Abstract
Rickettsiae are gram-negative, obligately intracellular bacteria responsible for arthropod-borne spotted fevers and typhus. Experimental studies have delineated a cluster of naturally rifampin-resistant spotted fever group species. We sequenced the 4,122- to 4,125-bp RNA polymerase β-subunit-encoding gene (rpoB) from typhus and spotted fever group representatives and obtained partial sequences for all naturally rifampin-resistant species. A single point mutation resulting in a phenylalanine-to-leucine change at position 973 of the Rickettsia conorii rpoB sequence and present in all the rifampin-resistant species was absent in all the rifampin-susceptible species. rpoB-based phylogenetic relationships among these rickettsial species yielded topologies which were in accordance with previously published phylogenies.
Rickettsiae are arthropod-borne, gram-negative, obligately intracellular bacteria (15). The genus Rickettsia has been divided into two groups, namely, the typhus group and the spotted fever group, on the basis of clinical presentation, immunological reactivity, intracellular location, and DNA G+C content (25). Phylogenetic appraisal based on a comparison of 16S rRNA genes (17, 26) has demonstrated that rickettsiae belong to the α-1 subgroup of the class Proteobacteria. Typhus group rickettsiae include Rickettsia prowazekii, the agent of epidemic typhus, and Rickettsia typhi, the agent of murine typhus. Phylogenetic approaches based on a comparison of sequences derived from the citrate synthase-encoding (18) and rOmpA-encoding (7) genes have resolved two principal subgroups within the spotted fever group (Table 1), the Rickettsia conorii subgroup and the Rickettsia massiliae subgroup, which also includes Rickettsia montanensis, Rickettsia aeschlimannii, Rickettsia rhipicephali, and the tick isolate Bar 29. Natural resistance to rifampin at MICs of 2 to 4 μg/ml is a phenotypic marker of the R. massiliae subgroup, whereas rickettsiae belonging to the R. conorii subgroup are naturally susceptible to rifampin (Rifs) at MICs of <1 μg/ml (16). Typhus group rickettsiae are naturally susceptible to rifampin (16), but rifampin-resistant (Rifr) strains of R. typhi (23) and R. prowazekii (14) have been selected in vitro after random mutagenesis. Alterations in the RNA polymerase β subunit resulting from mutations in rpoB (9, 10, 22) are the most common mechanisms for rifampin resistance (13). Indeed, amino acid substitutions in the RNA polymerase and rpoB point mutations have been demonstrated after in vitro selection of Rifr R. prowazekii (2, 14) and R. typhi (23). No data are available for naturally Rifr rickettsiae.
TABLE 1.
Rickettsial strains studied
| Species | Strain | Source | Geographical origin | Human disease |
|---|---|---|---|---|
| R. conorii | Moroccan, ATCC VR-141T | Unknown | Morocco | Mediterranean spotted fever |
| R. conorii | Seven (Malish) ATCC VR-613T | Unknown | South Africa | Mediterranean spotted fever |
| R. sibirica | 246, ATCC VR-151T | Dermacentor nuttali | Siberia | Siberian tick typhus |
| R. rickettsii | R(Bitteroot), ATCC VR-891T | Dermacentor andersoni | Montana | Rocky Mountain spotted fever |
| R. japonica | YM | Human | Japan | Oriental spotted fever |
| R. massiliae | Mtu1 | Rhipicephalus turanicus | Camargue, France | Not reported |
| Bar 29 | Bar 29 | Rhipicephalus sanguineus | Spain | Not reported |
| R. rhipicephali | 3-7-6 | Rhipicephalus sanguineus | Mississippi | Not reported |
| R. montanensis | M/5-6 | Microtus sp. | Ohio | Not reported |
| R. aeschlimanii | MC16 | Hyalomma | Morocco | Not reported |
| R. prowazekii | Brein L | Human | Poland | Epidemic typhus |
| R. typhi | Wilmington | Human | North Carolina | Murine typhus |
Therefore, we investigated the genetic basis for natural rifampin resistance in representatives of the typhus group and the two spotted fever subgroups of rickettsiae after amplification and sequencing of the rpoB gene. We derived phylogenetic relationships between the typhus group and spotted fever group rickettsiae and between Rifs and Rifr spotted fever group rickettsiae by sequence analysis of rickettsial rpoB. In addition, we confirmed the rifampin-based clustering of spotted fever group rickettsial species.
MATERIALS AND METHODS
Bacterial strains.
The rickettsial strains used in this study are listed in Table 1. All strains were cocultivated with Vero cells in minimum essential medium supplemented with 4% fetal calf serum and 2 mM glutamine at 32°C for 5 days. Cultures were checked for the absence of contamination by Mycoplasma species with a Mycoplasma Detection Kit (Boehringer Mannheim, Meylan, France). DNA was extracted by a standard phenol-chloroform procedure (19).
PCR amplification of rpoB.
Consensus PCR primers RC 1600D and RC 2030R were designed to hybridize to conserved regions of the gene identified following multiple alignments of the rplL, rpoB, and rpoC sequences available for the class Proteobacteria. Additional oligonucleotides were selected on the basis of data obtained from ongoing base sequence determinations (Table 2). All PCRs incorporated final concentrations of 2.5 × 10−2 U of Taq polymerase per μl, 1× Taq buffer, and 1.8 mM MgCl2 (Gibco BRL, Life Technologies, Cergy Pontoise, France); 120 μM each dATP and dTTP and 280 μM each dGTP and dCTP (Boehringer GmbH, Hilden, Germany); and 0.2 μM each primer (Eurogentec, Seraing, Belgium). PCR mixtures were subjected to the following thermal program: 35 cycles consisting of denaturation at 94°C for 10 s, primer annealing at 52°C for 20 s, and extension at 72°C for 50 s. Every program included predenaturation at 94°C for 90 s and a final elongation step at 72°C for 5 min. Sterile distilled water and noninfected Vero cell DNA were used as negative controls in each set of reactions. The success of amplification was assessed by UV illumination of the resolution of products by electrophoresis through an ethidium bromide-stained 0.8% agarose gel.
TABLE 2.
Oligonucleotide primers used for PCR amplification of rickettsial species
| Primer | Nucleotide sequence (5′-3′) | Positions relative to the open reading frame |
|---|---|---|
| RC 1600D | GCCAATTATCGCAGTTTATGG | 1,600–1,620 |
| RC 2030R | ACGATTTGCATCATCATTTTC | 2,030–2,010 |
| Bap 3850R | GCCCAACTTCCATTTCACC | 3,850–3,832 |
| Rick 1900D | GTAATGGTAGAACCGTATGAGG | 1,900–1,921 |
| Bap 355D | GAACAAGAAGTATATATGGG | 355–374 |
| Rc7 1700R | CGTAATGAGTCGGATGTACG | 1,700–1,681 |
| Bap rpIL 350D | TAGAAGATGCTGGAGC | 350–365 |
| Rc7 450D | ACACCAGGTGAGCGATGC | 450–467 |
| Bap rpoC 300R | AGTTCTATATGACCCAT | 300–284 |
| Rc7 3400D | GATGGAACTGTGGTAGATATCG | 3,400–3,421 |
Sequencing of rpoB.
Amplicons were purified for sequencing by use of a QIAquik Spin PCR Purification Kit (QIAGEN GmbH, Hilden, Germany) in accordance with the protocol of the supplier. Initial sequencing of PCR products was done with the same primers (Eurogentec) as those used for PCR. Subsequently, primers were chosen by comparison to the newly obtained sequences. Each base position was established at least three times in both the forward and the reverse directions. Sequencing reactions were carried out with the reagents in the ABI Prism dRhodamine Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer Applied Biosystems, Foster City, Calif.) in accordance with the manufacturer’s instructions and with the following program: 25 cycles of denaturation at 95°C for 20 s, primer annealing at 50°C for 10 s, and extension at 60°C for 4 min. Products of sequencing reactions were resolved by electrophoresis in a 0.2-mm 6% polyacrylamide denaturing gel and recorded with an ABI Prism 377 DNA Sequencer (Perkin Elmer Applied Biosystems) in accordance with the standard protocol of the supplier. The results obtained were processed into sequence data by use of sequence analysis software (Applied Biosystems), and partial sequences were combined into a single consensus sequence.
Partial rpoB amplification and sequencing in rickettsiae.
Since rpoB sequence analysis disclosed five nonsynonymous nucleotide positions in Rifs R. conorii and in Rifr R. massiliae and isolate Bar 29, we further investigated these codon positions by partial rpoB sequencing of Rifs and Rifr species. Fragments of rpoB were amplified and sequenced with internal primers by the same methods as those described above.
Sequence data analysis.
16S rRNA sequences and rpoB sequences of the nonrickettsial species studied were obtained from the GenBank database, as was the R. prowazekii (GenBank accession no. AF034531) rpoB sequence. Pairwise sequence comparisons for nucleic acid or peptide sequence homology were made with PC Gene software (Intelligenetics, Campbell, Calif.). The rpoB sequences were aligned by use of the multisequence alignment program CLUSTAL (8). Phylogenetic relationships were inferred from this alignment by use of programs within version 3.4 of the PHYLIP software package (6). A distance matrix was generated by use of DNADIST under the assumptions of Jukes and Cantor (11) and Kimura (12). Phylogenetic trees were derived from this matrix by neighbor joining. Maximum-likelihood analysis was done with DNAMLK and a molecular clock, and parsimony analysis was done with DNAPARS.
For alignment of multiple amino acid sequences, the parsimony method was done with PROTPARS; the distance matrix was calculated by use of PROTDIST Kimura or Dayhoff algorithms. This step was followed by the neighbor-joining method. Evaluation of individual node strength was done with the SEQBOOT bootstrapping method and 100 runs.
Levels of similarity between rpoB sequences and RNA polymerase β-subunit amino acid sequences were determined by use of the homology search function of DNASIS (Hitachi Software Engineering America, Ltd., Brisbane, Calif.).
Nucleotide sequence accession numbers.
The rpoB sequence data have been submitted to the DDBJ/EMBL/GenBank databases under the following accession numbers: R. conorii Moroccan. AF076435; R. conorii Seven, AF076434; R. massiliae, AF076433; isolate Bar 29, AF076436; R. prowazekii Brein L, AF076437; and R. typhi Wilmington AF083622.
RESULTS AND DISCUSSION
rpoB variability and genetic support of natural rifampin resistance in spotted fever group rickettsiae.
Complete rpoB sequences were determined for R. conorii Seven, R. conorii Moroccan, R. massiliae, isolate Bar 29, R. prowazekii, and R. typhi. Rickettsial rpoB consisted of a 4,122-bp open reading frame in the spotted fever group and a 4,125-bp open reading frame in the typhus group, with a G+C content of 35%, a low value consistent with that previously reported (24). A comparison of rpoB sequences showed five nonsynonymous mutations in Rifr isolate Bar 29 and R. massiliae versus Rifs R. conorii (Table 3). When these mutations were investigated with a larger representation of the Rifs subgroup and the more distant rickettsial species Rickettsia bellii, Phe973→Leu973 was found to be the only one common to the five members of the Rifs subgroup. This single point mutation, which appeared to be specific for the naturally rifampin-resistant subgroup, was not previously implicated in rifampin resistance in other bacteria.
TABLE 3.
Nonsilent rpoB sequence base positions divergent between rifampin-susceptible and rifampin-resistant spotted fever group Rickettsia species
| Strain | Amino acid/nucleotide at positiona:
|
Rifampin susceptibility | ||||
|---|---|---|---|---|---|---|
| 221 | 524 | 640 | 973 | 1031 | ||
| R. conorii | Thr/ACT | Ser/AGT | Ile/ATT | Phe/TTC | Ile/ATC | S |
| R. rickettsii | ND | Ser/AGT | Ile/ATT | Phe/TTC | Thr/ACC | S |
| R. sibirica | ND | Ser/AGT | Ile/ATT | Phe/TTC | Ile/ATC | S |
| R. japonica | ND | Ser/AGT | Val/GTT | Phe/TTT | Thr/ACC | S |
| R. massiliae | Ile/ATT | Asn/AAT | Val/GTT | Leu/TTA | Thr/ACC | R |
| R. rhipicephali | Ile/ATT | Asn/AAT | Val/GTT | Leu/TTA | Thr/ACC | R |
| Bar 29 | Ile/ATT | Asn/AAT | Val/GTT | Leu/TAA | Thr/ACC | R |
| R. montanensis | Ala/GCT | Ser/AGT | Val/GTT | Leu/TAA | Thr/ACC | R |
| R. aeschlimanii | Thr/ACT | Ser/AGT | Val/GTT | Leu/TAA | Thr/ACC | R |
Position numbering is that of the R. conorii Seven sequence. S, rifampin susceptible; R, rifampin resistant; ND, not determined.
In Escherichia coli, clusters of mutations were precisely determined (10): cluster I, encoding peptide amino acids 507 to 511 and 513 to 533; cluster II, encoding peptide amino acids 563 to 564 and 572; and cluster III, containing the single codon for Arg687. Clusters I and II comprise more than 90% of the mutations in Rifr E. coli (20, 21). A region homologous to clusters I and II has been reported to acquire mutations in Rifr Mycobacterium leprae (5, 9), Mycobacterium tuberculosis (22), Neisseria meningitidis (4), and Staphylococcus aureus (1). A unique point mutation changing Arg546 to Lys has been reported for Rifr R. prowazekii (2, 24), and three amino acid changes at residues 151, 201, and 271 have been described for Rifr R. typhi (23). It is noteworthy that all mutations reported to date were derived from rifampin-resistant isolates selected after a rifampin treatment failure in patients, in vitro selection, or directed mutagenesis. This study reports for the first time mutations associated with natural rifampin resistance among closely related bacterial species.
The clinical significance of these data remains hypothetical, since none of the five rifampin-resistant species has been isolated from patients. A randomized trial of 5-day rifampin versus 1-day doxycycline for treating Mediterranean spotted fever in Barcelona, Spain, revealed delayed apyrexia in the rifampin group, and the trial was stopped (3). No strain was isolated from patients during this trial; interestingly, however, the rpoB mutant isolate Bar 29 has been recovered from Rhipicephalus sanguineus ticks in the Barcelona area. The hypothesis that isolate Bar 29 was responsible for rifampin-resistant Mediterranean spotted fever in the Barcelona area remains to be confirmed by the isolation of this rickettsial species from any patient.
rpoB-based phylogenies of rickettsiae.
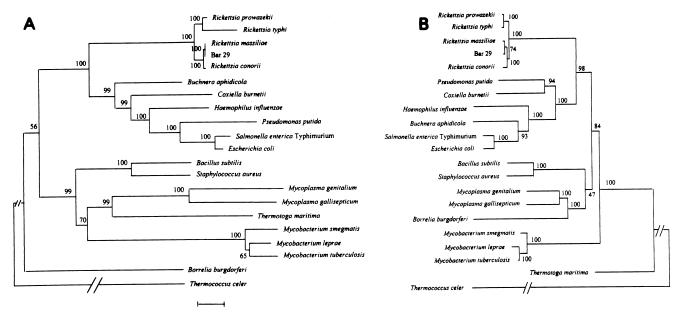
The sequences of rpoB of R. conorii Moroccan and Seven determined in this study were identical, as were those of R. prowazekii Brein L and Madrid E (14). Among the spotted fever group rickettsiae, the rpoB coding sequences were 97% similar, whereas among the typhus group rickettsiae, they were about 90% similar. A sequence similarity of about 92.8% was found between the spotted fever and typhus group rickettsiae, but only about 54.7 to 65.5% similarity was found between spotted fever group rpoB and rpoB from nonrickettsial bacteria. Dendrograms inferred from the entire rpoB alignment by neighbor-joining (Fig. 1) and parsimony methods yielded similar topologies with clusters of rickettsiae. The two typhus group species clustered together, as did the three spotted fever strains with bootstrap values of >80%. rpoB-based phylogenies supported the divergence of Rifr strains from other members of the spotted fever group, as previously indicated by partial gltA sequencing (16, 18) and sequence comparisons of rickettsial ompA (encoding an antigenic high-molecular-weight membrane protein) from all spotted fever group species (7).
FIG. 1.
Distance matrix trees derived from rpoB data (A) and from data for the 16S rRNA-encoding gene (B). The evolutionary distances were determined by the method of Kimura (12). These values were used to construct a dendrogram by the neighbor-joining method. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. The scale bar shows 5% differences in nucleotide sequences. The GenBank accession numbers for the 16S rRNA gene sequences included are as follows: Escherichia coli, M24996; Salmonella enterica serovar Typhimurium, X80681; Buchnera aphidicola, Z19056; Haemophilus influenzae, M35019; Pseudomonas putida, X93997; Coxiella burnetii, M21291; Rickettsia massiliae, L36214; isolate Bar 29, L36102; Rickettsia conorii Moroccan, L36105; Rickettsia typhi, L36221; Rickettsia prowazekii, M21789; Mycoplasma genitalium, X77334; Thermotoga maritima, M21774; Borrelia burgdorferi, X85204; Staphylococcus aureus, X68417; Bacillus subtilis, AF058766; Mycobacterium smegmatis, M12872; and Mycobacterium tuberculosis, X52917. Accession numbers for the nonrickettsial rpoB sequences included are as follows: Escherichia coli, V00339; Salmonella enterica serovar Typhimurium, X13854; Buchnera aphidicola, Z11913; Haemophilus influenzae, U68759; Pseudomonas putida, X15849; Coxiella burnetii, U86688; Mycoplasma genitalium, U39717; Thermotoga maritima, X61562; Borrelia burgdorferi, X71024; Staphylococcus aureus, X64172; Bacillus subtilis, D83789; Mycobacterium smegmatis, U24494; and Mycobacterium tuberculosis, L27989. The GenBank accession numbers for the rickettsial rpoB sequences are given in Materials and Methods.
ACKNOWLEDGMENTS
This work received financial support from BioMérieux S.A., La Balme Les Grottes, France.
We thank Christophe Mollet and Guy Vestris for technical assistance and Richard Birtles for reviewing the manuscript.
REFERENCES
- 1.Aubry-Damon H, Soussy C-J, Courvalin P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:2590–2594. doi: 10.1128/aac.42.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balayeva N M, Frovola O M, Genig V A, Nikolskaya V N. Some biological properties of antibiotic resistant mutants of Rickettsia prowazekii strain E induced by nitroso-guanidine. In: Kazàr J, editor. Rickettsiae and rickettsial diseases. Bratislava, Slovakia: Publishing House of the Slovak Academy of Sciences; 1985. pp. 85–91. [Google Scholar]
- 3.Bella F, Espero E, Uriz S, Serrano J A, Allegre M A, Tort J. Randomized trial of 5-day rifampin versus 1-day doxycycline therapy for MSF. J Infect Dis. 1991;164:433–434. doi: 10.1093/infdis/164.2.433. [DOI] [PubMed] [Google Scholar]
- 4.Carter P E, Abadi F J R, Yakubu D E, Pennington T H. Molecular characterization of rifampin-resistant Neisseria meningitidis. Antimicrob Agents Chemother. 1994;38:1256–1261. doi: 10.1128/aac.38.6.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole S T. Rifamycin resistance in mycobacteria. Res Microbiol. 1996;147:48–52. doi: 10.1016/0923-2508(96)80203-8. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. PHYLIP: Phylogeny Inference Package, version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 7.Fournier P-E, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol. 1998;48:839–849. doi: 10.1099/00207713-48-3-839. [DOI] [PubMed] [Google Scholar]
- 8.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 9.Honoré N, Cole S. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother. 1993;37:414–418. doi: 10.1128/aac.37.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 11.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 12.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 13.Ovchinnikov Y A, Monastyrskaya G S, Gubanov V V, Gurey S O, Chertov O Y, Modyanov T V, Grinkervich U A, Makarova I A, Marchenko T V, Polovnikova I N, Lipkin V M, Sverdlov E D. The primary structure of Escherichia coli RNA polymerase. Nucleotide sequence of the rpoB gene and amino-acid sequence of the β-subunit. Eur J Biochem. 1981;116:621–629. doi: 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- 14.Rachek L I, Tucker A M, Winkler H H, Wood D O. Transformation of Rickettsia prowazekii to rifampin resistance. J Bacteriol. 1998;180:2118–2124. doi: 10.1128/jb.180.8.2118-2124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raoult D, Roux V. Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev. 1997;10:694–719. doi: 10.1128/cmr.10.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolain J M, Maurin M, Vestris G, Raoult D. In vitro susceptibilities of 27 rickettsiae to 13 antimicrobials. Antimicrob Agents Chemother. 1998;42:1537–1541. doi: 10.1128/aac.42.7.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux V, Raoult D. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res Microbiol. 1995;146:385–396. doi: 10.1016/0923-2508(96)80284-1. [DOI] [PubMed] [Google Scholar]
- 18.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Severinov K, Soushko M, Goldfarb A, Nikiforov V. Rifampicin region revisited. New rifampicin-resistant and streptolydigin-resistant mutants in the β-subunit of Escherichia coli RNA polymerase. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 21.Singer M, Jin D J, Walter W A, Gross C A. Genetic evidence for the interaction between cluster I and cluster III rifampicin resistant mutations. J Mol Biol. 1993;231:1–5. doi: 10.1006/jmbi.1993.1251. [DOI] [PubMed] [Google Scholar]
- 22.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 23.Troyer J M, Radulovic S, Anderson S G E, Azad A F. Detection of point mutations in rpoB gene of rifampin-resistant Rickettsia typhi. Antimicrob Agents Chemother. 1998;42:1845–1846. doi: 10.1128/aac.42.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyeryar F J, Jr, Weiss E, Millar D B, Bozeman F M, Ormsbee R A. DNA base composition of rickettsiae. Science. 1973;180:415–417. doi: 10.1126/science.180.4084.415. [DOI] [PubMed] [Google Scholar]
- 25.Weiss E, Moulder J W. Rickettsiales Gieszczkiewicz 1939, 25AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 687–701. [Google Scholar]
- 26.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]