Abstract
Plasmid transfer is one important mechanism how antimicrobial resistance can spread between different species, contributing to the rise of multidrug resistant bacteria (MDRB) worldwide. Here were present whole genome sequencing (WGS) data of two MDRB isolates, an Escherichia coli and a Klebsiella quasipneumoniae, which were isolated from a single patient. Detailed analysis of long-read sequencing data identified an identical F2:A-:B- lncFII plasmid containing blaCTX-M-27 in both isolates, suggesting horizontal plasmid exchange between the two species. As the plasmid of the E. coli strain carried multiple copies of the resistance cassette, the genomic data correlated with the increased antimicrobial resistance (AMR) detected for this isolate. Our case report demonstrates how long-read sequencing data of MDRB can be used to investigate the role of plasmid mediate resistance in the healthcare setting and explain resistance phenotypes.
Keywords: long-read sequencing, blaCTX-M-27, plasmid transfer
1. Introduction
The spread of multidrug resistant bacteria (MDRB) has become a serious threat for healthcare systems worldwide as antimicrobial resistance (AMR) limits treatment options even for life-threatening infections [1]. Horizontal gene transfer enables bacteria to easily share antimicrobial resistance genes (ARG) between various species by exchanging AMR plasmids [2,3,4]. Yet, little is known about the impact horizontal plasmid transfer has on the transmission and epidemiology of AMR in the hospital setting. One limiting factor is the use of short-read sequencing, as the technology has difficulties to distinguish if ARGs are located on the chromosome or on a plasmid. However, since long-read sequencing technology has been introduced into routine microbiology laboratories, this information has become more readily available [5,6] and can help in the detection and analyzation of ARG movement by plasmid conjugation and other mobile genetic elements [6].
In this article, we wanted to show how long-read sequencing data can be used to identify plasmid-mediated multidrug resistance (MDR) in a routine clinical sample. Our data shows evidence of a plasmid transfer between an Escherichia coli and a Klebsiella quasipneumonia isolate. In addition, detailed analysis of the MDR plasmid revealed an “artificial” multicopy of the AMR cassette, leading to a significantly higher minimum inhibitory concentration (MIC) against antibiotics.
2. Materials and Methods
2.1. Bacterial Strain Collection
Anorectal swab samples were collected as part of routine surveillance for multiresistant Gram negative rods. Samples were cultivated on chromID® ESBL Agar (biomérieux, Marcy-l’Etoile, France), CHROMagar™ Acinetobacter (Mast Group, Paris, Frankreich), and Pseudomonas Cetrimide Agar (Thermo Fisher Scientific, Basingstoke, UK) and incubated at 36°C +/−1°C.
2.2. Identification of Bacteria and Antimicrobial Susceptibility Testing
Species identification was performed by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker, Bremen, Germany) with scores above 2.0. Antimicrobial susceptibility testing was performed using VITEK® 2 (biomérieux, Marcy-l’Etoile, France) and Etest® (biomérieux, Marcy-l’Etoile, France). MICs for antibiotics were interpreted according to the 2022 European Committee on Antimicrobial Susceptibility Testing (EUCAST, v.11.0) criteria.
2.3. Whole Genome Sequencing (WGS) and Data Analysis
Genomic DNA (gDNA) of bacterial isolates was extracted using the NEB Monarch Genomic Purification Kit (New England Biolabs, Ipswich, MA, USA). Both isolates were sequenced on a PacBio® Sequel IIe system (Pacific Biosciences, Menlo Park, CA, USA) as described previously [7]. Next, raw sequences were assembled de novo and analyzed using the SMRT® Link software suite v.9 with default parameters. Isolates were genotyped based on core genome multi-locus sequence typing (cgMLST) targets implemented in Ridom SeqSphere+ software version v.8.3.0 (Ridom GmbH, Münster, Germany) [8]. In addition, the multilocus sequence types (ST) were identified using the schemes for the respective species [9,10]. AMR genes and their location were determined using target gene sets for antimicrobial resistance based on the NCBI AMRFinderPlus [11]. To predict the plasmid replicon type the respective contigs were checked for complete circularization and uploaded to PlasmidFinder v.2.1 (24 January 2022: https://cge.cbs.dtu.dk/services/PlasmidFinder/) as well as pMLST 2.0 (24 January 2022: https://cge.cbs.dtu.dk/services/pMLST/) [12]. For annotation, we used the online Pipeline dfast (including HMM scan against TIGRFAM and RPSBLAST against COG database from NCBI) [13]. We performed BLAST analyses using BLASTn [14] and used BRIG Software (BLAST Ring Image Generator) [15] and progressive Mauve [16] for visualization.
3. Results
3.1. Species Identification and Antimicrobial Susceptibility Testing
During routine MDR screening upon intensive care unit (ICU) admission, two MDR isolates, an E. coli and a K. quasipneumoniae, were identified in a single patient. Growth on culture media was detected on chromID® ESBL Agar after 24 h with two different morphotypes. Both isolates showed phenotypically decreased antimicrobial susceptibility to various antibiotics, including cephalosporins, beta-lactam/beta-lactamase inhibitors and fluoroquinolones and were suspected to be extended spectrum beta-lactamase (ESBL) producers (Table 1). Etest® confirmed varying MICs for E. coli (Cefotaxime > 32 mg/L, Ceftazdime 12 mg/L, Ciprofloxacin 3 mg/L) and K. quasipneumoniae (Cefotaxime > 32 mg/L, Ceftazidime 1.5 mg/L, Ciprofloxacin 0.38 mg/L).
Table 1.
Antibiotic susceptibility of MDRB isolates. MIC = minimum inhibitory concentration; EUCAST = European Committee on Antimicrobial Susceptibility Testing; R = resistant; S = susceptible.
| Antibiotics | Classification by EUCAST | MIC (mg/L) | ||
|---|---|---|---|---|
| E.coli (1) | K. quasipneumoniae (2) | 1 | 2 | |
| Ampicillin | R | R | >=32 | >=32 |
| Amoxicillin | R | R | - | - |
| Amoxicillin/Sulbactam | R | R | 16 | 16 |
| Piperacillin/Tazobactam | S | S | <=4 | <=4 |
| Cefuroxime | R | R | >=64 | >=64 |
| Cefotaxime | R | R | >32 | >32 |
| Cefpodoxime | R | R | >=8 | >=8 |
| Ceftazidime | R | I | 12 | 1.5 |
| Ertapenem | S | S | <=0.5 | <=0.5 |
| Imipenem | S | S | <=0.25 | <=0.25 |
| Meropenem | S | S | <=0.25 | <=0 25 |
| Gentamicin | R | R | <=1 | <=1 |
| Ciprofloxacin | R | R | 3 | 0.38 |
| Levofloxacin | R | R | - | - |
| Moxifloxacin | R | R | >=8 | 2 |
| Tigecyclin | S | <=0.5 | - | |
| Trimethoprim/Sulfa | S | R | <=20 | >=320 |
3.2. Detection of ARG by WGS Data Analysis
As all MDR isolates are subjected to WGS as part of the routine hospital surveillance at the University Hospital Münster, both isolates were sequenced using long-read technology. Data analysis identified the E. coli isolate as multilocus sequence type (ST) 1722 and the K. quasipneumoniae isolate as ST138. Further data analysis for ARG identified blaCTX-M-27 in both isolates as well as numerous ARG such as blaLAP-2 and qnrS1. In addition, the K. quasipneumoniae carried the dfrA26 resistance gene matching phenotypic resistance to trimethoprim (Table 1 and Table 2).
Table 2.
ARGs identified by AMRFinderPlus.
| Class | Gene Symbol | Aligned Overlap (%) | Location |
|---|---|---|---|
| K. quasipneumoniae | |||
| BETA-LACTAM | blaOKP-B-2 | 100 | KqP_chromosome |
| FOSFOMYCIN | fosA | 100 | KqP_chromosome |
| PHENICOL/QUINOLONE | oqxB | 100 | KqP_chromosome |
| PHENICOL/QUINOLONE | oqxA | 100 | KqP_chromosome |
| BETA-LACTAM | blaTEM-1 | 100 | KqP_plasmid2 |
| SULFONAMIDE | sul2 | 100 | KqP_plasmid2 |
| TETRACYCLINE | tet(D) | 100 | KqP_plasmid2 |
| TRIMETHOPRIM | dfrA26 | 100 | KqP_plasmid2 |
| BETA-LACTAM | blaLAP-2 | 100 | KqP_plasmid3 |
| BETA-LACTAM | blaCTX-M-27 | 100 | KqP_plasmid3 |
| QUINOLONE | qnrS1 | 100 | KqP_plasmid3 |
| E. coli | |||
| FOSFOMYCIN | uhpT_E350Q | 100 | EC_chromosome |
| FOSMIDOMYCIN | cyaA_S352T | 100 | EC_chromosome |
| QUINOLONE | gyrA_S83L | 99.6 | EC_chromosome |
| AMINOGLYCOSIDE | aph(3’)-Ia | 100 | EC_plasmid1 |
| AMINOGLYCOSIDE | aph(6)-Id | 100 | EC_plasmid1 |
| AMINOGLYCOSIDE | aph(3’’)-Ib | 100 | EC_plasmid1 |
| BETA-LACTAM | blaTEM-1 | 100 | EC_plasmid1 |
| SULFONAMIDE | sul2 | 100 | EC_plasmid1 |
| TETRACYCLINE | tet(B) | 100 | EC_plasmid1 |
| BETA-LACTAM | blaLAP-2 | 100 | EC_plasmid2 |
| BETA-LACTAM | blaLAP-2 | 100 | EC_plasmid2 |
| BETA-LACTAM | blaLAP-2 | 100 | EC_plasmid2 |
| BETA-LACTAM | blaCTX-M-27 | 100 | EC_plasmid2 |
| BETA-LACTAM | blaCTX-M-27 | 100 | EC_plasmid2 |
| BETA-LACTAM | blaCTX-M-27 | 100 | EC_plasmid2 |
| QUINOLONE | qnrS1 | 100 | EC_plasmid2 |
| QUINOLONE | qnrS1 | 100 | EC_plasmid2 |
| QUINOLONE | qnrS1 | 100 | EC_plasmid2 |
3.3. Characterisation and Comparison of MDR Plasmids
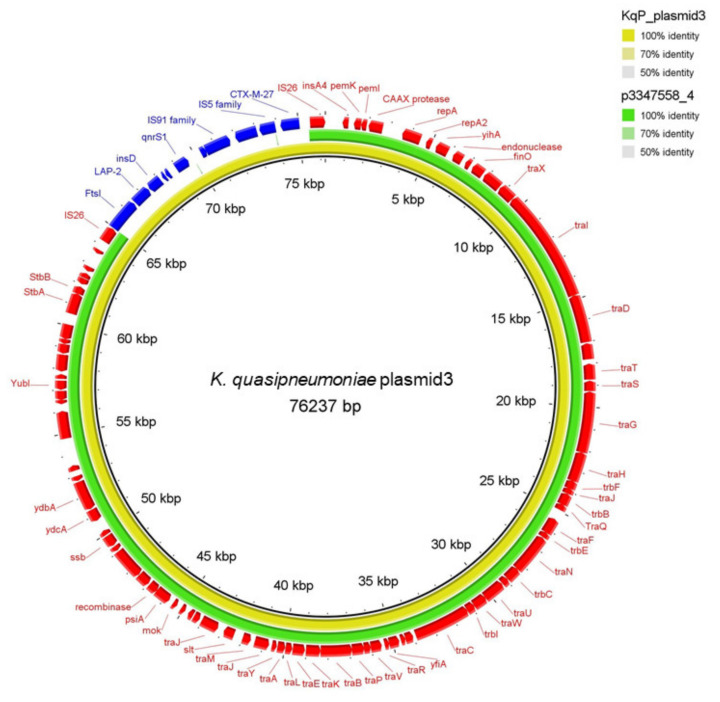
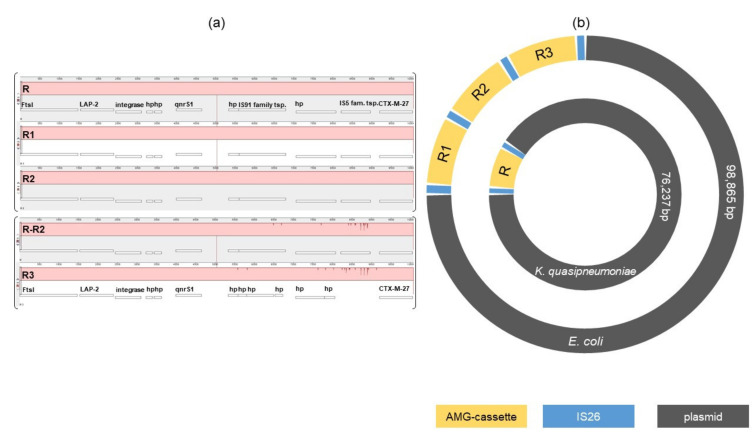
Next, we investigated if ESBL resistance genes were located on a plasmid or encoded on the chromosome. The web-tools PlasmidFinder predicted a IncFII plasmid and pMLST the FAB formula F2:A-:B for the contig that are carrying the blaCTX-M-27 alongside several ARG for both isolates. The plasmid length was 98,865 bp and 76,237 bp in the E. coli and K. quasipneumoniae isolates, respectively. The dfrA26 gene detected in the K. quasipneumoniae on the other hand was located on another plasmid. Both DNA sequences were annotated and showed 100% identical encoded proteins including a relaxome formed by TraI relaxase, TraM and TraY proteins and a type IV secretion system needed for self-conjugative plasmids [13,17] as well as the IncFII replicon (Figure 1). Visualization of the plasmids by BRIG using BLASTn alignment confirmed a 100% sequence match. The difference in total length of the plasmids is caused by the additional two copies of the AMR cassette (Figure 2b). The comparison using progressiveMauve alignment of the AMG-cassettes showed 100% identity of the K. quasipneumoniae AMG-cassette compared to the first two copies in the E. coli plasmid (Figure 2a). The third copy has some deletions and insertions leading to a frameshift causing the loss of the IS91 family transposase and the IS5 family transposase (Figure 2a). An online BLASTn search came up with several similar plasmids but without the AMR cassette. The nearest ancestor plasmid, calculated by BLASTn (Percent Query Coverage: 86%; Percent Identity: 99.95%, for BRIG visualisation see Figure 1), was isolated in 2020 in Switzerland (NCBI GenBank accession no. CP071077.1, plasmid p3347558_4) [18]. Overall, these results strongly suggest horizontal plasmid transfer has occurred between these two strains at one point in the past. Interestingly, the number of copies of the blaCTX-M-27 resistance cassette correlated with the increased MIC of cephalosporins and fluoroquinolones detected in the E. coli isolate compared to the K. quasipneumoniae isolate (Table 2 and Figure 2).
Figure 1.
Comparison of the K. quasipneumoniae plasmid and the reference plasmid p3347558_4 using BRIG. The outer Ring shows the annotated sequences by dfast: The AMG cassette is colored in blue, all other proteins are colored in red.
Figure 2.
(a): progessiveMauve alignment of the AMG-cassettes: R = AMG-cassette of the K. quasipneumoniae plasmid; R1-3 = AMG-cassettes of the E. coli plasmid; FtsI = peptidoglycan glycosyltransferase FtsI; LAP-2 = blaLAP-2; hp = hypotetical protein; qnrS1 = quinolone resistance protein qnrS1; tsp. = transpoase; CTX-M-27 = blaCTX-M-27. IS91 & IS5 family tsp. lost in R3 by frameshift because of insertions and deletions. (b): schematic blueprint of the two plasmids.
4. Discussion
Many reports support the view that plasmid transfer is a relevant mechanism how AMG spread within the hospital setting [19,20,21,22]. However, investigating plasmid transfer based on short-read sequencing data is challenging. Here we present data illustrating how long-read sequencing can be used as a high-resolution tool to monitor plasmids as part of the molecular surveillance of MDR isolates in the hospital setting. Our analysis strongly suggests an inter-species transfer of a self-conjugative F2:A-:B- plasmid between an E. coli and a K. quasipneumoniae isolate in vivo. While the exact time point and source of the plasmid transfer remains unclear, our observation demonstrates that this plasmid is capable to adapt to two different enterobacterial species. In fact, the plasmid described might possess a rather broad host range due to its ability to self-conjugate. Additionally, we could correlate the resistance phenotype of the MDR isolates with their genotype as sequencing data revealed three copies of the AMG cassette, explaining the increased MIC for cephalosporins and fluoroquinolones observed in the E. coli isolate. This example highlights the impact of duplication of AMG cassettes during or through plasmidal conjugations. The mechanisms how duplication of resistance cassettes occur in vivo and how this affects resistance phenotypes are not completely understood yet and will need further investigation as so far only few cases of clinical isolates have been described [23]. Medical records showed that the patient colonized with these MDR isolates has received prolonged treatment with cotrimoxazol/sultamicillin over several weeks because of recurred urinary tract infections (URTIs) prescribed by his general practitioner as well as piperacillin/tazobactam and meropenem twice while being admitted as inpatient in hospital. Fluoroquinolones were not used in the patient’s treatment. However, we must assume that even under selective pressure of a single antibiotic, the entire gene cassette is transferred and the ARG of the other antibiotic classes are co-selected, regardless of the antibiotic chosen previously. In the case of the isolate described this caused increased MIC of fluoroquinolones without using them in the patient’s treatment.
5. Conclusions
Taken together our data shows how analyzing long-read WGS data of routine MDR isolates can give insight into the transfer and function of plasmids in the hospital setting and can help to correlate genotype with resistance phenotype. Plasmid transfer and interaction with other mobile genetic elements in vivo are not completely understood yet but long-read WGS is a valuable tool to address this question and analyze MDR isolates in greater detail even in a routine laboratory setting.
Acknowledgments
We thank the sequencing team of the Institute of Hygiene and the clinical microbiology staff at University Hospital Münster for excellent technical assistance. We thank Natalie Effelsberg for scientific advice and critical review of the manuscript.
Author Contributions
Conceptualization, V.S. and A.M; methodology, A.M.; formal analysis, V.S., V.v.A. and F.S.; investigation, V.S. and V.v.A.; resources, F.S., A.M.; writing—original draft preparation, V.S. and V.v.A.; writing—review and editing, V.S., V.v.A., F.S. and A.M.; visualization, V.v.A. All authors have read and agreed to the published version of the manuscript.
Funding
V.S. was funded by an intramural research grant of the Medical Faculty of the Westfälische Wilhelms University Münster (IMF, SC 22 19 03).
We acknowledge support from the Open Access Publication Fund of the University of Münster.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Sample collection was part of routine surveillance and infection control activities carried out in accordance with the national recommendations of the Robert-Koch Institute, Germany. No additional patient data were collected for the purpose of this investigation. Formal consent was therefore not required.
Data Availability Statement
The sequence information presented in this study has been deposited under NCBI BioProject accession PRJNA802079.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Laxminarayan R., Duse A., Wattal C., Zaidi A.K.M., Wertheim H.F.L., Sumpradit N., Vlieghe E., Hara G.L., Gould I.M., Goossens H., et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 2.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Novais A., Viana D., Baquero F., Martinez-Botas J., Canton R., Coque T.M. Contribution of IncFII and broad-host IncA/C and IncN plasmids to the local expansion and diversification of phylogroup B2 Escherichia coli ST131 clones carrying blaCTX-M-15 and qnrS1 genes. Antimicrob. Agents Chemother. 2012;56:2763–2766. doi: 10.1128/AAC.06001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patino-Navarrete R., Rosinski-Chupin I., Cabanel N., Zongo P.D., Hery M., Oueslati S., Girlich D., Dortet L., Bonnin R.A., Naas T., et al. Specificities and Commonalities of Carbapenemase-Producing Escherichia coli Isolated in France from 2012 to 2015. mSystems. 2022;7:e01169-21. doi: 10.1128/msystems.01169-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zautner A.E., Bunk B., Pfeifer Y., Sproer C., Reichard U., Eiffert H., Scheithauer S., Gross U., Overmann J., Bohne W. Monitoring microevolution of OXA-48-producing Klebsiella pneumoniae ST147 in a hospital setting by SMRT sequencing. J. Antimicrob. Chemother. 2017;72:2737–2744. doi: 10.1093/jac/dkx216. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard A.E., Stoesser N., Wilson D.J., Sebra R., Kasarskis A., Anson L.W., Giess A., Pankhurst L.J., Vaughan A., Grim C.J., et al. Nested Russian Doll-Like Genetic Mobility Drives Rapid Dissemination of the Carbapenem Resistance Gene blaKPC. Antimicrob. Agents Chemother. 2016;60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Effelsberg N., Kobusch I., Linnemann S., Hofmann F., Schollenbruch H., Mellmann A., Boelhauve M., Kock R., Cuny C. Prevalence and zoonotic transmission of colistin-resistant and carbapenemase-producing Enterobacterales on German pig farms. One Health. 2021;13:100354. doi: 10.1016/j.onehlt.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Junemann S., Sedlazeck F.J., Prior K., Albersmeier A., John U., Kalinowski J., Mellmann A., Goesmann A., von Haeseler A., Stoye J., et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013;31:294–296. doi: 10.1038/nbt.2522. [DOI] [PubMed] [Google Scholar]
- 9.Diancourt L., Passet V., Verhoef J., Grimont P.A., Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C., Ochman H., et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I., Tyson G.H., Zhao S., Hsu C.H., McDermott P.F., et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019;63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A., Hasman H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS) Methods Mol. Biol. 2020;2075:285–294. doi: 10.1007/978-1-4939-9877-7_20. [DOI] [PubMed] [Google Scholar]
- 13.Che Y., Yang Y., Xu X., Brinda K., Polz M.F., Hanage W.P., Zhang T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. USA. 2021;118:e2008731118. doi: 10.1073/pnas.2008731118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alikhan N.F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Virolle C., Goldlust K., Djermoun S., Bigot S., Lesterlin C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes. 2020;11:1239. doi: 10.3390/genes11111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moser A.I., Keller P.M., Campos-Madueno E.I., Poirel L., Nordmann P., Endimiani A. A Patient with Multiple Carbapenemase Producers Including an Unusual Citrobacter sedlakii Hosting an IncC bla NDM-1- and armA-carrying Plasmid. Pathog. Immun. 2021;6:119–134. doi: 10.20411/pai.v6i2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aires-de-Sousa M., Ortiz de la Rosa J.M., Goncalves M.L., Costa A., Nordmann P., Poirel L. Occurrence of NDM-1-producing Morganella morganii and Proteus mirabilis in a single patient in Portugal: Probable in vivo transfer by conjugation. J. Antimicrob. Chemother. 2020;75:903–906. doi: 10.1093/jac/dkz542. [DOI] [PubMed] [Google Scholar]
- 20.Mathers A.J., Crook D., Vaughan A., Barry K.E., Vegesana K., Stoesser N., Parikh H.I., Sebra R., Kotay S., Walker A.S., et al. Klebsiella quasipneumoniae Provides a Window into Carbapenemase Gene Transfer, Plasmid Rearrangements, and Patient Interactions with the Hospital Environment. Antimicrob. Agents Chemother. 2019;63:e02513-18. doi: 10.1128/AAC.02513-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galani I., Panagea T., Chryssouli Z., Giamarellou H., Souli M. In vivo transmission of a plasmid containing the KPC-2 gene in a single patient. J. Glob. Antimicrob. Resist. 2013;1:35–38. doi: 10.1016/j.jgar.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Blake K.S., Choi J., Dantas G. Approaches for characterizing and tracking hospital-associated multidrug-resistant bacteria. Cell. Mol. Life Sci. 2021;78:2585–2606. doi: 10.1007/s00018-020-03717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuster C.F., Weber R.E., Weig M., Werner G., Pfeifer Y. Ultra-deep long-read sequencing detects IS-mediated gene duplications as a potential trigger to generate arrays of resistance genes and a mechanism to induce novel gene variants such as blaCTX-M-243. J. Antimicrob. Chemother. 2022;77:381–390. doi: 10.1093/jac/dkab407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence information presented in this study has been deposited under NCBI BioProject accession PRJNA802079.