Abstract
Aquatic environments are recognized as one of the main reservoirs for the emergence and dissemination of high-risk lineages of multidrug-resistant (MDR) bacteria of public health concern. However, the genomic characteristics of antibiotic-resistant Escherichia coli isolates from aquatic origins remain limited. Herein, we examined the antibiotic resistance and virulence genomic profiles of three E. coli recovered from surface water in northwest Mexico. Antimicrobial susceptibility testing, whole-genome sequencing (WGS), and in-depth in silico analysis were performed. Two E. coli exhibited MDR phenotypes. WGS-based typing revealed genetic diversity, and phylogenetic analysis corroborated a notable divergent relationship among the studied E. coli. One E. coli strain, harboring enterotoxigenic and extraintestinal pathogenic-associated virulence genes, was assigned to the ST4 lineage. MDR E. coli, belonging to the international high-risk clones ST410 and ST617, carried genes and mutations conferring resistance to aminoglycosides, β-lactams, quinolones, sulfonamides, tetracyclines, and trimethoprim. This study describes, for the first time, the detection and genomic profiling of high-risk lineages of E. coli ST410 and ST617 from surface water in Mexico. Additionally, our results underscore the role of surface water as a reservoir for critical pathogenic and MDR E. coli clones and the need for the surveillance and monitoring of aquatic environments via WGS from the One Health perspective.
Keywords: multidrug-resistance, environmental E. coli, aquatic environment, high-risk E. coli clones, ETEC O6, class 1 integron, whole-genome sequencing, agricultural drainage
1. Introduction
Antimicrobial resistance (AMR) has been a primary global public health issue in recent decades. AMR hinders clinical effectiveness and limits the therapeutic options for infectious diseases, leading to increased healthcare costs, treatment failure, morbidity, and mortality rates. At present, the estimated AMR-related death toll accounts for 700,000 annually, and it is projected to increase to 10 million deaths per year by 2050 unless action is taken [1]. AMR phenomenon represents a complex and multifaceted threat of growing concern to the human, animal, and environmental health due to the emergence, persistence, and spread of antibiotic resistance genes (ARGs) and their acquisition by clinically relevant bacteria in each interconnected sector [2].
The importance of the environment as a reservoir of antibiotic resistance and its role in the spread of potential ARGs has been widely recognized. Antibiotic resistance can arise either by mutational mechanisms or by resistance-conferring gene acquisition via horizontal gene transfer (HGT) events, that is, conjugation, transformation, or transduction. HGT is considered the most important factor in the recruitment and transmissibility of ARGs from the environmental gene pool to pathogenic bacteria [3].
Among all environmental compartments, the aquatic ecosystems are constantly subjected to anthropogenic contamination with antibiotics through the direct or indirect discharges of agricultural, aquaculture, domestic, hospital, and industrial effluents. The selective pressure exerted by antibiotic contamination impacts the resident bacterial communities towards the selection of antibiotic-resistant bacteria (ARB) and enrichment of drug-resistant genetic determinants [4]. Furthermore, aquatic environments have dynamic and distinct microbial composition patterns influenced by temporal and spatial disparities in physiochemical and biotic factors, including environmental stresses and nutrient composition [5]. Consequently, the aquatic milieus constitute an ideal reservoir for microbial genetic exchange, evolution, and dissemination of ARGs and ARB.
Among the ARB harboring transmissible resistance traits of greatest clinical relevance are extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, which have become a challenge for infection control because of their worldwide endemicity and multidrug-resistant (MDR) phenotypes [6,7]. Escherichia coli (E. coli) is a highly versatile member of the Enterobacteriaceae family that constitutes an important component of the commensal gastrointestinal microbiota of warm-blooded animals, including humans. However, its genome plasticity and HGT mechanisms have led to the evolution of this microorganism into highly adapted antibiotic-resistant and pathogenic clones [8].
Depending on the site of infection, pathogenic E. coli strains are classified as intestinal pathogenic E. coli (InPEC) or extraintestinal pathogenic E. coli (ExPEC) and further sub-categorized into distinct pathotypes based on clinical disease manifestations, virulence factors (VFs), and genetic background [9,10]. The long-term survival of E. coli has been described in secondary habitats, including food products, sediments, soil, and water [11,12]. Thus, environmental E. coli strains could be a source of opportunistic and MDR-related diseases of public health significance in animals and humans; therefore, such strains must be monitored and tracked.
Several studies have identified antibiotic-resistant and pathogenic E. coli strains in environmental water samples from Mexico [13,14,15,16,17]. Previous studies in Sinaloa, Mexico, have reported E. coli as a major biological pollutant of diverse aquatic resources, including agricultural drainage, drinking, and irrigation water [18,19,20]. Canizalez-Roman et al. [21] demonstrated multidrug-resistant InPEC strains in surface water samples from irrigation channels and river water across Sinaloa State, where they exhibited resistance to β-lactams, tetracycline, and trimethoprim/sulfamethoxazole.
To the best of our knowledge, genomic data describing the mechanisms underlying antibiotic resistance and virulence properties of E. coli from environmental water samples in Mexico are extremely limited. Therefore, this study aimed to examine, from a genomic perspective, the antibiotic resistance patterns and virulence determinants of E. coli isolates recovered from surface water of an agricultural drainage ditch that constantly receives irrigation water and domestic wastewater. In this study, we describe the genomic characteristics and determine the genetic diversity regarding the virulence-associated and antimicrobial resistance genes, plasmids, and prophage content of three antibiotic-resistant E. coli strains from the aquatic environment, two of which are international high-risk clones.
2. Materials and Methods
2.1. Bacterial Isolation and Identification
E. coli isolates ADD147, ADD167, and ADD183 were recovered from surface water samples in a longitudinal water quality assessment of a section (3.6 km) of the agricultural drainage ditch known as “La Michoacana” (30 km in total length) in the Sinaloa Valley (Navolato Municipality, Sinaloa, Mexico) during 2013. Overall, the microbiological and physicochemical water characteristics prevailing during sampling were in the following ranges for total coliforms (TC) (2.86 × 104−3.23 × 107 CFU/mL), fecal coliforms (FC) (5.45 × 103–1.77 × 107 CFU/mL), pH (7.8–8.01), temperature (21.51–24.76 °C), dissolved oxygen (DO) (8.65–21.24%), and salinity (0.54–0.64) [20]. The ditch has a semicircular profile (4 m wide × 2 m deep) on a plane landscape surrounded by crop fields and receives untreated agricultural and domestic wastewater discharge [22]. Bacterial isolation and species identification were conducted as previously described by Ahumada-Santos et al. [20]. All collected E. coli isolates were stored in brain heart infusion (BHI) broth containing 8% (v/v) DMSO at −80 °C. E. coli ADD147, ADD167, and ADD183 were selected based on preliminary phylogroups typing, antimicrobial resistance, and virulence profiles observed in testing for further study.
2.2. Antimicrobial Susceptibility Testing (AST)
The phenotypic antimicrobial resistance profile of E. coli isolates was determined using the disk diffusion method following the Clinical and Laboratory Standards Institute (CLSI) guidelines [23]. Twelve commercial antibiotic disks obtained from Diagnostic Research (ID), Mexico (PT-35 reference) spanning several antimicrobial classes were tested: aminoglycosides (amikacin (AK; 30 µg); gentamicin (GE; 10 µg); netilmicin (NET; 30 µg)), β-lactams (ampicillin (AM; 30 µg), carbenicillin (CB; 100 µg), cephalothin (CF; 30 µg), cefotaxime (CFX; 30 µg)), fluoroquinolones (ciprofloxacin (CPF; 5 µg), norfloxacin (NOF; 10 µg)), nitrofurans (nitrofurantoin (NF; 300 µg)), phenicols (chloramphenicol (CL; 30 µg)), and folate pathway antagonists (trimethoprim/sulfamethoxazole (SXT; 1.25/23.75 µg)). E. coli ATCC 25922 was used as a quality control strain. The inhibition zones were measured and interpreted according to the CLSI breakpoint criteria (M100 ED31:2021) [24]. The categorical interpretation of intermediate susceptibility was described as not susceptible. The isolates were classified as MDR strains if they exhibited non-susceptibility to at least one antimicrobial agent in three or more antimicrobial classes [25].
2.3. DNA Extraction, Whole-Genome Sequencing (WGS), and Read Preprocessing
For DNA extraction, each E. coli isolate was grown in Luria-Bertani (LB) broth at 37 ± 2 °C for 24 h. Genomic DNA (gDNA) was extracted using the ZymoBIOMICs DNA Miniprep Kit (Zymo Research Corp., Irvine, CA, USA) according to the manufacturer’s protocol. The purity, concentration, and integrity of gDNA were assessed by spectrophotometry (Nanodrop Lite, Thermo Fisher Scientific, Waltham, MA, USA) and electrophoresis on a 1.0% agarose gel. Paired-end DNA libraries were prepared using the Nextera XT Library Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions, and sequenced on the Illumina MiniSeq platform with a 2 × 150 bp approach. Sequence data quality was evaluated using the FastQC software v.0.11.9 [26]. Raw sequencing reads were quality trimmed and filtered using Cutadapt v2.4 [27] to remove adapters, low-quality bases (<Q30), and short reads (<50 bp). The processed reads were submitted to the KmerFinder v3.2 [28] web tool from the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/, accessed on 20 November 2021) for species confirmation.
The raw genome data were submitted to the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information (NCBI) under BioProject PRJNA715781 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA715781, accessed on 19 March 2021) and are accessible through individual SRA accession numbers: SRX10394959 (ADD147), SRX10394960 (ADD167), and SRX10394961 (ADD183).
2.4. Genome Assembly and Annotation
Following quality control and trimming, high-quality reads were assembled into contigs using SPAdes v3.15.1 [29] employing the “-- careful” option and different k-mer lengths (21, 33, 55, 77, 99, 107, and 117) and evaluated with QUAST v5.0.2 [30]. Due to the presence of small contigs after de novo assembly, a reference-guided scaffolding was performed using as a reference the best matching output from the KmerFinder tool. Therefore, the MeDuSa software [31] and the genome sequence of E. coli YD786 (RefSeq accession: GCF_001442495.1), WCHEC005784 (RefSeq accession: GCF_003051985.2), and 2014EL-1346-6 (RefSeq accession: GCF_002741195.1) were used for genome scaffolding of E. coli ADD147, ADD167, and ADD183, respectively. Scaffolds with lengths of less than 300 bp were excluded. The annotation was performed using Prokka v1.14 [32].
2.5. Serotyping, Phylogrouping, Multilocus Sequence Typing (MLST), Virulence, and Antimicrobial Resistance-Associated Genes
To perform the in silico molecular characterization, the genomes of E. coli sequenced here were analyzed using web tools provided by the CGE under default setting parameters (accessed on 21 November 2021). The O and H serotypes were defined using SerotypeFinder v2.0 [33]. The sequence type (ST) was determined following the seven-locus scheme proposed by Achtman [34] using MLST v2.0 (Escherichia coli #1 option) [35]. Virulence-associated genes (VAGs) and chromosomal point mutations along with acquired antimicrobial resistance determinants were identified and annotated with VirulenceFinder v2.0 [36] and ResFinder v4.1 [37], respectively. In addition, E. coli phylogroup classification based on the Clermont et al. scheme [38] was assessed using the ClermonTyping web tool [39] (http://clermontyping.iame-research.center/index.php, accessed on 21 November 2021).
2.6. Plasmid Replicons and Prophages Regions
Plasmid replicon identification was performed using PlasmidFinder v2.1 from the CGE using a minimum 90% nucleotide identity and 60% coverage threshold [40]. Prophage sequences in E. coli genomes were determined with Phage Search Tool Enhanced Release (PHASTER) (https://phaster.ca/, accessed on 21 November 2021). Prophage regions are predicted to be intact, questionable, and incomplete if the prophage sequence scores are ≥90, 70–90, and ≤70, respectively [41]. Only prophages identified as intact were used in the current analysis.
2.7. Phylogenetic Analysis
To place our three sequenced E. coli isolates into a global context, a collection of isolates with the same ST (i.e., ST4, ST410, and ST617) were selected from EnteroBase using the Achtman seven gene MLST scheme as a query (https://enterobase.warwick.ac.uk/species/index/ecoli, accessed on 2 March 2022) [42]. Phylogenetic analysis was performed using GrapeTree to construct a minimum spanning tree based on the core-genome MLST (cgMLST) V1 + Hierarchical Clustering (HierCC) V1 scheme from EnteroBase through the rapid neighbor-joining (RapidNJ) algorithm [43].
Because ST410, ST617, and ST4 phylogenetic trees comprised an excess of 200 isolates, refined subtrees were generated from clades containing E. coli ADD147, ADD167, and ADD183. Branch lengths were used to calculate the cgMLST allelic differences between closely related isolates. Metadata of E. coli isolates used for ST410, ST617, and ST4 subtree reconstruction are described in Supplementary Tables S1–S3, respectively.
3. Results
3.1. Genome Sequencing
E. coli ADD147, ADD167, and ADD183 were recovered in 2013 from the surface water of an agricultural drainage ditch and subjected to WGS. E. coli genomes were assembled de novo, yielding a coverage depth of 54.6× and exhibiting a genome length of 4.75 Mbp, 540 contigs, and 50.81% of GC content on average (Table 1). Because fragmented assemblies were obtained, the complete genome sequence of E. coli YD786 (GCF_001442495.1), WCHEC005784 (GCF_003051985.2), and 2014EL-1346-6 (GCF_002741195.1) were used as a reference to generate scaffolds. Hence, E. coli ADD147, ADD167, and ADD183 genome assemblies resulted in 47, 22, and 6 scaffolds, respectively (Table 1).
Table 1.
General features of antibiotic-resistant E. coli genomes from surface water of agricultural drainage in northwestern Mexico.
| Escherichia coli | |||
|---|---|---|---|
| ADD147 | ADD167 | ADD183 | |
| Length (bp) | 4,748,629 | 4,637,520 | 4,865,392 |
| GC (%) | 50.74 | 50.94 | 50.77 |
| Coverage depth (×) | 60 | 60 | 44 |
| Contigs | 594 | 523 | 504 |
| Scaffolds (>300 bp) | 47 | 22 | 6 |
| No. coding sequence (CDS) | 4440 | 4278 | 4553 |
| Ribosomal RNA (rRNA) | 8 | 8 | 11 |
| Transfer RNA (tRNA) | 81 | 81 | 80 |
| Serotype | ONT:H9 a | O101:H10 | O6:H16 |
| Phylogroup | C | A | A |
| Sequence type (ST) | ST410 | ST617 | ST4 |
| Clonal complex (CC) | CC23 | CC10 | CC10 |
a ONT, O-nontypable.
3.2. Phylogroups, Serotype, and Sequence Types
Based on the WGS data, in silico characterization was performed to gain insights into the genetic diversity of these E. coli isolates. In summary, two phylogenetic groups were identified. E. coli ADD167 and ADD183 were assigned to phylogroup A, whereas ADD147 was allocated to phylogroup C. The O:H typing predicted three dissimilar serotypes among the sequenced E. coli genomes, of which E. coli ADD147 was O-nontypable (ONT). Similarly, three distinct STs were detected, assigning E. coli ADD147, ADD167, and ADD183 to ST410, ST617, and ST4, respectively. Based on MLST data, both ST4 and ST617 were grouped within clonal complex 10 (CC10) (Table 1).
3.3. Virulence-Associated Genes (VAGs)
Whole-genome sequences were surveyed using the VirulenceFinder database (https://cge.cbs.dtu.dk/services/VirulenceFinder/, accessed on 21 November 2021) to identify the virulence potential of E. coli. Therefore, 14 different VAGs were detected, and the highest frequencies were for terC (tellurium ion resistance protein), followed by astA (EAST-1 heat-stable toxin), capU (hexosyltransferase homolog), gad (glutamate decarboxylase), and iss genes (increased serum survival protein). Each E. coli strain displayed a different combination of virulence genes (Table 2).
Table 2.
Phenotypic and genomic profiles of antibiotic-resistant E. coli isolates from surface water of agricultural drainage in northwestern Mexico.
| Isolate | Antimicrobial Resistance Profile a |
Acquired Antimicrobial Resistance Genes b | QRDR Mutations b |
Plasmid Replicons c |
Virulence Genes d |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-lactams | Aminoglycosides | Lincosamides | Multidrug Efflux System |
Phenicols | Sulfonamides | Tetracycline | Trimethoprim | |||||
| ADD147 | AM, CB, CPF, CL, GE, NET, NOF, STX | blaTEM-1B |
aadA1, aadA2, aph(3′)-Ia, aac(3)-IId |
lnu(F) | mdf(A) | floR, cmlA1 | sul2, sul3 | tet(A) | dfrA12 | gyrA-S83L+D87N; parC-S80I; parE-S458A | Col(pHAD28), IncFIB(AP001918), IncFIC(FII), IncX1 | gad, hra, lpfA, papC, terC |
| ADD167 | AM, CB, CF, CFX, CPF, GE, NF, NOF, STX | blaCTX-M-15 | aac(3)-IIa, aadA5, aph(3″)-Ib, aph(6)-Id | - | mdf(A) | - | sul1, sul2 | tet(B) | dfrA17 | gyrA-S83L+D87N; parC-S80I; parE-S458A | Col(MG828), Col(pHAD28) | astA, capU, gad, iss |
| ADD183 | CPF | - | - | - | mdf(A) | - | - | - | - | gyrA-S83L | ColRNAI, IncFII(pCoo) | astA, capU, eatA, gad, iss, kpsE, kpsM_K15, ltcA, ompT, terC, traT |
a Antimicrobial susceptibility testing by disk diffusion; AM, ampicillin; CB, carbenicillin; CF, cephalothin; CFX, cefotaxime; CPF, ciprofloxacin; CL, chloramphenicol; GE, gentamicin; NET, netilmicin; NF, nitrofurantoin; NOF, norfloxacin; STX, trimethoprim-sulfamethoxazole. A not susceptible interpretation result is highlighted in bold. b Information extracted from WGS data (ResFinder v4.1, https://cge.cbs.dtu.dk/services/ResFinder/, accessed on 21 November 2021). c Information extracted from WGS data (PlasmidFinder v2.1, https://cge.cbs.dtu.dk/services/PlasmidFinder/, accessed on 21 November 2021). d Information extracted from WGS data (VirulenceFinder v2.0, https://cge.cbs.dtu.dk/services/VirulenceFinder/, accessed on 21 November 2021).
Analysis of the distribution of VAGs indicated that strains under CC10 (ADD167 and ADD183) partially shared a virulence profile. However, E. coli ADD183 also displayed genes involved in capsular polysaccharide biosynthesis (kpsE, kpsM_K15), increased serum survival (traT), toxin production (ltcA, heat-labile enterotoxin A subunit), and protease production (eatA), namely, the serine protease autotransporter of Enterobacteriaceae (SPATE) EatA. A key feature of the enterotoxigenic E. coli (ETEC) pathotype, a member of the InPEC group, is the ability to express heat-labile (LT) and/or heat-stable (ST) enterotoxins [8]. Although further exploration is required, E. coli ADD183 was assigned to the ETEC pathotype according to the virulence profile predicted in this study.
3.4. Resistance Profile and Antimicrobial Resistance Genes (ARGs)
The phenotypic antimicrobial susceptibility was determined by disk diffusion according to CLSI standards. In summary, the phenotypic resistance profiles of E. coli ADD147 and ADD167 were comparable and cataloged both as MDR bacteria owing to their resistance to antibiotics from the β-lactam, fluoroquinolone, and folate pathway antagonists antimicrobial classes (Table 2) [25]. Both MDR E. coli strains showed resistance to ampicillin, carbenicillin, ciprofloxacin, norfloxacin, and trimethoprim/sulfamethoxazole. However, E. coli ADD147 further exhibited resistance to chloramphenicol, gentamicin, and netilmicin, whereas E. coli ADD167 was resistant to first- and third-generation cephalosporins (i.e., cephalothin and cefotaxime), suggesting a presumptive ESBL phenotype. In addition, E. coli ADD167 was not susceptible to gentamicin and nitrofurantoin. In contrast, ADD183 was sensitive to all antibiotics except ciprofloxacin, to which it remained not susceptible (Table 2).
Genome-based prediction of antimicrobial resistance determinants confirmed the previously observed phenotypic resistance profiles. We identified 21 different acquired genetic elements associated with resistance to β-lactams, aminoglycosides, lincosamides, phenicols, tetracyclines, and trimethoprim/sulfamethoxazole (Table 2). The multidrug transporter gene mdf(A) was found in all studied genomes. Two β-lactamase-encoding genes were identified: TEM-1-type narrow-spectrum β-lactamase (blaTEM-1B) and CTX-M-type ESBL (blaCTX-M-15). In total, eight allelic variants encoding aminoglycoside-modifying enzymes were detected (aadA1, aadA2, aadA5, aac(3)-IIa, aac(3)-IId, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id). Sulfonamide (sul1, sul2, and sul3) and trimethoprim (dfrA12 and dfrA17) resistance genes were present in two E. coli strains. Notably, the lincomycin resistance gene lnu(F) and the genes responsible for phenicol resistance (i.e., floR and cmlA1) were further observed in E. coli ADD147. In addition, two E. coli strains carried the genes tet(A) and tet(B) involved in tetracycline resistance. Nonetheless, neither lincosamide nor tetracycline antimicrobial classes were tested phenotypically. No carbapenem- and colistin-resistance-associated determinants were detected in the E. coli under study.
Further sequence analysis indicated that both MDR E. coli ADD147 and ADD167 possess a multidrug resistance region, which each houses a class 1 integron with ARGs associated. In strain ADD147, we identified a sul3-associated integron with a genetic structure dfrA12–orfF–aadA2–cmlA1–aadA1–qacH–tnp440–sul3, conferring resistance to trimethoprim, aminoglycoside, chloramphenicol, and sulfonamide, respectively. The chloramphenicol/florfenicol resistance gene floR was located upstream of the identified sul3-associated class 1 integron. In contrast, the class 1 integron found in E. coli ADD167 was flanked by regions co-harboring the ISEcp1-blaCTX-M-15-orf477Δ transposition unit and aac(3)-IIa genes (upstream), which confer resistance to β-lactams, including broad-spectrum cephalosporins and monobactams, and aminoglycosides, respectively; and the sul2–aph(3″)-Ib–aph(6)-Id gene array (downstream), involved in sulfonamide and aminoglycoside resistance. The genetic structure identified in this class 1 integron was the dfrA17–aadA5–qacEΔ1–sul1 gene cassette.
With regard to (fluoro)quinolone resistance, no plasmid-mediated quinolone resistance (PMQR) genes, such as qnr, qepA, or oqxAB, were observed. However, nucleotide sequence analysis of the quinolone resistance-determining region (QRDR) revealed point mutations leading to non-synonymous substitutions in the DNA gyrase and topoisomerase IV targets. In particular, double amino acid exchange was found in GyrA (S83L, D87N), whereas the S80I and S458A substitutions were recognized in ParC and ParE, respectively (Table 2). Additional chromosomal point mutations that confer antibiotic resistance have not been identified.
3.5. Detection of Plasmid Replicons and Prophages
WGS-based analysis using PlasmidFinder identified seven different replicons from both large and small plasmid groups, specifically, those relative to incompatibility F and X groups, and Col-like plasmids, respectively (Table 2). E. coli ADD147 concomitantly hosted two IncF-type replicons, IncFIB (AP001918) and IncFIC (FII), and the IncX-type (IncX1) and Col(pHAD28) plasmid groups, whereas E. coli ADD167 only harbored Col-like plasmids. E. coli ADD183 carried IncFII (pCoo) and ColRNAI plasmid replicons. We could not associate VAGs or ARGs with putative plasmids of E. coli ADD147 and ADD167. The putative IncFII plasmid (~125.3 kbp) from E. coli ADD183 contained several virulence-associated genes, including astA, capU, eatA, ltcA, and traT.
PHASTER analysis of the E. coli genomes identified 33 phage-related sequences. However, only seven prophage regions were predicted to be intact and considered for further analysis (Table 3); these showed homology to six different phages. Their length varied from 11 to 53 kb, of which the most frequent was the Enterobacteria phage P88.
Table 3.
Distribution of intact prophage regions of antibiotic-resistant E. coli isolates from surface water of agricultural drainage in northwestern Mexico.
| Isolate | Region | Length (kbp) | GC% | CDS | Phage (Hit Genes Counts) a |
|---|---|---|---|---|---|
| ADD147 | 1 | 11.5 | 48.4 | 18 | Enterobacteria phage VT2phi_272 (2) |
| ADD167 | 1 | 40.4 | 50.6 | 50 | Enterobacteria phage BP-4795 (9) |
| 2 | 15.1 | 54.5 | 21 | Escherichia phage pro483 (18) | |
| ADD183 | 1 | 40.2 | 49.9 | 44 | Enterobacteria phage P88 (21) |
| 2 | 53.7 | 51.6 | 45 | Enterobacteria phage P88 (27) | |
| 3 | 25.6 | 49.8 | 22 | Enterobacteria phage P4 (9) | |
| 4 | 10.3 | 54.2 | 15 | Escherichia phage SH2026Stx1 (3) |
a Phage with the highest number of CDS most similar to those in the region.
Predicted prophage regions were manually inspected to identify putative active phage signatures, that is, attachment site (att) sequences (attL and attR) and genes encoding structural proteins, DNA packaging and regulation, insertion into the host, and lysis. Prophages similar to VT2phi_272, pro483, and SH2026Stx1 lacked attachment sites, suggesting defective phages. Conversely, the prophages BP-4795, P88, and P4 possessed att sequences; however, the P4-like prophage cargo only contained integrase and a few structural-related proteins, lacking recombinase, terminase, transposase, and lysis module. Except for the iss gene encoded in the BP-4795 prophage, none of the predicted prophages carried further genetic elements implicated in virulence or antimicrobial resistance properties.
3.6. Phylogenetic Analysis
The EnteroBase search resulted in a total of 1522, 508, and 209 isolates available for phylogenetic analysis of E. coli ST410, ST617, and ST4 lineages, respectively, including ADD147, ADD167, and ADD183. All of them are derived from diverse isolation sources.
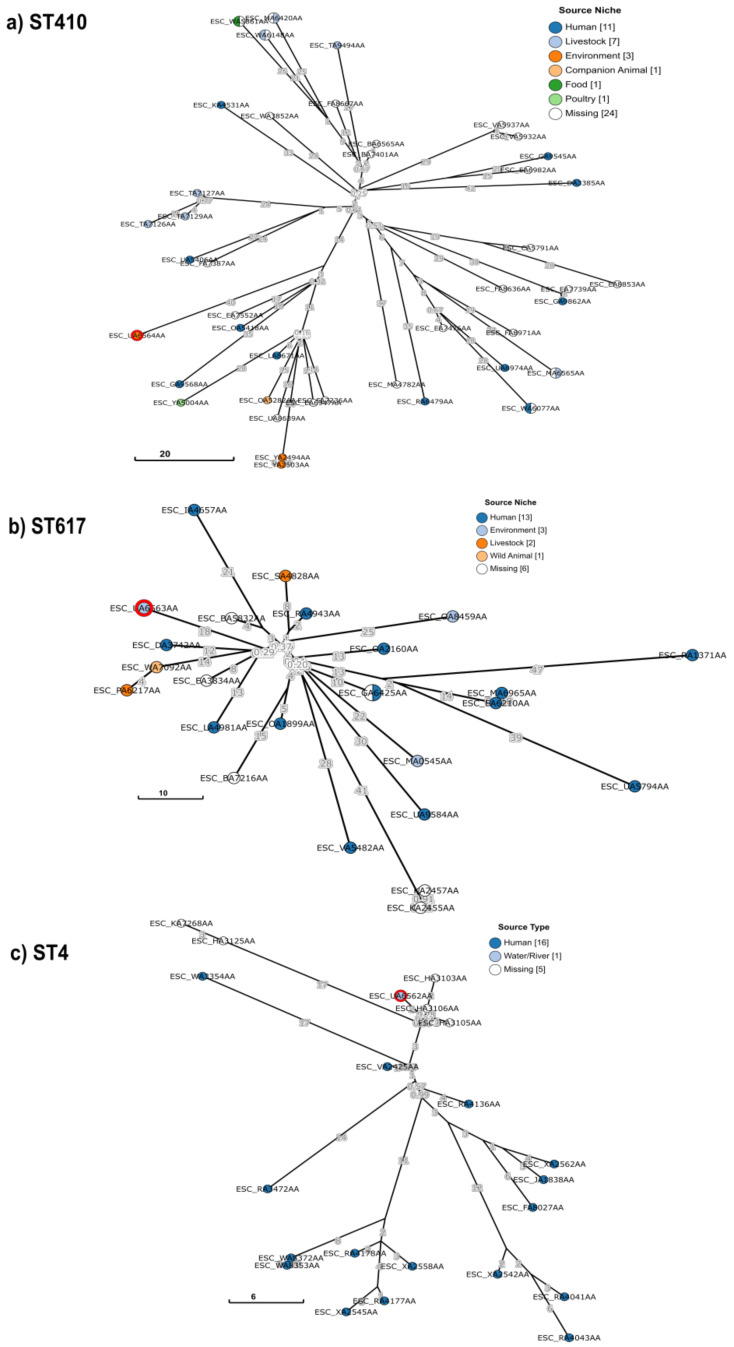
Overall, the global collection of ST410, ST617, and ST4 lineages was predominantly depicted by human-derived isolates, and to a lesser extent, by environment-sourced isolates. Of note, a significant number of isolates in each collection are from unknown origin (Supplementary Figures S1–S3). A similar trend was observed during the examination of refined subtrees. All three sequenced E. coli were clustered with closely related human-derived E. coli isolates (Figure 1a–c).
Figure 1.
GrapeTree phylogenetic subtrees of the ST410, ST617, and ST4 lineages based on core-genome multilocus sequence typing (cgMLST) distances. The phylogeny was reconstructed using the rapid neighbor-joining (RapidNJ) algorithm and the core-genome MLST (cgMLST) V1 + Hierarchical Clustering (HierCC) V1 scheme from EnteroBase. E. coli isolates from this study are highlighted with a red circle. Isolate ID is indicated as the EnteroBase Barcode. The nodes are color-coded according to the isolation source niche as indicated in the key and the numbers in brackets represent the number of isolates. Scale bar indicates the number of cgMLST allelic differences.
E. coli ADD147/ST410 was placed within a sub-cluster comprising companion animal, environmental, human, and poultry-sourced isolates showed 60–84 cgMLST allelic differences to E. coli ADD147 (Figure 1a). Notably, the twelve ST410 isolates belonging to this sub-clade were predicted like ONT:H9 serotype. E. coli ADD147 was more closely related to the unknown host source isolate ESC_EA7552AA from Vietnam (60 cgMLST allelic differences) and two human blood-sourced isolates ESC_OA5418AA and ESC_GA9568AA from Canada and Singapore (61–78 cgMLST allelic differences), respectively.
Similarly, E. coli ADD167/ST617 was located in a discrete cluster consisting of six isolates sourced from animal, environmental, and human origin showed 28–38 cgMLST allelic differences to E. coli ADD167 (Figure 1b). According to cgMLST analysis, the most closely related to E. coli ADD167 are the isolate ESC_EA3834AA, recovered from the hospital environment in Germany (26 cgMLST allelic differences), and the human blood-sourced isolate ESC_LA4981AA from the USA (31 cgMLST allelic differences).
Notably, according to EnteroBase and GrapeTree phylogenetic analysis of the global collection of ST4 lineage, E. coli ADD183 is the only water-sourced isolate reported. E. coli ADD183/ST4 was clustered with human-associated isolates showed 2–31 cgMLST allelic differences to E. coli ADD183 (Figure 1c). Furthermore, it was placed into a sub-clade containing five unknown host source E. coli isolates from the USA. The most closely related isolates to E. coli ADD183 were ESC_HA3106AA, ESC_HA3105AA, and ESC_HA3103AA differing only by two, three, and four cgMLST alleles, respectively.
4. Discussion
Aquatic environments are one of the main reservoirs and transmission sources for the dissemination of antibiotic resistance. This study constitutes the first approach for the genomic analysis of three antibiotic-resistant E. coli strains from surface water of agricultural drainage receiving raw irrigation water and domestic effluents in northwest Mexico (Sinaloa State). The objective of this genome interrogation was to determine the genetic diversity in terms of virulence features, antimicrobial resistance gene carriage, plasmids, and prophage content and to portray the potential risk that environmental E. coli could pose to animal or human health. Although the studied E. coli strains were isolated from the same area and period, they showed dissimilar genomic profiles. The resulting differences observed in the phenotypic and genomic profile could be partially explained by (1) the different evolutionary scenarios to which each E. coli strain was subjected [44]; (2) the accessory genome dynamics [45]; and (3) the environmental selective pressure imposed by the constant inflow of contaminated agricultural and domestic sewage on the studied agricultural drainage [22]. However, to address this knowledge gap, large-scale genomic epidemiological studies in the One Health framework are needed.
The main concern was the identification of international high-risk clones of E. coli carrying multiple acquired antibiotic resistance determinants in the agricultural drainage ditch in the Sinaloa Valley. In this area, intensive agriculture is practiced, and the drained water is subsequently reused for agricultural irrigation, livestock farming, and aquaculture purposes [20]. The “high-risk clone” designation has been used to describe bacterial lineages that enhance the dissemination of antibiotic resistance [46], and a prominent example of this is the ExPEC ST131, a successful globally disseminated clone associated with multiple antimicrobial resistance and enhanced pathogenicity and fitness features [47,48].
In this regard, the MDR TEM-1B-positive E. coli strain ADD147 belongs to the widespread ST410 group, a bacterial clone reported worldwide with a high cross-sectorial transmission between animals, humans, and the environment, and bearing diverse ARGs, mostly CTX-M-15, CMY-2, and NDM-5 ESBLs [49,50,51]. Although this isolate was devoid of ESBL genes, it harbored a sul3-associated class 1 integron with multiple resistance determinants with the dfrA12–orfF–aadA2–cmlA1–aadA1–qacH gene cassette array, a plasmid-borne integron structure first described in non-typhoidal Salmonella [52], and E. coli isolates from humans and animals [53,54,55,56]. However, we were not able to link the detected sul3-associated integron to a specific plasmid sequence.
The MDR CTX-M-15-positive E. coli strain ADD167 belongs to the ST617 lineage, a high-risk clone member of the internationally widespread CC10, which is related to clinical strains found in animal, environmental, food, and human samples worldwide and is predominantly associated with the CTX-M-15 and OXA-1 β-lactamases, and to a lesser extent, with CMY-2 [57,58,59,60,61]. Recent studies have reported the mobile colistin-resistant mcr-1 gene in E. coli ST617 from humans and food-producing animals, which threatens the effectiveness of polymyxins, one of the last-resort drug options for treating infections caused by multidrug- and carbapenem-resistant gram-negative bacteria [62,63]. No acquired resistance determinants for the last-resort treatment options were observed.
Based on the investigation of virulence factors, a low abundance of VAGs was predicted in the high-risk clones ADD147 (ST410) and ADD167 (ST617). Alternatively, strain ADD183 (O6:H16) belongs to the ETEC O6 group, the most common ETEC serogroup involved in multiple outbreaks and sporadic infections internationally, and typically carries both enterotoxins [64,65,66]. In contrast to what was reported in ETEC O6 strains [65,67], E. coli ADD183 harbored additional nonclassical virulence factors found in ETEC (eatA) and ExPEC (iss, kpsE, and kpsM_K15) strains, implying the potential of E. coli ADD183 to cause both intestinal and extraintestinal diseases. Therefore, further studies are needed to confirm this hypothesis.
Available information from previous studies shows that clinically sourced colistin-resistant E. coli belonging to ST410, ST617, and ST4 lineages have been identified worldwide. In particular, ST410 isolates have been reported in Asia (China and Vietnam), Europe (Italy), and America (Brazil), while ST617 isolates have been described in Asia (China, Korea, and Taiwan), and Europe (France and Italy). Alternatively, ST4 isolates have been reported in Arabia Saudi [68]. It is noteworthy that at present, there is no information regarding the prevalence or distribution of E. coli ST410 and ST617 lineages in Mexico, and the unique report of a single human-derived ETEC strain belonging to the ST4 lineage is by Saldaña-Ahuactzi et al. [69].
Although the determination of the origin or direct source was not part of this study, the phylogenetic analysis based on cgMLST elucidated the genetic relatedness and partially gave insight into the clinical relevance of the three E. coli strains studied here. Indeed, according to the former analysis, it is most likely that E. coli ADD147, ADD167, and ADD183 are human-derived strains that could be introduced into the agricultural drainage by the inflow of untreated domestic wastewater [20,22].
This study had several limitations. The results of the current analysis of a small number of E. coli isolates from surface water resources of the selected area cannot be extrapolated to aquatic ecosystems dedicated to agricultural practices at the regional or national level. Nonetheless, despite the limited number of E. coli isolates analyzed, our work highlights the need for extensive epidemiological and genomic studies on E. coli from the aquatic environment to understand the genetic diversity and the ongoing circulation of multidrug-resistant high-risk clones in the region.
5. Conclusions
To date, there is limited information regarding the genomic and molecular data describing the mechanisms underlying antibiotic resistance and virulence properties in E. coli from aquatic environments. To the best of our knowledge, this is the first report of genomic profiling of antibiotic-resistant E. coli strains from the surface water of agricultural drainage in Mexico. Our study highlights the identification of international high-risk clones ST410 and ST617 of E. coli carrying multiple antibiotic resistance determinants, including the widespread CTX-M-15 ESBL, in drained water that is subsequently reused for different practices, such as agricultural irrigation, livestock farming, and aquaculture. Moreover, the presence of enterotoxigenic E. coli strains harboring ExPEC-associated virulence factors was also evidenced. These results also underscore the role of surface water as a potential reservoir for critical pathogenic and multidrug-resistant E. coli clones and the urgent need for monitoring and tracking these bacterial populations to prevent their ongoing dissemination. Finally, the results of the present study will contribute to antibiotic resistance and virulence surveillance by employing WGS-based methodologies under the One Health concept.
Acknowledgments
We acknowledge the Consejo Nacional de Ciencia y Tecnología (CONACyT) of Mexico for the Doctoral degree scholarship granted to José Antonio Magaña-Lizárraga [No. 481143]. The authors would like to thank Karen Enciso-Ibarra and Julissa Enciso-Ibarra for their kind technical assistance in the preparation of DNA libraries and genomic sequencing.
Supplementary Materials
The following supporting information can be downloaded at:https://www.mdpi.com/article/10.3390/microorganisms10030662/s1, Figure S1: Neighbor-joining (RapidNJ) tree based on cgMLST distances of E. coli ST410 global collection; Figure S2: Neighbor-joining (RapidNJ) tree based on cgMLST distances of E. coli ST617 global collection; Figure S3: Neighbor-joining (RapidNJ) tree based on cgMLST distances of E. coli ST4 global collection; Table S1: Metadata associated with Escherichia coli ST410 genomes from EnteroBase used for cgMLST subtree phylogeny; Table S2: Metadata associated with Escherichia coli ST617 genomes from EnteroBase used for cgMLST subtree phylogeny; Table S3: Metadata associated with Escherichia coli ST4 genomes from EnteroBase used for cgMLST subtree phylogeny.
Author Contributions
Conceptualization, J.A.M.-L. and M.E.B.-F.; methodology, J.A.M.-L., B.G.-G. and M.E.B.-F.; software, J.A.M.-L.; validation, J.A.M.-L., B.G.-G. and M.E.B.-F.; formal analysis, J.A.M.-L.; investigation, J.A.M.-L.; resources, B.G.-G., J.G.R.-M., F.D.-V., I.F.V.-L. and M.E.B.-F.; data curation, J.A.M.-L.; writing—original draft preparation, J.A.M.-L.; writing—review and editing, B.G.-G., J.G.R.-M., F.D.-V., I.F.V.-L. and M.E.B.-F.; visualization, J.A.M.-L.; supervision, B.G.-G. and M.E.B.-F.; project administration, J.A.M.-L. and M.E.B.-F.; funding acquisition, M.E.B.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: NCBI BioProject PRJNA715781 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA715781 accessed on 8 February 2022) and SRA accession numbers: SRX10394959, SRX10394960, and SRX10394961.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. HM Government and Wellcome Trust; London, UK: 2016. [Google Scholar]
- 2.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Nisar M.A., Alvi R.F., Aslam M.A., Qamar M.U., Salamat M.K.F., et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berglund B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 2015;5:28564. doi: 10.3402/iee.v5.28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwu C.D., Korsten L., Okoh A.I. The incidence of antibiotic resistance within and beyond the agricultural ecosystem: A concern for public health. Microbiol. Open. 2020;9:e1035. doi: 10.1002/mbo3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manyi-Loh C., Mamphweli S., Meyer E., Okoh A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules. 2018;23:795. doi: 10.3390/molecules23040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance 2014. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 7.Castanheira M., Simner P.J., Bradford P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021;3:dlab092. doi: 10.1093/jacamr/dlab092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaper J.B., Nataro J.P., Mobley H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 9.Croxen M., Finlay B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 10.Leimbach A., Hacker J., Dobrindt U. Between Pathogenicity and Commensalism. Volume 358. Springer; Berlin/Heidelberg, Germany: 2013. E. coli as an All-Rounder: The Thin Line Between Commensalism and Pathogenicity; pp. 3–32. Current Topics in Microbiology and Immunology. [DOI] [PubMed] [Google Scholar]
- 11.Ishii S., Ksoll W.B., Hicks R.E., Sadowsky M.J. Presence and Growth of Naturalized Escherichia coli in Temperate Soils from Lake Superior Watersheds. Appl. Environ. Microbiol. 2006;72:612–621. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tymensen L., Pyrdok F., Coles D., Koning W., McAllister T.A., Jokinen C., Dowd S., Neumann N. Comparative accessory gene fingerprinting of surface water Escherichia coli reveals genetically diverse naturalized population. J. Appl. Microbiol. 2015;119:263–277. doi: 10.1111/jam.12814. [DOI] [PubMed] [Google Scholar]
- 13.Castillo F.Y.R., González F.J.A., Garneau P., Díaz F.M., Barrera A.L.G., Harel J. Presence of multi-drug resistant pathogenic Escherichia coli in the San Pedro River located in the State of Aguascalientes, Mexico. Front. Microbiol. 2013;4:147. doi: 10.3389/fmicb.2013.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosas I., Salinas E., Martínez L., Cruz-Córdova A., González-Pedrajo B., Espinosa N., Amábile-Cuevas C.F. Characterization of Escherichia coli Isolates from an Urban Lake Receiving Water from a Wastewater Treatment Plant in Mexico City: Fecal Pollution and Antibiotic Resistance. Curr. Microbiol. 2015;71:490–495. doi: 10.1007/s00284-015-0877-8. [DOI] [PubMed] [Google Scholar]
- 15.Delgado-Gardea M.C.E., Tamez-Guerra P., Gomez-Flores R., De La Serna F.J.Z.-D., La Vega G.E.-D., Nevárez-Moorillón G.V., Pérez-Recoder M.C., Sánchez-Ramírez B., Gonzalez-Horta C., Infante-Ramírez R. Multidrug-Resistant Bacteria Isolated from Surface Water in Bassaseachic Falls National Park, Mexico. Int. J. Environ. Res. Public Health. 2016;13:597. doi: 10.3390/ijerph13060597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Orgániz Á., Garza-Ramos U., Sampedro-Rosas M.L., González-González J., Nava-Faustino G., Toribio-Jiménez J. Patotipos y Resistencia a Antibióticos de Escherichia coli en Agua Residual. Rev. Int. Contam. Ambient. 2020;36:957–966. doi: 10.20937/RICA.53711. [DOI] [Google Scholar]
- 17.Amábile-Cuevas C., Arredondo-García J., Cruz A., Rosas I. Fluoroquinolone resistance in clinical and environmental isolates of Escherichia coli in Mexico City. J. Appl. Microbiol. 2010;108:158–162. doi: 10.1111/j.1365-2672.2009.04401.x. [DOI] [PubMed] [Google Scholar]
- 18.Chaidez C., Soto M., Martinez C., Keswick B. Drinking water microbiological survey of the Northwestern State of Sinaloa, Mexico. J. Water Health. 2008;6:125–129. doi: 10.2166/wh.2007.011. [DOI] [PubMed] [Google Scholar]
- 19.Cuevas O.L., Félix J.L., Edeza M.J., Quiroz C.C. Detección y resistencia a antibióticos de Escherichia coli y Salmonella en agua y suelo agrícola. Rev. Fitotec. Mex. 2009;32:119–126. [Google Scholar]
- 20.Ahumada-Santos Y.P., Báez-Flores M.E., Díaz-Camacho S.P., Uribe-Beltrán M.D.J., López-Angulo G., Vega-Aviña R., Chávez-Duran F.A., Montes-Avila J., Carranza-Díaz O., Möder M., et al. Spatiotemporal distribution of the bacterial contamination of agricultural and domestic wastewater discharged to a drainage ditch (Sinaloa, Mexico) Cienc. Mar. 2014;40:277–289. doi: 10.7773/cm.v40i4.2456. [DOI] [Google Scholar]
- 21.Canizalez-Roman A., Velazquez-Roman J., Valdez-Flores M.A., Flores-Villaseñor H., Vidal J.E., Muro-Amador S., Guadrón-Llanos A.M., Gonzalez-Nuñez E., Medina-Serrano J., Tapia-Pastrana G., et al. Detection of antimicrobial-resistance diarrheagenic Escherichia coli strains in surface water used to irrigate food products in the northwest of Mexico. Int. J. Food Microbiol. 2019;304:1–10. doi: 10.1016/j.ijfoodmicro.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Moeder M., Carranza-Diaz O., López-Angulo G., Vega-Aviña R., Chávez-Durán F.A., Jomaa S., Winkler U., Schrader S., Reemtsma T., Delgado-Vargas F. Potential of vegetated ditches to manage organic pollutants derived from agricultural runoff and domestic sewage: A case study in Sinaloa (Mexico) Sci. Total Environ. 2017;598:1106–1115. doi: 10.1016/j.scitotenv.2017.04.149. [DOI] [PubMed] [Google Scholar]
- 23.Performance Standards for Antimicrobial Disk Susceptibility Tests. 13th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 24.Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2021. [Google Scholar]
- 25.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 26.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Institute; Cambridge, UK: 2010. [Google Scholar]
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 28.Hasman H., Saputra D., Sicheritz-Ponten T., Lund O., Svendsen C.A., Frimodt-Møller N., Aarestrup F.M. Rapid Whole-Genome Sequencing for Detection and Characterization of Microorganisms Directly from Clinical Samples. J. Clin. Microbiol. 2014;52:139–146. doi: 10.1128/JCM.02452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosi E., Donati B., Galardini M., Brunetti S., Sagot M.-F., Lió P., Crescenzi P., Fani R., Fondi M. MeDuSa: A multi-draft based scaffolder. Bioinformatics. 2015;31:2443–2451. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- 32.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 33.Joensen K.G., Tetzschner A.M., Iguchi A., Aarestrup F.M., Scheutz F. Rapid and Easy in Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C.J., Ochman H., et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Ponten T., Ussery D.W., Aarestrup F.M., et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joensen K.G., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M., Aarestrup F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clermont O., Dixit O., Vangchhia B., Condamine B., Dion S., Bridier-Nahmias A., Denamur E., Gordon D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019;21:3107–3117. doi: 10.1111/1462-2920.14713. [DOI] [PubMed] [Google Scholar]
- 39.Beghain J., Bridier-Nahmias A., Le Nagard H., Denamur E., Clermont O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018;4:e000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carattoli A., Zankari E., Garcìa-Fernández A., Larsen M.V., Lund O., Villa L., Aarestrup F.M., Hasman H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Z., Alikhan N.-F., Mohamed K., Fan Y., the Agama Study Group. Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z., Alikhan N.-F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carriço J.A., Achtman M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mageiros L., Méric G., Bayliss S.C., Pensar J., Pascoe B., Mourkas E., Calland J.K., Yahara K., Murray S., Wilkinson T.S., et al. Genome evolution and the emergence of pathogenicity in avian Escherichia coli. Nat. Commun. 2021;12:765. doi: 10.1038/s41467-021-20988-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengtsson R.J., Dallman T.J., Allen H., De Silva P.M., Stenhouse G., Pulford C.V., Bennett R.J., Jenkins C., Baker K.S. Accessory Genome Dynamics and Structural Variation of Shigella from Persistent Infections. mBio. 2021;12:e00254-21. doi: 10.1128/mBio.00254-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodford N., Turton J., Livermore D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 47.Petty N.K., Ben Zakour N.L., Stanton-Cook M., Skippington E., Totsika M., Forde B.M., Phan M.-D., Moriel D.G., Peters K.M., Davies M., et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA. 2014;111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoesser N., Sheppard A., Pankhurst L., De Maio N., Moore C., Sebra R., Turner P., Anson L.W., Kasarskis A., Batty L., et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. mBio. 2016;7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaufler K., Semmler T., Wieler L.H., Wöhrmann M., Baddam R., Ahmed N., Müller K., Kola A., Fruth A., Ewers C., et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410—Another successful pandemic clone? FEMS Microbiol. Ecol. 2016;92:fiv155. doi: 10.1093/femsec/fiv155. [DOI] [PubMed] [Google Scholar]
- 50.Falgenhauer L., Imirzalioglu C., Ghosh H., Gwozdzinski K., Schmiedel J., Gentil K., Bauerfeind R., Kämpfer P., Seifert H., Michael G.B., et al. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int. J. Antimicrob. Agents. 2016;47:457–465. doi: 10.1016/j.ijantimicag.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 51.Roer L., Overballe-Petersen S., Hansen F., Schønning K., Wang M., Røder B.L., Hansen D.S., Justesen U.S., Andersen L.P., Fulgsang-Damgaard D., et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere. 2018;3:e00337-18. doi: 10.1128/mSphere.00337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antunes P., Machado J., Peixe L. Dissemination of sul3-Containing Elements Linked to Class 1 Integrons with an Unusual 3′ Conserved Sequence Region among Salmonella Isolates. Antimicrob. Agents Chemother. 2007;51:1545–1548. doi: 10.1128/AAC.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sáenz Y., Vinué L., Ruiz E., Somalo S., Martínez S., Rojo-Bezares B., Zarazaga M., Torres C. Class 1 integrons lacking qacE∆1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet. Microbiol. 2010;144:493–497. doi: 10.1016/j.vetmic.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 54.Moran R.A., Holt K.E., Hall R.M. pCERC3 from a commensal ST95 Escherichia coli: A ColV virulence-multiresistance plasmid carrying a sul3-associated class 1 integron. Plasmid. 2016;84–85:11–19. doi: 10.1016/j.plasmid.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Le-Vo H.-N., Tran P.T.-B., Le L., Matsumoto Y., Motooka D., Nakamura S., Jones J.W., Iida T., Cao V. Complex Class 1 Integron in a Clinical Escherichia coli Strain from Vietnam Carrying Both mcr-1 and blaNDM–1. Front. Microbiol. 2019;10:2472. doi: 10.3389/fmicb.2019.02472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zingali T., Reid C.J., Chapman T.A., Gaio D., Liu M., Darling A.E., Djordjevic S.P. Whole Genome Sequencing Analysis of Porcine Faecal Commensal Escherichia coli Carrying Class 1 Integrons from Sows and Their Offspring. Microorganisms. 2020;8:843. doi: 10.3390/microorganisms8060843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Báez J., Hernández-García M., Guamparito C., Díaz S., Olave A., Guerrero K., Cantón R., Baquero F., Gahona J., Valenzuela N., et al. Molecular Characterization and Genetic Diversity of ESBL-Producing Escherichia coli Colonizing the Migratory Franklin’s Gulls (Leucophaeus pipixcan) in Antofagasta, North of Chile. Microb. Drug Resist. 2015;21:111–116. doi: 10.1089/mdr.2014.0158. [DOI] [PubMed] [Google Scholar]
- 58.Rocha-Gracia R.C., Cortés-Cortés G., Lozano-Zarain P., Bello F., Martínez-Laguna Y., Torres C. Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 2015;203:315–319. doi: 10.1016/j.tvjl.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 59.Zong Z., Fenn S., Connor C., Feng Y., McNally A. Complete genomic characterization of two Escherichia coli lineages responsible for a cluster of carbapenem-resistant infections in a Chinese hospital. J. Antimicrob. Chemother. 2018;73:2340–2346. doi: 10.1093/jac/dky210. [DOI] [PubMed] [Google Scholar]
- 60.Nascimento T., Cantamessa R., Melo L., Fernandes M.R., Fraga E., Dropa M., Sato M.I., Cerdeira L., Lincopan N. International high-risk clones of Klebsiella pneumoniae KPC-2/CC258 and Escherichia coli CTX-M-15/CC10 in urban lake waters. Sci. Total Environ. 2017;598:910–915. doi: 10.1016/j.scitotenv.2017.03.207. [DOI] [PubMed] [Google Scholar]
- 61.Monte D.F., Sellera F.P., Fernandes M.R., Moura Q., Landgraf M., Lincopan N. Genome Sequencing of an Escherichia coli Sequence Type 617 Strain Isolated from Beach Ghost Shrimp (Callichirus major) from a Heavily Polluted Ecosystem Reveals a Wider Resistome against Heavy Metals and Antibiotics. Microbiol. Resour. Announc. 2019;8:e01471-18. doi: 10.1128/MRA.01471-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snesrud E., Ong A.C., Corey B., Kwak Y.I., Clifford R., Gleeson T., Wood S., Whitman T.J., Lesho E.P., Hinkle M., et al. Analysis of Serial Isolates of mcr-1-Positive Escherichia coli Reveals a Highly Active IS Apl1 Transposon. Antimicrob. Agents Chemother. 2017;61:e00056-17. doi: 10.1128/AAC.00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zając M., Sztromwasser P., Bortolaia V., Leekitcharoenphon P., Cavaco L.M., Ziȩtek-Barszcz A., Hendriksen R.S., Wasyl D. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated from Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019;10:1753. doi: 10.3389/fmicb.2019.01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin J., Yoon K.-B., Jeon D.-Y., Oh S.-S., Oh K.-H., Chung G.T., Kim S.W., Cho S.-H. Consecutive Outbreaks of Enterotoxigenic Escherichia coli O6 in Schools in South Korea Caused by Contamination of Fermented Vegetable Kimchi. Foodborne Pathog. Dis. 2016;13:535–543. doi: 10.1089/fpd.2016.2147. [DOI] [PubMed] [Google Scholar]
- 65.Pattabiraman V., Katz L.S., Chen J.C., McCullough A.E., Trees E. Genome wide characterization of enterotoxigenic Escherichia coli serogroup O6 isolates from multiple outbreaks and sporadic infections from 1975–2016. PLoS ONE. 2018;13:e0208735. doi: 10.1371/journal.pone.0208735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kartsev N.N., Fursova N.K., Pachkunov D.M., Bannov V.A., Eruslanov B.V., Svetoch E.A., Dyatlov I.A. Molecular Characterization of Enterotoxin-Producing Escherichia coli Collected in 2011–2012, Russia. PLoS ONE. 2015;10:e0123357. doi: 10.1371/journal.pone.0123357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwon T., Chung S.-Y., Jung Y.-H., Jung S.-J., Roh S.-G., Park J.-S., Kim C.-H., Kim W., Bak Y.-S., Cho S.-H. Comparative genomic analysis and characteristics of NCCP15740, the major type of enterotoxigenic Escherichia coli in Korea. Gut Pathog. 2017;9:55. doi: 10.1186/s13099-017-0204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dadashi M., Sameni F., Bostanshirin N., Yaslianifard S., Khosravi-Dehaghi N., Nasiri M.J., Goudarzi M., Hashemi A., Hajikhani B. Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: A systematic review. J. Glob. Antimicrob. Resist. 2021 doi: 10.1016/j.jgar.2021.10.022. in press. [DOI] [PubMed] [Google Scholar]
- 69.Saldaña-Ahuactzi Z., Cruz-Córdova A., Rodea G.E., Porta H., Navarro-Ocaña A., Eslava-Campos C., Cevallos M.A., Xicohtencatl-Cortes J. Genome Sequence of Enterotoxigenic Escherichia coli Strain FMU073332. Genome Announc. 2017;5:e01600-16. doi: 10.1128/genomeA.01600-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: NCBI BioProject PRJNA715781 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA715781 accessed on 8 February 2022) and SRA accession numbers: SRX10394959, SRX10394960, and SRX10394961.