Abstract
The cytochrome P450 (Cyp) proteins Cyp1A1 and Cyp1A2 are strongly induced in the mouse liver by the potent environmental toxicant 2, 3, 7, 8‐tetrachlorodibenzo‐p‐dioxin (TCDD), acting through the aryl hydrocarbon receptor (AHR). The induction of Cyp1A1 is localized within the centrilobular regions of the mouse liver at low doses of TCDD, progressing to pan‐lobular induction at higher doses. Even without chemical perturbation, metabolic functions and associated genes are basally zonated in the liver lobule along the central‐to‐portal axis. To investigate the mechanistic basis of spatially restricted gene induction by TCDD, we have developed a multiscale computational model of the mouse liver lobule with single‐cell resolution. The spatial location of individual hepatocytes in the model was calibrated from previously published high‐resolution images. A systems biology model of the network of biochemical signaling pathways underlying Cyp1A1 and Cyp1A2 induction was then incorporated into each hepatocyte in the model. Model simulations showed that a negative feedback loop formed by binding of the induced Cyp1A2 protein to TCDD, together with cooperative gene induction by the β‐catenin/AHR/TCDD transcription factor complex and β‐catenin, help produce the spatially localized induction pattern of Cyp1A1. Although endogenous WNT regulates the metabolic zonation of many genes, it was not a driver of zonal Cyp1A1 induction in our model. Conclusion: In this work, we used data‐driven computational modeling to identify the mechanistic basis of zonally restricted gene expression induced by the potent and persistent environmental pollutant TCDD. The multiscale model and derived results clarify the mechanisms of dose‐dependent hepatic gene induction responses to TCDD. Additionally, this work contributes to our broader understanding of spatial gene regulation along the liver lobule.
A multiscale liver lobule model was developed to analyze the mechanisms of zonal induction of cytochrome proteins P450 by TCDD. We found that a negative feedback loop formed by TCDD‐CYP1A2 binding and cooperation between transcription factors play key roles in the zonal induction of cytochrome proteins.

Abbreviations
- AHR
aryl hydrocarbon receptor
- APC
adenomatous polyposis coli
- BAT
β‐catenin/AHR/TCDD
- Cyp
cytochrome P450
- Cyp1A1
cytochrome P450 1A1
- Cyp1A2
cytochrome P450 1A2
- PBPK
physiologically based pharmacokinetic
- RNA‐seq
RNA‐sequencing
- TCDD
2, 3, 7, 8‐tetrachlorodibenzo‐p‐dioxin
2, 3, 7, 8‐tetrachlorodibenzo‐p‐dioxin (TCDD) is a potent environmental pollutant that binds the aryl hydrocarbon receptor (AHR) and induces multiple adverse responses in animals at high doses.( 1 , 2 ) As one of its early low‐dose effects, TCDD induces phase I drug‐metabolizing enzymes, specifically the hepatic cytochrome P450 proteins Cyp1A1 and Cyp1A2. Cyp1A1 is induced in centrilobular regions at low doses, with the induction pattern spreading to the whole lobule at high doses( 3 ) (Fig. 1A). Although transcriptional effects of TCDD in hepatocytes have been widely investigated, the mechanisms underlying zonal gene induction in the liver are poorly understood, hindering low‐dose extrapolation for hepatic dose‐response assessment of TCDD.( 4 , 5 )
FIG. 1.
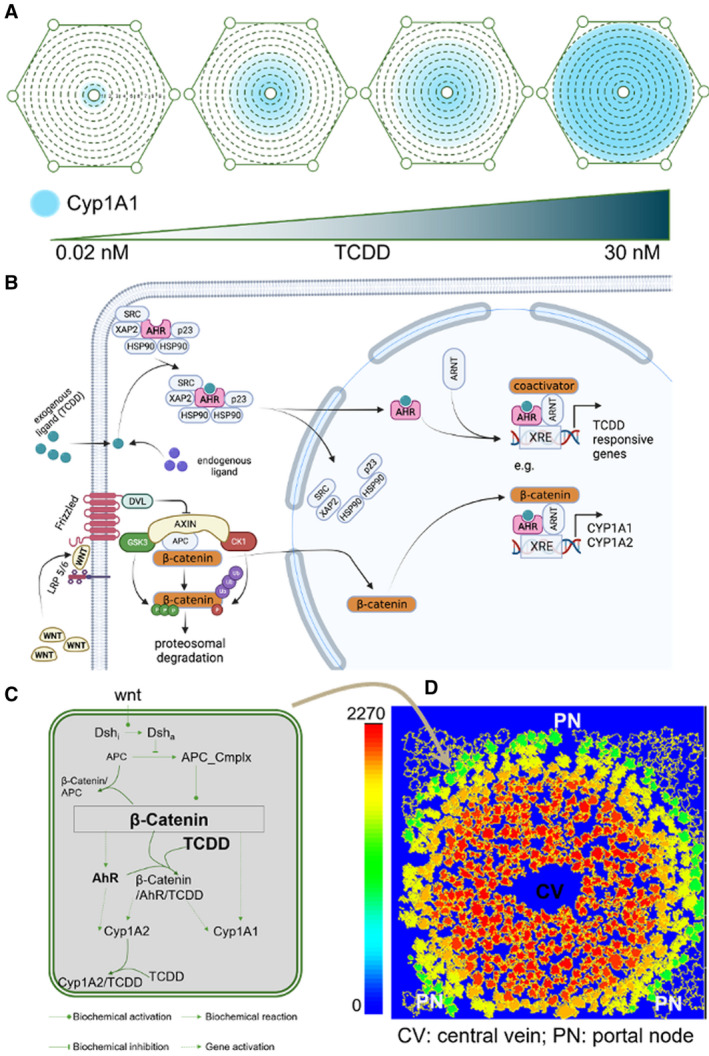
A multiscale computational model simulates hepatic zonal induction of Cyp1A1 by TCDD. (A) Zonal induction of Cyp1A1 in the mouse liver lobule by TCDD. The central‐to‐portal axis is segmented into nine zones following Halpern et al.(11) The area of the liver lobule expressing Cyp1a1 increases with increasing TCDD dose. (B) Schematic representation of the AHR and β‐catenin pathways and their intersection (made in BioRender [biorender.com]). (C) Intracellular model of the Wnt signaling pathway and downstream gene regulatory network for induction of Cyp1A1 by TCDD. (D) Multiscale virtual liver lobule model. The locations of the cells are calibrated from a single‐cell resolution image of the lobule in Halpern et al(11); the signaling pathway from (C) is implemented in each cell of the lobule model. The colors of the cells represent the expression level of Cyp1A1. Abbreviations: CV, central vein; and PV, portal vein.
Basal metabolic functions in liver lobules vary along the central‐to‐portal axis, in a process known as “metabolic zonation.”( 6 ) A key regulator of metabolic zonation is the WNT signaling pathway,( 7 , 8 , 9 , 10 ) to which nearly one third of zonated hepatic genes are functionally related.( 11 ) Multiple signaling components of the pathway, including Disheveled (DVL), adenomatous polyposis coli (APC) and β‐catenin, are zonally expressed along liver lobules.( 7 , 11 ) Prominently, APC has been described as the maintainer of metabolic zonation.( 7 ) In parallel, the WNT signaling pathway also regulates Cyp1A1 expression.( 12 , 13 , 14 , 15 ) Specifically, the WNT pathway component β‐catenin, acting as a transcription factor, controls the basal expression of both AHR and Cyp1A1. Preceding activation by WNT, β‐catenin is sequestered to the cytosol by the β‐catenin destruction complex composed of APC, Axin, CK1‐α, and GSK‐3β. This complex suppresses β‐catenin through sequestration (through Axin and APC), phosphorylation (through CK1‐α, and GSK‐3β), and subsequent proteasomal degradation.( 16 ) Before TCDD exposure, AHR is found in its inactive form in the cytosol, bound to its chaperone proteins—a dimer of 90‐kDa heat shock proteins, hepatitis B virus X–associated protein 2 (also known as AHR‐interacting protein), p23, and protein kinase SRC. TCDD‐liganded AHR disassociates from its chaperones, translocates to the nucleus, and binds to the AHR nuclear translocator protein. This heterodimeric transcription factor cooperates with activators and repressors to control gene expression by binding to dioxin response elements containing the core DNA motif 5'‐GCGTG‐3'.( 17 ) β‐Catenin is one such co‐activator, which binds to TCDD‐liganded AHR, forming a transcription factor complex that induces Cyp1A1 and Cyp1A2( 13 ) (Fig. 1B). Previous computational models of the WNT signaling pathway( 18 ) have captured different characteristics of the pathway: from detailed molecular models( 19 ) to simplified dynamic representations.( 20 , 21 ) A recent model investigated the role of the pathway in epithelial‐mesenchymal transition.( 22 ) A thermodynamic model based on mutant constructs of the Cyp1A1 promoter was recently proposed to explain cooperative interactions between the AHR transcription factor complex and individual binding sites on the Cyp1A1 promoter.( 23 ) These models, however, are either too complex to function as a subsystem within an integrated multiscale tissue model, or lack key molecular components involved in liver metabolic zonation.
Computational modeling is widely used in toxicology and pharmacology for dose‐response assessment. Physiologically based pharmacokinetic (PBPK) models that consider the liver as a homogeneous compartment have been developed for the Cyp1A1 gene induction system.( 24 ) Zonal induction of cytochrome genes was incorporated into a multi‐compartment geometric model of the liver lobule.( 25 , 26 ) Given the lack of single‐cell data at the time, both the cell signaling network and spatial resolution in these models were limited. Recently, “virtual tissues” have emerged as a versatile multiscale quantitative framework to simultaneously model the behaviors of intracellular signaling pathways, cells, tissues, and organs.( 27 , 28 , 29 , 30 , 31 ) Virtual tissues extend the concept of compartmental models down to the basic biological unit of life, a cell,( 28 , 29 ) and have been used for risk assessment, as well as simulation of physiological processes and tumor development.( 27 , 29 , 32 , 33 ) Recently, virtual liver lobule models have been used to simulate the physiological effects and metabolism of chemicals.( 34 , 35 ) Embedding systems biological models based on biochemical reactions within these spatial models allows us to study a physiological phenomenon at a resolution down to the molecular scale.( 36 ) Systems biology models have so far seen limited use in toxicology.( 37 ) Combining systems biology models with virtual tissues creates multiscale tissue models that can be used to investigate collective cellular behaviors and capture within‐tissue spatial heterogeneity.
To systematically investigate the zonal induction of cytochrome P450 genes by TCDD, we have developed a multiscale virtual model of a mouse liver lobule with embedded individual hepatocytes. WNT signaling molecules were modeled in the extracellular space of the tissue, while intracellular components of the WNT pathway were implemented in each hepatocyte. This virtual tissue model provides a platform to visualize zonated induction of Cyp1A1 in an intuitive way. The parameters of the WNT model were calibrated using previous experimental results, and were used the model to investigate the mechanisms of zonal induction of Cyp1A1 by TCDD. Our results indicate that binding between TCDD and the Cyp1A2 protein, zonal expression of β‐catenin, and cooperation among transcription factors (β‐catenin and β‐catenin/AhR/TCDD complex) together regulate zonal induction of Cyp1A1. In this work, we have explained the zonal induction of cytochrome proteins by TCDD using a systems biology approach. In addition to generating new hypotheses about hepatic gene induction by TCDD, the model suggests that a negative feedback loop architecture may be shared as a general mechanism of zonal gene expression in biological systems.
Materials and Methods
Biochemical Model of WNT Signal Pathway and TCDD‐Induced Gene Regulatory Network
The intracellular biochemical model includes key molecules regulating Cyp gene expression (Fig. 1B): WNT, DVL, APC, APC destructive complex, β‐catenin, and the protein complexes formed by these molecules. Considering the slow degradation of Disheveled and APC, we treated their concentrations as constant in the time frame of WNT signaling. Extracellular WNT molecules initiate the signaling cascade by activating Disheveled protein. Activated Disheveled protein then inhibits formation of the APC destructive complex.( 19 ) β‐Catenin is continuously synthesized and modeled by mass action kinetics. β‐Catenin degradation was modeled in two ways: degradation induced by the APC destructive complex, and basal first‐order degradation in accordance with mass action kinetics. β‐catenin reversibly binds with APC to form a protein complex. AHR, Cyp1A1, and Cyp1A2 proteins are continuously synthesized and degraded, with the degradation following first‐order mass action kinetics. Synthesis of these proteins is regulated by upstream β‐catenin and, when present, TCDD. Without TCDD, the WNT signaling cascade maintains homeostatic expression of AHR, Cyp1A1, and Cyp1A2. β‐Catenin induces both Cyp1A1 and AHR,( 12 , 13 , 23 ) which in turn acts as a transcription factor to induce Cyp1A1 and Cyp1A2 (Fig. 1B).( 13 )
TCDD significantly enhances the expression of Cyp1A1 and Cyp1A2,( 38 ) by forming a transcription factor complex along with AHR and β‐catenin (β‐catenin/AhR/TCDD [BAT]), which binds to sites in the promoters of Cyp1A1( 13 , 23 ) and Cyp1A2( 12 ) genes. Cyp1A2 protein in turn binds and sequesters TCDD. By limiting the amount of free TCDD, this reaction adds a negative feedback loop to the system, attenuating TCDD‐facilitated induction of Cyp1A1 and Cyp1A2.
The equations for this model are provided in the Supporting Materials.
PBPK Model for Estimating TCDD Concentration in the Liver Lobule
In the experiments from which the bulk RNA‐sequencing (RNA‐seq) data was collected to calibrate the tissue model, TCDD doses applied to the mice were in units of μg/kg. To estimate TCDD concentration in the liver lobule corresponding to different administered TCDD doses, we used a PBPK model. We have adapted a previously developed PBPK model of TCDD disposition for C57BL/6J mice( 24 ) to support multiple dosing every 4 days for 28 days, as described in Nault et al.( 39 ) For each of the doses used (0.01, 0.03, 0.1, 0.3, 1, 3, 10, or 30 μg/kg), the model was used to estimate the final TCDD liver concentration in nanometers at the end of the dosing regimen, corresponding to the time when RNA‐seq was performed in the original studies. These estimates of final TCDD concentrations in liver were used in the virtual tissue model (Supporting Fig. S1).
Numerical Simulations
Our multiscale model consists of two parts: a signaling pathway model and a single‐cell‐resolution liver lobule model. The signaling pathway model was developed in Jupyter notebook using Picotools( 40 ) and COPASI.( 41 ) The overall protein expression levels were adapted from the literature.( 42 ) Zonal gene‐expression levels were estimated based on single‐cell RNA‐seq data( 11 ) and immunofluorescence staining.( 7 ) TCDD‐induced gene‐expression values were adapted from a recent transcriptomic study.( 43 ) The experimental data used in this research are summarized in the Supporting Materials. The biochemical cell signaling model was exported from COPASI in SBML format and then incorporated into individual cells in CompuCell3D.( 44 ) All figures were plotted in Jupyter notebook using the Python Matplotlib (version 3.0.3) package and Prism (version 9) from GraphPad.
The liver lobule model was implemented in CompuCell3D (version 3.7.4) with script written in Python 3.7. CompuCell3D is a spatial and dynamical simulation environment for virtual tissues.( 44 ) The liver lobule model was developed based on the Cellular Potts Model.( 44 ) In this algorithm, the basic spatial unit is a lattice site, where each site can only be occupied by one cell. Based on their size, cells can occupy several lattice sites. The cells update their occupied sites based on an energy term, which the CompuCell3D algorithm tries to minimize.( 44 , 45 ) In our study, hepatocytes were fixed in place; hence we set the energy term very low to freeze each cell at its predefined location. The spatial locations and sizes of cells in the lobule, as well as their zone membership, were collected from single‐cell resolution images of hepatocytes in a mouse liver lobule. Additionally, the virtual liver lobule was segmented using concentric circles into nine zones with evenly distributed diameters, starting from the central vein and continuing to the portal area as in Halpern et al.( 11 ) Individual cells were assigned to different zones by distance from the central vein. CompuCell3D scripts and parameters are provided in the Supporting Materials.
Fitting of Parameters
The parameters in our biochemical cell signaling model were optimized to match a combination of modeling and experimental results in the literature. The parameters in the WNT signaling pathway model were optimized in COPASI using particle swarm algorithm to match the total β‐catenin nuclear concentration in previously published models.( 19 , 40 , 46 ) The parameters describing downstream gene regulation by TCDD were optimized to match published experimental data.( 41 ) All model parameters are provided in the Supporting Materials.
Results
The intracellular signaling model and virtual liver model together enabled us to analyze regulatory mechanisms of zonal induction of Cyp1A1 and Cyp1A2 by TCDD in the mouse liver lobule. We found that two factors are key regulators of this phenomenon: (1) the negative feedback loop formed by binding and sequestration of TCDD by Cyp1A2, and (2) cooperative gene induction by the transcription factors β‐catenin and BAT protein complex.
Modeling the Intracellular Signaling Pathway and Gene Regulatory Network
To capture key characteristics of the WNT signaling pathway, our simplified model (red, dashed line in Supporting Fig. S2) was compared with a detailed model from the literature (black, solid line in Supporting Fig. S2).( 19 , 46 ) Under 1‐nM WNT stimulation, our model responded slightly faster, and arrived at the same steady state as the detailed model (Supporting Fig. S2A). Next, we scanned the levels of WNT, APC, and Disheveled to evaluate their effects on steady‐state nuclear localization of β‐catenin. The results from our model were virtually identical to the detailed model (Supporting Fig. S2B‐D). Overall, our simplified model captured the major quantitative characteristics of the WNT signaling pathway. The detailed model( 19 ) was developed from experiments in Xenopus eggs, in which the protein expression levels are quite different from those in mammalian cells. Setting the concentrations of APC and Disheveled at the level of mammalian cells (APC, 10.28 nM; Disheveled, 2.05 nM),( 43 ) the temporal response of β‐catenin activation to reach steady state doubles compared with the detailed model. (Fig. 2A and Supporting Fig. S2A). We further scanned WNT and APC with concentrations around their expression levels in a mammalian cell. APC affects β‐catenin activation similarly to the detailed model (Fig. 2B and Supporting Fig. S2C). The effect of WNT, however, differs from the detailed model, as the nuclear localization of β‐catenin did not reach a plateau in our model due to the narrower expression range of WNT in mammalian cells (Fig. 2C and Supporting Fig. S2B).
FIG. 2.
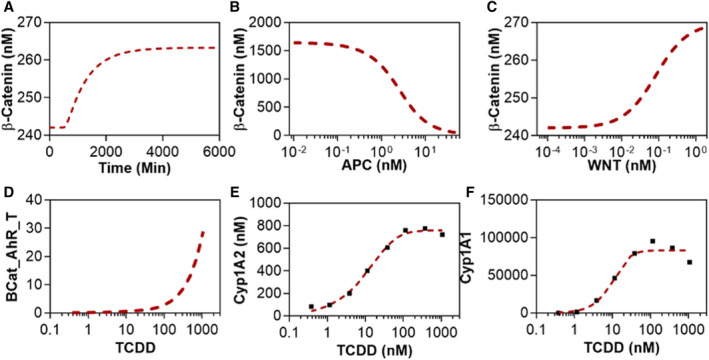
Calibration of the integrated intracellular model for TCDD induction of Cyp1A1. The dashed red lines show model simulations, while the black squares represent RNA‐seq data from the Gene Expression Omnibus (GSE62903 and GSE87543). (A) Dynamics of the WNT signaling pathway: time course of β‐catenin nuclear localization. (B,C) Effects of APC and WNT molecules on the steady‐state nuclear concentration of β‐catenin. (D‐F) Dose‐response curves of the β‐catenin/AHR/TCDD transcription factor complex, Cyp1A2, and Cyp1A1 with respect to TCDD concentration.
The second part of the intracellular model describes AHR and β‐catenin coordinately regulating Cyp1A1 and Cyp1A2 expression. The gene‐induction processes were modeled using the Hill equation.( 47 ) The expression levels of all genes were extracted from RNA‐seq data in the literature (GSE62903 and GSE87543).( 39 , 43 ) As per the experimental data, TCDD does not appear to significantly affect β‐catenin and AHR expression (Supporting Fig. S3). The nuclear concentration of the key transcription factor complex, BAT, is proportional to the concentration of TCDD (Fig. 2D). The steady‐state fold changes of Cyp1A1 and Cyp1A2 in our model were calibrated with experimental results (Fig. 2E,F). Cyp1A1 and Cyp1A2 showed a switch‐like response with respect to TCDD concentrations with a steeper slope for Cyp1A1 (Fig. 2E,F).( 48 ) The effective Hill coefficients of the dose‐response for Cyp1A1 and Cyp1A2 were 3.36 and 1.87, respectively. At high doses of TCDD (1,072 nM), the simulated expression of Cyp1A1 is higher than experimental results. This discrepancy might be a result of the toxic effects of TCDD reducing the physiological fitness of the cells in vivo. Overall, our intracellular biochemical model captured the experimentally observed behaviors of both the WNT signaling cascade and the TCDD‐induced gene induction pathway.
Modeling Basal Zonated Expression of Cyp1A1 and Cyp1A2
Low and intermediate doses of TCDD zonally induce Cyp1A1, with higher expression in the centrilobular zones.( 3 ) To model basal zonation in gene expression in the mouse liver, we segmented the lobule into nine zones along the central‐to‐portal axis with one to two cells in each zone, following Halpern et al.( 11 ) The zonated expression of key proteins in our biochemical model was extracted from multiple experimental data types, including single‐cell RNA‐seq( 11 ) and immunostaining( 7 ) results. APC messenger RNA (mRNA) expression increases exponentially from the central to portal area (Fig. 3A).( 7 ) Disheveled mRNA expression likewise increases from the central to portal area (Fig. 3B).( 11 ) Unphosphorylated β‐catenin (undergoing nuclear localization) presented a switch‐like expression pattern along the lobule in experimental studies (Fig. 3C).( 7 , 11 ) Based on the zonated expression of APC and Disheveled, our biochemical model could reproduce the switch‐like profile of β‐catenin nuclear localization (Fig. 3C), and the basal expression of its downstream genes (AHR, Cyp1A1, and Cyp1A2) (Fig. 3D‐F). Experimentally measured basal Cyp1A1 and Cyp1A2 expression was lower in the mid‐zonal regions of the lobule compared with our model simulations. Without TCDD stimulation, Cyp1A1 and Cyp1A2 show very low basal expression, which may be missed by single‐cell RNA‐seq because of its high dropout rate. To intuitively visualize the zonated gene expression, the biochemical model was implemented into each cell of the virtual liver lobule (Fig. 3G‐I). In this model, virtual hepatocyte locations are based on the spatial distribution of cells in a high‐resolution image of the liver lobule, adapted from Halpern et al.( 11 ) For each cell, the initial expression levels of the genes were defined as a function of cell location. The zonated expression of β‐catenin, AHR, Cyp1A1, and Cyp1A2 are illustrated along the lobular axis in the model (Fig. 3G‐I).
FIG. 3.
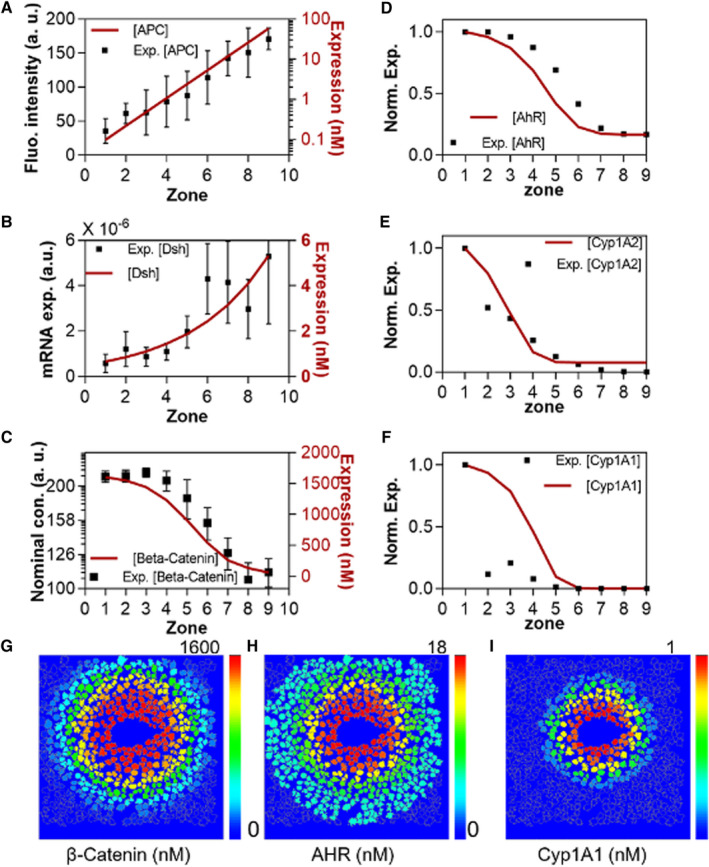
Calibration of metabolic zonation and basal expression of Cyp1A1 and Cyp1A2. (A‐C) Calibration of zonal protein expression profiles of APC, Disheveled, and β‐catenin with experimental data. APC and β‐catenin are calibrated with immunostaining data,(3) whereas Disheveled is calibrated with single‐cell RNA‐seq data.(11) (D‐F) Zonal expression profile of AHR gene regulatory network molecules: AHR, Cyp1A1, and Cyp1A2.(11) Red lines show simulated curves, while black squares represent experimental data. (G‐I) Zonated simulated concentration of β‐catenin, AHR, and Cyp1A1 along the virtual liver lobule.
Modeling Zonated Cyp1A1 and Cyp1A2 Induction by TCDD
TCDD induces Cyp1A1 in a switch‐like manner in individual hepatocytes.( 48 , 49 ) We stimulated the cells in our model with TCDD concentrations ranging from 0.38 nM to 1072 nM in the liver, corresponding to 0.01‐μg/kg to 30‐μg/kg doses administered by oral gavage. All cells in the model presented a switch‐like response to TCDD, though the shapes of the response curves varied across the zones of the lobule (Supporting Fig. S4). To characterize the zonal induction of Cyp1A1, we defined a “zonation index”: the percentage change in Cyp1A1 expression from zone 1 to zone 9 relative to its expression in zone 1 (Fig. 4C). Without TCDD stimulation, Cyp1A1 is zonally expressed at a very low level. In the presence of TCDD, Cyp1A1 expression increases both in the central and portal areas. The zonation index drops in a switch‐like manner with increasing TCDD dose. However, even at high TCDD doses, the Cyp1A1 gene is still zonally expressed (Fig. 4A,C). A critical regulator of Cyp1A1 induction is the transcription factor complex BAT. BAT concentration increases with the dose of TCDD, and is zonally distributed with highest levels observed in the centrilobular area (Fig. 4B). Unlike Cyp1A1, BAT does not attain pan‐lobular nuclear concentration in the TCDD concentration range we have simulated (Fig. 4B). The virtual liver lobule model enables us to visualize zonal gene induction intuitively, facilitating direct comparison to experimental results (Fig. 4E). The simulated zonation profiles of Cyp1A1 were aligned well with reported immunostaining results (right panels of Fig. 4E).( 47 )
FIG. 4.
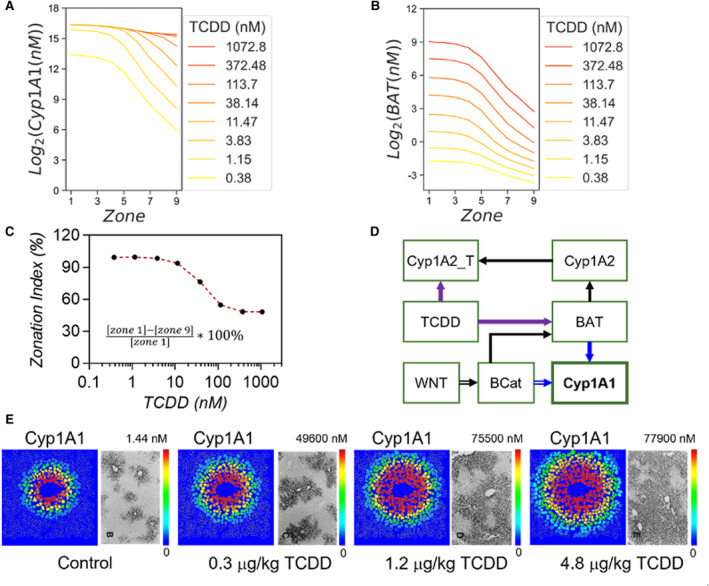
Simulation of zonal induction of Cyp1A1 by TCDD. (A,B) Zonal induction profiles of Cyp1A1 and BAT at various doses of TCDD. (C) The zonation index, representing the percentage changes in Cyp1A1 expression across the lobule, relative to the TCDD doses. (D)Simplified diagram of the signaling pathway network to highlight the three main regulators of zonal Cyp1A1 induction by TCDD: Wnt signaling, negative feedback induced by Cyp1A2 binding to TCDD, and the cooperation of regulatory transcription factors β‐catenin and the BAT complex. (E) Zonal induction of Cyp1A1 in the model compared with immunostaining results at different doses of TCDD. Immunostaining pictures reproduced from Tritscher et al.(3) Abbreviation: Cyp1A2_T, Cyp1A2‐TCDD molecular complex.
A simplified signaling network diagram summarizes the key factors regulating this dynamic process (Fig. 4D). The zonal induction of Cyp1A1 is affected by three components of this system: (1) the WNT signaling pathway (double line arrows), (2) the negative feedback loop formed by Cyp1A2‐TCDD interactions (purple arrows), and (3) the cooperation of transcription factors β‐catenin and BAT (blue arrows). The WNT signaling pathway regulates β‐catenin nuclear localization, which affects the concentration of BAT. BAT in turn induces Cyp1A2, which binds to TCDD (Cyp1A2_T in Fig. 4D), thus reducing the free pool of TCDD available to activate AHR and form the BAT complex. Furthermore, β‐catenin cooperates with BAT in regulating Cyp1A1. Next we evaluate the relative contributions of these three factors.
Zonated Cyp1A1 Induction Is Not Sensitive to the WNT Signaling Level in Our Model
The WNT signaling pathway regulates the basal metabolic zonation of gene expression along the liver lobule, as well as the nuclear localization of β‐catenin. Specifically, WNT2 is secreted by the sinusoidal endothelial cells around the central vein and maintains the zonal profiles of hepatic genes.( 2 )
To evaluate the effect of different WNT concentrations on zonal Cyp1A1 induction by TCDD, we scanned the WNT concentration (relative profile shown in Supporting Fig. S5; data from Halpern et al.( 11 )) along the central‐portal axis in our virtual liver lobule model. By setting the simulated WNT concentration to one of 0.05, 0.5, 5, 50, or 500 nM in zone 1, we set WNT concentrations in all other zones relative to zone 1, such that the relative WNT profile shown in Supporting Fig. S5 was maintained. This procedure enabled us to examine the differences in sensitivity of hepatocytes to WNT signaling, likely arising due to differences in WNT receptor concentrations across different zones. Our results show that in zones 1 through 4, changes in WNT levels had little relative effect on changes in BAT nuclear concentration (Fig. 5b), as β‐catenin was already maximally activated and concentrated in the nucleus even at low simulated WNT concentrations. From the midlobular zone 5 to the portal area (zone 9), the impact of WNT on changes in BAT nuclear concentration increased, resulting in a 20% increase of BAT nuclear concentration relative to basal nuclear concentration (Fig. 5B and Supporting Fig. S6A). However, because Cyp1A1 and β‐catenin are expressed at low levels in the portal area, the absolute changes in their expression and nuclear localization, respectively, were not significant when compared with their absolute expression and nuclear localization in the centrilobular area (Fig. 5A,B). The nuclear concentration of BAT, and expression profiles of Cyp1A1 and Cyp1A2, changed in a similar manner to β‐catenin nuclear concentration, as WNT expression was varied (Supporting Fig. S6C‐E). These results suggest that WNT receptors on hepatocytes in the centrilobular area might be saturated even at very low concentrations of WNT, whereas that might not be the case for hepatocytes in midzonal and periportal areas. With respect to expression of AHR, surprisingly, cells at midlobular zones 5 and 6 were the most sensitive to changes in WNT concentration (Supporting Fig. S6B). The effects of WNT concentration on Cyp1A1 zonal induction were mapped onto the virtual liver lobule (Fig. 5C).
FIG. 5.
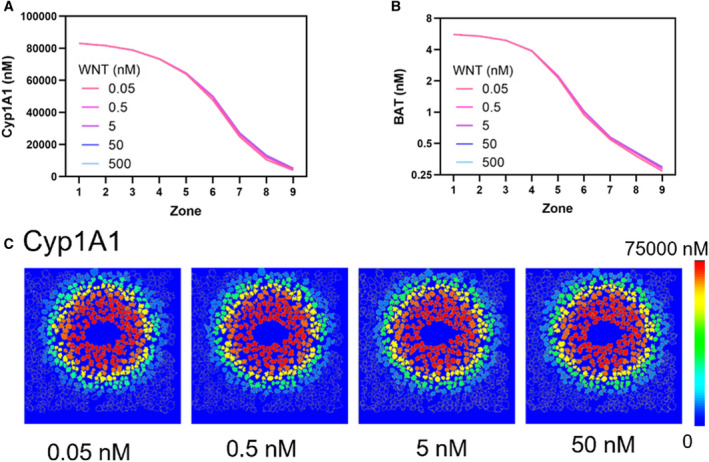
Zonal induction of Cyp1A1 in our signaling pathway model is not sensitive to the concentration of WNT. (A,B) Zonal simulated concentration profiles of Cyp1A1 (A) and BAT transcription factor complex (B) at different concentrations of WNT show minimal sensitivity to WNT. (C) Visualization of Cyp1A1 induction in the virtual liver lobule at different concentrations of WNT.
Zonated Cyp1A1 Induction Is Coordinately Regulated by Cyp1A2‐TCDD Binding and the Cooperation of Transcription Factors
β‐Catenin and the BAT complex cooperatively act as transcription factors inducing the Cyp1A1 gene (blue arrows in Fig. 4D).( 12 , 13 , 23 ) The concentration of BAT is affected by the binding of Cyp1A2 protein to TCDD, which effectively reduces the amount of free TCDD. Thus, zonal induction of Cyp1A1 by TCDD is coordinately regulated by (a) Cyp1A2‐TCDD binding and (b) BAT and β‐catenin cooperation.
To evaluate the comparative effects of mechanisms (a) and (b), we decoupled them in the model. In agreement with results in the previous section, WNT signaling pathway–induced metabolic zonation by itself could not generate the zonal induction of Cyp1A1 unless at least one of the two regulatory mechanisms (a) and (b) was present in the model (Fig. 6A‐D). Both Cyp1A1 and BAT were expressed uniformly along the liver lobule without any zonation when both the negative feedback produced by Cyp1A2‐TCDD binding (mechanism [a]) and TF cooperation between BAT and β‐catenin (mechanism [b]) were turned off in the model (Fig. 6A and Supporting Fig. S7). Either of the two mechanisms, Cyp1A2‐TCDD interaction or cooperation of transcription factors, by itself was able to reproduce zonal induction of Cyp1A1 (Fig. 6B,C). However, the effect of the negative feedback due to Cyp1A2‐TCDD binding on zonal gene induction was more significant (Fig. 6B,D).
FIG. 6.
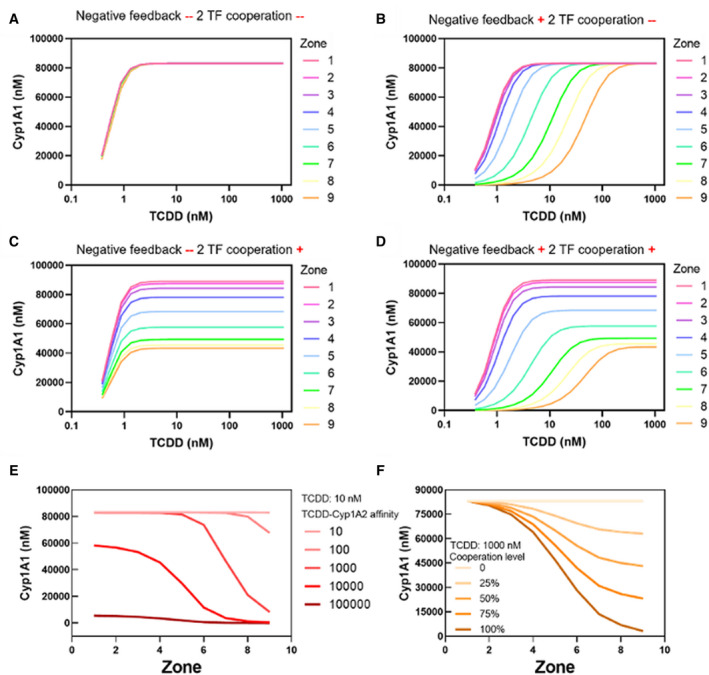
Negative feedback formed by binding of TCDD by the Cyp1A2 protein, and transcription factor cooperation among BAT and β‐catenin, together regulate the zonal induction of Cyp1A1. (A‐D) Zonal induction profiles of Cyp1A1 expression with four combinations of the two mechanisms. Zonal induction of Cyp1A1 was not observed unless at least one of the two regulatory mechanisms was present in the model. (E) Affinity of Cyp1A2 for TCDD strongly affects the zonal induction pattern of Cyp1a1. The lines indicate the forward reaction rate k10 of Cyp1A2 binding to TCDD (1/nMol*min). (F) Level of transcription factor cooperation regulates the zonal induction of Cyp1A1. Different lines represent different capabilities of β‐catenin to affect the transcriptional capacity of BAT (alpha in the model).
Without TCDD exposure, Cyp1A2 is minimally expressed. TCDD induces Cyp1A2 expression, which in turn binds to TCDD. This negative feedback reduces the pool of free TCDD available to form transcription factor complex BAT. The effect of this negative feedback loop on zonal Cyp1A1 induction was evaluated by scanning the forward reaction parameter k10 of the Cyp1A2‐TCDD binding interaction (Supporting Equation( 10 )), with the TCDD level set at 10 nM. The affinity of Cyp1A2 for TCDD strongly affects both the expression level and profile along the central‐to‐portal axis of Cyp1A1 (Fig. 6E) as well as other molecules (Supporting Fig. S8). As k10 increases, the concentration of BAT (Supporting Fig. S8A) and Cyp1A1 (Fig. 6E) decrease. Interestingly, cellular responses to alterations of k10 are zone‐specific. In the centrilobular region (zones 1‐3), the concentration of Cyp1A2‐TCDD is maintained at a very low level until the parameter k10 is larger than 100 1/(nMol*min). In the portal region (zones 7‐9), Cyp1A2‐TCDD increases with increasing k10 and is saturated at k10 = 1,000 (Supporting Fig. S8B). At steady state, due to the low concentration of TCDD relative to that of AHR, β‐catenin, and Cyp1A2, most of the TCDD molecules are either part of the BAT or the Cyp1A2‐TCDD complex. Thus, BAT concentration has an inverse profile compared to Cyp1A2‐TCDD (Supporting Fig. S8A,B). The switch‐like zonal gradient of BAT leads to similarly shaped zonal induction patterns for Cyp1A1 (Fig. 6E) and Cyp1A2 (Supporting Fig. S8C).
To evaluate the effect of transcription factor cooperation, we varied the capability of β‐catenin to affect the transcriptional capacity of BAT. The capability is defined as the percentage of BAT transcriptional capacity that can be affected by β‐catenin (alpha in the model). At a capability of 100%, BAT would not be able to induce Cyp1A1 without β‐catenin. A capability of 0%, on the other hand, would mean that β‐catenin has no effect on the transcriptional capacity of BAT. As this capability increases from 0% to 100%, the expression of Cyp1A1 drops more sharply from central to portal regions (Fig. 6F). A recent thermodynamic model predicted the capability of β‐catenin to affect the transcriptional power of BAT to be approximately 50% (Fig. 3 of Schulthess et al.( 23 )). Therefore, when tuning other parameters, alpha was set as 0.5 (Fig. 6F).
Discussion
We have described a single‐cell‐resolution virtual liver lobule model to investigate the determinants of AHR‐mediated zonal gene induction by TCDD. An intracellular model of the basal and inducible expression of Cyp proteins in the liver was developed and compared with published models.( 19 ) We used our model to investigate the mechanisms of zonal induction of Cyp1A1 by TCDD. The results indicated that binding and sequestration of TCDD by Cyp1A2, and cooperation of transcription factors β‐catenin and the β‐catenin‐AHR‐TCDD complex, together with basal metabolic zonation determine the zonal induction of Cyp1A1.
The WNT pathway plays a key role in regulating metabolic zonation in the liver. Specifically, APC has been implicated as a hepatic “zonation‐keeper.”( 7 ) Simulations from our minimal WNT signaling model aligned well with a previously published detailed model of the pathway. Acting downstream of WNT, the BAT complex acts as a transcription factor inducing Cyp family genes. Combining WNT and the downstream BAT complex, our model captured the dose‐response of Cyp1A1 and Cyp1A2 genes to TCDD stimulation (Figs. 3 and 4).
The role of the WNT signaling molecule in maintaining metabolic zonation has long been emphasized.( 7 , 10 , 50 ) Activating Beta‐catenin pathway via disrupting APC expression causes periportal genes to display a whole lobule expression. Activating the Beta‐catenin pathway by increasing the concentration of WNT molecules may not achieve the same results.( 7 ) Recently, endothelial cell–secreted WNT2 was confirmed to regulate β‐catenin expression in the liver.( 50 , 51 , 52 ) Knocking out WNT secreted by endothelial cells inhibited the expression of β‐catenin targeted genes, along with their zonal profiles. However, the cellular concentration of unphosphorylated β‐catenin was not reported in these studies.( 51 ) Surprisingly, our model simulations showed that the concentration of WNT does not significantly affect the zonal induction by TCDD of Cyp1A1. Other components of the WNT signaling pathway may limit the power of WNT in regulating the zonal induction of Cyp1A1 and Cyp1A2. WNT induces β‐catenin nuclear localization through regulation of APC and Disheveled. APC, a key player in the WNT signaling pathway, exhibits a gradient of increasing expression from the central to portal region,( 7 ) and Disheveled exponentially increases in the same direction (Fig. 3A,B).( 11 ) At the high end of WNT concentrations, in the centrilobular area, the low expression level of APC and Disheveled limits the regulative power of WNT molecules (Fig. 5A,B). Unphosphorylated β‐catenin maintains an expression plateau near the central region of the lobule.( 7 ) WNT signaling pathway parameters in our model were calibrated to reproduce the experimentally reported plateau of β‐catenin expression in the centrilobular region.
Binding and sequestration of TCDD by Cyp1A2 protein is a key determinant of the zonal induction of Cyp1A1 by TCDD. Basal metabolic zonation leads to zonal expression of β‐catenin, and TCDD has minimal effect on the expression of β‐catenin (Supporting Fig. S3)( 41 ) and AHR.( 13 , 23 ) By limiting the freely available TCDD pool, Cyp1A2‐TCDD binding serves as a negative feedback loop on the gene inductive effects of TCDD. β‐Catenin and AHR are basally synthesized. Thus, the freely available TCDD is the limiting factor determining the concentration of the active BAT complex. Although the concentration of β‐catenin varies significantly along the central‐to‐portal axis, the concentration of BAT is maintained at the same level along the central‐to‐portal axis without the Cyp1A2‐TCDD negative feedback loop (Supporting Fig. S7). In this biochemical system, Cyp1A2 competes with AHR and β‐catenin for TCDD. Due to this competition, more BAT is formed at the centrilobular area, where β‐catenin nuclear concentration is higher. As the affinity of Cyp1A2‐TCDD binding increases, BAT concentration drops. Considering the extreme cases, in which all TCDD binds either to Cyp1A2 or to AHR and β‐catenin, no zonation is observed in the level of the BAT transcription factor complex (Supporting Fig. S8). Thus, zonal induction of Cyp1A1 requires the affinity of Cyp1A2 for TCDD to be in a specific range. Our results show that switch‐like zonal expressions of Cyp1A1 and Cyp1A2 are most similar to experimentally observed patterns obtained at intermediate affinity of Cyp1A2 for TCDD (k10 = 10,000 (nmol * min)‐1) (), as reported elsewhere.( 23 ) The observation from our computational model that sequestration of TCDD by Cyp1A2 is required for zonal induction of Cyp1A1 has not been verified experimentally. Given the availability of Cyp1A2 null mice, however, the model’s prediction can be tested in future experiments.
Cooperation among transcription factors BAT and β‐catenin also affects the zonal induction of Cyp1A1 but to a lesser extent than the Cyp1A2‐TCDD negative feedback loop. Simulations based on a previous thermodynamic model( 23 ) support the contention that this transcription factor cooperation and basal metabolic zonation of β‐catenin contribute to zonal induction.( 23 ) β‐Catenin has one binding site in the Cyp1A1 gene promoter, whereas BAT has four binding sites.( 23 ) At TCDD concentrations producing saturated Cyp1A1 induction (3.83 nM), BAT concentration is much lower than that of β‐catenin. We therefore assume that the β‐catenin binding site at the promoter is always occupied. Without stimulation by TCDD, the capability of β‐catenin to induce Cyp1A1 is minimal. Thus, the role of β‐catenin as a transcription factor is primarily to augment the inductive capability of BAT. Without the Cyp1A2‐TCDD negative feedback loop, BAT exhibits a uniform pan‐lobular concentration profile, which would produce only a two‐fold difference in Cyp1A1 expression from the central to portal area (Fig. 6C). This difference is negligible compared with the more than 2,000‐fold change induced by BAT in the presence of the negative feedback loop (Fig. 6D).
Our multiscale virtual tissue model provides an intuitive way to visualize zonal gene expression in the liver lobule. This systems biology model integrates the molecular, cellular, and tissue‐level characteristics of the system. Through the work presented here, we demonstrated the utility of an integrated systems biology approach in elucidating the mechanisms determining the zonal expression and induction of a prototypical cytochrome P450 gene. Compared with prior compartmental liver models, our model offers a much higher spatial and biochemical resolution, enabling a dose‐response assessment from intracellular to tissue scale. Such a multiscale model can help identify emergent gene‐expression patterns at the tissue level. To reproduce tissue biology more faithfully, several factors can be added to further refine this model. First, intercellular communication can be integrated to mimic paracrine, juxtacrine, and endocrine aspects of cell signaling. Furthermore, non‐parenchymal cells types can be added to the model to represent more complex and diverse hepatic cell signaling networks.
Supporting information
Supplementary Material
Supported by the National Institute of Food and Agriculture and National Institute of Environmental Health Sciences (R01 ES031937).
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Boverhof DR, Burgoon LD, Tashiro C, Chittim B, Harkema JR, Jump DB, et al. Temporal and dose‐dependent hepatic gene expression patterns in mice provide new insights into TCDD‐mediated hepatotoxicity. Toxicol Sci 2005;85:1048‐1063. [DOI] [PubMed] [Google Scholar]
- 2. Bock KW, Kohle C. Ah receptor: Dioxin‐mediated toxic responses as hints to deregulated physiologic functions. Biochem Pharmacol 2006;72:393‐404. [DOI] [PubMed] [Google Scholar]
- 3. Tritscher AM, Goldstein JA, Portier CJ, McCoy Z, Clark GC, Lucier GW. Dose‐response relationships for chronic exposure to 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin in a rat tumor promotion model: quantification and immunolocalization of CYP1A1 and CYP1A2 in the liver. Can Res 1992;52:3436‐3442. [PubMed] [Google Scholar]
- 4. Conolly RB, Lutz WK. Nonmonotonic dose‐response relationships: mechanistic basis, kinetic modeling, and implications for risk assessment. Toxicol Sci 2004;77:151‐157. [DOI] [PubMed] [Google Scholar]
- 5. Dourson M, Becker RA, Haber LT, Pottenger LH, Bredfeldt T, Fenner‐Crisp PA. Advancing human health risk assessment: integrating recent advisory committee recommendations. Crit Rev Toxicol 2013;43:467‐492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gebhardt R, Matz‐Soja M. Liver zonation: novel aspects of its regulation and its impact on homeostasis. World J Gastroenterol 2014;20:8491‐8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, et al. APC tumor suppressor gene is the “zonation‐keeper” of mouse liver. Dev Cell 2006;10:759‐770. [DOI] [PubMed] [Google Scholar]
- 8. Torre C, Perret C, Colnot S. Transcription dynamics in a physiological process: beta‐catenin signaling directs liver metabolic zonation. Int J Biochem Cell Biol 2011;43:271‐278. [DOI] [PubMed] [Google Scholar]
- 9. Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol 2017;11:622‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: the role of Wnt/beta‐catenin signaling in liver zonation and beyond. Dev Dyn 2010;239:45‐55. [DOI] [PubMed] [Google Scholar]
- 11. Halpern KB, Shenhav R, Matcovitch‐Natan O, Tóth B, Lemze D, Golan M, et al. Single‐cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017;542:352‐356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerbal‐Chaloin S, Dume A‐S, Briolotti P, Klieber S, Raulet E, Duret C, et al. The WNT/β‐catenin pathway is a transcriptional regulator of CYP2E1, CYP1A2 and aryl hydrocarbon receptor gene expression in primary human hepatocytes. Mol Pharmacol 2014;86:624‐634. [DOI] [PubMed] [Google Scholar]
- 13. Braeuning A, Köhle C, Buchmann A, Schwarz M. Coordinate regulation of cxytochrome P450 1A1 expression in mouse liver by the aryl hydrocarbon receptor and the β‐catenin pathway. Toxicol Sci 2011;122:16‐25. [DOI] [PubMed] [Google Scholar]
- 14. Burke ZD, Reed KR, Phesse TJ, Sansom OJ, Clarke AR, Tosh D. Liver zonation occurs through a β‐catenin–dependent, c‐Myc–independent mechanism. Gastroenterology 2009;136:2316‐2324.e2313. [DOI] [PubMed] [Google Scholar]
- 15. Procházková J, Kabátková M, Bryja V, Umannová L, Bernatík O, Kozubík A, et al. The interplay of the aryl hydrocarbon receptor and β‐catenin alters both AhR‐dependent transcription and Wnt/β‐catenin signaling in liver progenitors. Toxicol Sci 2011;122:349‐360. [DOI] [PubMed] [Google Scholar]
- 16. Stamos JL, Weis WI. The β‐catenin destruction complex. Cold Spring Harbor Persp Biol 2013;5:a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rothhammer V, Quintana FJ. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol 2019;19:184‐197. [DOI] [PubMed] [Google Scholar]
- 18. Kofahl B, Wolf J. Mathematical modelling of Wnt/beta‐catenin signalling. Biochem Soc Trans 2010;38:1281‐1285. [DOI] [PubMed] [Google Scholar]
- 19. Lee E, Salic A, Krüger R, Heinrich R, Kirschner MW. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS Biol 2003;1:E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hartung N, Benary U, Wolf J, Kofahl B. Paracrine and autocrine regulation of gene expression by Wnt‐inhibitor Dickkopf in wild‐type and mutant hepatocytes. BMC Syst Biol 2017;11:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mirams GR, Byrne HM, King JR. A multiple timescale analysis of a mathematical model of the Wnt/beta‐catenin signalling pathway. J Math Biol 2010;60:131‐160. [DOI] [PubMed] [Google Scholar]
- 22. Gasior K, Hauck M, Wilson A, Bhattacharya S. A theoretical model of the Wnt signaling pathway in the epithelial mesenchymal transition. Theoretical Biol Med Model 2017;14:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schulthess P, Löffler A, Vetter S, Kreft L, Schwarz M, Braeuning A, et al. Signal integration by the CYP1A1 promoter—a quantitative study. Nucleic Acid Res 2015;43:5318‐5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hon‐Wing L, Ku RH, Paustenbach DJ, Andersenb ME. A physiologically based pharmacokinetic model for 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin in C57BL/6J and DBA/2J mice. Toxicol Lett 1988;42:15‐28. [DOI] [PubMed] [Google Scholar]
- 25. Andersen ME, Birnbaum LS, Barton HA, Eklund CR. Regional hepatic CYP1A1 and CYP1A2 induction with 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin evaluated with a multicompartment geometric model of hepatic zonation. Toxicol Appl Pharmacol 1997;144:145‐155. [DOI] [PubMed] [Google Scholar]
- 26. Andersen ME, Eklund CR, Mills JJ, Barton HA, Birnbaum LS. A multicompartment geometric model of the liver in relation to regional induction of cytochrome P450s. Toxicol Appl Pharmacol 1997;144:135‐144. [DOI] [PubMed] [Google Scholar]
- 27. Bhattacharya S, Shoda LKM, Zhang Q, Woods CG, Howell BA, Siler SQ, et al. Modeling drug‐ and chemical‐induced hepatotoxicity with systems biology approaches. Front Physiol 2012;3:462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drasdo D, Bode J, Dahmen U, Dirsch O, Dooley S, Gebhardt R, et al. The virtual liver: state of the art and future perspectives. Arch Toxicol 2014;88:2071‐2075. [DOI] [PubMed] [Google Scholar]
- 29. Shah I, Wambaugh J. Virtual tissues in toxicology. J Toxicol Environ Health Part B 2010;13:314‐328. [DOI] [PubMed] [Google Scholar]
- 30. Belmonte JM, Clendenon SG, Oliveira GM, Swat MH, Greene EV, Jeyaraman S, et al. Virtual‐tissue computer simulations define the roles of cell adhesion and proliferation in the onset of kidney cystic disease. Mol Biol Cell 2016;27:3673‐3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sluka JP, Fu X, Swat M, Belmonte JM, Cosmanescu A, Clendenon SG, et al. A liver‐centric multiscale modeling framework for xenobiotics. PLoS One 2016;11:e0162428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aslanidi O, Atia J, Benson AP, van den Berg HA, Blanks AM, Choi C, et al. Towards a computational reconstruction of the electrodynamics of premature and full term human labour. Prog Biophys Mol Biol 2011;107:183‐192. [DOI] [PubMed] [Google Scholar]
- 33. Ghaffarizadeh A, Heiland R, Friedman SH, Mumenthaler SM, Macklin P. PhysiCell: an open source physics‐based cell simulator for 3‐D multicellular systems. PLoS Comput Biol 2018;14:e1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White D, Coombe D, Rezania V, Tuszynski J. Building a 3D virtual liver: methods for simulating blood flow and hepatic clearance on 3D structures. PLoS One 2016;11:e0162215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu X, Sluka JP, Clendenon SG, Dunn KW, Wang Z, Klaunig JE, et al. Modeling of xenobiotic transport and metabolism in virtual hepatic lobule models. PLoS One 2018;13:e0198060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kitano H. Systems biology: a brief overview. Science 2002;295:1662. [DOI] [PubMed] [Google Scholar]
- 37. Leist M, Ghallab A, Graepel R, Marchan R, Hassan R, Bennekou SH, et al. Adverse outcome pathways: opportunities, limitations and open questions. Arch Toxicol 2017;91:3477‐3505. [DOI] [PubMed] [Google Scholar]
- 38. Poland A, Teitelbaum P, Glover E, Kende A. Stimulation of in vivo hepatic uptake and in vitro hepatic binding of [125I]2‐lodo‐3,7,8‐trichlorodibenzo‐p‐dioxin by the administration of agonist for the Ah receptor. Mol Pharmacol 1989;36:121‐127. [PubMed] [Google Scholar]
- 39. Nault R, Fader KA, Zacharewski T. RNA‐Seq versus oligonucleotide array assessment of dose‐dependent TCDD‐elicited hepatic gene expression in mice. BMC Genom 2015;16:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welsh CM, Fullard N, Proctor CJ, Martinez‐Guimera A, Isfort RJ, Bascom CC, et al. PyCoTools: a Python Toolbox for COPASI. Bioinformatics 2018;34:3702‐3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, et al. COPASI—a COmplex PAthway SImulator. Bioinformatics 2006;22:3067‐3074. [DOI] [PubMed] [Google Scholar]
- 42. Tan CW, Gardiner BS, Hirokawa Y, Layton MJ, Smith DW, Burgess AW. Wnt signalling pathway parameters for mammalian cells. PLoS One 2012;7:e31882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fader KA, Nault R, Kirby MP, Markous G, Matthews J, Zacharewski TR. Convergence of hepcidin deficiency, systemic iron overloading, heme accumulation, and REV‐ERBα/β activation in aryl hydrocarbon receptor‐elicited hepatotoxicity. Toxicol Appl Pharmacol 2017;321:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Swat MH, Thomas GL, Belmonte JM, Shirinifard A, Hmeljak D, Glazier JA. Multi‐scale modeling of tissues using CompuCell3D. In: Asthagiri AR, Arkin AP, eds. Methods in Cell Biology. Volume 110: Cambridge, MA: Academic Press; 2012:325‐366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Glazier JA, Graner F. Simulation of the differential adhesion driven rearrangement of biological cells. Phys Rev E 1993;47:2128. [DOI] [PubMed] [Google Scholar]
- 46. Benary U, Kofahl B, Hecht A, Wolf J. Modeling Wnt/β‐catenin target gene expression in APC and Wnt gradients under wild type and mutant conditions. Front Physiol 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauro HM. Enzyme kinetics for systems biology. Future Skill Software 2011. [Google Scholar]
- 48. Broccardo CJ, Billings RE, Chubb LS, Andersen ME, Hanneman WH. Single cell analysis of switch‐like induction of CYP1A1 in liver cell lines. Toxicol Sci 2004;78:287‐294. [DOI] [PubMed] [Google Scholar]
- 49. French CT, Hanneman WH, Chubb LS, Billings RE, Andersen ME. Induction of CYP1A1 in primary rat hepatocytes by 3,3′,4,4′,5‐pentachlorobiphenyl: evidence for a switch circuit element. Toxicol Sci 2004;78:276‐286. [DOI] [PubMed] [Google Scholar]
- 50. Yang J, Mowry LE, Nejak‐Bowen KN, Okabe H, Diegel CR, Lang RA, Williams BO, et al. Beta‐catenin signaling in murine liver zonation and regeneration: a Wnt‐Wnt situation! Hepatology 2014;60:964‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Preziosi M, Okabe H, Poddar M, Singh S, Monga SP. Endothelial Wnts regulate β‐catenin signaling in murine liver zonation and regeneration: a sequel to the Wnt‐Wnt situation. Hepatol Commun 2018;2:845‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leibing T, Géraud C, Augustin I, Boutros M, Augustin HG, Okun JG, et al. Angiocrine Wnt signaling controls liver growth and metabolic maturation in mice. Hepatology 2018;68:707‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


