Abstract
DWARF14 (D14) is an ɑ/β‐hydrolase and receptor for the plant hormone strigolactone (SL) in angiosperms. Upon SL perception, D14 works with MORE AXILLARY GROWTH2 (MAX2) to trigger polyubiquitination and degradation of DWARF53(D53)‐type proteins in the SUPPRESSOR OF MAX2 1‐LIKE (SMXL) family. We used CRISPR‐Cas9 to generate knockout alleles of the two homoeologous D14 genes in the Nicotiana benthamiana genome. The Nbd14a,b double mutant had several phenotypes that are consistent with the loss of SL perception in other plants, including increased axillary bud outgrowth, reduced height, shortened petioles, and smaller leaves. A ratiometric fluorescent reporter system was used to monitor degradation of SMXL7 from Arabidopsis thaliana (AtSMXL7) after transient expression in N. benthamiana and treatment with the strigolactone analog GR24. AtSMXL7 was degraded after treatment with GR245DS, which has the stereochemical configuration of natural SLs, as well as its enantiomer GR24 ent‐5DS. In Nbd14a,b leaves, AtSMXL7 abundance was unaffected by rac‐GR24 or either GR24 stereoisomer. Transient coexpression of AtD14 with the AtSMXL7 reporter in Nbd14a,b restored the degradation response to rac‐GR24, but required an active catalytic triad. We used this platform to evaluate the ability of several AtD14 mutants that had not been characterized in plants to target AtSMXL7 for degradation.
Keywords: CRISPR‐Cas9, phytohormones, proteolysis, signaling, transient expression
1. INTRODUCTION
Strigolactones (SLs) are a family of plant hormones derived from β‐carotene that have diverse functions in plants (Bouwmeester et al., 2021; Machin et al., 2020; Waters et al., 2017). SLs regulate axillary bud outgrowth (tillering), stem elongation, auxin transport, root elongation, leaf shape, leaf angle, leaf senescence, cambial growth, susceptibility to pathogenic microbes and root‐knot nematodes, stomatal closure responses, and drought tolerance (Agusti et al., 2011; Bu et al., 2014; Gomez‐Roldan et al., 2008; Kalliola et al., 2020; Kapulnik et al., 2011; Lahari et al., 2019; Lauressergues et al., 2015; Li et al., 2017, 2020; Marzec et al., 2016; Nasir et al., 2019; Ruyter‐Spira et al., 2011; Scaffidi et al., 2013; Shindo et al., 2020; Shinohara et al., 2013; Soundappan et al., 2015; Ueda & Kusaba, 2015; Umehara et al., 2008; Van Ha et al., 2014; Yamada et al., 2014). SLs are also exuded by roots into the rhizosphere, especially under nutrient‐poor conditions. There, SLs stimulate hyphal branching and metabolic activity of arbuscular mycorrhizal (AM) fungi, promoting beneficial symbiotic interactions with the host plant (Akiyama et al., 2005; Besserer et al., 2006, 2008; Kobae et al., 2018; Kretzschmar et al., 2012).
SL perception in flowering plants is mediated by the ɑ/β‐hydrolase DWARF14 (D14)/DECREASED APICAL DOMINANCE (DAD2)/RAMOSUS3 (RMS3) (Arite et al., 2009; de Saint Germain et al., 2016; Hamiaux et al., 2012; Waters et al., 2012). Upon activation by SL, D14 associates with the F‐box protein MORE AXILLARY GROWTH2 (MAX2)/DWARF3(D3), which acts as an adapter component of an SCF (Skp1–Cullin–F‐box) E3 ubiquitin ligase complex. Activated D14 also associates with a subset of proteins in the SUPPRESSOR OF MAX2 1‐LIKE (SMXL) family that are known as DWARF53 (D53) in rice and petunia, and SMXL6, SMXL7, and SMXL8 in Arabidopsis thaliana. This leads to the polyubiquitination of D53‐type SMXLs by SCFMAX2 and their degradation by the 26S proteasome (Jiang et al., 2013; Lee et al., 2020; Shabek et al., 2018; Soundappan et al., 2015; Wang, Wang, et al., 2015; Yao et al., 2016; Zhou et al., 2013).
A very similar signaling mechanism is used by karrikins (KARs), a class of plant growth regulators found in smoke. KAR signaling requires SCFMAX2 and an ancient paralog of D14 known as KARRIKIN INSENSITIVE2 (KAI2)/HYPOSENSITIVE TO LIGHT (HTL) (Nelson et al., 2011; Sun & Ni, 2011; Waters et al., 2012). In addition to mediating responses to KARs, KAI2 is hypothesized to perceive an unidentified, endogenous KAI2 ligand (KL) (Conn & Nelson, 2016; Sun et al., 2016). Upon activation, KAI2‐SCFMAX2 targets SMAX1 and its close paralog SMXL2 for degradation (Khosla, Morffy, et al., 2020; Stanga et al., 2013; Stanga et al., 2016; Wang et al., 2020; Zheng et al., 2020). This pathway regulates seed germination, hypocotyl/mesocotyl elongation, seedling responses to light, leaf shape, cuticle development, drought tolerance, root skewing, root hair density and elongation, and the capacity for AM symbiosis (Bunsick et al., 2020; Carbonnel et al., 2020; Choi et al., 2020; Gutjahr et al., 2015; Li et al., 2017, 2020; Nelson et al., 2009, 2010; Shen et al., 2007; Soundappan et al., 2015; Stanga et al., 2013, 2016; Sun & Ni, 2011; Swarbreck et al., 2019; Villaécija‐Aguilar et al., 2019; Zheng et al., 2020). In A. thaliana, SMAX1 and SMXL2 also associate with D14 and are targeted for degradation when a SL analog is applied, indicating some crosstalk between the SL and KAR signaling pathways may occur (Li et al., 2022; Wang et al., 2020).
D14 has highly conserved roles in angiosperms. This has been demonstrated through analysis of d14 mutants in petunia (Petunia hybrida), rice (Orzya sativa), A. thaliana, canola (Brassica napus), pea (Pisum sativum), barley (Hordeum vulgare), hexaploid wheat (Triticum aestivum), barrel medic (Medicago truncatula), and Lotus japonicus, as well as RNAi knockdown of D14 in soybean (Glycine max) hairy roots (Ahmad et al., 2020; Arite et al., 2009; Carbonnel et al., 2020; de Saint Germain et al., 2016; Hamiaux et al., 2012; Lauressergues et al., 2015; Liu et al., 2021; Marzec et al., 2016; Stanic et al., 2021; Waters et al., 2012). D14 orthologs from cotton (Gossypium hirsutum), poplar (Populus trichocarpa), and chrysanthemum (Dendranthema grandiflorum) have also been studied indirectly through cross‐species complementation of an Arabidopsis d14 (Atd14) mutant (Wang et al., 2019; Wen et al., 2015; Zheng et al., 2016). This approach enables in vivo analysis of gene function for species that have fewer genetic resources available or are less tractable to genetic studies than the major model plant systems (e.g., species lacking insertion/TILLING mutant collections and effective transformation methods). For example, this method has been used to identify SL receptors in root parasitic plants that arose from neofunctionalization of KAI2/HTL paralogs (Conn et al., 2015; de Saint Germain et al., 2021; Toh et al., 2015).
The utility of the cross‐species complementation approach is limited by the compatibility of the transgene product of interest with its noncognate cellular environment. For example, some MAX2 and KAI2/HTL transgenes from petunia; the bryophytes Selaginella moellendorffii, Marchantia polymorpha, and Physcomitrium (formerly Physcomitrella) patens; and the parasitic plant Striga hermonthica are nonfunctional or only partially functional in Arabidopsis (Conn et al., 2015; Drummond et al., 2011; Liu et al., 2014; Toh et al., 2015). This does not necessarily mean that these genes have reduced function in their native context; instead, the proteins they encode may not be able to interact well with Arabidopsis orthologs of their signaling partners (Khosla & Nelson, 2016). For example, at least two S. hermonthica KAI2/HTL proteins that are inactive in Arabidopsis can bind SL sensitively in vitro but cannot interact with Arabidopsis MAX2 (AtMAX2), or for that matter ShMAX2 (Wang et al., 2021). Conversely, S. hermonthica HTL7 (ShHTL7) causes Arabidopsis seed to germinate in the presence of picomolar SL. This response is several orders of magnitude lower than that conferred by ShHTL proteins with similar SL affinities in vitro and is likely due to the unusually high affinity of ShHTL7 for AtMAX2 (Toh et al., 2015; Tsuchiya et al., 2015; Uraguchi et al., 2018; Wang et al., 2021). Another disadvantage of the cross‐species complementation approach to investigate gene function is that it typically takes several generations to obtain homozygous transgenic lines that are suitable to study. Therefore, methods to evaluate plant gene function ex situ that are fast and also allow the cointroduction of compatible transgene partners are desirable.
We previously developed a ratiometric reporter system (pRATIO) that can monitor changes in the relative abundance of a transiently expressed target protein compared with a reference protein (Khosla, Rodriguez Furlan, et al., 2020). In this Gateway‐compatible system, a gene of interest (target) is translationally fused to a fluorescent or bioluminescent reporter. The target is cotranscribed with a reference gene that also encodes a fluorescent or bioluminescent reporter (Khosla, Rodriguez Furlan, et al., 2020). A modified “self‐cleaving” 2A peptide derived from foot‐and‐mouth disease virus is encoded between the two genes, causing the target and reference proteins to be translated separately. This approach enables multicistronic, stoichiometric expression in eukaryotes (Luke et al., 2010).
The pRATIO system has been applied successfully in Nicotiana benthamiana, a native Australian species that is closely related to tobacco (Nicotiana tabacum), to investigate degradation of Arabidopsis SMAX1, SMXL7, and KAI2 (Bally et al., 2018; Khosla, Morffy, et al., 2020; Khosla, Rodriguez Furlan, et al., 2020). When transiently expressed in N. benthamiana leaves, the Arabidopsis SMXL7 (AtSMXL7) ratiometric reporter is degraded after treatment with rac‐GR24, a racemic mixture of the synthetic SL analog GR245DS and its enantiomer, GR24ent‐5DS. In contrast, KAR1 or KAR2 treatments, which are expected to activate KAI2, do not affect AtSMXL7 stability (Khosla, Morffy, et al., 2020). This is consistent with prior studies that show degradation of rice D53 and Arabidopsis SMXL6, SMXL7, and SMXL8 in response to GR24, but not KAR1 treatments (Jiang et al., 2013; Wang, Wang, et al., 2015; Zhou et al., 2013). It also indicates that a receptor(s) in N. benthamiana is able to target AtSMXL7 for degradation in response to one or both components of rac‐GR24.
GR24‐induced degradation of D53‐type SMXLs is dependent on D14 in rice and Arabidopsis (Jiang et al., 2013; Samodelov et al., 2016; Wang, Wang, et al., 2015; Zhou et al., 2013). Genetic and evolutionary evidence also support the idea that AtSMXL7 is targeted by AtD14 and not AtKAI2 (Machin et al., 2020; Soundappan et al., 2015; Waters et al., 2015). Therefore, the AtSMXL7 degradation response is likely mediated by one or both of the two nearly identical D14 homoeologs found in the N. benthamiana genome. However, the possibility has been raised that there can be crosstalk between KAI2 and SMXL6, SMXL7, and SMXL8 in the regulation of Arabidopsis root skewing (Swarbreck et al., 2019). The N. benthamiana genome contains four KAI2 paralogs that are likely to encode functional proteins. Also, KAI2 proteins are not limited to KAR perception. Arabidopsis KAI2 is activated by GR24 ent‐5DS, whereas in parasitic plants, some evolutionarily “divergent” KAI2 (KAI2d) proteins are able to perceive GR245DS and natural SLs (Conn et al., 2015; de Saint Germain et al., 2021; Nelson, 2021; Toh et al., 2015; Tsuchiya et al., 2015). Therefore, it is not clear whether any NbKAI2 proteins might contribute to rac‐GR24‐induced degradation of AtSMXL7 in N. benthamiana.
We reasoned that transient coexpression in N. benthamiana could provide a way to rapidly evaluate the ability of D14 variants to target a SMXL7 ratiometric reporter for degradation. This could enable medium‐throughput screens for mutations that affect D14 signaling activity or its protein–protein interactions with SMXL7. Pairs of D14‐SMXL7 proteins from other species could also be evaluated as long as compatibility with N. benthamiana MAX2 is maintained. For this approach to be most effective, however, the endogenous SL receptors in N. benthamiana would need to be removed. Therefore, we used CRISPR‐Cas9 to knock out N. benthamiana D14a and D14b. We evaluate the combined roles of these genes in N. benthamiana shoot development. We also demonstrate that this mutant background can be used to analyze the capacity of Arabidopsis D14 variants to degrade AtSMXL7.
2. RESULTS
2.1. Knockout of two D14 genes in N. benthamiana with CRISPR‐Cas9
N. benthamiana is an allotetraploid that carries two D14 homoeologs in its genome (Figure 1a). The coding sequences of NbD14a and NbD14b are 97% identical at the nucleotide level, resulting in only three amino acid differences (Ala/Thr84, Leu/Ile119, and Ala/Thr257) between the 267‐aa proteins. We selected a pair of Cas9‐compatible gRNAs that would simultaneously target both NbD14 genes in each of their two exons (Figure 1). These gRNAs were cloned into an egg cell‐specific promoter‐controlled CRISPR‐Cas9 vector that was originally developed for use in A. thaliana (Wang, Xing, et al., 2015). This construct was stably introduced into wild‐type (wt) N. benthamiana through Agrobacterium tumefaciens‐mediated transformation. We identified a homozygous Nbd14a Nbd14b double mutant (hereafter, Nbd14a,b) in the T 0 generation and subsequently isolated a line free of the CRISPR‐Cas9 transgene. The Nbd14a‐1 allele is composed of two mutations: The first is a 14 bp deletion in exon 1 that results in a frameshift after Arg109 and premature truncation of the protein, and the second is a 1‐bp deletion in exon 2 (Figure 1b). The Nbd14b‐1 allele is a 1‐bp deletion at the same position in exon 2, which causes a frameshift after Cys209 (Figure 1b). This results in loss of the Asp217 and His246 residues of the catalytic triad in addition to many other amino acids. Both Asp217 and His246 are required for SL hydrolysis by Arabidopsis D14 in vitro, and the catalytic histidine residue is necessary for D14 activity in planta (Seto et al., 2019). Therefore, both d14 alleles are expected to cause a complete loss‐of‐function.
FIGURE 1.
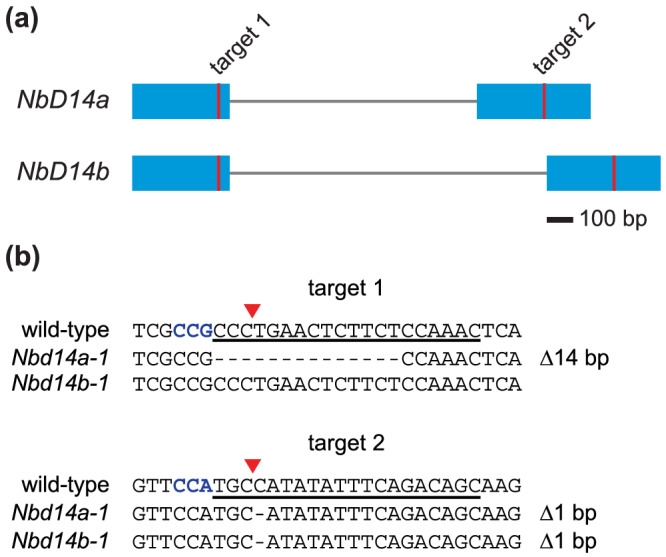
Mutation of two D14 genes in Nicotiana benthamiana with CRISPR‐Cas9. (a) Diagram of D14a and D14b genes in N. benthamiana. Blue boxes represent exons. Vertical red lines indicate gRNA target sites. (b) Sequences of Cas9‐induced frameshift alleles of NbD14a and NbD14b. gRNA target sequence is underlined. Protospacer adjacent motif (PAM) sequence (5′‐NGG‐3′) is indicated in bold blue font. Red triangles denote predicted Cas9 cleavage sites
2.2. N. benthamiana d14a,b has increased shoot branching and altered leaf shape
Loss‐of‐function mutations in D14 cause increased branching/tillering and semi‐dwarf stature in rice, barley, Arabidopsis, canola, petunia, pea, M. truncatula, and L. japonicus (Arite et al., 2009; Arumingtyas et al., 1992; Carbonnel et al., 2020; de Saint Germain et al., 2016; Hamiaux et al., 2012; Lauressergues et al., 2015; Marzec et al., 2016; Napoli & Ruehle, 1996; Stanic et al., 2021; Waters et al., 2012). To determine whether D14 has a similar role in N. benthamiana, we examined the shoot architecture of 5‐week‐old plants grown under greenhouse conditions. To our complete lack of surprise, Nbd14a,b plants had a more compact, “bushy” shoot architecture compared with wt (Figure 2a). Nbd14a,b plants had more axillary branches than wt (Figure 2b). We measured outgrowth of the first nine axillary buds of each plant and found that the three most basal axillary shoots were significantly longer in Nbd14a,b than wt (Figure 2a,c). At the younger, apical nodes, bud outgrowth was reduced, and there was no significant difference in the lengths of Nbd14a,b and wt axillary shoots. Consistent with the semi‐dwarf phenotype of d14 mutants in other angiosperms, we also observed that the height of Nbd14a,b plants was reduced significantly compared with wt (Figure 2a,d).
FIGURE 2.
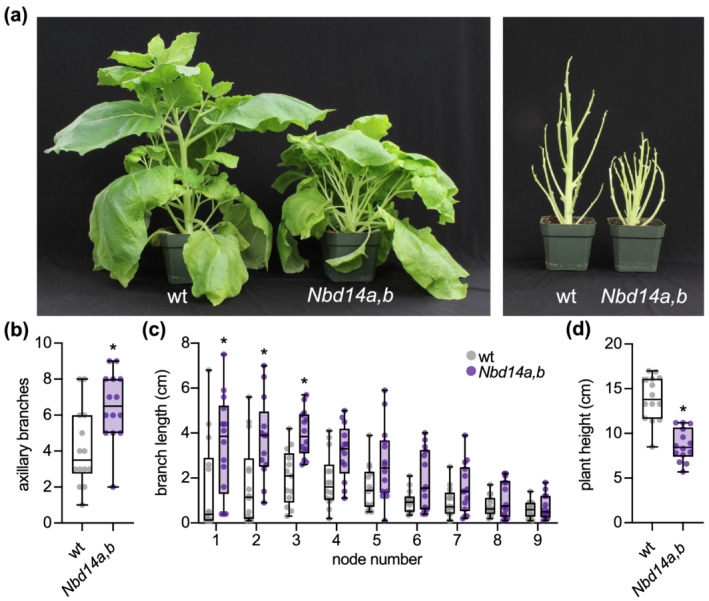
Nbd14a,b has more axillary bud outgrowth and reduced height. (a) Photographs of 10‐week‐old wt and Nbd14a,b N. benthamiana plants with (left) and without (right) leaves. (b) Total number of axillary branches > 1 cm and (c) axillary branch length of the first nine axillary nodes of 58‐day‐old wt and Nbd14a,b plants. (d) Primary shoot height of wt and 58‐day‐old Nbd14a,b. Box plots show median with 25th and 75th percentiles, and whiskers represent minimum and maximum values (n = 10–12 plants). For (b) and (d), *p < .05, unpaired t test with Welch correction. For (c), *p < .05, two‐way ANOVA with Bonferronis multiple comparisons test, comparing Nbd14a,b and wt at each node
The petioles of Arabidopsis d14 leaves have substantially reduced length compared with wt. In addition, the length and width of d14 leaves are reduced, resulting in a smaller and more rounded blade shape overall (Scaffidi et al., 2013; Soundappan et al., 2015; Waters et al., 2012). In M. truncatula, d14 leaflets have increased “solidity” due to increased but shallower serrations at the leaflet margin (Lauressergues et al., 2015). These observations led us to examine leaf morphology in Nbd14a,b. We measured the petiole length, blade length, and blade width of the subtending leaf for each of the first nine axillary buds (Figure 3). The petioles of the first five Nbd14a,b leaves were significantly shorter than wt (Figure 3a). Blade length and width were also reduced significantly for most of the five oldest Nbd14a,b leaves (Figure 3b,c). The most apical leaves of Nbd14a,b and wt had similar dimensions, at least at this age. Together, these data indicate that loss of NbD14a and NbD14b alters the shoot architecture and leaf morphology of N. benthamiana similarly to d14 mutants in other angiosperms.
FIGURE 3.
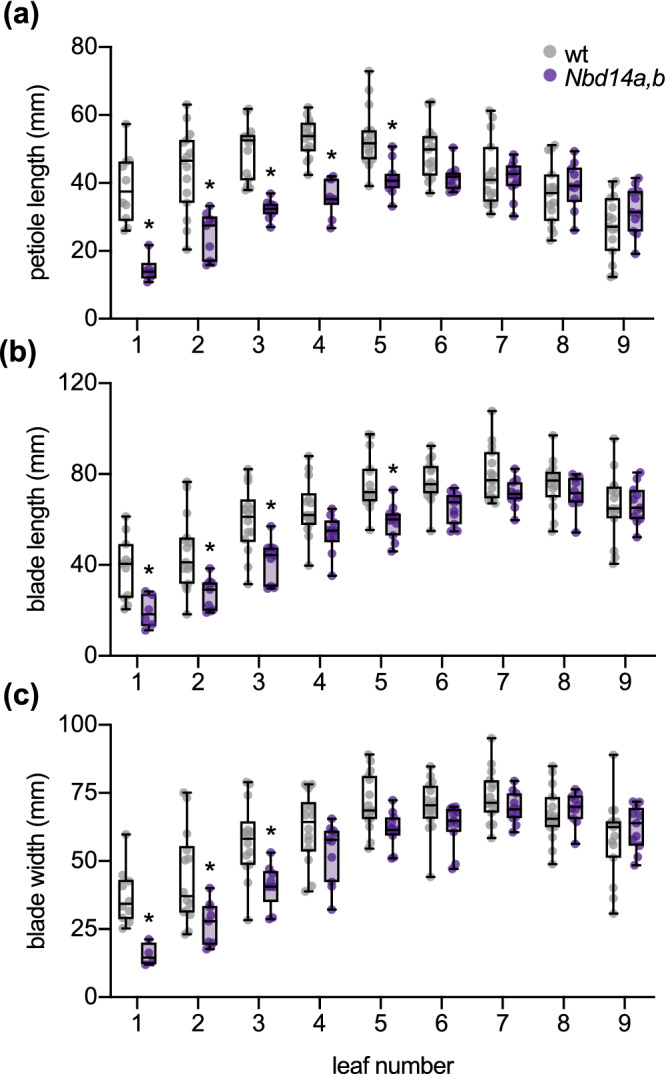
Nbd14a,b has smaller leaves and petioles. (a) Petiole lengths, (b) blade lengths, and (c) blade widths of the first nine leaves of 58‐day‐old wt and Nbd14a,b plants. Box plots show median with 25th and 75th percentiles, and whiskers represent minimum and maximum values (n = 10–12 plants). *p < .05, two‐way ANOVA with Bonferronis multiple comparisons test, comparing Nbd14a,b and wt at each leaf
2.3. GR24‐stimulated degradation of AtSMXL7 in N. benthamiana requires NbD14a,b
We previously used a ratiometric reporter system (pRATIO3212) to show that AtSMXL7 expressed in wt N. benthamiana leaves is degraded after treatment with rac‐GR24, but not KAR1 or KAR2 (Figure 4a; Khosla, Morffy, et al., 2020). To determine whether it is the GR245DS or GR24 ent‐5DS component of rac‐GR24 that triggers AtSMXL7 degradation, we tested the effects of optically pure compounds. Both 10‐μM GR245DS and GR24 ent‐5DS caused a statistically significant reduction in the ratio of fluorescence signals from the AtSMXL7‐mScarlet‐I target relative to the reference protein, Venus (Figure 4b). However, GR245DS caused a stronger decline of the SMXL7 reporter than GR24 ent‐5DS, at least within the 16‐h treatment period (p = .012, Student's two‐tailed t test).
FIGURE 4.
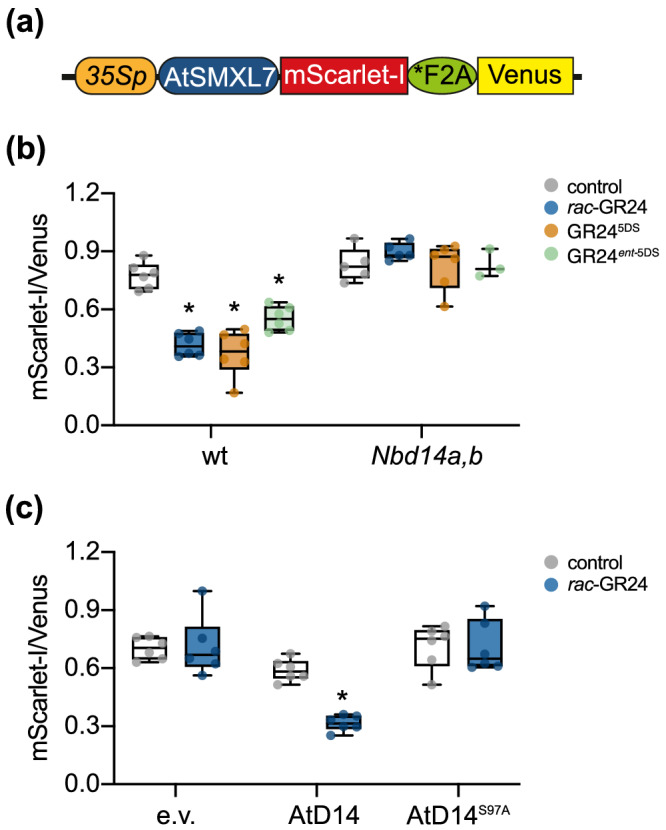
AtSMXL7 degradation in N. benthamiana is NbD14‐dependent. (a) Diagram of the ratiometric AtSMXL7 reporter expressed in pRATIO3212. mScarlet‐I is a fluorescent reporter protein translationally fused to the C‐terminus of AtSMXL7, *F2A is a modified “self‐cleaving” peptide, and Venus is a yellow fluorescent protein used for normalization. Diagram adapted from Khosla, Rodriguez Furlan, et al. (2020). (b) Background‐corrected AtSMXL7‐mScarlet‐I to Venus fluorescence in N. benthamiana wt and Nbd14a,b leaf discs after 16‐h treatment with solvent control (0.02% acetone [v/v]), or 10 μM rac‐GR24, GR245DS, or GR24 ent‐5DS. (c) Background‐corrected AtSMXL7‐mScarlet‐I to Venus fluorescence in Nbd14a,b leaf discs after 16‐h treatment with solvent control (0.02% acetone [v/v]), or 10‐μM rac‐GR24. pRATIO3212‐AtSMXL7 was coexpressed with pGWB415 empty vector (e.v.), AtD14, or AtD14S97A. Box plots show median with 25th and 75th percentiles, and whiskers represent minimum and maximum values (n = 3–6 leaf discs). *p < .05, two‐way ANOVA with (b) Dunnetts multiple comparisons test or (c) Bonferronis multiple comparisons test, comparing GR24 treatments with control for each genotype
We investigated whether GR24‐induced degradation of AtSMXL7 in N. benthamiana is D14‐dependent, or if other proteins such as KAI2 might also contribute. AtSMXL7‐mScarlet‐I abundance was not affected by treatment with rac‐GR24 or either of the purified GR24 stereoisomers in Nbd14a,b leaves (Figure 4b). This strongly suggests that AtSMXL7 degradation after GR24 treatment in N. benthamiana is only caused by NbD14 proteins. This result further implies that NbD14 proteins are more responsive to GR245DS, which shares a stereochemical configuration with naturally occurring SLs, than to GR24 ent‐5DS. Arabidopsis D14 shows a similar preference for GR245DS (Flematti et al., 2016; Samodelov et al., 2016; Scaffidi et al., 2014).
We next tested whether cotransformation of AtD14 could restore rac‐GR24‐induced degradation of the AtSMXL7 reporter to the Nbd14a,b mutant. As negative controls, we compared the effects of an empty vector (pGWB415) and an AtD14S97A mutant on AtSMXL7 degradation. Like many other ɑ/β‐hydrolases, D14 has a highly conserved Ser‐His‐Asp catalytic triad that is necessary for its enzymatic activity. AtD14S97A does not hydrolyze SL and is nonfunctional in plants (Abe et al., 2014; Hamiaux et al., 2012; Seto et al., 2019). Treatment with 10‐μM rac‐GR24 caused a statistically significant decrease in the AtSMXL7 target‐to‐reference ratio when 35S:AtD14 was cotransformed, but had no effect on samples co‐transformed with empty vector or 35S:AtD14 S97A (Figure 4c). This demonstrated that transient expression of AtD14 could rescue SL signaling in Nbd14a,b. It also raised the possibility that this approach could be used to evaluate the ability of different D14 variants to trigger SMXL7 degradation.
2.4. A rapid assay for the induction of AtSMXL7 degradation by AtD14 mutants
A recent study identified several amino acid substitutions that affect yeast two‐hybrid interactions of Petunia x hybrida DAD2 (PhDAD2, a D14 ortholog) with PhMAX2A and/or the SMXL7 ortholog PhD53A (Lee et al., 2020). An F135A substitution enhanced PhDAD2 interactions with PhD53A in the absence of rac‐GR24 but did not affect interactions with PhMAX2A. By contrast, N242I enhanced PhDAD2 interactions with PhMAX2A, but not PhD53A, in the absence of rac‐GR24. When these substitutions were combined, the PhDAD2 mutant protein showed enhanced interactions with both PhD53A and PhMAX2A that were not further stimulated by rac‐GR24. A third substitution, D166A, disrupted PhDAD2 interactions with PhMAX2A but not PhD53A (Lee et al., 2020). Based on these results, PhDAD2F135A and PhDAD2N242I might be expected to have hypersensitive responses to SL, whereas PhDAD2D166A may be insensitive to SL. Because these PhDAD2 variants were not tested in plants, however, it is possible that some of the altered interactions are specific to yeast two‐hybrid and do not translate to effects on SL signaling activity.
We reasoned that the Nbd14a,b mutant could provide a fast assay of D14 signaling activity that complements approaches such as yeast two‐hybrid. We synthesized amino acid substitutions in AtD14 that were equivalent to those previously characterized in PhDAD2: F136A, K243I, F136A/K243I, and D167A. We then tested the ability of these AtD14 variants to trigger degradation of an AtSMXL7 ratiometric reporter after 2‐, 10‐, 50‐, and 250‐nM rac‐GR24 treatments when coexpressed in Nbd14a,b leaves. AtD14F136A showed a similar response to rac‐GR24 as wt AtD14 (Figure 5a). AtD14K243I caused stronger degradation of the SMXL7 reporter than wt AtD14 at 10 nM and higher concentrations of rac‐GR24 (Figure 5b). The AtD14F136A/K243I double mutant showed a similarly enhanced response to rac‐GR24 as AtD14K243I (Figure 5c). It is notable that AtD14F136A/K243I responded positively to rac‐GR24, which was not expected from the yeast two‐hybrid assay of the equivalent DAD2 mutant (Lee et al., 2020). AtD14D167A did not trigger AtSMXL7 degradation as effectively as wt AtD14 (Figure 5d). In the presence of 1‐μM rac‐GR24, the relative fluorescence of AtSMXL7 reporter was reduced 16% by AtD14D167A compared with 43% by wt AtD14 (Figure S1). Because DAD2D166A appeared to have lost the rac‐GR24‐induced yeast two‐hybrid interaction with PhMAX2A, the observation of any rac‐GR24 response by AtD14D167A was also unexpected (Lee et al., 2020). These results support the potential for the Nbd14a,b mutant to serve as a platform for first‐pass screens of D14 variants in plants. This system may reveal changes to SL signaling activity that are not apparent in nonplant assays.
FIGURE 5.
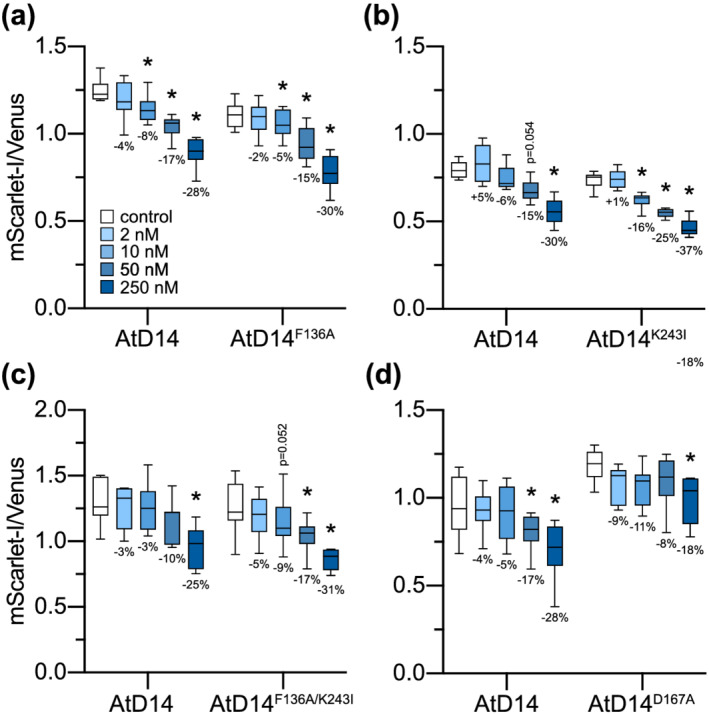
AtSMXL7 degradation is altered when coexpressed with AtD14 variants. Background‐corrected AtSMXL7‐mScarlet‐I to Venus fluorescence in Nbd14a,b leaf discs after 16‐h treatment with solvent control (0.025% acetone [v/v]), 2‐, 10‐, 50‐, or 250‐nM rac‐GR24. pRATIO3212‐AtSMXL7 was coexpressed with pGWB415‐AtD14 or (a) pGWB415‐AtD14F136A, (b) pGWB415‐AtD14K243I, (c) pGWB415‐AtD14F136A/K243I, or (d) pGWB415‐AtD14D167A. Box plots show median with 25th and 75th percentiles, and Tukeys whiskers (n = 6–8 independently transformed leaves, where each leaf value is the mean value of 3–6 discs). *p < .05, repeated‐measures two‐way ANOVA with Dunnetts multiple comparisons test, comparing GR24 treatments with control for each genotype. Percentages below box plots indicate the percent change in the mean mScarlet‐I/Venus ratio of each treatment compared with control
3. DISCUSSION
Here, we have shown that D14 proteins in N. benthamiana have similar functions in plant development and SL signaling as in other angiosperms. We did not investigate whether NbD14a and NbD14b have different roles in development or SL signaling. As these homoeologs are only distinguished at the protein level by three conservative amino acid substitutions, we anticipate that any differences in function would be due to unique expression patterns rather than protein activity. However, distinguishing their expression patterns would be challenging due to the very high nucleotide‐level similarity (97% identity) of NbD14a and NbD14b coding sequences.
The Nbd14a,b double mutant shows increased axillary bud outgrowth, reduced stature, and altered leaf morphology (Figures 2 and 3). The increase in axillary branch number in Nbd14a,b plants is primarily due to stronger outgrowth of buds at the most basal nodes (Figure 2c). This pattern of basitonic development, in which basal branches show more vigorous outgrowth than the apical branches, is also found in the SL‐insensitive or SL‐deficient decreased apical dominance (dad) mutants of P. hybrida, a related solanaceous species (Drummond et al., 2009; Hamiaux et al., 2012; Napoli & Ruehle, 1996; Snowden et al., 2005). Some Nbd14a,b leaves show decreased petiole length, leaf blade length, and leaf blade width (Figure 3). As with axillary bud outgrowth, these phenotypes are more pronounced in the smaller, older leaves found at basal nodes. By contrast, apical leaves of Nbd14a,b are similar to wt.
The similar developmental phenotypes of Nbd14a,b and SL‐insensitive mutants in other species implies that SL signaling is disrupted in this mutant. Indeed, we observed that Nbd14a,b has lost the ability to target an AtSMXL7 reporter for degradation in response to exogenous SL analogs (Figure 4). This strongly suggests that AtSMXL7 degradation is D14‐dependent and does not involve NbKAI2 protein(s). Importantly, the SL response of Nbd14a,b, as indicated by AtSMXL7 reporter degradation, could be rescued through transient expression of wt AtD14.
These results are consistent with the central role of D14 in SL signaling and developmental control that has been well‐established in other plants. The novel value of the Nbd14a,b mutant lies, however, in the utility of N. benthamiana as a medium for analyses of transiently expressed plant proteins. N. benthamiana is commonly used for transient expression experiments due to the simplicity of transformation and the robust transgene expression that can be achieved within a few days (Bally et al., 2018). We proposed that Nbd14a,b could provide a useful genetic background to perform preliminary analyses of the SL signaling activity of D14 variants. This may enable explorations of how D14 mutations affect ligand specificity or how D14 interactions with specific SMXL protein targets are achieved.
To test this idea, we examined the ability of several mutant forms of D14 to activate SMXL7 degradation. The AtD14S97A mutant showed no response to rac‐GR24, consistent with previous work that demonstrated this mutation abolishes SL hydrolysis and signaling (Figure 4c; Hamiaux et al., 2012; Abe et al., 2014; Seto et al., 2019). AtD14D167A had reduced SL signaling activity, presumably due to reduced interaction with MAX2 (Figure 5d). In contrast to the observation of abolished yeast two‐hybrid interactions between DAD2D166A and PhMAX2a, our data suggest that AtD14D167A has not completely lost the ability to work with MAX2 (Lee et al., 2020). Based on yeast two‐hybrid experiments with DAD2 mutants, AtD14F136A and AtD14K243I were expected to have enhanced or constitutive interactions with SMXL7 and MAX2, respectively (Lee et al., 2020). These mutations might be expected to cause hypersensitive SL responses by priming or enhancing SCFMAX2‐D14‐SMXL7 complex formation. We observed mildly enhanced responses to rac‐GR24 from AtD14K243I and AtD14F136A/K243I, but AtD14F136A appeared to have normal responses (Figure 5a–c). This might imply that interaction between SCFMAX2 and D14 is more of a limiting factor for formation of the SL signaling complex than interaction between D14 and SMXL7. Interestingly, the exquisitely sensitive, picomolar response to SL conferred by ShHTL7 is due to enhanced affinity for MAX2 rather than SL itself (Wang et al., 2021). Altogether, this assay enables a complementary analysis of D14 variants that reveals details of how they perform in plants, which may differ somewhat from in vitro or nonplant methods.
Assays that evaluate the activity of SL receptors can be useful tools to investigate the contributions of specific amino acids to SL recognition or signaling, or to screen for chemicals that affect SL signaling. To compare our approach and its utility with other alternatives, we discuss below the three major types of assays for SL receptors: (1) those that evaluate a SL receptor alone, (2) those that test protein–protein interactions between a SL receptor and MAX2 and/or its targets, and (3) those that report degradation of the SL receptor or its SMXL target protein(s), which are direct results of SL signaling.
3.1. In vitro assays for SL binding and hydrolysis by D14
A range of biochemical techniques have been used to evaluate the ability of D14 or KAI2 proteins to bind, hydrolyze, and be activated by SL. Isothermal calorimetry (ITC) and surface plasmon resonance are effective ways to measure the in vitro affinity (i.e., K d) of SL receptors for SL. These techniques have been used to study D14, several KAI2‐like proteins in Physcomitrella patens, and a set of 60 mutants of ShHTL7 (Bürger et al., 2019; Kagiyama et al., 2013; Pang et al., 2020). Yoshimulactone green (YLG) competition assays are another popular method to assess the affinity of a SL receptor for different ligands. In these in vitro assays, the SL analog YLG is hydrolyzed by the receptor, releasing a fluorescent byproduct. Compounds are tested for their ability to competitively interfere with YLG hydrolysis, producing a half‐maximal inhibitory concentration (IC50) value that generally corresponds with the receptor's affinity for the compound (Tsuchiya et al., 2015). The YLG competition assay was used to identify a D14 inhibitor from a chemical library of 800 compounds, as well as to test a set of binding pocket mutants of ShHTL7 (Uraguchi et al., 2018; Yoshimura et al., 2018). Hydrolysis of SL results in attachment of the methylbutenolide group from SL onto the catalytic His residue of the receptor (de Saint Germain et al., 2016; Yao et al., 2016). Formation of this “covalently linked intermediate molecule” (CLIM) can be tracked with liquid chromatography‐mass spectrometry and serves as another readout of a SL receptor's activity on a SL or SL analog. However, substrate‐binding and hydrolysis rates may correlate poorly with the receptor's signaling activity (Uraguchi et al., 2018). Differential scanning fluorimetry (DSF) or nano DSF have often been used to monitor shifts in the melting temperature or intrinsic fluorescence of SL receptors that are induced by potential ligands in vitro (Bürger et al., 2019; Hamiaux et al., 2012, 2018; Seto et al., 2019; Waters et al., 2015). These changes may indicate conformational changes in the SL receptor that correspond with its activation for downstream signal transduction.
In vitro assays for SL binding or activation of the SL receptor require purification of the protein of interest, which does not seem to pose a significant roadblock. A bigger issue is that these assays do not report how SL perception by the receptor affects downstream signaling events or incorporate the effects of in vivo factors (e.g., protein partners) on the receptor's ligand‐binding, hydrolysis, or signaling activities. For example, the presence of MAX2 is known to slow SL hydrolysis by D14 in vitro, and the affinity of D14 for D53/SMXL7 is enhanced by the presence of MAX2 (Shabek et al., 2018; Yao et al., 2016). This is also a potential weakness of in silico approaches such as molecular docking, pharmacophore modeling, and molecular dynamics simulations that model interactions between SL receptors in isolation and potential ligands (Bürger & Chory, 2020; Fukui et al., 2017; Lee et al., 2020; Mashita et al., 2016).
3.2. An in vivo assay for SL binding
A recent study has provided an exciting new approach to measure SL binding in vivo as well as in vitro. These SL biosensors incorporate a circularly permuted GFP (cpGFP) protein into an external loop joining alpha‐helices 6 and 7 of DAD2 or ShHTL7 (Chesterfield et al., 2020). SL induces conformational shifts in DAD2 or ShHTL7 that also affect the conformation of cpGFP, reducing its fluorescence. A second fluorescent protein fused to the biosensor enables ratiometric measurements of fluorescence that bypass problems with varying biosensor abundance. In tobacco protoplasts, the DAD2‐based biosensor can detect rac‐GR24 concentrations as low as 100 nM (Chesterfield et al., 2020). Because this system is sensitive to even single amino acid shifts in the placement of the cpGFP and must be fine‐tuned for each SL receptor, it may be better suited for screening for agonist or antagonist molecules of the receptor than evaluating the effects of receptor mutations. Also, while this system is able to provide valuable information on binding and detection of SLs by two SL receptors in vitro or in vivo, it does not address how perception affects downstream signaling.
3.3. Assays for SL‐induced protein–protein interactions
Other SL receptor assays have focused on protein–protein interactions that are induced by SL perception. One advantage of these approaches is that they can identify factors beyond ligand‐binding and hydrolysis that affect SL signaling. In vitro pulldowns, coimmunoprecipitation from plant tissue, size‐exclusion chromatography, and AlphaScreen are proven ways to assess interactions between SL receptors, MAX2/D3, and SMXL proteins (Jiang et al., 2013; Shabek et al., 2018; Wang et al., 2020, 2021; Yao et al., 2016, 2017; Zhao et al., 2015; Zhou et al., 2013). Generally, these are low‐throughput assays that can be quite challenging to perform due to difficulties in obtaining sufficient amounts of stable, soluble MAX2, and SMXL proteins. Yeast two‐hybrid or three‐hybrid assays that test interactions between SL receptors, MAX2, and/or SMXL proteins are better suited for testing the effects of SL receptor mutations or screening chemical libraries and have been successfully used for these purposes (Hamiaux et al., 2012; Lee et al., 2020; Nakamura et al., 2019; Seto et al., 2019; Toh et al., 2014; Wang et al., 2021). However, yeast‐based interaction assays may produce false‐positive results, at least for some KAI2 proteins (Wang et al., 2021; Yao et al., 2018).
3.4. Assays for SL‐induced proteolysis
The method we have described in this paper falls within a third class of assays that measure SL‐induced degradation of SL signaling components. These assays are relatively fast and highly specific readouts of SL signaling activity, whereas other downstream effects such as shoot branching, parasitic seed germination, or transcriptional responses may be affected by factors in addition to SLs. D14 is degraded after SL treatment in Arabidopsis and rice (Chevalier et al., 2014; Hu et al., 2017). An Arabidopsis line expressing a D14 fusion to luciferase has been developed as an in vivo assay for the ability of various SLs and SL analogs to induce D14 degradation (Sanchez et al., 2018). Most similar to our system are the ratiometric SL signaling sensors StrigoQuant and Strigo‐D2 (Samodelov et al., 2016; Song et al., 2022). StrigoQuant expresses Renilla luciferase and a SMXL6 fusion to firefly luciferase in a single transcript. These two reporters are separated during translation due to an intervening 2A peptide. StrigoQuant has been deployed in Arabidopsis protoplasts, where it can report SMXL6 degradation induced by rac‐GR24 or SLs at concentrations as low as 10 pM. Although achieving efficient protoplast isolation and transformation can be challenging, StrigoQuant offers the distinct advantage of being able to perform experiments with the many SL pathway mutants available for Arabidopsis. Coexpression of rice D14 with Strigoquant is able to restore SL‐induced SMXL6‐degradation responses to the Arabidopsis d14 mutant, demonstrating that it is feasible to test SL receptor variants with this system (Samodelov et al., 2016). Strigo‐D2 coexpresses from separate 35S promoters a SMXL6 C‐terminal domain (SMXL6‐D2) fused to the yellow fluorescent protein mVenus and a nuclear‐localized mCherry reference protein (Song et al., 2022). Overexpression of SMXL6‐D2‐mVenus has less negative impact on Arabidopsis growth and produces higher signal intensities than full‐length SMXL6‐mVenus. The Strigo‐D2 system provides a rapid (within 20 min) and sensitive readout of SL response in transgenic Arabidopsis plants. Responses to applied 5‐deoxystrigol concentrations as low as 5 nM can be detected, and a range of responsiveness can be observed in different plant tissues (Song et al., 2022). Putatively, the activity of D14 variants could be evaluated in the d14 Strigo‐D2 background through the generation of stable transgenic lines, although this may be time‐consuming.
3.5. Limitations of assays for SL‐induced proteolysis in N. benthamiana
We propose to use the Nbd14a,b‐pRATIO system we have established here as a complement to the techniques described above, but it has its own limitations. The success of this approach requires that heterologous proteins are compatible with endogenous N. benthamiana SL signaling components. For example, if NbMAX2 is not able to interact effectively with either the transiently coexpressed D14 or SMXL7 ratiometric reporter, the assays will fail. In cases where NbMAX2 is not compatible, it may be possible to co‐express a MAX2 clone derived from the species of interest. Another constraint to this approach is the presence of endogenous SLs that may activate some transiently expressed SL receptors prior to the application of an agonist. Receptors with high sensitivity to SL may still be identified by causing low SMXL7 reporter abundance pretreatment. However, adding a mutation that blocks SL biosynthesis to the Nbd14a,b line would be a useful way to eliminate background activation of D14 transgenes. Finally, it should be noted that our system is not suitable for studying the SL receptors in parasitic plants that mediate host perception. These SL receptors are neofunctionalized paralogs of KAI2 that target SMAX1 for degradation (Nelson, 2021). Thus, the endogenous KAI2 proteins that remain present in Nbd14a,b may confuse evaluations of SMAX1 reporter degradation by parasite SL receptors.
4. METHODS
4.1. Genes
NbD14a is found on Sol Genomics Network (SGN) scaffold Niben101Scf02153 (N. benthamiana Genome v1.0.1; Bombarely et al., 2012) and on N. benthamiana Sequencing Consortium (NbSC) scaffold NbLab330C11 (N. benthamiana v3.3; Naim et al., 2012). NbD14b is found on SGN scaffold Niben101Scf06949 and on NbSC scaffold NbLab330C03. AtD14 (AT3G03990) and AtSMXL7 (AT2G29970) have been previously described (Stanga et al., 2013; Waters et al., 2012).
4.2. Construction of CRISPR‐Cas9 constructs
20‐nt guide sequences were selected from the CRISPR‐P 2.0 database for the N. benthamiana genome (v0.4.4) to simultaneously target NbS00019774g0007 and NbS00024870g0006 with no mismatches (Bombarely et al., 2012; Liu et al., 2017). The next most likely off‐target sites (based on off‐score) for each guide selected had at least three mismatches and were located in intergenic or intron regions. The two guide sequences were cloned into the pHEE401E vector according to the simplified protocol for two gRNA expression cassettes for dicots (Wang, Xing, et al., 2015). Briefly, high‐fidelity PCR amplification of a pCBC‐DT1T2 template (containing a U6‐26 terminator and U6‐29 promoter) with overlapping primers was performed to add on a guide sequence and BsaI restriction site at each end. Primer sequences for NbD14‐DT1‐BsF, NbD14‐DT1‐F0, NbD14‐DT2‐R0, and NbD14‐DT2‐BsR are described in Table S1. After purification of the extended PCR fragment, GoldenGate cloning with BsaI and pHEE401E was performed. Electrocompetent Escherichia coli (strain DH5a) was transformed with the reaction product and selected on solid Luria Broth (LB) medium supplemented with 50‐mg/L kanamycin. Colony PCR was performed with U6‐26p‐F and U6‐29p‐R. Plasmids were purified from colonies positive for a successful vector insertion (726‐bp product), and both guide sequences were verified by Sanger sequencing with U6‐26p‐F and U6‐29p‐F. A. tumefaciens (strain GV3101) was transformed with the pHEE401E‐NbD14 construct and selected on solid LB medium supplemented with 50‐mg/L kanamycin, 25‐mg/L gentamicin, and 25‐mg/L rifampicin.
4.3. Stable transformation of N. benthamiana
Transformation of N. benthamiana with the pHEE401E‐NbD14 construct was performed by the Plant Transformation Facility at University of California, Davis. Newly expanded leaves from in vitro‐grown N. benthamiana plantlets were removed and cut into 1 cm2 pieces while suspended in a solution of A. tumefaciens adjusted to an OD600 of 0.1–0.2. Leaf pieces were transferred abaxial side down onto cocultivation medium consisting of Murashige and Skoog minimal organics with 8% (w/v) agar medium (MSO) supplemented with 30 g/L sucrose, 2.0 mg/L 6‐benzylaminopurine (BAP) and 200‐μM acetosyringone, pH 5.6–5.8, and incubated 2 days at 23°C in the dark. After 2 days, leaf pieces were transferred to shoot induction medium consisting of MSO medium supplemented with 30‐g/L sucrose, 2.0‐mg/L BAP, 400‐mg/L carbenicillin, 250‐mg/L cefotaxime, 25‐mg/L hygromycin and incubated for 10 days at 26°C under a 16‐h light (intensity 30 μM m2 s−1):8‐h dark photoperiod. After 10 days, tissue was subcultured every 21 days onto the same medium formulation until buds developed. Developing buds were then transferred to elongation medium consisting of MSO medium supplemented with 30‐g/L sucrose, 0.1‐mg/L BAP, 400‐mg/L carbenicillin, 250‐mg/L cefotaxime, and 25‐mg/L hygromycin, and the tissue was subcultured every 21 days until shoots developed. After shoots reached 3–4 cm in height, they were harvested and transferred to rooting medium consisting of 0.5× MSO medium supplemented with 15‐g/L sucrose, 0.2‐mg/L indole‐3‐butyric acid (IBA), 400‐mg/L carbenicillin, 250‐mg/L cefotaxime, and 25‐mg/L hygromycin for 14 days.
4.4. Plant growth conditions
Plants used in growth, branching, and leaf morphology assays were grown on soil in a greenhouse in Riverside, CA, from beginning of October 2019 through to mid December 2019. Plants were watered regularly every other day. Plants used in images for Figure 2a and SMXL7 degradation assays were grown on soil in a growth room under long day conditions (16 h white light at intensity 120 μM m2 s−1/8 h dark) at 22°C.
4.5. Genotyping
DNA was extracted from young leaf tissue using the DNAzol protocol (Molecular Research Center, Inc) and analyzed by PCR with Taq DNA polymerase (New England Biolabs). NbD14a was amplified with NbD14a,b‐F and NbD14a‐3'UTR‐R primers with the following thermal cycling conditions: 95°C for 3 min; 40 cycles of 95°C for 30 s, 59°C for 30 s, 68°C for 2 min; 68°C for 5 min. NbD14a,b‐F and NbD14a‐R were used for Sanger sequencing of purified PCR products. The first exon of NbD14b was amplified with NbD14a,b‐F and NbD14b‐Intron‐R, and the second exon was amplified with NbD14b‐Intron‐F and NbD14b‐R with the following thermal cycling conditions: 95°C for 3 min; 35 cycles of 95°C for 30 s, 52°C for 30 s, 68°C for 1 min; 68°C for 5 min. NbD14a,b‐F, NbD14b‐Intron‐R, NbD14b‐Intron‐F, and NbD14b‐R were used for Sanger sequencing of purified PCR products. The absence of the pHEE401E T‐DNA was validated using pHEE401EhygB‐F and pHEE401EhygB‐R primers for the hygromycin resistance gene with the following thermal cycling conditions: 95°C for 3 min; 40 cycles of 95°C for 30 s, 57°C for 30 s, 68°C for 45 s; 68°C for 5 min.
4.6. Plant growth, shoot branching, and leaf morphology assays
One‐week‐old seedlings were transplanted and grown for 51 days in the greenhouse. From the base of each plant and in developmental order, each branch at the base of the node was cut off and measured from the cutoff point to the meristematic zone, and the leaf was photographed. Each plant was measured from the soil level to the shoot apical meristem. Excised leaves were photographed and petiole length, leaf blade length, and leaf blade width were measured using ImageJ (NIH). Graphs and statistical analysis were performed in Prism (GraphPad).
4.7. SMXL7 degradation assays
GR245DS, GR24ent‐5DS, and rac‐GR24 were synthesized and purified by Dr Adrian Scaffidi and Dr Gavin Flematti (University of Western Australia). The ratiometric reporter for AtSMXL7, pRATIO3212‐SMXL7, is previously described (Khosla, Morffy, et al., 2020). pRATIO3212‐SMXL7 was transformed into an A. tumefaciens strain GV3101 that also carries a plasmid expressing p19, a suppressor of gene‐silencing. AtD14 variants were synthesized (Twist Bioscience), cloned into an ampicillin‐resistant pDONR220 entry vector, sequence‐verified, and cloned into the plant transformation vector pGWB415 (Nakagawa et al., 2007) using Gateway BP and LR cloning enzymes (Invitrogen). pGWB415‐D14 vectors were transformed into A. tumefaciens strain GV3101. Transient transformation of N. benthamiana and measurement of mScarlet‐I and Venus was performed as described previously in a detailed protocol (Khosla & Nelson, 2020), with the following modifications for the cell densities of A. tumefaciens cultures resuspended in infiltration media prior to injection in N. benthamiana leaves: Figure 4a, final OD600 = 0.6; Figure 4b, final OD600 = 1.2, with a 1:1 composition of pRATIO/p19:pGWB415 strains; Figure 5, final OD600 = 0.9, with an 8:1 composition of pRATIO/p19:pGWB415 strains. Graphs and statistical analysis were performed in Prism 9 (GraphPad).
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
Experiments were designed, carried out, and analyzed by ARFW, JAM, AK, and DCN. Figure preparation was made by ARFW. Manuscript preparation was performed by ARFW and DCN with contributions and final approval from all authors. Project design was performed by DCN. Funding to support the project was secured by DCN.
Supporting information
Figure S1. AtD14D167A is less sensitive to rac‐GR24 than wt AtD14.
Table S1. Primers used in this study
ACKNOWLEDGMENTS
We thank Dr Gavin Flematti and Dr Adrian Scaffidi (University of Western Australia) for supplying rac‐GR24 and purified GR24 enantiomers. We thank James Eckhardt and Claudia Sepulveda for providing assistance at early stages of the project. We also thank Dr David Tricoli and Bailey Van Bockern at the Plant Transformation Facility of the University of California, Davis for performing transformation of N. benthamiana. We gratefully acknowledge funding support from the US National Science Foundation (NSF) Research Traineeship (NRT) Program Grant DGE‐1922642 “Plants3D” to AW, and NSF grants IOS‐1740560 and IOS‐1856741 to DCN.
White, A. R. F. , Mendez, J. A. , Khosla, A. , & Nelson, D. C. (2022). Rapid analysis of strigolactone receptor activity in a Nicotiana benthamiana dwarf14 mutant. Plant Direct, 6(3), e389. 10.1002/pld3.389
The author(s) responsible for distribution of materials integral to the findings presented in this article is David C. Nelson (david.nelson@ucr.edu).
DATA AVAILABILITY STATEMENT
All biological resources developed in this study are available upon request and completion of a Material Transfer Agreement.
REFERENCES
- Abe, S. , Sado, A. , Tanaka, K. , Kisugi, T. , Asami, K. , Ota, S. , Kim, H. I. , Yoneyama, K. , Xie, X. , Ohnishi, T. , Seto, Y. , Yamaguchi, S. , Akiyama, K. , Yoneyama, K. , & Nomura, T. (2014). Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proceedings of the National Academy of Sciences of the United States of America, 111(50), 18084–18089. 10.1073/pnas.1410801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti, J. , Herold, S. , Schwarz, M. , Sanchez, P. , Ljung, K. , Dun, E. A. , Brewer, P. B. , Beveridge, C. A. , Sieberer, T. , Sehr, E. M. , & Greb, T. (2011). Strigolactone signaling is required for auxin‐dependent stimulation of secondary growth in plants. Proceedings of the National Academy of Sciences of the United States of America, 108(50), 20242–20247. 10.1073/pnas.1111902108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, M. Z. , Rehman, N. U. , Yu, S. , Zhou, Y. , Haq, B. U. , Wang, J. , Li, P. , Zeng, Z. , & Zhao, J. (2020). GmMAX2‐D14 and ‐KAI interaction‐mediated SL and KAR signaling play essential roles in soybean root nodulation. The Plant Journal, 101, 334–351. 10.1111/tpj.14545 [DOI] [PubMed] [Google Scholar]
- Akiyama, K. , Matsuzaki, K.‐I. , & Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435(7043), 824–827. 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- Arite, T. , Umehara, M. , Ishikawa, S. , Hanada, A. , Maekawa, M. , Yamaguchi, S. , & Kyozuka, J. (2009). d14, a strigolactone‐insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology, 50(8), 1416–1424. 10.1093/pcp/pcp091 [DOI] [PubMed] [Google Scholar]
- Arumingtyas, E. L. , Floyd, R. S. , Gregory, M. J. , & Mufert, I. C. (1992). Branching in Pisum: Inheritance and allelism tests with 17 ramosus mutants. Pisum Genetics, 24, 17–31. [Google Scholar]
- Bally, J. , Jung, H. , Mortimer, C. , Naim, F. , Philips, J. G. , Hellens, R. , Bombarely, A. , Goodin, M. M. , & Waterhouse, P. M. (2018). The rise and rise of Nicotiana benthamiana: A plant for all reasons. Annual Review of Phytopathology, 56(1), 405–426. 10.1146/annurev-phyto-080417-050141 [DOI] [PubMed] [Google Scholar]
- Besserer, A. , Bécard, G. , Jauneau, A. , Roux, C. , & Séjalon‐Delmas, N. (2008). GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiology, 148, 402–413. 10.1104/pp.108.121400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer, A. , Puech‐Pagès, V. , Kiefer, P. , Gomez‐Roldan, V. , Jauneau, A. , Roy, S. , Portais, J.‐C. , Roux, C. , Bécard, G. , & Séjalon‐Delmas, N. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biology, 4(7), e226. 10.1371/journal.pbio.0040226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely, A. , Rosli, H. G. , Vrebalov, J. , Moffett, P. , Mueller, L. A. , & Martin, G. B. (2012). A draft genome sequence of Nicotiana benthamiana to enhance molecular plant‐microbe biology research. Molecular Plant‐Microbe Interactions, 25(12), 1523–1530. 10.1094/MPMI-06-12-0148-TA [DOI] [PubMed] [Google Scholar]
- Bouwmeester, H. , Li, C. , Thiombiano, B. , Rahimi, M. , & Dong, L. (2021). Adaptation of the parasitic plant lifecycle: Germination is controlled by essential host signaling molecules. Plant Physiology, 185(4), 1292–1308. 10.1093/plphys/kiaa066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, Q. , Lv, T. , Shen, H. , Luong, P. , Wang, J. , Wang, Z. , Huang, Z. , Xiao, L. , Engineer, C. , Kim, T. H. , Schroeder, J. I. , & Huq, E. (2014). Regulation of drought tolerance by the F‐box protein MAX2 in Arabidopsis. Plant Physiology, 164, 424–439. 10.1104/pp.113.226837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunsick, M. , Toh, S. , Wong, C. , Xu, Z. , Ly, G. , McErlean, C. S. P. , Pescetto, G. , Nemrish, K. E. , Sung, P. , Li, J. D. , Scholes, J. D. , & Lumba, S. (2020). SMAX1‐dependent seed germination bypasses GA signalling in Arabidopsis and Striga. Nat Plants, 6, 646–652. 10.1038/s41477-020-0653-z [DOI] [PubMed] [Google Scholar]
- Bürger, M. , & Chory, J. (2020). In‐silico analysis of the strigolactone ligand‐receptor system. Plant Direct, 4(9), e00263. 10.1002/pld3.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger, M. , Mashiguchi, K. , Lee, H. J. , Nakano, M. , Takemoto, K. , Seto, Y. , Yamaguchi, S. , & Chory, J. (2019). Structural basis of Karrikin and non‐natural Strigolactone perception in Physcomitrella patens . Cell Reports, 26(4), 855, e5–865. 10.1016/j.celrep.2019.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel, S. , Das, D. , Varshney, K. , Kolodziej, M. C. , Villaécija‐Aguilar, J. A. , & Gutjahr, C. (2020). The karrikin signaling regulator SMAX1 controls Lotus japonicus root and root hair development by suppressing ethylene biosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 117(35), 21757–21765. 10.1073/pnas.2006111117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel, S , Torabi, S , Griesmann, M , Bleek, E , Tang, Y , Carbonnel, S. , Torabi, S. , Griesmann, M. , Bleek, E. , Tang, Y. , Buchka, S. , Basso, V. , Shindo, M. , Boyer, F.‐D. , Wang, T. L. , Udvardi, M. , Waters, M. T. , & Gutjahr, C. (2020). Lotus japonicus karrikin receptors display divergent ligand‐binding specificities and organ‐dependent redundancy. PLOS Genetics, 16(12), e1009249. 10.1371/journal.pgen.1009249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesterfield, R. J. , Whitfield, J. H. , Pouvreau, B. , Cao, D. , Alexandrov, K. , Beveridge, C. A. , & Vickers, C. E. (2020). Rational Design of Novel Fluorescent Enzyme Biosensors for direct detection of Strigolactones. ACS Synthetic Biology, 9(8), 2107–2118. 10.1021/acssynbio.0c00192 [DOI] [PubMed] [Google Scholar]
- Chevalier, F. , Nieminen, K. , Sánchez‐Ferrero, J. C. , Rodríguez, M. L. , Chagoyen, M. , Hardtke, C. S. , & Cubas, P. (2014). Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell, 26(3), 1134–1150. 10.1105/tpc.114.122903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Lee, T. , Cho, J. , Servante, E. K. , Pucker, B. , Summers, W. , Bowden, S. , Rahimi, M. , An, K. , An, G. , Bouwmeester, H. J. , Wallington, E. J. , Oldroyd, G. , & Paszkowski, U. (2020). The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nature Communications, 11, 2114. 10.1038/s41467-020-16021-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn, C. E. , Bythell‐Douglas, R. , Neumann, D. , Yoshida, S. , Whittington, B. , Westwood, J. H. , Shirasu, K. , Bond, C. S. , Dyer, K. A. , & Nelson, D. C. (2015). PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science, 349(6247), 540–543. 10.1126/science.aab1140 [DOI] [PubMed] [Google Scholar]
- Conn, C. E. , & Nelson, D. C. (2016). Evidence that KARRIKIN‐INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not Karrikin or Strigolactone. Frontiers in Plant Science, 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain, A. , Clavé, G. , Badet‐Denisot, M. A. , Pillot, J. P. , Cornu, D. , le Caer, J. P. , Burger, M. , Pelissier, F. , Retailleau, P. , Turnbull, C. , Bonhomme, S. , Chory, J. , Rameau, C. , & Boyer, F. D. (2016). An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nature Chemical Biology, 12(10), 787–794. 10.1038/nchembio.2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain, A. , Jacobs, A. , Brun, G. , Pouvreau, J.‐B. , Braem, L. , Cornu, D. , Clavé, G. , Baudu, E. , Steinmetz, V. , Servajean, V. , Wicke, S. , Gevaert, K. , Simier, P. , Goormachtig, S. , Delavault, P., & Boyer, F.‐D. (2021). A Phelipanche ramosa KAI2 protein perceives strigolactones and isothiocyanates enzymatically. Plant Communications, 2(5), 100166. 10.1016/j.xplc.2021.100166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, R. S. M. , Martínez‐Sánchez, N. M. , Janssen, B. J. , Templeton, K. R. , Simons, J. L. , Quinn, B. D. , Karunairetnam, S. , & Snowden, K. C. (2009). Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiology, 151(4), 1867–1877. 10.1104/pp.109.146720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, R. S. M. , Sheehan, H. , Simons, J. L. , Martínez‐Sánchez, N. M. , Turner, R. M. , Putterill, J. , & Snowden, K. C. (2011). The expression of Petunia Strigolactone pathway genes is altered as part of the endogenous developmental program. Frontiers in Plant Science, 2, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flematti, G. R. , Scaffidi, A. , Waters, M. T. , & Smith, S. M. (2016). Stereospecificity in strigolactone biosynthesis and perception. Planta, 243(6), 1361–1373. 10.1007/s00425-016-2523-5 [DOI] [PubMed] [Google Scholar]
- Fukui, K. , Yamagami, D. , Ito, S. , & Asami, T. (2017). A Taylor‐made design of Phenoxyfuranone‐type Strigolactone mimic. Frontiers in Plant Science, 8, 936. 10.3389/fpls.2017.00936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Roldan, V. , Fermas, S. , Brewer, P. B. , Puech‐Pagès, V. , Dun, E. A. , Pillot, J.‐P. , Letisse, F. , Matusova, R. , Danoun, S. , Portais, J.‐C. , & Bouwmeester, H. (2008). Strigolactone inhibition of shoot branching. Nature, 455(7210), 189–194. 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- Gutjahr, C. , Gobbato, E. , Choi, J. , Riemann, M. , Johnston, M. G. , Summers, W. , Carbonnel, S. , Mansfield, C. , Yang, S.‐Y. , Nadal, M. , Acosta, I. , Takano, M. , Jiao, W. B. , Schneeberger, K. , Kelly, K. A. , & Paszkowski, U. (2015). Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science, 350(6267), 1521–1524. 10.1126/science.aac9715 [DOI] [PubMed] [Google Scholar]
- Hamiaux, C. , Drummond, R. S. M. , Janssen, B. J. , Ledger, S. E. , Cooney, J. M. , Newcomb, R. D. , & Snowden, K. C. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Current Biology, 22(21), 2032–2036. 10.1016/j.cub.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Hamiaux, C. , Drummond, R. S. M. , Luo, Z. , Lee, H. W. , Sharma, P. , Janssen, B. J. , Perry, N. B. , Denny, W. A. , & Snowden, K. C. (2018). Inhibition of strigolactone receptors by N‐phenylanthranilic acid derivatives: Structural and functional insights. The Journal of Biological Chemistry, 293(17), 6530–6543. 10.1074/jbc.RA117.001154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Q. , He, Y. , Wang, L. , Liu, S. , Meng, X. , Liu, G. , Jing, Y. , Chen, M. , Song, X. , Jiang, L. , Yu, H. , Wang, B. , & Li, J. (2017). DWARF14, A receptor covalently linked with the active form of Strigolactones, undergoes Strigolactone‐dependent degradation in Rice. Frontiers in Plant Science, 8, 1935. 10.3389/fpls.2017.01935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Liu, X. , Xiong, G. , Liu, H. , Chen, F. , Wang, L. , Meng, X. , Liu, G. , Yu, H. , Yuan, Y. , Yi, W. , Zhao, L. , Ma, H. , He, Y. , Wu, Z. , Melcher, K. , Qian, Q. , Xu, H. E. , Wang, Y. , & Li, J. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature, 504(7480), 401–405. 10.1038/nature12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama, M. , Hirano, Y. , Mori, T. , Kim, S.‐Y. , Kyozuka, J. , Seto, Y. , Yamaguchi, S. , & Hakoshima, T. (2013). Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells, 18, 147–160. 10.1111/gtc.12025 [DOI] [PubMed] [Google Scholar]
- Kalliola, M. , Jakobson, L. , Davidsson, P. , Pennanen, V. , Waszczak, C. , Yarmolinsky, D. , Zamora, O. , Palva, E. T. , Kariola, T. , Kollist, H. , & Brosché, M. (2020). Differential role of MAX2 and strigolactones in pathogen, ozone, and stomatal responses. Plant Direct, 4, e00206. 10.1002/pld3.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik, Y. , Delaux, P.‐M. , Resnick, N. , Mayzlish‐Gati, E. , Wininger, S. , Bhattacharya, C. , Séjalon‐Delmas, N. , Combier, J.‐P. , Bécard, G. , Belausov, E. , Beeckman, T. , Dor, E. , Hershenhorn, J. , & Koltai, H. (2011). Strigolactones affect lateral root formation and root‐hair elongation in Arabidopsis. Planta, 233(1), 209–216. 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- Khosla, A. , Morffy, N. , Li, Q. , Faure, L. , Chang, S. H. , Yao, J. , Zheng, J. , Cai, M. L. , Stanga, J. P. , Flematti, G. R. , Waters, M. T. , & Nelson, D. C. (2020). Structure‐Function Analysis of SMAX1 Reveals Domains that Mediate its Karrikin‐Induced Proteolysis and Interaction with the Receptor KAI2. The Plant Cell, 32(8), 2639–2659. 10.1105/tpc.19.00752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla, A. , & Nelson, D. C. (2016). Strigolactones, super hormones in the fight against Striga. Current Opinion in Plant Biology, 33, 57–63. 10.1016/j.pbi.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Khosla, A. , & Nelson, D. C. (2020). Ratiometric measurement of protein abundance after transient expression of a transgene in Nicotiana benthamiana . Bio‐Protocol, 10(17), e3747. 10.21769/BioProtoc.3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla, A. , Rodriguez Furlan, C. , Kapoor, S. , Van Norman, J. M. , & Nelson, D. C. (2020). A series of dual reporter vectors for ratiometric analysis of protein abundance in plants. Plant Direct, 4(6), e00231. 10.1002/pld3.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobae, Y. , Kameoka, H. , Sugimura, Y. , Saito, K. , Ohtomo, R. , Fujiwara, T. , & Kyozuka, J. (2018). Strigolactone biosynthesis genes of Rice are required for the punctual entry of arbuscular mycorrhizal Fungi into the roots. Plant & Cell Physiology, 59(3), 544–553. 10.1093/pcp/pcy001 [DOI] [PubMed] [Google Scholar]
- Kretzschmar, T. , Kohlen, W. , Sasse, J. , Borghi, L. , Schlegel, M. , Bachelier, J. B. , Reinhardt, D. , Bours, R. , Bouwmeester, H. J. , & Martinoia, E. (2012). A petunia ABC protein controls strigolactone‐dependent symbiotic signalling and branching. Nature, 483(7389), 341–344. 10.1038/nature10873 [DOI] [PubMed] [Google Scholar]
- Lahari, Z. , Ullah, C. , Kyndt, T. , Gershenzon, J. , & Gheysen, G. (2019). Strigolactones enhance root‐knot nematode (Meloidogyne graminicola) infection in rice by antagonizing the jasmonate pathway. The New Phytologist, 224, 454–465. 10.1111/nph.15953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues, D. , André, O. , Peng, J. , Wen, J. , Chen, R. , Ratet, P. , Tadege, M. , Mysore, K. S. , & Rochange, S. F. (2015). Strigolactones contribute to shoot elongation and to the formation of leaf margin serrations in Medicago truncatula R108. Journal of Experimental Botany, 66(5), 1237–1244. 10.1093/jxb/eru471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. W. , Sharma, P. , Janssen, B. J. , Drummond, R. S. M. , Luo, Z. , Hamiaux, C. , Collier, T. , Allison, J. R. , Newcomb, R. D. , & Snowden, K. C. (2020). Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. The Journal of Biological Chemistry, 295(13), 4181–4193. 10.1074/jbc.RA119.011509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Martín‐Fontecha, E. S. , Khosla, A. , White, A. R. F. , Chang, S. , Cubas, P. , & Nelson, D. C. (2022). The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress. Plant Communications, 3(2), 100303. 10.1016/j.xplc.2022.100303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Nguyen, K. H. , Chu, H. D. , Ha, C. V. , Watanabe, Y. , Osakabe, Y. , Leyva‐González, M. A. , Sato, M. , Toyooka, K. , Voges, L. , Tanaka, M. , Mostofa, M. G. , Seki, M. , Seo, M. , Yamaguchi, S. , Nelson, D. C. , Tian, C. , Herrera‐Estrella, L. , & Tran, L. P. (2017). The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana . PLoS Genetics, 13(11), e1007076. 10.1371/journal.pgen.1007076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Nguyen, K. H. , Chu, H. D. , Watanabe, Y. , Osakabe, Y. , Sato, M. , Toyooka, K. , Seo, M. , Tian, L. , Tian, C. , Yamaguchi, S. , Tanaka, M. , Seki, M. , & Tran, L. P. (2020). Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE 2 in drought adaptation of Arabidopsis thaliana . The Plant Journal, 103, 111–127. 10.1111/tpj.14712 [DOI] [PubMed] [Google Scholar]
- Liu, H. , Ding, Y. , Zhou, Y. , Jin, W. , Xie, K. , & Chen, L.‐L. (2017). CRISPR‐P 2.0: An improved CRISPR‐Cas9 tool for genome editing in plants. Molecular Plant, 10(3), 530–532. 10.1016/j.molp.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Zhang, Y. , Matusova, R. , Charnikhova, T. , Amini, M. , Jamil, M. , Fernandez‐Aparicio, M. , Huang, K. , Timko, M. P. , Westwood, J. H. , Ruyter‐Spira, C. , van der Krol, S. , & Bouwmeester, H. J. (2014). Striga hermonthica MAX2 restores branching but not the very low Fluence response in the Arabidopsis thaliana max2 mutant. The New Phytologist, 202, 531–541. 10.1111/nph.12692 [DOI] [PubMed] [Google Scholar]
- Liu, R. , Hou, J. , Li, H. , Xu, P. , Zhang, Z. , & Zhang, X. (2021). Association of TaD14‐4D, a gene involved in Strigolactone signaling, with yield contributing traits in wheat. International Journal of Molecular Sciences, 22(7). 10.3390/ijms22073748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke, G. , Escuin, H. , De Felipe, P. , & Ryan, M. (2010). 2A to the fore ‐ research, technology and applications. Biotechnology & Genetic Engineering Reviews, 26(1), 223–260. 10.5661/bger-26-223 [DOI] [PubMed] [Google Scholar]
- Machin, D. C. , Hamon‐Josse, M. , & Bennett, T. (2020). Fellowship of the rings: A saga of strigolactones and other small signals. The New Phytologist, 225, 621–636. 10.1111/nph.16135 [DOI] [PubMed] [Google Scholar]
- Marzec, M. , Gruszka, D. , Tylec, P. , & Szarejko, I. (2016). Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare . Physiologia Plantarum, 158, 341–355. 10.1111/ppl.12460 [DOI] [PubMed] [Google Scholar]
- Mashita, O. , Koishihara, H. , Fukui, K. , Nakamura, H. , & Asami, T. (2016). Discovery and identification of 2‐methoxy‐1‐naphthaldehyde as a novel strigolactone‐signaling inhibitor. Journal of Pesticide Science, 41, 71–78. 10.1584/jpestics.D16-028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim, F. , Nakasugi, K. , Crowhurst, R. N. , Hilario, E. , Zwart, A. B. , Hellens, R. P. , Taylor, J. M. , Waterhouse, P. M. , & Wood, C. C. (2012). Advanced engineering of lipid metabolism in Nicotiana benthamiana using a draft genome and the V2 viral silencing‐suppressor protein. PLoS ONE, 7(12), e52717. 10.1371/journal.pone.0052717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. , Kurose, T. , Hino, T. , Tanaka, K. , Kawamukai, M. , Niwa, Y. , Toyooka, K. , Matsuoka, K. , Jinbo, T. , & Kimura, T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering, 104, 34–41. 10.1263/jbb.104.34 [DOI] [PubMed] [Google Scholar]
- Nakamura, H. , Hirabayashi, K. , Miyakawa, T. , Kikuzato, K. , Hu, W. , Xu, Y. , Jiang, K. , Takahashi, I. , Niiyama, R. , Dohmae, N. , Tanokura, M. , & Asami, T. (2019). Triazole Ureas covalently bind to Strigolactone receptor and antagonize Strigolactone responses. Molecular Plant, 12, 44–58. 10.1016/j.molp.2018.10.006 [DOI] [PubMed] [Google Scholar]
- Napoli, C. A. , & Ruehle, J. (1996). New mutations affecting meristem growth and potential in Petunia hybrida Vilm. The Journal of Heredity, 87(5), 371–377. 10.1093/oxfordjournals.jhered.a023016 [DOI] [Google Scholar]
- Nasir, F. , Tian, L. , Shi, S. , Chang, C. , Ma, L. , Gao, Y. , & Tian, C. (2019). Strigolactones positively regulate defense against Magnaporthe oryzae in rice (Oryza sativa). Plant Physiology and Biochemistry, 142, 106–116. 10.1016/j.plaphy.2019.06.028 [DOI] [PubMed] [Google Scholar]
- Nelson, D. C. (2021). The mechanism of host‐induced germination in root parasitic plants. Plant Physiology, 185(4), 1353–1373. 10.1093/plphys/kiab043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. C. , Flematti, G. R. , Riseborough, J.‐A. , Ghisalberti, E. L. , Dixon, K. W. , & Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 107(15), 7095–7100. 10.1073/pnas.0911635107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. C. , Riseborough, J.‐A. , Flematti, G. R. , Stevens, J. , Ghisalberti, E. L. , Dixon, K. W. , & Smith, S. M. (2009). Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiology, 149, 863–873. 10.1104/pp.108.131516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D. C. , Scaffidi, A. , Dun, E. A. , Waters, M. T. , Flematti, G. R. , Dixon, K. W. , Beveridge, C. A. , Ghisalberti, E. L. , & Smith, S. M. (2011). F‐box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 108(21), 8897–8902. 10.1073/pnas.1100987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, Z. , Zhang, X. , Ma, F. , Liu, J. , Zhang, H. , Wang, J. , Wen, X. , & Xi, Z. (2020). Comparative studies of potential binding pocket residues reveal the molecular basis of ShHTL receptors in the perception of GR24 in Striga. Journal of Agricultural and Food Chemistry, 68(45), 12729–12737. 10.1021/acs.jafc.0c04947 [DOI] [PubMed] [Google Scholar]
- Ruyter‐Spira, C. , Kohlen, W. , Charnikhova, T. , van Zeijl, A. , van Bezouwen, L. , de Ruijter, N. , Cardoso, C. , Lopez‐Raez, J. A. , Matusova, R. , Bours, R. , Verstappen, F. , & Bouwmeester, H. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiology, 155, 721–734. 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samodelov, S. L. , Beyer, H. M. , Guo, X. , Augustin, M. , Jia, K.‐P. , Baz, L. , Ebenhöh, O. , Beyer, P. , Weber, W. , al‐Babili, S. , & Zurbriggen, M. D. (2016). StrigoQuant: A genetically encoded biosensor for quantifying strigolactone activity and specificity. Science Advances, 2(11), e1601266. 10.1126/sciadv.1601266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, E. , Artuso, E. , Lombardi, C. , Visentin, I. , Lace, B. , Saeed, W. , Lolli, M. L. , Kobauri, P. , Ali, Z. , Spyrakis, F. , Cubas, P. , Cardinale, F. , & Prandi, C. (2018). Structure‐activity relationships of strigolactones via a novel, quantitative in planta bioassay. Journal of Experimental Botany, 69(9), 2333–2343. 10.1093/jxb/ery092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi, A. , Waters, M. T. , Ghisalberti, E. L. , Dixon, K. W. , Flematti, G. R. , & Smith, S. M. (2013). Carlactone‐independent seedling morphogenesis in Arabidopsis. The Plant Journal, 76, 1–9. 10.1111/tpj.12265 [DOI] [PubMed] [Google Scholar]
- Scaffidi, A. , Waters, M. T. , Sun, Y. K. , Skelton, B. W. , Dixon, K. W. , Ghisalberti, E. L. , Flematti, G. R. , & Smith, S. M. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiology, 165(3), 1221–1232. 10.1104/pp.114.240036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto, Y. , Yasui, R. , Kameoka, H. , Tamiru, M. , Cao, M. , Terauchi, R. , Sakurada, A. , Hirano, R. , Kisugi, T. , Hanada, A. , Umehara, M. , Seo, E. , Akiyama, K. , Burke, J. , Takeda‐Kamiya, N. , Li, W. , Hirano, Y. , Hakoshima, T. , Mashiguchi, K. , … Yamaguchi, S. (2019). Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nature Communications, 10, 191. 10.1038/s41467-018-08124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek, N. , Ticchiarelli, F. , Mao, H. , Hinds, T. R. , Leyser, O. , & Zheng, N. (2018). Structural plasticity of D3‐D14 ubiquitin ligase in strigolactone signalling. Nature, 563(7733), 652–656. 10.1038/s41586-018-0743-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H. , Luong, P. , & Huq, E. (2007). The F‐box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiology, 145(4), 1471–1483. 10.1104/pp.107.107227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, M. , Yamamoto, S. , Shimomura, K. , & Umehara, M. (2020). Strigolactones decrease leaf angle in response to nutrient deficiencies in rice. Frontiers in Plant Science, 11, 135. 10.3389/fpls.2020.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, N. , Taylor, C. , & Leyser, O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biology, 11, e1001474. 10.1371/journal.pbio.1001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden, K. C. , Simkin, A. J. , Janssen, B. J. , Templeton, K. R. , Loucas, H. M. , Simons, J. L. , Karunairetnam, S. , Gleave, A. P. , Clark, D. G. , & Klee, H. J. (2005). The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell, 17(3), 746–759. 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, C. , Zhao, J. , Guichard, M. , Shi, D. , Grossmann, G. , Schmitt, C. , Jouannet, V., & Greb, T. (2022). Strigo‐D2—a bio‐sensor for monitoring spatio‐temporal strigolactone signaling patterns in intact plants. Plant Physiology, 188(1), 97–110. 10.1093/plphys/kiab504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan, I. , Bennett, T. , Morffy, N. , Liang, Y. , Stanga, J. P. , Abbas, A. , Leyser, O. , & Nelson, D. C. (2015). SMAX1‐LIKE/D53 family members enable distinct MAX2‐dependent responses to Strigolactones and Karrikins in Arabidopsis. Plant Cell, 27(11), 3143–3159. 10.1105/tpc.15.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga, J. P. , Morffy, N. , & Nelson, D. C. (2016). Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta, 243(6), 1397–1406. 10.1007/s00425-015-2458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga, J. P. , Smith, S. M. , Briggs, W. R. , & Nelson, D. C. (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiology, 163, 318–330. 10.1104/pp.113.221259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic, M. , Hickerson, N. M. N. , Arunraj, R. , & Samuel, M. A. (2021). Gene‐editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola). Plant Biotechnology Journal, 19(4), 639–641. 10.1111/pbi.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X.‐D. , & Ni, M. (2011). HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de‐etiolation. Molecular Plant, 4, 116–126. 10.1093/mp/ssq055 [DOI] [PubMed] [Google Scholar]
- Sun, Y. K. , Flematti, G. R. , Smith, S. M. , & Waters, M. T. (2016). Reporter gene‐facilitated detection of compounds in Arabidopsis leaf extracts that activate the Karrikin signaling pathway. Frontiers in Plant Science, 7, 1799. 10.3389/fpls.2016.01799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck, S. M. , Guerringue, Y. , Matthus, E. , Jamieson, F. J. C. , & Davies, J. M. (2019). Impairment in karrikin but not strigolactone sensing enhances root skewing in Arabidopsis thaliana . The Plant Journal, 98(4), 607–621. 10.1111/tpj.14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh, S. , Holbrook‐Smith, D. , Stogios, P. J. , Onopriyenko, O. , Lumba, S. , Tsuchiya, Y. , Savchenko, A. , & McCourt, P. (2015). Structure‐function analysis identifies highly sensitive strigolactone receptors in Striga. Science, 350, 203–207. 10.1126/science.aac9476 [DOI] [PubMed] [Google Scholar]
- Toh, S. , Holbrook‐Smith, D. , Stokes, M. E. , Tsuchiya, Y. , & McCourt, P. (2014). Detection of parasitic plant suicide germination compounds using a high‐throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chemistry & Biology, 21(8), 988–998. 10.1016/j.chembiol.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Tsuchiya, Y. , Yoshimura, M. , Sato, Y. , Kuwata, K. , Toh, S. , Holbrook‐Smith, D. , Zhang, H. , McCourt, P. , Itami, K. , Kinoshita, T. , & Hagihara, S. (2015). PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science, 349(6250), 864–868. 10.1126/science.aab3831 [DOI] [PubMed] [Google Scholar]
- Ueda, H. , & Kusaba, M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiology, 169, 138–147. 10.1104/pp.15.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara, M. , Hanada, A. , Yoshida, S. , Akiyama, K. , Arite, T. , Takeda‐Kamiya, N. , Magome, H. , Kamiya, Y. , Shirasu, K. , Yoneyama, K. , Kyozuka, J. , & Yamaguchi, S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature, 455, 195–200. 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- Uraguchi, D. , Kuwata, K. , Hijikata, Y. , Yamaguchi, R. , Imaizumi, H. , Am, S. , Rakers, C. , Mori, N. , Akiyama, K. , Irle, S. , McCourt, P. , Kinoshita, T. , Ooi, T. , & Tsuchiya, Y. (2018). A femtomolar‐range suicide germination stimulant for the parasitic plant Striga hermonthica . Science, 362(6420), 1301–1305. 10.1126/science.aau5445 [DOI] [PubMed] [Google Scholar]
- Ha, C. V. , Leyva‐González, M. A. , Osakabe, Y. , Tran, U. T. , Nishiyama, R. , Watanabe, Y. , Tanaka, M. , Seki, M. , Yamaguchi, S. , Dong, N. V. , Yamaguchi‐Shinozaki, K. , Shinozaki, K. , Herrera‐Estrella, L. , & Tran, L. S. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences of the United States of America, 111, 851–856. 10.1073/pnas.1322135111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaécija‐Aguilar, J. A. , Hamon‐Josse, M. , Carbonnel, S. , Kretschmar, A. , Schmid, C. , Dawid, C. , Bennett, T. , & Gutjahr, C. (2019). SMAX1/SMXL2 regulate root and root hair development downstream of KAI2‐mediated signalling in Arabidopsis. PLoS Genetics, 15(8), e1008327. 10.1371/journal.pgen.1008327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Wang, B. , Jiang, L. , Liu, X. , Li, X. , Lu, Z. , Meng, X. , Wang, Y. , Smith, S. M. , & Li, J. (2015). Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53‐like SMXL repressor proteins for ubiquitination and degradation. Plant Cell, 27(11), 3128–3142. 10.1105/tpc.15.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Xu, Q. , Yu, H. , Ma, H. , Li, X. , Yang, J. , Chu, J. , Xie, Q. , Wang, Y. , Smith, S. M. , Li, J. , Xiong, G. , & Wang, B. (2020). Strigolactone and Karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis. Plant Cell, 32(7), 2251–2270. 10.1105/tpc.20.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Zhang, S. , Qiao, J. , Sun, Q. , Shi, Q. , Cai, C. , Mo, J. , Chu, Z. , Yuan, Y. , du, X. , Miao, Y. , Zhang, X. , & Cai, Y. (2019). Functional analysis of the GbDWARF14 gene associated with branching development in cotton. PeerJ, 7, e6901. 10.7717/peerj.6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Yao, R. , Du, X. , Guo, L. , Chen, L. , Xie, D. , & Smith, S. M. (2021). Molecular basis for high ligand sensitivity and selectivity of strigolactone receptors in Striga. Plant Physiology, 185, 1411–1428. 10.1093/plphys/kiaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.‐P. , Xing, H.‐L. , Dong, L. , Zhang, H.‐Y. , Han, C.‐Y. , Wang, X.‐C. , & Chen, Q.‐J. (2015). Egg cell‐specific promoter‐controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biology, 16, 144. 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, M. T. , Gutjahr, C. , Bennett, T. , & Nelson, D. C. (2017). Strigolactone signaling and evolution. Annual Review of Plant Biology, 68(1), 291–322. 10.1146/annurev-arplant-042916-040925 [DOI] [PubMed] [Google Scholar]
- Waters, M. T. , Nelson, D. C. , Scaffidi, A. , Flematti, G. R. , Sun, Y. K. , Dixon, K. W. , & Smith, S. M. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development, 139(7), 1285–1295. 10.1242/dev.074567 [DOI] [PubMed] [Google Scholar]
- Waters, M. T. , Scaffidi, A. , Moulin, S. L. Y. , Sun, Y. K. , Flematti, G. R. , & Smith, S. M. (2015). A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to Karrikins or Strigolactones. Plant Cell, 27(7), 1925–1944. 10.1105/tpc.15.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C. , Xi, L. , Gao, B. , Wang, K. , Lv, S. , Kou, Y. , Ma, N. , & Zhao, L. (2015). Roles of DgD14 in regulation of shoot branching in chrysanthemum (Dendranthema grandiflorum “Jinba”). Plant Physiology and Biochemistry, 96, 241–253. 10.1016/j.plaphy.2015.07.030 [DOI] [PubMed] [Google Scholar]
- Yamada, Y. , Furusawa, S. , Nagasaka, S. , Shimomura, K. , Yamaguchi, S. , & Umehara, M. (2014). Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta, 240, 399–408. 10.1007/s00425-014-2096-0 [DOI] [PubMed] [Google Scholar]
- Yao, J. , Mashiguchi, K. , Scaffidi, A. , Akatsu, T. , Melville, K. T. , Morita, R. , Morimoto, Y. , Smith, S. M. , Seto, Y. , Flematti, G. R. , Yamaguchi, S. , & Waters, M. T. (2018). An allelic series at the KARRIKIN INSENSITIVE 2 locus of Arabidopsis thaliana decouples ligand hydrolysis and receptor degradation from downstream signalling. The Plant Journal, 96(1), 75–89. 10.1111/tpj.14017 [DOI] [PubMed] [Google Scholar]
- Yao, R. , Ming, Z. , Yan, L. , Li, S. , Wang, F. , Ma, S. , Yu, C. , Yang, M. , Chen, L. , Chen, L. , Li, Y. , Yan, C. , Miao, D. , Sun, Z. , Yan, J. , Sun, Y. , Wang, L. , Chu, J. , Fan, S. , … Xie, D. (2016). DWARF14 is a non‐canonical hormone receptor for strigolactone. Nature, 536(7617), 469–473. 10.1038/nature19073 [DOI] [PubMed] [Google Scholar]
- Yao, R. , Wang, F. , Ming, Z. , Du, X. , Chen, L. , Wang, Y. , Zhang, W. , Deng, H. , & Xie, D. (2017). ShHTL7 is a non‐canonical receptor for strigolactones in root parasitic weeds. Cell Research, 27, 838–841. 10.1038/cr.2017.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura, M. , Sato, A. , Kuwata, K. , Inukai, Y. , Kinoshita, T. , Itami, K. , Tsuchiya, Y. , & Hagihara, S. (2018). Discovery of shoot branching regulator targeting strigolactone receptor DWARF14. ACS Central Science, 4, 230–234. 10.1021/acscentsci.7b00554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L.‐H. , Zhou, X. E. , Yi, W. , Wu, Z. , Liu, Y. , Kang, Y. , Hou, L. , de Waal, P. W. , Li, S. , Jiang, Y. , Scaffidi, A. , Flematti, G. R. , Smith, S. M. , Lam, V. Q. , Griffin, P. R. , Wang, Y. , Li, J. , Melcher, K. , & Xu, H. E. (2015). Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3‐ligase signaling effector DWARF3. Cell Research, 25(11), 1219–1236. 10.1038/cr.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. , Hong, K. , Zeng, L. , Wang, L. , Kang, S. , Qu, M. , Dai, J. , Zou, L. , Zhu, L. , Tang, Z. , Meng, X. , Wang, B. , Hu, J. , Zeng, D. , Zhao, Y. , Cui, P. , Wang, Q. , Qian, Q. , Wang, Y. , … Xiong, G. (2020). Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell, 32(9), 2780–2805. 10.1105/tpc.20.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, K. , Wang, X. , Weighill, D. A. , Guo, H.‐B. , Xie, M. , Yang, Y. , Yang, J. , Wang, S. , Jacobson, D. A. , Guo, H. , Muchero, W. , Tuskan, G. A. , & Chen, J. G. (2016). Characterization of DWARF14 genes in populus. Scientific Reports, 6(1), 21593. 10.1038/srep21593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. , Lin, Q. , Zhu, L. , Ren, Y. , Zhou, K. , Shabek, N. , Wu, F. , Mao, H. , Dong, W. , Gan, L. , Ma, W. , Gao, H. , Chen, J. , Yang, C. , Wang, D. , Tan, J. , Zhang, X. , Guo, X. , Wang, J. , … Wan, J. (2013). D14‐SCF(D3)‐dependent degradation of D53 regulates strigolactone signalling. Nature, 504(7480), 406–410. 10.1038/nature12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. AtD14D167A is less sensitive to rac‐GR24 than wt AtD14.
Table S1. Primers used in this study
Data Availability Statement
All biological resources developed in this study are available upon request and completion of a Material Transfer Agreement.


