Intracellular bacterial pathogens enter their hosts surrounded by a membrane-bound vacuole and use a panel of tricks to exploit or evade eukaryotic cell functions (9, 12). Chlamydia inhabits vesicles that do not fuse with lysosomes and remains within these parasitophorous vacuoles (termed inclusions) for the duration of its replication cycle. Although the biogenesis of these vacuoles is still poorly understood, it is becoming clear that the parasites which multiply within vacuoles modify those vesicles that arrest their maturation at discrete stages of the endocytic pathway, indicating more of a continuum along the endocyclic and lysosomal pathway than has been suspected in the past (18, 21, 30).
Chlamydia pneumoniae is the causative agent of about 10% of pneumonia cases in children. Chlamydia trachomatis is still a major cause of blindness in developing countries and is one of the most commonly encountered pathogens in sexually transmitted diseases. Chlamydia is a gram-negative bacterium and occurs in two forms. Well-shaped elementary bodies (EBs) are adapted to extracellular survival. The entry of EBs into host cells launches metabolic activity, i.e., the transformation of EBs into pleomorphic reticulate bodies (RBs), RB division by binary fission, back-differentiation of RBs to EBs, and EB release through the lysis of the host cells or by a process in which the inclusion membrane fuses with the plasma membrane. On infected monolayers, the cycle takes about 40 h. Electron micrographs highlight the multilayer structure of the cell envelope of EBs (44) and RBs (18, 36).
The bacterial cell wall peptidoglycan is a covalently closed, net-like polymer in which glycan strands made of alternating N-acetylglucosamine and N-acetylmuramic acid residues are cross-linked by peptides. In contrast to the vast majority of eubacteria, Chlamydia lacks detectable amounts of this essential polymer (20, 31). Yet, Chlamydia is susceptible to d-cycloserine, bacitracin, and penicillin, which are wall peptidoglycan inhibitors, and it produces three penicillin-binding proteins (PBPs) which are the molecular targets of penicillin action (4). This paradox is known as the chlamydial anomaly. In light of the results of the C. trachomatis genome sequencing project (40), Chopra et al. (8) have proposed that C. trachomatis has the information for the entire pathway of peptidoglycan synthesis. At variance with this view, we propose that in Chlamydia, a glycanless wall polymer whose synthesis is penicillin sensitive might substitute for a wall peptidoglycan.
PREDICTIVE STUDIES
This proposal relies, at least in part, on predictive studies involving 12 bacterial species whose genomes have also been sequenced. Escherichia coli, Haemophilus influenzae, Bacillus subtilis, Rickettsia prowazekii, Treponema pallidum, Mycobacterium tuberculosis, Helicobacter pylori, Borrelia burgdorferi, Aquifex aeolicus, and Synechocystis PCC6803 each possess a wall peptidoglycan. In contrast, Mycoplasma genitalium and Mycoplasma pneumoniae are wallless and peptidoglycanless bacteria. A search of similarity in amino acid sequence was carried out with the National Center for Biotechnology Information’s new gapped BLAST algorithm with the tblastn program (2). This program compares a protein query sequence against a nucleotide sequence database dynamically translated in all six reading frames (both strands). The probability that structural relatedness occurs by chance is expressed by an index P value. Values equal to or smaller than 10−3 are indicative of a statistically significant similarity.
LIPID II IN E. COLI
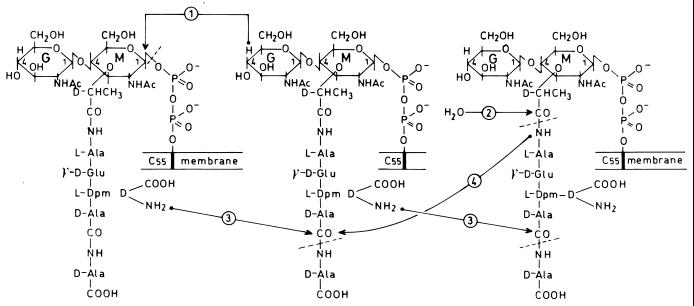
Lipid II (Fig. 1) is the immediate precursor of the wall peptidoglycan. Its synthesis involves an interchange of carriers that are compatible with the environments of the cell. In E. coli (42), UDP-N-acetylglucosamine is converted into UDP-N-acetylglucosamine-enolpyruvate by MurA and from this into UDP-N-acetylmuramic acid by MurB. The Ddl (ATP:ADP + Pi) ligase catalyzes the formation of a d-alanyl–d-alanine dipeptide, and the MurC, MurD, MurE, and MurF (ATP:ADP + Pi) ligases catalyze the formation of the UDP-N-acetylmuramoyl pentapeptide by sequential additions to UDP-N-acetylmuramic acid of l-alanine, d-glutamic acid, meso-diaminopimelic acid, and the preformed d-alanyl-d-alanine dipeptide. Then, MraY transfers the phospho-N-acetylmuramoyl pentapeptide from its uridylic carrier to the phosphate group of a membrane-anchored C55-isoprenoid alcohol phosphate, and in turn, MurG transfers the N-acetylglucosamine from its uridylic carrier to the lipid-linked N-acetylmuramic acid residue. Somehow, lipid II flips over the membrane bilayer, and the disaccharide pentapeptide moiety is exposed on the outer face of the membrane. In E. coli, ddl, murC, murD, murE, murF, murG, and mraY reside in a cluster (dcw) at the 2-min region of the chromosome.
FIG. 1.
Lipid II as the immediate biosynthetic precursor of wall peptidoglycan in E. coli and putative glycanless wall polypeptide in C. trachomatis. G, N-acetylglucosamine; M, N-acetylmuramic acid; Dpm, diaminopimelic acid; thick bar, transmembrane C55 lipid. Reaction 1, transglycosylase-catalyzed synthesis of a glycosidic bond. Reaction 2, N-acetylmuramoyl-l-alanine-catalyzed hydrolysis of a d-lactoyl–l-alanine bond. Reactions 3 and 4, acylserine transferase-catalyzed transpeptidations at the expense of the d-alanyl–d-alanine bond of pentapeptide units. The amino acceptors of the transfer reactions are the amino group at the D center of meso-diaminopimelic acid in reaction 3 and the amidase-released l-alanine amino group in reaction 4. The product of reactions 1 and 3 is the wall peptidoglycan. The product of reactions 2, 3, and 4 is the putative wall polypeptide.
LIPID II IN C. TRACHOMATIS
C. trachomatis possesses genes that are homologous to the E. coli ddl, murA, murB, murC, murD, murE, murF, and mraY genes. In particular, Table 1 gives the extent of homology between the MurA enolpyruvate transferases and MurB reductases which catalyze the first committed steps of lipid II synthesis and between the MraY transferases and MurG transglycosylases which catalyze the terminal steps of the pathway. The C. trachomatis ddl, murC, murD, murF, murG, and mraY genes cluster in a particular region of the genome (open reading frames [ORFs] D756 to D762), but murE, which encodes the meso-diaminopimelic-acid-adding enzyme, is outside the cluster (ORF D269). C. trachomatis has no l-Ala→d-Ala racemase-encoding gene, but it possesses dagA homologues for d-alanine-glycine permeases, suggesting that the synthesis of lipid II depends on an exogenous source of d-alanine (8). Peptidyl d-amino acids are present in rat liver tissues (32), and d-amino acids, including d-alanine, occur in tissues and body fluids of humans and other vertebrates (3, 22). Consistently, C. trachomatis is not susceptible to alaphosphine which is directed against the l-Ala→d-Ala racemase, and it is susceptible to d-cycloserine, which inhibits the Ddl ligase.
TABLE 1.
Homologous E. coli and C. trachomatis enzymes of the lipid II synthesis pathway
| Enzyme | Size (amino acids) of:
|
Pa | ||
|---|---|---|---|---|
| E. coli enzyme | C. trachomatis enzyme | Homologous segment | ||
| MurA | 419 | 444 | 400 | 10−62 |
| MurB | 342 | 290 | 200 | 10−6 |
| MraY | 360 | 335 | 230 | 10−60 |
| MurG | 355 | 352 | 320 | 10−32 |
P values smaller than 10−3 are indicative of significant amino acid sequence similarities.
PEPTIDOGLYCAN ASSEMBLY IN E. COLI
The assembly of the lipid II-transported disaccharide pentapeptide units into peptidoglycan and the remodelling of the polymer throughout the bacterial cell cycle are carried out by specialized transferases (14, 17). Glycosyltransferases catalyze glycan chain elongation (transglycosylation) by displacing the pyrophosphate linked to C-1 of N-acetylmuramic acid of a disaccharide unit by the 4-hydroxyl group of N-acetylglucosamine of another disaccharide unit (Fig. 1, reaction 1). Acylserine transferases working as transpeptidases catalyze peptide cross-linking between glycan strands (Fig. 1, reaction 3). The rupture of the d-alanyl–d-alanine bond at the carboxy end of a pentapeptide unit and the attack of the penultimate d-alanyl by the amino group at the D center of a meso-diaminopimelic acid of another peptide proceeds via the formation of a peptidyl enzyme in which the d-alanyl moiety is linked as an ester to a serine residue at the enzyme’s active site. Other acylserine transferases catalyze the hydrolytic breakdown of serine ester-linked peptidyl enzymes. The hydrolysis of the carboxy-terminal d-alanyl–d-alanine bonds by dd-carboxypeptidases limits the number of pentapeptides available for transpeptidation, and the hydrolysis of the interpeptide d-alanyl–(d)-meso-diaminopimelic bonds by dd-endopeptidases allows the wall peptidoglycan to undergo remodelling.
Penicillin is a mechanism-based inactivator of the dd(trans-, carboxy-, endo-) peptidases. The interaction produces stable serine ester-linked penicilloyl enzymes, and the inactivated enzymes can be detected as PBPs. The low-Mr PBPs are monofunctional dd-(carboxy-, endo-)peptidases. They do not seem to be essential. The high-Mr PBPs are, globally, the primary targets of penicillin action.
HIGH-MR PBPS
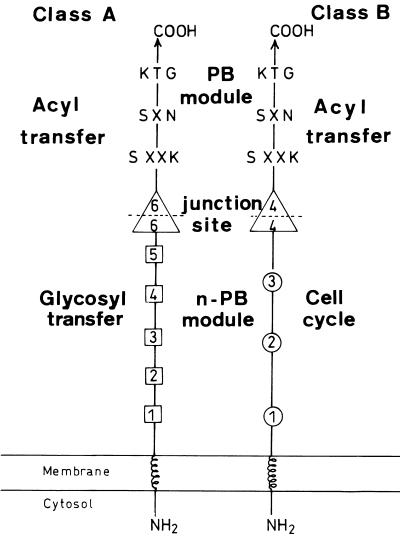
The high-Mr PBPs are multimodular (17). A transmembrane spanner is linked to the amino end of a non-penicillin-binding (n-PB) module which is linked to the amino end of an acylserine transferase PB module (Fig. 2). A conserved junction site links the n-PB and PB modules. The PB modules carry the three active-site-defining motifs SerXXLys (where Ser is the active serine residue and X is a variable amino acid residue), SerXAsn or an analogue, and LysThrGly or an analogue, which are characteristic of the penicilloyl serine transferases superfamily. Occasionally, adducts occur at various places along the polypeptide chains.
FIG. 2.
Modular design of the multimodular PBPs of classes A and B. Indicated are the positions of the five motifs of the transglycosylase n-PB module of class A PBPs (□) and the three motifs of the cell cycle n-PB module of class B PBPs (○). The intermodule junction sites (▵) are common to the PBPs of classes A and B.
In spite of these common structural features, the high-Mr PBPs fall into two classes, A and B, which are recognizable by the distinctive motifs borne by the n-PB modules of class A versus class B (17). The n-PB modules of the class A PBPs have an extended signature in the form of five motifs (Fig. 2). E. coli PBP 1a and PBP 1b of class A have been identified biochemically as transglycosylase (n-PB module)–transpeptidase (PB module) enzymes. They catalyze the conversion of lipid II into peptidoglycan in in vitro assays, and the conserved dicarboxylic amino acid residues Glu and Asp of motif 1 and Glu of motif 3 are important components of the transglycosylase catalytic center of the n-PB module (40a).
The n-PB modules of the class B PBPs have a less extended signature in the form of three motifs (Fig. 2). Motifs 1 to 3 of the class B PBP 2x of Streptococcus pneumoniae, whose structure has been determined (35), occupy positions that are likely to be sites of interaction between the n-PB and PB modules (17). Consistently, motifs 1 and 3 are important elements of the amino acid sequence folding information of the class B PBP 3 of E. coli (16). PBP 2x and PBP 3 each catalyze peptide bond formation (PB module) on properly structured thiolesters (1). However, PBP 3 does not perform transglycosylation on lipid II in in vitro assays. Consistently, the inactivation of the PBP 3-encoding ftsI (E. coli ftsI 63 mutant) does not induce a significant change in glycan chain lengths in the peptidoglycan of E. coli (23), indicating that the n-PB module of the class B PBPs fulfills functions other than glycan chain elongation in peptidoglycan synthesis. It has been proposed (17) that the class B PBPs perform peptide cross-linking (PB module) and that this activity is regulated by the associated n-PB module itself in interaction with components of morphogenetic networks involved in cell shape maintenance and cell septation.
PBPS IN C. TRACHOMATIS
C. trachomatis produces three PBPs of varying molecular masses (4). ORF D551 codes for a low-Mr PBP that is 343 amino acid residues long. ORFs D682 and D270 code for two high-Mr PBPs that are 1,080 and 647 amino acid residues long, respectively. Based on a long-lasting belief first formulated by Ishino et al. in the early 1980’s that the high-Mr PBPs are bifunctional transglycosylase-transpeptidase enzymes (24, 25), Chopra et al. (8) concluded that C. trachomatis has the required PBPs to manufacture a typical peptidoglycan from lipid II. The assignment of distinct functions to the class A and class B PBPs leads to a different conclusion. The two C. trachomatis high-Mr PBPs are both of class B, and the C. trachomatis genome has no ORFs that would code for proteins having the characteristic amino acid sequence signature of the transglycosylase (n-PB) module of the class A PBPs or the monofunctional transglycosylases known to be present in several bacterial species (39). Therefore, C. trachomatis does not synthesize a wall peptidoglycan because it lacks the required glycosyltransferases for glycan chain elongation from lipid II.
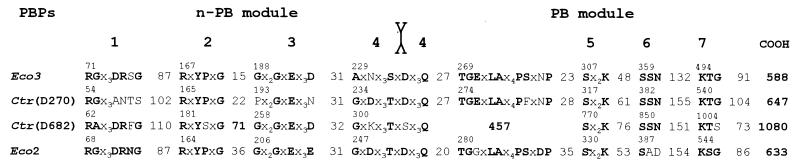
The identification of the class B-specific motifs borne by the two C. trachomatis high-Mr PBPs allows the n-PB and PB modules to be identified (Fig. 3). The motifs borne by the 647-amino-acid PBP occur with the expected spacing along the polypeptide chain. The motifs borne by the 1,080-amino-acid PBP occur in the correct order. However, this PBP has peculiar features. The n-PB module contains an extended polypeptide between motifs 2 and 3. A 457-amino-acid polypeptide is inserted downstream from the intermodule junction site. This insert is large enough to have its own fold, and it lacks amino acid sequence similarity with known proteins. Motif 7 of the PB module, LysThrSer, is somewhat unusual in that a serine residue substitutes for a glycine residue.
FIG. 3.
Occurrence of motifs characteristic of class B PBPs along amino acid sequences of the 647-amino-acid (ORF D270-encoded) PBP and the 1,080-amino-acid (ORFD682-encoded) PBP of C. trachomatis (Ctr). E. coli (Eco) PBP 3 and PBP 2 are the prototypes of PBPs of subclasses B3 and B2, respectively (17). The intermotif distances are in numbers of amino acid residues.
The hierarchical analysis of 34 high-Mr class B PBPs and their constitutive modules has led to several observations (17). The n-PB and PB modules of PBPs of gram-positive bacteria fall into subclasses B1, B4 (prototype: S. pneumoniae PBP 2x), and B5. The n-PB and PB modules of gram-negative bacteria fall into subclasses B2 (prototype: E. coli PBP 2) and B3 (prototype: E. coli PBP 3). An n-PB module of a given subclass is linked, almost invariably, to a PB module of the same subclass. In all likelihood, the PBPs of subclasses B1, B4, and B5 in the gram-positive bacteria are paralogs, i.e., they perform different functions; the PBPs of subclasses B2 and B3 in the gram-negative bacteria are also paralogs; and the PBPs of subclasses B4 and B5 (gram-positive bacteria) and the PBPs of subclasses B2 and B3 (gram-negative bacteria) may be orthologs, i.e., they perform similar functions. Few bacterial species do not obey these rules. The n-PB and PB modules of PBP VD of the gram-positive organism B. subtilis, which is involved in sporulation, belong to subclass B3. The PB module of PBB 2 of the gram-negative organism B. burgdorferi belongs to subclass B2, but the n-PB module to which the PB module is associated is an outlier distantly related to the same subclass.
As derived from predictive studies, the 647-amino-acid PBP of C. trachomatis belongs to subclass B3. The n-PB module, from motif 2 to the intermodule junction site, has structural relatedness with the n-PB module of B. subtilis PBP VD (P = 7 × 10−4), and the associated PB module has close similarity with the PB module of E. coli PBP 3 throughout the entire sequence (P = 5 × 10−33). In turn, the 1,080-amino-acid PBP of C. trachomatis belongs, most likely, to subclass B2. The n-PB module, from motif 1 to the intermodule junction site, has structural relatedness with the n-PB module of E. coli PBP 2 (P = 7 × 10−6), and the associated PB module, from motif 5 to motif 7, has structural relatedness with the PB modules of E. coli PBP 2 (P = 3 × 10−5) and B. burgdorferi PBP 2 (P = 1 × 10−7). One may note that in E. coli, the paralogous PBP 2 of subclass B2 and PBP 3 of subclass B3 perform different functions. They cannot substitute for each other.
A GLYCANLESS WALL POLYPEPTIDE IN CHLAMYDIA?
In the wall peptidoglycans of chemotype III found in a number of species of the family Micrococcaceae, the disaccharide and peptide units occur in the expected 1 to 1 molar ratio, but a large proportion of the N-acetylmuramic acid residues are not peptide substituted, and polypeptides consisting of peptides with the same amino acid sequences as the peptide units cross-link the glycan chains (13, 15). Admittedly, the presence of these polypeptides, which are made of peptide repeats, implies a tight coordination between N-acetylmuramoyl-l-alanine amidase and transpeptidase activities.
C. trachomatis possesses ORFs (D268, D601, and perhaps D759) which code for N-acetylmuramoyl-l-alanine amidases (8). Therefore, there is a possibility that the lipid II-transported l-Ala–γ-d-Glu–(l)-meso-diaminopimelic acid–(l)-d-Ala–d-Ala pentapeptides are released from their carrier by amidase action (Fig. 1, reaction 2) and then polymerized by the two class B PBPs into a cross-linked covalently closed wall polypeptide. d-Alanyl–d-alanine sequences could serve as carbonyl donors for two types of penicillin-sensitive transpeptidation reactions. Transpeptidation involving the l-alanine residue at the amino end of the amidase-released peptides as an acceptor would result in a head-to-tail assembly of linear polypeptide chains (Fig. 1, reaction 4). Transpeptidation involving the amino group at the D center of meso-diaminopimelic residues as the acceptor would create cross-linkages between linear polypeptide chains (Fig. 1, reaction 3).
This glycanless wall polypeptide, together with the lipoproteins to which it might be linked covalently (6), the lipopolysaccharides (37), and the highly disulfide cross-linked proteins (20) of the outer membrane, could provide Chlamydia with a cell envelope of sufficient mechanical strength. Moreover, the wall polypeptide could be remodelled throughout the chlamydial cell cycle by the low-Mr-PBP-catalyzed hydrolysis of the carboxy-terminal d-alanyl–d-alanine bonds of the pentapeptide units and the d-alanyl–(d)-meso-diaminopimelic acid cross-linkages at various places in the polymer.
LIPID II RECYCLING
In E. coli, the delivery of the disaccharide pentapeptide from lipid II generates an undecaprenyl pyrophosphate which is dephosphorylated, and then the C55 isoprenoid alcohol phosphate turns over the membrane bilayer so that the phosphate group faces the cytosol, thus allowing a new cycle to start. In Chlamydia, the C55 isoprenoid-pyrophosphate-disaccharide which results from the delivery of the pentapeptide might turn over the membrane bilayer, and a new cycle could start directly by ligase-catalyzed additions to the N-acetylmuramic acids of the lipid-borne disaccharide units of l-alanine, d-glutamic acid, meso-diaminopimelic acid, and the dipeptide d-alanyl–d-alanine. Attributing to the disaccharide moiety of lipid II the role of pentapeptide unit carrier would explain why N-acetylmuramic acid is not biochemically detected in Chlamydia or is detected in very small amounts. Alternatively, hydrolysis of the pyrophosphate bond might occur with the release of the C55 isoprenoid alcohol phosphate.
Bacitracin is a wall peptidoglycan inhibitor because it complexes the pyrophosphate group of lipid II before dephosphorylation occurs (38). Bacitracin also inhibits the synthesis of the outer membrane lipopolysaccharides whose polysaccharide chains are assembled on the same undecaprenyl pyrophosphate as that utilized in peptidoglycan synthesis and then are transferred as whole entities to the lipid A core of the molecule (37). In the absence of a typical wall peptidoglycan, the inhibition of the lipopolysaccharide synthesis might destabilize the chlamydial cell envelope. Following this view, the lipopolysaccharide of the outer membrane could be the target of bacitracin in Chlamydia.
CELL CYCLE PROTEINS
Cell cycle proteins channel PBP-catalyzed peptidoglycan assembly into wall expansion and septum formation in a cell cycle-dependent fashion. In recent years, the catalogue of these proteins has grown considerably. They are discussed in recent reviews (26, 33). Suffice it to say that in E. coli, a cell division dcw cluster at the 2-min region of the chromosome contains genes for the synthesis of lipid II and PBP 3 of subclass B3. It also contains genes encoding the cell division proteins MraZ (YABB), MraW (YABC), FtsL, FtsW, FtsQ, FtsA, and the ring-shaped FtsZ. A cell shape cluster at the 14-min region of the chromosome contains the gene encoding PBP 2 of subclass B2. It also contains genes encoding the low-Mr PBP5 and RodA.
Genes located outside these clusters are also devoted to cell division. Although essential to the process, some of them encode proteins which are not components of the morphogenetic apparatus itself. FtsK (encoded by a gene at 21 min) performs a septation function (N-terminal domain) and a chromosome partition function (C-terminal domain) (11, 43). ZipA (encoded by a gene at 52 min) is an integral membrane protein which interacts with the ring-shaped FtsZ (19). FtsH (encoded by a gene at 69 min) is a membrane-bound, ATP-dependent protease which degrades the heat shock transcription factor ς32 (41). FtsY (encoded by a gene at 78 min) is a functional homologue of a signal recognition particle protein involved in the reception and insertion of a subset of proteins at the plasma membrane (28). FtsN (encoded by a gene at 88 min) suppresses certain missense mutations in other fts genes (10).
In view of these advances, the question of which assortment of proteins in Chlamydia is involved in cell morphogenesis arises. To begin to solve the problem, one should note that the E. coli proteins are conserved, to various degrees, in those bacterial species that make wall peptidoglycan (Table 2, group A). FtsH, FtsY, the cell division MraW, FtsW, FtsZ, and the cell shape RodA are ubiquitous. MraW bears a putative S-adenosylmethionine-binding motif (7). FtsW and RodA are homologous integral membrane proteins with loops exposed on both faces of the plasma membrane (5). FtsZ is a GTPase related to eukaryotic tubulins. It localizes early at the division site, where it forms a ring-shaped structure that allows cell envelope constriction to take place (27, 29, 34).
TABLE 2.
Occurrence of E. coli protein homologues in bacterial species with known genomic data
| E. coli protein | Presence of homology with protein of:
|
Pc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group Aa
|
Group Bb
|
|||||||||||
| H. influenzae | B. subtilis | R. prowazekii | T. pallidum | M. tuberculosis | H. pylori | B. burgdorferi | A. aeolicus | Synechocystis PCC 6803 | M. pneumoniae | C. trachomatis | ||
| FtsH | + | + | + | + | + | + | + | + | + | + | + | <1 × 10−112 |
| FtsY | + | + | + | + | + | + | + | + | + | + | + | <9 × 10−44 |
| MraW | + | + | + | + | + | + | + | + | + | + | + | <1 × 10−32 |
| FtsW | + | + | + | + | + | + | + | + | + | − | + | <7 × 10−21 |
| RodA | + | + | + | + | + | + | + | + | + | − | + | <3 × 10−17 |
| FtsZ | + | + | + | + | + | + | + | + | + | + | − | <7 × 10−10 |
| FtsK | + | + | + | + | + | + | + | − | − | − | + | <4 × 10−87 |
| FtsA | + | + | + | + | − | + | + | + | − | − | − | <7 × 10−10 |
| MraZ | + | + | + | + | + | − | − | − | − | + | − | <7 × 10−8 |
| FtsN | + | + | − | − | − | − | − | − | − | − | − | <7 × 10−4 |
| FtsQ | + | − | − | − | − | − | − | − | − | − | − | 6 × 10−38 |
| FtsL | + | − | − | − | − | − | − | − | − | − | − | 1 × 10−5 |
| ZipA | + | − | − | − | − | − | − | − | − | − | − | 3 × 10−16 |
The bacterial species of group A have a wall peptidoglycan.
The bacterial species of group B are wall-less (M. pneumoniae) or wall peptidoglycan-less (C. trachomatis).
P values smaller than 10−3 are indicative of significant amino acid sequence similarities. When protein homologues occur in two or more bacterial species, the P value with the lowest significance is given.
Proteins functionally equivalent to the E. coli FtsK, FtsA, MraZ, FtsN, FtsQ, FtsL, and ZipA proteins (Table 2, group A) might also be ubiquitous, but then one has to assume that, depending on the proteins and the bacterial species, they have diverged so far from the corresponding E. coli proteins that similarity is marginal or almost nonexistent (P > 10−3). One may also note that, as result of diverging evolution, similarities between homologous proteins may be restricted to segments of the polypeptide chains only. Examples are given in Table 3.
TABLE 3.
Homologous E. coli and C. trachomatis proteinsa
| Protein |
E. coli
|
C. trachomatis
|
Homologous segment in the E. coli protein
|
P | |||
|---|---|---|---|---|---|---|---|
| Gene location (min) | Size (amino acids) | Gene location (ORF) | Size (amino acids) | NH2 | COOH | ||
| FtsZ | 2 | 383 | |||||
| MraW | 2 | 313 | D272 | 300 | 3 | 311 | 2 × 10−35 |
| FtsW | 2 | 414 | D760 | 403 | 48 | 397 | 1 × 10−28 |
| RodA | 14 | 370 | D726 | 379 | 47 | 363 | 2 × 10−21 |
| FtsK | 21 | 1,329 | D739 | 799 | 861 | 1,283 | 4 × 10−87 |
| FtsH | 69 | 644 | D841 | 913 | 140 | 593 | 1 × 10−142 |
| FtsY | 78 | 497 | D820 | 284 | 247 | 492 | 3 × 10−52 |
The homologies between the C. trachomatis and E. coli FtsK, FtsH, and FtsY proteins are limited to segments of the polypeptide chains.
The strict conservation of MraW, FtsW, RodA, and FtsZ among the peptidoglycan-containing bacterial species of group A is of particular significance when the wall-less mycoplasmas and the peptidoglycanless C. trachomatis are taken into consideration (Table 2, group B). The mycoplasmas do not synthesize lipid II. They produce MraW and FtsZ but lack FtsW and RodA, suggesting that FtsW and RodA might be connected to the flipping of lipid II through the membrane. C. trachomatis synthesizes lipid II. It produces MraW, FtsW, and RodA, and similarities with the corresponding E. coli proteins extend throughout the entire sequences (Table 3). But C. trachomatis lacks FtsZ, suggesting that in the bacterial species that manufacture a wall peptidoglycan, there is a link, direct or indirect, between FtsZ and the transglycosylase (n-PB module) of the class A PBPs. Consistently, the inactivation of FtsZ in E. coli (ftsZ 84 mutant) is associated with a significant change in the length distribution of glycan strands in newly synthesized peptidoglycan, with a shift from longer to shorter chain lengths (23).
CONCLUSIONS
Chlamydia is a peptidoglycanless bacterium because it does not have the information for the synthesis of class A PBPs (or monofunctional transglycosylases). The proposal that Chlamydia utilizes one or several N-acetylmuramoyl-l-alanine amidases and one bifunctional (cell cycle transpeptidase) PBP each of subclasses B2 and B3 to manufacture from lipid II a covalently closed, glycanless wall polypeptide made of peptide repeats whose synthesis is penicillin sensitive offers a clue to the chlamydial anomaly. This model has alternative possibilities. In particular, a combination of the identified enzymatic activities could lead to the synthesis of a wall polypeptide bearing few disaccharide units. Moreover, the question of what connections may exist between the presumed wall polypeptide, the presence of one additional domain in the PBP of subclass B2, the likely absence of a ring-shaped FtsZ-like protein, and the process of cell division in Chlamydia remains unanswered. These possibilities are presented here as a basis for future research on and an interpretation of a problem of great biological interest.
ACKNOWLEDGMENTS
This work was supported in part by the Belgian programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Services Fédéraux des Affaires Scientifiques, Techniques et Culturelles (PAI no. P4/03) and the Fonds de la Recherche Fondamentale Collective (contract no. 2.4534.95). C.G. is a Chercheur Qualifié of the Fonds National de la Recherche Scientifique.
We thank Martine Nguyen-Distèche and Jacques Coyette for their comments and suggestions.
REFERENCES
- 1.Adam M, Fraipont C, Rhazi N, Nguyen-Distèche M, Lakaye B, Frère J-M, Devreese B, Van Beeumen J, van Heijenoort Y, van Heijenoort J, Ghuysen J-M. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J Bacteriol. 1997;179:6005–6009. doi: 10.1128/jb.179.19.6005-6009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong D W, Gasper M, Lee S H, Zukowski J, Ercal N. d-Amino acid levels in human physiological fluids. Chirality. 1993;5:375–378. doi: 10.1002/chir.530050519. [DOI] [PubMed] [Google Scholar]
- 4.Barbour A G, Amano K-I, Hackstadt T, Perry L, Caldwell H D. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J Bacteriol. 1982;151:420–428. doi: 10.1128/jb.151.1.420-428.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. FtsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 6.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. New Compr Biochem. 1994;27:319–342. [Google Scholar]
- 7.Carrión M, Gómez M J, Ayala J A. Molecular analysis of the gene mraW of the dcw cluster of Escherichia coli. Workshop on structure, function and controls in microbial division. Inst Juan March Estud Investig. 1995;42:17–18. [Google Scholar]
- 8.Chopra I, Storey C, Falla T J, Pearce J H. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology. 1998;144:2673–2678. doi: 10.1099/00221287-144-10-2673. [DOI] [PubMed] [Google Scholar]
- 9.Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 10.Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draper G C, McLennan N, Begg K, Masters M, Donachie W D. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-del Portillo F, Finlay B B. The varied lifestyles of intracellular pathogens within eukaryotic vacuolar compartments. Trends Microbiol. 1995;3:373–380. doi: 10.1016/s0966-842x(00)88982-9. [DOI] [PubMed] [Google Scholar]
- 13.Ghuysen J-M. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- 14.Ghuysen J M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 15.Ghuysen J M, Bricas E, Lache M, Leyh-Bouille M. Structure of the cell walls of Micrococcus lysodeikticus. III. Isolation of a new peptide dimer Nα-[l-alanyl-γ-(α-d-glutamyl-glycine)]-l-lysyl-d-alanyl-Nα-[l-alanyl-γ-(α-d-glutamyl-glycine)]-l-lysyl-d-alanine. Biochemistry. 1968;7:1450–1460. doi: 10.1021/bi00844a030. [DOI] [PubMed] [Google Scholar]
- 16.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J-M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goffin C, Ghuysen J-M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 19.Hale C A, de Boer P A J. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatch T P. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzen R A, Scidmore M A, Rockey D D, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imai K, Fukushima T, Santa T, Homma H, Huang Y, Sakai K, Kato M. Distribution of free d-amino acids in tissues and body fluids of vertebrates. Enantiomer. 1997;2:143–145. [PubMed] [Google Scholar]
- 23.Ishidate K, Ursinus A, Höltje J V, Rothfield L. Analysis of the length distribution of murein glycan strands in ftsZ and ftsI mutants of Escherichia coli. FEMS Microbiol Lett. 1998;168:71–75. doi: 10.1111/j.1574-6968.1998.tb13257.x. [DOI] [PubMed] [Google Scholar]
- 24.Ishino F, Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981;101:905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- 25.Ishino F, Tamaki S, Spratt B G, Matsuhashi M. A mecillinam-sensitive peptidoglycan crosslinking reaction in Escherichia coli. Biochem Biophys Res Commun. 1982;109:689–696. doi: 10.1016/0006-291x(82)91995-7. [DOI] [PubMed] [Google Scholar]
- 26.Joseleau-Petit D, Vinella D, D’Ari R. Metabolic alarms and cell division in Escherichia coli. J Bacteriol. 1999;181:9–14. doi: 10.1128/jb.181.1.9-14.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Löwe J, Amos L A. Crystal structure of the bacterial cell division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 28.Luirink J, High S, Wood H, Giner A, Tollervey D, Dobberstein D. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature. 1992;359:741–743. doi: 10.1038/359741a0. [DOI] [PubMed] [Google Scholar]
- 29.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 30.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moulder J W. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect Agents Dis. 1993;2:87–89. [PubMed] [Google Scholar]
- 32.Nagata Y, Fujiwara T, Kawaguchi-Nagata K, Fukumori Y, Yamanaka T. Occurrence of peptidyl d-amino acids in soluble fractions of several eubacteria, archaea and eukaryotes. Biochim Biophys Acta. 1998;1379:76–82. doi: 10.1016/s0304-4165(97)00084-6. [DOI] [PubMed] [Google Scholar]
- 33.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nogales E, Downing K H, Amos L A, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 35.Pares S, Mouz N, Pétillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 36.Popov V L, Shatkin A A, Pankratova V N, Smirnova N S, von Bonsdorff C H, Ekman M R, Mörttinen A, Saikku P. Ultrastructure of Chlamydia pneumoniae in cell culture. FEMS Microbiol Lett. 1991;84:129–134. doi: 10.1016/0378-1097(91)90115-q. [DOI] [PubMed] [Google Scholar]
- 37.Reeves P. Biosynthesis and assembly of lipopolysaccharides. New Compr Biochem. 1994;27:281–318. [Google Scholar]
- 38.Siewert G, Strominger J L. Bacitracin: an inhibition of the dephosphorylation of lipid pyrophosphate, an intermediate in biosynthesis of the peptidoglycan of bacterial cell walls. Proc Natl Acad Sci USA. 1967;57:767–773. doi: 10.1073/pnas.57.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt B G, Zhou J, Taylor M, Merrick M J. Monofunctional biosynthetic peptidoglycan transglycosylases. Mol Microbiol. 1996;19:639–640. doi: 10.1046/j.1365-2958.1996.442924.x. [DOI] [PubMed] [Google Scholar]
- 40.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 40a.Terrak, M. T. K. Ghosh, J. van Heijenoort, J. Van Beeumen, M. Lampilas, J. Aszodi, J. A. Ayala, J. M. Ghuysen, and M. Nguyen-Distéche. The catalytic, glycosyl transferase and acyl transferase, modules of the cell wall peptidoglycan polymerizing penicillin-binding protein 1b of Escherichia coli. Mol. Microbiol., in press. [DOI] [PubMed]
- 41.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yomanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 43.Yu X-C, Weihe E K, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol. 1998;180:6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y X, Meng X M, Zhang L H, Su H, Li R D. Studies on the ultrastructure of envelope of elementary bodies of Chlamydia trachomatis. Sci Sin. 1980;23:1208–1215. [PubMed] [Google Scholar]