Abstract
Context
Data and studies based on exome sequencing for the genetic evaluation of short stature are limited, and more large-scale studies are warranted. Some factors increase the likelihood of a monogenic cause of short stature, including skeletal dysplasia, severe short stature, and small for gestational age (SGA) without catch-up growth. However, whether these factors can serve as predictors of molecular diagnosis remains unknown.
Objective
We aimed to explore the diagnostic efficiency of the associated risk factors and their exome sequences for screening.
Methods
We defined and applied factors that increased the likelihood of monogenic causes of short stature in diagnostic genetic tests based on next-generation sequencing (NGS) in 814 patients with short stature and at least 1 other factor.
Results
Pathogenic/likely pathogenic (P/LP) variants in genes, copy number variations, and chromosomal abnormalities were identified in 361 patients. We found P/LP variants among 111 genes, and RASopathies comprised the most important etiology. Short stature combined with other phenotypes significantly increased the likelihood of a monogenic cause, including skeletal dysplasia, facial dysmorphism, and intellectual disability, compared with simple severe short stature (<–3 SD scores). We report novel candidate pathogenic genes, KMT2C for unequivocal growth hormone insensitivity and GATA6 for SGA.
Conclusion
Our study identified the diagnostic characteristics of NGS in short stature with different risk factors. Our study provides novel insights into the current understanding of the etiology of short stature in patients with different phenotypes.
Keywords: short stature, whole exome sequencing, next generation sequencing
Children who are >2 SD below the population mean or the estimated familial target height are generally classified as having short stature and is a common reason for referrals to pediatric endocrinologists (1). Height in humans is influenced by hereditary, hormonal, nutritional, and environmental factors. Normal variations in adult height are largely attributed to the combined effects of various inherited genes. Thus, height is typically a polygenic trait (2-5). However, mutations in single genes can significantly affect height (6). Although several monogenic disorders can perturb growth, the role of genetic diagnostics in the evaluation of children with short stature has not reached a consensus.
With the use of next-generation sequencing (NGS) technology in clinical settings, genetic diagnostic strategies are playing increasingly important roles in determining the etiology and diagnosis of short stature. Genetic test algorithms might be useful for distinct diagnostic subgroups of patients with short stature (7). Exome sequencing has a high diagnostic yield for patients with short stature (8, 9). However, data and studies based on exome sequencing for the genetic evaluation of short stature are limited, and more large-scale studies are warranted.
Factors such as severe familial forms of isolated growth hormone deficiency (IGHD) or specific syndromic forms of multiple pituitary hormone deficiencies (MPHDs) increase the likelihood of a monogenic cause of short stature and severe short stature (<-3 SD compared with the population mean or midparental target height), body disproportion and/or skeletal dysplasia, and small for gestational age (SGA) without adequate catch-up growth (6, 10). However, these factors have not been rigorously validated as predictors or indicators for genetic diagnoses.
We collected samples from 814 patients with suspected monogenic short stature and analyzed 330 of them by whole-exome sequencing (WES) and 484 using an inherited disease panel (Fig. 1). We defined factors that increased the likelihood of a monogenic cause of short stature and considered them as indications for genetic diagnosis. We conducted an in-depth analysis of NGS data of patients with short stature and different phenotypes. Our study provides insights into the current understanding of the etiologies of short stature.
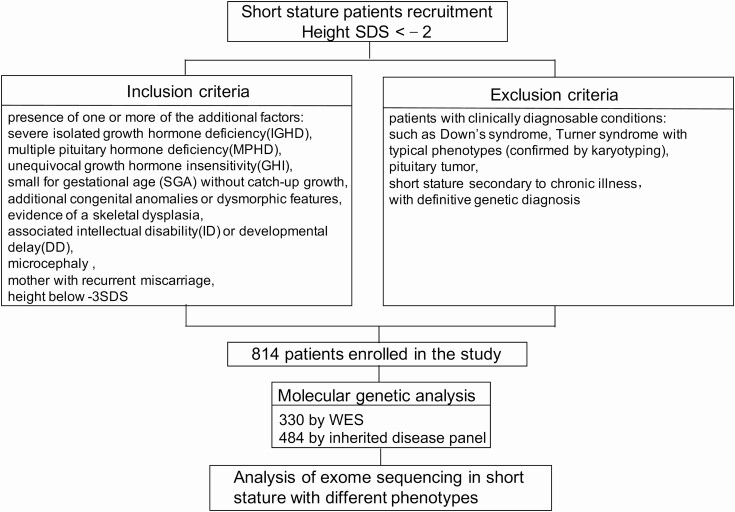
Figure 1.
Flowchart of patients recruitment and variants discovery approach. SDS, standard deviation score; WES, whole exome sequence.
Materials and Methods
Patient Referral
We screened pathogenic variants in 814 children with short stature who were followed up between July 2015 and March 2020 in the Department of Endocrinology and Metabolism at Shanghai Children’s Medical Center, Shanghai Jiaotong University School of Medicine and met the inclusion and exclusion criteria (Fig. 1) (Methods (11)).
The Ethics Committee of Shanghai Children’s Medical Center approved the study. Written informed consent was obtained from the parents of all participants.
Health Information and Clinical History
The documented medical history included birth status, feeding habits, growth, development, and a history of illness of the children and their family members. Physical examinations included facial features, height, weight, head circumference, seated height, arm span, and signs of sex development.
Serum peak growth hormone (GH) level upon provocation (2 independent provocation tests), and levels of insulin-like growth factor (IGF)-1 (12, 13), luteinizing hormone (LH), follicle-stimulating hormone (FSH), thyroid-stimulating hormone (TSH), adrenocorticotropic hormone (ACTH), and cortisol were determined using routine laboratory blood tests. Bone age was assessed by radiographic imaging and using the Greulich–Pyle Atlas method. Most patients were also assessed as needed by brain magnetic resonance imaging, echocardiography, gastrointestinal ultrasonography, and ultrasound of the urinary system.
Molecular Genetic Analysis
Peripheral blood samples were collected from the patients and their parents after obtaining written informed consent. Samples were analyzed by NGS and using the Agilent SureSelect capture technology (Agilent, Santa Clara, CA, USA), followed by either WES between 2018 and 2020 or an inherited disease panel (commercial version of Clearseq Inherited Disease panel from Agilent, part number: 5190-7519) comprising 2742 genes between 2015 and 2017. The captured libraries were sequenced using the Illumina HiSeq 2500 system (Illumina, San Diego, CA, USA) and reads were aligned to the Human Reference Genome (NCBI build37, hg 19) using Burrows–Wheeler aligner-maximum exact matches (14). Variants were called using the Genome Analysis Toolkit. All single nucleotide variants and indels were saved in variant call format files and annotated using Ingenuity Variant Analysis (Ingenuity Systems, Redwood City, CA, USA) and TGex (Translational Genomics Expert) platforms for variation filtering and interpretation (15). Briefly, all variants with a satisfactory sequencing depth and quality (average depth >150, 20× coverage >98%) were filtered according to a minor allele frequency of >0.01 in our in-house and genomAD exome (http://gnomad.broadinstitute.org/) databases (NGS sequencing data quality control metrics in Reference 11). The filtered variants were then sorted based on correlations between patient phenotypes and mutant genes using Ingenuity Variant Analysis and TGex. All suspected variants were confirmed by Sanger sequencing and validated using parental tests. Variants were manually classified according to the method recommended by the American College of Medical Genetics and Genomics (16).
CNVs were identified using open source CNVkit (17) software, which is a tool kit that can infer and visualize copy number from targeted DNA sequencing data. Previously aligned exome data (bam files) for sequencing variant screening were used again as input. Normal references for CNV identification were constructed based on sequencing data generated following the same protocol and experimental conditions from 10 normal males and 10 females who had no pathogenic CNVs, as validated by CMA. Individual CNVs were identified using default CNV kit settings. All CNVs identified using CNVkit were classified based on the CNV scoring metrics in ACMG/Clingen Technical Standards (18).
Statistical Analysis
Fisher’s exact test was carried out for categorical variables between groups. Results with P < .05 were considered statistically significant. All analyses were performed using Statistical Package for the Social Sciences for Windows (version 23.0,SPSS, Inc., Chicago, IL, USA).
Results
Demographic Data
The study involved 438 boys and 376 girls with a median age at diagnosis of 6.5 years (2 months to 17.68 years) and an average height SD of –3.043 (range –2.01 to –8.53).
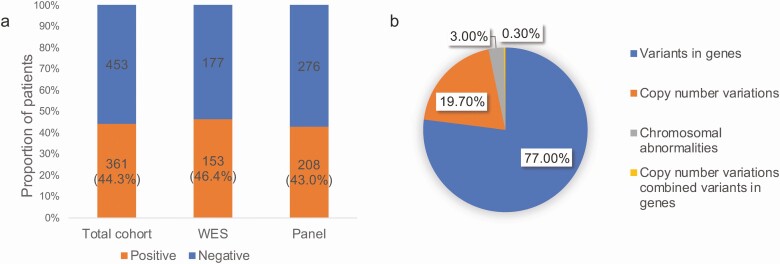
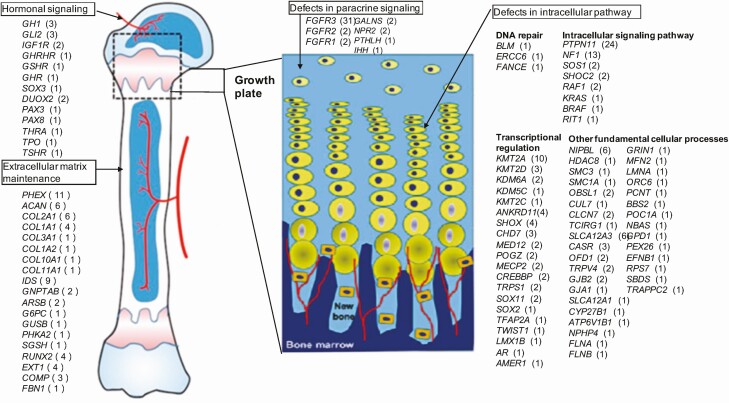
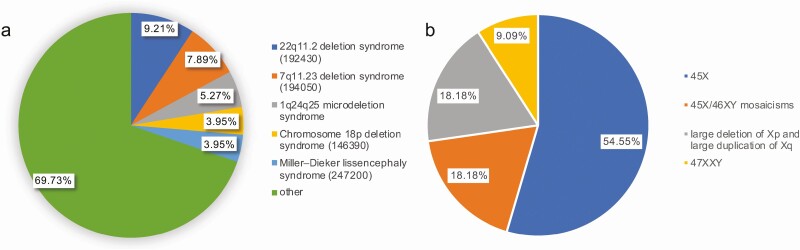
Among the 814 patients, samples of 330 and 484 with suspected monogenic short stature were respectively assessed using WES and the inherited disease panel. The P/LP variants in genes, CNVs, and chromosomal abnormalities were identified in 361 patients (Fig. 2). In addition, 279 patients harbored the P/LP variants distributed among 111 genes (Fig. 3), 72 had P/LP CNVs, and 11 had P/LP chromosomal abnormalities (Fig. 4).
Figure 2.
(A) In total, 44.3% (361/814) patients were identified with pathogenic/likely pathogenic (P/LP) variants; WES was 46.4% and that of the panel was 43.0%. (B) A total of 361 patients harbored P/LP variants, including 77.0% patients harboring variants in genes, 19.7% harboring copy number variations, 3.0% harboring chromosomal abnormalities, and 0.3% harboring number variations combined variants in genes. WES, whole exome sequence; Panel, inherited disease panel.
Figure 3.
A total of 279 patients were identified with pathogenic/likely pathogenic variants distributed among 111 genes; these genes were classified centered on the epiphyseal growth plate.
Figure 4.
(A) A total of 72 patients were identified with pathogenic/likely pathogenic copy number variations; 22q11.2 deletion syndrome was the most common copy number variation. (B) Eleven patients had pathogenic/likely pathogenic chromosomal abnormalities.
Analysis of Short Stature With Different Phenotypes
Table 1 shows the diagnostic efficiency of NGS in patients with short stature and various phenotypes.
Table 1.
The diagnostic efficiency of NGS in short stature patients with different phenotypes
| No. of patients | P/LP cases (%) | Variants in genes | CNVs | Chromosomal abnormalities | CNVs combined variants in genes | P | |
|---|---|---|---|---|---|---|---|
| severe IGHD | 16 | 4 (25%) | 4 | — | — | — | .121 |
| MPHD | 11 | 4 (36.4%) | 4 | — | — | — | <.001 |
| GHI | 39 | 8 (20.5%) | 6 | 2 | — | — | <.001 |
| SGA without catch-up growth | 87 | 21 (24.1%) | 11 | 9 | — | 1 | <.001 |
| Congenital anomalies or dysmorphic features | 387 | 217 (56.2%) | 162 | 45 | 10 | — | <.001 |
| Skeletal dysplasia | 235 | 152 (64.7%) | 146 | 6 | — | — | <.001 |
| Intellectual disability or developmental delay | 140 | 98 (70%) | 50 | 48 | — | — | <.001 |
| Microcephaly | 16 | 9 (56.3%) | 6 | 3 | — | — | .003 |
| Mother with recurrent miscarriage | 3 | 2 (66.7%) | / | 2 | — | — | .312 |
| Height below –3SD (none of the additional phenotypes) | 143 | 16 (11.2%) | 12 | 3 | 1 | — | (Ref.) |
P, Fisher’s exact test was carried out for categorical variables between different phenotypes and height below –3SD (none of additional phenotypes).
IGHD, isolated growth hormone deficiency; MPHD, multiple pituitary hormone deficiencies; GHI, unequivocal growth hormone insensitivity; SGA, small for gestational age; SDS, standard deviation scores; CNV, Copy number variation.
IGHD, MPHD, and GHI
Sixteen patients were diagnosed with severe IGHD based on clinical, laboratory, and imaging information, and a peak GH level on provocation was <3 ng/mL. The P/LP variants were detected in 4 (25%) of 16 patients. Among 11 patients diagnosed with MPHD, 4 (36.4%) harbored the P/LP variants (Table 2). Unequivocal growth hormone insensitivity (GHI) was diagnosed in 39 patients with short stature based on peak GH ≥7 μg/L and IGF-1 SDS ≤–2.0. Eight (20.5%) of the 39 patients had the P/LP variants (Table 3).
Table 2.
The phenotype and genotype analysis of patients with IGHD and MPHD
| Patient | Sex | Age (year) | Height (SDS) | GH peak (ng/mL) | Other pituitary hormone | Other phenotypes | MRI | Gene | Variation | Parental validation |
|---|---|---|---|---|---|---|---|---|---|---|
| 6135 | Male | 15.50 | –5.64 | 0.56 | Normal | / | Normal | GH1 |
NM_000515.4: c.242_243del p.(Ser81*) |
F/M |
| 6515 | Male | 3.92 | –3.17 | 0.01 | Normal | Cryptorchidism | Small pituitary size | GH1 |
NM_000515.4: c.291+1G>A p.? |
De novo |
| 10010 | Male | 2.83 | –8.54 | 0.06 | Normal | Big and protruding foreheads | Small pituitary size | GH1 |
NM_000515.4: [c.240del]/[Exon1-5 del] [p.(Ser81Glnfs*19)]/[p.?] |
F/M |
| 3973 | Male | 11.18 | –0.94 (<–2 SD the estimated familial target height) |
0.11 | Normal | Small penis, Mild learning difficulties |
Anterior pituitary hypoplasia | SOX3 |
NM_005634.2: c.424C>A p.(Pro142Thr) |
M |
| 5175 | Male | 2.56 | –5.3 | 0.45 | LH↓, FSH↓, TSH↓ | Micropenis, small testes |
Anterior pituitary hypoplasia | GLI2 |
NM_005270.4: c.3463_3464del p.(Asp1155Argfs*39) |
De novo |
| 5589 | Male | 2.25 | –5.75 | 0.04 | LH↓, FSH↓, TSH↓, ACTH↓ | Micropenis, small testes, polydactyly |
Anterior pituitary hypoplasia | GLI2 |
NM_005270.4: c.3137del p.(Gly1046Alafs*84) |
M |
| 6606 | Male | 5.90 | –4 | 0.52 | LH↓, FSH↓, TSH↓ | Micropenis, small testes, deafness, intellectual disability |
Anterior pituitary hypoplasia | GLI2 |
NM_005270.4: c.3640C>T p.(Gln1214*) |
M |
| 3969 | Male | 12.72 | –4.66 | 0.08 | TSH↓, ACTH↓ | Hematuria, normal renal function |
Anterior pituitary hypoplasia | NPHP4 |
NM_015102.4: c.3196C>T p.(Gln1066*) |
F/M |
25% (4/16) patients with severe IGHD were identified with pathogenic/likely pathogenic variants in 2 genes (GH1, SOX3); 36.36% (4/11) patients with MPHD were identified with pathogenic/likely pathogenic variants in 2 genes (GLI2, NPHP4).
Abbreviations: IGHD, isolated growth hormone deficiency; MPHD, multiple pituitary hormone deficiencies; SDS, standard deviation scores; F, paternal inheritance; M, maternal inheritance; F/M, inherited respectively from parents; LH, luteinizing hormone; FSH, follicle-stimulating hormone; TSH, thyroid-stimulating hormone; ACTH, adrenocorticotropic hormone.
Table 3.
20.51% (8/39) patients with unequivocal GHI were identified with pathogenic/likely pathogenic variants
| Patient | Sex | Age (year) | Height (SDS) | GH peak (ng/ml) | IGF–1 (SDS) | Other phenotypes | Variation | Parental validation |
|---|---|---|---|---|---|---|---|---|
| 4350 | Female | 10.08 | –3.37 | 19.71 | <–2SDS | CHD Facial dysmorphisms pectus excavatum |
PTPN11
NM_002834.3: c.1510A>G p.(Met504Val) |
NA |
| 8394 | Female | 8.58 | –4.35 | 13.06 | <–2SDS | CHD Facial dysmorphisms Pectus excavatum Amblyopia Deafness |
PTPN11
NM_002834.3: c.218C>T p.(Thr73Ile) |
De novo |
| 8953 | Male | 11.67 | –4.48 | 8.87 | <–2SDS | CHD Facial dysmorphisms Pectus excavatum Cryptorchidism |
PTPN11
NM_002834.3: c.923A>G p.(Asn308Ser) |
M |
| 8591 | Female | 12.33 | –3.54 | 10 | <–2SDS | CHD Webbed neck hp:0000465 |
PTPN11
NM_002834.3: c.188A>G p.(Tyr63Cys) |
De novo |
| 2221 | Male | 12.09 | –2.51 | 9.13 | <–2SDS | Subclinical hypothyroidism |
DUOX2
NM_014080.4: [c.3329G>A]/[c.1310G>C] [p.(Arg1110Gln)]/[p.(Gly437Ala)] |
F/M |
| 13165 | Female | 11.14 | –2.05 | 10.2 | <–2SDS | Primordial uterus Congenital spina bifida |
KMT2C
NM_170606.3: c.3841+1G>A p.? |
De novo |
| 5766 | Female | 11.25 | –3.52 | 9.84 | <–2SDS | / | dup(16)(q11.2)(over 300 kb) | NA |
| 7611 | Male | 8.33 | –3.09 | 10.73 | <–2SDS | CHD | del(22)(q11.21) [hg19(chr22:18 900 287 -21 245 501)] (over 2300 kb) |
NA |
Abbreviations: GHI, growth hormone insensitivity; CHD, congenital heart disease; F, paternal inheritance; M, maternal inheritance; F/M, inherited respectively from parents; NA, Not available.
SGA without catch-up growth
SGA without catch-up growth at the age of 2 years was diagnosed in 87 patients with short stature, including 45 and 42 with and without syndromic causes. The P/LP variants were detected in 21 (24.1%) of these patients; the P/LP cases for short children with and without syndromic causes were 14 (31.1%) of the 45 causes and 7 (16.7%) of the 42 causes (Table 4).
Table 4.
24.1% (21/87) SGA without catch-up growth after 2 years of birth were identified with pathogenic/likely pathogenic variants
| Patient | Sex | Age (year) | Height (SDS) | Phenotypes | Variation |
|---|---|---|---|---|---|
| 5341 | Female | 5.00 | –3.33 | SGA, CHD, facial dysmorphisms, development delay |
KMT2A
NM_001197104.1: c.11716C>T p.(Arg3906Cys) (het) (de novo) |
| 6533 | Female | 6.50 | –2 | SGA |
COL1A1
NM_000088.3: c.1171G>A p.(Asp391Asn) (het) (de novo) |
| 4042 | Male | 4.43 | –4.02 | SGA |
COL2A1
NM_001844.4: c.1016G>A p.(Gly339Asp) (het) (de novo) |
| 5621 | Female | 16.38 | –1.31 | SGA, cleft lip and palate, DSD, no olfactory bulb |
FGFR1
NM_023110.2: c.760C>Tp.(Arg254Trp) (het) (de novo) |
| WJ-584 | Male | 11.02 | –2.64 | SGA, facial dysmorphisms, microtia, absence of patella DSD |
ORC6
NM_014321.3: c.67A>G p.(Lys23Glu) (hom)(F/M) |
| WJ-656 | Male | 13.33 | –5.09 | SGA, facial dysmorphisms, microcephaly, development delay, acanthosis nigricans type 2 diabetes |
PCNT
NM_006031.5: [c.3103C>T]/[c.502C>T][p.(Arg1035*)]/[p.(Gln168*)] (compound heterozygote) (F/M) |
| 8816 | Male | 4.50 | –2.38 | SGA, CHD |
ANKRD11
NM_013275.5:c.3140_3143delp.(Gln1047Argfs*270) (het)(M) |
| 7290 | Male | 4.83 | –3.83 | SGA | RPS7 NM_021140.3:c.75+2T>Cp.? (het) (NA) |
| 9021 | Female | 7 | –2.4 | SGA facial dysmorphisms |
POC1A
NM_015426.4: c.981+1G>A p.? (hom)(F/M) |
| 9153 | Female | 3.92 | –2.3 | SGA |
CASR
NM_000388.3: c.3082C>T p.(Gln1028*) (het)(M) |
| 6500 | Female | 5.00 | –3.25 | SGA, DSD |
GHR
NM_000163.4: c.136+1G>A p.? (hom)(F/M) |
| 7500 | Male | 3.00 | –4.78 | SGA |
SOX11
NM_003108.3: c.425C>G p.(Ala142Gly)(het)(De novo) del(1)(q24.2-25.1)[hg19,(chr1:169 433 149-173 827 682)] (over 4300 kb) |
| 13921 | Female | 5.83 | –3.98 | SGA, IGF-1 >2 SD | IGF1Rgene deletion (whole gene) |
| 13693 | Male | 10.00 | –1.9 | SGA, intellectual disability | del(7)(q11.23)[hg19,(chr7:73 442 119-74 175 022)] (over 700 kb) |
| 10850 | Female | 7.67 | –5.8 | SGA, CHD, facial dysmorphisms, intellectual disability | del(18)(p11.31-p11.21)[hg19,(chr18:2 916 992-12 884 236)] (over 9900 kb) |
| 12721 | Female | 1.50 | –4.1 | SGA, facial dysmorphisms, development delay | del(7)(q36.1-q36.3)[hg19,(chr7:150 642 044-157 210 133)] (over 6500 kb) dup(18)(q23)[hg19,(chr18:77 439 801-77 514 510)] (over 200 kb) |
| 2882 | Female | 6.08 | –3.35 | SGA, CHD, facial dysmorphisms, intellectual disability, auricle deformity | del(9)(q21.11-q21.31)[hg19,(chr9:71000154-83236029)] (12236 kb) |
| 7767 | Female | 6.58 | –4.93 | SGA, CHD, intellectual disability | del(13)(q31.1-q32.1)[hg19,(79 314 118-96 544 277)] (17230 kb) |
| 7177 | Male | 7.00 | –1.9 | SGA | del(15)(q26.3)[hg19,(chr15:99 191 768-101 792 137)] (over 2600 kb) |
| 9951 | Female | 1.50 | –2.5 | SGA, facial dysmorphisms, development delay | del(16)(p13.11)[hg19,(chr16:15 737 124-16 317 328)] (over 500 kb) |
| 13727 | Female | 7.00 | –2.9 | SGA, development delay | dup(19)(p13.3)[hg19,(chr19:852 303-6 720 661)] (over 5800 kb) |
Abbreviations: SGA, small for gestational age; CHD, congenital heart disease; F, paternal inheritance; M, maternal inheritance; F/M, inherited respectively from parents; NA, Not available; het, heterozygote; hom, homozygote.
Congenital anomalies (dysmorphic features), skeletal dysplasia, intellectual disability/developmental delay (ID/DD), and microcephaly
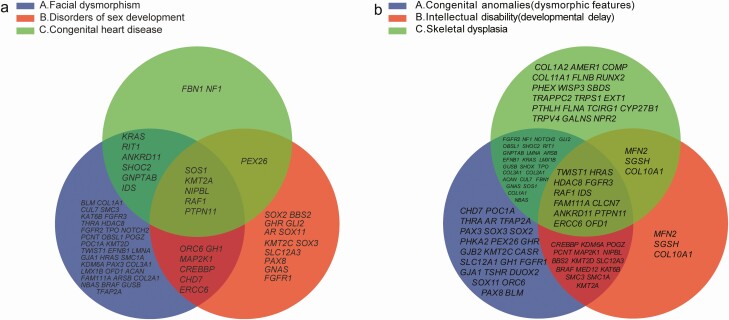
Among the 386 patients with short stature and congenital anomalies or dysmorphic features, the most prevalent were facial dysmorphism, disorders of sex development (DSD), and congenital heart disease (CHD) in 186 (48.2%), 96 (24.9%), and 93 (24.1%) of them, respectively. Figure 5A shows the intersection of pathogenic genes associated with these clinical features.
Figure 5.
The intersection of pathogenic genes associated with different clinical features. (A) In total, 70.4% (131/186) of the patients with facial dysmorphism were identified with P/LP variants related to 52 genes. Of the patients with disorders of sex development, 53.1% (51/96) were identified with P/LP variants related to 25 genes. Of the patients with congenital heart disease, 53.3% (49/92) were related to 14 genes. The intersection of pathogenic genes of these clinical features related to 5 genes, including PTPN11, RAF1, SOS1, NIPBL and KMT2A. (B) Patients with congenital anomalies or dysmorphic features were identified with pathogenic/likely pathogenic variants related to 76 genes (56.2%; 217/386). In total, 64.7% (152/235) of the patients with skeletal dysplasia had pathogenic/likely pathogenic variants related to 60 genes. Of the patients with intellectual disability or developmental delay, 70.0% (98/140) were identified with pathogenic/likely pathogenic variants related to 34 genes. The intersection of pathogenic genes of these clinical features related to 12 genes, including PTPN11, RAF1, HRAS, CLCN7, TWIST1, HDAC8, ANKRD11, OFD1, IDS, ERCC6, FAM111A, and FGFR3.
We identified the P/LP variants in 131 (70.4%) of 186 patients with facial dysmorphism (Table 1 (11)), in 16 (51.6%) of 31 with no other symptoms besides facial dysmorphism, and in 3 patients with these variants in the KMT2A gene. Among the 96 patients diagnosed with DSD, 70 and 26 were males and females, respectively, and the P/LP variants were detected in 51 (53.1% of them; Table 2 (11)). Thirty-nine male patients (46 XY) were diagnosed with cryptorchidism, and 26 (66.7%) of them harbored the P/LP variants (Table 3 (11)). Among 92 patients with CHD, 49 (53.3%) harbored P/LP variants (Table 4 (11)). Among 5 (20%) of 25 patients with short stature and CHD, P/LP variants were found in the NF1, PTPN11, and SHOC2 genes, and in 2 patients with 22q11.2 deletion syndrome (OMIM #611867).
Overall, 152 (64.7%) of 235 patients with skeletal dysplasia had P/LP variants. Pathogenic variants were identified in 59 genes and in 6 CNVs (Table 5 (11)). The P/LP variants detected in 98 (70.0%) of 140 patients with intellectual disability (ID) or developmental delay (DD) were related to 34 genes in 50 (51.0%) of these patients. Seven patients were diagnosed with Cornelia de Lange syndrome (OMIM #122470) related to variants in 4 genes (NIPBL, HDAC8, SMC1A, and SMC3). Five patients harbored the most common pathogenic variant of KMT2A (Table 6 (11)). We identified CNVs in 48 (48.48%) of 98 patients (Table 7 (11)). Figure 5B shows the intersections of pathogenic genes associated with congenital anomalies (dysmorphic features), skeletal dysplasia, and ID (DD). The P/LP variants were related to 6 genes and 4 CNVs in 9 (56.3%) of 16 patients with microcephaly (Table 8 (11)).
Short stature and maternal history of recurrent miscarriages
The mothers of 3 patients with short stature had experienced recurrent miscarriages. One of these patients had P/LP variants comprising a 2q37.3 deletion and a 9q34.3 duplication, and 1 had a 22q11.21 deletion.
Severe short stature (<–3 SD)
We diagnosed 364 patients with severe short stature (<–3 SD compared with the population mean or midparental target height) and 143 (39.3%) of them harbored P/LP variants. However, 143 of these patients had no other risk factors besides short stature (<–3 SD), whereas 16 (11.1%) of the 143 patients harbored the P/LP variants (Table 9 (11)).
Unexpected findings with short stature cases
We identified variants in genes (GATA6, PLCB4, and RYR1) that are not known to be related to short stature carried by patients 9990, 5260, and 9882 (Table 5). However, based on the type of variation, allele frequencies and other criteria, these variants could be classified into likely pathogenic groups. We assumed that these variants might contribute to our patients’ phenotypes, and the 3 genes could possibly be novel candidate genes responsible for short stature. However, due to the lack of evidence for certainty, we still regarded these situations as cases of uncertain diagnosis despite the pathogenicity classification.
Table 5.
Unexpected findings with short stature cases and novel candidate genes
| Patient | Sex | Age (year) | Height (SDS) | Phenotypes | Variation | ACMG category |
|---|---|---|---|---|---|---|
| 9990 | Male | 2 | –2.2 | SGA, CHD, type 1 diabetes |
GATA6
NM_005257.5: c.1366C>T p.(Arg456Cys) (het) (De novo) |
LP |
| 5260 | Male | 8 | –3.01 | facial asymmetry, development delay |
PLCB4
NM_000933.3: c.2980delA p.(Met994*) (het)(F) |
LP |
| 9882 | Male | 3.4 | –3.41 | pectus excavatum, scoliosis, cryptorchidism |
RYR1
NM_000540.2: c.7523G>A p.(Arg2508His) (het) (De novo) |
LP |
Pathogenic variants in genes that are not known to be related to short stature (GATA6, PLCB4, RYR1) were identified in patients 9990, 5260, and 9882.
Abbreviations: SGA, small for gestational age; CHD, congenital heart disease; F, paternal inheritance; het, heterozygote; LP, likely pathogenic.
Discussion
Growth is regulated by several genetic factors, but some individuals with significantly short stature harbor single-gene mutations that considerably affect height (19, 20). To accurately identify the etiology of short stature is challenging because extensive etiological heterogeneity and clinical complexity are involved. We identified factors that increased the likelihood of a monogenic cause of short stature and considered them as indications for genetic tests (Fig. 1). We applied NGS to samples from 814 patients with suspected monogenic short stature and at least 1 of the factors listed in Fig. 1. We identified 361 patients with P/LP variants by NGS in our study, and the P/LP variants were distributed among 111 genes; RASopathies caused by mutations in genes of the Ras–MAPK pathway comprised the most important etiology of short stature in our cohort (Fig. 3). The CNVs diagnosed using NGS mostly caused 22q11.2 and 7q11.23 deletion syndromes. Our patients were of short stature with a risk factor, and the diagnosis yield for monogenic diseases was higher than that in the general group of children with short stature.
Genetic defects of the GH–IGF-1 axis have been associated with severe IGHD and MPHD (21). Our findings showed that variants in GH1 constitute a major cause of severe IGHD. Variants in GLI2 were detected in 3 of 11 patients with MPHD. Serum peak GH level on provocation in positive IGHD and MPHD patients was <1 ng/mL.
Classical GHI originally described by Laron et al. in 1966 (22, 23) and called Laron-type dwarfism or Laron syndrome (OMIM #262500) is caused by a defect in the GH receptor (GHR) gene, resulting in extreme GH resistance and an associated IGF-1 deficiency (24). This rare and extreme phenotype became synonymous with a diagnosis of GHI. During the past 20 years, the GHI categories have been expanded to include mild or moderate GHI and several other congenital and acquired conditions associated with it (25). Among our patients with GHI, 20.51% harbored pathogenic variants, of which PTPN11 was the most common. Studies have suggested that the constitutively activated RAS–MAPK pathway in Noonan syndrome (OMIM #163590) and other RASopathies can lead to inhibition of the JAK/STAT pathway, relatively low levels of IGF-I, and subsequently short stature (26). The most common mutation affects PTPN11, which encodes the cytoplasmic SH2 domain-containing protein tyrosine phosphatase 2 (SHP-2). This enzyme dephosphorylates STAT5b, consequently activating mutations of PTPN11 and downregulating STAT5b activity, while activating the MAPK pathway. The growth response to GH is lower in individuals who are PTPN11 variant-positive than those who are negative (27). Our findings suggested that GHI is most likely caused by variants in PTPN11. We identified a patient with GHI pathogenic variants of KMT2C. KMT2C encodes a histone methyltransferase that regulates gene transcription by modifying chromatin structure. A heterozygous mutation in KMT2C is associated with Kleefstra syndrome-2 (OMIM #617768), which is a rare genetic syndrome with delayed psychomotor development, variable intellectual disability, and mild dysmorphic features. Some patients have short stature, but the involvement of the GH-IGF-1 axis is unknown (28-30). Our findings suggested that the limited growth of patients with a heterozygous mutation in KMT2C can be attributed to an IGF-1 deficiency.
The process of human fetal growth is regulated by fetal and maternal genetic factors that affect the intrauterine environment to ensure effective nutrient exchange between the mother and fetus via the placenta. Small for gestational age has been defined either as being below the tenth percentile for weight at a given gestational age or as having a birth length or weight SD < 2.0 (below the 2.3 percentile) (31). Among the causes of SGA are maternal health and obstetric factors, placental insufficiency, and fetal genetic factors. Among children with idiopathic SGA, ~85% catch up to the third percentile of length by the age of 2 years (32, 33). Children without catch-up growth require further evaluation, especially a subset with progressive postnatal growth failure. The diagnostic yield of NGS in SGA in the present study was 21 (24.1%) of 87, among whom 13 (14.9%) and 8 (9.2%) had P/LP variants in genes and CNVs which was below that of the total cohort (361/814; 44.3%) (P < .05). Imprinted genes in the placenta are important for the control of fetal growth (34, 35). A recent study of 269 patients with SGA with short stature reported a diagnostic yield of 107 (39.78%) of the 269 patients by comparative genomic hybridization combined with methylation analysis, and 32.34% (87/269) patients were diagnosed with imprinting disorders and 7.44% (20/269) were CNVs (35). The diagnostic power of exome sequencing in SGA is limited; further methylation analysis can be an effective approach to diagnose SGA, and environmental causes for SGA should be considered.
One patient with SGA, CHD, and diabetes harbored pathogenic variants in GATA6, which encodes GATA-binding protein 6 and has not yet been associated with short stature. GATA6 belongs to a small family of zinc finger transcription factors that play important roles in the regulation of cellular differentiation and organogenesis during development in vertebrate. The GATA6 phenotypic spectrum includes neonatal-, childhood-, and adult-onset diabetes; exocrine pancreatic insufficiency; pancreatic agenesis or hypoplasia; various cardiac malformations, hypothyroidism, hypopituitarism and pituitary agenesis; intestinal malrotation; hernias; colonic perforation; structural kidney abnormalities; neurocognitive deficits; and seizures (36-38). Two patients with pathogenic variants in GATA6 had intrauterine growth restriction (39, 40). Thus, GATA6 may be a candidate pathogenic gene for SGA without catch-up growth.
RASopathies were the most important etiology of short stature in patients with CHD (Table 4 (11)). The P/LP variants were detected in 20% of the short stature patients who presented with no other symptoms except CHD, and 22q11.2 deletion syndrome was the most common pathogenic variant. The clinical presentation of 22q11.2 deletion syndrome varies by age, and clinical complexity might pose challenges in accurate diagnoses (41). Next-generation sequencing should facilitate the earlier detection and increased recognition of 22q11.2 deletion syndrome.
We detected P/LP variants in 51 (53.1%) of the 96 patients with short stature and DSD. Thirty-nine males (46 XY) had cryptorchidism and 26 (66.7%) of the 39 patients harbored the P/LP variants. Cryptorchidism (OMIM #219250) is 1 of the most frequent congenital birth defects in boys and appears in 2% to 4% of full-term male births (42). Maldescent testicles can be an isolated event or result from a variety of syndromes (syndromic cryptorchidism) and other nonsyndromic diseases (nonsyndromic cryptorchidism) (43-45). Data from 50 studies have associated cryptorchidism with 44 syndromes, as well as genomic loci include 38 protein-coding genes and 22 structural variations containing microdeletions and microduplications (46). Our findings suggest that short stature combined with cryptorchidism considerably increases the likelihood of a monogenic cause of short stature.
Geneticists identified facial dysmorphism in 186 patients in our cohort, and we detected P/LP variants related to 52 genes in 131 (70.4%) of the patients. Many syndromes have recognizable facial features, and Face2gene has achieved a high diagnostic rate in genetic diseases based on facial images (47). Our findings suggested that short stature combined with facial dysmorphism indicates a need for genetic testing. The P/LP variants were detected in 16 (51.6%) of the 31 patients who presented with no other symptoms except facial dysmorphism. Three patients harbored the P/LP variants in KMT2A. Wiedemann–Steiner syndrome (OMIM #605130) is a rare genetic disorder characterized by facial gestalt, neurodevelopmental delay, skeletal anomalies, and growth retardation, which is caused by variations in KMT2A (48). Most patients exhibited suggestive features, but characteristics were less obvious in others (49). Wiedemann–Steiner syndrome is an important consideration for short stature alone with facial dysmorphism.
In our study, 152 (64.7%) of the 235 patients with skeletal dysplasia harbored the P/LP variants related to 59 genes and 6 CNVs (Table 5 (11)). Skeletal dysplasia features, mainly attributable to variants in protein-coding genes, rarely involve structural variations. MFN2, RYR1, and PLCB4 have not been associated with short stature in previous reports; patient phenotypes, types of variations, allele frequencies, and other criteria could classify variants into P/LP groups. Variants in MFN2 or RYR1 lead to a slow, progressive development of neuromuscular disorders, and clinical manifestations include skeletal deformities (50, 51). Pathogenic variants in PLCB4 are associated with auriculocondylar syndrome (OMIM #602483), which is mainly characterized by micrognathia, a small mandibular condyle, facial asymmetry, and question mark–shaped ears. It is a rare disease that segregates in an autosomal dominant pattern in most of the families described in the literature with evident intrafamilial variability (52, 53).
Both DD and ID affect 1% to 3% of children and a genetic etiology is involved in approximately 50% of those affected (54). Our findings suggested that DD and ID combined with short stature increased the likelihood of a monogenic cause, and structural variations containing microdeletions and microduplications were major causes of these conditions. Cornelia de Lange, Wiedemann–Steiner, and Williams–Beuren (OMIM #194050) syndromes are common pathologies (Table 6 and Table 7 (11)).
Microcephaly is defined as a head circumference of >2 SD below the mean for gender and age. Growth retardation accompanied by microcephaly is mainly associated with microcephalic primordial dwarfism such as Cornelia de Lange, MOPD I (OMIM #210710), MOPD II (OMIM #210720), Seckel (OMIM #210600), and Meier–Gorlin (OMIM #224690) syndromes (20). Our findings showed an extremely high positive diagnostic yield for microcephaly with mental retardation, and syndromes associated with abnormal DNA repair, such as Bloom (OMIM #210900) and Cockayne (OMIM #216400, #133540) syndromes, should be recognized (Table 8 (11)).
A recent study diagnosed a pathological cause of severe short stature (<–3 SD compared with the population mean) in 76% and 71% of girls and boys investigated, but a genetic cause of severe short stature was not determined (55). For severe short stature without other symptoms, genetic defects affecting paracrine factors in the growth plate (FGFR3, GNAS, and IHH), genetic defects affecting the cartilage extracellular matrix (ACAN), genetic defects affecting the GH–IGF-1–IGF-1R axis (GHRHR, GHSR, and IGF1R), and Wiedemann–Steiner syndrome (KMT2A) with fewer characteristics should be carefully analyzed.
In conclusion, NGS combined with risk factor screening significantly increased the diagnostic yield of patients with short stature. The diagnostic power of exome sequencing in children with SGA is limited, and adding methylation studies can be an effective approach to diagnose children with SGA. Variants in PTPN11 might comprise the main etiology of mild GHI, and further investigation should target the effectiveness of recombinant human growth hormone (rhGH) therapy for patients with Noonan syndrome and IGF-1 therapy may be an appropriate therapy for these patients. Short stature with facial features indicates the possibility of a genetic etiology, even if accompanied by a single symptom. Some of the patients in this study harbored the P/LP variants in GATA6, RYR1, and PLCB4 that have not yet been associated with short stature. Based on phenotypes, types of variations, allele frequencies, and other criteria, gene variants can be classified into P/LP groups. Short stature might also be a non-primary component of a few syndromic disorders, and WES presents a higher diagnostic yield than short stature panels for these conditions.
Limitations
Our study had some limitations. This study was performed in 1 institute with a large referral population, which could have created a selection bias that likely increased the diagnostic yield of WES in this study. Some children with short stature may have been already diagnosed either clinically or genetically and hence were ineligible for the study, such as those with achondroplasia (OMIM #100800). Some patients were not assessed using WES and rare CNVs are difficult to diagnose using NGS. Although CNV detection based on read-depth information from WES data has been widely adopted in clinical practical, the discovery rate of rare and nonrecurrent CNVs still largely depends on principle of the algorithm, quality of the raw sequencing data, and number of samples in the same batch (56). Future research should further expand the survey sample and improve testing methods.
Acknowledgments
We thank all patients and their families for participating in this project.
Glossary
Abbreviations
- ACTH
adrenocorticotropic hormone
- CHD
congenital heart disease
- CNV
copy number variation
- DSD
disorders of sex development
- FSH
follicle-stimulating hormone
- GH
growth hormone
- GHI
growth hormone insensitivity
- IGF
insulin-like growth factor
- IGHD
isolated growth hormone deficiency
- LH
luteinizing hormone
- MPHD
multiple pituitary hormone deficiency
- NGS
next-generation sequencing
- P/LP
pathogenic/likely pathogenic
- SGA
small for gestational age
- TSH
thyroid-stimulating hormone
- WES
whole-exome sequencing.
Financial Support: This study was supported by the Science and Technology Commission of Shanghai Municipality (Shanghai Clinical Research center for Children’s Rare Diseases 20MC1920400), Shanghai health and Family Planning Commission (20204Y0346), Pudong New Area Science and Technology Development Fund (PKJ2018-Y46), the National Science Foundation for Young Scientists of China (81900722), and Key project of Chongqing Kewei Joint Medical research project (2018ZDXM008).
Additional Information
Disclosure Summary: The authors declared no conflicts of interest.
Data Availability
Data are available from the corresponding author on reasonable request.
Ethics Declaration: The Ethics Committee at Shanghai Children’s Medical Center approved the study. Written informed consent was obtained from the parents of all participants.
References
- 1. Rogol AD, Hayden GF. Etiologies and early diagnosis of short stature and growth failure in children and adolescents. J Pediatr. 2014;164(5 Suppl):S1-14.e6. [DOI] [PubMed] [Google Scholar]
- 2. Hirschhorn JN, Lettre G. Progress in genome-wide association studies of human height. Horm Res. 2009;71(Suppl 2):5-13. [DOI] [PubMed] [Google Scholar]
- 3. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hemani G, Yang J, Vinkhuyzen A, et al. Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs. Am J Hum Genet. 2013;93(5):865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marouli E, Graff M, Medina-Gomez C, et al. ; EPIC-InterAct Consortium; CHD Exome+ Consortium; ExomeBP Consortium; T2D-Genes Consortium; GoT2D Genes Consortium; Global Lipids Genetics Consortium; ReproGen Consortium; MAGIC Investigators . Rare and low-frequency coding variants alter human adult height. Nature. 2017;542(7640):186-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dauber A, Rosenfeld RG, Hirschhorn JN. Genetic evaluation of short stature. J Clin Endocrinol Metab. 2014;99(9):3080-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wit JM, Kiess W, Mullis P. Genetic evaluation of short stature. Best Pract Res Clin Endocrinol Metab. 2011;25(1):1-17. [DOI] [PubMed] [Google Scholar]
- 8. Hauer NN, Popp B, Schoeller E, et al. Clinical relevance of systematic phenotyping and exome sequencing in patients with short stature. Genet Med. 2018;20(6):630-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo MH, Shen Y, Walvoord EC, et al. Whole exome sequencing to identify genetic causes of short stature. Horm Res Paediatr. 2014;82(1):44-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collett-Solberg PF, Ambler G, Backeljauw PF, et al. Diagnosis, genetics, and therapy of short stature in children: a Growth Hormone Research Society International Perspective. Horm Res Paediatr. 2019;92(1):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X , Yao R, Chang G, et al. Data from: Clinical profiles and genetic spectra of 814 Chinese children with short stature. figshare. Dataset. Deposited August 17 2021. ProMED-mail website. 10.6084/m9.figshare.14617449.v8 [DOI]
- 12. Xu SS, Gu XF, Pan H, et al. Reference values for serum IGF-1 and IGFBP-3 in children and adolescents. J Clin Pediatrics. 2009;27(12):1105–10. [Google Scholar]
- 13. Juul A, Dalgaard P, Blum WF, et al. Serum levels of insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) in healthy infants, children, and adolescents: the relation to IGF-I, IGF-II, IGFBP-1, IGFBP-2, age, sex, body mass index, and pubertal maturation. J Clin Endocrinol Metab. 1995;80(8):2534-2542. [DOI] [PubMed] [Google Scholar]
- 14. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu X, Li N, Xu Y, et al. Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: the China experience. Genet Med. 2018;20(9):1045-1053. [DOI] [PubMed] [Google Scholar]
- 16. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riggs ER, Andersen EF, Cherry AM, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22(2):245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lui JC, Nilsson O, Baron J. Recent research on the growth plate: Recent insights into the regulation of the growth plate. J Mol Endocrinol. 2014;53(1):T1-T9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wit JM, Oostdijk W, Losekoot M, van Duyvenvoorde HA, Ruivenkamp CA, Kant SG. MECHANISMS IN ENDOCRINOLOGY: novel genetic causes of short stature. Eur J Endocrinol. 2016;174(4):R145-R173. [DOI] [PubMed] [Google Scholar]
- 21. Rohayem J, Drechsel H, Tittel B, Hahn G, Pfaeffle R, Huebner A. Long-term outcomes, genetics, and pituitary morphology in patients with isolated growth hormone deficiency and multiple pituitary hormone deficiencies: a single-centre experience of four decades of growth hormone replacement. Horm Res Paediatr. 2016;86(2):106-116. [DOI] [PubMed] [Google Scholar]
- 22. Laron Z. Laron syndrome (primary growth hormone resistance or insensitivity): the personal experience 1958-2003. J Clin Endocrinol Metab. 2004;89(3):1031-1044. [DOI] [PubMed] [Google Scholar]
- 23. Laron Z, Pertzelan A, Mannheimer S. Genetic pituitary dwarfism with high serum concentration of growth hormone–a new inborn error of metabolism? Isr J Med Sci. 1966;2(2):152-155. [PubMed] [Google Scholar]
- 24. Eshet R, Laron Z, Pertzelan A, Arnon R, Dintzman M. Defect of human growth hormone receptors in the liver of two patients with Laron-type dwarfism. Isr J Med Sci. 1984;20(1):8-11. [PubMed] [Google Scholar]
- 25. Wit JM, de Luca F. Atypical defects resulting in growth hormone insensitivity. Growth Horm IGF Res. 2016;28:57-61. [DOI] [PubMed] [Google Scholar]
- 26. De Rocca Serra-Nédélec A, Edouard T, Tréguer K, et al. Noonan syndrome-causing SHP2 mutants inhibit insulin-like growth factor 1 release via growth hormone-induced ERK hyperactivation, which contributes to short stature. Proc Natl Acad Sci U S A. 2012;109(11):4257-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Binder G. Noonan syndrome, the Ras-MAPK signalling pathway and short stature. Horm Res. 2009;71(Suppl 2):64-70. [DOI] [PubMed] [Google Scholar]
- 28. Faundes V, Newman WG, Bernardini L, et al. ; Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study; Deciphering Developmental Disorders (DDD) Study . Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am J Hum Genet. 2018;102(1):175-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kleefstra T, Kramer JM, Neveling K, et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet. 2012;91(1):73-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koemans TS, Kleefstra T, Chubak MC, et al. Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLoS Genet. 2017;13(10):e1006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clayton PE, Cianfarani S, Czernichow P, Johannsson G, Rapaport R, Rogol A. Management of the child born small for gestational age through to adulthood: a consensus statement of the International Societies of Pediatric Endocrinology and the Growth Hormone Research Society. J Clin Endocrinol Metab. 2007;92(3):804-810. [DOI] [PubMed] [Google Scholar]
- 32. Hokken-Koelega AC, De Ridder MA, Lemmen RJ, Den Hartog H, De Muinck Keizer-Schrama SM, Drop SL. Children born small for gestational age: do they catch up? Pediatr Res. 1995;38(2):267-271. [DOI] [PubMed] [Google Scholar]
- 33. Karlberg J, Albertsson-Wikland K. Growth in full-term small-for-gestational-age infants: from birth to final height. Pediatr Res. 1995;38(5):733-739. [DOI] [PubMed] [Google Scholar]
- 34. Stalman SE, Solanky N, Ishida M, et al. Genetic analyses in small-for-gestational-age newborns. J Clin Endocrinol Metab. 2018;103(3):917-925. [DOI] [PubMed] [Google Scholar]
- 35. Fuke T, Nakamura A, Inoue T, et al. Role of imprinting disorders in short children born SGA and Silver-Russell syndrome spectrum. J Clin Endocrinol Metab. 2021;106(3):802-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allen HL, Flanagan SE, Shaw-Smith C, et al. ; International Pancreatic Agenesis Consortium . GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet. 2011;44(1):20-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Franco E, Shaw-Smith C, Flanagan SE, Shepherd MH, Hattersley AT, Ellard S; International NDM Consortium . GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013;62(3):993-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bui PH, Dorrani N, Wong D, Perens G, Dipple KM, Quintero-Rivera F. First report of a de novo 18q11.2 microdeletion including GATA6 associated with complex congenital heart disease and renal abnormalities. Am J Med Genet A. 2013;161A(7):1773-1778. [DOI] [PubMed] [Google Scholar]
- 39. Yau D, De Franco E, Flanagan SE, Ellard S, Blumenkrantz M, Mitchell JJ. Case report: maternal mosaicism resulting in inheritance of a novel GATA6 mutation causing pancreatic agenesis and neonatal diabetes mellitus. Diagn Pathol. 2017;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bonnefond A, Sand O, Guerin B, et al. GATA6 inactivating mutations are associated with heart defects and, inconsistently, with pancreatic agenesis and diabetes. Diabetologia. 2012;55(10):2845-2847. [DOI] [PubMed] [Google Scholar]
- 41. McDonald-McGinn DM, Sullivan KE, Marino B, et al. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klonisch T, Fowler PA, Hombach-Klonisch S. Molecular and genetic regulation of testis descent and external genitalia development. Dev Biol. 2004;270(1):1-18. [DOI] [PubMed] [Google Scholar]
- 43. Hadziselimovic F, Hadziselimovic NO, Demougin P, Oakeley EJ. Decreased expression of genes associated with memory and x-linked mental retardation in boys with non-syndromic cryptorchidism and high infertility risk. Mol Syndromol. 2014;5(2):76-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hadziselimovic F. Involvement of fibroblast growth factors and their receptors in epididymo-testicular descent and maldescent. Mol Syndromol. 2016;6(6):261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barthold JS, Wang Y, Kolon TF, et al. Pathway analysis supports association of nonsyndromic cryptorchidism with genetic loci linked to cytoskeleton-dependent functions. Hum Reprod. 2015;30(10):2439-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Urh K, Kolenc Ž, Hrovat M, Svet L, Dovč P, Kunej T. Molecular mechanisms of syndromic cryptorchidism: data synthesis of 50 studies and visualization of gene-disease network. Front Endocrinol (Lausanne). 2018;9:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gurovich Y, Hanani Y, Bar O, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25(1):60-64. [DOI] [PubMed] [Google Scholar]
- 48. Li N, Wang Y, Yang Y, et al. Description of the molecular and phenotypic spectrum of Wiedemann-Steiner syndrome in Chinese patients. Orphanet J Rare Dis. 2018;13(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baer S, Afenjar A, Smol T, et al. Wiedemann-Steiner syndrome as a major cause of syndromic intellectual disability: a study of 33 French cases. Clin Genet. 2018;94(1):141-152. [DOI] [PubMed] [Google Scholar]
- 50. Züchner S. MFN2 hereditary motor and sensory neuropathy. 2005 Feb 18 [updated 2020 May 14]. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews® [Internet]. Seattle, WA: University of Washington, Seattle; 1993–2020. [PubMed] [Google Scholar]
- 51. Lawal TA, Todd JJ, Witherspoon JW, et al. Ryanodine receptor 1-related disorders: an historical perspective and proposal for a unified nomenclature. Skelet Muscle. 2020;10(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nabil A, El Shafei S, El Shakankiri NM, et al. A familial PLCB4 mutation causing auriculocondylar syndrome 2 with variable severity. Eur J Med Genet. 2020;63(6):103917. [DOI] [PubMed] [Google Scholar]
- 53. Romanelli Tavares VL, Zechi-Ceide RM, Bertola DR, et al. Targeted molecular investigation in patients within the clinical spectrum of Auriculocondylar syndrome. Am J Med Genet A. 2017;173(4):938-945. [DOI] [PubMed] [Google Scholar]
- 54. Han JY, Lee IG. Genetic tests by next-generation sequencing in children with developmental delay and/or intellectual disability. Clin Exp Pediatr. 2020;63(6):195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kärkinen J, Miettinen PJ, Raivio T, Hero M. Etiology of severe short stature below -3 SDS in a screened Finnish population. Eur J Endocrinol. 2020;183(5):481-488. [DOI] [PubMed] [Google Scholar]
- 56. Yao R, Zhang C, Yu T, et al. Evaluation of three read-depth based CNV detection tools using whole-exome sequencing data. Mol Cytogenet. 2017;10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the corresponding author on reasonable request.