Summary
We used a machine learning approach to analyze the within-gene distribution of missense variants observed in hereditary conditions and cancer. When applied to 840 genes from the ClinVar database, this approach detected a significant non-random distribution of pathogenic and benign variants in 387 (46%) and 172 (20%) genes, respectively, revealing that variant clustering is widespread across the human exome. This clustering likely occurs as a consequence of mechanisms shaping pathogenicity at the protein level, as illustrated by the overlap of some clusters with known functional domains. We then took advantage of these findings to develop a pathogenicity predictor, MutScore, that integrates qualitative features of DNA substitutions with the new additional information derived from this positional clustering. Using a random forest approach, MutScore was able to identify pathogenic missense mutations with very high accuracy, outperforming existing predictive tools, especially for variants associated with autosomal-dominant disease and cancer. Thus, the within-gene clustering of pathogenic and benign DNA changes is an important and previously underappreciated feature of the human exome, which can be harnessed to improve the prediction of pathogenicity and disambiguation of DNA variants of uncertain significance.
Keywords: medical genetics, mutation, molecular pathology, variant clustering, pathogenicity prediction, functional domains, gain-of-function, loss-of-function, missense, human genome
Graphical abstract

Introduction
It has been previously noted that mechanisms associated with the pathogenesis of missense variants often correlate with the three-dimensional structure of proteins1, 2, 3 and that, for some disease-associated genes, mutations appear to cluster within specific regions.4, 5, 6, 7, 8, 9, 10, 11, 12 More systematic analyses have identified DNA sub-regions intolerant to missense variants13,14 and domains in protein families enriched in variants associated with disease.15,16 In addition, within many genes, pathogenic missense variants tend to cluster within specific domains or regions of the encoded proteins, whereas most loss-of-function variants do not10,15 with the exception of the penultimate and last exons where premature termination codons can escape nonsense-mediated decay (NMD).17 This information has been used empirically to estimate the pathogenic potential of newly detected variants for individual genes. Surprisingly perhaps, the same information has never been systematically considered to determine clusters of benign and deleterious variants across the entire human exome, or to score the pathogenicity of DNA changes on the same scale.
Identifying variants that underlie Mendelian phenotypes and cancer represents a major challenge in genetic medicine. Achieving this goal relies critically on developing the capability to recognize the one or a few mutations that have a clinical impact among the hundreds of thousands of benign variants which are normally present in an individual’s genome.18,19 Next generation sequencing (NGS) is now being applied routinely in clinical diagnostic settings,20 making the prediction of pathogenic DNA variants, and particularly of missense variants, a crucial element not only for medical research, but also in the context of patient diagnosis and disease management. The American College of Medical Genetics and Genomics (ACMG) has provided guidelines for the classification of variants into five categories: pathogenic, likely pathogenic, variants of uncertain significance (VUSs), likely benign, and benign, thereby establishing a terminology that has been widely adopted by the molecular genetics community.21 Recently this approach has been validated by using a Bayesian classification framework.22 However, these criteria still leave many rare missense variants classified as VUSs,23 resulting in an impasse at the diagnostic level. Many in silico tools have been developed over the past few years to help with this problem, delineating features that are typical of variants with benign versus pathogenic outcomes. Such features are generally uni-dimensional and include evolutionary conservation at the nucleotide or amino acid sequence levels, the structure of the protein, and the severity of the amino acid substitution in terms of change in hydrophobicity, charge, and size. Some of these tools exploit these features per se,24, 25, 26, 27, 28, 29, 30 whereas others take advantage of machine learning approaches and are trained on sets of pathogenic versus benign DNA changes.31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 Finally, meta-predictors offer an optimized combination of existing tools, such as those mentioned above,46, 47, 48, 49, 50, 51, 52, 53, 54, 55 although they sometimes suffer from circularity issues that are prone to producing falsely optimistic results (e.g., Grimm et al.56).
In this work, we address the topic of within-gene distribution of pathogenic and benign variants, reveal significant clustering of missense changes across the entire coding genome, and use this information to build a positional score. This value is then integrated with other unsupervised independent features (to avoid any circularity issues) to yield a pathogenicity score (MutScore) that has high discriminatory power for variants of uncertain significance.
Material and methods
Computation of the positional score
First, we extracted missense variants reported in ClinVar as of November 21, 2020 as pathogenic and likely pathogenic (PLP) and as benign and likely benign (BLB). Since some genes have a reduced number of BLB missense variants within ClinVar, we first computed the maximal allele frequency (AF) in gnomAD (maximum of gnomAD_exomes_AF and gnomAD_genomes_AF from dbNFSP4.0) of all reported PLP variants (missense, LoF, and others) for every gene. We then termed this value max-AF-PLP (maximum allelic frequency of PLP variants per gene) and extracted variants having a higher frequency than this value within the gnomAD dataset (r.2.1.1, PASS variants only), since variants occurring more frequently than max-AF-PLP can be considered as likely benign (ACMG criteria BS1). This allowed the addition of 354,888 BLB variants to our analysis and the building of the positional score as well as the amino acid change score (see below, Table S1).
To build our positional score, we used a random forest approach for each transcript, considered individually, having at least ten PLP (arbitrary threshold) and one BLB (the random forest needs at least one negative observation) variants in the training set (see below for details of the set). We used the randomForest function from the R package randomForest (v.4.6.14), with default parameters, except for the ntree value, which was set to 1,000. We considered as positive cases the amino acid positions of PLP missense variants and as negative cases all BLB missense variants (including common gnomAD variants with AF higher than max-AF-PLP) from the training set. Then, for every variant, we selected the score from the isoform in which the highest number of PLP variants was found and defined this value as the positional score. The positional score was set to zero for variants that were present only in transcripts with fewer than ten missense PLP variants in the training set.
Cluster analysis
We computed clusters of missense PLP variants in every transcript of every annotated gene by taking regions of consecutive positional scores higher than 0.05 and containing 5 or more PLP missense variants. A clustering score was also computed, defined as the minimum value between the fraction of PLP missense variants located inside detected clusters (over the total number of PLP variants; this is indicative of the precision of the clustering) and the fraction of the transcript not covered by clusters (over the total length of the transcript; this is indicative of the density of the clustering). Hence, the resulting score ranges between 0 and 1, with higher values indicating higher clustering. We then performed a permutation test for every transcript to evaluate the significance of the clustering score. More precisely, for each transcript, we ran 1,000 simulations by randomly assigning new amino acids positions for PLP and BLB and computing the resulting clustering score. With the resulting scores of these simulations, we could derive a p value and considered as significant those transcripts for which a p value below 0.05 with false discovery rate (FDR) correction was obtained.
Genes were also classified according to their clustering scores. Specifically, they were defined as having low, medium, or high PLP clustering if their best-performing isoform had scores below 0.33, between 0.33 and 0.66, or above 0.66, respectively.
A similar analysis was performed for the cluster analysis of BLB variants, by taking regions of consecutive positional scores lower than 0.05 and containing 10 or more BLB missense variants.
Finally, genes were classified according to the mode of inheritance of their associated phenotype, according to the OMIM database.57 More precisely, genes were included in the autosomal-dominant class if all of their associated phenotypes were labeled in OMIM as “autosomal dominant (AD)” and not “autosomal recessive (AR)” or “somatic mutation (SMu),” whereas they were included in the autosomal-recessive class if all their associated phenotypes were labeled as “autosomal recessive.” For the somatic class, we selected genes with at least one phenotype linked to the label “somatic mutation” and no occurrences of “autosomal recessive.” Genes linked to phenotypes having non-Mendelian or uncertain inheritance (OMIM entries beginning with a question mark or within curly brackets) were excluded from our analyses.
Selection of existing features
To build our model, we first selected all unsupervised scores available through dbNFSP4.058 that were also relevant to coding variants, namely SIFT,24 SIFT4G,59 LRT,29 PROVEAN,25 GERP++RS,27 phyloP100way, phyloP30way, and phyloP17way,26 phastCons100way, phastCons30way, and phastCons17way,28 SiPhy29way,30 dbscSNV-ADA, and dbscSNV-RF.60 Proportion expressed across transcripts (pext) scores61 were downloaded separately from the gnomAD database, since they are not available within dbNFSP4.0. The maximal and the mean score for every variant was then computed, yielding two features: pext-max and pext-mean. In order to replace missing values, we computed the median value of all missense mutations from dbNFSP4.0 for each feature, except for dbscSNV-ADA, dbscSNV-RF, and pext scores, for which missing values were simply zeroed.
Computation of the amino acid score
The amino acid change score was computed as follows. First, for every possible missense variant (e.g., Arg > Trp), we computed the number of genes containing this change as PLP in at least one instance (value a) as well as the number of genes containing this change as BLB, again at least once (value b). We then defined the amino acid score as being a/(a+b), for all missense changes. This approach was adopted so as to avoid any potential bias from genes with numerous identical changes, such as for instance Gly > Xaa substitutions in collagen triple helices.62
Annotation of variants
All variants from the training set (see below), the testing sets (see below), gnomAD, and ClinVar were annotated by using ANNOVAR63 (based on RefSeq genes) with data from the dbNFSP v.4.0 database and with custom-made tables for scores that were not present in this dataset. These tables included data from ClinPred,52 dbscSNV-ADA and -RF,60 as well as CONDEL.48
Training set
For the training set, we used variants reported in ClinVar as of November 21, 2020. The VCF file was downloaded from the ClinVar website. PLP variants were defined as annotated either as pathogenic or likely pathogenic and BLB variants as annotated benign or likely benign (Table S1). Variants with conflicting interpretations (CI) and variants of uncertain significance (VUSs) were discarded from the training set.
We excluded ClinPred from comparison with MutScore for the training set and testing sets since it uses directly allelic frequency (AF) from gnomAD as a feature, and therefore it is biased against AF, since most BLB variants have higher AF than PLP variants. By stratifying the analysis by AF, we could indeed show that the performance of ClinPred is lower for very rare variants (Figure S1).
Computation of the prediction score (MutScore)
MutScore was built using a random forest approach (randomForest function from the R package randomForest, default parameters, except ntree = 1,000; increasing the number of trees did not improve the performance of the model) with selected existing and novel features, as described above (n = 18; SIFT, SIFT4G, LRT, PROVEAN, GERP++RS, phyloP100way, phyloP30way, phyloP17way, phastCons100way, phastCons30way, phastCons17way, SiPhy29way, dbscSNV-ADA, dbscSNV-RF, pext-mean, pext-max, amino acid change score, and positional score) on the training set. The ratio of pathogenic out-of-box (OOB) votes over the total number of votes outputted by the random forest model was taken as the score for the variants of the training set for all further analysis. The score for other variants was obtained as the resulting probability of the model (R function “predict”).
10-fold cross-validation on the training set
The training set used to build MutScore (PLP and BLB variants from ClinVar) was split randomly into ten equal parts. Iteratively, nine parts were considered. These parts constituted the training subset of the cross-validation, whereas the remaining tenth was taken as a validation subset. This subdivision was used to compute the amino acid change score, the positional score, and finally the “MutScore.” Subsequently, performance was assessed on both the ten training and the ten validation subsets. AUCs were computed for all permutations, yielding ten values for the training subsets and ten values for the validation subsets. These two groups of AUCs were then compared using an unpaired t test (R function “t.test”), with unequal variance (Figure S2).
Testing sets
For testing set 1, we used ClinVar PLP and BLB variants that were present in the database on September 19, 2021. We excluded variants that were present in the training set (entries present in ClinVar up to November 21, 2020), that were used to train the positional score, or that were present in testing set 3 (see below). This procedure yielded 1,867 PLP and 459 BLB variants (Table S1).
We further stratified PLP variants from the testing set 1 according to the star-based classification system in ClinVar: zero stars (no assertion criteria provided), one star (criteria provided, single submitter), two stars (criteria provided, multiple submitters, no conflicts), three stars (reviewed by expert panel), and four stars (practice guideline). An additional stratification included their molecular origin, i.e., germline only (CLNORIGIN = 1) or de novo (CLNORIGIN = 32 or 33). To establish the performance of the tested tools on these subsets of variants, we used only BLB variants for genes also carrying retained PLP variants, to avoid type 2 circularity.56
For testing set 2, we used as PLP variants all disease-causing missense mutations (DM) from the HGMD database (v.2020.2) that were added since 2017 and were neither in ClinVar nor were included in the training set or another testing set (n = 14,327). Since the HGMD database does not contain BLB variants, we used all missense variants from gnomAD that were (1) absent from the training set, (2) not used to train the positional score, (3) absent from both HGMD and ClinVar, and (4) present only in genes for which we defined at least one PLP variant. In order to have a similar number of BLB and PLP variants, we used an AF threshold of >0.000177 in gnomAD (maximum value of exome and genome subsets), resulting in the selection of 13,248 BLB variants (Table S1). For the analysis of AUCs as a function of time (Figure S3B), we used only HGMD variants published during the year considered.
For testing set 3, we used variants from the DoCM database as PLP variants, excluding variants from the training set and variants used to build the positional and amino acid change features (n = 205). Since the DoCM database only contains pathogenic variants, BLB variants were selected according to the same criteria described for testing set 2 (n = 207, Table S1).
For all testing sets, AUCs of MutScore were compared to other tools using the DeLong test in the “roc.test” function from the package pROC (v1.17.0.1) with default arguments, except for “method=delong.”
Analysis of VUS and CI variants
VUSs and CI variants (ClinVar, dataset of November 21, 2020) in the 3,663 genes with at least one PLP variant in our training set were selected and annotated with MutScore, VEST4, and REVEL (Table S1). Two thresholds were computed for each score to reclassify such variants as likely pathogenic (LP), VUS, or likely benign (LB). Specifically, a variant was reclassified as LP if its score was above the value for which 95% of variants from the training set with that score (or higher) were indeed PLPs. Similarly, a variant was classified as LB if its score was below the value for which 95% of variants from the training set with that score (or lower values) were BLB.
Analytical and graphical software
All the analyses outlined above were performed with R (v.4.0.3) and the following packages: gridExtra (v.2.3), MASS (v.7.3.53), pROC (v.1.17.0.1), dplyr (v.1.0.4), randomForest (v.4.6.14), caTools (v.1.18.1), rpart.plot (v.3.0.9), rpart (v.4.1.15), and stringr (v.1.4.0). The figures resulting from these analyses were also obtained by the use of the same software.
MutLand and MutScore-batch online apps
Plots were performed in R (v.4.0.3), using the following packages: MASS (v.7.3-53.1) shiny (v.1.6.0), UniProt.ws (v.2.30.0), drawProteins (v.1.10.0), inlmisc (v.0.5.2), DT (v.0.17), shinythemes (v.1.2.0), waiter (v.0.2.0), and caTools (v.1.18.1). We computed the data obtained from ClinVar, gnomAD, conservation scores within dbNFSP4.0, as well as pext scores from gnomAD to build a graphical representation of the mutational landscape for genes with at least one PLP variant in ClinVar (n = 3,663). UniProt information about regions and domains was obtained with the UniProt.ws and drawProteins packages.
Results
Detection of intragenic variant clustering
We set out to investigate whether the clustering of missense lesions could be a general feature of the entire human morbid genome, rather than a phenomenon limited to a few specific loci, families of genes/proteins, or conserved domains. We selected all pathogenic or likely pathogenic (PLP) missense DNA variants reported in the ClinVar database,64 the largest public database assessing the pathogenicity of known human variants, as a reference set for disease-causing mutations in the human genome, whereas benign and likely benign substitutions (BLB) were extracted from both ClinVar and gnomAD65 (see Material and methods for details on variant selection). We then determined a “positional score,” based on a random forest model, for each transcript of every gene annotated in the RefSeq database (n = 52,630)66 using the position of amino acids affected by PLP and by BLB missense variants as a feature. This process allowed us to associate a score for the likelihood of pathogenicity for every single codon of every known transcript. Following the selection of disease genes harboring sufficient (10 or more) PLP variants to potentially allow the detection of clustering (840 genes), we identified 3,854 regions in 740 genes with multiple PLP variants and positional scores above a minimal threshold (as defined in the Material and methods). We then computed the clustering score, a parameter assessing both the precision and the density of variant clustering within a given transcript, and subdivided these 740 genes into three classes (with low, medium, or high clustering), as a function of this score (Figures 1A and 1B). The biological significance of such clustering was then assessed by means of a permutation test, and 387 genes were found to have at least one transcript with a p value below 0.05 after FDR correction, indicating that statistically significant clustering of pathogenic missense mutations occurred in more than 40% of all well-characterized disease genes (Figures 1A and 1B, Table S2). In addition, more than 79% of significantly clustered genes had at least one transcript with medium or high clustering (Figure 1B; examples of one gene for each class, Figures 2A–2C; global overview, Figure S4). Interestingly, a large majority of the genes that were associated only with autosomal-dominant phenotypes or with the presence of clinically relevant somatic mutations showed significant clustering (65.9% and 79.6% for the autosomal-dominant and the somatic classes, respectively, Figures S5B and S5C, Table S3), whereas corresponding significant values were obtained for a minority of genes linked to autosomal recessive-only phenotypes (19.7%), with 2.6% of them displaying high clustering (Figure S5A, Table S3).
Figure 1.
Genome-wide quantification of clustering of PLP and BLB missense variants
(A and C) Plots of clustering precision (defined as the proportion of variants of a given transcript located inside clusters) versus clustering density (defined as the proportion of a given transcript not covered by clusters) for all transcripts with at least ten PLP and one BLB missense variants in ClinVar. Dots indicate individual transcripts (some genes may be represented by more than one dot). Squares indicate clustering score thresholds (at 0.33 and 0.66 units) used to define three categories: low, medium, and high clustering. Transcripts with non-significant clustering scores (see Material and methods) are marked in dark gray. Rectangles with yellow dots depict schematically examples of transcripts with dense or precise clustering.
(B and D) Pie charts of the same data depicted in (A) and (C) for the transcript of a given gene with the highest clustering score. Genes for which no clusters were detected, in any of their transcripts, are also shown (in light gray).
Figure 2.
Examples of transcripts with significant clustering score p values
Shown are (A) low PLP clustering (COL3A1, GenBank: NM_000090.3); (B) medium PLP clustering (BRAF, GenBank: NM_004333.6); (C) high PLP clustering (KRT6A, GenBank: NM_005554.4); and (D) high PLP clustering and low BLB clustering (NSD1, GenBank: NM_022455.4). Variants were extracted from ClinVar, November 21, 2020 release.
The same analysis was performed for BLB variants. Within the 851 genes considered (i.e., carrying 10 or more BLB missense changes), 10,136 enriched regions in 490 of them were identified, including 172 (20.2% of the total) displaying a significant clustering score (Figures 1C and 1D, Table S2). However, only 9 genes (1.1%) exhibited high clustering of BLB variants (Figure 1D). The majority of transcripts showed precise but sparse BLB clustering (bottom right, Figure 1C), as for example NSD1, which also displays high PLP clustering (Figure 2D). Sub-classification of BLB variants according to recessive versus dominant and somatic occurrence resulted in a trend similar to that observed for PLP variants (Figures S5D–S5F, Table S3).
Implementation of MutScore
We developed MutScore with the goal of exploiting the biological information contained in this patterning of variants to predict the pathogenicity of DNA changes, and potentially obtaining additional knowledge on the molecular mechanisms underlying pathogenicity. In addition, we aimed at an improved classification of VUSs, which in whole-genome sequencing (WGS) or whole-exome sequencing (WES) studies are mostly represented by low-frequency missense substitutions.
In addition to the positional score described above, we also calculated an “amino acid change score,” which is intended to approximate the likelihood of a particular amino acid substitution (e.g., Arg > Trp) to result in a pathological phenotype (see Material and methods for details on both scores). Moreover, we collected 16 additional existing features from published predictive tools, all of which originated from unsupervised approaches,24, 25, 26, 27, 28, 29, 30,59, 60, 61 to avoid artificially inflating the resulting predictive power (Figure 3A).
Figure 3.
Outline of the procedures used to build MutScore and MutLand and importance of different features within the training set
(A) Framework representing the different steps followed to generate MutScore and MutLand (dark green, database; light green, variant set; light blue, computation; dark blue, scores; orange, graphical output).
(B) Ranking of features in the model based on mean decrease in accuracy.
(C) Ranking of features in the model based on mean decrease in Gini index.
Finally, we built another random forest model that considered these 18 features, trained on a set containing only two discrete classes: PLP (36,966 variants) and BLB (29,066 variants), as assessed by ClinVar in its release of November 21, 2020 (Table S1). The importance of each feature was computed as either mean decrease in accuracy (Figure 3B) or decrease in Gini index (mean decrease in impurity, Figure 3C). In both cases, the positional score as well as the PROVEAN25 and SIFT24 scores were found to be the three most important features (Figures 3B and 3C, Table S4). The least important features were represented by the output of two splicing predictors, probably because only a small minority of missense variants are predicted to alter splicing.
To generate a final score for pathogenicity (MutScore), the model was applied to all single-nucleotide substitutions resulting in a missense change, for all possible RefSeq genes (range: 0–1, representing the likelihood for a given missense variant to be pathogenic). A 10-fold cross-validation step was also performed to check whether overfitting had occurred during training. The areas under the curve (AUCs) from the validation sets from this cross-validation procedure (average = 0.949) were not statistically different from the corresponding training sets (average = 0.950) (two-tailed unpaired t test, p = 0.701, Figure S2), showing no overfitting of the model on the training data.
Testing sets
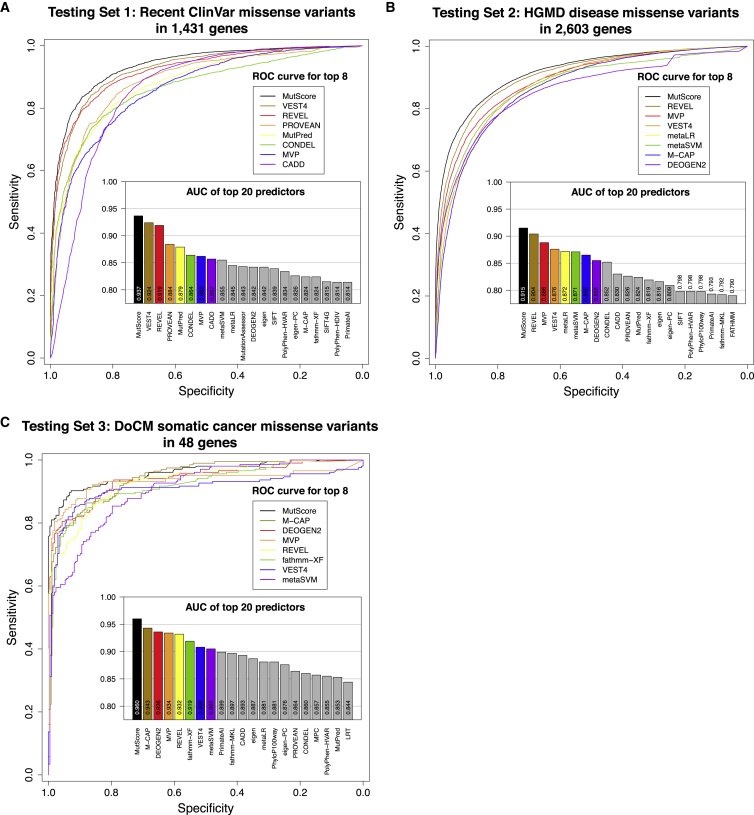
To determine the real predictive power of our tool, we evaluated its performance with respect to different sets of data that were not used in the training process. Specifically, we considered recent ClinVar entries, logged between November 22, 2020 and September 19, 2021 (testing set 1), the HGMD database67 (testing set 2), and the database of cancer variants DoCM68 (testing set 3).
For testing set 1, including 5,021 PLP and 2,035 BLB variants, MutScore had an AUC of 0.937 (Figure 4A) which was the highest value, at a statistically significant level, across all tools tested (p < 0.05, DeLong test). The closest competitors were REVEL and VEST4, with AUCs of 0.924 and 0.919, respectively. For testing set 2, PLP variants were selected to correspond to all disease-causing mutations (DM) from the HGMD database that were present in neither testing set 1 nor in the training set. In addition, to avoid using variants that were used to train other tools (such as REVEL and VEST4), we deliberately selected only PLP variants from HGMD that were added after such tools were published, i.e., in 2017 or at a later date. For BLB entries, we selected variants from the gnomAD database that did not overlap with previous analyses (see Material and methods for details). This resulted in the selection of 14,327 PLP and 13,248 BLB missense variants in 2,603 different genes. Again, MutScore exhibited a significantly higher performance than other tools (p < 0.05, DeLong test, Figure 4B, Table S5) with an AUC of 0.915 compared to 0.904 for REVEL, the second-best predictor. No other predictor achieved an AUC above 0.900. For testing set 3, we selected somatic cancer variants from the DoCM database, a curated set of somatic variants with established relevance to cancer biology. More precisely, we considered all variants from this database that were not present in the training set and were not used to build the model as PLP entries, whereas BLB entries were selected according to the same procedures described for testing set 2. This resulted in the identification of 205 PLP and 207 BLB variants. Once more, MutScore had the highest AUC value, in a statistically significant way (AUC = 0.960, p < 0.05, DeLong test), followed by M-CAP (AUC = 0.943, Figure 4C, Table S5).
Figure 4.
Performance of MutScore and other tools with respect to different testing sets
Shown are (A) testing set 1: recent ClinVar variants (PLPs versus BLBs); (B) testing set 2: recent variants from the HGMD database (from 2017 onward; PLPs) versus frequent gnomAD variants (BLBs); and (C) testing set 3: variants from the DoCM database (PLPs) versus frequent gnomAD variants (BLBs). ROC curves for the top-8 predictors and histograms of AUCs for the top-20 predictors are also shown.
It is interesting to note that the performance of some predictors appeared to decrease when evaluating more recent variants, and in particular when HGMD data prior to 2017 versus post-2017 (testing set 2) were used (Figure S3A). This could be due to certain tools having been overfitted on data used to train them versus data that were completely naive to them (Figure S3B), a conclusion supported by the observation that untrained tools (SIFT, PROVEAN, GERP, PhyloP, etc.) retained their power on old versus new entries (Figure S3A). More specifically, the performance bias toward older entries by trained versus untrained tools was significant (average differences between new and old entries: −0.0024 and −0.0170 for untrained and trained tools, respectively; p value = 5.8 × 10−5, by t test, bilateral with unequal variance).
Performance on subsets of testing set 1 from ClinVar
We wished to test whether MutScore performance would reflect the actual pathogenicity of variants as assessed by curated experimental evidence. ClinVar attributes a score ranging from zero to four stars to every entry, depending upon the number of submissions and on data supporting its pathogenicity. For instance, it was recently shown that the tool Rhapsody69 classified PLP variants with zero stars (i.e., no assertion criteria provided) less accurately than other variants. MutScore’s AUCs correlated well with the number of stars attributed by ClinVar to PLP variants, with values of 0.908 for variants with zero stars, 0.946 for variants with one star (i.e., criteria provided, single submitter), 0.963 for variants with two stars (i.e., criteria provided, multiple submitters, no conflicts), and 0.967 for variants with three stars (i.e., reviewed by expert panel). Since there were too few missense variants with four stars (i.e., practice guideline) to assess, we could not compute performance for such a small dataset. Other prediction tools displayed the same trend (Figure S6, Table S5). MutScore had a significantly higher AUC with 0.946 for the PLP variants with zero stars, which represents more than 66% of all PLP variants.
We also investigated the performance of MutScore according to ClinVar’s defined origin of PLP missense variants: strictly germline versus de novo. In this test, MutScore had the highest AUCs for germline and de novo variants (0.938 and 0.908, respectively, Figures S7A and S7B, Table S5). Again, all differences were statistically significant (p < 0.05, DeLong test), except for the comparison with VEST4 on de novo variants. MutScore also displayed a lower decrease in AUC for de novo variants compared to germline variants when compared to VEST4 and REVEL (Table S5). This can be explained by the fact that both de novo and somatic missense mutations tend to exert their influence via dominant gain-of-function mechanisms and hence usually only affect specific portions of a protein (e.g., a kinase domain). Traditional tools consider amino acid residue conservation, but usually do not take into account mutational clustering or positional information, and therefore their predictive power may be less efficient with respect to that of MutScore.
Finally, since the positional score appears to be the most important feature of our model (Figure 3B), we evaluated MutScore’s performance for genes that were previously well characterized from a mutational standpoint (HCGs [highly characterized genes], with a positional score > 0) versus genes that were not (PCGs [poorly characterized genes], positional score = 0). MutScore displayed the highest AUC for both HCGs and PCGs for testing set 1 (ClinVar, Figures S8A and S8B) as well as for testing set 2 (HGMD, Figures S8C and S8D), although the difference with respect to other top-performing tools for PCGs was only marginal (≤0.006 units, overall) and statistically not significant (Table S5). Testing set 3 was not used since it did not contain sufficient PLP variants in PCGs. As expected, performance for HCGs was higher than for PCGs in all cases.
Variants of uncertain significance (VUSs) and with conflicting interpretation (CI)
As a next step, we investigated the ability of MutScore, as well as of the two predictors that displayed the best performance in previous tests (VEST4 and REVEL), to re-classify ambiguous variants. Following the reassessment of all VUSs and conflicting interpretation (CI) variants from ClinVar, MutScore succeeded in reclassifying 54.7% of VUSs and 61.7% of CI variants, compared to 35.5% and 33.9% of VUSs and 40.7% and 37.3% of CI variants for VEST4 and REVEL, respectively (Figures 5 and S9). In particular, MutScore had an edge in redirecting VUSs and CI variants toward BLB variants (ACMG classes 2 and 1), compared to other predictors (Figure 5, left column and Figure S9). Again, this was probably a consequence of the positional score, which allows for a better assessment of variants with respect to their presence within mutational clusters or outside of them, whereas existing algorithms evaluate variants independently of such regional information.70 We can assume that many variants were reclassified as BLB by virtue of their presence outside pathogenic clusters.
Figure 5.
Distribution of MutScore, VEST4, and REVEL scores of ClinVar’s PLP, BLB, VUSs and CI variants
MutScore provides a better separation of all classes of variant and allows for an improved re-classification of VUSs and CI variants (see Figure S9 as well). Red, PLP variants; blue, BLB variants; light blue, VUSs/CIs reclassified as likely benign; light orange, VUSs/CIs not reclassified; light red, VUSs/CIs reclassified as likely pathogenic; black lines, thresholds for reclassification. The thresholds used to reclassify variants are defined in the Material and methods, and correspond specifically to 0.140 and 0.730 for MutScore, 0.187 and 0.819 for VEST4, and 0.086 and 0.682 for REVEL for BLB and PLP variants, respectively.
MutLand and MutScore-batch
To provide a visual representation of our results in individual genes, as well as to facilitate the scoring of newly identified variants, we created an interactive, web-accessible interface displaying data from ClinVar, gnomAD, and UniProt, conservation scores from other tools, PLP and BLB clusters, as well as the output from MutScore. As an example, Figure 6 shows the MutLand output for KCNQ2, which is known to harbor clusters of PLP missense variants in specific transmembrane segments, in the pore loop, and in some intracellular helices.10,71 In the MutLand representation of this gene, clustering of PLP missense variants can be easily identified, whereas PLP LoF variants do not cluster. Many VUSs are scattered along the entire protein sequence. Of these, those affecting amino acids in the C-terminal portion, located outside of PLP clusters and with a lower MutScore value, might then be reclassified as likely benign. To allow the user to interrogate multiple MutScore values simultaneously, we also created a separate web interface, MutScore-batch.
Figure 6.
Example of a MutLand graphical output
Here we display the output for KCNQ2 (GenBank: NM_172107.4), listing various attributes of this gene/protein, including MutScore. Variants were extracted from ClinVar, release of November 21, 2020.
Discussion
It has long been known that the pattern of pathogenic variants within a limited number of genes72 or gene classes follows a non-random and biological function-driven distribution,9,10,73 such as variants in the triple-helical region of collagen74,75 or in the BRCT region of BRCA1.76,77 Here we find that clustering of pathogenic missense variants occurs in almost half of all human genes, genome-wide, and that for about 18% of them such clustering is highly delimited. This also applies to benign missense changes, irrespective of their allelic frequency, as clusters of BLB variants are detectable in approximately 20% of all genes associated with a hereditary condition. Based on the distribution of MutScore values in relation to protein regions and protein classes, it is reasonable to assume that this clustering of DNA variants is mainly shaped by pathogenic mechanisms occurring at the protein level. For instance, in humans the majority of dominant mutations lead to gain-of-function or dominant-negative events, usually affecting amino acid residues located within specific domains or regions of a given protein.15,78,79 This phenomenon is clearly reflected in our finding that PLP variants associated with dominant conditions are mostly identified in clusters, and is even more pronounced for somatic variants involved in cancer (Figure S5). By contrast, most recessive missense mutations act via loss-of-function (or reduced function) mechanisms and in general this type of DNA changes is more dispersed along the protein sequence.72 In particular, recessive PLP missense variants tend to cluster less and, if they do, to generate larger and more loosely defined regions. The mirror effect of these events is often visible for BLB variants: in genes associated with dominant conditions, BLB variants tend to cluster anywhere in the gene other than in regions with PLP clusters. Conversely, in genes linked to recessive conditions, BLB clustering is in general absent.
We reasoned that the clustering or dispersion of missense variants might be used to infer the pathogenicity of newly observed DNA changes, similar to the use of evolutionary conservation across species. For this purpose, we developed MutScore, a predictor tool, and its graphical interface, MutLand, to enable the easy visualization of mutational landscapes. In addition to existing unsupervised dimensions, MutScore integrates two novel features into its final predictive model: a positional score and an amino acid change score. Furthermore, it builds heavily on curated information of clinically relevant DNA variants, as defined by ClinVar. The positional score in particular, essentially defining regions of a gene in which pathogenic mutations or benign variants are likely (or unlikely) to occur, appears to be the most important feature, conferring upon MutScore an edge over existing algorithms. Interestingly, the integration of as many benign variants as possible within the model also appears to be very important, since it allows for the determination of a “harmlessness threshold” that is gene specific and improves prediction even more.
When tested with real data from three independent datasets (HGMD and recent ClinVar data for constitutional disorders, as well as DoCM for cancer), MutScore performed markedly better than existing tools in discriminating between pathogenic and non-pathogenic variants. This performance allowed for a high rate of disambiguation of VUSs, a key issue in current NGS-based genetic diagnostics. One limitation of our scoring approach lies in the fact that prediction is partly dependent on the positional score, and therefore on information pertaining to existing pathogenic variants, such as the number of entries in ClinVar and their quality and accuracy. This explains why MutScore’s performance is still uneven with respect to different genes in the human genome and essentially matches that of other meta-predictors for poorly characterized genes. As more and more variants are identified and their clinical implications are assessed, both information on variant clustering and MutScore’s performances are likely to increase substantially over time. Furthermore, integration of data on the tridimensional structure of proteins, e.g., from the AlphaFold project,80 may help the future development of MutScore and improve even further its predictive power.
In conclusion, our analysis of the regional distribution of missense variants within human disease genes reveals that extensive clustering is common, both for pathogenic and benign DNA changes. This observation may help in clarifying protein structure and function and will hopefully prime further research into mechanisms of selection and evolution. Moreover, the AI-driven integration of clustering information in MutScore allows for more accurate pathogenicity scoring and the disambiguation of variants of uncertain significance. Together with its graphical interface MutLand and with MutScore-batch, this tool promises to become a useful instrument in genetic medicine and could be used as a stepping stone for new research projects aiming to define further key properties of the morbid human genome.
Acknowledgments
We would like to thank Ms. Sitta Föhr for her careful revision of this manuscript. This work was supported by the Swiss National Science Foundation (grants #176097 and #204285 to C.R.). Research performed by D.N.C. and P.D.S. was partly supported by QIAGEN Inc.
Declaration of interests
D.N.C. and P.D.S. acknowledge QIAGEN Inc. for their financial support through a License Agreement with Cardiff University. The other authors do not declare any conflicts of interest.
Published: February 3, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.01.006.
Data and code availability
MutScore values can be retrieved from https://iob-genetic.shinyapps.io/mutscore/ or from https://iob-genetic.shinyapps.io/mutscore-batch and they can be used under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. For commercial uses of MutScore, please contact the authors.
The code used to generate the MutScore and the analyses presented here is available at https://github.com/mquinodo/mutscore, except for the part using ANNOVAR, which can be found at https://annovar.openbioinformatics.org/en/latest/.
Web resources
ClinPred, https://sites.google.com/site/clinpred
gnomAD, (downloads), https://gnomad.broadinstitute.org/downloads
MutScore-batch, https://iob-genetic.shinyapps.io/mutscore-batch
Supplemental information
References
- 1.Tokheim C., Bhattacharya R., Niknafs N., Gygax D.M., Kim R., Ryan M., Masica D.L., Karchin R. Exome-scale discovery of hotspot mutation regions in human cancer using 3D protein structure. Cancer Res. 2016;76:3719–3731. doi: 10.1158/0008-5472.CAN-15-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medina-Carmona E., Betancor-Fernández I., Santos J., Mesa-Torres N., Grottelli S., Batlle C., Naganathan A.N., Oppici E., Cellini B., Ventura S., et al. Insight into the specificity and severity of pathogenic mechanisms associated with missense mutations through experimental and structural perturbation analyses. Hum. Mol. Genet. 2019;28:1–15. doi: 10.1093/hmg/ddy323. [DOI] [PubMed] [Google Scholar]
- 3.Iqbal S., Pérez-Palma E., Jespersen J.B., May P., Hoksza D., Heyne H.O., Ahmed S.S., Rifat Z.T., Rahman M.S., Lage K., et al. Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc. Natl. Acad. Sci. USA. 2020;117:28201–28211. doi: 10.1073/pnas.2002660117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Althari S., Najmi L.A., Bennett A.J., Aukrust I., Rundle J.K., Colclough K., Molnes J., Kaci A., Nawaz S., van der Lugt T., et al. Unsupervised clustering of missense variants in HNF1A using multidimensional functional data aids clinical interpretation. Am. J. Hum. Genet. 2020;107:670–682. doi: 10.1016/j.ajhg.2020.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dietz H.C., Saraiva J.M., Pyeritz R.E., Cutting G.R., Francomano C.A. Clustering of fibrillin (FBN1) missense mutations in Marfan syndrome patients at cysteine residues in EGF-like domains. Hum. Mutat. 1992;1:366–374. doi: 10.1002/humu.1380010504. [DOI] [PubMed] [Google Scholar]
- 6.Kamburov A., Lawrence M.S., Polak P., Leshchiner I., Lage K., Golub T.R., Lander E.S., Getz G. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc. Natl. Acad. Sci. USA. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talbot K., Ponting C.P., Theodosiou A.M., Rodrigues N.R., Surtees R., Mountford R., Davies K.E. Missense mutation clustering in the survival motor neuron gene: a role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum. Mol. Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 8.Wang C.M., Dixon P.H., Decordova S., Hodges M.D., Sebire N.J., Ozalp S., Fallahian M., Sensi A., Ashrafi F., Repiska V., et al. Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region. J. Med. Genet. 2009;46:569–575. doi: 10.1136/jmg.2008.064196. [DOI] [PubMed] [Google Scholar]
- 9.Geisheker M.R., Heymann G., Wang T., Coe B.P., Turner T.N., Stessman H.A.F., Hoekzema K., Kvarnung M., Shaw M., Friend K., et al. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 2017;20:1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lelieveld S.H., Wiel L., Venselaar H., Pfundt R., Vriend G., Veltman J.A., Brunner H.G., Vissers L.E.L.M., Gilissen C. Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 2017;101:478–484. doi: 10.1016/j.ajhg.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waring A.A.J. 2020. Exploration of rare-missense variant clustering in Mendelian disease-genes. PhD thesis, University of Oxford. [Google Scholar]
- 12.Buljan M., Blattmann P., Aebersold R., Boutros M. Systematic characterization of pan-cancer mutation clusters. Mol. Syst. Biol. 2018;14:e7974. doi: 10.15252/msb.20177974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayeck T.J., Stong N., Wolock C.J., Copeland B., Kamalakaran S., Goldstein D.B., Allen A.S. Improved pathogenic variant localization via a hierarchical model of sub-regional intolerance. Am. J. Hum. Genet. 2019;104:299–309. doi: 10.1016/j.ajhg.2018.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silk M., Petrovski S., Ascher D.B. MTR-Viewer: identifying regions within genes under purifying selection. Nucleic Acids Res. 2019;47(W1):W121–W126. doi: 10.1093/nar/gkz457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Palma E., May P., Iqbal S., Niestroj L.M., Du J., Heyne H.O., Castrillon J.A., O’Donnell-Luria A., Nürnberg P., Palotie A., et al. Identification of pathogenic variant enriched regions across genes and gene families. Genome Res. 2020;30:62–71. doi: 10.1101/gr.252601.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coban-Akdemir Z., White J.J., Song X., Jhangiani S.N., Fatih J.M., Gambin T., Bayram Y., Chinn I.K., Karaca E., Punetha J., et al. Baylor-Hopkins Center for Mendelian Genomics Identifying genes whose mutant transcripts cause dominant disease traits by potential gain-of-function alleles. Am. J. Hum. Genet. 2018;103:171–187. doi: 10.1016/j.ajhg.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pottinger T.D., Puckelwartz M.J., Pesce L.L., Robinson A., Kearns S., Pacheco J.A., Rasmussen-Torvik L.J., Smith M.E., Chisholm R., McNally E.M. Pathogenic and uncertain genetic variants have clinical cardiac correlates in diverse biobank participants. J. Am. Heart Assoc. 2020;9:e013808. doi: 10.1161/JAHA.119.013808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark M.M., Stark Z., Farnaes L., Tan T.Y., White S.M., Dimmock D., Kingsmore S.F. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom. Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavtigian S.V., Greenblatt M.S., Harrison S.M., Nussbaum R.L., Prabhu S.A., Boucher K.M., Biesecker L.G., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018;20:1054–1060. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kryukov G.V., Pennacchio L.A., Sunyaev S.R. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am. J. Hum. Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garber M., Guttman M., Clamp M., Zody M.C., Friedman N., Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–i62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L., Edwards K.J., Day I.N., Gaunt T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 34.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raimondi D., Tanyalcin I., Ferté J., Gazzo A., Orlando G., Lenaerts T., Rooman M., Vranken W. DEOGEN2: prediction and interactive visualization of single amino acid variant deleteriousness in human proteins. Nucleic Acids Res. 2017;45(W1):W201–W206. doi: 10.1093/nar/gkx390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 37.Sundaram L., Gao H., Padigepati S.R., McRae J.F., Li Y., Kosmicki J.A., Fritzilas N., Hakenberg J., Dutta A., Shon J., et al. Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 2018;50:1161–1170. doi: 10.1038/s41588-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gulko B., Hubisz M.J., Gronau I., Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Q., Hu Y., Sun J., Cheng Y., Cheung K.H., Zhao H. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci. Rep. 2015;5:10576. doi: 10.1038/srep10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shihab H.A., Rogers M.F., Gough J., Mort M., Cooper D.N., Day I.N., Gaunt T.R., Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34:511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pejaver V., Mooney S.D., Radivojac P. Missense variant pathogenicity predictors generalize well across a range of function-specific prediction challenges. Hum. Mutat. 2017;38:1092–1108. doi: 10.1002/humu.23258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter H., Douville C., Stenson P.D., Cooper D.N., Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Favalli V., Tini G., Bonetti E., Vozza G., Guida A., Gandini S., Pelicci P.G., Mazzarella L. Machine learning-based reclassification of germline variants of unknown significance: The RENOVO algorithm. Am. J. Hum. Genet. 2021;108:682–695. doi: 10.1016/j.ajhg.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y.F. Unified inference of missense variant effects and gene constraints in the human genome. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008922. e1008922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alirezaie N., Kernohan K.D., Hartley T., Majewski J., Hocking T.D. ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am. J. Hum. Genet. 2018;103:474–483. doi: 10.1016/j.ajhg.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda J.I., Nanatsue K., Yamagishi R., Ito M., Haga N., Hirata H., Ogi T., Ohno K. InMeRF: prediction of pathogenicity of missense variants by individual modeling for each amino acid substitution. NAR Genom Bioinform. 2020;2:a038. doi: 10.1093/nargab/lqaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi H., Zhang H., Zhao Y., Chen C., Long J.J., Chung W.K., Guan Y., Shen Y. MVP predicts the pathogenicity of missense variants by deep learning. Nat. Commun. 2021;12:510. doi: 10.1038/s41467-020-20847-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimm D.G., Azencott C.A., Aicheler F., Gieraths U., MacArthur D.G., Samocha K.E., Cooper D.N., Stenson P.D., Daly M.J., Smoller J.W., et al. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 2015;36:513–523. doi: 10.1002/humu.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X., Li C., Mou C., Dong Y., Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103. doi: 10.1186/s13073-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 60.Jian X., Boerwinkle E., Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cummings B.B., Karczewski K.J., Kosmicki J.A., Seaby E.G., Watts N.A., Singer-Berk M., Mudge J.M., Karjalainen J., Satterstrom F.K., O’Donnell-Luria A.H., et al. Genome Aggregation Database Production Team. Genome Aggregation Database Consortium Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. doi: 10.1038/s41586-020-2329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butterfield R.J., Foley A.R., Dastgir J., Asman S., Dunn D.M., Zou Y., Hu Y., Donkervoort S., Flanigan K.M., Swoboda K.J., et al. Position of glycine substitutions in the triple helix of COL6A1, COL6A2, and COL6A3 is correlated with severity and mode of inheritance in collagen VI myopathies. Hum. Mutat. 2013;34:1558–1567. doi: 10.1002/humu.22429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stenson P.D., Mort M., Ball E.V., Chapman M., Evans K., Azevedo L., Hayden M., Heywood S., Millar D.S., Phillips A.D., Cooper D.N. The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 2020;139:1197–1207. doi: 10.1007/s00439-020-02199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ainscough B.J., Griffith M., Coffman A.C., Wagner A.H., Kunisaki J., Choudhary M.N., McMichael J.F., Fulton R.S., Wilson R.K., Griffith O.L., Mardis E.R. DoCM: a database of curated mutations in cancer. Nat. Methods. 2016;13:806–807. doi: 10.1038/nmeth.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponzoni L., Peñaherrera D.A., Oltvai Z.N., Bahar I. Rhapsody: predicting the pathogenicity of human missense variants. Bioinformatics. 2020;36:3084–3092. doi: 10.1093/bioinformatics/btaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiang J., Yang J., Chen L., Chen Q., Yang H., Sun C., Zhou Q., Peng Z. Reinterpretation of common pathogenic variants in ClinVar revealed a high proportion of downgrades. Sci. Rep. 2020;10:331. doi: 10.1038/s41598-019-57335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J., Kim E.C., Chen C., Procko E., Pant S., Lam K., Patel J., Choi R., Hong M., Joshi D., et al. Identifying mutation hotspots reveals pathogenetic mechanisms of KCNQ2 epileptic encephalopathy. Sci. Rep. 2020;10:4756. doi: 10.1038/s41598-020-61697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turner T.N., Douville C., Kim D., Stenson P.D., Cooper D.N., Chakravarti A., Karchin R. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet. 2015;24:5995–6002. doi: 10.1093/hmg/ddv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sivley R.M., Dou X., Meiler J., Bush W.S., Capra J.A. Comprehensive analysis of constraint on the spatial distribution of missense variants in human protein structures. Am. J. Hum. Genet. 2018;102:415–426. doi: 10.1016/j.ajhg.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parkin J.D., San Antonio J.D., Pedchenko V., Hudson B., Jensen S.T., Savige J. Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum. Mutat. 2011;32:127–143. doi: 10.1002/humu.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiu Y., Mekkat A., Yu H., Yigit S., Hamaia S., Farndale R.W., Kaplan D.L., Lin Y.S., Brodsky B. Collagen Gly missense mutations: Effect of residue identity on collagen structure and integrin binding. J. Struct. Biol. 2018;203:255–262. doi: 10.1016/j.jsb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams R.S., Chasman D.I., Hau D.D., Hui B., Lau A.Y., Glover J.N. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J. Biol. Chem. 2003;278:53007–53016. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- 77.Lee M.S., Green R., Marsillac S.M., Coquelle N., Williams R.S., Yeung T., Foo D., Hau D.D., Hui B., Monteiro A.N., Glover J.N. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res. 2010;70:4880–4890. doi: 10.1158/0008-5472.CAN-09-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Veitia R.A., Caburet S., Birchler J.A. Mechanisms of Mendelian dominance. Clin. Genet. 2018;93:419–428. doi: 10.1111/cge.13107. [DOI] [PubMed] [Google Scholar]
- 79.Li Y., Zhang Y., Li X., Yi S., Xu J. Gain-of-function mutations: an emerging advantage for cancer biology. Trends Biochem. Sci. 2019;44:659–674. doi: 10.1016/j.tibs.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MutScore values can be retrieved from https://iob-genetic.shinyapps.io/mutscore/ or from https://iob-genetic.shinyapps.io/mutscore-batch and they can be used under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. For commercial uses of MutScore, please contact the authors.
The code used to generate the MutScore and the analyses presented here is available at https://github.com/mquinodo/mutscore, except for the part using ANNOVAR, which can be found at https://annovar.openbioinformatics.org/en/latest/.