Abstract
The RET proto-oncogene encodes a receptor tyrosine kinase whose alterations are responsible for various human cancers and developmental disorders, including thyroid cancer, non-small cell lung cancer, multiple endocrine neoplasia type 2, and Hirschsprung’s disease. RET receptors are physiologically activated by glial cell line-derived neurotrophic factor (GDNF) family ligands that bind to the coreceptor GDNF family receptor α (GFRα). Signaling via the GDNF/GFRα1/RET ternary complex plays crucial roles in the development of the enteric nervous system, kidneys, and urinary tract, as well as in the self-renewal of spermatogonial stem cells. In addition, another ligand, growth differentiation factor-15 (GDF15), has been shown to bind to GFRα-like and activate RET, regulating body weight. GDF15 is a stress response cytokine, and its elevated serum levels affect metabolism and anorexia-cachexia syndrome. Moreover, recent development of RET-specific kinase inhibitors contributed significantly to progress in the treatment of patients with RET-altered cancer. This review focuses on the broad roles of RET in development, metabolic diseases, and cancer.
Keywords: RET protooncogene, GDNF family ligands, cancer, Hirschsprung’s disease, kidney development, body weight control
1. Introduction
We identified RET (Rearranged during Transfection) as an oncogene activated by DNA rearrangement in 1985.1) RET encodes a transmembrane tyrosine kinase with a unique extracellular domain that consists of four cadherin-like domains and a cysteine-rich region with 16 cysteine residues in a stretch of 120 amino acids (Fig. 1).2–4) Alternative splicing in the 3′ region produces three different isoforms (1072, 1106, and 1114 amino acids) with short (9 amino acids, RET9), intermediate (43 amino acids, RET43), and long (51 amino acids, RET51) carboxyl-terminal tails, respectively,5) and the expression level of RET43 was reported to be low compared with RET9 and RET51. As observed for cadherin, Ca2+ ions bind to the cadherin-like domains, and these are required for RET activation by GDNF and neurturin (NRTN).4,6)
Figure 1.
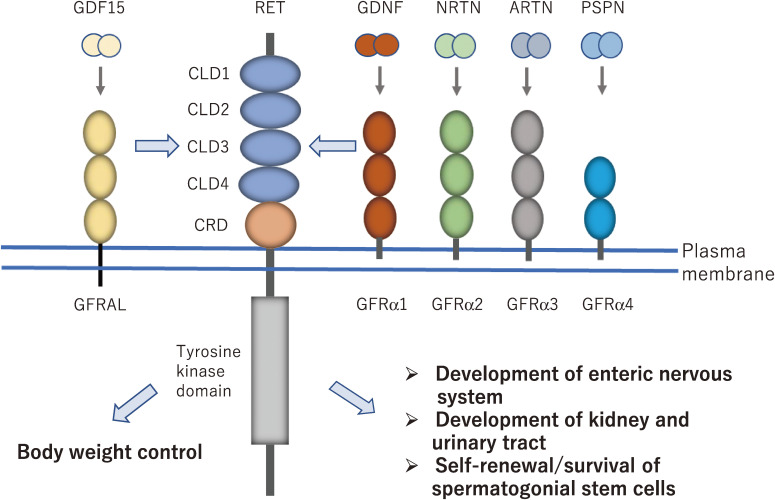
RET activation by GDNF family ligands (GFLs) that bind to coreceptor GDNF family receptor α (GFRα). GDNF, NRTN, ARTN and PSPN preferentially bind to GFRα1, GFRα2, GFRα3 and GFRα4, respectively. GDNF/GFRα1/RET signaling complex is essential for the development of the enteric nervous system, kidney and urinary tract, and self-renewal/survival of spermatogonial stem cells. GDF15/GFRAL/RET complex plays a crucial role in body weight control. GDNF, glial cell line-derived neurotrophic factor; NRTN, neurturin; ARTN, artemin; PSPN, persephin; GDF15, growth differentiation factor-15; GFRAL, GDNF family receptor α-like; CLD, cadherin-like domain; CRD, cysteine-rich domain.
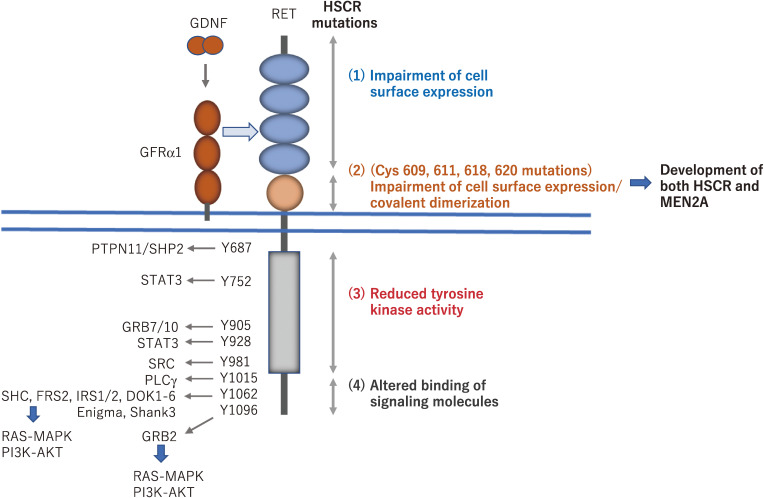
Following RET activation by GDNF family ligands (GFLs), specific tyrosine residues in its intracellular domain are autophosphorylated. At least 14 of the 18 tyrosine residues in the intracellular domain can be phosphorylated, and some of them represent docking sites for key adaptor proteins, leading to the activation of important signaling pathways (Fig. 2).4,7,8) For example, phosphorylated tyrosine 1062 mediates binding of the adaptor proteins SHC and FRS2, which are responsible for activation of the RAS/MAPK and/or PI3K/AKT signaling pathways.9–11) Phosphorylated tyrosine 1015 mediates binding of phospholipase C γ (PLCγ), resulting in the activation of protein kinase C.12) These pathways play important roles in cell migration, proliferation, survival, and differentiation.
Figure 2.
Intracellular signaling pathways activated by RET via phosphotyrosines and RET mutations in Hirschsprung’s disease (HSCR). Phosphorylated tyrosines in the intracellular domain of RET interact with a wide range of adaptor proteins, which leads to the activation of downstream signaling pathways, including the RAS/MAPK and PI3K-AKT pathways. For example, phosphotyrosine 1062 represents a multifunctional docking site for SHC, FRS2, and DOK family proteins. Missense mutations identified in HSCR patients are distributed along the whole coding sequence of the RET gene. Based on functional analyses of mutant RET, the HSCR mutations are classified as follows. (1) Most mutations in the RET extracellular domain impair its cell surface expression, most likely due to misfolding of the RET protein. (2) Mutations of Cys609, 611, 618, and 620 can result in the development of both HSCR (loss-of-function) and multiple endocrine neoplasia type 2A (MEN2A) (gain-of-function) phenotypes that are caused by impaired cell surface expression of RET in ENCDCs and covalent dimerization in thyroid C cells, respectively. (3) Mutations in the tyrosine kinase domain almost completely or partially disturb the RET kinase activity, resulting in the impairment of RAS/MAPK, PI3K/AKT, and/or PLCγ signaling pathways. (4) Mutations in the carboxyl-terminal tail alter the binding of adaptor proteins such as SHC.
RET has been shown to be a causative gene for a variety of human diseases.5) RET activating point mutations are responsible for the development of the hereditary cancer syndrome multiple endocrine neoplasia type 2 (MEN2), which develops into medullary thyroid carcinoma (MTC) and pheochromocytoma.13–16) RET activation by gene rearrangement is found in papillary thyroid carcinoma (PTC), non-small cell lung carcinoma (NSCLC), salivary gland intraductal carcinoma, and other cancers.17–24) In addition, RET-inactivating point mutations or deletions lead to the development of Hirschsprung’s disease (HSCR),25,26) which is a congenital malformation characterized by aganglionosis of variable length of the distal gastrointestinal tract. To date, the molecular mechanisms through which RET mutations lead to disease development have been extensively studied.
2. RET activation by GDNF family ligands
In 1993, GDNF was purified and cloned as a neurotrophic factor that enhances the survival of midbrain dopaminergic neurons.27) GDNF is a distant member of the transforming growth factor-β (TGF-β) superfamily, and three other proteins of GFLs, including NRTN, artemin (ARTN), and persephin (PSPN) were identified.28) These four family ligands show approximately 40% amino acid identity with each other and can activate RET kinase. However, GFLs cannot bind to RET directly, but GPI-anchored coreceptors named GFRα1–4 are necessary for their binding.28–32) GDNF, NRTN, ARTN, and PSPN bind preferentially to GFRα1, GFRα2, GFRα3, and GFRα4, respectively (Fig. 1), although crosstalk occurs to a certain extent between the ligand and coreceptor pairs.8) Formation of the GFL-GFRα-RET 2:2:2 ternary complex results in activation of intracellular signaling, which supports the survival and differentiation of various neurons, including peripheral sensory and autonomic neurons as well as central motor and dopaminergic neurons.28) Recent cryo-EM analysis demonstrated that the extracellular region of RET is folded and packed in a ‘C-clamp’ shape, which is stabilized by extensive inter-domain interactions. Because of this unique C-clamp shape, two RET molecules are recruited onto dimeric GFL-GFRα complexes, and the two cysteine-rich domains of RET are brought into close proximity, thereby promoting dimer formation and activation of the activity of the intracellular tyrosine kinase domain.33–35)
More recently, another member of the TGF-β superfamily, GDF15 (also known as MIC-1) was found to bind to GFRα-like (GFRAL) with high affinity and then activate RET (Fig. 1).36) The significance of this signaling complex is described in detail in a separate section.
3. Role of GDNF/GFRα1/RET signaling in the development of the enteric nervous system and Hirschsprung’s disease
Gdnf-, Gfrα-, and Ret-deficient mice share phenotypes characterized by a lack of enteric neurons in the whole gastrointestinal tract and kidney agenesis or dysgenesis.37–42) This finding clearly revealed the importance of signaling via GDNF/GFRα1/RET multicomponent receptors in development (Fig. 1). We and others found that phosphorylated tyrosine 1062 in the RET carboxyl-terminal tail represents a docking site for several adaptor proteins such as SHC and FRS2 and is important for activation of the RAS/MAPK and PI3K-AKT pathways (Fig. 2).5) Ret mutant mice in which tyrosine 1062 was replaced with phenylalanine exhibited severe defects of enteric neurons in the intestine and small kidneys, indicating a crucial role of signaling via tyrosine 1062 in organogenesis.43,44)
The enteric nervous system (ENS) originates from the neural crest, mostly at the vagal level. Neural crest-derived cells invade the foregut and begin their long rostrocaudal migration toward the end of the colon.45) In addition to extensive migration, establishment of the ENS requires controlled cell proliferation, differentiation and network formation by differentiated enteric neurons. During embryogenesis, migrating enteric neural crest-derived cells (ENCDCs) express RET and GFRα1. Gdnf mRNA is expressed in the mesenchyme of the gut and is abundant in the stomach on embryonic day 9.5 and extends to the cecum on embryonic day 10.5, suggesting a role of GDNF as a chemoattractant for ENCDC migration.
RET is a major causative gene for HSCR (prevalence: one in 5000 live births),25,26) which is a congenital malformation of the ENS that lacks enteric neurons mainly in the distal gastrointestinal tract. Based on the length of the aganglionic segment, HSCR is classified into two groups: short-segment HSCR (patients with aganglionosis as far as the rectosigmoidal junction) and long-segment HSCR (patients with aganglionosis beyond the rectosigmoid junction). RET mutations were found in approximately 50% of patients with familial HSCR and 10–20% of sporadic cases.45) Notably, there is a clear association between RET mutations and long-segment HSCR, total colonic aganglionosis, and total intestinal aganglionosis. A variety of missense, nonsense, and frameshift mutations have been identified along the entire coding sequence of RET, but meta-analysis data showed that RET mutations in HSCR are more commonly found in exon 10 (7.55%), 13 (11.32%), and 15 (7.55%).46) These mutations are inactivating and abrogate RET signaling, which is responsible for the migration and proliferation of ENCDCs during embryogenesis. Biochemical and cell biological analyses have elucidated various mechanisms through which RET missense mutations cause HSCR (Fig. 2).47) (1) Mutations in the RET extracellular domain impair its cell surface expression, most likely due to misfolding of the RET protein.48–50) (2) Mutations of Cys609, 611, 618, and 620 can result in the development of both HSCR (loss-of-function) and multiple endocrine neoplasia type 2A (MEN2A) (gain-of-function) that are caused by impaired cell surface expression of RET in ENCDCs and covalent dimerization in thyroid C cells, respectively (described below in the section on RET mutations in cancer).5) (3) Mutations in the tyrosine kinase domain almost completely or partially disturb RET kinase activity, resulting in the impairment of RAS/MAPK, PI3K/AKT, and/or PLCγ signaling pathways.51–53) (4) Mutations in the carboxyl-terminal tail alter the binding of adaptor proteins such as SHC.54,55)
In addition to the coding sequence, non-coding regions of the RET gene play a pivotal role in the development of HSCR. A common intronic enhancer polymorphism (RET +3, or rs2435357) was identified, which is a risk factor for HSCR and impairs RET expression.56) This common polymorphism might also interact with other genetic alterations, modulating the HSCR phenotype and may explain the failure to identify coding sequence mutations in the majority of HSCR cases, even in patients from families showing linkage to RET.
Nrtn- and Gfrα2-deficient mice showed a reduced number of myenteric neurons in the small intestine and a drastic reduction in cholinergic innervation in the salivary and lacrimal glands. GDNF and NRTN are considered rare susceptibility HSCR genes, sometimes in conjugation with RET mutations.57,58)
4. Role of GDNF/GFRα1/RET signaling in the development of kidney and renal anomaly
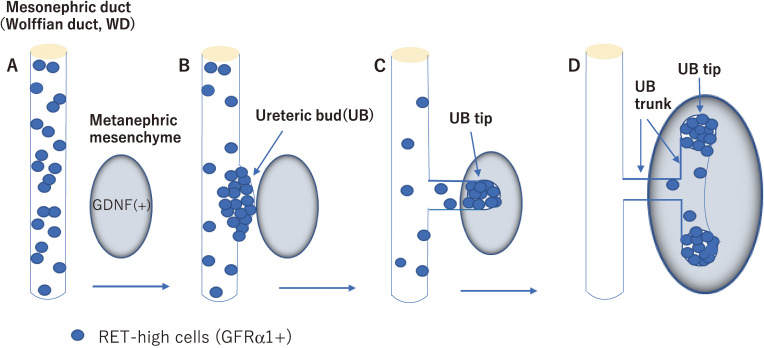
RET-mediated GDNF signaling has been shown to be essential for kidney development.59) During early development, RET and GFRα1 are expressed along the Wolffian duct (WD), whereas GDNF is expressed in the metanephric mesenchyme (MM) adjacent to the caudal portion of the WD (Fig. 3A). The ureteric bud (UB) emerges from the caudal portion of the WD, invaginates into the MM and begins to branch repeatedly. Of note, RET is highly expressed in a particular region of the WD where UB formation occurs (Fig. 3B) and subsequently at the tips of branching UB in the MM (Fig. 3C). During the whole process of kidney development, the UB tips appear to be formed entirely by RET-expressing cells that respond to GDNF in the MM, whereas the UB trunk largely consists of Ret-negative cells (Fig. 3D),60,61) indicating that RET expression is fine-tuned during UB branching. In the absence of Gdnf, Ret, or Gfrα1 gene, the most frequent consequence is a failure of UB formation, resulting in renal agenesis or dysgenesis and ureter defects (no ureters, small ureters, abnormally connected ureters, etc.).37–42) Several Ret-mutant mice showed decreased UB branching by affecting the RAS/MAPK, PI3K/AKT, and/or PLCγ pathways.8,43,62) In addition, the transcription factors ETV4 and ETV5 were identified as key components of a gene network downstream of RET that promotes branching morphogenesis.63,64)
Figure 3.
RET-dependent cell movement during ureteric bud (UB) formation and branching. RET-positive cells (blue) are initially dispersed along the mesonephric duct (Wolffian duct, WD) (A). RET-positive cells start to move to form the primary UB and the ventral mesonephric duct is depleted of RET-positive cells (B). As the UB grows out, RET-positive cells form the UB tips while RET-negative cells follow and form the UB trunk (C, D).
A previous report showed that heterozygous RET mutations were found in approximately 30% of a series of 29 human fetuses with bilateral or unilateral renal agenesis (BRA or URA) and one heterozygous GDNF mutation in a fetus with URA.65) However, another analysis of a large series of 105 cases, including 90 fetuses with either BRA or URA and contralateral renal hypodysplasia or multicystic dysplastic kidney reported only seven potential mutations in the RET coding sequence (6.6%) and no mutation in GDNF.66)
Congenital anomalies of the kidney and urinary tract (CAKUT) account for 40–50% of chronic renal diseases in children. CAKUT covers a wide range of structural malformations that result from a defect in the morphogenesis of the kidney and/or urinary tract. One study of 122 living patients, encompassing various CAKUT, found RET or GDNF variations in approximately 5% of patients.67) Moreover, in a large cohort of 749 individuals from 650 families with CAKUT, the coding exons of the 17 known dominant CAKUT-causing genes were analyzed. Among them, 37 different heterozygous mutations in 12 out of 17 genes examined were detected in 47 patients from 41 of the 650 families (6.3%), in which only three mutations in RET (0.5%) were included.68) These findings indicated that RET mutations are less commonly associated with CAKUT than expected.
5. Role of GDF15/GFRAL/RET signaling in body weight control
GDF15 is a member of the TGF-β family and is associated with body weight regulation.36) The administration of GDF15 to obese mice reduced food intake and body weight, demonstrating its anti-obesity effects.69) GDF15 is a stress-induced hormone and its plasma levels increase with age, intense exercise, obesity, smoking, and pregnancy. Higher levels are also observed in various human diseases, including cardiovascular disease, chronic kidney disease, diabetes, many advanced cancers, and serious infections (Fig. 4).70–72)
Figure 4.
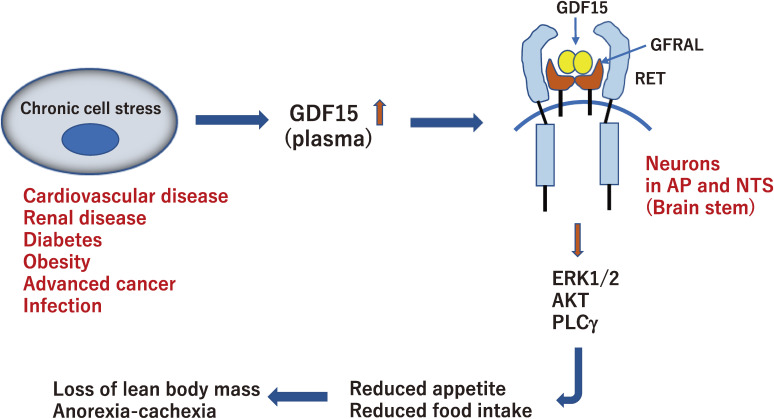
Action of GDF15 under conditions of chronic stress. Circulating GDF15 increases in response to various cellular stresses and crosses the blood-brain barrier. GDF15 binds to GFRAL and activates RET in neurons in the hindbrain area postrema (AP) and nucleus of the solitary tract (NTS). This central pathway leads to reduced food intake, causing loss of lean body mass and anorexia-cachexia syndrome.
In 2017, the receptor for GDF15 was identified by four pharmaceutical company research laboratories as GFRAL, the expression of which is detected in neurons of the hindbrain area postrema (AP) and nucleus of the solitary tract (NTS), leading to a decrease in food intake and body weight in mice.73–76) More notably, GDF15-GFRAL requires RET tyrosine kinase as a signaling receptor for body weight regulation. GDF15 binding to GFRAL initiates RET phosphorylation, resulting in the activation of ERK1/2, AKT, PLCγ, and FOS (Fig. 4). The co-expression of RET and GFRAL in AP and NTS was confirmed by in situ hybridization analysis.
In homeostatic conditions, animals use hypothalamic neural circuits to maintain body weight by integrating metabolic and hormonal signals from the periphery. However, under stress conditions, they use an alternative neuronal pathway for metabolic changes.36,77) Elevated GDF15 plasma levels are observed in various chronic human diseases and are associated with body weight loss. In advanced cancers, the circulating levels of GDF15 increase by up to 10–100-fold and induce anorexia-cachexia syndrome (Fig. 4). Cachexia is defined as a metabolic syndrome associated with extreme involuntary wasting of lean body mass with or without loss of fat mass. GDF15 is now considered the main actor in cachexia observed in patients with cancer. However, there are no approved drugs for this condition. Recently, a monoclonal antibody that targets GFRAL and inhibits RET signaling has been developed. The antibody prevented the GDF15-driven interaction of RET with GFRAL and cancer-related cachexia by reversing excessive lipid oxidation in tumor-bearing mice.78) Further clinical trials of drugs targeting the GDF15-GFRAL pathway will shed light on the treatment of patients with cancer-related cachexia.
6. RET rearrangements in cancer
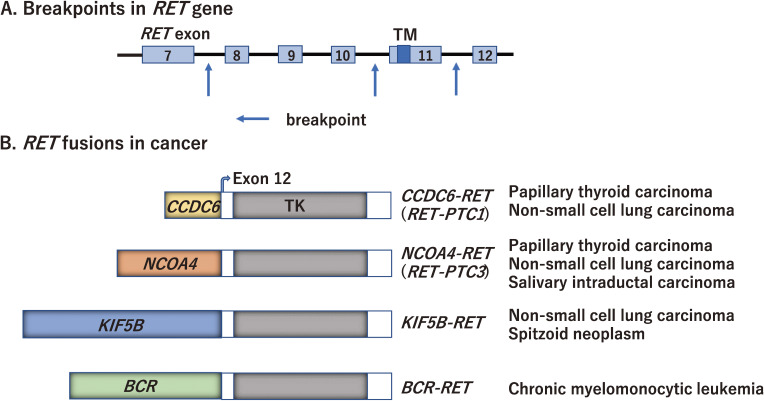
Since RET was discovered as an oncogene in 1985,1) a variety of RET rearrangements and point mutations have been identified in human cancers.79) Somatic RET rearrangements involve the 3′ sequence of RET, which contains the tyrosine kinase domain, and the 5′ sequence of various partner genes that contain dimerization domains such as the coiled-coil domain. RET breakpoints often occur within intron 11 and less frequently within introns 7 and 10 (Fig. 5A). To date, more than 35 genes have been reported to form fusion genes with RET.79)
Figure 5.
RET rearrangement in human cancer. A. Breakpoints in the RET gene. The breakpoints of RET often occur within intron 11 and less frequently within introns 7 and 10 (indicated by arrows). B. Representative RET rearrangement identified in human cancers. PTC, papillary thyroid carcinoma; TM, transmembrane domain; TK, tyrosine kinase domain.
RET fusion has been detected in 5–35% of adult PTCs, in which rearrangement with the CCDC6 gene has most frequently been observed (named RET/PTC1) (Fig. 5B).17–19) The other 5′ partner genes for RET fusion in PTC include PRKAR1A, NCOA4, GOLGA5, TRIM24, TRIM33, KTN1, and RFG9. The prevalence of RET rearrangements is much higher (50–80%) in radiation-induced PTCs following the Chernobyl radioactive fallout or the atomic bomb in Japan.80–83) The highest frequency of rearrangement was found in post-Chernobyl children. A significant predominance of RET fusion with the NCOA4 gene (named RET/PTC3) (Fig. 5B) over RET/PTC1 was observed in PTCs of the first post-Chernobyl decade. RET, CCDC6, and NCOA4 are located on the long arm of chromosome 10; thus, both RET/PTC1 and RET/PTC3 are induced by paracentric inversion. It is notable that RET rearrangements and BRAF mutations are largely mutually exclusive in PTCs. In addition to PTC, RET rearrangements were detected at a much lower prevalence in other types of thyroid cancer, such as follicular thyroid, anaplastic thyroid, and medullary thyroid carcinomas.79)
RET rearrangements are found in a portion (1–2%) of NSCLC cases with KIF5B-RET fusion being the most commonly identified (Fig. 5B).20–23) Because KIF5B is located on the short arm of chromosome 10, the KIF5B-RET fusion is created by pericentric inversion. CCDC6, NCOA4, TRIM33, and CUX1 are also partner 5′ genes for RET fusion in NSCLCs.84) Patients with RET fusion-positive NSCLCs have shown unique clinicopathological characteristics; they are young (<60 years old), female, and non-smoking patients.
Next-generation DNA and/or RNA sequencing approaches are used to identify less frequent RET rearrangements in a wide variety of cancer types. These include colorectal,85) breast,86) ovarian,87) chronic myelomonocytic leukemia88) and spitzoid tumors (Fig. 5B).89) Large-scale analyses have revealed that RET fusion can be detected in 0.2% of colorectal cancers (6/3117 cases)85) and 0.1% of breast cancers (8/9693 cases).86) Recently, a high frequency of RET rearrangements (>40%) has been detected in salivary intraductal carcinomas, including NCOA4-RET (Fig. 5B) and TRIM27-RET,24,90) suggesting that its detection is useful for diagnosing a particular type of salivary carcinoma.
7. RET mutations in cancer
Germline RET-activating mutations give rise to a hereditary cancer syndrome, MEN2.13–16) Based on the clinical phenotypes, MEN2 is classified into three subtypes: MEN2A, MEN2B, and familial medullary thyroid carcinoma (FMTC). MEN2A is the most common subtype, which is characterized by MTC in all patients combined with the development of pheochromocytoma and parathyroid hyperplasia/adenoma (hyperparathyroidism) in ∼50% and ∼20% of the patients, respectively. Lichen amyloidosis is occasionally observed in patients with MEN2A. MEN2B is a more aggressive subtype with early onset of MTC. In addition to the development of MTC and pheochromocytoma, MEN2B patients display mucosal neuroma, hyperganglionosis of the intestine, thickening of the corneal nerve, and marfanoid habitus, but not hyperparathyroidism. FMTC is the most indolent subtype of MEN2 and usually develops MTC in the later stages of life. FMTC is now considered a variant of MEN2A.91)
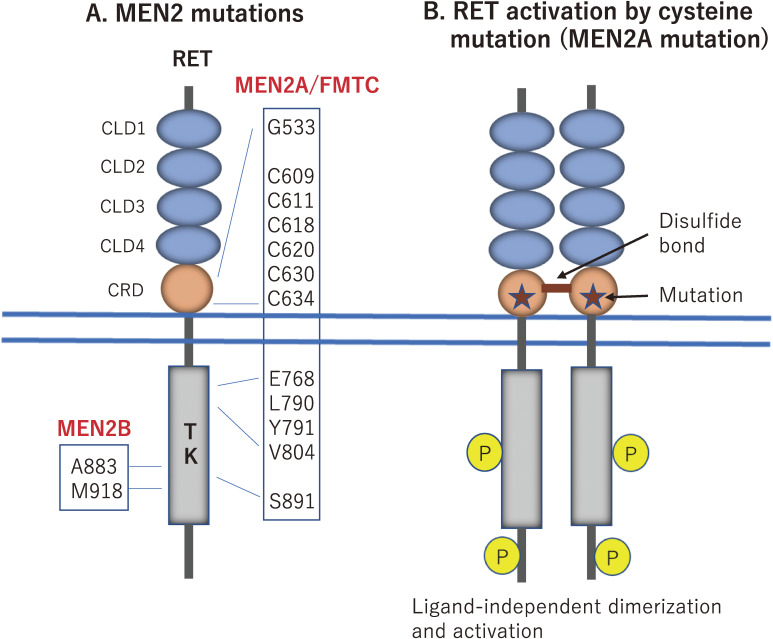
The majority of MEN2A mutations (>95%) have been identified in one of six cysteine residues (codons 609, 611, 618, and 620 in exon 10 and codons 630 and 634 in exon 11) in the cysteine-rich region of the RET extracellular domain (Fig. 6A). Among these, Cys634 mutations have been found in ∼85% of the patients.92) The same cysteine mutations in the RET extracellular domain also cause the FMTC phenotype with a high frequency of ∼60% for Cys609, 611, 618, 620, or 630 substitutions and a lower frequency of ∼30% for Cys634 substitutions.93) We and Santoro et al. demonstrated that cysteine substitutions result in ligand-independent constitutive activation of RET through the formation of an aberrant intermolecular disulfide bond between two mutant RET (Fig. 6B).94,95) In addition, the transforming activity of RET with Cys609, 611, 618, or 620 mutations is considerably lower than that of RET with Cys634 mutation, due to the impaired cell surface expression of the former four mutants.96) This may explain why the Cys609, 611, 618, or 620 mutations predispose to the development of indolent FMTC rather than MEN2A. Of notes, RET cysteine mutations affecting Cys609, 611, 618, and 620 can develop both MEN2A/FMTC (gain-of-function) and HSCR (loss-of-function) phenotypes, which are caused by RET covalent dimerization in thyroid C cells and impaired cell surface expression in ENCDCs, respectively (Fig. 2).97)
Figure 6.
Germline RET mutations in MEN2. A. The majority of MEN2A mutations (>95%) are identified in one of six cysteine residues (codons 609, 611, 618, and 620 in exon 10 and codons 630 and 634 in exon 11) in the cysteine-rich domain (CRD) of the RET extracellular region. In addition to cysteine substitutions, FMTC mutations are frequently found at noncysteine residues in both the extracellular and intracellular regions. The M918 mutation is detected in >95% of MEN2B patients. B. Mechanism of RET activation by cysteine mutations. When a cysteine residue is replaced with another amino acid in MEN2A/FMTC (indicated by stars), mutant RET proteins form ligand-independent covalent dimerization, resulting in constitutive activation.
Moreover, Gly533Cys (G533C) (exon 8 in the extracellular domain),98) Glu768Asp (E768D), Leu790Phe (L790F), Tyr791Phe (Y791F), Val804Met/Leu (V804M/L), and Ser891Ala(S891A) substitutions (exons 13–15 in the kinase domain) have been reported in some families with FMTC and/or MEN2A (Fig. 6A).47,91)
Two specific mutations, Met918Thr (M918T) and Ala883Phe (A883F), are associated with the development of MEN2B (Fig. 6A).15,16,99) The M918T mutation is found in >95% of patients and the A883F mutation in less than 4% of patients. These substitutions may induce conformational changes in the kinase domain that increase kinase activity and ATP binding and may alter substrate specificity.100–103) However, the mechanisms by which different RET mutations in MEN2A and MEN2B induce distinct clinical phenotypes remain elusive.
According to data published in a public database in 2015 (Catalog of Somatic Mutations in Cancer), somatic RET mutations have been identified with a high frequency (>40%) in sporadic MTC patients. The M918T mutation is most frequent in these patients, and other less common somatic mutations are also observed at residues C634, C630, A883, and others.79) A recent study using next-generation sequencing uncovered the presence of RET mutations in a variety of cancers at a low frequency, including colorectal carcinoma,85,104) breast carcinoma,86) endometrial and ovarian carcinoma, skin melanoma, Merkel cell carcinoma, and paraganglioma.87)
8. Therapeutic application of RET kinase inhibitors to RET mutation-positive tumors
A variety of multiple tyrosine kinase inhibitors (MTKIs) have been used in clinical trials to treat RET-mutation-positive tumors. These MTKIs include vandetanib, cabozantinib, sorafenib, sunitinib, ponatinib, lenvatinib, alectinib, and RXDX-105, which target several tyrosine kinases.105) Each molecule has distinct inhibitory activities against various targets. The clinical efficacy of MTKIs in RET-altered cancers is limited as shown by the lower overall objective response ratios (ORRs: 16–53% in RET-rearranged NSCLCs) and the shorter progression-free survival (PFS: 2.3–7.3 months in RET-rearranged NSCLCs). The limited efficacy of MTKIs is at least partially attributed to the off-target activity of these molecules.79,105) In addition, while cabozantinib and vandetanib can effectively inhibit the activity of the RET M918T mutant, they fail to block the activity of the gatekeeper mutants RET V804M and V804L.106,107)
Thus, more selective RET inhibitors are expected to achieve higher potency and lower toxicity. Recently, two RET-specific inhibitors, pralsetinib (BLU-667) and selpercatinib (LOXO-292), have been developed to inhibit wild-type RET and a broad spectrum of RET mutants, including those carrying the M918T mutation, the gatekeeper mutations V804L and V804M, and the CCDC6-RET and KIF5B-RET rearrangements.79,108–110) The use of these inhibitors improved ORR and the median duration of response, and revealed greater benefit for RET-altered MTC and NSCLC. For example, in the patients with RET-fusion-positive NSCLC who had previously received platinum-based chemotherapy, selpercatinib showed an ORR of 64% and a median duration of response of 17.5 months. Sixty-three percent of the responses were ongoing at a median follow-up of 12.1 months.110) Notably, both inhibitors also exhibited anti-tumor activity in patients with brain metastasis.111,112) Selpercatinib has been approved (September, 2021) and implemented (December 2021) for RET-rearranged NSCLC in Japan.
9. Conclusions
Over the past 30 years, a wide range of RET kinase functions have been studied, including roles in development, neurological disorders, and cancer. Recently, RET activation by GDF15 has been shown to play a pivotal role in the regulation of appetite and body weight during stress conditions, opening up new horizons in RET research. Further studies on GDF15/GFRAL-RET signaling will promote our understanding of cachexia metabolic signatures in patients with cancer and the development of therapeutic interventions to improve their outcomes. In addition, although not discussed in this review, the roles of RET in self-renewal and/or survival of spermatogonial stem cells113–115) and hematopoietic stem cells,116) in intestinal immunity,117,118) and in neuropathic pain119) are also being actively studied. Research on RET kinases continues to have a profound impact on a wide range of fields of life science.
The development of RET kinase inhibitors has been actively pursued, and many clinical studies are in progress. Selective RET inhibitors, selpercatinib and pralsetinib, have demonstrated remarkable clinical efficacy and safety, and are beneficial to patients with advanced RET-altered cancers. RET is also expressed in different neuronal cells, including dopaminergic neurons, motor neurons, sympathetic neurons, and parasympathetic neurons. Thus, it is possible that RET-dependent signaling plays a role in neurodegenerative diseases, such as Parkinson’s disease and amyotrophic lateral sclerosis. RET agonists and their appropriate delivery to the nervous system will contribute significantly to the development of new therapeutic strategies for such diseases.
Acknowledgements
The author thanks all laboratory members and research collaborators for insightful discussion. Work at the author’s laboratory is partly supported by Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. I would like to thank Editage (www.editage.com) for English language editing.
Non-standard abbreviation list
- AP
area postrema
- ARTN
artemin
- BRA
bilateral renal agenesis
- CAKUT
congenital anomalies of the kidney and urinary tract
- ENCDC
enteric neural crest-derived cell
- ENS
enteric nervous system
- FMTC
familial medullary thyroid carcinoma
- GDF15
growth differentiation factor-15
- GDNF
glial cell line-derived neurotrophic factor
- GFL
GDNF family ligand
- GFRα
GDNF family receptor α
- GFRAL
GDNF family receptor-α like
- HSCR
Hirschsprung’s disease
- MEN2
multiple endocrine neoplasia type2
- MM
metanephric mesenchyme
- MTC
medullary thyroid carcinoma
- MTKI
multiple tyrosine kinase inhibitors
- NRTN
neurturin
- NSCLC
non-small cell lung carcinoma
- NST
nucleus of the solitary tract
- ORR
objective response ratio
- PFS
progression-free survival
- PLCγ
phospholipaseC γ
- PSPN
persephin
- PTC
papillary thyroid carcinoma
- RET
Rearranged during Transfection
- TGF-β
transforming growth factor-β
- UB
ureteric bud
- URA
unilateral renal agenesis
- WD
Wolffian duct
Profile
Masahide Takahashi was born in Gifu Prefecture, Japan, in 1954 and graduated from Nagoya University School of Medicine in 1979. He majored in Pathology at Nagoya University Graduate School of Medicine and received his PhD degree in 1983. He subsequently studied as a research fellow at Dana-Farber Cancer Institute and Harvard Medical School in Boston between 1983 and 1985. After returning to Japan, he worked as a researcher at Aichi Cancer Center Research Institute in Nagoya between 1985 and 1990, and he moved to Nagoya University as an Assistant Professor. He became Professor of Pathology at Nagoya University in 1996. He was appointed as Dean of Nagoya University School of Medicine between 2012 and 2017, and Trustee and Vice President of Nagoya University between 2017 and 2020. He became Director and Professor of the International Center for Cell and Gene Therapy at Fujita Health University in 2020. His pioneering work included the discovery of the RET protooncogene, mutations in which are responsible for various human cancers and developmental diseases. His research group has elucidated the mechanisms of disease development caused by RET mutations. For his accomplishments, he has received The Japan Pathology Award (2001), The Chunichi Cultural Award (2010), Medical Award of The Japan Medical Association (2019), Princess Takamatsu Cancer Research Fund Prize (2020), and Medal with Purple Ribbon from the Government of Japan (2020).
References
- 1).Takahashi M., Ritz J., Cooper G.M. (1985) Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 42, 581–588. [DOI] [PubMed] [Google Scholar]
- 2).Takahashi M., Buma Y., Iwamoto T., Inaguma Y., Ikeda H., Hiai H. (1988) Cloning and expression of the ret proto-oncogene encoding a tyrosine kinase with two potential transmembrane domains. Oncogene 13, 571–578. [PubMed] [Google Scholar]
- 3).Iwamoto T., Taniguchi M., Asai N., Ohkusu K., Nakashima I., Takahashi M. (1993) cDNA cloning of mouse ret proto-oncogene and its sequence similarity to the cadherin superfamily. Oncogene 8, 1087–1091. [PubMed] [Google Scholar]
- 4).Ibáñez C.F. (2013) Structure and physiology of the RET receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol. 5, a009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Takahashi M. (2001) The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 12, 361–373. [DOI] [PubMed] [Google Scholar]
- 6).Nozaki C., Asai N., Murakami H., Iwashita T., Iwata Y., Horibe K., et al. (1998) Calcium-dependent Ret activation by GDNF and neurturin. Oncogene 16, 293–299. [DOI] [PubMed] [Google Scholar]
- 7).Kawamoto Y., Takeda K., Okuno Y., Yamakawa Y., Ito Y., Taguchi R., et al. (2004) Identification of RET autophosphorylation sites by mass spectrometry. J. Biol. Chem. 279, 14213–14224. [DOI] [PubMed] [Google Scholar]
- 8).Kawai K., Takahashi M. (2020) Intracellular RET signaling pathways activated by GDNF. Cell Tissue Res. 382, 113–123. [DOI] [PubMed] [Google Scholar]
- 9).Asai N., Murakami H., Iwashita T., Takahashi M. (1996) A mutation at tyrosine 1062 in MEN2A-Ret and MEN2B-Ret impairs their transforming activity and association with shc adaptor proteins. J. Biol. Chem. 271, 17644–17649. [DOI] [PubMed] [Google Scholar]
- 10).Hayashi H., Ichihara M., Iwashita T., Murakami H., Shimono Y., Kawai K., et al. (2000) Characterization of intracellular signals via tyrosine 1062 in RET activated by glial cell line-derived neurotrophic factor. Oncogene 19, 4469–4475. [DOI] [PubMed] [Google Scholar]
- 11).Kurokawa K., Iwashita T., Murakami H., Hayashi H., Kawai K., Takahashi M. (2001) Identification of SNT/FRS2 docking site on RET receptor tyrosine kinase and its role for signal transduction. Oncogene 20, 1929–1938. [DOI] [PubMed] [Google Scholar]
- 12).Borrello M.G., Alberti L., Arighi E., Bongarzone I., Battistini C., Bardelli A., et al. (1996) The full oncogenic activity of Ret/ptc2 depends on tyrosine 539, a docking site for phospholipace C-g. Mol. Cell. Biol. 16, 2151–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Mulligan L.M., Kwok J.B.J., Healey C.S., Elsdon M.J., Eng C., Gardner E., et al. (1993) Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature 363, 458–460. [DOI] [PubMed] [Google Scholar]
- 14).Donis-Keller H., Dou S., Chi D., Carlson K.M., Toshima K., Lairmore T.C., et al. (1993) Mutations in the RET protooncogene are associated with MEN 2A and FMTC. Hum. Mol. Genet. 2, 851–856. [DOI] [PubMed] [Google Scholar]
- 15).Hofstra R.M.W., Landsvater R.M., Ceccherini I., Stulp R.P., Stelwagen T., Luo Y., et al. (1994) A mutation in the RET proto-oncogene associated with multiple endocrine neoplasia type 2B and sporadic medullary thyroid carcinoma. Nature 367, 375–376. [DOI] [PubMed] [Google Scholar]
- 16).Carlson K.M., Dou S., Chi D., Scavarda N., Toshima K., Jackson C.E., et al. (1994) Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc. Natl. Acad. Sci. U.S.A. 91, 1579–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Grieco M., Santoro M., Berlingieri M.T., Melillo R.M., Donghi R., Bongarzone I., et al. (1990) PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell 60, 557–563. [DOI] [PubMed] [Google Scholar]
- 18).Nikiforov Y.E. (2002) RET/PTC rearrangement in thyroid tumors. Endocr. Pathol. 13, 3–16. [DOI] [PubMed] [Google Scholar]
- 19).Romei C., Ciampi R., Elisei R. (2016) A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat. Rev. Endocrinol. 12, 192–202. [DOI] [PubMed] [Google Scholar]
- 20).Kohno T., Ichikawa H., Totoki Y., Yasuda K., Hiramoto M., Nammo T., et al. (2012) KIF5B-RET fusion in lung cancer. Nat. Med. 18, 375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Takeuchi K., Soda M., Togashi Y., Suzuki R., Sakata S., Hatano S., et al. (2012) RET, ROS and ALK fusions in lung cancer. Nat. Med. 18, 378–381. [DOI] [PubMed] [Google Scholar]
- 22).Lipson D., Capelletti M., Yelensky R., Otto G., Parker A., Jarosz M., et al. (2012) Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat. Med. 18, 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Ju Y.S., Lee W.-C., Shin J.-Y., Lee S., Bleazard T., Won J.-K., et al. (2012) A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 22, 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Skálová A., Vanecek T., Uro-Coste E., Bishop J.A., Weinreb I., Thompson L.D.R., et al. (2018) Molecular profiling of salivary gland intraductal carcinoma revealed a subset of tumors harboring NCOA4-RET and novel TRM27-RET fusions: A report of 17 cases. Am. J. Surg. Pathol. 42, 1445–1455. [DOI] [PubMed] [Google Scholar]
- 25).Romeo G., Ronchetto P., Luo Y., Barone v., Seri M., Ceccherini I., et al. (1994) Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung’s disease. Nature 367, 377–378. [DOI] [PubMed] [Google Scholar]
- 26).Edery P., Lyonnet S., Mulligan L.M., Pelet A., Dow E., Abel L., et al. (1994) Mutations of the RET protooncogene in Hirschsprung’s disease. Nature 367, 378–380. [DOI] [PubMed] [Google Scholar]
- 27).Lin L.F., Doherty D.H., Lile J.D., Bektesh S., Collins F. (1993) GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science 260, 1130–1132. [DOI] [PubMed] [Google Scholar]
- 28).Airaksinen M.S., Saarma M. (2002) The GDNF family: signalling, biological functions and therapeutic value. Nat. Rev. Neurosci. 3, 383–394. [DOI] [PubMed] [Google Scholar]
- 29).Jing S., Wen D., Yu Y., Holst P.L., Luo Y., Fang M., et al. (1996) GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell 85, 1113–1124. [DOI] [PubMed] [Google Scholar]
- 30).Treanor J.J., Goodman L., de Sauvage F., Stone D.M., Poulsen K.T., Beck C.D., et al. (1996) Characterization of a multicomponent receptor for GDNF. Nature 382, 80–83. [DOI] [PubMed] [Google Scholar]
- 31).Klein R.D., Sherman D., Ho W.-H., Stone D., Bennett G.L., Moffat B., et al. (1997) A GPI-linked protein that interacts with Ret to form a candidate neurturin receptor. Nature 387, 717–721. [DOI] [PubMed] [Google Scholar]
- 32).Buj-Bello A., Adu J., Pinon L.G., Horton A., Thompson J., Rosenthal A., et al. (1997) Neurturin responsiveness requires a GPI-linked receptor and the Ret receptor tyrosine kinase. Nature 387, 721–724. [DOI] [PubMed] [Google Scholar]
- 33).Bigalke J.M., Aibara S., Roth R., Dahl G., Gordon E., Dorbéus S., et al. (2019) Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain. Sci. Adv. 5, eaau4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Li J., Shang G., Chen Y.J., Brautigam C.A., Liou J., Zhang X., et al. (2019) Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands. eLife 8, e47650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Adams S.E., Purkiss A.G., Knowles P.P., Nans A., Briggs D.C., Borg A., et al. (2021) A two-site flexible clamp mechanism for RET-GDNF-GFRa1 assembly reveals both conformational adaptation and strict geometric spacing. Structure 29, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Breit S.N., Brown D.A., Tsai V.W.-W. (2021) The GDF15-GFRAL pathway in health and metabolic disease: Friend or foe? Annu. Rev. Physiol. 83, 127–151. [DOI] [PubMed] [Google Scholar]
- 37).Schuchardt A., D’Agati V., Larsson-Blomberg L., Costantini F., Pachnis V. (1994) Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380–383. [DOI] [PubMed] [Google Scholar]
- 38).Sánchez M.P., Silos-Santiago I., Frisén J., He B., Lira S.A., Barbacid M. (1996) Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382, 70–73. [DOI] [PubMed] [Google Scholar]
- 39).Pichel J.G., Shen L., Sheng H.Z., Granholm A.C., Drago J., Grinberg A., et al. (1996) Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382, 73–76. [DOI] [PubMed] [Google Scholar]
- 40).Moore M.W., Klein R.D., Fariñas I., Sauer H., Armanini M., Phillips H., et al. (1996) Renal and neuronal abnormalities in mice lacking GDNF. Nature 382, 76–79. [DOI] [PubMed] [Google Scholar]
- 41).Cacalano G., Fariñas I., Wang L.-C., Hagler K., Forgie A., Moore M., et al. (1998) GFRα1 is an essential receptor component for GDNF in the developing nervous system and kidney. Neuron 21, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Enomoto H., Araki T., Jackman A., Heuckeroth R.O., Snider W.D., Johnson E.M., Jr., et al. (1998) GFRa1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron 21, 317–324. [DOI] [PubMed] [Google Scholar]
- 43).Jijiwa M., Fukuda T., Kawai K., Nakamura A., Kurokawa K., Murakumo Y., et al. (2004) A targeting mutation of tyrosine 1062 in ret causes a marked decrease of enteric neurons and renal hypoplasia. Mol. Cell. Biol. 24, 8026–8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Wong A., Bogni S., Kotka P., de Graaff E., D’Agati V., Costantini F., et al. (2005) Phosphotyrosine 1062 is critical for the in vivo activity of the Ret9 receptor tyrosine kinase isoform. Mol. Cell. Biol. 25, 9661–9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Lake J.I., Heuckeroth R.O. (2013) Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G1–G24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Tomuschat C., Puri P. (2015) RET gene is a major risk factor for Hirschsprung’s disease: a meta-analysis. Pediatr. Surg. Int. 31, 701–710. [DOI] [PubMed] [Google Scholar]
- 47).Takahashi M., Kawai K., Asai N. (2020) Roles of the RET proto-oncogene in cancer and development. JMA J. 3, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Iwashita T., Murakami H., Asai N., Takahashi M. (1996) Mechanisms of Ret dysfunction by Hirschsprung mutations affecting its extracellular domain. Hum. Mol. Genet. 5, 1577–1580. [DOI] [PubMed] [Google Scholar]
- 49).Carlomagno F., De Vita G., Berlingieri M.T., de Franciscis V., Melillo R.M., Colantuoni V., et al. (1996) Molecular heterogeneity of RET loss of function in Hirschsprung’s disease. EMBO J. 15, 2717–2725. [PMC free article] [PubMed] [Google Scholar]
- 50).Cosma M.P., Cardone M., Carlomagno F., Colantuoni V. (1998) Mutations in the extracellular domain cause RET loss of function by a dominant negative mechanism. Mol. Cell. Biol. 18, 3321–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Pasini B., Borrello M.G., Greco A., Bongarzone I., Luo Y., Mondellini P., et al. (1995) Loss of function effect of RET mutations causing Hirschsprung disease. Nat. Genet. 10, 35–40. [DOI] [PubMed] [Google Scholar]
- 52).Pelet A., Geneste O., Edery P., Pasini A., Chappuis S., Attie T., et al. (1998) Various mechanisms cause RET-mediated signaling defects in Hirschsprung’s disease. J. Clin. Invest. 101, 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Iwashita T., Kurokawa K., Qiao S., Murakami H., Asai N., Hashimoto M., et al. (2001) Functional analysis of RET with Hirschsprung’s mutations affecting its kinase domain. Gastroenterology 121, 24–33. [DOI] [PubMed] [Google Scholar]
- 54).Geneste O., Bidaud C., De Vita G., Hofstra R.M.W., Tartare-Deckert S., Buys C.H.C.M., et al. (1999) Two distinct mutations of the RET receptor causing Hirschsprung’s disease impair the binding of signaling effector to a multifunctional docking site. Hum. Mol. Genet. 8, 1989–1999. [DOI] [PubMed] [Google Scholar]
- 55).Ishiguro Y., Iwashita T., Murakami H., Asai N., Iida K., Goto H., et al. (1999) The role of amino acids surrounding tyrosine 1062 in Ret in specific binding of the Shc phosphotyrosine-binding domain. Endocrinology 140, 3992–3998. [DOI] [PubMed] [Google Scholar]
- 56).Emison E.S., MaCallion A.S., Kashuk C.S., Bush R.T., Grice E., Lin S., et al. (2005) A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434, 857–863. [DOI] [PubMed] [Google Scholar]
- 57).Manié S., Santoro M., Fusco A., Billaud M. (2001) The RET receptor: function in development and dysfunction in congenital malformation. Trends Genet. 17, 580–589. [DOI] [PubMed] [Google Scholar]
- 58).Sergi C.M., Caluseriu O., McColl H., Eisenstat D. (2017) Hirschsprung’s disease: clinical dysmophology, genes, micro-RNA, and future perspectives. Pediatr. Res. 81, 177–191. [DOI] [PubMed] [Google Scholar]
- 59).Costantini F., Shakya R. (2006) GDNF/Ret signaling and the development of the kidney. BioEssays 28, 117–127. [DOI] [PubMed] [Google Scholar]
- 60).Chi X., Michos O., Shakya R., Riccio P., Enomoto H., Lichit J.D., et al. (2009) Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell 17, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Michos O. (2009) Kidney development: from ureteric bud formation to branching morphogenesis. Curr. Opin. Genet. Dev. 19, 484–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Davis T.K., Hoshi M., Jain S. (2014) To bud or not to bud: the RET perspective in CAKUT. Pediatr. Nephrol. 29, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Lu B.C., Cebrian C., Chi X., Kuure S., Kuo R., Bates C.M., et al. (2009) Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat. Genet. 41, 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Riccio P., Cebrian C., Zong H., Hippenmeyer S., Costantini F. (2016) Ret and Etv4 promote directed movements of progenitor cells during renal branching morphogenesis. PLoS Biol. 14, e1002382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Skinner M.A., Safford S.D., Reeves J.G., Jackson M.E., Freemerman A.J. (2008) Renal aplasia in humans is associated with RET mutations. Am. J. Hum. Genet. 82, 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Jeanpierre C., Mace G., Parisot M., Moriniere V., Pawtowsky A., Benabou M., et al. (2011) RET and GDNF mutations are rare in fetuses with renal agenesis or other severe kidney development defects. J. Med. Genet. 48, 497–504. [DOI] [PubMed] [Google Scholar]
- 67).Chatterjee R., Ramos E., Hoffman M., VanWinkle J., Martin D.R., Davis T.K., et al. (2012) Traditional and targeted exome sequencing reveals common, rare and novel functional deleterious variants in RET-signaling complex in a cohort of living US patients with urinary tract malformations. Hum. Genet. 131, 1725–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Hwang D.-W., Dworschak G.C., Kohl S., Saisawat P., Vivante A., Hilger A.C., et al. (2014) Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 85, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Johnen H., Lin S., Kuffner T., Brown D.A., Tsai V.W., Bauskin A.R., et al. (2007) Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat. Med. 13, 1333–1340. [DOI] [PubMed] [Google Scholar]
- 70).Tsai V.W.W., Husaini Y., Sainsbury A., Brown D.A., Breit S.N. (2018) The MIC-1/GDF15-GFRAL pathway in energy homeostasis: Implications for obesity, cachexia, and other associated diseases. Cell Metab. 28, 353–368. [DOI] [PubMed] [Google Scholar]
- 71).Wollert K.C., Kempf T., Wallentin L. (2017) Growth differentiation factor 15 as a biomarker in cardiovascular disease. Clin. Chem. 63, 140–151. [DOI] [PubMed] [Google Scholar]
- 72).Luan H.H., Wang A., Hilliard B.K., Carvalho F., Rosen C.E., Ahasic A.M., et al. (2019) GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell 178, 1231–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Hsu J.Y., Crawley S., Chen M., Ayupova D.A., Lindhout D.A., Higbee J., et al. (2017) Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 550, 255–259. [DOI] [PubMed] [Google Scholar]
- 74).Mullican S.E., Lin-Schmidt X., Chin C.N., Chavez J.A., Furman J.L., Armstrong A.A., et al. (2017) GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat. Med. 23, 1150–1157. [DOI] [PubMed] [Google Scholar]
- 75).Yang L., Chang C.C., Sun Z., Madsen D., Zhu H., Padkjær S.B., et al. (2017) GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat. Med. 23, 1158–1166. [DOI] [PubMed] [Google Scholar]
- 76).Emmerson P.J., Wang F., Du Y., Liu Q., Pickard R.T., Gonciarz M.D., et al. (2017) The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med. 23, 1215–1219. [DOI] [PubMed] [Google Scholar]
- 77).Ahmed D.S., Isnard S., Lin J., Routy B., Routy J.-P. (2021) GDF15/GFRAL pathway as a metabolic signature for cachexia in patients with cancer. J. Cancer 12, 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Suriben R., Chen M., Higbee J., Oeffinger J., Ventura R., Li B., et al. (2020) Antibody-mediated inhibition of GDF15-GFRAL activity reverses cancer cachexia in mice. Nat. Med. 26, 1264–1270. [DOI] [PubMed] [Google Scholar]
- 79).Subbiah V., Yang D., Velcheti V., Drilon A., Meric-Bernstam F. (2020) State-of-the-art strategies for targeting RET-dependent cancers. J. Clin. Oncol. 38, 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80).Ricarte-Filho J.C., Li S., Garcia-Rendueles M.E., Montero-Conde C., Voza F., Knauf J.A., et al. (2013) Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J. Clin. Invest. 123, 4935–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81).Rabes H.M., Demidchik E.P., Sidorow J.D., Lengfelder E., Beimfohr C., Hoelzel D., et al. (2000) Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-chernobyl papillary thyroid carcinomas: biological, phenotypic, and clinical implications. Clin. Cancer Res. 6, 1093–1103. [PubMed] [Google Scholar]
- 82).Elisei R., Romei C., Vorontsova T., Cosci B., Veremeychik V., Kuchinskaya E., et al. (2001) RET/PTC rearrangements in thyroid nodules: studies in irradiated and not irradiated, malignant and benign thyroid lesions in children and adults. J. Clin. Endocrinol. Metab. 86, 3211–3216. [DOI] [PubMed] [Google Scholar]
- 83).Hamatani K., Eguchi H., Ito R., Mukai M., Takahashi K., Taga M., et al. (2008) RET/PTC rearrangements preferentially occurred in papillary thyroid cancer among atomic bomb survivors exposed to high radiation dose. Cancer Res. 68, 7176–7182. [DOI] [PubMed] [Google Scholar]
- 84).Kohno T., Nakaoku T., Tsuta K., Tsuchihara K., Matsumoto S., Yoh K., et al. (2015) Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl. Lung Cancer Res. 4, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Le Rolle A.-F., Klempner S.J., Garrett C.R., Seery T., Sanford E.M., Balasubramanian S., et al. (2015) Identification of characterization of RET fusion in advanced colorectal cancer. Oncotarget 6, 28929–28937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Paratala B.S., Chung J.H., Williams C.B., Yilmazel B., Petrosky W., Williams K., et al. (2018) RET rearrangement are actionable alterations in breast cancer. Nat. Commun. 9, 4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Kato S., Subbiah V., Marchlik E., Elkin S.K., Carter J.L., Kurzrock R. (2017) RET aberration in diverse cancers: Next-generation sequencing of 4871 patients. Clin. Cancer Res. 23, 1988–1997. [DOI] [PubMed] [Google Scholar]
- 88).Ballerini P., Struski S., Cresson C., Prade N., Toujani S., Deswarte C., et al. (2012) RET fusion genes are associated with chronic myelomonocytic leukemia and enhance monocytic differentiation. Leukemia 26, 2384–2389. [DOI] [PubMed] [Google Scholar]
- 89).Wiesner T., He J., Yelensky R., Esteve-Puig R., Botton T., Yeh I., et al. (2014) Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat. Commun. 5, 3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Skálová A., Ptáková N., Santana T., Agaimy A., Ihrler S., Uro-Coste E., et al. (2019) NCOA4-RET and TRIM27-RET are characteristic gene fusions in salivary intraductal carcinoma, including invasive and metastatic tumors: Is “intraductal” correct? Am. J. Surg. Pathol. 43, 1303–1313. [DOI] [PubMed] [Google Scholar]
- 91).Mulligan L.M. (2014) RET revisited: expanding the oncogenic portfolio. Nat. Rev. Cancer 14, 173–186. [DOI] [PubMed] [Google Scholar]
- 92).Mulligan L.M., Eng C., Healey C.S., Clayton D., Kwok J.B., Gardner E., et al. (1994) Specific mutations of the RET proto-oncogene are related to disease phenotype in MEN 2A and FMTC. Nat. Genet. 6, 70–74. [DOI] [PubMed] [Google Scholar]
- 93).Eng C., Clayton D., Schuffenecker I., Lenoir G., Cote G., Gagel R.F., et al. (1996) The relationship between specific RET proto-oncogene mutations and disease phenotype in multiple endocrine neoplasia type 2. JAMA 276, 1575–1579. [PubMed] [Google Scholar]
- 94).Asai N., Iwashita T., Matsuyama M., Takahashi M. (1995) Mechanism of activation of the ret proto-oncogene by multiple endocrine neoplasia 2A mutations. Mol. Cell. Biol. 15, 1613–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Santoro M., Carlomagno F., Romano A., Bottaro D.P., Dathan N.A., Grieco M., et al. (1995) Activation of RET as a dominant transforming gene by germline mutations of MEN2A and MEN2B. Science 267, 381–383. [DOI] [PubMed] [Google Scholar]
- 96).Ito S., Iwashita T., Asai N., Murakami H., Iwata Y., Sobue G., et al. (1997) Biological properties of Ret with cysteine mutations correlate with multiple endocrine neoplasia type 2A, familial medullary thyroid carcinoma, and Hirschsprung’s disease phenotype. Cancer Res. 57, 2870–2872. [PubMed] [Google Scholar]
- 97).Takahashi M., Iwashita T., Santoro M., Lyonnet S., Lenoir G.M., Billaud M. (1999) Co-segregation of MEN2 and Hirschsprung’s disease: the same mutation of RET with both gain and loss-of-function? Hum. Mutat. 13, 331–336. [DOI] [PubMed] [Google Scholar]
- 98).Castro M.R., Thomas B.C., Richards M.L., Zhang J., Morris J.C. (2013) Multiple endocrine neoplasia type 2A due to an exon 8 (G533C) in a large North American kindred. Thyroid 23, 1547–1552. [DOI] [PubMed] [Google Scholar]
- 99).Smith D.P., Houghton C., Ponder B.A. (1997) Germline mutation of RET codon 883 in two cases of de novo MEN 2B. Oncogene 15, 1213–1217. [DOI] [PubMed] [Google Scholar]
- 100).Songyang Z., Carraway K.L., 3rd, Eck M.J., Harrison S.C., Feldman R.A., Mohammadi M., et al. (1995) Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature 373, 536–539. [DOI] [PubMed] [Google Scholar]
- 101).Murakami H., Iwashita T., Asai N., Shimono Y., Iwata Y., Kawai K., et al. (1999) Enhanced phosphatidylinositol 3-kinase activity and high phosphorylation state of its downstream signalling molecules mediated by ret with the MEN 2B mutation. Biochem. Biophys. Res. Commun. 262, 68–75. [DOI] [PubMed] [Google Scholar]
- 102).Gujral T.S., Singh V.K., Jia Z., Mulligan L.M. (2006) Molecular mechanisms of RET receptor-mediated oncogenesis in multiple endocrine neoplasia 2B. Cancer Res. 66, 10741–10749. [DOI] [PubMed] [Google Scholar]
- 103).Plaza-Menacho I., Barnouin K., Goodman K., Martínez-Torres R.J., Borg A., Murray-Rust J., et al. (2014) Oncogenic RET kinase domain mutations perturb the autophosphorylation trajectory by enhancing substrate presentation in trans. Mol. Cell 53, 738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104).Mendes Oliveira D., Grillone K., Mignogna C., De Falco V., Laudanna C., Biamonte F., et al. (2018) Next-generation sequencing analysis of receptor-type tyrosine kinase genes in surgically resected colon cancer: identification of gain-of-function mutations in the RET proto-oncogene. J. Exp. Clin. Cancer Res. 37, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Drilon A., Hu Z.I., Lai G.G.Y., Tan D.S.W. (2018) Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat. Rev. Clin. Oncol. 15, 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106).Dagogo-Jack I., Stevens S.E., Lin J.J., Nagy R., Ferris L., Shaw A.T., et al. (2018) Emergence of a RET V804M gatekeeper mutation during treatment with vandetanib in RET-rearranged NSCLC. J. Thorac. Oncol. 13, e226–e227. [DOI] [PubMed] [Google Scholar]
- 107).Huang Q., Schneeberger V.E., Luetteke N., Jin C., Afzal R., Budzevich M.M., et al. (2016) Preclinical modeling of KIF5B-RET fusion lung adenocarcinoma. Mol. Cancer Ther. 15, 2521–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Subbiah V., Gainor J.F., Rahal R., Brubaker J.D., Kim J.L., Maynard M., et al. (2018) Precision targeted therapy with BLU-667 for RET-driven cancers. Cancer Discov. 8, 836–849. [DOI] [PubMed] [Google Scholar]
- 109).Wirth L.J., Sherman E., Robinson B., Solomon B., Kang H., Lorch J., et al. (2020) Efficacy of selpercatinib in RET-altered thyroid cancers. N. Engl. J. Med. 383, 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110).Drilon A., Oxnard G.R., Tan D.S.W., Loong H.H.F., Johnson M., Gainor J., et al. (2020) Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N. Engl. J. Med. 383, 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111).Subbiah V., Gainor J.F., Oxnard G.R., Tan D.S.W., Owen D.H., Cho B.C., et al. (2021) Intracranial efficacy of selpercatinib in RET fusion-positive non-small cell lung cancers on the LIBRETTO-001 trial. Clin. Cancer Res. 27, 4160–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Mehta G.U., Barone A.K., Bradford D., Larkins E., Kim J., Pai-Scherf L., et al. (2021) US Food and Drug Administration regulatory updates in neuro-oncology. J. Neurooncol. 53, 375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Meng X., Lindahl M., Hyvönen M.E., Parvinen M., de Rooij D.G., Hess M.W., et al. (2000) Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493. [DOI] [PubMed] [Google Scholar]
- 114).Jijiwa M., Kawai K., Fukihara J., Nakamura A., Hasegawa M., Suzuki C., et al. (2008) GDNF-mediated signaling via RET tyrosine 1062 is essential for maintenance of spermatogonial stem cells. Genes Cells 13, 365–374. [DOI] [PubMed] [Google Scholar]
- 115).Takashima S., Kanatsu-Shinohara M., Tanaka T., Morimoto H., Inoue K., Ogonuki N., et al. (2015) Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports 4, 489–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116).Fonseca-Pereira D., Arroz-Madeira S., Rodrigues-Campos M., Barbosa I.A., Domingues R.G., Bento T., et al. (2014) The neurotrophic factor receptor RET drives haematopoietic stem cell survival and function. Nature 514, 98–101. [DOI] [PubMed] [Google Scholar]
- 117).Veiga-Fernandes H., Coles M.C., Foster K.E., Patel A., Williams A., Natarajan D., et al. (2007) Tyrosine kinase receptor RET is a key regulator of Peyer’s patch organogenesis. Nature 446, 547–551. [DOI] [PubMed] [Google Scholar]
- 118).Ibiza S., García-Cassani B., Ribeiro H., Carvalho T., Almeida L., Marques R., et al. (2016) Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defense. Nature 535, 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119).Mahato A.K., Sidorova Y.A. (2020) RET receptor tyrosine kinase: Role in neurodegeneration, obesity, and cancer. Int. J. Mol. Sci. 21, 7108. [DOI] [PMC free article] [PubMed] [Google Scholar]