Summary
Non-obstructive azoospermia (NOA) is a severe and frequent cause of male infertility, often treated by testicular sperm extraction followed by intracytoplasmic sperm injection. The aim of this study is to improve the genetic diagnosis of NOA, by identifying new genes involved in human NOA and to better assess the chances of successful sperm extraction according to the individual’s genotype. Exome sequencing was performed on 96 NOA-affected individuals negative for routine genetic tests. Bioinformatics analysis was limited to a panel of 151 genes selected as known causal or candidate genes for NOA. Only highly deleterious homozygous or hemizygous variants were retained as candidates. A likely causal defect was identified in 16 genes in a total of 22 individuals (23%). Six genes had not been described in man (DDX25, HENMT1, MCMDC2, MSH5, REC8, TDRKH) and 10 were previously reported (C14orf39, DMC1, FANCM, GCNA, HFM1, MCM8, MEIOB, PDHA2, TDRD9, TERB1). Seven individuals had defects in genes from piwi or DNA repair pathways, three in genes involved in post-meiotic maturation, and 12 in meiotic processes. Interestingly, all individuals with defects in meiotic genes had an unsuccessful sperm retrieval, indicating that genetic diagnosis prior to TESE could help identify individuals with low or null chances of successful sperm retrieval and thus avoid unsuccessful surgeries.
Keywords: genetics of male infertility, non-obstructive azoospermia, whole-exome sequencing, genetic diagnosis
Introduction
Male infertility is considered by the World Health Organization (WHO) as a global health concern affecting more than 50 million couples worldwide. Non-obstructive azoospermia (NOA), defined by the complete absence of spermatozoa in the ejaculate, even after centrifugation of the semen sample followed by microscopic examination of the pellet,1 occurs in approximately 1% of men and 10% of infertile men.2 It is a highly heterogeneous condition with a broad genetic basis. Spermatogenesis is a highly complex process comprising three main successive steps: spermocytogenesis that allows the proliferation and growth of spermatogonia, meiosis (I and II) which produces haploid cells, and spermiogenesis during which round spermatids undergo numerous biochemical and morphological changes including chromatin compaction, acrosome biogenesis, and flagellum assembly and elongation, to produce mature spermatozoa. Defects in any of these complex and specialized processes can prevent the production of sperm cells and induce NOA, which is expected to be extremely genetically heterogeneous. Consistent with these observations, mutations in more than 600 genes were shown to decrease fertility in animal models3 and 2,274 genes present an elevated expression in testis, including 474 genes that are detected only in testis.4 Currently, only two genetic analyses are performed routinely for NOA-affected individuals: a karyotype which identifies sex chromosome anomalies, in particular Klinefelter syndrome (47 XXY) and various translocations, and the search for microdeletions in the AZF region. Using these tests, a diagnosis is obtained for approximately 20% of the studied individuals5 suggesting that the overwhelming majority of affected individuals remains undiagnosed.
Regardless of the diagnosis results or lack thereof, most couples want to initiate a pregnancy if possible with their own gametes. For NOA-affected individuals, the only option to achieve intra-couple pregnancy is to perform testicular sperm extraction (TESE) followed by in vitro fertilization (IVF) performed using intracytoplasmic sperm injection (ICSI), but success rates remains low and sperm cells are found in only 30%–50% of cases.5 The whole TESE-ICSI procedure is invasive, lengthy, psychologically wounding, and expensive and should be avoided when the chances of success are too low. Renouncing TESE is a difficult decision but it is the physician’s responsibility to propose a medical procedure only if the expected gain overweighs the risks. In the absence of a precise diagnosis, the practitioner has no predictive information and in many cases, a TESE-ICSI will be unsuccessfully performed rather than moving faster toward the alternative procedures of sperm donation or adoption. In addition, infertile men, and by extension their relatives and potential offspring, are at higher risk of related adverse health issues requiring preventive follow up or specific treatments. This is well established for Klinefelter syndrome, Kallmann syndrome, or primary ciliary dyskinesia but increasing evidence also points to important links between cancer risks and infertility.6 Genetic diagnosis is therefore essential, especially when associated with a prognostic value, to provide informed counselling to individuals undergoing TESE and/or endocrine therapy.7
In the past decade, the overwhelming development of high-throughput sequencing and in particular the use of whole-exome sequencing (WES) has permitted the identification of many pathological genes in the field of male infertility.8,9 Recent cohort studies or familial studies also demonstrated the relevance of WES and highlighted the involvement of addition genes in the etiology of NOA.10, 11, 12 In view of the extreme genetic heterogeneity of NOA and the increasing affordability of the technique, we believe that WES should now be included in the panel of genetic techniques proposed to infertile men.
Here, we recruited 96 infertile men with idiopathic NOA who had undergone TESE, and we performed WES on each one. To facilitate data analysis and to focus on high-confidence gene defects, we defined a list including 151 candidate genes and we considered only highly deleterious homo or hemi-zygous variants.
Subjects and methods
Study subjects
A total of 96 unrelated men originating from North Africa affected by primary infertility and displaying non-syndromic non-obstructive azoospermia were recruited and treated at the “Clinique les Jasmins” in Tunis, Tunisia according to the established routine protocol. All individuals had a normal karyotype and no microdeletion of the Y chromosome. None declared to have any other health defect. In particular, they did not present anosmia, any disorder of sex development, or abnormal secondary sex characteristics. Details on all individuals are provided in Table S1.
Informed consent was obtained from all the individuals participating in the study according to local protocols and the principles of the Declaration of Helsinki. The study was approved by local ethics committees, and samples were then stored in the Fertithèque collection declared to the French Ministry of health (DC-2015-2580) and the French Data Protection Authority (DR-2016-392).
All individuals had two sperm collections realized at least 2 months apart, to establish the diagnosis of azoospermia. Semen was collected by masturbation after 3 days of sexual abstinence. Semen samples were incubated for 30 min at 37°C for liquefaction and then centrifuged at 3,000 × g for 15 min.
Tissue samples collection and histological examination
All samples were obtained during routine therapeutic testicular sperm extraction (TESE) from all individuals recruited in the study. In all cases, a micro-TESE procedure was performed using a 6-fold magnifying loupe as described previously.13
Tissues were fixed by immersion in 4% paraformaldehyde (PFA) for 14 h, embedded in paraffin, and sectioned (5 μm). For histological analysis, after being deparaffinized, slides were stained with hematoxylin and eosin. The colored sections were analyzed under light microscope.
Gene panel design and predictive outcome
Two independent reviewers conducted a literature search in Pubmed using the following keywords: (azoospermia gene) OR (non-obstructive azoospermia gene) OR (genetics of male infertility) OR (meiotic gene). The MGI database describing the phenotypes of KO mice was also interrogated using the keyword “azoospermia.” The search was performed on several occasions, the last search occurring on July 1, 2021.
Whole-exome sequencing and variants filtering
Genomic DNA was isolated from EDTA blood using the DNeasy Blood & Tissue Kits from QIAGEN SA. Genetic data were obtained from various sequencing centers, in particular Novogene and Integragen. Coding regions and intron/exon boundaries were sequenced after enrichment using SureSelect Human All Exon V6 –from Agilent.
An alignment-ready GRCh38 reference genome (including ALT, decoy, and HLA) was produced using “run-gen-ref hs38DH” from Heng Li’s bwakit package (https://github.com/lh3/bwa). The exomes were analyzed using a bioinformatics pipeline developed in-house. The pipeline consists of two modules, both distributed under the GNU General Public License v3.0 and available on github.
The first module (URL of this primary module in indicated in the web resources) takes FASTQ files as input and produces a single merged GVCF file per variant-caller, as follows. Adaptors are trimmed and low-quality reads filtered with fastp 0.20.0 (FASTP),14 reads are aligned with BWA-MEM 0.7.17 (BWA-MEM),15 duplicates are marked using samblaster 0.1.24 (SAMBLASTER),16 and BAM files are sorted and indexed with samtools 1.9 (SAMTOOLS). SNVs and short indels are called from each BAM file using strelka 2.9.10 (STRELKA)17 and GATK 4.1.8.1 to produce individual GVCF files from each variant-caller. These are finally merged with mergeGVCFs.pl to obtain a single multi-sample GVCF per caller. Using several variant-callers allows to compensate for each caller’s flaws.
The second module (URL of this secondary module in indicated in the web resources) takes each merged GVCF as input and produces annotated analysis-ready TSV files. This is achieved by performing up to 15 streamlined tasks, including the following. Low-quality variant calls (DP < 10, GQ < 20, or less than 15% of reads supporting the ALT allele) are discarded. Variant Effect Predictor v10418 is used to annotate the variants and predict their impact, allowing to filter low-impact (MODIFIER) variants and/or prioritize high-impact ones (e.g., stop-gain or frameshift variants). Variants with a minor allele frequency greater than 1% in gnomAD v.2.0 or 3% in 1000 Genomes Project phase 3 are filtered. Additional information can be found in Arafah et al.19 Copy number variants (CNVs) were searched using the ExomeDepth software package as previously reported.20,21
Sanger verification of candidate variants
Candidate variants were subjected to Sanger verification using an Applied Biosystems 3500XL Genetic Analyzer. Analyses were performed using SeqScape software (Applied Biosystems). Sequences of primers used and expected product sizes are summarized in Table S2.
Results
Established gene panel
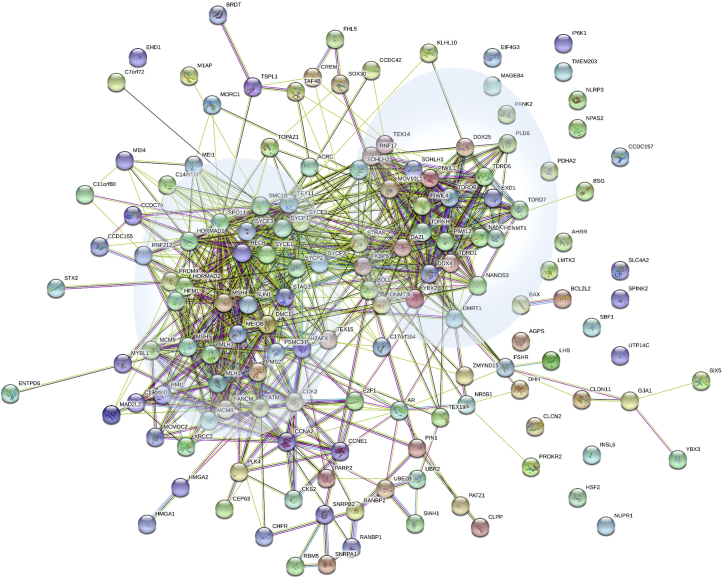
An initial search using selected keywords selected 20,595 manuscripts. Based on titles and abstracts, we excluded all manuscripts describing genes related to other spermatic phenotypes, genetic associations and risk factors, chromosome anomalies, AZF deletions, and CNVs affecting multiple genes and syndromic NOA, leading to the selection of approximately 1,000 manuscripts, which were analyzed in detail. From these eligible publications, we extracted the gene names, inheritance pattern, the testicular histological information when available, animal models if studied, functional tests if performed, the ethnicity and geographic origin of the investigated individuals, sporadic cases or familial cases, and nature of the identified variants. Data from the MGI database (http://www.informatics.jax.org/) describing the phenotypes of KO mice using the keyword “azoospermia” yielded 313 hits. The described phenotypes were analyzed and correlated with the characteristics of generated knock-out mice (conditional/spontaneous mutation etc.). Candidate genes were retained only if strong evidence was available from the studied documents. Overall a total of 151 genes were selected as likely implicated in NOA (Table S3). This list is available as an open evolving resource that can be updated by all actors of the field in a free access URL (see the link in the web resources). A STRING analysis was performed on all the selected gene products (Figure 1), highlighting numerous interactions between many of the selected proteins.
Figure 1.
STRING analysis of physical and functional protein-protein interactions between the proteins encoded by the 151 candidate genes including in the NOA panel
Analysis performed using STRING: https://string-db.org/. Two main clusters are highlighted in blue circles, loosely regrouping meiotic genes (left) and PIWI and DNA repair genes (right).
Exome sequencing
After variant filtering, each person carried on average 62 homozygous and 1,497 heterozygous variants. Overall, among these variants 3.3% were classified by Variant Effect Predictor as having a HIGH impact (alleles with such variants are not expected to produce a functional protein). In addition, 7.2% of variants were classified as having a likely deleterious effect (L-HIGH): this includes missense variants predicted as deleterious by at least three methods among SIFT, PolyPhen, CADD, mutationTaster, or REVEL, as well as splice-region variants predicted deleterious by both ada and rfmethods from dbscSNV.22 On average, each person carried 2.1 homozygous HIGH variants and 3.6 homozygous L-HIGH variants passing all previous filters. We identified a total of 25 heterozygous HIGH variants and 47 L-HIGH heterozygous variants in one of the 151 candidate genes. At this stage, as the confirmation of the segregation of the variants was not possible (to establish if the variants were de novo, mother transmitted, or localized in trans), we did not investigate the potentially dominant transmission nor did we consider compound heterozygous variants. Overall we identified 22 subjects (23%) with a homozygous or hemizygous HIGH (n = 14) or L-HIGH (n = 8) variant in one of the 151 candidate genes (Table 1). We focused on the variants that present a high probability of being responsible for the individuals’ NOA.
Table 1.
List of all candidate variants identified
| Gene | Subject | Variant | Type of variant | MAF, gnomAD |
|---|---|---|---|---|
| Loss-of-function (LoF) variants in previously reported candidate genes | ||||
| C14orf39 | P0142 | NM_174978.3: c.204_205del; p.His68GlnfsTer2 | frameshift | 4.1E−06 |
| FANCM | P0138 | NM_020937.4: c.5791C>T; p.Arg1931Ter | stop gained | 1.0E−03 |
| GCNA | P0137 | NM_052957.5: c.1507del; p.Glu504LysfsTer11 | frameshift | 0.0E+00 |
| HFM1 | P0369 | NM_001017975.6: c.3588+1G>A | splice donor | 0.0E+00 |
| MEIOB | P0074 | NM_001163560.3: c.1118_1121del; p.Phe373SerfsTer6 | frameshift | 0.0E+00 |
|
TDRD9 |
P0080 | NM_153046.3: c.3483_3484dup; p.Ser1162IlefsTer3 | frameshift | 0.0E+00 |
| P0279 | NM_153046.3: c.720_723del; p.Ser241ProfsTer4 | frameshift | 7.7E−05 | |
| Deleterious missense variants previously reported in NOA candidate genes | ||||
| MCM8 | P0370 | NM_001281520.2: c.482A>C; p.His161Pro | missense | 0.0E+00 |
| P0281 | NM_001281520.2: c.482A>C; p.His161Pro | missense | 0.0E+00 | |
|
PDHA2 |
P0253 | NM_005390.5: c.679A>G; p.Met227Val | missense | 6.0E−05 |
| P0144 | NM_005390.5: c.679A>G; p.Met227Val | missense | 6.0E−05 | |
| Loss-of-function (LoF) variants in novel strong candidate genes (no mutations described in NOA-affected men) | ||||
| DDX25 | P0283 | NM_013264.5: c.1129C>T; p.Arg377Ter | stop gained | 1.6E−05 |
| HENMT1 | P0272 | NM_001102592.2: c.456C>G; p.Tyr152Ter | stop gained | 0.0E+00 |
| MCMDC2 | P0085 | NM_173518.5: c.1795C>T; p.Arg599Ter | stop gained | 1.3E−04 |
| MSH5 | P0355 | NM_172166.4: c.537+1G>A | splice donor | 0.0E+00 |
| P0321 | ENST00000375703.7: c.1015_2508del | CNV (homo) | 0.0E+00 | |
| REC8 | P0088 | NM_001048205.2: c.860_861del; p.Pro287ArgfsTer74 | frameshift | 0.0E+00 |
| TDRKH | P0110 | NM_001083965.2: c.1003A>T; p.Lys335Ter | stop gained | 0.0E+00 |
| Novel deleterious missense variants in NOA candidate genes | ||||
| DMC1 | P0352 | NM_007068.4: c.364A>G; p.Thr122Ala | missense | 0.0E+00 |
| P0082 | NM_007068.4: c.860C>A; p.Pro287His | missense | 0.0E+00 | |
| HENMT1 | P0109 | NM_001102592.2: c.226G>A; p.Gly76Arg | missense | 8.0E−06 |
| TERB1 | P0145 | NM_001136505.2: c.733G>A; p.Gly245Arg | missense | 6.5E−06 |
All variants were homozygous and considered to be most likely deleterious and responsible for the studied phenotype.
Identified gene variants
A total of seven individuals had a homozygous loss-of-function variant (mainly frameshift variants) in six genes (C14orf39 [MIM: 617307], FANCM [MIM: 609644], GCNA [MIM: 300369], HFM1 [MIM: 615684], MEIOB [MIM: 617670], TDRD9 [MIM: 617963]) previously described to be associated with NOA23, 24, 25, 26, 27, 28, 29, 30 (Table 1). In addition, two individuals harbored the same homozygous missense variants in MCM8 (MIM: 608187): GenBank: NM_001281520.2: c.482A>C (p.His161Pro). Similarly, two other persons carried the same homozygous variants in PDHA2. Both these genes have previously been associated with NOA.31,32 Overall, for these 11 individuals we can consider that a near certain diagnosis has been obtained.
An additional seven individuals were identified with a homozygous loss-of-function variant in six genes whose function was described as critical for spermatogenesis (Table 1). For each of these genes (DDX25 [MIM: 607663], HENMT1 [MIM: 612178], MCMDC2 [MIM: 617545], MSH5 [MIM: 603382], REC8 [MIM: 608193], TDRKH [MIM: 609501]), knock out mouse models have been produced which all exhibit male sterility.33, 34, 35, 36, 37, 38, 39, 40 Some of these genes have been described to be linked with female infertility (MSH5),41 but no deleterious variants have yet been described in men with NOA. We can therefore consider that the data presented here confirm the direct link of these genes with NOA. Last, four individuals carried missense variants in genes already described to induce NOA (DMC1 [MIM: 602721], HENMT1 [MIM: 612178], and TERB1 [MIM: 617332]). DMC1 homozygous missense variants have been described in male and female infertile siblings and KO mice present with male and female sterility.42,43 Here, two individuals carried a distinct homozygous missense variant in DMC1. The two DMC1 variants identified, GenBank: NM_007068.4: c.364A>G (p.Thr122Ala) and c.860C>A (p.Pro287His), are not found in gnomAD and are respectively predicted as pathogenic by 9 and 11 out of 12 prediction software (see varsome in web resources). Similarly, the HENMT1 and TERB1 variants are very rare in gnomAD (2 occurrences) and are respectively predicted as pathogenic by 9/12 and 7/11 prediction software. The presence of all variants was confirmed by Sanger sequencing (Figure S1), and all variants were submitted to Clinvar (see web resources) under the reference SUB10823688.
Overall, we obtained a high-confidence diagnosis for 18 individuals (HIGH impact variants or L-HIGH variants in genes identified in two individuals) and a likely diagnosis for 4 additional individuals (missense variant not previously published).
Histological phenotype and TESE outcome
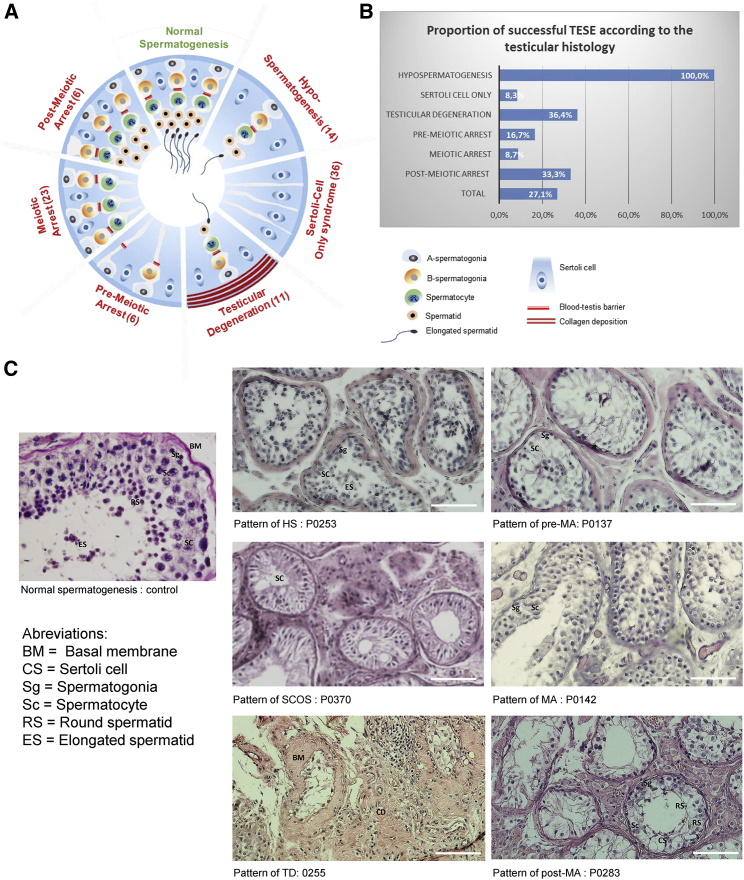
All subjects underwent surgery to perform a testicular sperm extraction (TESE). In all cases one part of the extracted tissues was fixed and colored for histological analysis and the other part was dilacerated for attempts to extract living spermatozoa. Fixed slides were carefully examined to obtain a histological diagnosis. Overall, 14 individuals were classified as having hypospermatogenesis, 36 a Sertoli cell only syndrome, 11 a testicular degeneration, 6 a pre-meiotic arrest, 23 a meiotic arrest, and 6 a post-meiotic arrest (Figure 2A). All individuals with hypospermatogenesis had a positive TESE (100%), subjects with testicular degeneration had 36.6% positive TESE, those with post-meiotic arrest 33.3%, Sertoli-cell only syndrome 8.3%, and pre-meiotic and meiotic arrest had 16.7 and 8.7%, respectively (Figures 2B and 2C). The positive TESE results might seem in conflict with the histological diagnoses of Sertoli-cell only syndrome and pre-meiotic and meiotic arrest. This can be explained by the fact that the tubules might be unevenly affected and some of the histological results might not be representative of the whole testis environment.
Figure 2.
Definition and schematic representation of the different NOA sub-phenotypes illustrated by subjects’ histological sections
(A) The testicular biopsies were categorized into different histopathological patterns, indication of the total number of individuals with each anomaly.
Normal spermatogenesis: the seminiferous tubules are lined by a thin basement membrane and the germinal epithelium shows normal progression from spermatogonia to spermatozoa along with spermatocytes and spermatids.
Hypospermatogenesis (HS): the germinal epithelium shows all the stages of germ cells but their number is reduced.
Sertoli cell only syndrome: the tubules contain only Sertoli cells and no germ cells.
Testicular degeneration: including both seminiferous tubule hyalinization and degenerating tubules. Hyalinized tubules are characterized by a thickened basement membrane, smaller diameter devoid of epithelial cells, and collagen deposition. Degenerating tubules are characterized by hypocellularity.
Germ cell maturation arrest (pre-meiotic arrest [PreMA], meiotic arrest [MA], and post-meiotic arrest [Post-M]): at a specific cell stage the process of spermatogenesis is arrested.
Mixed pattern (not shown): there is variation in the histopathological pattern in the same testicular biopsy.
(B) Proportion of individuals with successful TESE according to the testis histology.
(C) Examples of testis sections from individuals with different testicular defects.
Genotype phenotype correlation
Among the 22 diagnosed persons, 12 (55%) had gene defects altering a gene described to be critical for meiosis (C14orf39, DMC1, HFM1, MCM8, MCMDC2, MEIOB, MSH5, REC8, TERB1) (Table 2). The histological diagnosis was concordant for 10 individuals, evidencing a meiotic arrest. For all these individuals, diagnosed with genetic defect in a meiotic gene, the TESE was negative (Table 2). Only two individuals with a missense variant in the meiotic MCM8 gene were scored as having a Sertoli-cell only syndrome. Seven individuals had defects in genes described to be involved in the Piwi pathway or in DNA repair (FANCM, GCNA, HENMT1, TDRD9). Among these individuals, TESE was positive for 2 individuals (29%). Last, three individuals had defects in genes involved in post-meiotic maturation (DDX25, PDHA2) and one (33%) had a positive TESE.
Table 2.
Predicted gene function and clinical information on affected individuals
| Gene | Function | Patient | Left/right testis volume (mL) | FSH (IU/L) | Testis histology | Sperm retrieval |
|---|---|---|---|---|---|---|
| Genes involved in meiosis | ||||||
| C14orf39 | synaptonemal complex | P0142 | 10–15/10–15 | N/A | MeA | NEG |
| DMC1 | chiasma strand invasion | P0352∗ | >15/>15 | N/A | MeA | NEG |
| P0082∗ | 10–15/10–15 | 9 | MeA | NEG | ||
| HFM1 | chiasma strand invasion | P0369 | 5–10/5–10 | 5.16 | MeA | NEG |
| MCM8 | chiasma synthesis and stabilization | P0370 | 5–10/5–10 | 16.7 | SCOS | NEG |
| P0281 | <5/<5 | 27 | SCOS | NEG | ||
| MCMDC2 | meiotic recombination | P0085 | 5–10/5–10 | N/A | MeA | NEG |
| MEIOB | chiasma synthesis and stabilization | P0074 | 10–15/10–15 | 2.16 | MeA | NEG |
| MSH5 | chiasma synthesis and stabilization | P0355 | 10–15/10–15 | 6.08 | MeA | NEG |
| P0321 | >15/>15 | 9.65 | MeA | NEG | ||
| REC8 | meiotic recombination | P0088 | 5–10/5–10 | 12.9 | MeA | NEG |
| TERB1 | assembly of a meiotic telomere complex | P0145∗ | 10–15/10–15 | 4.23 | MeA | NEG |
| Genes involved in Piwi pathway and/or DNA repair | ||||||
| FANCM | DNA repair pathway | P0138 | 5–10/5–10 | N/A | TD | NEG |
| GCNA | regulator of genome stability | P0137 | >15/>15 | 2.24 | PrMeA | NEG |
| HENMT1 | component of the piwi pathway | P0272 | 10–15/10–15 | 3.29 | HS | POS |
| P0109 | – | – | MeA | NEG | ||
| TDRD9 | Piwi pathway, transposon silencing | P0080 | <5/<5 | 16 | HS | POS |
| P0279 | 10–15/10–15 | 4.6 | MeA | NEG | ||
| TDRKH | component of the piwi pathway | P0110 | 5–10/5–10 | 27.6 | MeA | NEG |
| Genes involved in post-meiotic maturation | ||||||
| DDX25 | post-transcriptional regulation | P0283 | 5–10/5–10 | N/A | PoMA | NEG |
| PDHA2 | carbohydrate oxidation | P0253 | 10–15/10–15 | 2.87 | HS | POS |
| P0144 | 10–15/10–15 | 2.4 | MeA | NEG | ||
Indicated testis histology are: Sertoli cell only (SCOS), testicular degeneration (TD), hypospermatogenesis (HS), spermatogenic arrest : pre-meiotic (PrMeA), meiotic (MeA), post-meiotic (PoMA). Sperm retrieval is either positive (POS) or negative (NEG).
Discussion
The recent development of high-throughput sequencing methods permitted the identification of an exponential number of candidate genes in all genetic disorders. The field of male infertility also benefited from these advances and the past years saw the identification and characterization of numerous infertility genes. For example, we identified SPINK2 as a cause for post-meiotic arrest44 and a recent cohort study focusing on NOA-affected individuals with spermatogenic maturation arrest (MA) identified 5 candidate genes and a likely genetic defect in 23 out of 147 individuals (16%).10 Here we performed WES analysis on 96 NOA-affected individuals showing different testicular histologies; we identified 7 candidate genes and identified a likely genetic defect in 22 out of 96 individual (23%). For men with NOA, the benefit of a clear molecular diagnosis following exome sequencing has however been limited as the genetic diagnosis only rarely has a direct impact on the subject’s proposed treatment. Clear guidelines associated with the predicted treatment outcome are necessary to turn WES into a useful tool for the routine diagnosis and care of NOA-affected individuals.
Exome sequencing is a powerful tool in the framework of infertility diagnosis
Following exome sequencing, using our list of candidate genes and strict guidelines designed to be integrated into an automated bioinformatics process, we obtained a high-confidence diagnosis for 19% of the analyzed persons and an additional very likely diagnosis for 4%. Currently, the only genetic tests carried out routinely for NOA-affected individuals are a karyotype and the search for microdeletions in the AZF region of the Y chromosome. These analyses yield a diagnosis for less than 20% of the tested individuals.5 In this study, we see that exome sequencing combined with our proposed analysis method permits clinicians to double the diagnosis efficiency for this pathology. Here, all subjects were recruited at the Clinique les Jasmins in Tunis and originated from North Africa. We can estimate that half of the included individuals were born to related parents and this certainly contributed to this high diagnosis efficiency. The yield of diagnosis of a genetically more heterogeneous population might therefore be lower but diagnosis efficiency should improve rapidly for all individuals with the publication of new studies which will permit researchers to quickly enrich the list of candidate genes. Here, we presented only the variants affecting the genes from a limited list of genes likely linked with NOA. Despite all our efforts, this list is likely incomplete and may contain mistakes, but more importantly, considering how fast the field is moving, it will certainly be incomplete by the time this article is printed. However, this list is available online and we aim to keep it updated, to serve as a shared tool for the analysis of WES data in the context of NOA. Variants in other genes not included in the list of candidate genes appeared interesting but were not discussed in this manuscript, as additional work has to be carried out to confirm or infirm their implication in NOA. To that end we generated four knock-out mice lines using CRISPR-Cas9 technology to gain additional information on their function. Also, we were very restrictive in the selection of the candidate variants. As we did not have the possibility to perform familial studies to assess the segregation of the variants, we only considered homozygous or hemizygous variants and did not select the individuals with two deleterious variants in the same candidate genes as we could not confirm that the variants were bi-allelic. Several variants did not fulfill our selection criteria but appeared interesting. This was the case for potential splice variants, not affecting consensus acceptor or donor site, but appearing as good candidates necessitating further functional validation.
Interestingly, some of the identified genes have been described to be linked with female infertility and primary ovarian insufficiency (C14orf39,29,45 HFM1,46 MCM8,32 MEIOB,47 MSH5,41 DMC142) highlighting common mechanisms between male and female meiosis. As many genes have been described to induce both male and female infertility, it might be relevant to produce a large list of candidate genes regrouping genes described to be associated with either and both male and female fertility.
Overall, the use of a well-established candidate gene list and strict criteria for variant selection permits a quick and easy analysis of WES results, allowing a rapid clinical diagnosis. As highlighted before, the list of candidate genes is expected to be in constant evolution, so WES appears much preferable to a targeted sequencing approach that would very rapidly become obsolete and would not permit the discovery of new candidate genes.
Histological phenotype and identified gene defects
Careful examination of histological results permitted to identify 6 types of testicular defects. The most frequent defects were SCOS observed in 36 individuals and meiotic arrest observed in 23 (Figure 2). There was a relatively good correlation between the histological phenotype and the outcome of TESE. All individuals with hypospermatogenesis had a positive TESE (100%); results were fair for individuals with testicular degeneration (36.6% positive TESE) and post-meiotic arrest (33.3%) and poor for Sertoli-cell only syndrome (8.3%) and pre-meiotic and meiotic arrest (16.7 and 8.7%, respectively). In some cases, this classification was difficult to make as different tubules might present slightly different anomalies and some persons might have several overlapping phenotypes (Table S2). Histological classification of testicular tissues has always been recognized as challenging. A large study performed on 1,418 samples show a matched phenotypic classification between right and left testicular biopsies in 81.2% of SCO cases whereas a matched classification was only obtained for 25%–67.7% of post-meiotic arrests.48 This illustrates that multiple sampling is necessary to obtain an overall view of the spermatogenesis. Here, for simplification, we retained the observed majoritarian defect. Interestingly, a diagnosis was obtained for 12 out of 22 individuals (55%) identified as having a meiotic arrest. Conversely, diagnosis efficiency was low for the other testis histological categories and a likely variant was identified in 6% of SCOS-affected individuals, in 9% of testicular degeneration, in 21% of hypospermatogenesis, and in 17% of individuals with pre- and post-meiotic arrest. One of the main reasons to explain the predominance of individuals with a genetic diagnosis among the subjects scored with a meiotic arrest might be that meiosis has been extensively studied and many meiotic genes have been identified and are present in our list of candidate genes.
We tried to group the identified genes by function. Nine of the identified genes were described to have a direct function in meiotic processes, five a role in piwi pathways or DNA repair (Table 2). Only two genes described to be involved in post-meiotic processes were identified. It is interesting to note that we observed genes regrouped in these broad functions following the string analysis of the 151 candidate genes (Figure 1). There was a relatively good correlation between the histological phenotype and what was expected from the presumed gene function. Nine out of the 16 identified genes are described to be directly involved in meiosis (Table 2) and 10 out of the 12 individuals concerned had a testis histology phenotype of meiotic arrest. The two remaining individuals had variants in MCM8, involved in chiasma synthesis and chromosomal pairing stabilization and were scored to have SCOS. Conversely, three individuals scored to have a meiotic arrest had variants in HENMT1, TDRD9, and TDRKH, three genes known to be involved in the PIWI pathway. These small discrepancies were also illustrated by the fact that several persons with defects in the same genes (HENMT1, TDRD9, and PDHA2) had a different histological subphenotype and a different TESE outcome. For PDHA2, two individuals had the same mutation and one had a hypospermatogenesis and a positive TESE, the other a meiotic arrest and a negative TESE. Overall these observations highlight the limit of the histological classification but also the difficulty of predicting the effects of different (or even identical) gene defects. Variable expressivity and penetrance of defects in a same gene or even of the very same genetic defects are well-known caveats of human genetics.49 Here we see that individuals with the same gene defect (as observed for PDHA2) can present a different testicular phenotype and TESE outcome. It is not possible to know if these discrepancies are due to the testicular tissue heterogeneity and to the skill of the surgeon on the day of the biopsies, or to the variable expressivity or penetrance of the gene defect. The main goal for performing a genetic diagnosis for NOA-affected individuals is to obtain for the concerned persons a reliable negative diagnosis that may prevent a useless TESE procedure. Genes such as PDHA2, which do not provide a clear TESE prognostic, would be classified as uncertain and would not counter-indicate TESE.
It is interesting to note that all 12 individuals with defects in meiotic genes had a negative TESE indicating that a negative prognostic can be reached with a good degree of confidence, in particular for meiotic genes. In a recent study, Krausz and colleagues studied NOA-affected men with a maturation arrest and a negative TESE.10 Interestingly, they identified a total of 12 candidate genes of which 9 can be considered to be meiotic genes (MEI1, MEIOB, MSH4, RAD51L1, SHOC1, STAG3, SYCE1, TERB1, TEX11), thus confirming the strong link between defects in meiotic genes and negative TESE.
Conclusion
We used here a simple strategy based on a list of candidate genes to facilitate the interpretation of WES data and the identification of causal gene defects associated with a prediction of their consequence on spermatogenesis. Our results indicate that a clinical diagnosis interpreted in light of other clinical arguments can provide a strong argument against TESE. This demonstrates that exome sequencing can help the clinician to provide his patient with a more precise and relevant information which will help them adopt the most appropriate course of action, avoiding, in some instances, hopeless surgical procedures.
Acknowledgments
This work was funded by the French National Research Agency (ANR), projects MASFLAGELLA (ANR-19-CE17-0014), FLAGEL-OME (ANR-19-CE17-0014), and the INSERM (Institut National de la Santé et de la Recherche Médicale) and Bettencourt Foundation.
Declaration of interests
The authors declare no competing interests.
Published: February 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.01.011.
Data and code availability
All identified variants were submitted to Clinvar under the reference SUB10823688.
Details of the exome analysis pipeline used in the study is available on github (URL provided in the web resources section).
Web resources
gnomAD Browser, https://gnomad.broadinstitute.org/
MGI database, http://www.informatics.jax.org/
OMIM, https://www.omim.org/
Varsome, https://varsome.com
TIMC primary, https://github.com/ntm/grexome-TIMC-Primary
TIMC secondary, https://github.com/ntm/grexome-TIMC-Secondary
Supplemental information
References
- 1.Aziz N. The importance of semen analysis in the context of azoospermia. Clinics (São Paulo) 2013;68(Suppl 1):35–38. doi: 10.6061/clinics/2013(Sup01)05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tournaye H., Krausz C., Oates R.D. Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol. 2017;5:544–553. doi: 10.1016/S2213-8587(16)30040-7. [DOI] [PubMed] [Google Scholar]
- 3.Matzuk M.M., Lamb D.J. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 5.Tüttelmann F., Werny F., Cooper T.G., Kliesch S., Simoni M., Nieschlag E. Clinical experience with azoospermia: aetiology and chances for spermatozoa detection upon biopsy. Int. J. Androl. 2011;34:291–298. doi: 10.1111/j.1365-2605.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberg M.L., Li S., Behr B., Pera R.R., Cullen M.R. Relationship between semen production and medical comorbidity. Fertil. Steril. 2015;103:66–71. doi: 10.1016/j.fertnstert.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Okutman O., Rhouma M.B., Benkhalifa M., Muller J., Viville S. Genetic evaluation of patients with non-syndromic male infertility. J. Assist. Reprod. Genet. 2018;35:1939–1951. doi: 10.1007/s10815-018-1301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell M.J., Metzler-Guillemain C., Toure A., Coutton C., Arnoult C., Ray P.F. Single gene defects leading to sperm quantitative anomalies. Clin. Genet. 2017;91:208–216. doi: 10.1111/cge.12900. [DOI] [PubMed] [Google Scholar]
- 9.Ray P.F., Toure A., Metzler-Guillemain C., Mitchell M.J., Arnoult C., Coutton C. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin. Genet. 2017;91:217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 10.Krausz C., Riera-Escamilla A., Moreno-Mendoza D., Holleman K., Cioppi F., Algaba F., Pybus M., Friedrich C., Wyrwoll M.J., Casamonti E., et al. Genetic dissection of spermatogenic arrest through exome analysis: clinical implications for the management of azoospermic men. Genet. Med. 2020;22:1956–1966. doi: 10.1038/s41436-020-0907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oud M.S., Houston B.J., Volozonoka L., Mastrorosa F.K., Holt G.S., Alobaidi B.K.S., deVries P.F., Astuti G., Ramos L., Mclachlan R.I., et al. Exome sequencing reveals variants in known and novel candidate genes for severe sperm motility disorders. Hum. Reprod. 2021;36:2597–2611. doi: 10.1093/humrep/deab099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakhro K.A., Elbardisi H., Arafa M., Robay A., Rodriguez-Flores J.L., Al-Shakaki A., Syed N., Mezey J.G., Abi Khalil C., Malek J.A., et al. Point-of-care whole-exome sequencing of idiopathic male infertility. Genet. Med. 2018;20:1365–1373. doi: 10.1038/gim.2018.10. [DOI] [PubMed] [Google Scholar]
- 13.Bouker A., Halouani L., Kharouf M., Latrous H., Makni M., Marrakchi O., Zouari R., Fourati S. Step-by-step loupes-mTESE in non-obstructive azoospermic men, a retrospective study. Basic Clin. Androl. 2019;29:11. doi: 10.1186/s12610-019-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at arXiv. 2013 arXiv:1303.3997. [Google Scholar]
- 16.Faust G.G., Hall I.M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S., Scheffler K., Halpern A.L., Bekritsky M.A., Noh E., Källberg M., Chen X., Kim Y., Beyter D., Krusche P., Saunders C.T. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 18.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arafah K., Lopez F., Cazin C., Kherraf Z.-E., Tassistro V., Loundou A., Arnoult C., Thierry-Mieg N., Bulet P., Guichaoua M.-R., Ray P.F. Defect in the nuclear pore membrane glycoprotein 210-like gene is associated with extreme uncondensed sperm nuclear chromatin and male infertility: a case report. Hum. Reprod. 2021;36:693–701. doi: 10.1093/humrep/deaa329. [DOI] [PubMed] [Google Scholar]
- 20.Kherraf Z.-E., Amiri-Yekta A., Dacheux D., Karaouzène T., Coutton C., Christou-Kent M., Martinez G., Landrein N., Le Tanno P., Fourati Ben Mustapha S., et al. A Homozygous Ancestral SVA-Insertion-Mediated Deletion in WDR66 Induces Multiple Morphological Abnormalities of the Sperm Flagellum and Male Infertility. Am. J. Hum. Genet. 2018;103:400–412. doi: 10.1016/j.ajhg.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plagnol V., Curtis J., Epstein M., Mok K.Y., Stebbings E., Grigoriadou S., Wood N.W., Hambleton S., Burns S.O., Thrasher A.J., et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–2754. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasak L., Punab M., Nagirnaja L., Grigorova M., Minajeva A., Lopes A.M., Punab A.M., Aston K.I., Carvalho F., Laasik E., et al. GEMINI Consortium Bi-allelic Recessive Loss-of-Function Variants in FANCM Cause Non-obstructive Azoospermia. Am. J. Hum. Genet. 2018;103:200–212. doi: 10.1016/j.ajhg.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin H., Ma H., Hussain S., Zhang H., Xie X., Jiang L., Jiang X., Iqbal F., Bukhari I., Jiang H., et al. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet. Med. 2019;21:62–70. doi: 10.1038/s41436-018-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arafat M., Kleiman S.E., AbuMadighem A., Zeadna A., Levitas E., Vardi I.H., Barda S., Lehavi O., Hauser R., Lunenfeld E., et al. Pathogenic variations in Germ Cell Nuclear Acidic Peptidase (GCNA) are associated with human male infertility. Eur. J. Hum. Genet. 2021;29:1781–1788. doi: 10.1038/s41431-021-00946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy J.J., Wyrwoll M.J., Mcfadden W., Malcher A., Rotte N., Pollock N.C., Munyoki S., Veroli M.V., Houston B.J., Xavier M.J., et al. GEMINI Consortium Variants in GCNA, X-linked germ-cell genome integrity gene, identified in men with primary spermatogenic failure. Hum. Genet. 2021;140:1169–1182. doi: 10.1007/s00439-021-02287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arafat M., Har-Vardi I., Harlev A., Levitas E., Zeadna A., Abofoul-Azab M., Dyomin V., Sheffield V.C., Lunenfeld E., Huleihel M., Parvari R. Mutation in TDRD9 causes non-obstructive azoospermia in infertile men. J. Med. Genet. 2017;54:633–639. doi: 10.1136/jmedgenet-2017-104514. [DOI] [PubMed] [Google Scholar]
- 28.Tang D., Lv M., Gao Y., Cheng H., Li K., Xu C., Geng H., Li G., Shen Q., Wang C., et al. Novel variants in helicase for meiosis 1 lead to male infertility due to non-obstructive azoospermia. Reprod. Biol. Endocrinol. 2021;19:129. doi: 10.1186/s12958-021-00815-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan S., Jiao Y., Khan R., Jiang X., Javed A.R., Ali A., Zhang H., Zhou J., Naeem M., Murtaza G., et al. Homozygous mutations in C14orf39/SIX6OS1 cause non-obstructive azoospermia and premature ovarian insufficiency in humans. Am. J. Hum. Genet. 2021;108:324–336. doi: 10.1016/j.ajhg.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y., Li Y., Murtaza G., Zhou J., Jiao Y., Gong C., Hu C., Han Q., Zhang H., Zhang Y., et al. Whole-exome sequencing of consanguineous families with infertile men and women identifies homologous mutations in SPATA22 and MEIOB. Hum. Reprod. 2021;36:2793–2804. doi: 10.1093/humrep/deab185. [DOI] [PubMed] [Google Scholar]
- 31.Yıldırım Y., Ouriachi T., Woehlbier U., Ouahioune W., Balkan M., Malik S., Tolun A. Linked homozygous BMPR1B and PDHA2 variants in a consanguineous family with complex digit malformation and male infertility. Eur. J. Hum. Genet. 2018;26:876–885. doi: 10.1038/s41431-018-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouali N., Francou B., Bouligand J., Imanci D., Dimassi S., Tosca L., Zaouali M., Mougou S., Young J., Saad A., Guiochon-Mantel A. New MCM8 mutation associated with premature ovarian insufficiency and chromosomal instability in a highly consanguineous Tunisian family. Fertil. Steril. 2017;108:694–702. doi: 10.1016/j.fertnstert.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Tsai-Morris C.-H., Sheng Y., Lee E., Lei K.-J., Dufau M.L. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim S.L., Qu Z.P., Kortschak R.D., Lawrence D.M., Geoghegan J., Hempfling A.-L., Bergmann M., Goodnow C.C., Ormandy C.J., Wong L., et al. HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse. PLoS Genet. 2015;11:e1005620. doi: 10.1371/journal.pgen.1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guiraldelli M.F., Eyster C., Wilkerson J.L., Dresser M.E., Pezza R.J. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 2013;9:e1003383. doi: 10.1371/journal.pgen.1003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNairn A.J., Rinaldi V.D., Schimenti J.C. Repair of Meiotic DNA Breaks and Homolog Pairing in Mouse Meiosis Requires a Minichromosome Maintenance (MCM) Paralog. Genetics. 2017;205:529–537. doi: 10.1534/genetics.116.196808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vries S.S., Baart E.B., Dekker M., Siezen A., de Rooij D.G., de Boer P., te Riele H. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 1999;13:523–531. doi: 10.1101/gad.13.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H., Beasley M.D., Warren W.D., van der Horst G.T.J., McKay M.J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Saxe J.P., Chen M., Zhao H., Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finsterbusch F., Ravindranathan R., Dereli I., Stanzione M., Tränkner D., Tóth A. Alignment of Homologous Chromosomes and Effective Repair of Programmed DNA Double-Strand Breaks during Mouse Meiosis Require the Minichromosome Maintenance Domain Containing 2 (MCMDC2) Protein. PLoS Genet. 2016;12:e1006393. doi: 10.1371/journal.pgen.1006393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo T., Zhao S., Zhao S., Chen M., Li G., Jiao X., Wang Z., Zhao Y., Qin Y., Gao F., Chen Z.J. Mutations in MSH5 in primary ovarian insufficiency. Hum. Mol. Genet. 2017;26:1452–1457. doi: 10.1093/hmg/ddx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He W.-B., Tu C.-F., Liu Q., Meng L.-L., Yuan S.-M., Luo A.-X., He F.-S., Shen J., Li W., Du J., et al. DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J. Med. Genet. 2018;55:198–204. doi: 10.1136/jmedgenet-2017-104992. [DOI] [PubMed] [Google Scholar]
- 43.Pittman D.L., Cobb J., Schimenti K.J., Wilson L.A., Cooper D.M., Brignull E., Handel M.A., Schimenti J.C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 44.Kherraf Z.-E., Christou-Kent M., Karaouzene T., Amiri-Yekta A., Martinez G., Vargas A.S., Lambert E., Borel C., Dorphin B., Aknin-Seifer I., et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol. Med. 2017;9:1132–1149. doi: 10.15252/emmm.201607461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou D., Yao C., Xu B., Luo W., Ke H., Li Z., Qin Y., Guo T. Variations of C14ORF39 and SYCE1 identified in idiopathic premature ovarian insufficiency and nonobstructive azoospermia. J. Clin. Endocrinol. Metab. 2021 doi: 10.1210/clinem/dgab777. Published online October 26, 2021. [DOI] [PubMed] [Google Scholar]
- 46.Wang J., Zhang W., Jiang H., Wu B.-L., Primary Ovarian Insufficiency Collaboration Mutations in HFM1 in recessive primary ovarian insufficiency. N. Engl. J. Med. 2014;370:972–974. doi: 10.1056/NEJMc1310150. [DOI] [PubMed] [Google Scholar]
- 47.Caburet S., Todeschini A.-L., Petrillo C., Martini E., Farran N.D., Legois B., Livera G., Younis J.S., Shalev S., Veitia R.A. A truncating MEIOB mutation responsible for familial primary ovarian insufficiency abolishes its interaction with its partner SPATA22 and their recruitment to DNA double-strand breaks. EBioMedicine. 2019;42:524–531. doi: 10.1016/j.ebiom.2019.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulze W., Thoms F., Knuth U.A. Testicular sperm extraction: comprehensive analysis with simultaneously performed histology in 1418 biopsies from 766 subfertile men. Hum. Reprod. 1999;14(Suppl 1):82–96. doi: 10.1093/humrep/14.suppl_1.82. [DOI] [PubMed] [Google Scholar]
- 49.Maya I., Sukenik-Halevy R., Basel-Salmon L., Sagi-Dain L. Ten points to consider when providing genetic counseling for variants of incomplete penetrance and variable expressivity detected in a prenatal setting. Acta Obstet. Gynecol. Scand. 2020;99:1427–1429. doi: 10.1111/aogs.13963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All identified variants were submitted to Clinvar under the reference SUB10823688.
Details of the exome analysis pipeline used in the study is available on github (URL provided in the web resources section).