J mice were randomized in three groups, vehicle control (VC) and NNK group received corn oil with oral gavage (0.1 ml) while NNK + DAS received DAS dissolved in corn oil (100 μM/0.1 ml). After 2 weeks of priming, NNK and NNK + DAS groups received single dose of NNK (24 μM/0.1 ml) while VC group received glyceryl trioctonoate by intraperitoneal injection (shown by hollow arrow). Animals were sacrificed 2- and 4-week postinjection (shown by solid arrow). All the five mouse lung lobes were excised and snapped frozen in liquid nitrogen followed by storage at –80°C. Body weight of the animals: Body weights of the mice in each group (VC, NNK, NNK + DAS) were monitored once every week and represented as a mean weight of the group at each time point.
Metabolite extraction and LCMS analysis of lung tissues
Lung tissues from three groups (VC, NNK and NNK + DAS) with two time points of week 2 and week 4 were subjected to organic extraction of cellular metabolites for LC–MS analysis. Briefly, 30 mg tissue will be weighed and pulverized with Yttria Grinding ball using CryoMill at 20 Hz for 2 min to ensure completely homogenized tissue. All the preparation will be done on ice. 40:40:20 methanol, acetonitrile and water with formic acid will be used for the organic extraction of the metabolites. Post 20 min incubation, samples will be centrifuged at 14 000g for 10 min and steps will be repeated. Final supernatant will be transferred to the tube containing 0.08 times volume of 15% Ammonium bicarbonate for analysis using LCMS as reported previously (22). Briefly, LC separation was performed on a XBridge BEH Amide column (2.1 mm × 150 mm, 2.5 μm particle size, 130 Å pore size; Waters) coupled with a Waters XBridge BEH XP VanGuard cartridge (2.1 mm × 5 mm, 2.5 μm particle size, 130 Å pore size) guard column. The column over temperature was set to 25°C. The solvent A prepared by water/acetonitrile (95:5, v/v) with 20 mm NH3AC and 20 mm NH3OH at pH 9; and solvent B prepared by acetonitrile/water (80:20, v/v) with 20 mm NH3AC and 20 mm NH3OH at pH 9 in the following solvent B percentages over time: 0 min, 100%; 3 min, 100%; 3.2 min, 90%; 6.2 min, 90%; 6.5 min, 80%; 10.5 min, 80%; 10.7 min, 70%; 13.5 min, 70%; 13.7 min, 45%; 16 min, 45%; 16.5 min, 100%. The flow rate was set to 300 μl/min with an injection volume 5 μl. The column temperature was set at 25°C. MS scans were obtained in both positive and negative ion mode with a resolution of 70 000 at m/z 200, in addition to an automatic gain control target of 3 × 106 and m/z scan range of 72–1000. The data was analyzed in three independent tissues from each group (VC, NNK and NNK + DAS) from two time points (weeks 2 and 4). The data was analyzed for statistical and pathway analysis using metaboAnalyst 5.0 software.
Methyl-seq library preparation
Methyl-seq library preparation was performed using Agilent SureSelect Mouse Methyl-Seq kit as described in the manufacturer’s protocol (Methyl-seq protocol, Version X, August 2017). 3 μg of DNA was used in the library preparation. 550 ng of adaptor ligated DNA was used for the hybridization capture and the indexed library was prepared. The following changes were made to minimize the loss of DNA in the process of enzymatic reactions. AMPure XP beads (Beckman Coulter, USA) were incubated with DNA reaction mix for 10 min at room temperature prior to pelleting by magnetization. AMPure XP beads were then washed twice with 80% ethanol and dried at 37°C for 5 min. DNA was dissolved for 10 min at 37°C. AMPure beads were retained in the solution for adenylation and end-repair reaction. An equal volume of binding buffer was added to this reaction mix at 1:1 ratio to enable the AMPure XP beads to rebind to DNA, incubated at room temperature for 10 min. DNA was further purified using AMPure XP beads followed by concentration and size of DNA fragments were determined by Agilent Bioanalyzer 2100.
RNA-seq sample preparation and next-generation sequencing analysis
Total RNA was extracted from snap-frozen lung tissue of all the experimental groups using the AllPrep DNA/RNA Mini Kit (Qiagen, CA, USA). The quality and quantity of the extracted RNA samples were determined with an Agilent 2100 Bioanalyzer. The library was constructed using the SureSelectXT RNA Direct Library Preparation kit according to the manufacturer’s manual. Samples were sequenced on the Illumina HiSeq 2500 instrument with 150 bp paired end reads, to a minimum depth of 30–40 million reads per sample.
Computational analyses of RNA-seq and Methyl-seq next-generation sequencing data
Cufflinks v2.2.1 (24) program cuffdiff was used to calculate expression levels, using the UCSC gene annotations and default parameters, as previously described (25). Data quality was checked using the FastQC 0.11.2 software. The reads were compared with standard mouse genome (mm9) provided by the UCSC database and treated by HISAT2. Further, differential gene expression analysis was performed by the DESeq2 of R package as previously (23,24). For statistical analysis, two comparisons (VC with NNK and NNK with NNK + DAS) were analyzed by p value and false discovery rate as described by Storey et al.
Reactome pathway analysis
The gene expression levels with log2 fold changes ≥0.3 were investigated by the Reactome Pathway Analysis software. The top 20 canonical pathways associated with NNK and DAS treatment were identified.
Validation by quantitative PCR
The mRNAs significantly modulated by NNK and NNK + DAS treatment at week 2 and week 4 were determined using quantitative PCR (qPCR) analysis. RNA (previously used for RNA-seq library preparation) was reversed-transcribed by TaqMan and analyzed by the QuantStudio 5 Real-time PCR system using SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA). The relative fold change was normalized to the internal standard gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
Statistical analysis
Statistical analysis between groups was performed by Student’s t test. P value <0.05, and false discovery rate (FDR) <10% were considered significant, except as otherwise stated. Metabolites were analyzed using analysis of variance (ANOVA) and P < 0.05 was considered significant. All the data was analyzed with at least three animals in each group per time point.
Results
Effect of NNK and DAS treatment on body weight of A/J mice
There were no signs of toxicity and no mortality was observed in any of the groups of A/J mice. At the time of sacrifice, no gross changes in the kidney, spleen, liver, stomach or intestine were seen at varying treatment periods. Final body weights were found to be constant at the termination time points in all the groups as compared to its initial probably suggesting lack of toxicity in the all the treated groups (Figure 1C).
Effect of DAS on cellular metabolites in NNK induced lung tissues of A/J mice
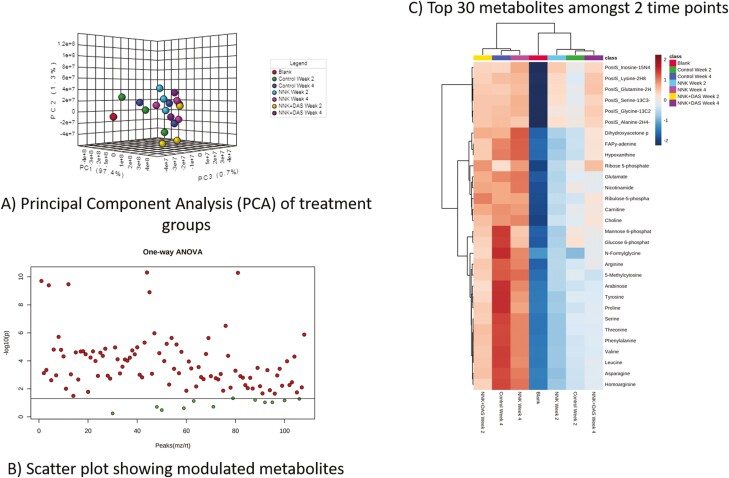
Cellular metabolites are known to modulate the different cellular process including epigenetic reprogramming and consequentially transcriptomic regulation (25–28). Hence, metabolomic landscape using LC–MS analysis from all three groups (VC, NNK and NNK + DAS) was studied. Principle component analysis (PCA) plot demonstrated the independent distribution of samples (Control, NNK and NNK + DAS) of both week 2 and week 4 (Figure 2A). All the NNK-treated as well as NNK + DAS-treated samples were clustered together as compared to their respective controls. Many metabolites were significantly detected in the lung in comparison of both time points in all three groups, VC, NNK and NNK + DAS showing stronger differences amongst the compared groups (Figure 2B). We next analyzed the top modulated cellular metabolites in comparing the groups of the two time points, week 2 and week 4. Top deregulated metabolites include the Tricarboxylic acid (TCA) cycle metabolites including Glutamate and glutamine, Amino acid metabolism derivatives like Arginine, tyrosine, alanine, lysine, serine and glucose metabolism metabolites like Glucose-6-phosphate (Figure 2C). Interestingly the metabolites are mainly downregulated in NNK at week 2 group but upregulated in the NNK-treated week 4 group, demonstrating differential metabolic rewiring induced by NNK. Similarly, metabolites in NNK + DAS at week 2 and week 4 were modulated as compared to their respective NNK-treated controls. Also, week 2 and week 4 showed different level of metabolites in NNK versus NNK + DAS group anticipating differential role of same metabolites in the cancer progression.
Figure 2.
Metabolite profiling of A/J mice lung tissues treated with NNK and NNK + DAS using LC−MS. (A) Principal component analysis (PCA) plot in which each dot demonstrating a sample in each group (VC, NNK and NNK + DAS) at two time points (week 2 and week 4). (B) Scatter plot showing the modulated metabolites in the overall comparison (VC versus NNK and NNK versus NNK + DAS at week 2 and week 4) using one-way ANOVA (*P < 0.05). Each red dot represents one metabolite with P < 0.05 while the green dot represents metabolite without statistical significance. X-axis of the plot represents peak. (C) Heatmap showing top 30 differentially modulated metabolites in each group with upregulated and downregulated metabolites in each group.
Importantly, NNK with the parental origin of Nicotine, various nicotine derivatives including nicotinamide, methyl-nicotinamide and nicotinic acid were predominantly formed in NNK-treated mice lung tissue (Supplementary Figure 1A). The NNK and NNK + DAS showed significant upregulation of Nicotine and its derivative at all the time points as compared to its respective control groups. The exact position of the nicotine derivative, nicotinic acid and nicotinamide which involves in the metabolic pathway of nicotinate and nicotinamide pathway are shown in (Supplementary Figure 1B).
DAS induces the methylation changes in NNK-treated lung tissues of A/J mice
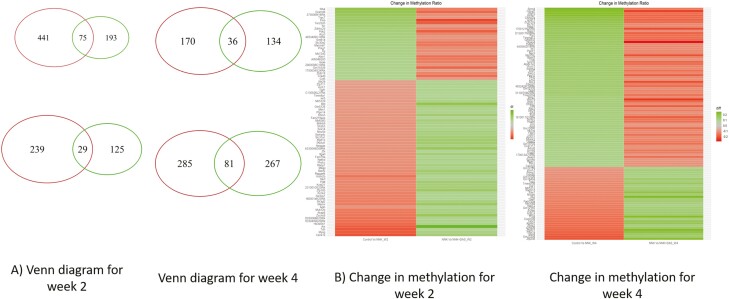
Epigenetics as one of the regulatory mechanisms of transcription, global CpG methylation was evaluated in lung tissues isolated from A/J mice from all three groups (VC, NNK and NNK + DAS). Percent of methylated CpG by regions in the gene is shown in Supplementary Fgure 2A. Interestingly, maximum % of DMRs are associated with distal intergenic regulatory region followed by promotor region of the gene (Supplementary Figure 2A). Further, distribution of differentially methylated regions (DMRs) in the gene region showed that the body of the gene regions and downstream region of the gene followed by promotor region with the highest counts of DMRs (represented on Y-axis) by number of CpG (represented on X-axis) as compared to other regions of the gene (Supplementary Figure 2B). At week 2, number of DMRs in each group’s comparison (VC versus NNK and NNK versus NNK + DAS) and their overlap identified 516 genes to be hypomethylated in VC versus NNK while 268 genes were hypermethylated in NNK versus NNK + DAS (Figure 3A). Similarly, 154 genes were hypermethylated in NNK versus NNK + DAS while 268 genes were hypomethylated in VC versus NNK comparison (Figure 3A). The methylation difference of threshold of >0.1 was used to identify the DMRs and top differentially methylated genes in both comparisons of VC versus NNK and NNK versus NNK + DAS are represented in the heatmap (Figure 3B). In week 2, hypermethylated genes in VC versus NNK showed hypomethylated in NNK versus NNK + DAS. The complete list of genes differentially methylated in both the comparisons in week 2 is represented in Supplementary Table 1. Genes including transient receptor potential cation channel subfamily C member 7 (TRPC7) involved in the proliferative process in lung cancer, pyruvate dehydrogenase kinase isoform 2 (PDK2), ataxin 1 (ATXN1) known to be involved in checkpoint regulation pathways, showed hypermethylation in VC versus NNK comparison while hypomethylated in the NNK versus NNK + DAS comparison. Importantly, hypomethylated genes includes NFKB repressing factor (NKRF), SRY-box transcription factor 14 (SOX14) transcription factor gene which regulates p53 signaling in the lung carcinogenesis process were hypomethylated, demonstrating the early epigenetic changes at week 2.
Figure 3.
Regulation of differentially methylated regions (DMRs) in NNK-treated lung tissues and effect of DAS in A/J mice model. (A) Venn diagram demonstrating the hypermethylated and hypomethylpriated genes in VC versus NNK & NNK versus NNK + DAS groups at week 2 and week 4. Genes with q < 0.05 and log2(fold change) >1 or <−1 were considered for the given analysis. (B) Heatmap showing the genes with differentially methylated regions (DMRs) in both VC versus NNK and NNK versus NNK + DAS group in both week 2 and week 4 time points.
Week 4 change in methylation demonstrated most of the genes hypermethylated in VC versus NNK comparison was hypomethylated in the NNK versus NNK + DAS comparison (Figure 3B). The genes like dual specificity phosphatase 5 (DUSP5), epithelial cellular adhesion molecule (EpCAM), adenylate cyclase 6 (ADCY6), Ras-related C3 botulinum toxin substrate 3 (Rac3), which regulates downstream pathways in the lung carcinogenesis were hypermethylated by NNK treatment while hypomethylated with DAS treatment. Potassium calcium-activated channel subfamily N member 1 (KCNN1), proteasome 20S subunit alpha 3 (PSMA3), homologous recombination DNA repair pathway gene (Rad51D) were hypomethylated by NNK treatment but hypermethylated by DAS. The differentially methylated genes in both the comparisons at week 4 are represented in Supplementary Table 2.
DAS induces transcriptomic changes in NNK-treated lung tissues of A/J mice
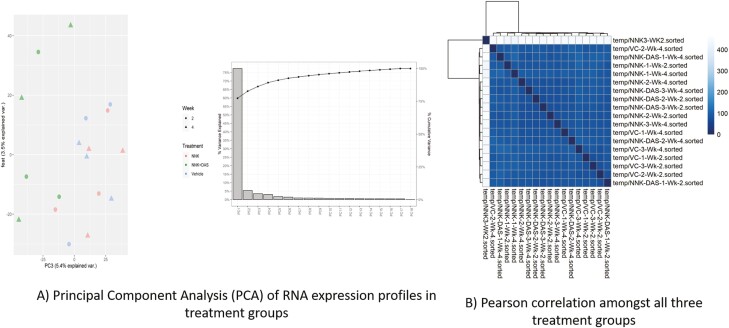
To understand the global transcriptomic changes in the lung tissues treated with NNK and DAS, RNA-seq analysis were performed using RNA isolated from total lung tissues. The initial principal component analysis (PCA) demonstrated the sample distribution of all three groups (VC, NNK and NNK + DAS) week 2 as well as week 4 in Figure 4A. Samples in each group showed to be independently standing out with the minimal overlap featuring the biologically differential expression amongst the three independent groups. We next compared the distribution of raw count in both the time points (week 2 and week 4) for all the samples in three groups. The comparison demonstrated the similarity in the distribution of raw count with both upper and lower quartiles amongst the samples of all three groups (VC, NNK and NNK + DAS) in Figure 4B.
Figure 4.
Effect of DAS on RNA-seq analysis in NNK-treated A/J mice model. (A) Principal component analysis (PCA) of RNA expression profiles between VC, NNK and NNK + DAS groups at week 2 and week 4 time points. (B) Pearson correlation amongst all three groups at two time points clustered by Euclidean distance.
The global view of the differential genes was represented using MA plot, with the log2 fold change on the Y-axis over the mean of normalized counts on X-axis. The distribution of gene expression with variation has been represented by blue dots while the gray dots represent the distribution of unchanged expression within the comparison. At week 2, comparison between the differential gene expression showed the NNK induced lowering of log fold gene expression as compared to its respective controls including VC as well as NNK + DAS (Supplementary Figure 3). While at week 4, the NNK induced distribution of changed gene expression (blue dots) seems to be predominant as compared to its vehicle control. However, the effect of DAS treatment on NNK induced change on gene expression remains marginal (Supplementary Figure 3).
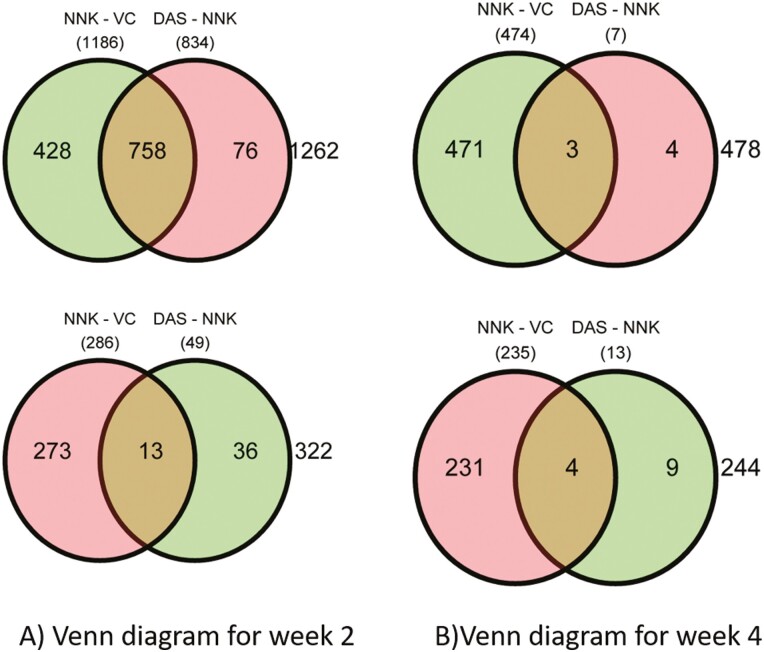
Venn diagram demonstrated, at week 2 the 1186 genes were upregulated while 286 genes were downregulated in NNK versus VC (Figure 5A). Similarly, 834 genes were downregulated while 49 genes were upregulated in NNK versus NNK + DAS treatment. Transcription factors like FOXO1 (FKHR), one of the fork head family of transcription factors, Fas (CD95) and its ligand FasL signal apoptosis and are involved in tissue homeostasis, Prostaglandin E synthase 2 (PTGES2) involving in inflammation were shown to be top upregulated genes (Supplementary Table 3). Cyclin-dependent kinase-9 (CDK-9), chemokine ligand-13 (CXCL-13), FRAT Regulator of WNT Signaling Pathway 1 (FRAT-1) shown to be downregulated by NNK (Supplementary Table 4). At week 4, 474 genes were upregulated while 253 genes were downregulated in NNK versus VC comparison (Figure 5B). While seven genes were downregulated while 13 genes were upregulated in NNK versus NNK + DAS comparison. The cross section represented the common genes in the indicated comparisons. Junctophilin-2 (JPH2), serine protease inhibitor (Serpina-1b), transthyretin (TTR) shown to be upregulated by NNK treatment (Supplementary Table 5). While NOTCH Regulated Ankyrin Repeat Protein (NRARP), Phosphogluconate Dehydrogenase (PGD), T-Complex 11 Like 2 (TCP11L2), Serine/threonine-protein kinase, Unc-51 like autophagy activating kinase (ULK1) shown to be downregulated (Supplementary Table 6). Further qPCR validation of the representative gene list (Supplementary Figure 4) at both the weeks demonstrating the week 4, NNK versus NNK + DAS showed marginal modulation of the genes (up or downregulation) as compared to either of the comparisons suggesting DAS has more impact in week 2 than in week 4.
Figure 5.
Overview of early changes in differentially expressed genes (DEGs) between NNK and NNK + DAS treatment in A/J mice model: Venn diagram demonstrating number of overexpressed (green) and downregulated (pink circle) genes in both VC versus NNK and NNK versus NNK + DAS groups at (A) week 2 and (B) week 4. Genes with q < 0.05 and threshold of log2(fold change) >1 or <–1 were considered for the given analysis.
NNK modulates canonical biochemical pathways in lung tissues of A/J mice
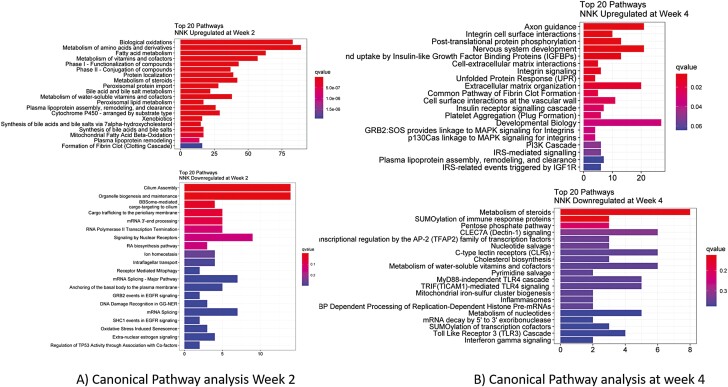
To dissect the transcriptome, the pathway analysis using Reactome was performed. Top 20 pathways upregulated and downregulated at week 2 showed significantly upregulated pathways include Phase I and Phase II drug metabolism pathways, and metabolic pathways including steroids, lipids, vitamins and co-factors and mitochondrial oxidative pathways (Figure 6A). Interestingly, downregulated pathways include crucial metabolic pathway for lung cancer progression including GRB2 events and SHC-1 events in EGFR signaling, regulation of TP53 association, estrogen signaling and oxidative stress induced senescence (Figure 6A). At week 4, upregulation of various signaling pathways driving the carcinogenesis process includes integrin signaling, PI3K cascade, IRS mediated signaling pathway, MAPK signaling pathways (Figure 6B). Majority of the downregulated pathways include various immunological response pathways including SUMOylation of immune response proteins, MyD88 independent TLR4 cascade, inflammasome, TLR3 cascade, interferon gamma signaling pathways. Other biochemical pathways including regulation of AP-2 transcription factor family are also upregulated.
Figure 6.
Differential pathway analysis between NNK and NNK + DAS treatment in A/J mice: Reactome pathway analysis showing the top 20 canonical pathways upregulated by the NNK as compared to VC while downregulation by DAS treatment at (A) week 2, (B) week 4.
Discussion
Lung cancer survival statistics are very poor considering the survival ranking among the poorest of all cancers despite the addition of the latest targeted therapies and immunotherapies. But early detection has proven effective in the prevention of severity in the disease management. Clinically, low dose computed tomography (LD-CT), chest X-ray, bronchoscopy along with detection of circulating biomarkers and liquid biopsies in target population has proven effective modes of early detection. However, to date, no early screening approach has been shown to decrease mortality from the disease. Remarkably, no reliable circulating biomarkers of lung cancer are available to aid in either early diagnosis or monitoring of disease recurrence. Efforts to identify such biomarkers are ongoing and involve the application of novel technologies such as gene expression analysis and mass spectrometry–based proteomics.
Various mouse models have been developed to understand the role of specific gene/pathway in the progression of lung cancer. These models are generally developed based on frequently occurring mutations in the crucial genes including oncogenes and tumor suppressor genes. Tobacco carcinogens induced lung tumors in mice are similar in morphology, histopathology, and molecular characteristics to human adenocarcinomas. NNK is tobacco-specific carcinogen (TSCs) from nicotine-derived nitrosamine class of carcinogen responsible for K-Ras point mutation in codon 12 of the A/J mice inducing primary lung tumors (29). NNK induced A/J mouse lung cancer model for the primary lung adenoma development is one of the most used models to study the early developmental changes in lung cancer (9,30,31). The selection of dose of NNK was pertinent with the earlier studies conducted for the long-term carcinogenesis using 3 μM of NNK for eight weeks for the tumor as an end point (32). While non-toxic dose of DAS was selected consistent with the studies with oraganosulphur compounds showing modulation of the biomarkers including xenobiotic metabolism markers; CYPs in the in vivo system (33,34). Various stages in the progression of lung cancer including hyperplasia, adenoma are consistent with the human lung cancer progression, with respect to histology as well as molecular changes (35). Early molecular changes including immunological manifestation as well as hyperproliferative molecular changes in the initial stages of NNK induced lung lesions have been demonstrated previously (36,37). Our current study, for the first time, demonstrated the sequential metabolomic, transcriptomic and epigenomic changes in the NNK induced lung before the tumorigenesis process.
NNK metabolic products are responsible for the genomic as well as metabolomic changes in the cells, responsible for the initiation process of carcinogenesis. Earlier study demonstrated effect of nicotine and its methylated metabolite, N-methyl-nicotine (M-nicotine), on human luteal cells by measuring release of progesterone and prostaglandins (PGs) from cultured cells and by testing gene expression of vascular endothelial growth factor (VEGF), an angiogenic factor strictly involved in luteal pathophysiology. Nicotine and M-nicotine can induce a sort of luteal insufficiency by inhibiting progesterone release, likely through modulation of the PG system (38). Also, when Nicotinic acid hydrazide, acidic derivative of nicotine when administered through oral administration shown to induce lung adenomas and adenocarcinomas (39). In the given study, our LC–MC metabolome data demonstrated the modulation of nicotine metabolism pathway as one of the major pathways in the lung carcinogenesis process. Nicotine and its derivative including methyl-nicotine and nicotinic acid have shown to be upregulated in NNK-treated A/J lung tissues. The direct interaction of these metabolites either with early events such as the genomic rearrangements or the later events like with the signaling kinases remains to be deciphered.
Cancer cell metabolism is highly linked with the epigenetic regulation of transcription including DNA methylation, histone acetylation and chromatin remodeling (40). SERBP1, RNA-binding protein functioning as a central regulator of cancer metabolism and indirect modulator of epigenetic regulation in Glioblastoma (41). Our study shown, metabolites potentially regulate one of the important mechanisms, that is, epigenetic regulation of the gene expression including CpG methylation. DAS has shown to modulate the CpG methylation in distal intergenic region as well as promotor region of genes including like Dual Specificity Phosphatase 5 (DUSP5) and homologous recombination DNA repair pathway gene (Rad51D). DUSP5 is a key regulator of various pathways including upregulation of MAPK by epigenetic regulation of miRNAs (42). Moreover, DUSP5 is a direct target of p53, it represents a mechanism by which p53 might negatively regulate cell-cycle progression by downregulating mitogen- or stress-activated protein kinases (43). Our study further shows the early-stage methylation regulation of transcriptional modulation of DUSP5 protein in NNK induced in vivo A/J mice model. As reported by others, adenylate cyclase overexpression has been responsible for the metastatic potential of the lung cancer cells (44). As an early event, our study indicated it is hypermethylated in the NNK induced lung tissue as compared to its respective vehicle control, depicting these genes can be a part of late event in the process of carcinogenesis.
The garlic derived organosulphur compounds have shown to protect against the various processes in carcinogenesis progression including stem cell phenotype restoring thyroid-specific gene expression in thyroid carcinoma cells (45), exerts antitumor efficacy via activation of MAPK pathway (46) and effect against acute pancreatitis with associated lung injury (47). Diallyl trisulfide (DATS) has shown to suppress the viability of human lung cancer cell lines H358 and H460 by causing G2-M phase cell-cycle arrest and apoptotic cell death (48). Diallyl disulphide (DADS) suppressed the fibronectin induced invasion and migration potential of A549 cells by the upregulation of the epithelial markers, E-cadherin and cytokeratin-18, and the downregulation of the mesenchymal markers, N-cadherin and vimentin, and the transcription factors, snail, slug and twist (49). In our study, we found that DAS modulated the transcription regulation of crucial genes including FOXO1 (FKHR), fork head family of transcription factors, Fas (CD95) and its ligand FasL signal apoptosis and are involved in tissue homeostasis, Prostaglandin E synthase 2 (PTGES2). Previously, FOXO1 expression and localization has been shown to be correlated with nodal involvement in progression of the disease depicting it as favorable prognostic factor in non-small cell lung cancer (NSCLC) (50). Previously, it was shown that when normal human lung fibroblasts, exposed to cigarette smoke constituents, elicit COX-2 expression with consequent prostaglandin synthesis, thus creating a proinflammatory environment, responsible for epithelial transformation (51). Biomarker of inflammation inducing prostaglandin E-2 (PGE-2) have shown to be overexpressed in lung carcinogenesis is consistent with our finding showing the earlier phenomenon in the sequential event (52).
Dissecting the drivers in the sequential lung carcinogenesis, we further studied Reactome based canonical pathways involved in these early events in the NNK induced lung carcinogenesis. Metabolic enzymes facilitating the activities of NNK are the main constituents through which NNK-specific metabolic pathways and subsequent NNK activation occur. Cytochrome pigment 450 (CYP450) enzymes belonging to the CYP multigene family, for example, catalyze the hydroxylation of NNK in the oxidative metabolism pathway (53). Our study demonstrated the significant upregulation of the NNK metabolism Phase I and Phase II drug metabolism pathways at week 2 post carcinogen treatment. Importantly, at week 2, GRB2 events and SHC-1 events in EGFR signaling, regulation of TP53 association, estrogen signaling, and oxidative stress pathways shown to be downregulated outlining signaling kinases are late events, post week 2 in the process. This is context, integrin signaling, PI3K cascade, IRS mediated signaling pathway, MAPK signaling pathways showed to be modulated at week 4 post carcinogen treatment illustrating the week 4 is the point at which extensive network of signaling kinases plays pivotal role in the driving process of carcinogenesis.
In summary, we have for the first time demonstrated the correlation of metabolomic, epigenomic and transcriptomic analyses in NNK induced sequential events in lung carcinogenesis events. Our study demonstrated the differential cellular metabolic rewiring, expression of genes demonstrating modulation of transcriptional regulators and signaling kinases at week 2 and week 4 postcarcinogen treatment. We further show that CpG methylation with differentially methylated regions in the genes responsible for the expression/repression events of the early carcinogenesis. We also deciphered the preventive effects of diallyl sulphide administration on these molecular changes in the early events in the process. The correlation of significantly different metabolites with the aforementioned transcriptomic and epigenomic changes in all three groups (VC, NNK and NNK + DAS) suggesting the sequential driving cause during the initiation and promotion stages of NNK induced lung carcinogenesis. These biomarkers will be important in understanding of lung cancer carcinogenesis, potential prevention and treatment strategy in smoking induced lung cancer in human.
Supplementary Material
Acknowledgements
The authors are grateful to Dr Kong’s laboratory for their incredible support and technical assistance.
Glossary
Abbreviations
- DNMTs
NA methyl transferases
- HDACs
istone deacetylases
- NNK
nicotine-derived nitrosamine ketone
- PCA
principal component analysis
- TSCs
tobacco-specific carcinogen
Funding
This work was supported in part by institutional funds, P30 ES005022 and by AT009152 from the National Center for Complementary and Integrative Health (NCCIH) to Dr Kong.
Conflict of Interest Statement: All authors declare no conflict of interest.
References
- 1. Marquette, C.H., et al. ; AIR project Study Group. (2020) Circulating tumour cells as a potential biomarker for lung cancer screening: a prospective cohort study. Lancet. Respir. Med., 8, 709–716. [DOI] [PubMed] [Google Scholar]
- 2. Shoji, F., et al. (2020) Complete blood cell count-derived inflammatory biomarkers in early-stage non-small-cell lung cancer. Ann. Thorac. Cardiovasc. Surg., 26, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang, Y.H., et al. (2020) Identifying circulating miRNA biomarkers for early diagnosis and monitoring of lung cancer. Biochim. Biophys. Acta. Mol. Basis Dis., 1866, 165847. [DOI] [PubMed] [Google Scholar]
- 4. Bennett, C.W., et al. (2016) Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget, 7, 71013–71035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pisanic, T.R.2nd, et al. (2015) DREAMing: a simple and ultrasensitive method for assessing intratumor epigenetic heterogeneity directly from liquid biopsies. Nucleic Acids Res., 43, e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sestini, S., et al. (2015) Circulating microRNA signature as liquid-biopsy to monitor lung cancer in low-dose computed tomography screening. Oncotarget, 6, 32868–32877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boldry, E.J., et al. (2020) Effects of 2-phenethyl isothiocyanate on metabolism of 1,3-butadiene in smokers. Cancer Prev. Res. (Phila)., 13, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang, Q., et al. (2021) Oral administration of EGCG solution equivalent to daily achievable dosages of regular tea drinkers effectively suppresses miR483-3p induced metastasis of hepatocellular carcinoma cells in mice. Food Funct., 12, 3381–3392. [DOI] [PubMed] [Google Scholar]
- 9. Hudlikar, R.R., et al. (2017) Polymeric black tea polyphenols (PBPs) inhibit benzo(a)pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung carcinogenesis potentially through down-regulation of p38 and Akt phosphorylation in A/J mice. Mol. Carcinog., 56, 625–640. [DOI] [PubMed] [Google Scholar]
- 10. García-Rodríguez, M.D.C., et al. (2021) Antigenotoxic effects of (−)-epigallocatechin-3-gallate (EGCG) and its relationship with the endogenous antioxidant system, 8-hydroxydeoxyguanosine adduct repair (8-OHdG), and apoptosis in mice exposed to chromium(VI). J. Toxicol. Environ. Health. A, 84, 331–344. [DOI] [PubMed] [Google Scholar]
- 11. Chai, Y.S., et al. (2020) Curcumin regulates the differentiation of naïve CD4+T cells and activates IL-10 immune modulation against acute lung injury in mice. Biomed. Pharmacother., 125, 109946. [DOI] [PubMed] [Google Scholar]
- 12. Hudlikar, R.R., et al. (2019) Dose-related modulatory effects of polymeric black tea polyphenols (PBPs) on initiation and promotion events in B(a)P and NNK-induced lung carcinogenesis. Nutr. Cancer, 71, 508–523. [DOI] [PubMed] [Google Scholar]
- 13. Zhang, Y., et al. (2020) Phytochemicals of garlic: promising candidates for cancer therapy. Biomed. Pharmacother., 123, 109730. [DOI] [PubMed] [Google Scholar]
- 14. Ghasemi, S., et al. (2021) Epigenetic targeting of cancer stem cells by polyphenols (cancer stem cells targeting). Phytother. Res., 35, 3649–3664. [DOI] [PubMed] [Google Scholar]
- 15. Yang, Y., et al. (2019) DNA methylome and transcriptome alterations and cancer prevention by triterpenoid ursolic acid in UVB-induced skin tumor in mice. Mol. Carcinog., 58, 1738–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang, X.H., et al. (2015) Gene expression profiling signatures for the diagnosis and prevention of oral cavity carcinogenesis-genome-wide analysis using RNA-seq technology. Oncotarget, 6, 24424–24435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thakur, C., et al. (2019) Connections between metabolism and epigenetics in cancers. Semin. Cancer Biol., 57, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Varier, R.A., et al. (2011) Histone lysine methylation and demethylation pathways in cancer. Biochim. Biophys. Acta, 1815, 75–89. [DOI] [PubMed] [Google Scholar]
- 19. Elstrom, R.L., et al. (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res., 64, 3892–3899. [DOI] [PubMed] [Google Scholar]
- 20. Strohecker, A.M., et al. (2014) Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy, 10, 384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qu, Z., et al. (2021) Exposure to a mixture of cigarette smoke carcinogens disturbs gut microbiota and influences metabolic homeostasis in A/J mice. Chem. Biol. Interact., 344, 109496. [DOI] [PubMed] [Google Scholar]
- 22. Chen, L., et al. (2020) HNF4 regulates fatty acid oxidation and is required for renewal of intestinal stem cells in mice. Gastroenterology, 158, 985–999.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang, Y., et al. (2019) UVB drives different stages of epigenome alterations during progression of skin cancer. Cancer Lett., 449, 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hudlikar, R.R., et al. (2021) Epigenomic, transcriptomic, and protective effect of carotenoid fucoxanthin in high glucose-induced oxidative stress in Mes13 kidney mesangial cells. Chem. Res. Toxicol., 34, 713–722. [DOI] [PubMed] [Google Scholar]
- 25. Wallace, D.C., et al. (2010) Energetics, epigenetics, mitochondrial genetics. Mitochondrion, 10, 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cyr, A.R., et al. (2011) The redox basis of epigenetic modifications: from mechanisms to functional consequences. Antioxid. Redox Signal., 15, 551–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janke, R., et al. (2015) Metabolism and epigenetics. Annu. Rev. Cell Dev. Biol., 31, 473–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu, R., et al. (2020) Redox signaling, mitochondrial metabolism, epigenetics and redox active phytochemicals. Free Radic. Biol. Med. doi: 10.1016/j.freeradbiomed.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hisamoto, A., et al. (2007) Point mutation of K-ras gene in cisplatin-induced lung tumours in A/J mice. Lung Cancer, 58, 15–20. [DOI] [PubMed] [Google Scholar]
- 30. Hecht, S.S., et al. (1994) Lung tumor induction in A/J mice by the tobacco smoke carcinogens 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and benzo[a]pyrene: a potentially useful model for evaluation of chemopreventive agents. Carcinogenesis, 15, 2721–2725. [DOI] [PubMed] [Google Scholar]
- 31. Rioux, N., et al. (1998) Prevention of NNK-induced lung tumorigenesis in A/J mice by acetylsalicylic acid and NS-398. Cancer Res., 58, 5354–5360. [PubMed] [Google Scholar]
- 32. Woo, J.K., et al. (2009) Liposomal encapsulation of deguelin: evidence for enhanced antitumor activity in tobacco carcinogen-induced and oncogenic K-ras-induced lung tumorigenesis. Cancer Prev. Res. (Phila)., 2, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hong, J.Y., et al. (1992) Inhibitory effects of diallyl sulfide on the metabolism and tumorigenicity of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in A/J mouse lung. Carcinogenesis, 13, 901–904. [DOI] [PubMed] [Google Scholar]
- 34. Kim, S.H., et al. (2015) Diallyl disulfide prevents cyclophosphamide-induced hemorrhagic cystitis in rats through the inhibition of oxidative damage, MAPKs, and NF-κB pathways. Biomol. Ther., 23, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gordon, W., et al. (2013) The tobacco carcinogen nitrosamine induces a differential gene expression response in tumour susceptible A/J and resistant C3H mouse lungs. Eur. J. Cancer, 49, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Razani-Boroujerdi, S., et al. (2007) Early manifestations of NNK-induced lung cancer: role of lung immunity in tumor susceptibility. Am. J. Respir. Cell Mol. Biol., 36, 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang, G., et al. (1997) Characterization of early pulmonary hyperproliferation and tumor progression and their inhibition by black tea in a 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis model with A/J mice. Cancer Res., 57, 1889–1894. [PubMed] [Google Scholar]
- 38. Miceli, F., et al. (2005) Effects of nicotine on human luteal cells in vitro: a possible role on reproductive outcome for smoking women. Biol. Reprod., 72, 628–632. [DOI] [PubMed] [Google Scholar]
- 39. Toth, B. (1981) Nicotinic acid hydrazide carcinogenesis in mice. Oncology, 38, 106–109. [DOI] [PubMed] [Google Scholar]
- 40. Morrison, A.J. (2021) Cancer cell metabolism connects epigenetic modifications to transcriptional regulation. FEBS J. doi: 10.1111/febs.16032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kosti, A., et al. (2020) The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation. Genome Biol., 21, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Du, M., et al. (2020) microRNA-95 knockdown inhibits epithelial-mesenchymal transition and cancer stem cell phenotype in gastric cancer cells through MAPK pathway by upregulating DUSP5. J. Cell. Physiol., 235, 944–956. [DOI] [PubMed] [Google Scholar]
- 43. Ueda, K., et al. (2003) Dual-specificity phosphatase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene, 22, 5586–5591. [DOI] [PubMed] [Google Scholar]
- 44. Tan, M., et al. (2013) Overexpression of adenylate cyclase-associated protein 1 is associated with metastasis of lung cancer. Oncol. Rep., 30, 1639–1644. [DOI] [PubMed] [Google Scholar]
- 45. Zhang, L., et al. (2021) Diallyl trisulphide, a H2 S donor, compromises the stem cell phenotype and restores thyroid-specific gene expression in anaplastic thyroid carcinoma cells by targeting AKT-SOX2 axis. Phytother. Res., 35, 3428–3443. [DOI] [PubMed] [Google Scholar]
- 46. Jiang, X., et al. (2017) Garlic-derived organosulfur compound exerts antitumor efficacy via activation of MAPK pathway and modulation of cytokines in SGC-7901 tumor-bearing mice. Int. Immunopharmacol., 48, 135–145. [DOI] [PubMed] [Google Scholar]
- 47. Mathan Kumar, M., et al. (2020) Protective effect of diallyl disulfide against cerulein-induced acute pancreatitis and associated lung injury in mice. Int. Immunopharmacol., 80, 106136. [DOI] [PubMed] [Google Scholar]
- 48. Xiao, D., et al. (2009) Diallyl trisulfide selectively causes Bax- and Bak-mediated apoptosis in human lung cancer cells. Environ. Mol. Mutagen., 50, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Das, B., et al. (2019) Diallyl disulphide suppresses the cannonical Wnt signaling pathway and reverses the fibronectin-induced epithelial mesenchymal transition of A549 lung cancer cells. Food Funct., 10, 191–202. [DOI] [PubMed] [Google Scholar]
- 50. Maekawa, T., et al. (2009) Expression and localization of FOXO1 in non-small cell lung cancer. Oncol. Rep., 22, 57–64. [PubMed] [Google Scholar]
- 51. Martey, C.A., et al. (2004) Cigarette smoke induces cyclooxygenase-2 and microsomal prostaglandin E2 synthase in human lung fibroblasts: implications for lung inflammation and cancer. Am. J. Physiol. Lung Cell. Mol. Physiol., 287, L981–L991. [DOI] [PubMed] [Google Scholar]
- 52. Yoshimatsu, K., et al. (2001) Inducible prostaglandin E synthase is overexpressed in non-small cell lung cancer. Clin. Cancer Res., 7, 2669–2674. [PubMed] [Google Scholar]
- 53. Akopyan, G., et al. (2006) Understanding tobacco smoke carcinogen NNK and lung tumorigenesis. Int. J. Oncol., 29, 745–752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.