Abstract
The children of related parents show increased risk of early mortality. The Native American genome typically exhibits long stretches of homozygosity, and Latin Americans are highly heterogeneous regarding the individual burden of homozygosity, the proportion and the type of Native American ancestry. We analysed nationwide mortality and genome-wide genotype data from admixed Chileans to investigate the relationship between common causes of child mortality, homozygosity and Native American ancestry. Results from two-stage linear-Poisson regression revealed a strong association between the sum length of runs of homozygosity (SROH) above 1.5 Megabases (Mb) in each genome and mortality due to intracranial non-traumatic haemorrhage of foetus and newborn (5% increased risk of death per Mb in SROH, P = 1 × 10−3) and disorders related to short gestation and low birth weight (P = 3 × 10−4). The major indigenous populations in Chile are Aymara–Quechua in the north of the country and the Mapuche–Huilliche in the south. The individual proportion of Aymara–Quechua ancestry was associated with an increased risk of death due to anencephaly and similar malformations (P = 4 × 10−5), and the risk of death due to Edwards and Patau trisomy syndromes decreased 4% per 1% Aymara–Quechua ancestry proportion (P = 4 × 10−4) and 5% per 1% Mapuche–Huilliche ancestry proportion (P = 2 × 10−3). The present results suggest that short gestation, low birth weight and intracranial non-traumatic haemorrhage mediate the negative effect of inbreeding on human selection. Independent validation of the identified associations between common causes of child death, homozygosity and fine-scale ancestry proportions may inform paediatric medicine.
Introduction
Runs of homozygosity (ROHs), i.e. continuous stretches of homozygous alleles originated by inheritance of identical-by-descent haplotypes from both parents, are universally common in humans (1). ROHs reflect the demographic histories of both individuals and populations: while recent parental relatedness translates into long ROH, shared parental ancestry tens to hundreds generations ago is reflected into many shorter homozygous stretches (2). The fraction of the autosomal genome in ROH of length above 1.5 Megabases (Mb) shows a good correlation (r = 0.86) with inbreeding coefficients estimated from pedigrees and ROHs are typically used to improve the accuracy of homozygosity mapping of recessive Mendelian diseases (3). However, studies on the relationship between ROH and the risk of polygenic and multifactorial diseases have long been neglected (4). Several case-control studies have been published on the effects of the length and the number of ROHs on various complex diseases, in particular adult and childhood cancers, often with inconsistent results (5–16). Most of these studies have been conducted in outbred populations of European origin, although Native American genomes show on average long stretches of homozygosity and Latin Americans are highly heterogeneous regarding the individual burden of homozygosity. Large studies on the association between ROH and quantitative traits have produced more consistent results. In a recent meta-analysis, Clark et al. (17) investigated 100 phenotypes in more than 1.4 million individuals and found 32 phenotypes associated with ROH length. In particular, the authors replicated a previously reported decrease in height with increasing ROH (17). As height is in turn associated with several complex diseases, including diabetes, heart disease and cancer, the study also suggests that homozygosity may influence the individual susceptibility to complex, multi-factorial diseases (18–22).
The demographic history of the Americas has been shaped by extensive admixture between indigenous Native Americans, Europeans and Africans (23). Latin Americans are a highly heterogeneous ethnic group and the distribution of ancestry proportions varies greatly both between and within Latin American regions and countries (24). Latin Americans also show specific disease prevalences and an overall increased risk of diabetes and asthma, as well as stomach and gallbladder cancers (25–28). Lately, research has been focused on disease susceptibility according to the individual proportions of Native American, European and African ancestry (29–31). However, there are more than 400 indigenous groups in Latin America (32) and most studies neglected so far the influence of the type of Native American ancestry, which may have a considerable impact on disease risk (33).
In Chile, the four major indigenous populations are the Aymara and Quechua in the northern highlands and the Mapuche and Huilliche in the south of the country. Genetic studies in Chile benefit from the geography as the country is very long and narrow, bounded by the Pacific Ocean in the west and by the Andes in the east, allowing investigation of gradual changes from north to south. Disease-specific mortality rates generally vary throughout the country, the Northern and Southern Native American components are regionally separated and the proportion of African ancestry is low (3%). The present study builds on previous research that found important differences between Aymara–Quechua and Mapuche–Huilliche ancestry in relation to the leading causes of death in Chile (33). Considering individual differences in ancestry proportions and differentiating between fine-scale types of ancestry will become increasingly important for personalized disease prevention. We incorporate an additional layer of genetic variability—the individual burden of ROH—and assess the association between child mortality, fine-scale Native American ancestry proportions and ROH length. Since the offspring of first cousins’ marriages shows a 4–5% increased childhood mortality, we only examine the most common causes of child mortality, where the effect of ROH would be strong enough to be detected in our analyses (34,35).
Here, we take advantage of genome-wide genotype data from 1786 admixed Chileans and nationwide mortality data from 2007 to 2017 to investigate the relationship between ROH, genetic ancestry and child mortality in Chile. We use the PLINK software to calculate the sum length of runs of homozygosity (SROH) above 1.5 Mb in each genome and the ADMIXTURE software to estimate the individual proportions of Aymara–Quechua, Mapuche–Huilliche, European and African ancestry (36,37). We assess the relationship between ROH, genetic ancestry and regionally aggregated mortality data by multiple linear regression to estimate the expected regional SROH and ancestry proportions, followed by multiple Poisson regression to quantify the association between regional mortality rates and the expected SROH and ancestry proportions.
Results
Figure 1 depicts the estimated SROH, Aymara–Quechua and Mapuche–Huilliche ancestry proportions for each of the 15 Chilean regions (A–C). The central region ‘Metropolitana de Santiago’ showed the lowest average SROH (19.7 Mb). The highest SROH was observed in the southern De La Araucanía (39.0 Mb), De Magallanes y de la Antártica Chilena (30.7 Mb) and De Aisén del General Carlos Ibáñez del Campo (28.2 Mb) regions, followed by the most northern Chilean region (De Arica y Parinacota, 27.9 Mb). The estimated Aymara–Quechua ancestry proportions ranged from 4.7% in the southern De Los Ríos region to 29.6% in the northern De Tarapacá region. Overall, the estimated Mapuche–Huilliche ancestry proportions were higher than the Aymara–Quechua proportions, ranging from 25.3% (De Tarapacá region) to 49.9% (De Los Ríos regions). Regions with a high SROH generally showed high Native American ancestry proportions too.
Figure 1.

Maps of Chile representing the estimated regional SROH (A), Aymara–Quechua ancestry (B) and Mapuche–Huilliche ancestry proportions (C). D shows the first (African versus non-African) versus second (Native American versus European) principal components of genetic variability, the first versus third (Aymara–Quechua versus Mapuche–Huilliche) principal components are shown in E. Reference populations are represented in green (Africans), beige (Europeans), red (Mapuche-Huilliche) and blue (Aymara-Quechua). Study participants are shown in a white-to-black scale representing individual estimated SROH.
The estimated sum length of the genome in ROH above 1.5 Mb for each study individual is also plotted against the first three principal components of genetic variability in Figs. 1D and E. The estimated individual SROH ranged from 5.6 to 112.3 Mb with a mean equal to 30.7 Mb. The first principal component distinguished between African and non-African ancestry, the second principal component separated the European from the Native American ancestry and the increase in SROH with increasing proportions of Native American ancestry was striking (D). The third principal component separated the Aymara–Quechua and the Mapuche–Huilliche subcomponents of Chilean Native American ancestry, and individuals with large proportions of Aymara–Quechua ancestry clearly presented large fractions of the genome in ROH (E).
Table 1 shows the SROH distribution in the study according to the proportions of Aymara–Quechua and Mapuche–Huilliche ancestry, gender, age (four categories of almost equal size), educational level (three categories), socio-economic status (six categories, from the lowest D/E stratum [semi- and un-skilled manual occupations, unemployed and lowest grade occupations] to the highest ABC1 category representing the high middle class) and salary (four groups).
Table 1.
Distribution of the SROH above 1.5 Mb according to the ancestry proportions and other sociodemographic characteristics of study participants
| Variable | Level | N | SROH (Mb) | |||
|---|---|---|---|---|---|---|
| Pval | Estimate | 95% CI | ||||
| Intercept | Ref. | 1786 | 0.007 | −9.53 | −16.42 | −2.65 |
| Aymara–Quechua ancestry | per 1% | 1786 | 1.5 × 10−97 | 0.93 | 0.85 | 1.01 |
| Mapuche–Huilliche ancestry | per 1% | 1786 | 9.9 × 10−31 | 0.86 | 0.71 | 1.00 |
| Gender | Male | 1089 | 1.6 × 10−13 | Ref. | ||
| Female | 697 | −10.48 | −13.26 | −7.69 | ||
| Age | <24 years | 387 | 0.05 | 0.30 | −3.74 | 4.34 |
| 24–26 years | 455 | 3.55 | −0.31 | 7.41 | ||
| 27–32 years | 487 | 3.70 | 0.34 | 7.07 | ||
| >32 years | 457 | Ref. | ||||
| Educational level | Primary/secondary school | 1279 | 0.92 | Ref. | ||
| Technical | 57 | 1.43 | −6.00 | 8.87 | ||
| University/postgrade | 450 | −0.14 | −3.40 | 3.12 | ||
| Socio-economic status | E | 33 | 0.31 | 2.12 | −7.59 | 11.84 |
| D | 453 | −1.51 | −5.90 | 2.88 | ||
| C3 | 504 | Ref. | ||||
| C2 | 150 | 1.36 | −3.76 | 6.47 | ||
| ABC1 | 27 | 11.65 | 1.18 | 22.1 | ||
| Missing | 619 | −0.79 | −5.61 | 4.03 | ||
| Salary | <350 000$ | 198 | 0.39 | −0.31 | −5.76 | 5.15 |
| 350 000–450 000$ | 231 | 3.46 | −1.38 | 8.31 | ||
| 450 000$+ | 514 | Ref. | ||||
| Missing | 843 | 0.60 | −4.22 | 5.42 | ||
Results are based on a multiple linear regression model that simultaneously included all the listed variables. Ref.: Reference, N: Number of individuals, Pval: Global probability value, CI: Confidence interval.
The reference SROH (intercept) was −9.53 Mb (95%CI: −16.42 to −2.65) in the fitted multiple linear regression model. The SROH increased by 0.93 Mb (95%CI: 0.85–1.01) for each 1% increase in the proportion of Aymara–Quechua ancestry, and 0.86 Mb (95%CI: 0.71–1.00) for every 1% increase in the proportion of Mapuche–Huilliche ancestry. Males showed a lower SROH than females (−10.48 Mb, 95%CI: −13.26 to −7.69). This result was due to regional differences in the recruitment of women and men: while 60% of the women were recruited in the most northern Chilean region (De Arica y Parinacota), which showed the highest proportion of Aymara–Quechua ancestry and in turn a high SROH, the proportion of men recruited in this region was only 31% (Supplementary Material, Table S1). Individuals in the highest socio-economic category (ABC1) showed the highest SROH (+11.65 Mb compared with individuals in the C3 category, 95%CI: 1.18–22.1), but differences in SROH by age, educational level, socio-economic status and salary did not reach the 5% level of statistical significance.
As a proof of principle, in addition to potential associations with common causes of child death, we also checked the well-established, negative association between SROH and height in the investigated cohort. As expected, Chileans in the fourth SROH quartile (33.5–502 Mb) were on average shorter (−1.32 cm, 95%CI: −2.29 to −0.35) than Chileans in the first SROH quartile (0–17.5 Mb, Supplementary Material, Table S2), adding plausibility to the rest of our findings.
After examining the geographical distribution of SROH and ancestry proportions, the association between regionally aggregated mortality rates, SROH and ancestry proportions was quantified by multiple Poisson regression. To facilitate the interpretability of results, standardized mortality ratios (SMRs) are reported per Mb in SROH, and per 1% proportion of Aymara–Quechua and Mapuche–Huilliche ancestry. Only perinatal conditions (first character P or Q in the code of the 10th version of the International Classification of Diseases—ICD10) causing at least 200 deaths before the age of 5 years in Chile between 2007 and 2017 were considered, resulting in 23 investigated ICD10-categories.
Table 2 shows the estimated SMR for certain conditions originating in the perinatal period and congenital malformations, deformations and chromosomal abnormalities. Bold type is used to highlight SMR with associated probability values lower than 0.002 (0.05/23), and therefore significant after accounting for multiple testing. SROH was associated with an increased mortality due to disorders related to short gestation and low birth weight, not elsewhere classified (ICD-10 code P07, 3% risk increase per Mb in SROH, 95%CI: 1–4%) and to intracranial non-traumatic haemorrhage of foetus and newborn (ICD-10 code P52, 5% risk increase per Mb in SROH, 95%CI: 2–9%). The proportion of Aymara–Quechua ancestry was associated with an increased mortality due to respiratory distress of newborn (ICD-10 code P22, 3% increased risk per 1% ancestry proportion, 95%CI: 1–5%) and to anencephaly and similar malformations (ICD-10 code Q00, 5% increased risk per 1% ancestry proportion, 95%CI: 3–7%), and with a decreased mortality due to Edwards syndrome and Patau syndrome (ICD-10 code Q91, 4% decreased risk per 1% ancestry proportion, 95%CI: −6 to −2%). The proportion of Mapuche–Huilliche ancestry was associated with an increased mortality due to other congenital malformations of brain (ICD-10 code Q04, 7% increased risk per 1% ancestry proportion, 95%CI: 2–12%) and with a decreased mortality due to Edwards syndrome and Patau syndrome (ICD-10 code Q91, 5% decreased risk per 1% ancestry proportion, 95%CI: −8 to −2%).
Table 2.
Number of deaths before the age of 5 years and SMRs due to common childhood diseases per Mb in SROH, 1% proportion of Aymara–Quechua and 1% proportion of Mapuche–Huilliche genetic ancestry
| ICD | Description | Deaths | SROH (Mb) | Aymara–Quechua ancestry (%) | Mapuche–Huilliche ancestry (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pval | SMR | 95% CI | Pval | SMR | 95% CI | Pval | SMR | 95% CI | |||||||
| P | Certain conditions originating in the perinatal period | ||||||||||||||
| P01 | Foetus and newborn affected by maternal complications of pregnancy | 466 | 0.80 | 1.00 | 0.97 | 1.04 | 0.76 | 1.00 | 0.97 | 1.04 | 0.55 | 0.99 | 0.95 | 1.03 | |
| P02 | Foetus and newborn affected by complications of placenta, cord and membranes | 416 | 0.22 | 0.98 | 0.94 | 1.01 | 0.003 | 1.05 | 1.02 | 1.08 | 0.48 | 1.02 | 0.97 | 1.06 | |
| P07 | Disorders related to short gestation and low birth weight, n. e. c. | 2025 | 0.0003 | 1.03 | 1.01 | 1.04 | 0.004 | 0.98 | 0.96 | 0.99 | 0.04 | 1.02 | 1.00 | 1.04 | |
| P21 | Birth asphyxia | 638 | 0.66 | 0.99 | 0.97 | 1.02 | 0.07 | 1.02 | 1.00 | 1.05 | 0.19 | 1.02 | 0.99 | 1.05 | |
| P22 | Respiratory distress of newborn | 983 | 0.14 | 1.02 | 0.99 | 1.04 | 0.001 | 1.03 | 1.01 | 1.05 | 0.81 | 1.00 | 0.97 | 1.03 | |
| P26 | Pulmonary haemorrhage originating in the perinatal period | 335 | 0.38 | 0.98 | 0.95 | 1.02 | 0.37 | 0.98 | 0.95 | 1.02 | 0.07 | 1.04 | 1.00 | 1.08 | |
| P28 | Other respiratory conditions originating in the perinatal period | 844 | 0.63 | 0.99 | 0.97 | 1.02 | 0.32 | 0.99 | 0.97 | 1.01 | 0.21 | 0.98 | 0.95 | 1.01 | |
| P36 | Bacterial sepsis of newborn | 630 | 0.66 | 0.99 | 0.96 | 1.02 | 0.74 | 1.00 | 0.98 | 1.03 | 0.81 | 1.00 | 0.96 | 1.03 | |
| P52 | Intracranial non-traumatic haemorrhage of foetus and newborn | 479 | 0.001 | 1.05 | 1.02 | 1.09 | 0.12 | 0.98 | 0.95 | 1.01 | 0.28 | 0.98 | 0.94 | 1.02 | |
| P77 | Necrotizing enterocolitis of foetus and newborn | 663 | 0.05 | 1.03 | 1.00 | 1.06 | 0.01 | 0.96 | 0.94 | 0.99 | 0.02 | 0.96 | 0.92 | 0.99 | |
| Q | Congenital malformations, deformations and chromosomal abnormalities | ||||||||||||||
| Q00 | Anencephaly and similar malformations | 649 | 0.14 | 0.98 | 0.95 | 1.01 | 4.4 × 10 −5 | 1.05 | 1.03 | 1.07 | 0.01 | 1.04 | 1.01 | 1.07 | |
| Q04 | Other congenital malformations of brain | 275 | 0.08 | 0.96 | 0.93 | 1.00 | 0.08 | 1.03 | 1.00 | 1.07 | 0.002 | 1.07 | 1.02 | 1.12 | |
| Q20 | Congenital malformations of cardiac chambers and connections | 226 | 0.61 | 1.01 | 0.96 | 1.07 | 0.07 | 0.96 | 0.91 | 1.00 | 0.32 | 0.97 | 0.91 | 1.03 | |
| Q21 | Congenital malformations of cardiac septa | 264 | 0.34 | 0.98 | 0.93 | 1.02 | 0.58 | 1.01 | 0.97 | 1.05 | 0.31 | 1.03 | 0.98 | 1.08 | |
| Q23 | Congenital malformations of aortic and mitral valves | 244 | 0.60 | 1.01 | 0.96 | 1.06 | 0.02 | 0.94 | 0.90 | 0.99 | 0.26 | 0.97 | 0.91 | 1.02 | |
| Q24 | Other congenital malformations of heart | 1101 | 0.66 | 1.00 | 0.98 | 1.03 | 0.07 | 1.02 | 1.00 | 1.04 | 0.17 | 1.02 | 0.99 | 1.04 | |
| Q25 | Congenital malformations of great arteries | 409 | 0.51 | 1.01 | 0.98 | 1.05 | 0.41 | 0.99 | 0.95 | 1.02 | 0.74 | 0.99 | 0.95 | 1.04 | |
| Q60 | Renal agenesis and other reduction defects of kidney | 501 | 0.70 | 0.99 | 0.96 | 1.03 | 0.52 | 0.99 | 0.96 | 1.02 | 0.70 | 0.99 | 0.95 | 1.03 | |
| Q79 | Congenital malformations of the musculoskeletal system, n. e. c. | 542 | 0.36 | 0.98 | 0.95 | 1.02 | 0.52 | 1.01 | 0.98 | 1.04 | 0.61 | 1.01 | 0.97 | 1.05 | |
| Q89 | Other congenital malformations, not elsewhere classified | 608 | 0.22 | 1.02 | 0.99 | 1.04 | 0.12 | 1.02 | 0.99 | 1.05 | 0.15 | 1.02 | 0.99 | 1.06 | |
| Q90 | Down syndrome | 390 | 0.63 | 0.99 | 0.96 | 1.03 | 0.80 | 1.00 | 0.97 | 1.04 | 0.50 | 1.01 | 0.97 | 1.06 | |
| Q91 | Edwards syndrome and Patau syndrome | 908 | 0.21 | 1.02 | 0.99 | 1.05 | 0.0004 | 0.96 | 0.94 | 0.98 | 0.002 | 0.95 | 0.92 | 0.98 | |
| Q99 | Other chromosome abnormalities, n. e. c. | 327 | 0.63 | 1.01 | 0.97 | 1.05 | 0.34 | 1.02 | 0.98 | 1.05 | 0.45 | 1.02 | 0.97 | 1.06 | |
n.e.c.: not elsewhere classified. Bold represents associated probability values smaller than 0.05/23 investigated diseases = 0.002.
To examine the robustness of the identified associations, we inspected visually the relationship between gender-specific mortality rates, SROH and ancestry, and performed gender-stratified regression analyses. Figure 2A shows the scatterplot for SROH and disorders related to short gestation and low birth weight, not elsewhere classified (ICD-10 code P07). Results are represented in red for young girls and blue for young boys. The corresponding estimated SMRs are shown in Supplementary Material, Table S3. Opposite associations were noticed for young girls (1% risk decrease) and young boys (3% risk increase per Mb in SROH). Results for young girls strongly depended on the data from De Maule region, which showed the highest average SROH (76.2 Mb). However, effect estimates for males and females were even more different after exclusion of this high-leverage observation (6% risk decrease for young girls, data not shown).
Figure 2.

Estimated SROH versus mortality rate due to disorders related to short gestation and low birth weight, not elsewhere classified (ICD-10 code P07, A); intracranial non-traumatic haemorrhage of foetus and newborn (ICD-10 code P52, B). Estimated Aymara–Quechua ancestry proportion versus mortality rate due to respiratory distress of newborn (ICD-10 code P22, C), anencephaly and similar malformations (ICD-10 code Q00, D) and Edwards syndrome and Patau syndrome (ICD-10 code Q91, E). Estimated Mapuche–Huilliche ancestry proportion versus mortality rate due to Edwards syndrome and Patau syndrome (ICD-10 code Q91, F). Observations and regression lines are shown in blue for young boys and red for young girls.
In contrast, consistent associations in young girls and boys were found for SROH and intracranial non-traumatic haemorrhage of foetus and newborn (ICD-10 code P52, Fig. 2B, the estimated SMR for young girls was 1.01 after exclusion of the De Maule region, data not shown). Visual inspection of the remaining scatter plots revealed no apparent outliers or leverage points, and gender-stratified analyses revealed undifferentiated associations between Aymara–Quechua ancestry and respiratory distress of newborn (ICD-10 code P22, Fig. 2C, 4% increased mortality risk for young girls and 3% increased risk for young boys per 1% ancestry proportion) and between Aymara–Quechua ancestry and anencephaly and similar malformations (ICD-10 code Q00, Fig. 2D, 5% increased risk per 1% ancestry proportion for both young girls and boys). Consistent associations were also observed for Aymara–Quechua ancestry and Edwards syndrome and Patau syndrome (ICD-10 code Q91, Fig. 2E, 3% decreased risk per 1% ancestry proportion for both young girls and boys), Mapuche–Huilliche ancestry and Edwards syndrome and Patau syndrome (Fig. 2F, 5% decreased risk for young girls and 3% decreased risk for young boys per 1% ancestry proportion) and Mapuche–Huilliche ancestry and other congenital malformations of brain (ICD-10 code Q04, Supplementary Material, Fig. S1, 2% increased risk for young girls and 4% increased risk for young boys per 1% ancestry proportion).
Discussion
The present study is, to our knowledge, the first one to investigate the relationship between common causes of childhood mortality, homozygosity—quantified as the total genome length in ROH above 1.5 Mb—and the individual proportions of two different types of Native American ancestry. A strength of the study was the large regional variability in SROH (from 19.7 to 39.0 Mb) combined with the differences in child mortality among Chilean regions, which potentially translated into a higher statistical power to investigate the formulated hypothesis than analysing similar data from European-ancestry cohorts in high-income countries. The simultaneous consideration of SROH, Aymara–Quechua and Mapuche–Huilliche ancestry in the multiple Poisson regression models was also a novelty, which aimed at disentangling the role of the two distinct genetic components on early disease risk, allowing at the same time to adjust SROH effect estimates for the potential effect of a poorer access to the health system during pregnancy with increasing proportions of Native American ancestry.
As the burden of homozygosity, cultural and socioeconomic factors are closely related, potential confounding effects may be complex and difficult to disentangle. The importance of accounting for social confounding was demonstrated in a large meta-analysis that examined full-sibling data: SROH differences between siblings are entirely due to Mendelian segregation and thus independent of cultural or socioeconomic confounding. On average across all investigated traits, SROH effect estimates based on sibships were 22% smaller than population-based estimates, possibly reflecting the contribution of non-genetic confounders (17). The present study was based on genetically admixed Chileans, who showed continuous gradients of homozygosity and ancestry, likely making our results less prone to sociocultural confounding than the comparisons of separated groups (children with related versus unrelated parents, or children of Native American compared with European ancestry).
The reported findings relied on individual-level genotype and aggregated mortality data. In order to reduce any ecological bias, we examined in the first stage of our analyses the potential confounding effects of age, gender, educational level, socioeconomic status and salary, and investigated in the second stage the relationship between rare outcomes (child deaths due to a specific cause) and regional estimates of SROH, Aymara–Quechua and Mapuche–Huilliche ancestry proportions. In agreement with previous studies, we found a negative relationship between SROH and height (17), but gender was the only investigated covariate that showed an effect on SROH, likely reflecting the regional differences in the recruitment of women and men in the study.
Native Americans show the highest burden of ROH across the globe: 67% of Native American genomes have at least one chromosomal homozygous segment above 10 Mb in length as a result of recent inbreeding and close kin unions, which were widespread in the central Andes in the Late Intermediate Period (~1000–1470 AD). The present results confirm the association between SROH and Native American ancestry proportions—as a benchmark for D and E in Fig. 1, the average inbreeding coefficient for second cousins translates into ~0.0156 × 3 Gb = 46.9 Mb. We also attempted to identify SROH differences according to the type of Native American ancestry, but the difference between SROH increase per 1% proportion in Aymara–Quechua compared with 1% proportion in Mapuche–Huilliche ancestry did not reach statistical significance (overlapping 95%CIs). Some individuals with average proportions of Aymara–Quechua and Mapuche–Huilliche ancestry also displayed a high SROH (middle part of Fig. 1E) and the increased SROH found in the highest socio-economic group (ABC1, +11.7 Mb compared with individuals in the C3-category) substantiates the previously reported assortative mating in Chileans (38,39).
We detected a strong association between SROH and intracranial non-traumatic haemorrhage of foetus and newborn—as an indicative value, please note that the 5.44% increased mortality rate per Mb in SROH predicts a 5.44 × 46.9 = 255% increased mortality risk for second cousin marriages. The statistical analysis of aggregate data in our study did not reveal other relationships between SROH and the investigated, common types of congenital malformations, deformations and chromosomal abnormalities leading to child death. This result was rather unexpected since several studies have reported a higher incidence of birth defects in the offspring of first cousins compared with non-consanguineous parents. On the other hand, a large prospective survey following over 20 000 pregnant women in South India did not find any relationship between the parental degree of consanguinity and congenital anomalies or foetal development. It is important to mention here that we did not directly investigate the incidence, but the mortality caused by these disorders.
Inbreeding depression is the result of both differential fertility and mortality before reaching reproductive age. Advances in medicine and public health have reduced differences in reproductive success through family planning and the reduction in infant mortality, particularly in high-income regions. The relationship between SROH and human fertility has received some attention in research, but the potential impact of homozygosity on child mortality has long been neglected. The present study focuses on this second component. Disorders related to short gestation and low birth weight constitute the most common cause of childhood mortality in Chile (n = 2025 deaths during a period of 11 years in this study). Interestingly, they were associated with SROH in overall and young boys’ analyses, but not among young girls, possibly mediating a negative effect of homozygosity on human male selection. Also the association between SROH and intracranial non-traumatic haemorrhage of foetus and newborn showed a stronger association for young boys than for young girls.
Investigating possible associations between common causes of infant death and fine-scale Native American ancestry, we found that each 1% increase in the proportion of Aymara–Quechua ancestry was associated with a 3% increased mortality risk due to respiratory distress of newborn, possibly with a stronger association effect for young girls than young boys. Interestingly, we identified previously an association between Aymara ancestry and mortality due to other interstitial pulmonary diseases, which represent the third cause of adult death due to respiratory diseases in Chileans after pneumonia and chronic obstructive pulmonary disease (33). Aymara–Quechua ancestry was also associated with an increased risk of child death due to anencephaly and similar malformations (ICD-10 code Q00), and Mapuche–Huilliche ancestry was associated with congenital malformations of brain (ICD-10 code Q04), which could point to a different completion of death certificates in the north and south of Chile. Anencephaly is known to be influenced by multiple risk factors including folic acid deficit, mutations in the genes of the folate pathway and exposure to a variety of toxins and contaminants. Folic acid supplementation could be particularly indicated to pregnant women with large proportions of Native American ancestry, and the high levels of arsenic in drinking-water and copper mining in the north of Chile could contribute to the identified association with Aymara–Quechua ancestry.
We also identified a consistent, negative association between the two investigated types of Native American ancestry and child mortality due to Edwards syndrome (trisomy 18) and Patau syndrome (trisomy 13). The stronger associations found for females in gender-stratified analyses likely reflect the higher prevalence of Edwards syndrome in female offspring. The risk of trisomy in the offspring increases with maternal age and data were not available to adjust for this potential confounder in our analyses. However, we found no association with infant death due to the more common Down syndrome. Whole and mosaic trisomy syndromes are not inherited, but partial trisomy can be transmitted from parents who carry a balanced translocation. A lower frequency of genetic rearrangements between chromosomes 18/13 and other chromosomes in Native Americans than Europeans could also contribute to the observed negative association.
In summary, the identified associations between SROH, disorders related to short gestation and low birth weight, and intracranial non-traumatic haemorrhage of foetus and newborn; Aymara–Quechua ancestry, respiratory distress of newborn, anencephaly and similar malformations, and Edwards syndrome and Patau syndrome; and Mapuche–Huilliche ancestry, other congenital malformations of brain, and Edwards syndrome and Patau syndrome warrant further investigation using individual-level data to clarify their relevance to population genetics and public health. From the point of view of population genetics, independent validation of the identified associations with SROH may improve our understanding of the link between homozygosity and human selection. The present results may also have important implications for health care and disease prevention. For illustration, the difference in Aymara–Quechua ancestry proportion between the two regions De Tarapacá and De Los Ríos was 25%, which translates into a differential mortality rate due to respiratory distress of newborn of 25 × 3% = 75%.
Materials and Methods
Ethics approval
Ethics approval was obtained from the Medical Faculties of the Universidad de Chile (approval #123-2012) and the Pontificia Universidad Católica de Chile (#11-159), and from Universidad de Tarapacá and University College London as described in Ruiz-Linares et al. (13). Written informed consent was obtained from all study participants. Structured questionnaires applied to volunteers of the aggregate-data are available upon request.
Study participants and associated sociodemographic data
The study comprised 1786 individuals who were recruited from 2010 to 2013; 39% of the study participants were women and the median age was 27 years. About two-thirds of the subjects were professional soldiers, with a relatively large proportion of men born in the south of Chile (Del Maule, Del Biobío and De la Araucanía regions). On the contrary, 60% of the women were recruited from the northern region De Arica y Parinacota, which was also the region, where the most participants were recruited from (42%), followed by Metropolitana de Santiago (18%), and Del Bío-Bío (9%). Since the majority of soldiers are typically recruited from among the middle classes, with minorities from the social elite and the lowest socioeconomic groups, this dataset represented the general Chilean population quite well. Part of the study collective has been described before by Ruiz-Linares et al. (24) and Lorenzo Bermejo et al. (33).
Aggregated mortality data
Aggregated mortality data were obtained from the Chilean Department of Statistics and Health information (www.deis.cl). Causes of death were grouped according to the 10th version of the International Classification of Diseases (www.who.int/classifications/icd/, ICD10). We considered the first three characters of the ICD10 code and investigated perinatal conditions (first character P and Q). Only diseases causing at least 200 deaths before the age of 5 years in Chile between 2007 and 2017 were considered, resulting in 23 investigated ICD10-categories. From 2007 to 2017, Chile was divided into 15 regions, which are the country’s first-level administrative division.
Genotyping and quality control
Blood samples were collected by certified phlebotomists and trained nurses. DNA was extracted following standard laboratory procedures. Genotypes were available for 730 525 single nucleotide polymorphism (SNP) markers from the Illumina’s Human610-Quad beadchip. Intentional duplicates, samples with sex information inconsistent with genotype data and related individuals with shared identity by descent proportion ≥0.25 were removed.
For the calling of ROH, variants with a minor allele frequency (MAF) under 5% and more than 3% missing genotypes across all individuals were excluded, leaving 593 740 SNPs for the subsequent ROH calling. For the genetic principal component and ancestry analyses, variants with an MAF under 5% and more than 5% missing genotypes were excluded. Genetic variants were filtered to only include autosomal polymorphisms, variants with a missing call rate under 5%, and variants without adenine-thymine or guanine-cytosine alleles to avoid DNA strand flipping. In addition, SNPs were pruned based on linkage disequilibrium at r2 higher than 0.2. After merging the study samples with the reference samples, more than 35 000 SNPs were used for the ROH and ancestry analyses.
Genetic principal component analysis and estimation of ancestry proportions
Genetic principal component analyses were conducted using the eigenstrat function available at www.popgen.dk/software/index.php/Rscripts (40). The ADMIXTURE software (version 1.3) was used for supervised estimation of individual African, European, Northern Native Chilean and Southern Native Chilean ancestry components relying on reference individuals (37). Surrogates of African and European ancestry were 108 Yorubans in Ibadan, Nigeria, and 99 Utah residents with Northern and Western European ancestry as well as 107 individuals from Iberian populations in Spain from the 1000 Genome Project (41). The major indigenous peoples in Chile are the Aymara and Quechua in the northern regions and the Mapuche and Huilliche in the southern regions of Chile; 8 Aymara and 9 Mapuche individuals were selected as reported in Lorenzo et al. and complemented with 22 Aymara, 40 Quechua and 4 Huilliche from Reich et al., and 32 Huilliche from Lindo et al. (33,42–44).
ROH calling
ROHs were called using the free, open-source whole genome association analysis toolset PLINK (version 1.07) (36). PLINK parameters were fixed according to the recommendations of the ROHgen consortium to enhance the comparability of results as follows: homozyg-window-snp 50; homozyg-snp 50; homozyg-kb 1500; homozyg-gap 1000; homozyg-density 50; homozyg-window-missing 5 and homozyg-window-het 1.
Statistical analyses
Data were analyzed using the R software environment for statistical computing and graphics (version 3.6.0). The relationship between the aggregated mortality data, SROH and Native American ancestry proportions was investigated in two stages.
First, the expected regional SROH, Aymara–Quechua and Mapuche–Huilliche ancestry were estimated by multiple linear regression using the models
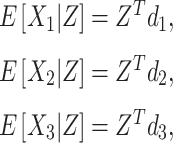 |
where the SROH (represented by 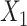 ), the Aymara–Quechua ancestry proportion (represented by
), the Aymara–Quechua ancestry proportion (represented by 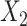 ) and the Mapuche–Huilliche ancestry proportion (represented by
) and the Mapuche–Huilliche ancestry proportion (represented by 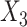 ) depended on the product of a design matrix
) depended on the product of a design matrix 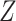 times a fixed-effect vector
times a fixed-effect vector 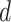 , which included the intercept and the response variables gender and region. Each model was run 15 times, altering each time the reference region.
, which included the intercept and the response variables gender and region. Each model was run 15 times, altering each time the reference region.
In the second stage, the association between regional mortality rates and expected SROH, Aymara–Quechua and Mapuche–Huilliche ancestry proportions was quantified by multiple Poisson regression using the underlying model
 |
where the response variable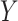 represents the disease-specific mortality rate accumulated over eleven years (from 2007 to 2017), considering the intercept, gender and region as explanatory variables (region-level design matrix
represents the disease-specific mortality rate accumulated over eleven years (from 2007 to 2017), considering the intercept, gender and region as explanatory variables (region-level design matrix 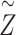 multiplied by the fixed-effect vector
multiplied by the fixed-effect vector 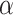 ;
; 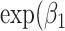 ) was used to estimate the SMR per 1 Mb increase in SROH,
) was used to estimate the SMR per 1 Mb increase in SROH, 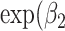 ) the SMR per 1% increase in Aymara–Quechua ancestry proportion and
) the SMR per 1% increase in Aymara–Quechua ancestry proportion and 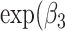 ) the SMR per 1% increase in Mapuche–Huilliche ancestry proportion.
) the SMR per 1% increase in Mapuche–Huilliche ancestry proportion.
Availability of data and code
The Source code in R to reproduce all the results described is provided as Supplementary Material, and the necessary input files are available at www.biometrie.uni-heidelberg.de/ StatisticalGenetics/Software_and_Data
Conflict of Interest statement
The authors declare that they have no conflicts of interest.
Funding
European Union’s Horizon 2020 research and innovation programme grant 825741. The funder had no role in the design of the study, the collection, analysis or interpretation of the data, or in writing the manuscript.
Supplementary Material
Contributor Information
Fabienne Koenigstein, Statistical Genetics Research Group, Institute of Medical Biometry, Heidelberg University, Heidelberg, Germany.
Felix Boekstegers, Statistical Genetics Research Group, Institute of Medical Biometry, Heidelberg University, Heidelberg, Germany.
James F Wilson, Centre for Global Health Research, Usher Institute, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, Scotland; MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh, Scotland.
Macarena Fuentes-Guajardo, Departamento de Tecnología Médica, Facultad de Ciencias de la Salud, Tarapacá University, Arica, Chile.
Rolando Gonzalez-Jose, Instituto Patagónico de Ciencias Sociales y Humanas, Centro Nacional Patagónico, CONICET, Puerto Madryn, Argentina.
Gabriel Bedoya, Instituto de Biología, Grupo Genmol, Universidad de Antioquía, Medellín, Colombia.
Maria Cátira Bortolini, Instituto de Biociências, Universidad Federal do Rio Grande do Sul, Puerto Alegre, Brazil.
Victor Acuña-Alonzo, National Institute of Anthropology and History, Mexico City, Mexico.
Carla Gallo, Laboratorios de Investigación y Desarrollo, Facultad de Ciencias y Filosofía, Universidad Peruana Cayetano Heredia, Lima, Perú.
Andres Ruiz Linares, Ministry of Education Key Laboratory of Contemporary Anthropology and Collaborative Innovation Center of Genetics and Development, School of Life Sciences and Human Phenome Institute, Fudan University, Shanghai, China; Aix-Marseille Université, CNRS, EFS, ADES, Marseille, France; Department of Genetics, Evolution and Environment, UCL Genetics Institute, University College London, London, UK.
Francisco Rothhammer, Instituto de Alta Investigación, Tarapacá University, Arica, Chile.
Justo Lorenzo Bermejo, Statistical Genetics Research Group, Institute of Medical Biometry, Heidelberg University, Heidelberg, Germany.
References
- 1. Broman, K.W. and Weber, J.L. (1999) Long homozygous chromosomal segments in reference families from the Centre d'Etude du polymorphisme humain. Am. J. Hum. Genet., 65, 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ceballos, F.C., Joshi, P.K., Clark, D.W., Ramsay, M. and Wilson, J.F. (2018) Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet., 19, 220–234. [DOI] [PubMed] [Google Scholar]
- 3. McQuillan, R., Leutenegger, A.L., Abdel-Rahman, R., Franklin, C.S., Pericic, M., Barac-Lauc, L., Smolej-Narancic, N., Janicijevic, B., Polasek, O., Tenesa, A. et al. (2008) Runs of homozygosity in European populations. Am. J. Hum. Genet., 83, 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rudan, I., Campbell, H., Carothers, A.D., Hastie, N.D. and Wright, A.F. (2006) Contribution of consanguinuity to polygenic and multifactorial diseases. Nat. Genet., 38, 1224–1225. [DOI] [PubMed] [Google Scholar]
- 5. Enciso-Mora, V., Hosking, F.J. and Houlston, R.S. (2010) Risk of breast and prostate cancer is not associated with increased homozygosity in outbred populations. Eur. J. Hum. Genet., 18, 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomsen, H., MIDS, F., Woltmann, A., Johansson, R., Eyfjörd, J.E., Hamann, U., Manjer, J., Enquist-Olsson, K., Henriksson, R., Herms, S. et al. (2015) Inbreeding and homozygosity in breast cancer survival. Sci. Rep., 5, 16467. 10.1038/srep16467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bacolod, M.D., Schemmann, G.S., Wang, S., Shattock, R., Giardina, S.F., Zeng, Z., Shia, J., Stengel, R.F., Gerry, N., Hoh, J. et al. (2008) The signatures of autozygosity among patients with colorectal cancer. Cancer Res., 68, 2610. 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spain, S.L., Cazier, J.B., CORGI Consortium, Houlston, R., Carvajal-Carmona, L. and Tomlinson, I. (2009) Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res., 69, 7422–7429. [DOI] [PubMed] [Google Scholar]
- 9. Siraj, A.K., Khalak, H.G., Sultana, M., Al-Rasheed, M., Bavi, P., Al-Sanea, N., Al-Dayel, F., Uddin, S., Alkuraya, F.S. and Al-Kuraya, K.S. (2013) Colorectal cancer risk is not associated with increased levels of homozygosity in Saudi Arabia. Genet. Med., 14, 720–728. [DOI] [PubMed] [Google Scholar]
- 10. McWhirter, R.E., Thomson, R.J., Marthick, J.R., Rumbold, A.R., Brown, M.A., Taylor-Thomson, D., Maypilama, E.L., Condon, J.R. and Dickinson, J.L. (2014) Runs of homozygosity and a cluster of vulvar cancer in young Australian aboriginal women. Gynecol. Oncol., 133, 421–426. [DOI] [PubMed] [Google Scholar]
- 11. Loveday, C., Sud, A., Litchfield, K., Levy, M., Holroyd, A., Broderick, P., Kote-Jarai, Z., Dunning, A.M., Muir, K., Peto, J. et al. (2019) Runs of homozygosity and testicular cancer risk. Andrology., 7, 555–564. [DOI] [PubMed] [Google Scholar]
- 12. Hosking, F.J., Papaemmanuil, E., Sheridan, E., Kinsey, S.E., Lightfoot, T., Roman, E., Irving, J.A., Allan, J.M., Taylor, M., Tomlinson, I.P. et al. (2010) Genome-wide homozygosity signatures and childhood acute lymphoblastic leukemia risk. Blood, 115, 4472–4477. [DOI] [PubMed] [Google Scholar]
- 13. Thomsen, H., Inacio da Silva Filho, M., Fuchs, M., Ponader, S., Pogge von Strandmann, E., Eisele, L., Herms, S., Hoffmann, P., Engert, A., Hemminki, K. and Försti, A. (2016) Evidence of inbreeding in Hodgkin lymphoma. PLoS One, 11, e0154259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sud, A., Cooke, R., Swerdlow, A.J. and Houlston, R.S. (2015) Genome-wide homozygosity signature and risk of Hodgkin lymphoma. Sci. Rep., 5, 14315. 10.1038/srep14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orloff, M.S., Zhang, L., Bebek, G. and Eng, C. (2012) Integrative genomic analysis reveals extended germline homozygosity with lung cancer risk in the PLCO cohort. PLoS One, 7, e31975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomsen, H., Chen, B., Figlioli, G., Elisei, R., Romei, C., Cipollini, M., Cristaudo, A., Bambi, F., Hoffmann, P., Herms, S. et al. (2016) Runs of homozygosity and inbreeding in thyroid cancer. BMC Cancer, 16, 227. 10.1186/s12885-016-2264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark, D.W., Okada, Y., Moore, K.H.S., Mason, D., Pirastu, N., Gandin, I., Mattsson, H., Barnes, C.L.K., Lin, K., Zhao, J.H. et al. (2019) Associations of autozygosity with a broad range of human phenotypes. Nat. Commun., 10, 4957. 10.1038/s41467-019-12283-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawlor, D., Ebrahim, S. and Davey, S.G. (2002) The association between components of adult height and type II diabetes and insulin resistance: British Women's heart and health study. Diabetologia, 45, 1097–1106. [DOI] [PubMed] [Google Scholar]
- 19. Lawlor, D.A., Taylor, M., Davey Smith, G., Gunnell, D. and Ebrahim, S. (2004) Associations of components of adult height with coronary heart disease in postmenopausal women: the British women’s heart and health study. Heart, 90, 745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith, G.D., Hart, C., Upton, M., Hole, D., Gillis, C., Watt, G. and Hawthorne, V. (2000) Height and risk of death among men and women: aetiological implications of associations with cardiorespiratory disease and cancer mortality. J. Epidemiol. Community Health, 54, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gunnell, D., Okasha, M., Smith, G.D., Oliver, S.E., Sandhu, J. and Holly, J.M. (2001) Height, leg length, and cancer risk: a systematic review. Epidemiol. Rev., 23, 313–342. Epub August 24, 2002 PubMed PMID: 12192740. [DOI] [PubMed] [Google Scholar]
- 22. Ceballos, F.C., Hazelhurst, S., Clark, D.W., Agongo, G., Asiki, G., Boua, P.R., Xavier Gómez-Olivé, F., Mashinya, F., Norris, S., Wilson, J.F. and Ramsay, M. (2020) Autozygosity influences cardiometabolic disease-associated traits in the AWI-gen sub-Saharan African study. Nat. Commun., 11, 5754. 10.1038/s41467-020-19595-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mörner, M. (1967) Race mixture in the history of Latin America. Little, Brown, Boston. [Google Scholar]
- 24. Ruiz-Linares, A., Adhikari, K., Acuña-Alonzo, V., Quinto-Sanchez, M., Jaramillo, C., Arias, W., Fuentes, M., Pizarro, M., Everardo, P., de Avila, F. et al. (2014) Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet., 10, e1004572. 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eslick, G.D. (2010) Epidemiology of gallbladder cancer. Gastroenterol. Clin. N. Am., 39, 307–330. [DOI] [PubMed] [Google Scholar]
- 26. Arnold, M., Moore, S.P., Hassler, S., Ellison-Loschmann, L., Forman, D. and Bray, F. (2014) The burden of stomach cancer in indigenous populations: a systematic review and global assessment. Gut, 63, 64–71. [DOI] [PubMed] [Google Scholar]
- 27. Pino-Yanes, M., Thakur, N., Gignoux, C.R., Galanter, J.M., Roth, L.A., Eng, C., Nishimura, K.K., Oh, S.S., Vora, H., Huntsman, S. et al. (2015) Genetic ancestry influences asthma susceptibility and lung function among Latinos. J. Allergy Clin. Immunol., 135, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu, H., Huff, C.D., Yamamura, Y., Wu, X. and Strom, S.S. (2015) The relationship between native American ancestry, body mass index and diabetes risk among Mexican-Americans. PLoS One, 10, e0141260. 10.1371/journal.pone.0141260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bryc, K., Durand, E.Y., Macpherson, J.M., Reich, D. and Mountain, J.L. (2015) The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet., 96, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carter-Pokras, O.D. and Gergen, P.J. (1993) Reported asthma among Puerto Rican, Mexican-American, and Cuban children, 1982 through 1984. Am. J. Public Health, 83, 580–582. Epub April 01, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salari, K., Choudhry, S., Tang, H., Naqvi, M., Lind, D., Avila, P.C., Coyle, N.E., Ung, N., Nazario, S., Casal, J. et al. (2005) Genetic admixture and asthma-related phenotypes in Mexican American and Puerto Rican asthmatics. Genet. Epidemiol., 29, 76–86. [DOI] [PubMed] [Google Scholar]
- 32. San Sebastian, M. and Hurtig, A.K. (2007) Review of health research on indigenous populations in Latin America, 1995-2004. Salud Publica Mex., 49, 316–320. [DOI] [PubMed] [Google Scholar]
- 33. Lorenzo Bermejo, J., Boekstegers, F., Gonzalez Silos, R., Marcelain, K., Baez Benavides, P., Barahona Ponce, C., Muller, B., Ferreccio, C., Koshiol, J., Fischer, C. et al. (2017) Subtypes of native American ancestry and leading causes of death: Mapuche ancestry-specific associations with gallbladder cancer risk in Chile. PLoS Genet., 13, e1006756. 10.1371/journal.pgen.1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wright, A.F. and Hastie, N.D. (2001) Complex genetic diseases: controversy over the Croesus code. Genome Biol., 2, comment2007.1. 10.1186/gb-2001-2-8-comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bittles, A.H., Mason, W.M., Greene, J. and Rao, N.A. (1991) Reproductive behavior and health in consanguineous marriages. Science (New York, N.Y.), 252, 789–794. [DOI] [PubMed] [Google Scholar]
- 36. Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M.A., Bender, D., Maller, J., Sklar, P., de Bakker, P.I., Daly, M.J. and Sham, P.C. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alexander, D.H., Novembre, J. and Lange, K. (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res., 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cohn, P., Rothhammer, F. and Cruz-Coke, R. (1985) Correlation between genetic structure and social class in Chile. Rev. Med. Chil., 113, 470–471. [PubMed] [Google Scholar]
- 39. Valenzuela, C.Y. (2011) Human sociogenetics. Biol. Res., 44, 393–404. [PubMed] [Google Scholar]
- 40. Price, A.L., Patterson, N.J., Plenge, R.M., Weinblatt, M.E., Shadick, N.A. and Reich, D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 41. Genomes Project, C., Auton, A., Brooks, L.D., Durbin, R.M., Garrison, E.P., Kang, H.M., Korbel, J.O., Marchini, J.L., McCarthy, S., McVean, G.A. et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reich, D., Patterson, N., Campbell, D., Tandon, A., Mazieres, S., Ray, N., Parra, M.V., Rojas, W., Duque, C., Mesa, N. et al. (2012) Reconstructing native American population history. Nature, 488, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de la Fuente, C., Avila-Arcos, M.C., Galimany, J., Carpenter, M.L., Homburger, J.R., Blanco, A., Contreras, P., Cruz Davalos, D., Reyes, O., San Roman, M. et al. (2018) Genomic insights into the origin and diversification of late maritime hunter-gatherers from the Chilean Patagonia. Proc. Natl. Acad. Sci. U. S. A., 115, E4006–E4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lindo, J., Haas, R., Hofman, C., Apata, M., Moraga, M., Verdugo, R.A., Watson, J.T., Viviano Llave, C., Witonsky, D., Beall, C. et al. (2018) The genetic prehistory of the Andean highlands 7000 years BP though European contact. Sci. Adv., 4, eaau4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


