Abstract
Hepatocellular carcinoma (HCC), the sixth most common cancer worldwide, has an incidence rate equal to mortality. Over 80% of HCC cases occur within a high‐risk population, mainly patients with both cirrhosis and chronic hepatitis B or C. With a 5‐year survival rate ranging from <16% for advanced HCC to >90% for early stage HCC, there is a high medical need for the early detection of HCC. In this study, we systematically evaluated biomarkers mentioned in international guidelines and peer‐reviewed literature for HCC surveillance and diagnosis with the aim of identifying combinations that display high sensitivity and specificity for early stage HCC. Fifty biomarkers were measured in the first sample panel, panel A (n = 110), and subjected to univariate analysis. Of these, 35 biomarkers (38 assays) from panel A and an additional 13 biomarkers from the literature were prioritized for subsequent multivariate evaluation with lasso regression and exhaustive search of two‐ to four‐biomarker combinations (panel B). Panel B included 1,081 samples from patients with HCC (n = 308) or with chronic liver diseases (n = 740). Among all patients, 61.0% had hepatitis B, 32.9% had hepatitis C, and 60.5% had cirrhosis; 40.6% of patients with HCC had early stage cancer. Protein induced by vitamin K absence‐II (PIVKA‐II; also known as des‐gamma‐carboxy prothrombin [DCP]) and alpha‐fetoprotein (AFP) demonstrated the best clinical performance, both individually and in combination, and the addition of a third biomarker (Lens culinaris agglutinin‐reactive fraction of AFP [AFP‐L3], cartilage oligomeric matrix protein [COMP], insulin‐like growth factor‐binding protein 3 [IGFBP3], or matrix metalloproteinase 3 [MMP3]) further increased sensitivity for the detection of both early stage and all‐stage HCC. The addition of age and sex to the three‐biomarker panel resulted in an improved diagnostic performance. Conclusion: The combination of AFP and PIVKA‐II, with either IGFBP3, COMP or MMP3, plus age and sex, demonstrated the best performance for the detection of early‐ and all‐stage HCC. These novel panels performed similar to that of the GALAD score (sex [gender], age, plus serum levels of AFP, AFP‐L3 and DCP [PIVKA‐II]), a promising screening tool developed for HCC detection.

Abbreviations
- AASLD
American Association for the Study of Liver Diseases
- AFP
alpha‐fetoprotein
- AFP‐L3
Lens culinaris agglutinin‐reactive fraction of alpha‐fetoprotein
- ASH
alcoholic steatohepatitis
- AUC
area under the curve
- BCLC
Barcelona Clinic Liver Cancer
- COMP
cartilage oligomeric matrix protein
- DCP
des‐gamma‐carboxyprothrombin (also known as PIVKA‐II)
- EASL
European Association for the Study of the Liver
- GALAD
sex (gender), age, Lens culinaris agglutinin‐reactive fraction of alpha‐fetoprotein, alpha‐fetoprotein, des‐gamma‐carboxy prothrombin
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HE4
human epididymis protein 4
- IGFBP3
insulin‐like growth factor‐binding protein 3
- JSH
Japanese Society of Hepatology
- MMP3
matrix metalloproteinase 3
- NASH
nonalcoholic steatohepatitis
- PIVKA‐II
protein induced by vitamin K absence‐II (also known as DCP)
- ROC
receiver operating curve
- USG
ultrasonography
Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide and the third leading cause of cancer‐related mortality (~800,000 deaths/year).( 1 , 2 , 3 , 4 ) HCC usually develops in patients with cirrhosis and is triggered by continuous cycles of inflammation and repair in the liver, which may be caused by chronic viral infection (hepatitis B virus [HBV], hepatitis C virus [HCV]), or alcoholic or nonalcoholic steatohepatitis (ASH or NASH).( 5 , 6 , 7 , 8 , 9 ) HCC is typically diagnosed at advanced stages, leaving limited treatment options and resulting in a dismal 5‐year survival rate of <16%; however, early detection and prompt treatment significantly increases survival.( 9 ) The average overall survival ranges from 80 months for early stage HCC (stage A; single nodule or three nodules <3 cm) to 15 months for stage C (portal invasion, nodal invasion, and metastases); patients with stage D (severe liver damage and performance status >2) have a dismal overall survival of approximately 4 months.( 10 , 11 ) Current guidelines for HCC management recommend surveillance of high‐risk patients every 6 months using ultrasonography (USG); however, its effectiveness in the detection of early stage HCC is limited by operator skill and expertise and patient characteristics.( 9 , 12 , 13 , 14 , 15 , 16 ) USG displays 51% sensitivity and 91% specificity for early stage HCC diagnosis; magnetic resonance imaging (MRI) and computed tomography (CT) display higher sensitivity (83.7% and 62.5%, respectively) and similar specificity (89.1% and 87.5%, respectively), but both are more costly with exposure to radiation and/or contrast agents and are therefore infrequently employed for HCC surveillance.( 12 , 13 , 17 , 18 , 19 ) The measurement of the tumor biomarker alpha‐fetoprotein (AFP) has a reported sensitivity of 41%‐65% and specificity of 80%‐90% for the detection of HCC at any stage (20 ng/mL cutoff), but its use remains controversial.( 12 ) AFP is included in the recommendations by the Japanese Society of Hepatology (JSH), Asia‐Pacific Association for the Study of the Liver, and American Association for the Study of Liver Diseases (AASLD) but not the European Association for the study of the Liver (EASL).( 20 ) Two other biomarkers, des‐gamma‐carboxyprothrombin (DCP; also known as protein induced by vitamin K absence II [PIVKA‐II]), and Lens culinaris agglutinin‐reactive fraction of AFP (AFP‐L3), while routinely used for HCC screening in Japan, are not widely used outside Japan and are not included in other guidelines. However, both have been approved as risk markers of HCC by the US Food and Drug Administration in the United States and have a European Conformity marking in Europe.( 19 , 21 ) The addition of age and sex (gender) to biomarkers AFP, AFP‐L3%, and DCP (also known as PIVKA‐II) in a diagnostic algorithm, the GALAD score, has resulted in a specificity of 81.6%‐93.3% and sensitivity of 80.2%‐85.6%, depending on where the analysis was undertaken, for the detection of early stage HCC. This is superior to any biomarker used alone.( 22 , 23 ) Despite these promising developments, novel serologic tests have not yet been incorporated into the guidelines for the surveillance of patients at high risk.( 24 , 25 , 26 ) The development of a serologic biomarker panel that can be applied to populations with diverse liver disease etiologies and complement the limitations of USG to improve the detection of early stage HCC is essential. The aim of this study was to systematically and comprehensively evaluate the sensitivity and specificity of a curated collection of biomarkers in a large sample cohort and to determine the biomarkers and biomarker combinations best suited for detection of early stage HCC.
Patients and Methods
Biomarker Selection
Biomarker candidates were included in this study if they were mentioned in the following international guidelines: AASLD, National Comprehensive Cancer Network, European Society for Medical Oncology–European Society of Digestive Oncology, EASL–European Organization for Research and Treatment of Cancer, British Society of Gastroenterology, National Institute for Health and Care Excellence, JSH, Liver Cancer in China guidelines, and Liver Cancer Association–National Cancer Center Korea Practice guidelines.( 14 , 15 , 26 , 27 , 28 , 29 , 30 , 31 ) An additional PubMed literature search was performed using the term “HCC diagnosis, screening, and surveillance biomarkers.” All biomarker candidates were then evaluated in identical sample panels, permitting head‐to‐head comparison between biomarkers, and a multivariate analysis of all potential biomarker combinations was undertaken.
Study Cohort
The study recruited patients at high risk for developing HCC, diagnosed with either cirrhotic liver disease (independent of etiology), noncirrhotic NASH, chronic HBV infection, chronic HCV infection, and liver fibrosis (stage ≥F3)/cirrhosis or lesions suspicious for HCC based on imaging or untreated HCC. Patients were ineligible for the study if they were <18 years old, were receiving cancer treatment, had a history of other malignancy (other than liver cancer), had received immunomodulatory treatment within the last 6 months, or did not have sonography information within <4 weeks. Before the start of any cancer‐related therapy, a single blood draw was taken and each patient’s diagnosis was documented. Six samples from patients who took anticoagulants (warfarin/phenprocoumon) were excluded from PIVKA‐II analysis because the PIVKA‐II level was shown to significantly increase after warfarin administration.( 32 )
Recruited patients were split into two cohorts, panels A and B. Biomarkers were assessed in a step‐wise approach, allowing head‐to‐head comparison of single biomarker performance in the first cohort (panel A) and then a multivariate analysis of possible biomarker combinations in the second cohort (panel B).
Sample Preparation, Storage, and Measurement
Blood serum and ethylene diamine tetraacetic acid‐plasma samples were collected before surgery, percutaneous ethanol injection, chemotherapy, or radiotherapy. Samples were stored at −70°C until analysis; repeated freeze–thaw cycles were avoided. A combination of two radiologic methods (USG, CT, or MRI) was used to confirm HCC diagnosis in cirrhotic liver. Biopsy and histopathologic analysis was mandatory in noncirrhotic liver. Biomarkers were analyzed using mouse microsomal triglyceride transfer protein enzyme‐linked immunosorbent assay, integrated mutation profiling of actionable cancer targets (IMPACT),( 33 ) cobas e 601, cobas c 311, Fujifilm Micro Total Analysis System Wako, or Lumipulse immunoassays, according to the instructions of the manufacturers (Supporting Tables S1 and S2).
Data Analyses
Univariate receiver operating curve (ROC) analysis was used in panel A to assess the area under the curve (AUC) of each biomarker for distinguishing all‐stage HCC versus benign liver disease controls (Fig. 1). Biomarkers were prioritized and selected based on the clinical performance of AUC ≥ 0.7.
FIG. 1.
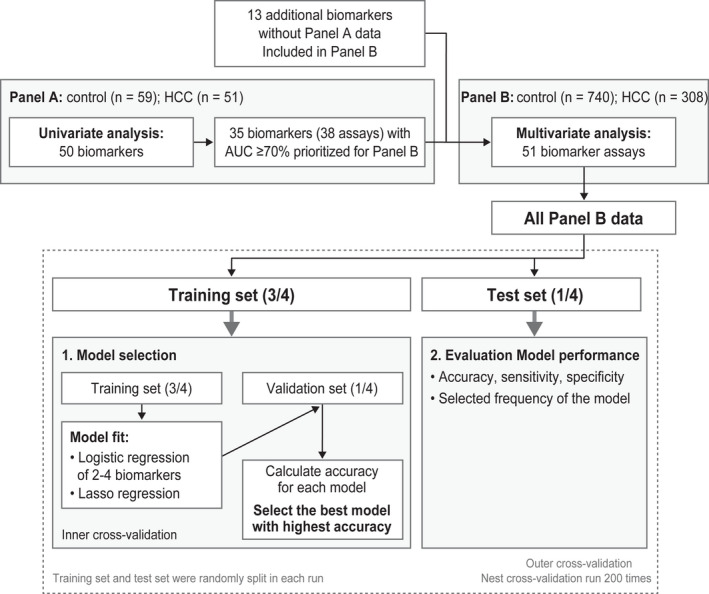
Biomarker analysis strategy. Abbreviations: AUC, area under the curve; HCC, hepatocellular carcinoma.
Selected biomarkers were analyzed with multivariate models in the second cohort (panel B). The aim of the multivariate analysis was to identify the best performing panel of biomarkers that could separate early stage HCC or all‐stage HCC from benign controls with high accuracy. Multivariate analyses were performed using two methods: 1) lasso regression (no fixed panel size) and 2) exhaustive search with logistic regression (fixed panel size of two to four biomarkers). In lasso regression, the number of selected biomarkers was determined by the hyperparameter “lambda” that controlled the number of features for each model. The lasso regression optimized the best model by maximizing the diagnostic accuracy while minimizing the number of biomarkers. In the exhaustive search, logistic regression models based on all possible combinations of two to four biomarkers were evaluated and compared. All possible two‐biomarker combinations were analyzed among the 41 biomarkers. Next, one or two possible biomarkers were added to the best performing two‐biomarker combination, PIVKA‐II + AFP, to search for three‐ and four‐biomarker combinations, e.g., PIVKA‐II + AFP + X and PIVKA‐II + AFP + X + Y. Estimation of diagnostic accuracy was done by a nested cross‐validation, with the data split randomly into training (3/4) and test (1/4) sets. The optimal model was selected using an inner cross‐validation of the training set and tested on the remaining data set. This procedure was repeated 100 times. The test data in the outer loop were used to evaluate the clinical performance of the optimal model. The nested cross‐validation procedure provided an estimate of the stability and a robust estimate of the performance (to avoid overfitting) for each model (Fig. 1). The performance of AUC, sensitivity at 90% specificity, and specificity at 90% sensitivity were reported for each model for detecting early stage and all‐stage HCC. The acceptance criteria (target performance) for the detection of early stage HCC (Barcelona Clinic Liver Cancer [BCLC] stages 0 and A) were preset at 90% sensitivity with specificity >70% or at 90% specificity with sensitivity >70%; this was also used as a reference for all‐stage HCC. The best performing biomarker combinations identified in the panel plus age and sex were then compared with the GALAD algorithm.
In addition, the combination of the selected biomarkers using AND/OR rules was performed and compared with the models of the same biomarkers, using logistic regression from the multivariate analyses. In the AND rule analysis, the overall three‐biomarker combination was considered positive if all three biomarkers were positive and negative if any one biomarker was negative. In the OR rule analysis, the overall three‐biomarker combination was considered positive if any of the three biomarkers were positive and negative if all three biomarkers were negative. The cutoffs used in this analysis were prespecified.
Results
Blood samples were obtained from 1,322 patients between 2014 and 2016 at seven centers in four countries (China, Germany, Spain, and Thailand) (Fig. 2).
FIG. 2.
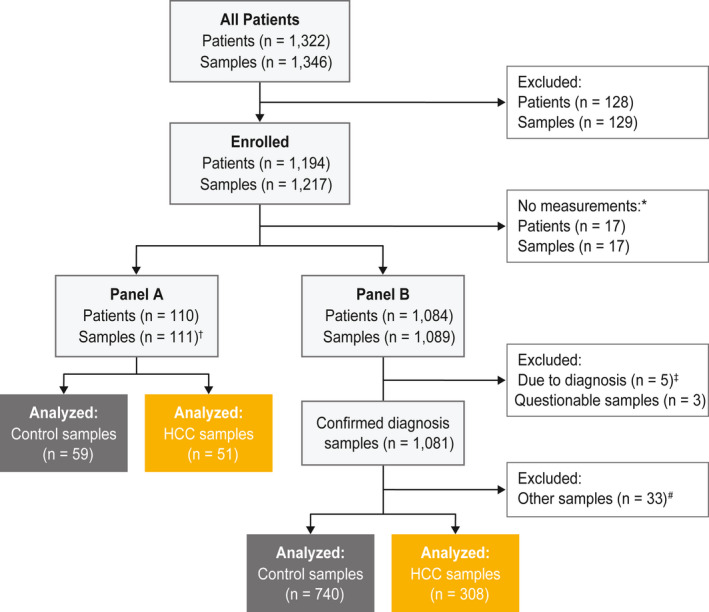
Study design and sample selection. Highlighted boxes show which samples were included in the analysis. *No measurements as a sample were taken on visit 2. †One sample from two visits and only the sample from visit one was measured. ‡Five samples were excluded due to a suspicious diagnosis at visit one or a second replicate diagnosis at visit two. #Patients had CCC, mixed HCC/CCC, or variable HCC diagnosis and were excluded from the analysis. Abbreviation: CCC, cholangiocellular carcinoma; HCC, hepatocellular carcinoma.
In panel A, 50 biomarkers were measured in 110 samples from 110 patients; 72.7% were men, 31.8% had cirrhosis, 17.3% and 16.4% had chronic HBV and HCV infection, respectively, 2.7% had ASH, and 0.9% had NASH (Table 1; Fig. 3). Of all biomarkers evaluated in panel A, 35 biomarkers had AUC ≥ 0.7 and were prioritized for multivariate evaluation. With the addition of two further assays for PIVKA‐II and one assay for carboxylesterase 1, 38 assays were selected; a further 13 biomarkers (detected with mainly commercially available, in vitro, diagnostic assays) were included without panel A data (Supporting Tables S1 and S2). In total, 51 biomarker assays were measured in panel B. Biomarkers were excluded for multivariate analysis if the same biomarkers were replicated on Fujifilm Micro Total Analysis System Wako, Lumipulse, or IMPACT platforms.
TABLE 1.
Summary of patient demographic for samples analyzed in Panel A and Panel B
| Demographics | ||
|---|---|---|
| Panel A | Control (n = 59) | HCC (n = 51) |
| Mean age, years | 54.4 | 61 |
| Gender, n (%) | ||
| Female | 25 (42.4) | 5 (9.8) |
| Male | 34 (57.6) | 46 (90.2) |
| Cirrhosis, n (%) | 0 (0.0) | 35 (68.6) |
| Panel B | Control (n = 740) | HCC (n = 308) |
| Mean age, years | 55.5 | 60.8 |
| Gender, n (%) | ||
| Female | 323 (43.7) | 65 (21.1) |
| Male | 417 (56.4) | 243 (78.9) |
| Race, n (%) | ||
| African black | 2 (0.3) | 0 (0.0) |
| Asian | 638 (86.2) | 248 (80.5) |
| Caucasian/white | 92 (12.4) | 55 (17.9) |
| Mixed | 7 (0.9) | 3 (1.0) |
| Other | 1 (0.1) | 2 (0.6) |
| Cirrhosis, n (%) | 395 (53.4) | 239 (77.6) |
ASH, alcoholic steatohepatitis; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; n.a., not applicable; NASH, non‐alcoholic steatohepatitis.
FIG. 3.
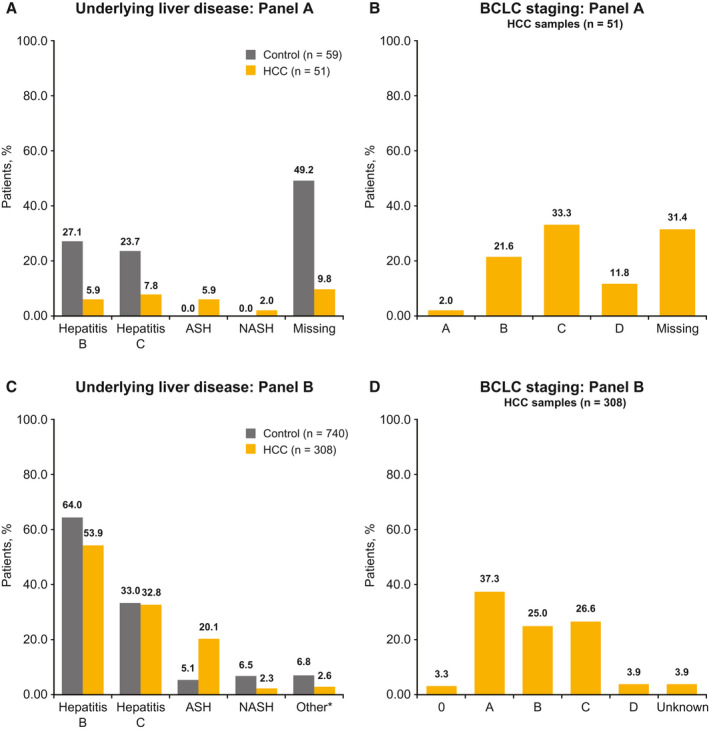
Baseline disease characteristics. (A,B) Percentage of patients included in panel A. (C,D) Percentage of patients included in panel B. *Other comprised adenoma, focal nodular hyperplasia, and hemangioma. Abbreviations: ASH, alcoholic steatohepatitis; BCLC, Barcelona clinic liver cancer; HCC, hepatocellular carcinoma; NASH, non‐alcoholic steatohepatitis.
Panel B analysis was performed on 1,089 samples from 1,084 patients and, after excluding samples with suspicious diagnosis and questionable serum/plasma description, the final sample population consisted of 1,081 samples (740 controls, 308 HCC, and 33 with other diagnoses) (Fig. 2). Among controls and patients with HCC, 61% had HBV (63.9% controls and 53.9% patients with HCC), 32.9% had HCV (33% controls and 32.8% patients with HCC); 60.5% had cirrhosis (53.4% controls and 77.6% patients with HCC); 5.3% had noncirrhotic NASH (6.5% controls and 2.3% patients with HCC), and 9.5% had ASH (5.1% controls and 20.1% patients with HCC) (Table 1; Fig. 3). Controls were comprised of patients with chronic liver disease, such as cirrhotic liver disease, noncirrhotic NASH, ASH, and HBV or HCV. Among patients with HCC, 40.6% had BCLC stage 0 or A, 25.0% stage B, 26.6% stage C, and 3.9% stage D; stage information was missing for 3.9% of patients (Table 1; Fig. 3).
Although the univariate ROC analysis showed that no single biomarker fulfilled clinical requirements for the detection of early stage or all‐stage HCC, PIVKA‐II and AFP displayed the best individual clinical performance for early stage HCC versus controls, with AUC of 79.0% (PIVKA‐II) and 83.4% (AFP) (Fig. 4). The 20 best performing biomarkers from panel B are shown in Table 2.
FIG. 4.
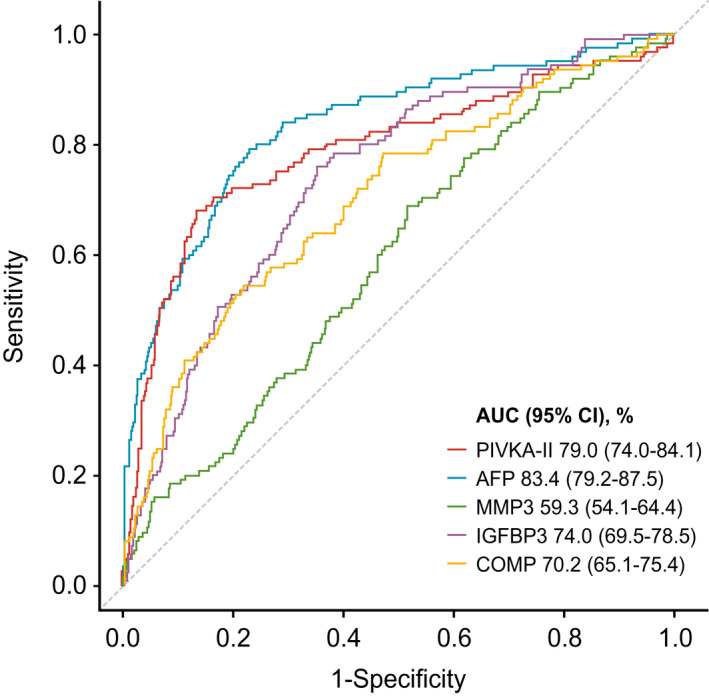
ROC curves showing univariate analyses of early stage HCC versus controls with chronic liver diseases of PIVKA‐II, AFP, MMP3, IGFBP3, and COMP. Abbreviation: CI, confidence interval. Abbreviations: AFP, alpha‐fetoprotein; AUC, area under the curve; COMP, cartilage oligomeric matrix protein; HCC, hepatocellular carcinoma; IGFBP3, Insulin‐like growth factor‐binding protein 3; MMP3, matrix metalloproteinase‐3; PIVKA‐II, protein induced by vitamin k absence‐II; ROC, receiver operating characteristics.
TABLE 2.
Top 20 individual biomarkers that showed best performance in the univariate analysis for detection of early stage HCC (panel B)
| Biomarker | AUC % | HCC sensitivity %* |
|---|---|---|
| AFP cobas e 601 | 83.4 | 53.6 |
| PIVKA‐II cobas e 601 | 79.0 | 56.0 |
| GPC3‐N | 78.2 | 36.8 |
| AFP‐L3% Fujifilm Micro Total Analysis System Wako | 77.8 | 47.2 |
| AFP‐L3 cobas e 601 | 76.5 | 48.8 |
| HE4 | 76.5 | 34.4 |
| IGFBP3 | 74.0 | 30.4 |
| HGF | 72.5 | 30.4 |
| MMP2 | 72.4 | 26.4 |
| TIMP1 | 71.1 | 30.4 |
| Ang2 | 71.0 | 28.8 |
| GGT2 | 70.9 | 28.0 |
| COMP | 70.2 | 36.0 |
| GP73 | 70.1 | 31.2 |
| sAxl | 70.1 | 26.4 |
| Cyfra 21‐1 | 69.7 | 22.4 |
| DKK1 | 68.9 | 21.6 |
| CEA | 68.8 | 25.6 |
| IGF1 | 68.7 | 22.0 |
| IL6 | 68.3 | 28.0 |
| CA 19‐9 | 68.1 | 32.0 |
Abbreviations: Ang2, angiopoietin‐2; CA 19‐9, cancer antigen 19‐9; CEA, carcinoembryonic antigen; Cyfra 21‐1, cytokeratin 19 fragments; DKK1, dickkopf‐related protein 1; GGT2, gamma‐glutamyltransferase‐2; GP73, Golgi membrane protein 1; GPC3, glypican‐3; HGF, hepatocyte growth factor; IGF1, insulin‐like growth factor 1; IL6, interleukin‐6; sAxl, soluble Axl; TIMP1, tissue inhibitor of metalloproteinase‐1.
Sensitivity of biomarkers was assessed at 90% specificity.
Multivariate analysis using lasso regression was performed using 41 biomarkers to identify parsimonious models that met the target performance of 70% sensitivity at 90% specificity or 70% specificity at 90% sensitivity. Lasso regression with no fixed panel size (number of variables in the model not specified) identified PIVKA‐II and AFP as the best combination of biomarkers for the identification of both all‐stage and early stage HCC versus controls (Supporting Table S3; Supporting Fig. S1). The addition of either gamma‐glutamyltransferase 2, human epididymis protein 4 (HE4), osteoprotegerin (OPG), or insulin‐like growth factor binding protein 3 (IGFBP3) to PIVKA‐II + AFP for all‐stage HCC (Supporting Fig. S1A) and HE4, cartilage oligomeric matrix protein (COMP), carcinoembryonic antigen, matrix metalloproteinase 3 (MMP3), or OPG to PIVKA‐II + AFP for early stage HCC (Supporting Fig. S1B) enhanced diagnostic value. No further improvement in clinical performance was observed with models containing more than three biomarkers. We then undertook an exhaustive search of all combinations of two to four biomarkers (approximately 1,600 combinations) to separate early stage HCC from controls (125 vs. 740) and all‐stage HCC from controls (308 vs. 740). The exhaustive search determined that only PIVKA‐II + AFP met all criteria for the identification of all‐stage HCC versus controls, with no enhanced detection noted on the addition of a third biomarker (Supporting Table S4). However, for early stage HCC versus controls, none of the two‐biomarker panels met the acceptance criteria and only the three‐biomarker panels, composed of PIVKA‐II + AFP with either MMP3, IGFBP3, or COMP, met the acceptance criteria (Table 3). Notably, however, the third‐selected biomarker (IGFBP3, COMP, or MMP3) was not consistently selected in the multivariate analyses (Supporting Table S4).
TABLE 3.
Clinical performance of the biomarker combinations showing 90% specificity/sensitivity in the multivariate analysis, with and without age and gender, for all‐ and early‐stage HCC
| All‐stage HCC (n = 308) vs controls (n = 734)* | Early‐stage HCC (n = 125) vs controls (n = 734)* | |||||
|---|---|---|---|---|---|---|
| AUC, % | Sensitivity at 90% specificity, % | Specificity at 90% sensitivity, % | AUC, % | Sensitivity at 90% specificity, % | Specificity at 90% sensitivity, % | |
| Biomarker Combination Without Age and Gender (Sex) | ||||||
| AFP+PIVKA‐II+MMP3 | 94.4 | 83.8 | 84.7 | 89.1 | 65.6 | 72.9 |
| AFP+PIVKA‐II+IGFBP3 | 94.3 | 85.4 | 85.8 | 88.5 | 68.0 | 74.3 |
| AFP+PVKA‐II+COMP | 94.4 | 84.1 | 85.6 | 89.0 | 67.2 | 74.1 |
| GALAD (cobas e 601 by Roche) | 94.1 | 84.4 | 82.3 | 88.1 | 68.0 | 69.1 |
| GALAD (Fujifilm Micro Total Analysis System WakoTM) | 94.5 | 81.8 | 84.4 | 89.8 | 64.8 | 77.1 |
| Biomarker Combination With Age and Gender (Sex) | ||||||
| AFP+PIVKA‐II+MMP3 | 95.3 | 84.4 | 85.9 | 90.7 | 67.2 | 75.1 |
| AFP+PIVKA‐II+IGFBP3 | 95.3 | 87.0 | 85.8 | 90.6 | 71.2 | 77.4 |
| AFP+PVKA‐II+COMP | 95.4 | 85.7 | 85.5 | 90.9 | 72.8 | 80.2 |
| GALAD (cobas e 601 by Roche) | 95.2 | 86.0 | 84.1 | 90.5 | 71.2 | 77.7 |
| GALAD (Fujifilm Micro Total Analysis System WakoTM) | 94.5 | 81.8 | 84.4 | 89.8 | 64.8 | 77.1 |
AFP, alpha‐fetoprotein; AFP‐L3, Lens culinaris agglutinin‐reactive fraction of AFP; AUC, area under the curve; COMP, cartilage oligomeric matrix protein; GALAD, Gender, age, AFP‐L3, AFP, and PIVKA‐II; HCC, hepatocellular carcinoma; HE4, epididymal protein 4; IGFBP3, Insulin‐like growth factor‐binding protein 3; IL6, interleukin 6; MMP3, matrix metalloproteinase‐3; PIVKA‐II, protein induced by vitamin k absence‐II.
*Samples with missing measurements were excluded from the multivariate analysis.
The combination of the selected biomarkers PIVKA‐II + AFP plus a third biomarker (either MMP3, IGFBP3, or COMP) using AND/OR rules did not show better clinical performance compared with the logistic regression models. None of the AND/OR combinations reached the target sensitivity and specificity (data not shown).
The addition of age and sex to the three‐biomarker panels further enhanced diagnostic performance (Table 3). The HCC detection capability of the novel biomarker panel AFP + PIVKA‐II with either COMP, MMP3, or IGFBP3, plus age and sex, was compared with that of the GALAD score (−10.08 + 0.09 × Age + 1.67 × Sex + 2.34 × log10[AFP] + 0.04 × AFP‐L3% + 1.33 × log10[PIVKA‐II]), where Sex = 1 for men and 0 for women( 23 , 41 ) (Fig. 5). The performance of the novel biomarker panels was similar to the GALAD score. The addition of age and sex to the novel three‐biomarker panels enhanced specificity at the high‐sensitivity region; similar results were obtained for two versions of the GALAD score, using assays from different manufacturers, and for the detection of all‐stage and early stage HCC (AUC > 95% and >90%, respectively) (Table 3; Supporting Table S5; Supporting Fig. S2).
FIG. 5.
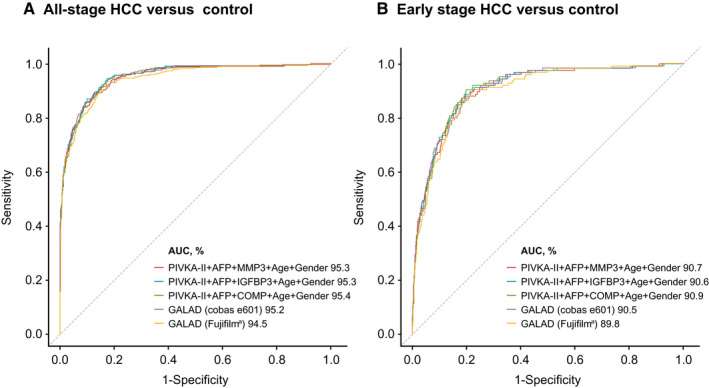
Results of combined biomarker panels plus age and gender (sex). (A) For all‐stage HCC. (B) For early stage HCC. aFujifilm, Fujifilm Micro Total Analysis System Wako. Abbreviations: AFP, alpha‐fetoprotein; AUC, area under the curve; COMP, cartilage oligomeric matrix protein; GALAD, Gender, age, AFP‐l3, AFP, and PIVKA‐II; IGFBP3, Insulin‐like growth factor‐binding protein 3; MMP3, matrix metalloproteinase‐3.
Discussion
In this study, we described the most comprehensive analysis to date of a large panel of serum biomarkers that have been proposed for surveillance of HCC. Through univariate and multivariate analysis, we identified candidate biomarker combinations that were compared with AFP alone and with the GALAD model.
In summary, none of the evaluated individual biomarkers alone met the clinical requirements for the detection of early stage HCC with high sensitivity or high specificity. However, the combination of AFP + PIVKA‐II, along with a third biomarker (AFP‐L3, IGFBP3, COMP, or MMP3) plus age and sex demonstrated the best performance for the detection of early stage and all‐stage HCC. Further, when compared with the GALAD score, which has been shown to be superior to AFP for the detection of early stage and all‐stage HCC, the assessed three‐biomarker panels comprised of AFP + PIVKA‐II and either COMP, IGFBP‐3, or MMP3, plus age and sex, demonstrated similar sensitivity and specificity.( 23 , 34 , 35 ) Importantly, the assessed three‐biomarker panels did not reach the requirements for high sensitivity without taking into account age and sex.
The assessment of serum AFP for HCC surveillance among individuals at high risk is established in guidelines throughout Asia, but its use remains controversial elsewhere.( 12 , 26 , 30 , 31 ) AFP has been inconsistently incorporated into guidelines. For example, it is recommended for surveillance in JSH guidelines but not in EASL or AASLD guidelines.( 26 ) Although multiple studies have highlighted the value of AFP, either in combination with USG or other known biomarkers, in the detection of both early stage and all‐stage HCC, its utility is limited due to elevations in patients with chronic hepatitis (15%‐58%) and liver cirrhosis (11%‐47%) and due to the fact that levels are not elevated in up to 50% of patients with HCC.( 12 ) These limitations have driven the search for novel biomarkers that could improve early stage HCC surveillance.
Although many promising candidates have been evaluated, none have yet been adopted into clinical practice.( 36 , 37 ) The assessment of these biomarkers is hampered by a lack of head‐to‐head data in large well‐characterized cohorts, with most studies including only a small number of biomarkers. We therefore developed a panel of biomarkers intended to be as comprehensive as possible by curating a list of those described in HCC guidelines and in peer‐reviewed literature at the time the study was initiated. We undertook a systematic assessment of their univariate performance and then applied multivariate analysis followed by an exhaustive search to identify the best performing combinations from this large panel.
Our data show that none of the tested biomarkers, including previously extensively studied AFP, PIVKA‐II, glypican‐3, dickkopf‐related protein 1, and Golgi membrane protein 1, had sufficient sensitivity as single markers for routine use in HCC surveillance. The sensitivity for early stage HCC of the two best performing individual markers, AFP and PIVKA‐II, increased from 53.6% and 56.0%, respectively, to ~70% when used in combination. This is in accordance with studies reporting individual AFP and PIVKA‐II sensitivities of 41%‐65% and 28%‐89%, respectively, compared with a sensitivity of 78.3% when used in combination for the detection of all‐stage HCC.( 13 , 38 , 39 , 40 )
Additionally, our comprehensive analysis has identified COMP, IGFBP3, and MMP3 as biomarkers that further enhance the clinical performance of AFP + PIVKA‐II, when used as a three‐biomarker panel, for the detection of both early stage and all‐stage HCC. Taken together, our data and the results from previous studies emphasize the need for additional HCC‐specific biomarkers that could further enhance the diagnostic value of AFP.
The GALAD score has shown great potential sensitivity for the detection of early stage HCC and may be particularly well suited to the surveillance of patients with NASH in whom the performance of USG is limited.( 22 , 23 , 35 , 41 ) In this study, several biomarker combinations containing AFP + PIVKA‐II, with the addition of either COMP, IGFBP3, or MMP3, plus age and sex, displayed similar sensitivities and specificities to those of the GALAD score for the detection of early stage and all‐stage HCC. The success of both the three‐biomarker panels analyzed and the GALAD model relies on taking into account specific risk factors, such as male sex and advanced age, when determining the HCC risk for patients with chronic liver diseases.( 42 ) GALAD also allows the cutoff to be adjusted to customize the sensitivity and specificity of the biomarker panels according to local patient demographics and prevalence of HCC, for which substantial regional differences have been described.( 14 , 43 , 44 ) All three novel three‐biomarker panels fulfilled the acceptance criteria and performed at a similar level as the GALAD score for early stage HCC detection; however, the third biomarker in each novel panel was inconsistently selected in the exhaustive search process and therefore require independent confirmation. Importantly, the performance of the novel three‐biomarker panels should be viewed cautiously as the performance was based on nested cross‐validation from a single study, which may overestimate performance, whereas the clinical performance of the GALAD algorithm has been validated in several independent studies.( 22 , 23 , 24 , 35 , 41 ) Limitations of this study are the patient population, which was comprised mainly of patients with liver disease due to chronic hepatitis, which could have influenced the biomarkers selected in the multivariate analysis. However, multiple studies assessing GALAD in a greater proportion of patients with chronic liver disease with nonviral etiology have yielded similar results.( 22 , 23 , 35 , 41 ) Details on etiology were also missing from a number of patients in panel A. Additionally, the majority of included patients with HCC identified as Asian and few women were enrolled; however, this is reflective of the higher incidence of HCC in both Asian and male populations.( 4 ) In order to validate the clinical performance of the biomarker combinations, this study had a phase 2 case‐control design and included patients with varying baseline and disease characteristics, such as sex, age, liver disease etiology, and the presence of cirrhosis. Ongoing phase 3 studies are expected to provide additional evidence in support of biomarker combinations in longitudinal studies.( 45 , 46 ) Another limitation of the current study was using different assay formats that may have detected different analyte pools. However, in this study, the clinical performance of AFP, PIVKA‐II, and AFP‐L3 across all the assay formats was shown to be comparable.
In conclusion, we assessed the largest number of biomarkers for HCC surveillance to date and identified combinations of serum biomarkers that may be of use in the early diagnosis of patients at risk. These assays are rapid, easy to perform, noninvasive, and less dependent on operator skill than USG. Prospective studies to validate such biomarker panels are urgently needed. Currently, there are several large ongoing studies collecting samples for this purpose, including the Hepatocellular Carcinoma Early Detection Strategy study( 45 ) and the Texas Hepatocellular Carcinoma Consortium Cohort study.( 46 ) While a single‐center case‐control study found that the addition of USG provided only minimal improvement in the performance of the GALAD score,( 43 ) USG results were scored categorically (either positive or negative) when numerical/continuous variables, e.g., number and size of nodules, may have been more informative. Larger, prospective, multicenter studies are required to formally assess this combinational approach, ideally using real‐world screening cohorts. Thus, the potential for further improving detection rates for early stage HCC by combining biomarker panels with individual modalities, such as USG, may be of interest for future research.
Supporting information
Supplementary Material
Acknowledgment
We thank the patients and their families for their contribution to the study. We thank Marcus‐Rene Lisy and Christine Anna Muth of Roche Diagnostics for the development of the PIVKA‐II and AFP‐L3 cobas assays, Vinzent Rolny of Roche Diagnostics for his help with the biostatistical analysis, and Friedemann Krause for his help with the study design. We also thank Carmen K.M. Chan, Stephen L. Chan, Charing C.N. Chong, Paul B.S. Lai, Pete Tse, Grace L.H. Wong, John Wong, and Vincent W.S. Wong from the Chinese University of Hong Kong. COBAS is a trademark of Roche. All other product names and trademarks are the property of their respective owners. Editorial support was provided by Nichola Cruickshanks of inScience Communications, Springer Healthcare Ltd., United Kingdom, and was funded by Roche Diagnostics.
Supported by Roche Diagnostics GmbH.
Potential conflict of interest: HL‐YC reports consultancy fees from Abbvie, Arbutus Biopharma, ContraVir, Gilead Sciences, GRAIL, MedImmune, Janssen Pharmaceutica, Intellia therapeutics, Roche, Vir Biotechnology, Aligos Therapeutics, Vaccitech and VenatoRx Pharmaceuticals and speaker’s bureau participation for Gilead Sciences and Roche Diagnostics. TP reports speaker’s bureau participation for Bristol‐Myers Squibb, Gilead Science, Bayer, Abbott and Eisai and MSD and research grant/contracts from Gilead Science, Roche Diagnostics, Janssen, Fibrogen and VIR. YH, MSdL and DM are employees of Roche Diagnostics. MSdL has a patent for a new binding agent and assay for PIVKA‐II pending. BK, JIEM, MBM, ST, TT and WS have nothing to disclose.
References
Author names in bold designate shared co‐first authorship.
- 1. Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am 2015;24:1‐17. [DOI] [PubMed] [Google Scholar]
- 2. Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet 2003;362:1907‐1917. [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization Cancer. https://www.who.int/news‐room/fact‐sheets/detail/cancer. Published September 21, 2021. Accessed September, 2021.
- 4. World Health Organization; International Agency for Research on Cancer . Liver. https://gco.iarc.fr/today/data/factsheets/cancers/11‐Liver‐fact‐sheet.pdf Published December 2020. Accessed January, 2021.
- 5. Axley P, Ahmed Z, Ravi S, Singal AK. Hepatitis C virus and hepatocellular carcinoma: a narrative review. J Clin Transl Hepatol 2018;6:79‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee WY, Bachtiar M, Choo CCS, Lee CG. Comprehensive review of hepatitis B virus‐associated hepatocellular carcinoma research through text mining and big data analytics. Biol Rev Camb Philos Soc 2019;94:353‐367. [DOI] [PubMed] [Google Scholar]
- 7. Kumar R, Goh BG, Kam JW, Chang PE, Tan CK. Comparisons between non‐alcoholic steatohepatitis and alcohol‐related hepatocellular carcinoma. Clin Mol Hepatol 2020;26:196‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int 2016;36:317‐324. [DOI] [PubMed] [Google Scholar]
- 9. Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol 2015;21:10573‐10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Llovet JM, Fuster J, Bruix J; Barcelona‐Clínic Liver Cancer Group . The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl 2004;10(Suppl 1.):S115‐S120. [DOI] [PubMed] [Google Scholar]
- 11. Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE study. Liver Int 2015;35:2155‐2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song P, Tang W, Kokudo N. Serum biomarkers for early diagnosis of hepatocellular carcinoma. Transl Gastrointest Cancer 2014;3:103‐105. [Google Scholar]
- 13. Bialecki ES, Di Bisceglie AM. Diagnosis of Hepatocellular Carcinoma. HPB (Oxford) 2005;7:26‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer . EASL‐EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908‐943. Erratum in: J Hepatol 2012;56:1430. [DOI] [PubMed] [Google Scholar]
- 15. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723‐750. [DOI] [PubMed] [Google Scholar]
- 16. Omata M, Cheng A‐L, Kokudo N, Kudo M, Lee JM, Jia J, et al. Asia‐Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int 2017;11:317‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Attwa MH, El‐Etreby SA. Guide for diagnosis and treatment of hepatocellular carcinoma. World J Hepatol 2015;7:1632‐1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta‐analysis. Gastroenterology 2018;154:1706‐1718.e1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gish RG. Early detection of hepatocellular carcinoma through surveillance using biomarkers. Gastroenterol Hepatol (N Y) 2014;10:121‐123. [PMC free article] [PubMed] [Google Scholar]
- 20. Purcell Y, Copin P, Paulatto L, Pommier R, Vilgrain V, Ronot M. Hepatocellular carcinoma surveillance: Eastern and Western perspectives. Ultrasonography 2019;38:191‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Food and Drug Administration . AFP‐L3% immunological test systems ‐ class II special controls guidance document guidance for industry and FDA staff. Published October 4, 2005. Accessed November 1, 2021.
- 22. Best J, Bilgi H, Heider D, Schotten C, Manka P, Bedreli S, et al. The GALAD scoring algorithm based on AFP, AFP‐L3, and DCP significantly improves detection of BCLC early stage hepatocellular carcinoma. Z Gastroenterol 2016;54:1296‐1305. [DOI] [PubMed] [Google Scholar]
- 23. Berhane S, Toyoda H, Tada T, Kumada T, Kagebayashi C, Satomura S, et al. Role of the GALAD and BALAD‐2 serologic models in diagnosis of hepatocellular carcinoma and prediction of survival in patients. Clin Gastroenterol Hepatol 2016;14:875‐886.e6. [DOI] [PubMed] [Google Scholar]
- 24. Vogel A, Cervantes A, Chau I, Daniele B, Llovet J, Meyer T, Nault JC, et al.; ESMO Guidelines Committee . Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann Oncol 2018;29(Suppl. 4):iv238‐iv255. Erratum in: Ann Oncol 2019;30:871‐873. [DOI] [PubMed] [Google Scholar]
- 25. Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358‐380. [DOI] [PubMed] [Google Scholar]
- 26. No authors listed . Clinical practice guidelines for hepatocellular carcinoma differ between Japan, United States, and Europe. Liver Cancer 2015;4:85‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. National Comprehensive Cancer Network . NCCN clinical practice guidelines in oncology: hepatobiliary cancers. Version 4.2019. https://www.nccn.org/professionals/physician_gls/pdf/hepatobiliary.pdf. Published December 2019. Accessed December 2019.
- 28. Kokudo N, Takemura N, Hasegawa K, Takayama T, Kubo S, Shimada M, et al. Clinical practice guidelines for hepatocellular carcinoma: The Japan Society of Hepatology 2017 (4th JSH‐HCC guidelines) 2019 update. Hepatol Res 2019;49:1109‐1113. [DOI] [PubMed] [Google Scholar]
- 29. Ryder SD; British Society of Gastroenterology . Guidelines for the diagnosis and treatment of hepatocellular carcinoma (HCC) in adults. Gut 2003;52(Suppl. 3):iii1‐iii8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korean Liver Cancer Association; National Cancer Center . 2018 Korean Liver Cancer Association‐National Cancer Center Korea Practice Guidelines for the Management of Hepatocellular Carcinoma. Gut Liv 2019;13:227‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou J, Sun HC, Wang Z, Cong WM, Wang JH, Zeng MS, et al. Guidelines for diagnosis and treatment of primary liver cancer in China (2017 edition). Liver Cancer 2018;7:235‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee W, Chung H‐J, Kim S, Jang S, Park C‐J, Chi H‐S, et al. PIVKA‐II is a candidate marker for monitoring the effects of the oral anticoagulant warfarin. Clin Biochem 2010;43:1177‐1179. [DOI] [PubMed] [Google Scholar]
- 33. Claudon A, Vergnaud P, Valverde C, Mayr A, Klause U, Garnero P. New automated multiplex assay for bone turnover markers in osteoporosis. Clin Chem 2008;54:1554‐1563. [DOI] [PubMed] [Google Scholar]
- 34. Caviglia GP, Abate ML, Petrini E, Gaia S, Rizzetto M, Smedile A. Highly sensitive alpha‐fetoprotein, Lens culinaris agglutinin‐reactive fraction of alpha‐fetoprotein and des‐gamma‐carboxyprothrombin for hepatocellular carcinoma detection. Hepatol Res 2016;46:E130‐E135. [DOI] [PubMed] [Google Scholar]
- 35. Best J, Bechmann LP, Sowa J‐P, Sydor S, Dechêne A, Pflanz K, et al. GALAD score detects early hepatocellular carcinoma in an international cohort of patients with nonalcoholic steatohepatitis. Clin Gastroenterol Hepatol 2020;18:728‐735.e4. [DOI] [PubMed] [Google Scholar]
- 36. Rich N, Singal AG. Hepatocellular carcinoma tumour markers: current role and expectations. Best Pract Res Clin Gastroenterol 2014;28:843‐853. [DOI] [PubMed] [Google Scholar]
- 37. De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R. Novel biomarkers in hepatocellular carcinoma. Dig Liver Dis 2018;50:1115‐1123. [DOI] [PubMed] [Google Scholar]
- 38. Song P, Gao J, Inagaki Y, Kokudo N, Hasegawa K, Sugawara Y, et al. Biomarkers: evaluation of screening for and early diagnosis of hepatocellular carcinoma in Japan and China. Liver Cancer 2013;2:31‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi JY, Jung SW, Kim HY, Kim M, Kim Y, Kim DG, et al. Diagnostic value of AFP‐L3 and PIVKA‐II in hepatocellular carcinoma according to total‐AFP. World J Gastroenterol 2013;19:339‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xing H, Zheng Y‐J, Han J, Zhang H, Li Z‐L, Lau W‐Y, et al. Protein induced by vitamin K absence or antagonist‐II versus alpha‐fetoprotein in the diagnosis of hepatocellular carcinoma: a systematic review with meta‐analysis. Hepatobiliary Pancreat Dis Int 2018;17:487‐495. [DOI] [PubMed] [Google Scholar]
- 41. Johnson PJ, Pirrie SJ, Cox TF, Berhane S, Teng M, Palmer D, et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol Biomarkers Prev 2014;23:144‐153. [DOI] [PubMed] [Google Scholar]
- 42. Corey KE, Gawrieh S, deLemos AS, Zheng H, Scanga AE, Haglund JW, et al. Risk factors for hepatocellular carcinoma in cirrhosis due to nonalcoholic fatty liver disease: a multicenter, case‐control study. World J Hepatol 2017;9:385‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang JD, Addissie BD, Mara KC, Harmsen WS, Dai J, Zhang N, et al. GALAD score for hepatocellular carcinoma detection in comparison with liver ultrasound and proposal of GALADUS score. Cancer Epidemiol Biomarkers Prev 2019;28:531‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Borges KA, Dai J, Parikh ND, Schwartz M, Nguyen MH, Roberts LR, et al. Rationale and design of the Hepatocellular Carcinoma Early Detection Strategy study: a multi‐center longitudinal initiative of the National Cancer Institute's Early Detection Research Network. Contemp Clin Trials 2019;76:49‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feng Z, Marrero JA, Khaderi S, Singal AG, Kanwal F, Loo N, et al. Design of the Texas Hepatocellular Carcinoma Consortium Cohort Study. Am J Gastroenterol 2019;114:530‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


