Abstract
Background:
Coronaviruses (CoVs) are single-stranded, polyadenylated, enveloped RNA of positive polarity with a unique potential to alter host tropism. This has been exceptionally demonstrated by the emergence of deadly virus outbreaks of the past: Severe Acute Respiratory Syndrome (SARS-CoV) in 2003 and Middle East Respiratory Syndrome (MERS-CoV) in 2012.
Summary:
The 2019 outbreak by the new cross-species transmission of SARS-CoV-2 has put the world on alert. CoV infection is triggered by receptor recognition, membrane fusion, and successive viral entry mediated by the surface Spike (S) glycoprotein. S protein is one of the major antigenic determinants and the target for neutralizing antibodies. It is a valuable target in antiviral therapies because of its central role in cell-cell fusion, viral antigen spread, and host immune responses leading to immunopathogenesis. The receptor-binding domain of S protein has received greater attention as it initiates host attachment and contains major antigenic determinants. However, investigating the therapeutic potential of fusion peptide as a part of the fusion core complex assembled by the heptad repeats 1 and 2 (HR1 and HR2) is also warranted. Along with receptor attachment and entry, fusion mechanisms should also be explored for designing inhibitors as a therapeutic intervention.
Key message:
In this article, we review the S protein function and its role in mediating membrane fusion, spread, tropism, and its associated pathogenesis with notable therapeutic strategies focusing on results obtained from studies on a murine β-Coronavirus (m-CoV) and its associated disease process.
Keywords: Cell-cell fusion, Coronavirus, Fusion peptide, Murine CoV (m-CoV), Pathogenesis, SARS-CoV, SARS-CoV-2, Spike protein, Virus-host attachment protein
Introduction
The new coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus, SARS-CoV-2. It is about 80% genetically identical to SARS-CoV and displays clinical symptoms similar to those reported in previous outbreaks of Coronaviruses (CoVs), namely, SARS-CoV and MERS-CoV.1–8 The severity of CoV infections depends on virus-mediated tissue damage as well as the antiviral immune inflammation, which are influenced by viral tropism, infectivity, virus spread, and specificity of host responses, all of which are majorly regulated by the CoV Spike (S) protein. S protein is a multidomain Class I viral fusion protein that exerts its effect stepwise, by a concerted action of individual domains.9–11 Considering the highly infectious nature of CoVs and their associated lethality and ability to emerge from zoonotic hosts, SARS-CoV-2 has once again conjured the entire scientific community in an urgency to develop effective therapeutics against CoVs.
The vast reservoir of research contributed by Corona virologists over the years where Mouse Hepatitis Virus (MHV), a group 2-β-CoV, has been an epicenter sheds light on the importance of S protein in controlling the pathogenic properties of CoV. Murine-CoV (m-CoV) infection in mice is the best-explored experimental animal model for studying respiratory, enteric, hepatic, as well as neurological illness because of their differential organ tropism like the distinct tropism showed by the strains of human-CoVs (H-CoVs).10, 12–21 However, it is difficult to explore the disease mechanisms in humans, and the generation of an animal model is necessary where the function of individual genes can be studied. In this context, discontinuous transcription process producing subgenomic mRNA in CoV, despite their enormous genome size, has proved advantageous for developing a reverse genetic system for CoVs wherein each protein could be targeted separately to understand their contribution to pathogenic properties.22–25 A battery of recombinant strains created by adopting a reverse genetics tool has largely demonstrated that the S protein is a major determinant of pathogenic properties associated with m-CoV.
S protein of SARS-CoV-2 and SARS-CoV share about 74% to 76% amino acid identity in their receptor-binding domain (RBD). Still, they show significant differences in their ability to infect and transmit in humans.2, 4, 26, 27 Both the SARS strains identify ACE2 receptor, SARS-CoV-2 binds to it with 10 to 20 folds more affinity.8,11,27–30 An interesting question that stems from this observation is, can S protein alone contribute to the higher virulence of SARS-CoV-2? A lack of a suitable animal model to study H-CoV pathology makes it challenging to answer this question. However, owing to the high similarity in recombination potential and replication kinetics of a fellow β -CoV, MHV, we can draw parallels between H-CoV and m-CoV S proteins to gain a better understanding of the unanswered questions related to virus entry, replication kinetics, pathogenesis, host immune responses, and virus persistence.5, 31–33 This review presents insights into the contribution of S protein in the resultant immune inflammation and pathogenesis based on the notable contributions over the last three decades with particular emphasis on the presence of two consecutive prolines in the fusion peptide of m-CoV Spike. Coronavirus induced cell-cell fusion mechanism offers a potential target for antiviral therapy. This review will discuss how knowledge from m-CoV studies holds lessons to understand SARS-CoV pathogenesis.
CoV Virions and Genome
The family name Corona is because of the outward radial projection of S protein, which gives the virion a crown-like structure. CoV virions can be spherical or pleiomorphic, with typical sizes ranging from 60 to 200 nm.20, 34, 35 Their genome is an internal 5' capped single strand of helical RNA with a length of 26 to 32 kb, complexed with Nucleocapsid (N) protein encoded by gene seven in MHV and nine in SARS. The genome comprises six to ten open reading frames (ORFs) where the ORF1 forms 2/3rd of the genome coding for nonstructural polyproteins with helicase, polymerase, and replicase functions. The latter one-third of the genome encodes structural proteins in the order Hemagglutinin Esterase (HE), S, the envelope protein (E), transmembrane glycoprotein (M), and nucleocapsid phosphoprotein (N). ORF2 may or may not be functional in all strains of CoVs; in certain MHV strains it encodes a 65 kD HE protein that forms smaller spikes on the surface, but their function in the life cycle is not well understood. The genome is packaged within a lipid envelope containing S, E, and M encoded by genes 3, 5, and 6, respectively.20, 34, 35 The M and E proteins are essential for virus assembly and S protein functions in virus entry. Some CoV strains also have an Internal protein (I) of unknown function. 36 Figure 1A and B depict an overview of the virion structure and genome organization.
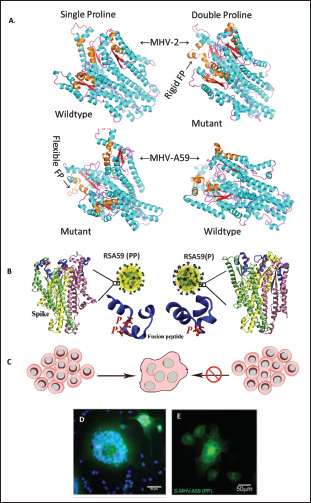
Figure 1. CoV Virion, Genome Organization and Functional Domains of Spike Protein. (A) Pictorial Representation of the Generalized Structure of the CoV virion. CoV is Somewhat Pleomorphic Containing an Internal Helical Positive-Stranded RNA Genome Complexed with Nucleocapsid Phosphoprotein (N), Surrounded by the Viral Envelope Containing Spike (S), Envelope (E) and Membrane (M) Proteins as Specified in the Text. (B) Schematic Organization of the Genes in the m-CoV and SARS-CoV. The CoV Single-Stranded Genome Encodes for Several Nonstructural Proteins (yellow), and Four Structural Proteins: Spike (S; Blue), Envelope (E; Orange), Membrane (M; Green), and Nucleocapsid (N; Purple). Some MHV Strains Also Encode a Hemagglutinin Esterase Protein (HE; Grey). (C) and (D) Line Diagram Showing the Spike Protein Organized into Two Subunits S1 and S2 Post Cleavage Between the Domain Linker Regions. Each Functionally Important Domain of the Protein is Marked and Numbered as per MHV-A59 and MHV-2 (C); SARS-CoV-2 and SARS-CoV Spike Sequence Location (D). The Percentage Identity and Similarity Between the Two Proteins are Indicated for Different Segments.
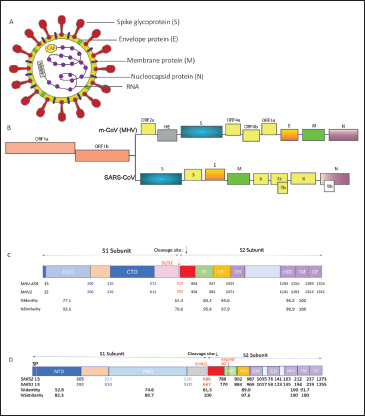
Source: Figure Designed by Fareeha Saadi and Debnath Pal.
Spike Function in Attachment, Fusion, and Entry into the Host Cell
S protein is a large, multifunctional, highly glycosylated type I transmembrane protein incorporating 21 to 35 N-glycosylation sites.37–40 They assemble into trimers on the viral surface to form the characteristic “corona” and mediate virus-host attachment through the ectodomain. The ectodomain of all CoV S proteins share the same two domains: An N-terminal S1 domain responsible for binding the receptor on the host surface and a C-terminal S2 domain responsible for fusion of their envelope with the host cell membrane to deliver their nucleocapsid. 41 As a Class-I fusion protein, S protein has the typical α-helical secondary structure and contains characteristic two heptad repeats comprising of repetitive heptapeptide with hydrophobic residues in the S2 subunit that aid the formation of coiled-coil structure to participate in the fusion process. 9 The characteristic postfusion structures of the HR have been solved for SARS-CoV and m-CoV, they form the characteristic six-helix bundle. Their functional roles in MHV and SARS-CoV were confirmed by mutating key residues and by inhibition experiments using HR2 peptides.42–45
The S1 contains two subdomains, an N-terminal domain (NTD) and a C-terminal domain (CTD), both of which can function as receptor binding domains (RBDs) in different strains of CoVs.4,8,46–53 For example, MHV uses the S1 NTD as the RBD to bind to the carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) in contrast to its close relatives SARS-CoVs and MERS-CoV who have the RBD in their S1 CTDs.8, 54–57 After the binding of the spike with the receptor, S2 triggers the fusion process either by entering the host cell directly or after internalization through endocytosis. 58 Additionally, S protein is known to become fusion competent by cleavage.58, 59 Some β- and all γ-CoVs spike have a furin cleavage site between the S1 and S2 domains, which is typically recognized and cleaved by a Golgi-resident host protease, Furin.7, 60, 61 Interestingly, within the β-genera, two closely related m-CoV species, MHV-2 and MHV-A59, display different cleavage requirements for infectivity. 62 MHV-A59 strain shows pH-dependent and independent fusion processes. 63 Whereas, in MHV-2, the entry is shown to be dependent on the endosomal proteases, Cathepsin B, and L that are activated at low pH. 64 In SARS-CoV also, the fusion process is dependent on Cathepsin L and the proteolytic processing of the spike in sequential cleavage events. 65 The requirement of endosomal proteases for efficient entry by MHV-2 and SARS-CoV could be attributed to their uncleaved spike.
Kathryn Holmes and Larry Sturman, 66 in very early studies, showed that MHV-A59 S protein is proteolytically cleaved by furin into S1 and S2 subunits at RRAHR (Arginine-Arginine-Alanine-Histidine-Arginine) during the virus attachment to the host membrane. This cleavage property was later shown to be essential for cell-cell fusion in MHV-A59 infection. Recent studies have shown that SARS-CoV-2 S protein also contains a furin recognition site between S1 and S2 subunits, which was absent in SARS-CoV. 11 However, SARS-CoV shows trypsin mediated sequential cleavage of Spike at two discrete locations. 67 The first cleavage (R667) takes place at the S1/S2 boundary by Cathepsin.64, 67 Later, another cleavage site (S2’) was recognized within the S2 domain as a second cleavage event at R797 that exposed the fusion peptide required for entry. Reports also suggest that a cleavage mediated by Elastase at T795, next to the fusion peptide, can significantly alter the cleavage activation in SARS-CoV. 68 Additionally, the S proteins of SARS-CoV use the transmembrane protease/serine subfamily member 2 (TMPRSS2) colocalized with ACE2 on the cell surface to enter the host cell.69, 70 TMPRSS2 family proteases are majorly found in the respiratory tract and lungs which can explain their tropism in the lung tissues. It is recently shown that SARS-CoV-2 entry also employs the TMPRSS2 activity and hence the similarity in tropism. 4 The two cleavage events, one by furin and second by TMPRSS2 might increase the infectivity of SARS-CoV-2. 26
Overview of m-CoVs and H-CoVs Belonging to the Genera β-Coronavirus
m-CoV, SARS-CoV, MERS-CoV, and SARS-CoV-2 belong to the family Coronaviridae, order Nidovirales, and genus β-coronavirus. The other three genera in the family Coronaviridae are α-coronavirus, γ-coronavirus, and δ-coronavirus. Among the four genera, α and β-coronaviruses have majorly emerged to cause several gastrointestinal, respiratory, hepatic, and CNS illness in mammals. Whereas γ-coronaviruses are known to infect only avian species, and δ-coronaviruses infect both mammalian and avian species. 10
Five naturally occurring strains of m-CoV have been identified and thoroughly studied. MHV infects many vertebrate hosts and induces several diseases oscillating in severity. MHV-1 causes respiratory disease. MHV-2 is purely hepatotropic and causes severe hepatitis and lethality, whereas MHV-3 is a neurotropic strain that results in hepatitis, vasculitis, initial ependymitis, meningitis, and encephalitis but not white matter lesions.15, 19, 71 Temperature-sensitive JHM strain causes severe encephalitis and demyelination but is unable to infect the liver and is highly lethal. It infects nonneuronal cells and has a special affinity toward astrocytes.21, 72 MHV-A59 is dual hepato-neurotropic.12, 19, 73
So far, seven humans CoVs have been identified. NL63 and 229E (α-CoVs) and OC43, HKU1 (β-CoVs) cause mild common cold symptoms whereas, SARS-CoV, MERS-CoV(β-CoVs) are known to cause acute respiratory syndrome and the newly emerged SARS-CoV-2 (β-CoV) with its high zoonotic potential causes severe acute respiratory syndrome associated with heightened lethality.1, 10 The periodical emergence of the new CoVs among humans can be attributed to their great genetic diversity, high pervasiveness, and recurrent genetic recombination. The risk of cross-species transmission increases further because of the increase in the human-animal interaction, thus enabling the viruses to select the most favorable receptors in the host. While m-CoVs are known to infect cells in the hepatic and central nervous system primarily, SARS-CoV2, SARS-CoV, and MERS-CoV primarily target the respiratory system and present with diffuse alveolar damage.3, 35, 74 Major clinical manifestations of SARS-CoV-2 causing COVID-19 are like the previous outbreaks, including cough, pneumonia, RNAemia, respiratory distress, cardiac injury, and the serious ground-glass opacities in both lungs preceding death.75, 76 Interestingly, SARS-CoV-2 rapidly translocates to the lower respiratory tract and other organs, including the liver, gastrointestinal tract, kidney, CNS, and cardiac muscles leading to multiorgan failure, all of which can be attributed to the appropriate choice of vastly expressed host receptor by the virus.4, 8, 30, 77
Comparison of S Protein Between m-CoV; MHV-A59 and MHV-2 and H-CoV; SARS-CoV and SARS-CoV-2
MHV-A59 (gene bank accession number 9629812, RefSeq: NC_001846.1) and MHV-2 (gene bank accession number: AF201929) show 94% to 98% sequence homology in their replicase genes, 83% to 95% sequence homology of genes 2a, 3, 5b, 6, and 7. However, considering the significant differences in their cleavage and fusion properties, both of which rely on S protein, the S gene identities were compared (Figure 1). As discussed above, the major difference between the two strains is cleavability and fusogenicty. MHV-2 lacks the furin cleavage site, unlike MHV-A59, and their cleavage depends on Cathepsins. Both strains’ cleavage signals showed a 64.3% sequence identity. Their fusion peptides are 83.3% identical. Like MHV-2, SARS-CoV (NCBI Accession: NP_828851.1) also depends on Cathepsins for their spike cleavage into S1 and S2. When compared with SARS-CoV-2 (NCBI Accession: QIK02964.1, RefSeq: YP_009724390.1), which shares a furin cleavage site similar to MHV-A59, they were 61.5% identical in their cleavage signals and 78.9% identical in the fusion peptide. We also compared the RBD, HR1, HR2, and TM domains; their respective identities and sequence similarities are denoted in Figure 1.
Corollary Between SARS-CoV-2 and m-CoV Disease Kinetics
Intracranial (i.c.) inoculation of MHV-A59 and RSA59 in mice is used as extensively studied prototypic animal model to study virus-induced inflammation of the brain, spinal cord, optic nerve, and retinal ganglionic cells of the eye. Acute disease symptoms during three to seven days postinfection (p.i.) are characterized by robust host immune responses accompanied by pathological consequences including meningitis, encephalitis, and myelitis with or without hepatitis. Infectious viral particles are cleared within the first 10 to 14 days; however, at this time, mice begin to develop demyelination, either clinical or accompanied by chronic hind limb paralysis. Demyelination and consecutive axonal loss reach its peak at day 30 p.i. when the infectious particle is completely cleared, and innate immune inflammation is majorly resolved, this represents the second or chronic disease phase. On day 30 p.i., only viral RNA persists at a low level in the spinal cord.18, 19, 78 An intermediate stage may be present, which is governed by the continuous infiltrating T cells into the CNS and helps connect the acute host-inflammatory responses with the chronic anti-inflammatory responses. This phase, however, is less explored.
Likewise, SARS-CoV-2 infection is divided into three phases. 79 The first phase is represented by the initial infection of the mucosal membranes and respiratory tract and dissemination of virus into the peripheral blood. During the second pneumonia phase, the virus replicates profusely and causes an upregulation of host-inflammatory responses. If the antiviral immune response is proficiently active, and the virus replication is reduced, the patient enters the final recovery phase; otherwise, the uncontrolled cytokine storm results in lethality.28, 80 Results from case reports have shown that the timely detection of virus RNA is crucial as the viral titer lies within the detection limit only for a short duration of time, i.e., in between the first two phases and drops down drastically by the second week.79, 80 Figure 2 depicts the characteristic neuroinflammatory pathology induced by m-CoV MHV-A59, and a comparative schematic of viral replication with disease kinetics of m-CoV and SARS-CoV-2.
Figure 2. Disease Kinetics and Pathological Manifestations of m-CoV MHVA59 and Its Corollary with SARS-COV-2 (A) Shows the Kinetics of MHV-A59 Viral RNA, Infectious Particles, and Acute Stage Neuroinflammation with the Onset of Demyelination and Chronic Stage Peak of Demyelination Disease Following Intracranial Inoculation. (B) Shows the Proposed Kinetics of SARS-CoV-2 Viral RNA, Infectious Particles, and Disease Course with the Evidence of Cytokine Release Syndrome. Infected Humans Succumb on 21 Days p.i. Upon Inefficient Anti-inflammatory Responses (red dash line) or Enter the Recovery Phase Upon the Development of Herd Immunity.
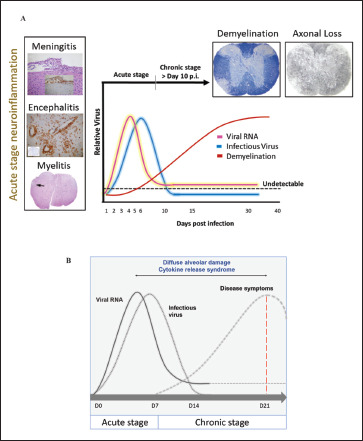
Source: Figure designed by Fareeha Saadi.
The studies we have at our disposal about SARS-CoV-2 infection mostly tell us about the disease kinetics extending up to 21 days. However, m-CoV has significantly shed light on the fact that major disabling chronic pathology of demyelination and axonal loss associated with MHV-A59 and RSA59 reaches its peak rather silently when the infectious viral particles go below the detection limit, acute inflammation is resolved, and only viral RNA persists at a low level. 81 So, the question is, do the same phenomena exist in the patients recovered from COVID-19? Though it is too early to comment on cases of COVID-19 patients, mounting evidence indicates a possibility. A study showed that patients affected with SARS-CoV-2 presented with neurological symptoms like headache (8%) and confusion (9%). 82 Also, several studies have shown that SARS-CoV and MERS-CoV can infect and replicate in the CNS of both humans and animal models, especially the brainstem.3,74,83–86 While many animal models of SARS-CoV were tested, none were able to mimic the disease symptoms like humans, but MHV-1 produced a SARS-CoV like lethal disease in mouse and has provided few insights into the host cytokine responses against the virus. 87 Still, there is a shortage of studies directly correlating the replication of the virus with the resultant pathology as the disease caused by both SARS-CoV in humans and MHV-1 in mice are milder than SARS-CoV-2. Moreover, the neurological manifestations of SARS-CoV-2 infection is finding more relevance by the day and therefore the consequences of SARS-CoV-2 infection can be analyzed from the eye of m-CoV, MHV-A59 to uncover aspects of disease pathology that may be instrumental in designing therapeutic strategies.
Spike Controls m-CoV Pathogenesis
While a vast range of studies was identifying and emphasizing on the spike function in infection and associated pathology, less was clear about the role of S protein in CoVviral antigen spread, cell-cell spread, and associated pathogenesis. The study on natural recombinants obtained by co-infection of MHV-2 and LA-7 strains opened avenues for RNA recombination, where distinct mutations could be linked to altered pathologies, if any, in a measure to understand the role of distinct spike domains in the disease outcome. 88 Paul Masters and group developed the first reverse genetic system for MHV using RNA recombination.25, 89 This has been extensively used to exchange specifically the structural and nonstructural genes among the MHV strains and to introduce single amino acid substitutions. 90 The large genome size of the CoVs made it difficult to use the reverse genetics approach as a tool to design the whole genome infectious cDNA clone. Still, in the last few years, full-length infectious clones for many coronaviruses, including MHV, TGEV (transmissible gastroenteritis coronavirus), NL63, and SARS-CoV have been developed.91–94 Very recently, a complete genome clone of SARS-CoV-2 and a SARS-CoV-2 mNeonGreen reporter virus has been generated. 95
Using targeted RNA recombination, two isogenic recombinants of MHV, RSA59, and RSMHV2 (background is from demyelinating strain MHV-A59) that differed only in the S gene were generated. RSA59 had the spike from MHV-A59, and RSMHV2 had the spike from the parental nondemyelinating MHV-2 strain.96, 97 The replication, virulence, and spread of the recombinants were like the parental spike strains (Figure 3).81, 96, 98 These recombinant viruses have served as suitable tools to study the role of Spike mediated pathogenesis which has been thoroughly reviewed previously.18, 19 From the perspective of neuropathogenesis, the major highlights from the comparative studies are that spike gene mediated entry in the brain or the induction of encephalitis may not be the restricting factors for causing demyelination as both strains induced acute stage neuroinflammation but only RSA59 caused chronic stage demyelination. Other functions of spike like cell tropism, virus transport along the axons associated with virus spread, fusion, and persistence may attribute to the differential chronic stage pathologies between the two strains. RSA59 takes advantage of the synaptically linked neuronal and glial cells circuits to travel from the neuronal body centripetally along the axons to spread from brain to the brainstem during day 3 p.i. It reaches the spinal cord grey matter by day 5 upon releasing at the nerve endings and quickly translocate to the white matter microglia and oligodendrocytes by day 7 p.i. as a function of its cell-cell fusion (Figure 4).99, 100, 101, 102 RSA59 was also capable of retrograde axonal transport from the optic nerve into the retinal ganglionic cells and caused optic nerve inflammation and demyelination. Additionally, RSA59 induces a persistent activation of CD11b+ microglia/macrophages in the demyelinating regions which showed a mechanism of macrophage-mediated myelin stripping in RSA59 infection (Figure 4).99, 19 RSMHV2 and RSA59 also differ in their cleavage signal site. As mentioned before, RSA59 S is cleaved posttranslationally into S1 and S2 subunits, but RSMHV2 S protein does not and is unable to cause fusion in vitro.103, 104 Penn 97-1, a recombinant strain with S1 from demyelinating strain MHV-A59 and S2 domain from MHV-2 was nonfusogenic and did not induce demyelination, demonstrating the role of S2 domain in the fusogenic property of MHV-A59.16, 105 Thus, variable regions of the S2 domain or differences in the cleavage signal site and fusion domain between RSA59 and RSMHV2 could be candidates to explain the differential axonal transport and demyelination potential.
Figure 3. Differential Neuropathological Outcomes of m-CoV Spike Protein Recombinants, Demyelinating Strain RSA59 and Nondemyelinating Strain RSMHV2. Serial 5 µm Thick Brain Sagittal Sections From RSA59 (C, E, G) and RSMHV2 (D, F, H) Infected Mice at Day 7 p.i. Immunostained for leukocyte common antigen (LCA) or CD45 (C-F) or CD11b (G, H) are Shown. LCA Staining Shows Acute Encephalitis in RSA59 (C, E) and RSMHV2 (D, F) Infected Mouse Brain Characterized by the Presence of Inflammatory Infiltrates Throughout the Parenchyma. Serial 5 µm Thick Cross-Sections From RSA59 and RSMHV2 Infected Mouse Spinal Cord at Day 30 p.i. (Peak of Demyelination) were Stained with luxol fast blue (LFB) for Myelin (I, J) or LCA (K, L). Schematic of the Genomic Organization Depicts That RSA59/DM Strain (A) and RSMHV2/NDM Strain (B) are Isogenic Except for the Spike Gene but Show Significant Differences in the Chronic Stage Pathology. DM Contains the Spike Gene From Parental Demyelinating Strain MHV-A59, and NDM Contains the Spike Gene From Parental Nondemyelinating Strain MHV-2, Both Strains Contain Heterologous Enhanced Green Fluorescent Protein (EGFP). The Expression of EGFP Helps to Trace the Virus Particle in Real-Time in Situ. The Majority of LCA+ Inflammatory Cells Stained Positive for the Macrophage/Microglia Marker CD11b in Both RSA59 (G) and RSMHV2 (H) Infected Mouse Brain, Which is Characteristic of Neuroinflammation and Encephalitis. Large Demyelinating Lesions (I) with LCA+ Cells (K) were Observed in RSA59 Infected Mouse Spinal Cord White Matter. In Contrast, Normal Myelin Staining (J) was Observed in RSMHV2 Infected Mouse Spinal Cord with Only Rare Scattered LCA+ Cells in the Grey Matter (L). C and D are Laser-scanned (Aperio) Images of Glass Slides. Original Magnifications for E–H are 100x. Original Magnification for I–L is 40x.
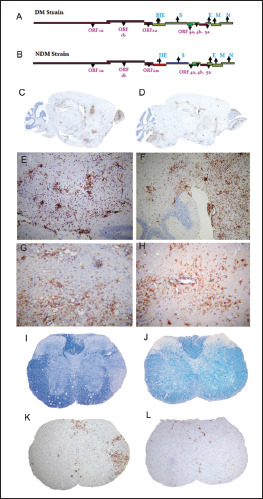
Source: Figure adapted from Das Sarma et al. 99
Figure 4. Spike Regulates the Trafficking of RSA59 in the White Matter and Subsequent Neuropathology. Infected Mouse Spinal Cord at Day 6/7 p.i. were analyzed for the Spread of the Virus. EGFP Fluorescence (Green) Identifies Virus-infected Cells of Infected Mice Spinal Cords. Immunolabeling With Neuronal Cytoskeletal Markers Shows the Co-localization of RSA59-EGFP with NFM in (A) and Synaptophysin in (E). Insets Show Proof of Transport of RSA59 Along the Neuronal Cytoskeleton to Get Released at the Nerve End (Synaptophysin, E) to Infect the Oligodendrocytes in the White Matter. RSA59 Infected Spinal Cord, Showed the Viral Antigen Translocation Denoted by EGFP Expression From Grey to White Matter (B). The Area of EGFP-positive Cells From A is Marked in a Corresponding LFB-stained Section (C). In Contrast, RSMHV2 is Predominantly Restricted to the Grey Matter and Grey-white Matter Junction, as Shown by EGFP-positive Cells (F). The Area of EGFP-positive Cells From D is Marked in the Corresponding LFB-stained Section (G). Ventral Horn White Matter (D) Reveal Olig2 Positive Nuclei Surrounded by EGFP Positive Cytoplasm (Arrows) Demonstrating RSA59 Infection of White Matter Oligodendrocytes. (H) Demonstrates EGFP Fluorescence in the Axonal Pattern with Surrounding Co-localization of PLP in the Myelin Sheaths Showing Axonal Transport of RSA59. Thirty days p.i. Spinal Cords were Stained for Toluidine Blue (1 µm Thick) to Delineate the Preservation of Myelin, Axons, and Axon-myelin Coherence (I, J, P, and Q). Sections Showed That Axon-Myelin Coherence is Disrupted in Demyelinated Plaques of RSA59 Infected Mouse Spinal Cord (I, J). In RSMHV2 Infected Mouse Spinal Cord, Myelin Remains Relatively Preserved with Rare Examples of Early Axonal Degeneration Characterized by Loss of the Central Axon and Collapse of the Myelin Sheath (P, Q). Representative Foci of Demyelination and Axonal Injury were Selected From the Toluidine Blue-stained Sections and Processed for Ultrastructural Studies by High-resolution TEM. Demyelinating Plaque in RSA59 Infected Mouse Spinal Cord (K–M), Showed Activated Macrophages, Complete Axonal Degeneration with Only Residual Empty Vacuoles, Extensive Loss of Myelin, Hypomyelination, Naked Axons with No Myelin Sheath at all. In Contrast, RSMHV2 Infected Mouse Spinal Cords Show Only Rare Examples of Early Axonal Degeneration with Collapsed Myelin (R). K is Magnified Further in M and N. (M) Shows a Macrophage (Upper Portion of the Figure) is Observed Surrounding an Intact Axon (Lower Portion of the Figure) with Uncompacted Myelin. Myelin Figures are Observed Within the Cytoplasm of this Macrophage Indicative of Prior Engulfment of Myelin. (O) Shows a Higher Magnification Image Demonstrating Close Apposition of the Macrophage Cell Membrane and an Outer Layer of Uncompacted Myelin where the Macrophage has Formed a Pseudopodium Showing Evidence of Myelin Stripping. The Axoplasm and Axolemma are Intact, and the Adjacent Myelin Shows Vesiculation Indicative of Myelin Degeneration. These Images Demonstrate a Macrophage-mediated Myelin Stripping as a Mechanism of Demyelination in RSA59 Infection. Mechanistically, the Role of the Presence of Activated Microglia/Macrophage within the Demyelinating Plaques of RSA59 Infected Spinal Cords, as Shown in Figure 3, is Clarified Here. Original Magnification for A, D, E is 630x, B is 40×, C and G is 20×, F, I, P is 100×, H is 200×, J and Q is 1000×, M is 3000×, K is 5000×, L and R is 6000x, N is 15000x and O is 40000x.
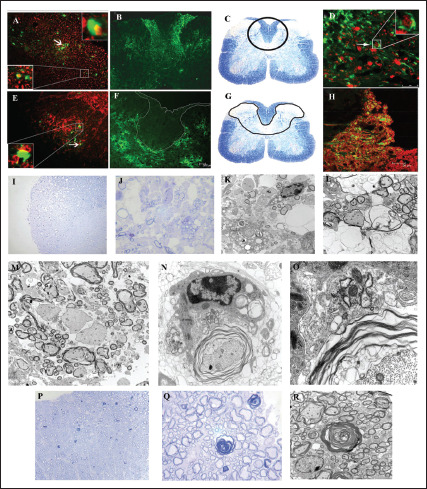
Source: Figure Adapted From (Das Sarma et al., 2009; Das Sarma et al., 2008; Kenyon et al. 2015).99, 100, 101
Delineating the Minimum Essential Motif in the S2 Domain of Spike Protein Responsible for Fusogenicity and Demyelination
The strategy for inhibiting the fusion of CoV S2 domain is based on the current understanding of the conformational transition that takes place during the fusion event. 34 The basis for this transition is the formation of the six-helix bundle viral fusion core driven by hydrophobic interactions between HR1 and HR2. 9 Thus, all efforts of researchers have been directed in disrupting the formation of the fusion core, and this has been facilitated by the fact that HR regions are the better-conserved regions of the S protein and more amenable for peptide design. The identification of the double-proline site opens a new possibility of mimetic peptide design with the potential to disrupt Spike fusogenicity.
In 1990, for the first time, it was indicated that MHV-A59 contains an internal FP (929–944) that could be the fusion domain considering its hydrophobicity and location adjacent to the heptad repeat domains. 106 Previous studies had shown that substitution of 936 methionine residue with lysine (M936K) or leucine (M936L) in the fusion domain did not affect fusion, but the substitution of 938 proline with lysine (P938K) partially impaired the fusogenicity. 107 The fusion domain of MHV-A59 contains two consecutive central proline (938, 939), but that of the MHV-2 (nonfusogenic and nondemyelinating) strain contains only one proline (976). A series of 3-dimensional in silico and biophysical nuclear magnetic resonance (NMR) studies were carried out on MHV-2 and MHV-A59 Spike fusion domains in their wild-type and mutant states which revealed that the fusion peptide might contribute in a significant way toward the fusogenicity of MHV-A59(Figure 5). 62 The double proline fusion peptides were found to be markedly rigid than single proline. The presence of two proline in the Spike of MERS-CoV, SARS-CoV and HCoV-HKU1 were also shown to stabilize the S protein in the prefusion state. 108 Recently, as a proof of concept for the use of S-2P as a vaccine for SARS-CoV-2 it was shown that an mRNA vaccine expressing the MERS S-2P elicits a protective response in mice. Similarly, SARS-CoV-2 S-2P expressing mRNA served as a potent immunogen inducing both neutralizing activity and CD8 T cell responses in mice. It protects mice from both upper and lower airway SARS-CoV-2 infections. The mRNA vaccine has shown promising results in phase II clinical trials and is soon expected to be in the phase III efficacy evaluation phase. 109 The S-2P design has been used in several vaccine strategies including the
Figure 5. Two Consecutive Prolines in the Centre of the Fusion Peptide of m-CoV Regulate Fusogenicity. (A) Diagram Showing the Illustration of MHV-2 and MHV-A59 Spike Fusion Domain Structures in the Trimeric Quaternary State. Frames From a 1µS Long Molecular Dynamics Simulation Trajectory were Randomly Drawn to Depict the Shown Structures. The Fusion Peptides are Marked in Brown. The Single Proline Containing Fusion Peptides Showed more Flexibility than the Double Proline Cases in the Trajectory. (B) Shows the Illustration of Spike of RSA59 (PP) Wildtype Recombinant and RSA59 (P) Single Proline Mutant of RSA59. Only RSA59(PP) can Form Syncytia (C, D). Spike Alone can Traffic to the Cell Surface and Cause Syncytia Formation was Shown by the Transient Overexpression of Spike of MHV-A59 (S-MHV-A59 [PP]) in HeLa Cells (E) by YFP.
95% effective, BNT162b2 by BioNTech/Pfizerand 94% effective mRNA1273 by Moderna/NIAID, the first two vaccines approved by the FDA upon exercising their emergency authority protocols.110–112
In earlier studies, the role of two Proline infusogenicity was demonstrated by generating a mutant by targeted RNA recombination, RSA59 (P), which had one deleted proline. RSA59 (PP), i.e., the wild-type recombinant MHV-A59 strain and RSA59 (P), were compared in a series of in vitro and in vivo experiments. RSA59(P) had significantly reduced fusion and syncytia formation. 62 Both the PP ad P mutants have the same cytoplasmic tail which has the retention signal for controlling protein trafficking, therefore, RSA59 (P) slower replication and syncytia formation could not be only linked to the movement of S Protein to the cell surface but a combinatorial outcome of low replication, destabilized S protein structure and inefficient trafficking (Figure 5).62, 113 RSA59 (P) showed limited ability to spread across the brain parenchyma; viral antigen staining was mostly restricted to the site of infection and in the meninges and it induced significantly low demyelination pathology compared to the wild-type. Additionally, RSA59(P) showed significantly less inflammation of the optic nerve and induced no damage to the RGC. The virus did transport retrogradely, but its reduced ability toward cell-cell fusion did not allow it to move until the retina and infect RGCs. 114 The proline thus plays an essential role in fusion, which further affects the replication, spread across the brain and spinal cord, retrograde transport to the optic nerve and RGC, and the ability to cause demyelination.
These reports, for the first-time, unraveled the role of two proline in the backbone of S protein fusion peptide in m-CoV pathogenesis. M-CoV fusion mechanism offers a potential therapeutic target. Dissecting the minimum essential required for fusogenicity is a significant advancement for designing a mimetic peptide to restrict CoV infection.
S2 and Not S1 Domain of Spike Can Serve as Better Pan-CoV Drug Target
Despite considerable progress in the understanding of spike protein functions, it may still be limited in our ability to predict the effect of mutations that Spike, and the virus genome might acquire. Therefore, insights from decade-long studies on other CoVs and understandings from them hold a compelling case for improving our understanding of the current COVID-19 pandemic and assess its pathology and course of evolution.
Given the rapid expected evolution of the SARS-CoV-2, it is imperative to ask which of its two domains, S1 or S2, is likely to evolve faster. This can be answered from sequence comparisons with other existing members of the β-coronaviruses. The receptor-binding domain is most variable across the CoVs, and this has also been corroborated by the limited success we have achieved in drugs’ discovery against the receptor-binding domain as target.10, 11 Antibodies capable of recognizing the SARS-CoV receptor binding domain have failed to recognize the SARS-CoV-2 receptor binding domain, although they share >74% identity, thereby showing low cross-reactivity. 30 In comparison, the fusion domain (S2) has more conserved regions, and studies in m-CoV have demonstrated the importance of specific residues, including our illustration of the high significance of two prolines in the fusion peptide and Spike associated disease pathology.62, 114–116 The fusion domain thus promises to be a better cross-functional success as the potential pan-coronavirus therapeutic target. However, the receptor-binding domain contains most antigenic determinants because of its larger surface exposure in the prefusion state. Another aspect of Spike that strengthens our premise for S2 as a therapeutic target is based on the profound structural and functional similarities it shares within all class I viral membrane fusion proteins. In all cases, S2 is in a metastable state that exists in prefusion and postfusion conformations. The prefusion structures are similarly triggered for state change through comparable conformational rearrangement, folding to a highly similar six-helix bundle post-fusion structure with exposed fusion peptides. The complexity and intricacy of the fusion mechanism appear to be specialized and inherited, although its independent evolution in viruses cannot be completely ruled out. Spike evolution is justifiable; therefore, an intriguing question because a Spike from neurotropic strain MHV-JHM exists that can mediate receptor-independent virus entry into cells that do not express its receptor. 117 This suggests that receptor binding need not be essential for a virus to fuse, infect, and replicate, and its sole purpose is cell-recognition. However, such a Spike with S2 alone may be inefficient for the fusion.
It has been shown that the HR1/HR2 regions of class I viral fusion proteins of enveloped viruses like HIV, respiratory syncytial virus (RSV), 21 Ebola virus, 100 paramyxoviruses SV5, 101 Nipah virus, 22 and murine hepatitis virus (MHV) 23 serve as efficient drug targets. Likewise, the heptad region in the spike S2 domain is highly conserved and peptide sequences based on these segments have been most tested for inhibiting CoVs. During SARS-CoV 2003 outbreak, three independently developed peptides based on the heptad repeat two region were found to inhibit CoV fusion.118–120 Later another three peptides based on the same region were specifically found to block the six-helix bundle formation. 121 SARS-CoV-2 not only binds to the ACE2 receptor with higher affinity, but it also displays a significantly higher capability of membrane fusion compared to SARS-CoV which is elucidated by the formation of syncytium in culture. Thus, suggesting that the fusion machinery of SARS-CoV-2 can serve as an important drug target. This could be partly attributed to the presence of few alterations in the SARS-CoV-2 HR1 region. The HR2 peptide sequence is identical in both SARS-CoV and SARS-CoV-2 and the mutations in HR1 might have resulted in rendering the virus fusogenic. Recent study showed that peptides targeted against the HR1 domain were effective against SARS-CoV-2 and other H-CoVs. 29 Nonheptad repeats regions-based peptides have also been developed for fusion inhibition study, albeit to a limited degree. Two designed peptides upstream of the S1/S2 and S2’ cleavage sites have elicited anti-fusogenic activity likely because of interference with S1/S2 cleavage and conformational restriction for fusion. 121 Our discovery of surface-exposed double proline containing rigid segments in the S2 domain could offer a new premise for structure-based peptide design for CoV fusion inhibition. These segments could also be presented as antigens to raise antibodies and, therefore, also suitable for vaccine design. There is a need to do clinical tests using few peptides discovered – for proof of pan-COVID utility.
m-CoV Spike in Inflammation and Immunity
The integration of reverse genetics, molecular biology, and pathology has significantly contributed toward the understanding of S protein function in CoV biology, to the extent of deciphering a minimum essential motif required for entry and fusogenicity. However, Spike function is not limited to mediating entry into the cell, and it is known to regulate immunopathogenesis, which is the critical regulator of virus infection.18, 19, 26 In fact, the interaction between the virus and the innate immune system determines the outcome of the disease. While the early control of virus replication elicits a strong pro-inflammatory, Type I interferon response, it also helps promote the development of an anti-inflammatory response.122–124 The balance between these two contrasting yet complementary states of immunity is central for reinstating tissue homeostasis.125, 126 A shift toward either side can be detrimental and result in immune pathology and tissue damage.
MHV infection causes neuroinflammation as a result of pronounced activation of CNS resident immune cells, microglia, and astrocytes.72, 127 Microglia, upon activation, take the characteristic activated phenotype and start expressing microglia/macrophage-specific protein Iba1(ionized calcium-binding adaptor molecule 1), which promotes ruffling and phagocytosis.99, 19, 128, 129 Both RSA59 and RSMHV2 trigger innate immune responses predominated by chemokines like CXCL10, CXCL9, CCL5, and CCL12 and CD molecules during the acute stage. Antiviral host responses are associated with perforins and genes involved in interferon (IFN) gamma signaling. The inflammatory responses gradually decline in RSMHV2 infection after virus clearance. However, RSA59 chronic disease is represented with persistent CD11b+ microglia within the demyelinating lesions, and the production of microglia-associated inflammatory mediators. These differences could be attributed to the ability of RSA59 to translocate through the axons and evade the robust immune responses targeted to clear the virus completely. However, parallelly, regulatory, and anti-inflammatory cytokine IL-10 significantly controls the enlargement of tissue lesions in an attempt to reduce the chronic pathology.130–133 IFN responses can promote phagolysosomes maturation and autophagy in the persistently activated microglia/macrophages, which can engulf the myelin sheath, leading to demyelination. 99
Further, virus-specific T cell effector functions are essential to eliminate the infectious virus load during most acute infections.134–136 Control of m-CoV spread requires the functioning of both CD4 and CD8 T cells, were CD8 T cells are the primary effectors but require support functions from CD4 T cells.20, 137 A recent study in CD4-/- mice showed impaired virus clearance, despite the presence of functional CD8 T cells, demonstrating the importance of CD4 T cells for the efficient functioning of CD8 T responses. 138 While T cells are the helpers for the development of intact adaptive responses, their functions are acutely regulated by CNS resident immune cells to reduce their cytolytic effects. 139 However, a deviation from the optimum control of CNS cells can significantly dampen the T cell functions that may sometimes result in the establishment of persistent virus infection. 139 In a recent study, we have shown a critical role of CD4 T cells in RSA59 clearance and resultant pathology. The demyelination pathology is significantly more severe in the CD4-/- mice where phagocytic M2 phenotypic microglia/macrophages are found in abundance in the demyelinating plaques.
The studies on Spike mutants have served as a valuable tool in the elucidation of the Spike mediated mechanisms of intracellular spread and the processes that specifically allow CoVs to evade the immune system to establish successful disease pathologies. Figure 6 presents an illustration of the kinetics of immune cell infiltration and pathogenesis upon RSA59 infection.
Figure 6. Encephalitic and Demyelinating m-CoV Infection and Kinetics of Immune Cell Infiltration and Pathogenesis. Following Intracranial Inoculation of MHV-RSA59, into C57BL/6 Mice the Virus Migrates into the Brain Parenchyma and Infects the CNS Resident Cells, Namely, Astrocytes, Microglia, Oligodendrocytes, and Neurons. The First Seven Days are Marked by Rapid Replication of Virus and Heightened Innate Immune Response with Characteristic Neuroinflammation and the Production of Proinflammatory Signals Such as TNF-α, IL-6, CCl5 and CXCL10 Released by Activated Glial Cells Causing a Cytokine Storm. These Proinflammatory Signals Cause a Surge of Peripheral Innate Immune cells into the CNS, Majorly, Neutrophils, Macrophages and Natural Killer (NK) Cells. The Levels Of Neutrophils Decline as Early as Day 5, However, Enhanced Proinflammatory Signals Lead to a Shift in the Adaptive Response Wherein the Virus-specific CD4+ and CD8+ T Cells That Secrete IFN-γ, Infiltrate and Accumulate in the CNS. The T Cells Migrate in Response to Widespread Virus Replication Throughout the Brain Parenchyma and Its Transport to the Brain Stem and the Spinal Cord White Matter (Indicated in Grey Shaded Rectangle) Upon Crossing the Grey-white Matter Junction. Further, During Early Chronic Infection Stage (Days 10–15 Post Infection) the Infectious Virus Particles Start to Decline Particularly in Response to CXCL10, CCl5 and IFN-γ, and Goes Below the Detection Limit. As the Virus is Cleared From the Glial Cells, CD8+ T Cells Population Declines Drastically. CD4+ T Cells are Present in Significant Numbers Even Beyond Day 16 Showing Their Probable Association with Anti-inflammatory Response in Consort with the Microglia. Activated Microglia are Still Present in Large Numbers Around the Dead and Sick Neurons in the Demyelinating Regions at the Peak of Demyelination and Axonal Loss (Day 30).
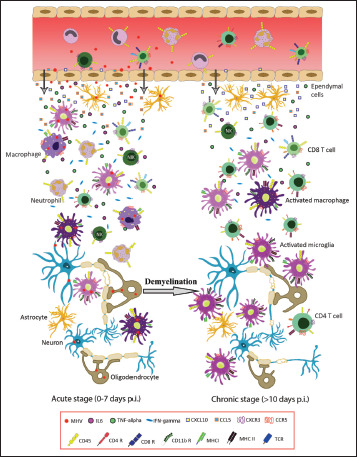
Source: Figure Designed by Fareeha Saadi.
Insights From m-CoV Help to Understand the Underlying Mechanisms of Immune Inflammation in SARS-CoV-2
As the infection kinetics of MHV-A59 and SARS-CoV-2 are quite similar, it is not surprising to note that the immune induction is also similar in the two strains based on the current literature.140, 141 SARS-CoV-2 infection initially replicates in the airway epithelium and induces virus-associated pyroptosis in the infected cells.1, 140–142 This highly inflammatory type of cell death program triggers a whole chain of pro-inflammatory responses like m-CoV, starting with IL1β and other chemokines secretion, which is followed by the infiltration of activated macrophages and establishment of a local inflammatory niche.28, 77, 141 Many cytokines and chemokines, mostly of the TH1 response, are identified in the plasma of the patients suffering COVID-19 during the acute infection, including, but not limited to, IL6, IFNγ, MCP1, and IP-10.141, 143 In most individuals, these innate immune responses clear the virus, and patients recover. But dysregulation of these tightly controlled host responses can trigger a cytokine release storm (CRS) which is represented by increasing levels of IL-6, IL-2, IL-7, IL-10, granulocyte colony-stimulating factor (G-CSF), IP-10, MCP1, macrophage inflammatory protein 1α (MIP1α), and tumor necrosis factor (TNF). 28 Inflammatory monocytes derived activated macrophages persist in the lung tissues like the activated glial cell persistence in RSA59 infection that secrete significant levels of cytokines like MCP1, IP-10, and MIP1α. 144
Interestingly, the adaptive immune system, including CD4 T cells and B cell responses, occurs concomitantly after the first stage of the infection, i.e., after the first week in SARS-CoV-2, as is also seen in m-CoV.145, 146 While CD8 T cells are required for direct killing of the virus, CD4 T cells are essential for activating both CD8 and B cells as well as cytokine production. Not much is known about the role of CD4 T cells in the COVID-19 pathology. Still, there are pieces of evidence from case reports which show the presence of T cells in the lungs and lymphopenia, indicating their putative role in the attempt to prevent tissue damage.141, 147–149
The main reason behind the extreme morbidity associated with COVID-19 is the imbalance between this see-saw where proinflammation is so high it outweighs the protective anti-inflammatory response. It has been demonstrated in SARS-CoV that virus infection downregulates the expression of ACE2, which has shown to play essential roles in lung injury and the regulation of the renin-angiotensin system (RAS).75, 150 RAS dysregulation disturbs the blood pressure and fluid/electrolyte balance and induces a heightened inflammatory response along with increased vascular permeability. Now, SARS-CoV-2's binding to the receptor is 10 to 20 folds higher, and it is not unlikely to hypothesize that it causes a much more significant downregulation in ACE2, which results in the inefficiency of RAS and dysregulation between the pro and anti-inflammatory responses which results in a cytokine storm.27–30 This highly pro-inflammatory microenvironment and permeable vasculature invite an ever-increasing number of inflammatory cells into the lungs, which ultimately clogs the airways resulting in lethality. SARS-CoV emerged, caused a few hundred deaths, and disappeared, but it seems that SARS-CoV-2 is here to stay. The efficiency of human-human transmission and an enormous virus load is still beyond the control and scope of the current database of knowledge that has been generated in years. JHMV, a highly neurovirulent CoV, shows similar uncontrolled acute inflammation state, whereas less as 10PFU of virus creates havoc in the CNS of the mice so much so that results in the death of mice at ten days p.i.5, 151, 152 Though this virus and its recombinant strains have generated enormous amounts of results to understand CoV biology, it is considered unsuitable for the study of virus persistence. MHV-2, on the other hand, triggers a robust innate immunity in the acute stage, which helps clear the virus and bring back homeostasis. 129 It does not cause any kind of progressive illness. MHV-A59, because of its mild nature has proven to be an appropriate model to study the development of a protective host response that efficiently balances the innate antiviral responses at the same time, the virus persists in the spinal cord, silently causing chronic pathology without any visible symptoms. SARS-CoV was reported to infect the brains of patients, especially the neurons, and upon intranasal inoculation in animal models could also infect the thalamus and the brain stem regions of the brain.3, 153–156 In fact, another study showed MERS-CoV tropism only in the brainstem upon low dose intranasal inoculation, indicating that the infection in the brain and not lungs led to significant morbidity in mice. 157 The mechanism of the CNS invasion is not clear, but they probably take the synaptic route, as shown in other CoVs.101, 86 A neurotropic propensity is common to many CoVs.155, 158, 159 Furthermore, some CoVs can spread trans-synaptically to the brain from the mechanoreceptors and chemoreceptors in the lung and lower respiratory airways. 160 As SARS-CoV and SARS-CoV-2, are quite similar and specific neurological symptoms have been demonstrated in COVID-19 patients, the potential invasion of SARS-CoV2 in the brain might probably lead to the acute respiratory failure of patients.160–165
COVID induced by m-CoV and H-CoV share similarities in disease, including acute immune responses, virus persistence, and the development of adaptive protective immunity. Their comparative study to investigate therapeutics is an open forum.
Discussion and Conclusions
SARS-CoV-2 has infected millions of people worldwide to date, implying that it has presented itself with a golden opportunity to diversify further. 166 With a low fidelity of the virally encoded RNA-dependent-RNA-polymerase, 167 its large size of genome and replication strategy is a boon for frequent homologous recombination. This process enables the exchange of genetic material during co-infection. Persistent infection can further lead to the accumulation of adaptive mutations. Therefore, there is a high possibility of the virus becoming more lethal and jump to more species and cause more devastation. In such a scenario arresting Spike mediated fusion, either during virus-host attachment or cell-cell spread, can be the key to early containment of infection. Else because of the significant role of the spike in tissue tropism, its modification through evolution would mean altered cell and tissue tropism, and additional association with other viral and host factors may accelerate the alteration of virus pathogenicity. Such rapid change may overwhelm our capacity for surveillance. Recent studies showed that SARS-CoV-2 is actively evolving. D614 G mutation within the viral spike protein (S1 CTD) has rendered the virus more infective than the wild-type virus, though virus’s response to antibodies was unaltered.168–171 While this ensures that vaccines currently under development can be effective against the new strain, it also raises serious concern to address the COVID-19 pandemic and future coronaviruses, by investigating their entry and fusion mechanisms in consort, which can guide new therapeutic efforts. Many other aspects of the viral fusion reaction are also potential targets for modulation. These include the inhibition of the proteases that cleave the Spike, small molecules, or lipids that alter the lipid composition or ionic environment and antibodies that recognize the S domains. The research community is poised to see what novel therapeutics can address CoV infections.
Several treatment options for COVID-19 are emerging at an accelerated rate. Many vaccines candidates have been developed. Some have even cleared the trials and approved by the FDA. The first two mRNA vaccines delivered by nanoparticle showed exceptional results so far. 171 A third adenovirus-based vaccine 172 from United Kingdom also made to the market and is showing 91% effectivity. While vaccine development is the need of the hour to control COVID-19 pandemic and return to the prepandemic normal state, our long-term goal should be addressing the pan-COVID question and that demands the development of effective broad-spectrum therapeutics which cannot be achieved without considering in detail the host immune response. It is becoming increasingly evident from the current data on SARS-CoV-2 that host responses contribute a great deal to the morbidity. Therefore, we need to consider the host responses alongside Spike functions to be fully prepared for future outbreaks. Cellular factors like the production of reactive oxygen species have been associated with many virus infections, including CoVs. An imbalance between reactive oxygen species and antioxidants’ generation leads to a dysregulation of oxidative stress, endoplasmic reticulum stress and unfolded protein response pathways. 173 Deprivation of antioxidant mechanisms and oxidative damage to the tissues is relevant to the aging process, which could be one of the reasons for making the aged more susceptible to SARS-CoV-2 infection.2,4,77,160,174,175 Also, nonstructural proteins have shown to affect tropism and pathogenesis by regulating the rate of virus replication either by interacting with cell type-specific factors or with components of the immune response.176–181 But Spike alone can as well alter fusion, entry, cell-cell fusion, pathogenesis, host immune response, and virus persistence. This was corroborated from an excellent mouse model system of RSA59/RSMHV2 infection, demonstrating demyelination pathology because of either pathogenic immune outcome, direct virus-induced cytopathic effects, or their combined outcome for RSA59, and weak effect for RSMHV2, the strains differing only in their Spike. When immunity clears the virus and prevents subsequent pathology, it also supports a paradigm where cell-mediated immunity affects pathology indicated by microglia-mediated myelin stripping and the inability of T cell effector functions to completely clear the virus which results in persistently active immune responses.18, 19, 138, 182 This establishes that inflammation is a double-edged sword and provides useful clues for hypothesizing SAR-CoV-2 immune response. The unique nature of S glycoprotein suggests a need to focus on identifying the domains of Spike that interact with the specific host cellular pathways and use the information for therapeutic intervention. Health threats from CoVs are constant and understanding the antiviral host responses targeted against Spike is essential for controlling the virus with a specific goal to preserve global health and establish economic stability.
Acknowledgments
m-CoV research in an experimental animal model has provided a timely and appropriate platform to take a step toward writing a comprehensive review of the importance of S protein in COVID biology. This work has been a collaborative effort for two decades between the Indian Institute of Science Education and Research Kolkata (IISER-K), India; Indian Institute of Sciences (IISc), India; University of Pennsylvania, USA; Thomas Jefferson University, USA and University of Colorado Denver, USA. The joint endeavor has been supported and nurtured by the Indo-U.S. Science and Technology Forum (IUSSTF) on several occasions. M-CoV research was generously funded by the Department of Biotechnology (DBT), India, Council for Scientific and Industrial Research (CSIR), India; National Multiple Sclerosis Society, USA; M. E. Groff Surgical Medical Research and Education Charitable Trust; and Lindback Foundation Career Enhancement Award, USA. The authors would like to convey their special gratitude to Dr Manmeet Singh, Mr Saurav Saswat Rout, Ms Debanjana Chakravarty, Ms Lucky Sarkar, and Dr Rahul Basu for their significant contribution toward understanding the role of Spike fusogenicity in the progression of neuropathogenesis. The authors would also like to express their gratefulness to Dr Dhriti Chatterjee, Dr Kaushiki Biswas, Dr Abhinoy Kishore, Dr Subhajit Das Sarma, Dr Afaq Hussain, Mr Soumya Kundu, Mr Abhishek Bose, Mr Sourodip Sengupta, Ms Mithila Kamble, Ms Vaishali Mulchandani, Mr Saurav, Mr Safiriyu Abass A, Dr Mahua Maulik, and Ms Soma Nag who have contributed immensely to the generation of a large volume of work on m-CoV research over the years. Authors would like to thank Dr Kenneth S Shindler, Dr Lawrence C Kenyon, and Dr Randall J Cohrs for their long-standing collaboration on m-CoV research. The success of this work has been primarily dependent on the collaborative efforts of the national and international m-COVID team and many researchers whose names are listed in the references.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Jayasri Das Sarma  https://orcid.org/0000-0002-3980-4060
https://orcid.org/0000-0002-3980-4060
Authors’ Contribution
This review was conceptualized by JDS. Literature survey was conducted by FS in consultation with JDS. FS and JDS wrote the original manuscript where DP contributed significantly to reviewing the in silico part of the review.
Statement of Ethics
Most of the m-CoV work discussed in this review either adopted from our published work or from other studies adhered to the experimental procedures and animal care and use in accordance with good animal ethics approved by the Institutional Animal Care and use Committee.
References
- 1.Lu R, Zhao X, Li J. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020; 395(10224): 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J.SARS-CoV-2: An emerging coronavirus that causes a global threat. Int J Biol Sci 2020; 16(10): 1678–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu J, Gong E, Zhang B. et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med 2005; 202(3): 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M, Kleine-Weber H, Schroeder S. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181(2): 271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S and Dandekar AA.. Immunopathogenesis of coronavirus infections: Implications for SARS. Nat Rev Immunol 2005; 5(12): 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P, X-L Yang, X-G Wang. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579(7798): 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss SR.Forty years with coronaviruses. J Exp Med 2020; 217(5): e20200537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letko M, Marzi A, Munster V.. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020; 5: 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosch BJ, van der Zee R, de Haan CA. et al. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J Virol 2003; 77(16): 8801–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F.Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 2016; 3(1): 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrapp D, Wang N, Corbett KS. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020; 367(6483): 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavi E, Gilden DH, Wroblewska Z. et al. Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology 1984; 34(5): 597–603. [DOI] [PubMed] [Google Scholar]
- 13.Stohlman SA, Weiner LP.. Chronic central nervous system demyelination in mice after JHM virus infection. Neurology 1981; 31(1): 38–44. [DOI] [PubMed] [Google Scholar]
- 14.Fazakerley JK, Buchmeiert MJ.. Pathogenesis of virus-induced demyelination. Adv Virus Res 1993; 42: 249–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tardieu M, Boespflug O, Barbé T.. Selective tropism of a neurotropic coronavirus for ependymal cells, neurons, and meningeal cells. J Virol 1986; 60(2): 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das Sarma J, Fu L, Hingley ST. et al. Mouse hepatitis virus type-2 infection in mice: An experimental model system of acute meningitis and hepatitis. Exp Mol Pathol 2001; 71(1): 1–12. [DOI] [PubMed] [Google Scholar]
- 17.Tardieu M, Goffinet A, Harmant-van Rijckevorsel G. et al. Ependymitis, leukoencephalitis, hydrocephalus, and thrombotic vasculitis following chronic infection by mouse hepatitis virus 3 (MHV 3). Acta Neuropathol 1982; 58(3): 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das Sarma J.Microglia-mediated neuroinflammation is an amplifier of virus-induced neuropathology. J Neurovirol 2014; 20(2): 122–136. [DOI] [PubMed] [Google Scholar]
- 19.Das Sarma J.A mechanism of virus-induced demyelination. Interdiscip Perspect Infect Dis 2010; 2010: 109239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender SJ, Weiss SR.. Pathogenesis of murine coronavirus in the central nervous system. J Neuroimmune Pharmacol 2010; 5(3): 336–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleming JO, Trousdale MD, el-Zaatari FA. et al. Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies. J Virol 1986; 58(3): 869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong YS, Makino S.. Evidence for coronavirus discontinuous transcription. J Virol 1994; 68(4): 2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sola I, Almazán F, Zúñiga S. et al. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2015; 2(1): 265–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawicki SG, Sawicki DL, Siddell SG.. A contemporary view of coronavirus transcription. J Virol 2007; 81(1): 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masters PS.Reverse genetics of the largest RNA viruses. Adv Virus Res 1999; 53: 245–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou X, Liu Y, Lei X. et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020; 11(1): 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang J, Ye G, Shi K. et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020; 581(7807): 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coperchini F, Chiovato L, Croce L. et al. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 2020; 53: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia S, Liu M, Wang C. et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 2020; 30(4): 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang Y, Wu L. et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020; 181(4): 894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu K and Baric RS.. Evidence for variable rates of recombination in the MHV genome. Virology 1992; 189(1): 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham RL, Baric RS.. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J Virol 2010; 84(7): 3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fehr AR, Coronaviruses: Perlman S.. An overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, eds. Coronaviruses: Methods and protocols . New York, NY: Springer New York; 2015: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang T, Bidon M, Jaimes JA. et al. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir Res 2020; 178: 104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Wit E, van Doremalen N, Falzarano D. et al. SARS and MERS: Recent insights into emerging coronaviruses. Nat Rev Microbiol 2016; 14(8): 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer F, Peng D, Hingley ST. et al. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J Virol 1997; 71(2): 996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bas T, Gao GY, Lvov A. et al. Post-translational N-glycosylation of type I transmembrane KCNE1 peptides: Implications for membrane protein biogenesis and disease. J Biol Chem 2011; 286(32): 28150–28159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oostra M,, de Haan CAM,, de Groot RJ. et al. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J Virol 2006; 80(5): 2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masters PS.The molecular biology of coronaviruses. Adv Virus Res 2006; 66: 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J Alsaadi EA, Jones IM.. Membrane binding proteins of coronaviruses. Future Virol 2019; 14(4): 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delmas B and Laude H.. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol 1990; 64(11): 5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Supekar VM, Bruckmann C, Ingallinella P. et al. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc Natl Acad Sci 2004; 101(52): 17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Cole DK, Lou Z. et al. Construct design, biophysical, and biochemical characterization of the fusion core from mouse hepatitis virus (a coronavirus) spike protein. Protein Expr Purif 2004; 38(1): 116–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.W-E Chan, C-K Chuang, S-H Yeh. et al. Functional characterization of heptad repeat 1 and 2 mutants of the spike protein of severe acute respiratory syndrome coronavirus. J Virol 2006; 80(7): 3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Z, Matthews AM, Weiss SR.. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J Virol 1999; 73(10): 8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H-X Lin, Feng Y, Wong G. et al. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD–ACE2 receptor interaction. J Gen Virol 2008; 89(4): 1015–1024. [DOI] [PubMed] [Google Scholar]
- 47.Delmas B, Gelfi J, L'Haridon R. et al. Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 1992; 357(6377): 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Godet M, Grosclaude J, Delmas B. et al. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol 1994; 68(12): 8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann H, Pyrc K, van der Hoek L. et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci 2005; 102(22): 7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babcock GJ, Esshaki DJ, Thomas WD Jr.. et al. . Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol 2004; 78(9): 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, Moore MJ, Vasilieva N. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426(6965): 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong SK, Li W, Moore MJ. et al. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem 2004; 279(5): 3197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shang J, Ye G, Shi K. et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020; 581(7807): 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng G, Sun D, Rajashankar KR. et al. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci 2011; 108(26): 10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wessner DR, Shick PC, Lu JH. et al. Mutational analysis of the virus and monoclonal antibody binding sites in MHVR, the cellular receptor of the murine coronavirus mouse hepatitis virus strain A59. J Virol 1998; 72(3): 1941–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miura HS, Nakagaki K, Taguchi F.. N-terminal domain of the murine coronavirus receptor CEACAM1 is responsible for fusogenic activation and conformational changes of the spike protein. J Virol 2004; 78(1): 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan K, Zelus BD, Meijers R. et al. Crystal structure of murine sCEACAM1a [1,4]: A coronavirus receptor in the CEA family. EMBO J 2002; 21(9): 2076–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heald-Sargent T and Gallagher T.. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 2012; 4(4): 557–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Colman PM, Lawrence MC.. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol 2003; 4(4): 309–319. [DOI] [PubMed] [Google Scholar]
- 60.Walls AC, Y-J Park, Tortorici MA. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020; 181(2): 281–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Q, Qiu Y, J-Y Li. et al. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol Sin 2020; 35(3): 337–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh M, Kishore A, Maity D. et al. A proline insertion-deletion in the spike glycoprotein fusion peptide of mouse hepatitis virus strongly alters neuropathology. J Biol Chem 2019; 294(20): 8064–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eifart P, Ludwig K, Bottcher C. et al. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J Virol 2007; 81(19): 10758–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pu Y and Zhang X.. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J Virol 2008; 82(16): 8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsuyama S, Ujike M, Morikawa S. et al. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci USA 2005; 102(35): 12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frana MF, Behnke JN, Sturman LS. et al. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: Host-dependent differences in proteolytic cleavage and cell fusion. J Virol 1985; 56(3): 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belouzard S, Chu VC, Whittaker GR.. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. . Proc Natl Acad Sci 2009; 106(14): 5871–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belouzard S, Madu I, Whittaker GR.. Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain. J Biol Chem 2010; 285(30): 22758–22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuyama S, Nagata N, Shirato K. et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 2010; 84(24): 12658–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shulla A, Heald-Sargent T, Subramanya G. et al. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 2011; 85(2): 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarma JD, Fu L, Hingley ST. et al. Mouse hepatitis virus type-2 infection in mice: An experimental model system of acute meningitis and hepatitis. Exp Mol Pathol 2001; 71(1): 1–12. [DOI] [PubMed] [Google Scholar]
- 72.Bergmann CC, Lane TE, Stohlman SA.. Coronavirus infection of the central nervous system: Host-virus stand-off. Nat Rev Microbiol 2006; 4(2): 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dandekar AA, Wu GF, Pewe L. et al. Axonal damage is T cell mediated and occurs concomitantly with demyelination in mice infected with a neurotropic coronavirus. J Virol 2001; 75(13): 6115–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding Y, He L, Zhang Q. et al. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: Implications for pathogenesis and virus transmission pathways. J Pathol 2004; 203(2): 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuba K, Imai Y, Rao S. et al. Lessons from SARS: Control of acute lung failure by the SARS receptor ACE2. J Mol Med (Berl) 2006; 84(10): 814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, Liu L, Zhang D. et al. SARS-CoV-2 and viral sepsis: Observations and hypotheses. Lancet 2020; 395(10235): 1517–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu Y, Cheng Y, Wu Y.. Understanding SARS-CoV-2-mediated inflammatory responses: From mechanisms to potential therapeutic tools. Virol Sin 2020; 35(3): 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butchi NB, Hinton DR, Stohlman SA. et al. Ifit2 deficiency results in uncontrolled neurotropic coronavirus replication and enhanced encephalitis via impaired alpha/beta interferon induction in macrophages. J Virol 2014; 88(2): 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin L, Lu L, Cao W. et al. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect 2020; 9(1): 727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Liu SM, Yu XH. et al. Coronavirus disease 2019 (COVID-19): Current status and future perspectives. Int J Antimicrob Agents 2020; 55(5): 105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Das Sarma J, Fu L, Tsai JC. et al. demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J Virol 2000; 74(19): 9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu J, Gong E, Zhang B. et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med 2005; 202(3): 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu J, Zhong S, Liu J. et al. Detection of severe acute respiratory syndrome coronavirus in the brain: Potential role of the chemokine mig in pathogenesis. Clin Infect Dis 2005; 41(8): 1089–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Netland J, Meyerholz DK, Moore S. et al. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol 2008; 82(15): 7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCray PB, Pewe L, Wohlford-Lenane C. et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 2007; 81(2): 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li K, Wohlford-Lenane C, Perlman S. et al. Middle east respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis 2016; 213(5): 712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Albuquerque N, Baig E, Ma X. et al. Murinehepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J mice. 2006; 80(21): 10382–10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keck JG, Soe LH, Makino S. et al. RNA recombination of murine coronaviruses: Recombination between fusion-positive mouse hepatitis virus A59 and fusion-negative mouse hepatitis virus 2. J Virol 1988; 62(6): 1989–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koetzner CA, Parker MM, Ricard CS. et al. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol 1992; 66(4): 1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuo L, G-J Godeke, Raamsman MJB. et al. retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: Crossing the host cell species barrier. J Virol 2000; 74(3): 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yount B, Curtis KM, Fritz EA. et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA 2003; 100(22): 12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.González JM, Pénzes Z, Almazán F. et al. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by insertion of an intron. J Virol 2002; 76(9): 4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yount B, Denison MR, Weiss SR. et al. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 2002; 76(21): 11065–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Donaldson EF, Yount B, Sims AC. et al. Systematic assembly of a full-length infectious clone of human coronavirus NL63. J Virol 2008; 82(23): 11948–11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xie X, Muruato A, Lokugamage KG. et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Das Sarma J, Scheen E, S-h Seo. et al. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J Neurovirol 2002; 8(5): 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ontiveros E, Kuo L, Masters PS. et al. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 2001; 289(2): 230–238. [DOI] [PubMed] [Google Scholar]
- 98.Phillips JJ, Chua MM, Lavi E. et al. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: The murine coronavirus spike protein is a major determinant of neurovirulence. J Virol 1999; 73(9): 7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Das Sarma J, Kenyon LC, Hingley ST. et al. Mechanisms of primary axonal damage in a viral model of multiple sclerosis. J Neurosci 2009; 29(33): 10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Das Sarma J, Iacono K, Gard L. et al. Demyelinating and nondemyelinating strains of mouse hepatitis virus differ in their neural cell tropism. J Virol 2008; 82(11): 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kenyon LC, Biswas K, Shindler KS. et al. Gliopathy of demyelinating and non-demyelinating strains of mouse hepatitis virus. Front Cell Neurosci 2015; 9: 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shindler KS, Chatterjee D, Biswas K. et al. Macrophage-mediated optic neuritis induced by retrograde axonal transport of spike gene recombinant mouse hepatitis virus. J Neuropathol Exp Neurol 2011; 70(6): 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamada YK, Takimoto K, Yabe M. et al. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology 1997; 227(1): 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Biswas K, and Das Sarma J.. Effect of microtubule disruption on neuronal spread and replication of demyelinating and nondemyelinating strains of mouse hepatitis virus. J Virol 2014; 88(5): 3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das Sarma J, Fu L, Hingley ST. et al. Sequence analysis of the S gene of recombinant MHV-2/A59 coronaviruses reveals three candidate mutations associated with demyelination and hepatitis. J Neurovirol 2001; 7(5): 432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chambers P, Pringle CR, Easton AJ.. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol 1990; 71( Pt 12): 3075–3080. [DOI] [PubMed] [Google Scholar]
- 107.Luo Z and Weiss SR.. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 1998; 244(2): 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pallesen J, Wang N, Corbett KS. et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA 2017; 114(35): E7348–E7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Corbett KS, Edwards DK ,Leist SR. et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020; 586, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jackson LA, Anderson EJ, Rouphael NG. et al. An mRNA Vaccine against SARS-CoV-2 - preliminary report. N Engl J Med 2020; 383(20): 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mulligan MJ, Lyke KE, Kitchin N. et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020; 586(7830): 589–593. [DOI] [PubMed] [Google Scholar]
- 112.Dai L and Gao GF.. Viral targets for vaccines against COVID-19. Nat Rev Immunol 2021; 21(2): 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaufman G, Liu P, Leibowitz JL.. Identification of novel functional regions within the spike glycoprotein of MHV-A59 based on a bioinformatics approach. Virus Res 2014; 189: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rout SS, Singh M, Shindler KS. et al. One proline deletion in the fusion peptide of neurotropic mouse hepatitis virus (MHV) restricts retrograde axonal transport and neurodegeneration. J Biol Chem 2020. May 15; 295(20): 6926–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xia S, Zhu Y, Liu M. et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol 2020; 17: 765–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Madu IG, Roth SL, Belouzard S. et al. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol 2009; 83(15): 7411–7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Miura TA, Travanty EA, Oko L. et al. The spike glycoprotein of murine coronavirus mhv-jhm mediates receptor-independent infection and spread in the central nervous systems of Ceacam1a−/− mice. J Virol 2008; 82(2): 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bosch BJ, Martina BE, Van Der Zee R. et al. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci U S A 2004; 101(22): 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu S,, Xiao G,, Chen Y. et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004; 363(9413): 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yuan K, Yi L, Chen J. et al. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem Biophys Res Commun 2004; 319(3): 746–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.B-J Zheng, Guan Y, M-L Hez. et al. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir Ther 2005; 10(3): 393–403. [PubMed] [Google Scholar]
- 122.Stetson DB,, Medzhitov R.Type I Interferons in Host Defense. Immunity 2006; 25(3): 373–381. [DOI] [PubMed] [Google Scholar]
- 123.Teijaro JR.Type I interferons in viral control and immune regulation. Curr Opin Virol 2016; 16: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Murira A and Lamarre A.. Type-I interferon responses: From friend to foe in the battle against chronic viral infection. Front Immunol 2016; 7(609) 2016 Dec 19; 7: 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cicchese JM, Evans S Hult C. et al. Dynamic balance of pro- and anti-inflammatory signals controls disease and limits pathology. Immunol Rev 2018; 285(1): 147–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tisoncik JR, Korth MJ, Simmons CP. et al. into the eye of the cytokine storm. Microbiol Mol Biol Rev 2012; 76(1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Streit WJ,, Kincaid-Colton CA.The brain's immune system. Sci Am 1995; 273(5): 54–5, 8–61. [DOI] [PubMed] [Google Scholar]
- 128.Streit WJ, Mrak RE, Griffin WST.. Microglia and neuroinflammation: A pathological perspective. J Neuroinflammation 2004; 1(1): 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chatterjee D, Addya S, Khan RS. et al. Mouse hepatitis virus infection upregulates genes involved in innate immune responses. PLoS One 2014; 9(10): e111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Savarin C and Bergmann CC.. Fine tuning the cytokine storm by IFN and IL-10 following neurotropic coronavirus encephalomyelitis. Front Immunol 2018; 9: 3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Savarin C and Bergmann CC.. Viral-induced suppression of self-reactive T cells: Lessons from neurotropic coronavirus-induced demyelination. J Neuroimmunol 2017; 308: 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Puntambekar SS, Hinton DR, Yin X. et al. Interleukin-10 is a critical regulator of white matter lesion containment following viral induced demyelination. Glia 2015; 63(11): 2106–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.de Aquino MT, Kapil P, Hinton DR. et al. IL-27 limits central nervous system viral clearance by promoting IL-10 and enhances demyelination. J Immunol 2014; 193(1): 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hufford MM, Kim TS, Sun J. et al. The effector T cell response to influenza infection. Curr Top Microbiol Immunol 2015; 386: 423–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Griffin DE.The immune response in measles: Virus control, clearance and protective immunity. Viruses 2016; 8(10): 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Savarin C, Bergmann CC, Hinton DR. et al. Differential regulation of self-reactive CD4+ T cells in cervical lymph nodes and central nervous system during viral encephalomyelitis. Front Immunol 2016; 7(370). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bergmann CC, Parra B, Hinton DR. et al. Perforin and gamma interferon-mediated control of coronavirus central nervous system infection by CD8 T cells in the absence of CD4 T cells. J Virol 2004; 78(4): 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chakravarty D, Saadi F, Kundu S. et al. CD4 deficiency causes poliomyelitis and axonal blebbing in murine coronavirus induced neuroinflammation. J Virol 2020. Jul 1; 94(14): e00548–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miller KD, Schnell MJ, Rall GF.. Keeping it in check: Chronic viral infection and antiviral immunity in the brain. Nat Rev Neurosci 2016; 17(12): 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tay MZ, Poh CM, Rénia L. et al. The trinity of COVID-19: Immunity, inflammation and intervention. Nat Rev Immunol 2020; 20, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang C, Wang Y, Li X. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395(10223): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.S-Y Fung, K-S Yuen, Z-W Ye. et al. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: Lessons from other pathogenic viruses. Emerg Microbes Infect 2020; 9(1): 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Conti P, Ronconi G, Caraffa A. et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-inflammatory strategies. J Biol Regul Homeost Agents 2020. March-April; 34(2): 327–331. [DOI] [PubMed] [Google Scholar]
- 144.Merad M and Martin JC.. Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat Rev Immunol 2020; 20: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Quinti I, Lougaris V, Milito C. et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol 2020; 146(1): 211–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Biswas K, Chatterjee D, Addya S. et al. Demyelinating strain of mouse hepatitis virus infection bridging innate and adaptive immune response in the induction of demyelination. Clin Immunol 2016; 170: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wen W, Su W, Tang H. et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov 2020; 6(1): 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tan M, Liu Y, Zhou R, et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology 2020; 160(3), 261–268. https://doi.org/10.1111/imm.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jin Y, Yang H, Ji W. et al. Virology, epidemiology, pathogenesis, and control of COVID-19. Viruses 2020. Mar 27; 12(4): 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Glowacka I, Bertram S, Herzog P. et al. Differential down regulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol 2010; 84(2): 1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lane TE, Hosking MP.. The pathogenesis of murine coronavirus infection of the central nervous system. Crit Rev Immunol 2010; 30(2): 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu MT, Lane TE.. Chemokine expression and viral infection of the central nervous system: Regulation of host defense and neuropathology. Immunol Res 2001; 24(2): 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Netland J, Meyerholz DK, Moore S. et al. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virology 2008; 82(15): 7264–7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hwang CS.Olfactory neuropathy in severe acute respiratory syndrome: Report of A case. Acta Neurol Taiwan 2006; 15(1): 26–28. [PubMed] [Google Scholar]
- 155.Bohmwald K, Gálvez NMS, Ríos M. et al. Neurologic alterations due to respiratory virus infections. Front Cell Neurosci 2018; 12: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.To KF, Tong JH, Chan PK. et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: An in-situ hybridization study of fatal cases. J Pathol 2004; 202(2): 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li K, Wohlford-Lenane C, Perlman S. et al. Middle east respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. Int J Infect Dis 2016; 213(5): 712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Desforges M, Le Coupanec A, Stodola JK. et al. Human coronaviruses: Viral and cellular factors involved in neuroinvasiveness and neuropathogenesis. Virus Res 2014; 194: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tao X, Garron T, Agrawal AS. et al. Characterization and demonstration of the value of a lethal mouse model of middle east respiratory syndrome coronavirus infection and disease. J Virol 2016; 90(1): 57 Jul; 51(5): 384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Delgado-Roche L and Mesta F.. Oxidative stress as key player in severe acute respiratory syndrome coronavirus (SARS-CoV) infection. Arch Med Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Conde Cardona G, Quintana Pájaro LD, Quintero Marzola ID. et al. Neurotropism of SARS-CoV 2: Mechanisms and manifestations. J Neurol Sci 2020; 412: 116824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Filatov A, Sharma P, Hindi F. et al. Neurological complications of coronavirus disease (COVID-19): Encephalopathy. Cureus 2020; 12(3): e7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chen N, Zhou M, Dong X. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet (London, England). 2020; 395(10223): 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gandhi S, Srivastava AK, Ray U. et al. Is the collapse of the respiratory center in the brain responsible for respiratory breakdown in covid-19 patients? ACS Chem Neurosci 2020. May 20; 11(10): 1379–1381. [DOI] [PubMed] [Google Scholar]
- 165.Brun G, J-F Hak, Coze S. et al. COVID-19—White matter and globus pallidum lesions. Demyelination or small-vessel vasculitis? Neurol Neuroimmunol Neuroinflamm 2020; 7(4): e777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Singh T and Murugan S.. Anatomy of the COVID-19 pandemic: An epidemiologists view point. Ann Neurosci 2020; 27(2): 47–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li X, Wang W, Zhao X. et al. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol 2020; 92(5): 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Korber B, Fischer WM, Gnanakaran S. et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182(4): 812–827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Eaaswarkhanth M, Al Madhoun A, Al-Mulla F.. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int J Infect Dis 2020; 96: 459–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Grubaugh ND, Hanage WP, Rasmussen AL.. Making sense of mutation: What D614G means for the COVID-19 pandemic remains unclear. Cell 2020; 182(4): 794–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Samrat SK, Tharappel AM, Li Z. et al. Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res 2020; 288: 198141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Folegatti PM, Ewer KJ, Aley PK. et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020; 396(10249): 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ozgur R, Uzilday B, Iwata Y. et al. Interplay between the unfolded protein response and reactive oxygen species: A dynamic duo. J Exp Bot 2018; 69(14): 3333–3345. [DOI] [PubMed] [Google Scholar]
- 174.Romano AD, Serviddio G, de Matthaeis A. et al. Oxidative stress and aging. J Nephrol 2010; 23 (Suppl 15): S29–36. [PubMed] [Google Scholar]
- 175.Nikolich-Zugich J, Knox KS, Rios CT. et al. SARS-CoV-2 and COVID-19 in older adults: What we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience 2020; 42(2): 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.da Silva SJR, da Silva CTA, Mendes RPG. et al. Role of nonstructural proteins in the pathogenesis of SARS-CoV-2. J Med Virol 2020. Sep; 92(9): 1427–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Law AHY, Lee DCW, Cheung BKW. et al. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J Virol 2007; 81(1): 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Züst R, Cervantes-Barragán L, Kuri T. et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: Implications for the rational design of coronavirus vaccines. PLoS Pathog 2007; 3(8): e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Narayanan K, Ramirez SI, Lokugamage KG. et al. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res 2015; 202: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Narayanan K, Huang C, Lokugamage K. et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 2008; 82(9): 4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lokugamage KG, Narayanan K, Nakagawa K. et al. Middle east respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol 2015; 89(21): 10970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wu GF, Dandekar AA, Pewe L. et al. CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J Immunol 2000; 165(4): 2278. [DOI] [PubMed] [Google Scholar]


