Abstract
Knowledge about the impact of prior severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of the elderly on mRNA vaccination response is needed to appropriately address the demand for additional vaccinations in this vulnerable population. Here, we show that octogenarians, a high-risk population, mount a sustained SARS-CoV-2 spike-specific immunoglobulin G (IgG) antibody response for 15 months following infection. This response boosts antibody levels 35-fold upon receiving a single dose of BNT162b2 mRNA vaccine 15 months after recovery from coronavirus disease 2019 (COVID-19). In contrast, antibody responses in naive individuals boost only 6-fold after a second vaccine. Spike-specific angiotensin-converting enzyme 2 (ACE2) antibody binding responses in the previously infected octogenarians following two vaccine doses exceed those found in a naive cohort after two doses. RNA sequencing (RNA-seq) demonstrates activation of interferon-induced genetic programs, which persist only in the previously infected. A preferential increase of specific immunoglobulin G heavy chain variable (IGHV) clonal transcripts that are the basis of neutralizing antibodies is observed only in the previously infected nuns.
Keywords: SARS-CoV-2, BNT162b2 vaccine, octogenarian, naive versus COVID-19-recovered individuals, PBMC, cytokine, antibody, neutralization, Omicron, immune transcriptome, IGHV clones
Graphical abstract
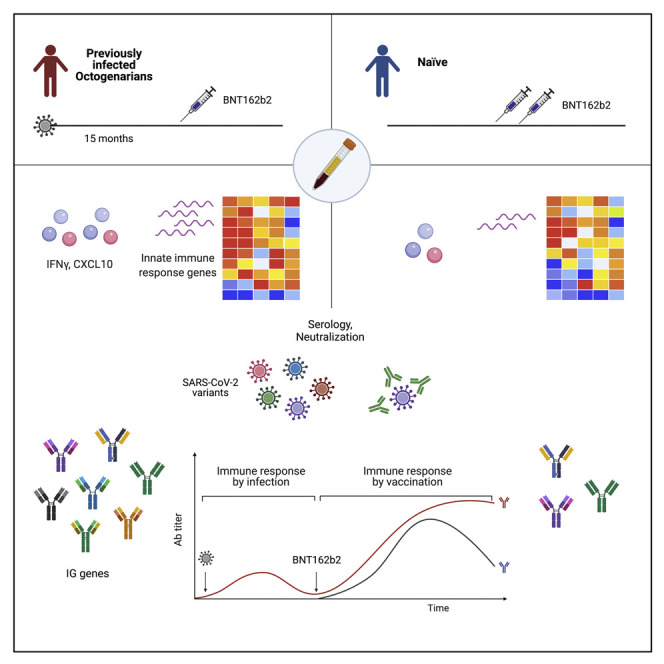
A data-driven approach for optimizing vaccination strategies in the very old population is needed. Lee et al. demonstrate that octogenarians mount a sustained antibody response following COVID-19 infection that is boosted upon receiving a single dose of BNT162b2 mRNA vaccine more than 1 year after recovery from COVID-19.
Introduction
With the persistence of the coronavirus disease 2019 (COVID-19) epidemic, the question of when and to whom to offer additional vaccinations emerged as a discussion point across geographic areas (Dolgin, 2021a, 2021b). A point of agreement is the vulnerability of the aged to COVID-19 morbidity and mortality (Covino et al., 2021). There is a body of information on vaccination response both before and after COVID-19 disease in younger, and generally healthy, populations, with median ages ranging from 32 to 47 years old (Ebinger et al., 2021; Goel et al., 2021; Krammer et al., 2021; Lozano-Ojalvo et al., 2021; Sokal et al., 2021; Wang et al., 2021). While a large-scale study has revealed high and comparable efficacy of the BNT162b2 mRNA COVID-19 vaccine in young and older adults (Polack et al., 2020), at least in the short term, there are limited data available for those in the 8th and higher decades of life (Hyams et al., 2021).
The need for a data-driven approach for optimizing vaccination strategies in the very old population is 4-fold. One, this is a group that has disproportionately experienced death due to COVID-19; two, aging of the immune system can be associated with functional declines and poor vaccine responses titers (Bartholomeus et al., 2018; Gonçalves et al., 2019; Sokal et al., 2021); three, this population has a higher prevalence of co-morbidities, many of which are specifically linked to COVID-19 morbidity and mortality, than younger populations; and four, this population is not infrequently domiciled in close living quarters, facilitating disease transmission.
Tyrol, Austria was an epicenter early in the COVID-19 pandemic, whereby an outbreak in the ski resort of Ischgl in February 2020 led to a seroprevalence of 42% (Borena et al., 2021; Lee et al., 2021b). Another outbreak took place in a nearby convent with widespread transmission among a group of nuns (median 81 years old). In June 2021, 15 months following the outbreak, the community of nuns was offered the opportunity to receive the BNT162b2 mRNA COVID-19 vaccine. Following informed consent, and with the use of deidentified samples, the immune response of this cohort was characterized both before and after receipt of a single dose of the BNT162b2 mRNA COVID-19 vaccine. The primary comparison group was a public community of men and women slightly younger from a neighboring town (median 58 years old). Within the community of nuns, the largest group were women with significant co-morbidity who experienced mild-moderate COVID-19 symptoms. Studied for secondary comparison were two smaller groups, five severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected nuns with significant underlying co-morbidity housed in a subacute care setting (median 86 years old) and five nuns who did not experience COVID-19 infection (median age 73 years old). The five SARS-CoV-2-infected nuns received two doses of BNT162b2 in January 2021, 10 months following the COVID-19 outbreak in the convent, and the five noninfected nuns received two doses of BNT162b2 in April 2021.
Here, we use serology and transcriptome analyses to explore and understand the immune response of individuals in their 8th and higher decade of life who received one versus two vaccination doses, without and with significant co-morbidity and/or previous infection, with our primary comparative control being a more commonly studied slightly younger naive population who received two doses of BNT162b2. Specifically, the sequencing depth in our study facilitated the identification of an expanded immune response in the previously infected nuns following a single vaccination.
Results
Antibody response in octogenarians receiving an mRNA vaccine 15 months after documented SARS-CoV-2 infection and a second dose after 20 months
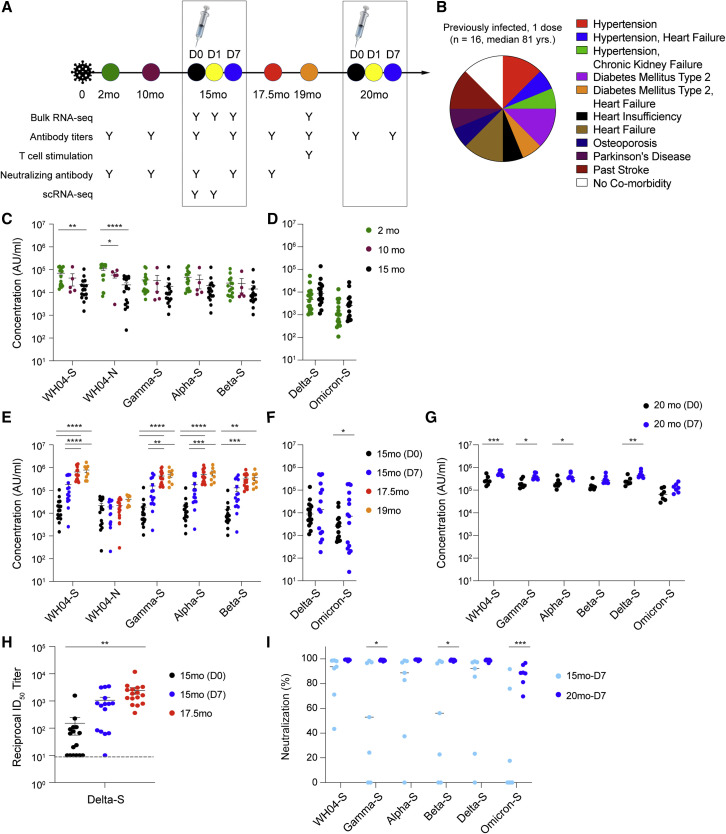
To address the deficit of information in the aged, we investigated the immune response of a group of octogenarian non-cloistered nuns (n = 16; median 81 years old; Figures 1A and 1B; Table S1) exhibiting normal age-related co-morbidities before and after a single BNT162b2 vaccination 15 months following documented COVID-19 disease. Comparative immune response data to vaccination from uninfected community-dwelling members, with and without co-morbidity (n = 14; median 58 years old; Figures 2A and 2B; Table S1), were analyzed simultaneously with ancillary data following a two-vaccination course from octogenarian nuns (n = 5; median 86 years old) previously infected with SARS-CoV-2 (Figures 3A and 3B; Table S1) with age-related co-morbidities requiring chronic subacute care and naive nuns (n = 5; median 73 years old; Figures 3C and 3D). Four out of the 16 previously infected nuns reported mild post-vaccine symptoms, fatigue and joint pain, after the single vaccine dose (Table S1). No symptoms were reported by the 14 naive individuals after their first and second vaccination (Table S1). We show here that this group of 16 octogenarians demonstrated stable antibody endpoint titers against the spike protein of four variants (WH04, Alpha, Beta, and Gamma) from 2 to 15 months following disease (Figure 1C; Table S2), with a significant boost following a single mRNA vaccination that persisted through a 4-month follow-up period (Figure 1E). Antibody titers against the Delta and Omicron spike protein were lower, and a significant induction of anti-Omicron spike was observed at day 7 post-vaccination (Figures 1D–1F). Endpoint titers exceeded those generated by a second vaccine response in a slightly younger community-dwelling group, used as a comparator (Figures 2C and 2D; Table S2). The increase of anti-spike titers for WHO4, Alpha, Beta, Gamma, and Delta, but not Omicron, in the naive cohort was significant (Figures 2C and 2D). Antibody levels for individuals with and without defined co-morbidities are presented for the previously infected cohort (Figure S2) and the naive community-dwelling group (Figure S3).
Figure 1.
Antibody response of an octogenarian population elicited by two vaccine doses 15 and 20 months after SARS-CoV-2 infection
(A) Sixteen nuns (median age 81 years old) received one dose of the BNT162b2 vaccine 15 months after recovering from COVID-19 and a second dose after another 5 months. Blood was collected over a period of 20 months as indicated by the colored circles.
(B) Co-morbidities. The co-morbidities of each individual are listed in Table S1.
(C–G) Plasma IgG antibody binding the SARS-CoV-2 RBD (spike) from different variants and the N protein within 15 months after infection (C and D), after the first dose vaccination (E and F), and after the second vaccination (G). p values are from two-way ANOVA with Tukey’s multiple comparisons test (C, E, and G) and unpaired t test with Welch’s correction (D and F). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (2 mo, n = 16; 10 mo, n = 5; 15 mo [D0], n = 16; 15 mo [D7], n = 15; 17.5 mo, n = 16; 19 mo, n = 8; 20 mo [D0], n = 7; 20 mo [D7], n = 7).
(H) Neutralizing antibody response to Delta (B.1.617.2). p values are from one-way ANOVA with Tukey’s multiple comparisons test. ∗∗p < 0.01. The individual data are listed in Table S2. Colors in dot plots were matched to the ones in timeline in (A) (15 mo [D0], n = 16; 15 mo [D7], n = 15; 17.5 mo, n = 16).
(I) Blocking of interactions between ACE2 and SARS-CoV-2 spike (RBD) interactions was tested by competition ELISA. p values are from two-way ANOVA with Sidak’s multiple comparisons test. ∗p < 0.05 and ∗∗∗p < 0.001 (15 mo [D7], n = 7; 20 mo [D7], n = 7).
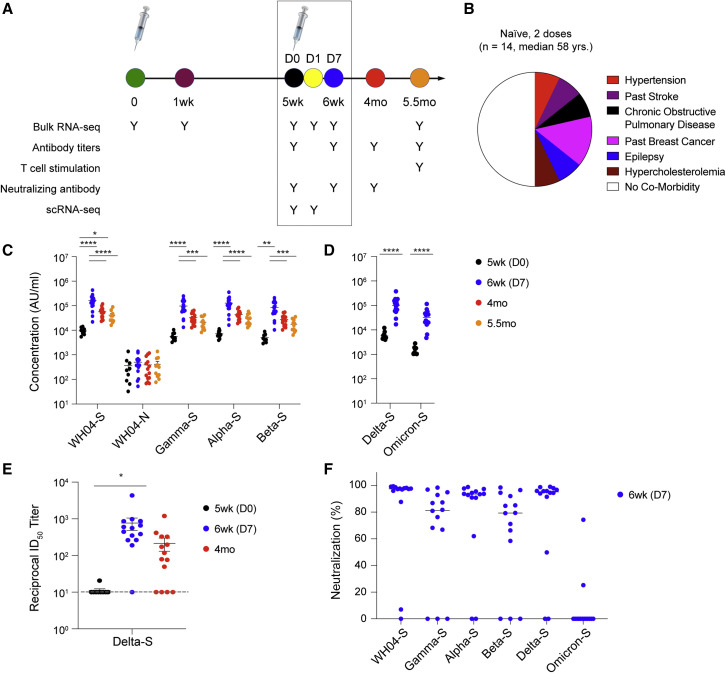
Figure 2.
Antibody response of a naive community after two vaccine doses
(A) Fourteen naive persons (median age 58 years old) received two doses of the BNT162b2 vaccine. Blood was collected over a period of 5.5 months as indicated by the colored circles.
(B) Co-morbidities. The co-morbidities of the individuals are listed in Table S1.
(C and D) Plasma IgG antibody binding the SARS-CoV-2 RBD (spike) from different variants and the N protein. p values are from two-way ANOVA with Tukey’s multiple comparisons test (C) and unpaired t test with Welch’s correction (D). ∗p < 0.05, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (5wk [D0], n = 9; 6wk [D7], n = 14; 4 mo, n = 14; 5.5 mo, n = 11).
(E) Neutralizing antibody response to the Delta variant (B.1.617.2). p values are from one-way ANOVA with Tukey’s multiple comparisons test. ∗p < 0.05. The individual data are listed in Table S2. Colors in dot plots were matched to ones in timetable in (A) (5wk [D0], n = 9; 6wk [D7], n = 14; 4 mo, n = 14).
(F) Blocking of interactions between ACE2 and SARS-CoV-2 spike (RBD) interactions was tested by competition ELISA (6wk [D7], n = 14).
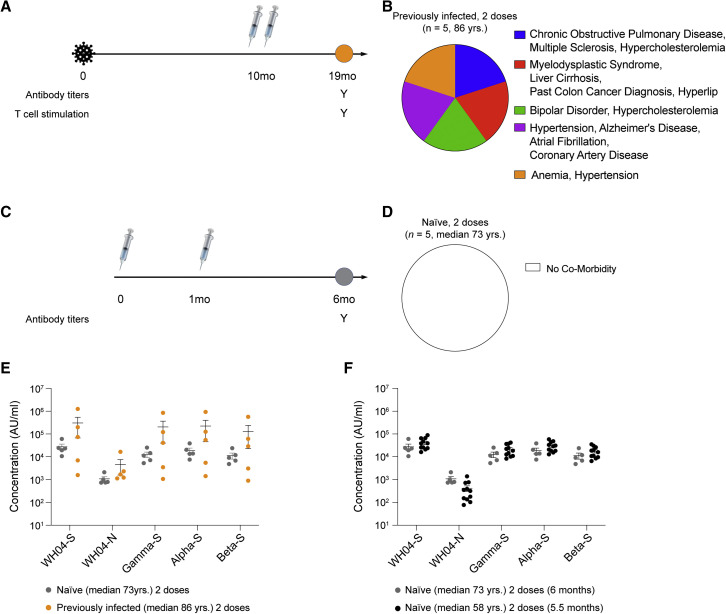
Figure 3.
Antibody response of convalescent and naive octogenarians after two vaccine doses
(A and B) Five nuns (median age 86 years old) with underlying co-morbidities received two doses of the BNT162b2 vaccine 10 months after recovering from COVID-19.
(C and D) Five naive nuns (median age 73 years old) with no underlying co-morbidities received two doses of the BNT162b2 vaccine.
(E) Plasma IgG antibody binding to the SARS-CoV-2 RBD (spike) from different variants and the N protein (naive two doses, n = 5; previously infected two doses, n = 5).
(F) Comparison of antibody titers between two naive groups. p values are from two-way ANOVA with Sidak’s multiple comparisons test. The individual data are listed in Table S2. Colors in dot plots were matched to ones in timetable in (A) and (C) (naive two doses [median 73 years], n = 5; naive two doses [median 58 years], n = 14).
A current discussion centers around whether a one- or two-dose vaccination schedule in individuals previously infected with SARS-CoV-2 might be superior in different circumstances, such as distinct age groups. The octogenarian nuns were offered a second vaccination in November 2021, and seven nuns received a dose of BNT 5 months after the first dose, 20 months after their SARS-CoV-2 infection (Figure 1A). Antibodies titers were measured just prior to the second vaccination (D0), and a significant increase was observed for all spikes, exception for Beta and Omicron, at day 7 post-vaccination (D7) (Figure 1G; Table S2).
Antibody response in naive and COVID-19-recovered octogenarians receiving a two-dose vaccination 10 months after SARS-CoV-2 infection
Next, we analyzed antibody levels in five nuns (median 86 years old) who had received two doses of BNT162b2 10 months following initial infection (Figures 3A and 3B) and in five SARS-CoV-2-naive nuns (median 73 years old; Figures 3C and 3D) who had received the standard two-dose vaccination series. Antibody titers in this previously infected group and in this naive group were measured 9 months and 5 months post-vaccination, respectively. While the previously infected group was characterized by high frequency of co-morbidities, with all individuals having more than one co-morbidity, the naive group had no co-morbidities. The anti-spike antibody levels were compared, and there were no significant differences in response comparing the two groups following completion of the vaccination series (Figure 3E; Table S2). We also directly compared the anti-spike antibody levels in the two different SARS-CoV-2 naive populations (median 58 years and 73 years) after receiving two doses and found no significant differences (Figure 3F; Table S2).
Duration of neutralizing antibody responses between previously infected and naive individuals following vaccination
At this point in the pandemic, a critical question is whether antibodies induced by either natural infection or vaccination can neutralize current variants. In vitro neutralization activity against Delta (B.1.617.2) was determined using a pseudovirus neutralization assay. Prior to receiving the BNT162b2 vaccine (15 months [D0]), the reciprocal 50% inhibitory dilution (ID50) geometric mean titer (GMT) in the previously infected cohort was 152, with 6/16 showing no detectable neutralizing antibodies against Delta (Figure 1H). Seven days (15 months [D7]) following the single vaccine dose, the reciprocal ID50 GMT had risen ∼7-fold to 1,060 and the number of non-responders had decreased to 1/15 (one sample was not collected). During the 2.5 months following the vaccination, all 16 individuals had detectable neutralizing antibody responses against Delta, and the reciprocal ID50 GMT had further risen ∼2.3-fold to 2,440 on average from 15 months (day 7 after vaccination) to 17.5 months, resulting in an ∼16-fold increase (Figure 1H). In contrast, and as expected, community-dwelling members with no prior history of infection had no detectable neutralizing antibody at the time of receiving the second vaccine dose (5 weeks [D0]; reciprocal ID50 GMT of 11; five samples not collected), with a single exception that had low levels of neutralizing antibody (Figure 2E). By day 7 after the second dose (6 weeks [D7]), the reciprocal ID50 GMT in this group was 763, an ∼68-fold increase from a week prior (5 weeks [D0]). However, in contrast to the previously infected octogenarians, the naive vaccinated group experienced an ∼3.5-fold decrease (reciprocal ID50 GMT of 223) in neutralizing antibody levels from 6 weeks to 4 months (4 mo, Figure 2E). By 4 months, 4/14 naive vaccinated individuals had no detectable neutralizing antibody against Delta. Overall, the fold change elicited by the second dose, from 5 weeks (D0) to 4 months, in the naive individuals was significantly higher, ∼20-fold, than in the previously infected cohort after the first dose, ∼16-fold. However, at 17.5 months (2.5 months after the single vaccination), the previously infected group had a larger magnitude of neutralizing antibody than the naive group after receiving their second dose.
Next, we determined the degree to which antibodies induced in the previously infected octogenarians after one or two vaccinations can prevent binding of the Omicron spike protein to the angiotensin-converting enzyme 2 (ACE2) receptor. For this, we used the ACE2 binding inhibition assay (Ebinger et al., 2021). ACE2 binding inhibition approached 100% for the ancestral variant and Alpha, Beta, Gamma, and Delta after the second vaccination (Figure 1I). A highly significant increase of ACE2 binding inhibition was also observed for the Omicron variant after the second vaccination (Figure 1I). In contrast, little ACE2 binding inhibition for Omicron was obtained in the naive group after the second vaccination (Figure 2F).
Immediate immune response to BNT162b2 mRNA vaccination
Early responses to vaccination are elevated levels of interferons and other cytokines. To gauge the early vaccine response, we measured serum levels of a panel of cytokines in the previously infected nuns prior to and after the single vaccination and prior to and after the second vaccination in the naive group (Figures S4A–S4D; Table S3). Out of the 10 cytokines measured, a significant increase of circulating interferon γ (IFN-γ) and CXCL10 was observed in both groups within 1 day following vaccination. CXCL10 (IP-10) is regulated by IFN-γ (Lee et al., 2021a) and is rapidly induced following vaccination and viral infections, and it has been identified as a biomarker reflecting COVID-19 severity (Huang et al., 2005; Laing et al., 2020; Sobolev et al., 2016). While IFN-γ levels in both groups returned to baseline levels at day 7, CXCL10 levels had returned to baseline in the naive group but remained significantly elevated in the COVID-19 recovered group, suggesting an extended inflammatory response. Levels of the other cytokines tested (interleukin-2 [IL-2], IL-4, IL-6, IL-8, IL-10, IL-15, IL-16, IL-1b, tumor necrosis factor alpha [TNF-α], and VEGF) were unchanged in the COVID-19 recovered group (Table S3). In contrast, in the naive group, IL-16 levels declined post-vaccination and IL-8 levels increased at day 7 post-vaccination (Figure S4E).
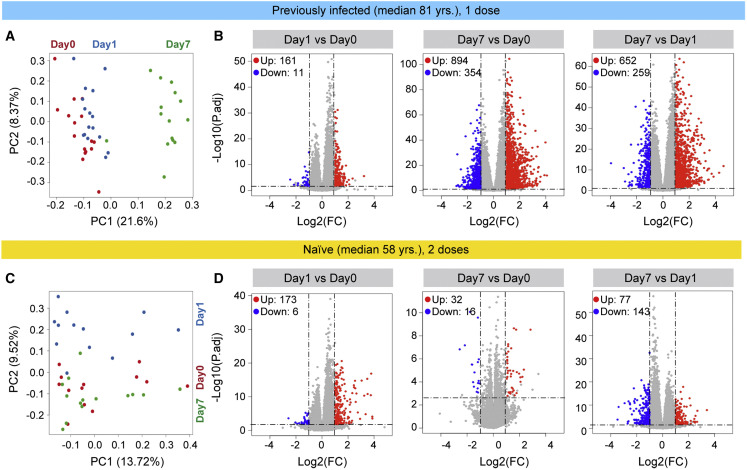
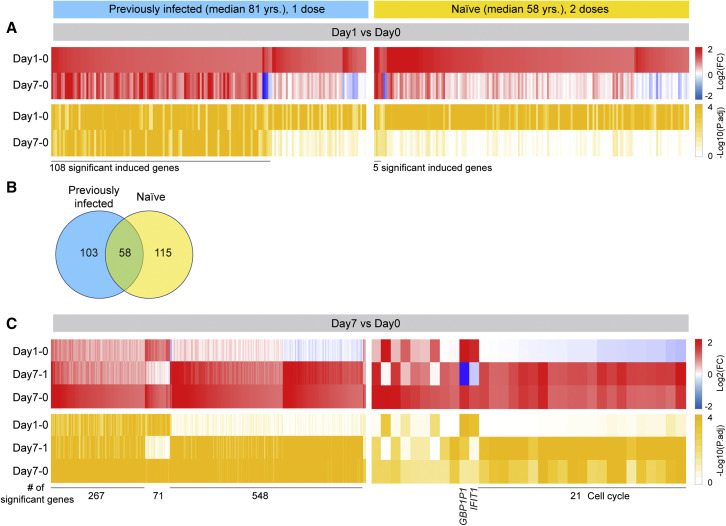
To further understand the molecular differences to the vaccine response between the two cohorts, we performed bulk RNA sequencing (RNA-seq) on buffy coats from the 16 previously infected nuns prior to and after the first vaccination and from the 14 naive subjects prior to and after the first and second dose (Figure 4 ; Tables S4, S5, S6, S7, S8, S9, and S10). RNA-seq was conducted on 115 samples with an average sequencing depth of 240 million reads per sample (Table S1). The greatest transcriptome differences in the previously infected nuns were observed at day 7 post-vaccination (Figure 4A). A total of 161 genes were induced at least 2-fold within 1 day of the vaccination (Figure 4B; Table S4), 894 genes were induced by day 7 (Table S5), and 652 genes were activated between day 1 and day 7 (Table S6). Gene set enrichment analysis (GSEA) demonstrated that the genes activated within 1 day after the vaccination were enriched in immune-response, IFN, and JAK-STAT pathways (Figures S5A–S5C). The naive group exhibited a distinctly different transcriptome response (Figures 4C, 4D, and S5D–S5F). While expression of 173 genes was induced within 1 day post-vaccination (Figure 4D; Table S7), only 32 genes were elevated at day 7 as compared with day 0 (Figure 4D; Table S8). A total of 77 genes were activated between days 1 and 7 (Table S9). As expected, the genes activated at day 1 were part of IFN and cytokine pathways (Figure S5D). We also investigated the immune transcriptomes prior to and after the primary vaccination of the naive group, and very few induced genes related to chemokine and cytokine signaling were identified (Figure S6; Table S10).
Figure 4.
Immune transcriptomes following vaccination
(A) Principal-component analysis (PCA) of transcriptomes generated prior to (day 0) and after the vaccination days 1 (day 1) and 7 (day 7) from the 16 previously infected octogenarians. The variation in the global gene expression profiles across the three time points is shown. Principal components 1 (PC1) and 2 (PC2), which represent the greatest variation in gene expression, are shown. Red dots: day 0; blue dots: day 1, and green dots: day 7 (day 0, n = 16; day 1, n = 16; day 7, n = 15).
(B) Volcano plots of differentially expressed genes (DEGs) comparing day 1 versus day 0, day 7 versus day 0, and day 7 versus day 1 in the convalescents. DEGs (adjusted p value [p adj.] < 0.05) with a log2 fold change (FC) of more than 1 or less than −1 are indicated in red and blue, respectively. Non-significant DEGs are indicated in gray. The numbers of upregulated and downregulated genes are listed in Tables S4, S5, and S6.
(C) PCA of transcriptomes from the SARS-CoV-2 antigen-naive cohort prior to (day 0) and day 1 and day 7 after the second vaccination. Red dots: day 0, blue dots: day 1, and green dots: day 7 (day 0, n = 13; day 1, n = 13; day 7, n = 14).
(D) Volcano plot of DEGs comparing day 1 versus day 0, day 7 versus day 0, and day 7 versus day 1. The numbers of upregulated and downregulated genes are listed in Tables S7, S8, and S9. Red dots indicate significant upregulation; blue dots indicate significant downregulation (p adj. < 0.05).
Differential transcriptomes between the previously infected and naive individuals
To further understand the stark differences to the first vaccination in the previously infected group and the second vaccination in the naive cohort, we dug deeper and analyzed the longitudinal expression of the genes activated at day 1 post-vaccination. Out of the 161 genes activated in the previously infected population, expression of 108 genes was still significantly activated at day 7 (Figure 5A; Table S11). Forty percent of these genes are part of IFN and virus-response pathways. In contrast, out of the 173 genes induced in the naive population at day 1, only five genes (IFI44, IFI44L, RSAD2, IFIT1, and GBP1P1) were expressed at elevated levels at day 7 (Figure 5A; Table S12). These findings suggest a prolonged vaccine-induced transcriptomic response in the previously infected individuals.
Figure 5.
Comparison of immune transcriptomes between COVID-19 recovered and SARS-CoV-2 naive cohorts after BNT162b2 vaccination
(A) Heatmaps showing log2 FC (top, red) and corresponding p adj. (bottom, ocher) of significantly upregulated genes between day 0 and day 1 in COVID-19 recovered (left) and SARS-CoV-2 naive (right) cohorts. The genes specifically activated between day 0 and day 1 and still induced by day 7 are listed in Tables S11 and S12.
(B) Venn diagram displays the number of significantly induced genes between day 0 and day 1 in both cohorts. GSEA analysis is in Figure S5.
(C) Heatmaps showing log2 FC (top) and corresponding p adj. (bottom) of genes significantly activated at day 7 in COVID-19 recovered (left) and SARS-CoV-2-naive (right) cohorts. The genes induced between day 0 and day 7 fall into different categories based on their activation pattern (Tables S11 and S12).
A direct comparison of genes induced in both groups at day 1 post-vaccination identified 58 immune-relevant genes shared between the previously infected and SARS-CoV-2 naive population (Figures 5B and S7; Tables S11 and S12). The 115 genes preferentially activated in the naive population are part of IFN signaling, and the 103 genes differentially expressed in the previously infected population are enriched for transcription factors.
Unlike in the naive population, extensive persistent transcriptome changes were observed in the previously infected, and additional gene classes were activated between days 1 and 7 post-vaccination (Figures 5C and S8; Table S11). While a total of 548 genes were induced at least 2-fold in the previously infected cohort, only 21 genes were induced in the naive population between day 1 and 7 post-vaccination.
T cell activation
For an initial exploration of T cell immunity, a standard clinical laboratory assay of T cell stimulation was performed in serum 4 months after vaccination. Data showed that the previously infected cohort with a single vaccine dose demonstrated a significantly higher proportion of definitively positive tests to COVID-19 spike protein (13 out of 16) than the naive group (1 out of 14; 22%; Figure S9; Table S13). Three out of the five previously infected nuns (median 86 years old age) demonstrated positive after the second vaccination.
Antibody germline repertoire
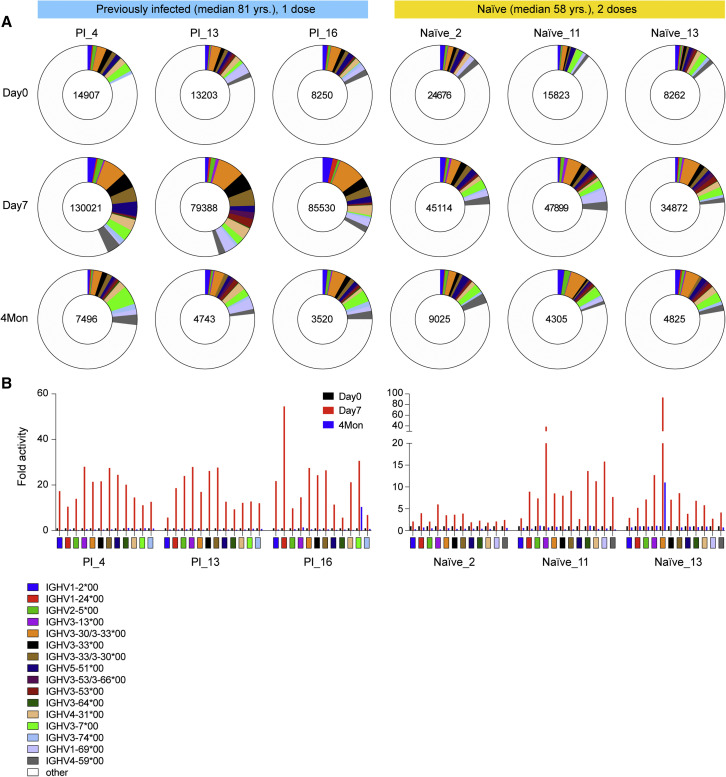
The preferential increase in anti-spike antibodies and neutralizing antibodies in the previously infected nuns upon receiving the first dose (Figure S10) begged the question about the expression profiles of specific germline variable gene classes. With a sequencing depth exceeding 240 million reads per sample, we determined the range of immunoglobulin heavy-chain variable region gene (IGHV), immunoglobulin kappa variable region gene (IGKV), and immunoglobulin lambda variable region gene (IGLV) usage in three previously infected nuns after the one vaccination and in three naive persons after the second vaccination (Figures 6 and S11; Table S14). RNA-seq was conducted prior to the vaccination (D0) and at 7 days (D7) and 4 months after the vaccination. IGHV genes used in rearrangements of high-level CDR3 revealed the use of a broad range of germlines in both cohorts, with a larger breadth in the previously infected persons. In addition, a preferential expansion of transcripts from specific germline genes occurred in the previously infected individuals (Figure 6A; Table S15). Most notably, the IGHV1-2, IGHV1-24, IGHV2-5, IGHV3-13, IGHV3-30, IGHV3-33, IGHV5-51, IGHV3-53/3–66, IGHV4-31, and IGHV3-7 clonal transcripts increased more than 15-fold within 1 week after the single vaccination of the previously infected group, exceeding that observed in the naive group (Figure 6B; Table S15). While similar patterns have been observed by others (Andreano et al., 2021b), there are also differences that could be explained by the sequencing depth. Notably, while elevated usage of IGHV2-5 in a recent study was restricted to previously infected individuals (Andreano et al., 2021b), we observed some induction also in the naive group. At 4 months post-vaccination, expression of all transcript classes that were elevated at day 7 had returned to levels seen prior to the vaccination.
Figure 6.
SARS-CoV-2-RBD-specific B cell memory
(A) Pie chart shows the distribution of antibody sequences of three convalescent and three naive individuals prior to the vaccination (day 0) and after 7 days (day 7) and 4 months (Table S15). The number of sequences analyzed for each individual is shown in the inner circle. Sizes of pie slices are proportional to the number of clonally related sequences. Persisting clones (same Integrative Genomics Viewer [IGV] genes) in both time points are shown as colored slices. White indicates sequences isolated at single time point.
(B) Induction fold activity of specific variable gene classes identified in the three convalescent and three naive individuals prior to the vaccination and after 7 days (Table S15). The color code in (A) and (B) are identical.
Germline IGHV3-53/IGHV3-66, IGHV5-51, and IGHV3-30/IGHV3-33 are the basis of neutralizing antibodies, targeting the spike protein receptor-binding domain (RBD), produced after SARS-CoV-2 infection (Andreano et al., 2021b; Andreano and Rappuoli, 2021; Zhang et al., 2021; Zost et al., 2020). Transcripts from these genes are preferentially elevated in the previously infected nuns that have a higher level of neutralizing antibodies as compared with the naive group with lower levels of neutralizing antibodies (Figures 1H, 1I, 2E, and 2F). IGHV1-24, which is preferentially induced in the previously infected nuns after a single vaccine has also been identified in extremely potent monoclonal antibodies from COVID-19 recovered patients (Andreano et al., 2021a). The diversity of B cell receptor (BCR) repertoires was also measured using the widely used Chao1 biodiversity index that is sensitive to changes in rare species (Chao, 1984). It significantly increased at D7, both in the recently infected group after the first vaccination and in the naive group after the second dose (Figure S12).
Lastly, we analyzed the CDR3 sequences for shared characteristics in both cohorts. We found nine CDR3 sequences induced in both groups between days 0, 7, and 46 in the previously infected group and at day 39 in the naive group (Table S16). The induction levels were higher in the previously infected group compared with the naive group. Although median of CDR3 amino acid length is similar in both groups, the previously infected group shows more diversity compared with the naive group, and longer ones were detected distinctively in individual’s repertoire (Table S16).
The spike (S) protein is the major surface antigen of SARS-CoV-2, and it uses its RBD to engage the host receptor ACE2 for viral entry (Zhou et al., 2020). RBD-targeting antibodies can neutralize SARS-CoV-2 by blocking ACE2 binding. In previous work, 294 SARS-CoV-2 RBD-targeting antibodies with information on IGHV gene usage have been described (Yuan et al., 2020). Here, we have identified isoforms of IGHV3-30 and IGHV3-33 as well as isoforms of IGHV3-53 and IGHV3-66 that are frequently used in these antibodies (Figure 6). The prevalence of IGHV3-53 has been recognized in COVID-19 patients (Yan et al., 2021; Yuan et al., 2020).
Discussion
In this real-world study, we provide evidence that a single dose of the mRNA vaccine BNT162b2 elicits a strong immune response in an octogenarian population after receiving a single vaccination 15 months after a documented infection with SARS-CoV-2 and recovery from COVID-19. Aging is associated with a decline of the immune system, commonly referred to as immunosenescence, and increased chronic low-grade systemic inflammation, also referred to as inflammaging (Zost et al., 2020), has been associated with a poor vaccine response (Lozano-Ojalvo et al., 2021). However, our data demonstrate that the immune response to BNT162b2 in this previously infected elderly population (median 81 years old) exceeds that of a younger naive cohort (median 59 years old) receiving a two-dose regimen.
The optimal window for providing the booster vaccine to individuals previously infected with SARS-CoV-2 has not been precisely defined and may be age dependent. Recent studies have investigated the immune response in younger populations recovered from COVID-19 (Andreano et al., 2021a; Hyams et al., 2021; Wang et al., 2021). In general, the immune responses, including spike-specific immunoglobulin G (IgG) antibody levels, in individuals younger than 50 years having received booster doses within 1–6 months after the original SARS-CoV-2 infection were similar to those seen after two doses of vaccine in individuals of similar age without prior infection (Ebinger et al., 2021; Goel et al., 2021; Hyams et al., 2021). While our previously infected population has a median age of 81 years, other studies use different definitions of elderly, ranging from 61 years (Abu Jabal et al., 2021) to >71 years (Anderson et al., 2020). A large-scale clinical study provided evidence that natural immune protection that develops after a SARS-CoV-2 infection followed by a single vaccination provides considerably more protection against the SARS-CoV-2 Delta variant than two doses of the Pfizer-BioNTech mRNA (BNT162b2) vaccine in SARS-CoV-2-naive individuals (Planas et al., 2021). It remains unclear whether one or two vaccine doses are needed for previously infected individuals. Different countries are advocating different approaches with some dictating one booster and others two. Although a single vaccine dose appears to result in a solid immune response in COVID-19 recovered individuals, studies with two vaccine doses are emerging (Ebinger et al., 2021; Goel et al., 2021). Third booster vaccinations are now available for SARS-CoV-2-naive individuals (https://www.cdc.gov/coronavirus/2019-ncov/vaccines/booster-shot.html), and its effectiveness in reducing transmission and severe disease was demonstrated in individuals 60 years and older who were fully vaccinated with the standard two-dose regimen at least 5 months prior (Bar-On et al., 2021).
The rise in antibody titers with vaccinations was preceded by a robust induction of IFN-γ pathway genes that exceeded the response induced by a two-vaccination course in the uninfected, slightly younger comparative community group. The strong, prolonged immune response in the previously infected elderly that had received a single vaccination more than 1 year after infection greatly exceeded that observed after the booster vaccination in naive individuals. This was seen both in the antibody evolution and in transcriptomes. Exploring the genomic immune responses after vaccination through RNA-seq approaches can identify transcriptional signatures associated with effective antibody production, but published data are limited to younger age individuals (Andreano and Rappuoli, 2021; Arunachalam et al., 2021; Lee et al., 2021b). Sequencing depth is a consideration for interpretation of such studies, and a sequencing depth of more than 240 million reads per sample permitted the identification of specific gene signatures in the elderly after a single vaccine dose. This also allowed the identification of specific IGHV germline classes, including IGHV1-69, IGHV1-24, IGHV1-2, and IGHV3-53, that are preferentially expressed in some of the previously infected octogenarians after a single vaccine but induced less in the naive individuals. These hepatitis C virus (HCV) genes are used by several potent neutralizing antibodies (Andreano et al., 2021a). While the expression of some IGHV germline classes, such as IGHV3-30 and IGHV3-53, is specifically elevated in our octogenarian cohort after the single dose, expression in younger COVID-19 recovered individuals with a median age of 41 years was independent of the vaccination status (Wang et al., 2021). Longitudinal antibody measurements in our elderly cohort, especially after receiving additional BNT162b2 doses, will provide a better understanding of the need, timing, and value of specific vaccination regimen in this vulnerable population.
The fast-spreading SARS-CoV-2 Omicron variant has a propensity of immune evasion, and breakthrough infections are common (Kuhlmann et al., 2022). While a third dose of BNT162b2 augments the magnitude of the antibody response to Omicron (Kotaki et al., 2022; Muik et al., 2022; Sievers et al., 2022), questions remained about the efficacy of previous SARS-CoV-2 infection followed by a two-dose mRNA vaccination in the elderly. Our study demonstrated previous infection followed by two doses of BNT162b2 about 1.5 years later resulted in a strong Omicron neutralization based on an ACE2 binding inhibition assay, far exceeding that seen in naive individuals receiving two BNT162b2 doses. A recent study demonstrated that previously infected and vaccinated persons display residual neutralization of Omicron (Cele et al., 2022), possibly accounting for the milder disease seen in many individuals.
Results from this real-world study are encouraging for vaccine efficacy in previously infected individuals in their 80s and beyond. While the optimal window between previous infection and a booster shot is not known, our study demonstrates that a 15-month gap between infection and the first vaccination did not negatively interfere with the immune response but resulted in robust production of antibodies, qualitatively and quantitatively exceeding that of naive individuals who received two doses. Similarly, a larger interval between vaccinations followed by a breakthrough infection correlated with increased neutralization activity against SARS-CoV-2 variants (Miyamoto et al., 2022; Sidik, 2022). This has practical implications for health care professionals making decisions on the need for booster vaccinations.
Limitations of the study
This elderly previously infected cohort was from a narrowly defined geographic area and included only one gender (females). The SARS-CoV-2 antigen-naive population was from a narrowly defined geographic area. The study was confined to the BNT162b2 mRNA vaccine.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human immune cells | Austria, Tyrol | N/A |
| Human serum | Austria, Tyrol | N/A |
| Critical commercial assays | ||
| Maxwell RSC simply RNA Blood Kit | Promega | Cat# ASB1380 |
| V-PLEX Custom Human Biomarkers | Meso Scale Discovery | Cat# K151A9H |
| V-PLEX SARS-CoV-2 Panel 17 (IgG) Kit | Meso Scale Discovery | Cat# K15429U |
| V-PLEX SARS-CoV-2 Panel 23 (IgG) Kit | Meso Scale Discovery | Cat# K15567U |
| V-PLEX SARS-CoV-2 Panel 23 (ACE2) Kit | Meso Scale Discovery | Cat# K15570U |
| ELISpot Path: Human IFN-γ | Mabtech | Cat# 3420-4AST-P1-1 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina, | Cat# RS-20020595 |
| Luciferase assay | Promega | E4550 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE190747 |
| Human reference genome UCSC, hg19 | UCSC Genome Browser | http://hgdownload.soe.ucsc.edu/downloads.html#mouse |
| Experimental models: Cell lines | ||
| HEK293T/17 | ATCC | CRL-11268 |
| HEK293T-ACE2 | Michael Farzan and Huihui Mu, Scripps Research | N/A |
| Recombinant DNA | ||
| Spike_SARS-CoV-2 B.1.617.2 | Vaccine Research Center, NIH (Corbett et al., 2021a) | N/A |
| VRC5601: pHR’CMV Luc | Naldini et al., 1996 | N/A |
| VRC5602: pCMV ΔR8.2 | Naldini et al., 1996 | N/A |
| VRC9260: TMPRSS2 | Vaccine Research Center, NIH (DiPiazza et al., 2021) | N/A |
| Software and algorithms | ||
| MSD DISCOVERY WORKBENCH analysis software | N/A | https://www.mesoscale.com/en/products_and_services/software |
| Trimmomatic (version 0.36) | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| STAR (2.5.4a) | Dobin et al., 2013 | https://anaconda.org/bioconda/star/files?version=2.5.4a |
| HTSeq | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| R (3.6.3) | N/A | https://www.R-project.org/ |
| Bioconductor | Huber et al., 2015 | https://www.bioconductor.org/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| RUVSeq | Risso et al., 2014 | https://bioconductor.org/packages/release/bioc/html/RUVSeq.html |
| Dplyr | N/A | https://CRAN.R-project.org/package=dplyr |
| ggplot2 | Wickham, 2009 | https://ggplot2.tidyverse.org/ |
| GSEA | N/A | https://www.gsea-msigdb.org/gsea/msigdb |
| MiXCR | Bolotin et al., 2015 | https://mixcr.readthedocs.io/en/master/ |
| Chao1 | Chao, 1984 | https://immunarch.com/articles/web_only/v6_diversity.html |
| PRISM GraphPad (9.0.0) | N/A | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Lothar Hennighausen (lotharh@nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Study population, study design and recruitment
Sixteen COVID-19 recovered volunteers who were infected with SARS-CoV-2 and developed COVID-19 in the spring of 2020 and 14 SARS-CoV-2 naive healthy volunteers and (Data S1 and Table S1) were recruited for the study under informed consent. Recruitment and blood sample collection took place between January and August 2021. This study was approved (EK Nr: 1064/2021) by the Institutional Review Board (IRB) of the Office of Research Oversight/Regulatory Affairs, Medical University of Innsbruck, Austria, which is responsible for all human research studies conducted in the State of Tyrol (Austria). The investigators do not need to have an affiliation with the University of Innsbruck. A waiver of informed consent was obtained from the Institutional Review Board (IRB) of the Office of Research Oversight/Regulatory Affairs, Medical University of Innsbruck (https://www.i-med.ac.at/ethikkommission/). Written informed consent was obtained from all subjects. This study was determined to impose minimal risk on participants. All methods were carried out in accordance with relevant guidelines and regulations. All research has been have been performed in accordance with the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/). In addition, we followed the ‘Sex and Gender Equity in Research – SAGER – guidelines’ and included sex and gender considerations where relevant.
Method details
Quantification of immunoproteins
Serum samples from all participants were collected from their blood. After thawing, serum samples were centrifuged for 3 min at 2000 g to remove particulates prior to sample preparation and analysis. The electrochemiluminescence V-PLEX assay (Meso Scale Discovery, MD) was used to measure proinflammatory proteins (IFN-γ, IL-1b, IL-2, IL-4, IL-6, IL-8, IL-10 and TNF-α), cytokines (IL-15, IL16 and VEGF) and chemokine (CXCL10). Serum samples were diluted 2-fold and measured in duplicate. The cytokines concentration was determined with the electrochemiluminescent labels whilst the plate is inserted into the MSD instrument (MESO QUICKPLEX SQ 120). All samples were assayed in duplicate. High and low controls were used to assess variance between plates. The inter-assay coefficient of variations was <10%. The results were analyzed using MSD DISCOVERY WORKBENCH analysis software.
Antibody assay
End-point binding IgG antibody titers to various SARS-CoV-2–derived antigens were measured using the Meso Scale Discovery (MSD) platform. SARS-CoV-2 spike, nucleocapsid, Alpha, Beta and Gamma spike subdomains were assayed using the V-plex multispot COVID-19 serology kits (K15429U and K15567U). Plates were coated with the specific antigen on spots in the 96 well plate and the bound antibodies in the samples (1:50000 dilution) were then detected by anti-human IgG antibodies conjugated with the MSD SULPHO-TAG which is then read on the MSD instrument which measures the light emitted from the tag.
ACE2 binding inhibition (Neutralization) ELISA
The V-PLEX COVID-19 ACE2 Neutralization kit (Meso Scale Discovery, K15570U) was used to quantitatively measure antibodies that block the binding of ACE2 to its cognate ligands (SARS-CoV-2 and variant spike subdomains). Plates were coated with the specific antigen on spots in the 96 well plate and the bound antibodies in the samples (1:10 dilution) were then detected by Human ACE2 protein conjugated with the MSD SULPHO-TAG which is then read on the MSD instrument which measures the light emitted from the tag.
Lentiviral pseudovirus neutralization
Pseudotyped lentiviral reporter viruses were produced as previously described (Corbett et al., 2020; Wu et al., 2021). Briefly, HEK293T/17 cells (ATCC CRL-11268) were transfected with the following: [1] a plasmid encoding S protein from Wuhan-Hu-1 strain (GenBank no. MN908947.3) with a p.Asp614Gly mutation (D614G) or a plasmid encoding B.1.617.2 S protein that was altered via site-directed mutagenesis (Delta), [2] a plasmid encoding luciferase reporter, [3] a plasmid encoding the lentivirus backbone, and [4] a plasmid encoding the human transmembrane protease serine 2 (TMPRSS2) gene. Serum, in duplicate, were tested for neutralizing activity against the pseudoviruses by quantification of luciferase activity in relative light units. The percentage of neutralization was normalized, with luciferase activity in uninfected cells defined as 100% neutralization and luciferase activity in cells infected with pseudovirus alone as 0% neutralization. Titers were calculated using a log (agonist) versus normalized-response (variable slope) nonlinear regression model in GraphPad and are reported as the serum dilution required to achieve 50% (50% inhibitory dilution [ID50]) neutralization. The input dilution of serum is 1:20, thus, 20 is the lower limit of quantification. Samples that do not neutralize at the limit of detection at 50% are plotted at 10, and that value was used for geometric mean calculations.
T-cell activation assay
T-cell reactivity to SARS-CoV-2 peptides was measured by an ELISPOT assay using a human IFN-γ kit (Mabtech, Nacka Strand, Sweden) according to manufacturer’s instructions. After washing of ELISPOT plate with sterile PBS, conditioning of plate with medium for 30 min and subsequent removal of medium, buffy coat cells were added to wells. Buffy coat cells were stimulated by adding each, SARS-CoV-2 S1 peptide and SARS-CoV-2 N, M, O-peptide mix, followed by incubation in a humified incubator at 37°C and 5% CO2. Anti-CD-28 was added to each well in a concentration of 1μg mL-1 to enhance stimulation. Anti CD3 mAbs served as positive control. Detection of stimulated T-cells was done by adding PBS plus 0.5 fetal calf serum (PBS/0.5% FCS) - containing detection antibodies to each well and incubation for 2 h at room temperature after removal of cells and washing with sterile PBS. Subsequent incubation and washing using PBS, streptavidin- ALP diluted in PBS/0.5% FCS was added. After another incubation of 1h at room temperature and washing with PBS, substrate solution was added to each well. After incubation at room temperature till emergence of distinct spots, color development was stopped by thorough washing with tap water. The plate was dried before counting spots by using an AID ELISPOT reader system. Normalized reads were obtained by subtraction of the negative control wells. Results were presented as spot forming colonies per million immune cells in percent.
Extraction of the buffy coat and purification of RNA
Whole blood was collected, and total RNA was extracted from the buffy coat and purified using the Maxwell RSC simply RNA Blood Kit (Promega) according to the manufacturer’s instructions. The concentration and quality of RNA were assessed by an Agilent Bioanalyzer 2100 (Agilent Technologies, CA).
mRNA sequencing (mRNA-seq) and data analysis
The Poly-A containing mRNA was purified by poly-T oligo hybridization from 1 mg of total RNA and cDNA was synthesized using SuperScript III (Invitrogen, MA). Libraries for sequencing were prepared according to the manufacturer’s instructions with TruSeq Stranded mRNA Library Prep Kit (Illumina, CA, RS-20020595) and paired-end sequencing was done with a NovaSeq 6000 instrument (Illumina) yielding 200–350 million reads per sample.
The raw data were subjected to QC analyses using the FastQC tool (version 0.11.9) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). mRNA-seq read quality control was done using Trimmomatic (Bolger et al., 2014) (version 0.36) and STAR RNA-seq (Dobin et al., 2013) (version STAR 2.5.4a) using 150 bp paired-end mode was used to align the reads (hg19). HTSeq (Anders et al., 2015) (version 0.9.1) was to retrieve the raw counts and subsequently, Bioconductor package DESeq2 (Love et al., 2014) in R (https://www.R-project.org/) was used to normalize the counts across samples and perform differential expression gene analysis. Additionally, the RUVSeq (Risso et al., 2014) package was applied to remove confounding factors. The data were pre-filtered keeping only genes with at least ten reads in total. The visualization was done using dplyr (https://CRAN.R-project.org/package=dplyr) and ggplot2 (Wickham, 2009). The genes with log2 fold change >1 or < -1 and adjusted p-value (pAdj) < 0.05 corrected for multiple testing using the Benjamini-Hochberg method were considered significant and then conducted gene enrichment analysis (GSEA, https://www.gsea-msigdb.org/gsea/msigdb).
For T- or B-cell receptor repertoire sequencing analysis, trimmed fastq files from bulk RNA-seq were aligned against human V, D and J gene sequences using the default settings with MiXCR (Bolotin et al., 2015, 2017). CDR3 sequence and the rearranged BCR/TCR genes were identified. The diversity of BCR/TCR genes was investigated by the Chao1 index (Chao, 1984).
Quantification and statistical analysis
Differential expression gene (DEG) identification used Bioconductor package DESeq2 in R. p-values were calculated using a paired, two-side Wilcoxon test and adjusted p-value (pAdj) corrected using the Benjamini–Hochberg method. Genes with log2 fold change >1 or < -1, pAdj <0.05 and without 0 value from all sample were considered significant. For significance of each GSEA category, significantly regulated gene sets were evaluated with the Kolmogorov-Smirnov statistic. p-values of cytokines were calculated using two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli on GraphPad Prism software (version 9.0.0). A value of ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 was considered statistically significant.
Ethics statement
This study was approved (EK Nr: 1064/2021) by the Institutional Review Board (IRB) of the Office of Research Oversight/Regulatory Affairs, Medical University of Innsbruck, Austria, which is responsible for all human research studies conducted in the State of Tyrol (Austria) regardless of whether, or not, the investigators have an affiliation with the University of Innsbruck.
Acknowledgments
This work was supported by the Intramural Research Programs (IRPs) of the National Institute of Diabetes and Digestive and Kidney Diseases, USA and the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, USA. Our gratitude goes to the participants who contributed to this study to advance our understanding of COVID-19 vaccination. We thank Yuhai Dai from the NIDDK clinical core for helping antibody assay, Nikolaus Wick from the Speziallabor Wick for conducting T cell activation assay, and Lingshu Wang, Wei Shi, and Eun Sung Yang from the NIAID for providing the plasmids used to make the lentiviruses. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). RNA sequencing was conducted in the NIH Intramural Sequencing Center, NISC (https://www.nisc.nih.gov/contact.htm).
Author contributions
H.K.L., L.K., and L.H. designed the study. L.K.; L.K., Sr.; S.K.; and B.P. recruited patients and collected material. H.K.L. analyzed RNA-seq data. J.I.M., A.P.W., S.B.-B., and N.J.S. conducted lentiviral pseudovirus neutralization assay. M.W. conducted antibody assays. H.K.L., L.K., P.A.F., and L.H. analyzed data. H.K.L. and L.K. administrated the project. L.H. supervised the project. H.K.L., L.K., J.I.M., P.A.F., and L.H. wrote the paper. All authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: March 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110680.
Supplemental information
Data and code availability
-
•
RNA-seq data from this study were generated in the laboratory of the last author and deposited under the accession GSE190747 in the Gene Expression Omnibus (GEO).
-
•
Cytokine data displayed in Figure S4 and antibody data shown in Figures 1C–1I, 2C–2F and 3E–3F are listed in Data S1, Tables S2 and S3. Analyzed RNA-seq data in Figures 4 and 5, and Figures S5–S8 are listed in Data S1, Tables S4–12. T-cell activation data shown in Figure S9 are listed in Data S1, Table S13. Analyzed immunoglobulin genes in Figures 6, S10 and S11 are listed in Data S1, Tables S14–S16.
-
•
This paper does not report original code.
-
•
Additional Supplemental Items are available from Mendeley Data at https://data.mendeley.com/datasets/smhwct443j/1.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abu Jabal K., Ben-Amram H., Beiruti K., Batheesh Y., Sussan C., Zarka S., Edelstein M. Impact of age, ethnicity, sex and prior infection status on immunogenicity following a single dose of the BNT162b2 mRNA COVID-19 vaccine: real-world evidence from healthcare workers, Israel, December 2020 to January 2021. Euro Surveill. 2021;26:2100096. doi: 10.2807/1560-7917.ES.2021.26.6.2100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e1816. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Paciello I., Piccini G., Manganaro N., Pileri P., Hyseni I., Leonardi M., Pantano E., Abbiento V., Benincasa L., et al. Hybrid immunity improves B cells and antibodies against SARS-CoV-2 variants. Nature. 2021;600:530–535. doi: 10.1038/s41586-021-04117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Rappuoli R. Immunodominant antibody germlines in COVID-19. J. Exp. Med. 2021;218:e20210281. doi: 10.1084/jem.20210281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Freedman L., Kalkstein N., Mizrahi B., Alroy-Preis S., Ash N., Milo R., et al. BNT162b2 vaccine booster dose protection: a nationwide study from Israel. medRxiv. 2021 doi: 10.1101/2021.08.27.21262679. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeus E., De Neuter N., Meysman P., Suls A., Keersmaekers N., Elias G., Jansens H., Hens N., Smits E., Van Tendeloo V., et al. Transcriptome profiling in blood before and after hepatitis B vaccination shows significant differences in gene expression between responders and non-responders. Vaccine. 2018;36:6282–6289. doi: 10.1016/j.vaccine.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Davydov A.N., Frenkel F.E., Fanchi L., Zolotareva O.I., Hemmers S., Putintseva E.V., Obraztsova A.S., Shugay M., et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol. 2017;35:908–911. doi: 10.1038/nbt.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Borena W., Bánki Z., Bates K., Winner H., Riepler L., Rössler A., Pipperger L., Theurl I., Falkensammer B., Ulmer H., et al. Persistence of immunity to SARS-CoV-2 over time in the ski resort Ischgl. EBioMedicine. 2021;70:103534. doi: 10.1016/j.ebiom.2021.103534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covino M., Russo A., Salini S., De Matteis G., Simeoni B., Della Polla D., Sandroni C., Landi F., Gasbarrini A., Franceschi F. Frailty assessment in the emergency department for risk stratification of COVID-19 patients aged ≥80 years. J. Am. Med. Dir. Assoc. 2021;22:1845–1852.e1. doi: 10.1016/j.jamda.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. COVID vaccine immunity is waning - how much does that matter? Nature. 2021;597:606–607. doi: 10.1038/d41586-021-02532-4. [DOI] [PubMed] [Google Scholar]
- Dolgin E. Is one vaccine dose enough if you've had COVID? What the science says. Nature. 2021;595:161–162. doi: 10.1038/d41586-021-01609-4. [DOI] [PubMed] [Google Scholar]
- Ebinger J.E., Fert-Bober J., Printsev I., Wu M., Sun N., Prostko J.C., Frias E.C., Stewart J.L., Van Eyk J.E., Braun J.G., et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat. Med. 2021;27:981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves E., Bonduelle O., Soria A., Loulergue P., Rousseau A., Cachanado M., Bonnabau H., Thiebaut R., Tchitchek N., Behillil S., et al. Innate gene signature distinguishes humoral versus cytotoxic responses to influenza vaccination. J. Clin. Invest. 2019;129:1960–1971. doi: 10.1172/JCI125372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.J., Su I.J., Theron M., Wu Y.C., Lai S.K., Liu C.C., Lei H.Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyams C., Marlow R., Maseko Z., King J., Ward L., Fox K., Heath R., Tuner A., Friedrich Z., Morrison L., et al. Effectiveness of BNT162b2 and ChAdOx1 nCoV-19 COVID-19 vaccination at preventing hospitalisations in people aged at least 80 years: a test-negative, case-control study. Lancet Infect. Dis. 2021;21:1539–1548. doi: 10.1016/S1473-3099(21)00330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaki R., Adachi Y., Moriyama S., Onodera T., Fukushi S., Nagakura T., Tonouchi K., Terahara K., Sun L., Takano T., et al. SARS-CoV-2 Omicron-neutralizing memory B-cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci. Immunol. 2022:eabn8590. doi: 10.1126/sciimmunol.abn8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Srivastava K., Alshammary H., Amoako A.A., Awawda M.H., Beach K.F., Bermudez-Gonzalez M.C., Bielak D.A., Carreno J.M., Chernet R.L., et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 2021;384:1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann C., Mayer C.K., Claassen M., Maponga T., Burgers W.A., Keeton R., Riou C., Sutherland A.D., Suliman T., Shaw M.L., et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet. 2022;399:625–626. doi: 10.1016/S0140-6736(22)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Jung O., Hennighausen L. JAK inhibitors dampen activation of interferon-stimulated transcription of ACE2 isoforms in human airway epithelial cells. Commun. Biol. 2021;4:654. doi: 10.1038/s42003-021-02167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Knabl L., Pipperger L., Volland A., Furth P.A., Kang K., Smith H.E., Knabl L., Sr., Bellmann R., et al. Immune transcriptomes of highly exposed SARS-CoV-2 asymptomatic seropositive versus seronegative individuals from the Ischgl community. Sci. Rep. 2021;11:4243. doi: 10.1038/s41598-021-83110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Ojalvo D., Camara C., Lopez-Granados E., Nozal P., Del Pino-Molina L., Bravo-Gallego L.Y., Paz-Artal E., Pion M., Correa-Rocha R., Ortiz A., et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naive and COVID-19 recovered individuals. Cell Rep. 2021;36:109570. doi: 10.1016/j.celrep.2021.109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Arashiro T., Adachi Y., Moriyama S., Kinoshita H., Kanno T., Saito S., Katano H., Iida S., Ainai A., et al. Vaccination-infection interval determines cross-neutralization potency to SARS-CoV-2 Omicron after breakthrough infection by other variants. medRxiv. 2022 doi: 10.1016/j.medj.2022.02.006. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Lui B.G., Wallisch A.K., Bacher M., Muhl J., Reinholz J., Ozhelvaci O., Beckmann N., Guimil Garcia R.C., Poran A., et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik S.M. Immunity against Omicron from breakthrough infection could be a matter of timing. Nature. 2022 doi: 10.1038/d41586-022-00004-x. [DOI] [PubMed] [Google Scholar]
- Sievers B.L., Chakraborty S., Xue Y., Gelbart T., Gonzalez J.C., Cassidy A.G., Golan Y., Prahl M., Gaw S.L., Arunachalam P.S., et al. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 2022;14:eabn7842. doi: 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev O., Binda E., O'Farrell S., Lorenc A., Pradines J., Huang Y., Duffner J., Schulz R., Cason J., Zambon M., et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat. Immunol. 2016;17:204–213. doi: 10.1038/ni.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Barba-Spaeth G., Fernandez I., Broketa M., Azzaoui I., de La Selle A., Vandenberghe A., Fourati S., Roeser A., Meola A., et al. mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants. Immunity. 2021;54:2893–2907.e2895. doi: 10.1016/j.immuni.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595:426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer; 2009. Ggplot2 : Elegant Graphics for Data Analysis. [Google Scholar]
- Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., He P., Huang X., Luo K., Zhang Y., Yi H., Wang Q., Li F., Hou R., Fan X., et al. Germline IGHV3-53-encoded RBD-targeting neutralizing antibodies are commonly present in the antibody repertoires of COVID-19 patients. Emerg. Microb. Infect. 2021;10:1097–1111. doi: 10.1080/22221751.2021.1925594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ju B., Ge J., Chan J.F., Cheng L., Wang R., Huang W., Fang M., Chen P., Zhou B., et al. Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2. Nat. Commun. 2021;12:4210. doi: 10.1038/s41467-021-24514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data from this study were generated in the laboratory of the last author and deposited under the accession GSE190747 in the Gene Expression Omnibus (GEO).
-
•
Cytokine data displayed in Figure S4 and antibody data shown in Figures 1C–1I, 2C–2F and 3E–3F are listed in Data S1, Tables S2 and S3. Analyzed RNA-seq data in Figures 4 and 5, and Figures S5–S8 are listed in Data S1, Tables S4–12. T-cell activation data shown in Figure S9 are listed in Data S1, Table S13. Analyzed immunoglobulin genes in Figures 6, S10 and S11 are listed in Data S1, Tables S14–S16.
-
•
This paper does not report original code.
-
•
Additional Supplemental Items are available from Mendeley Data at https://data.mendeley.com/datasets/smhwct443j/1.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.