Abstract
Age at depressive onset (AAO) corresponds to unique symptomatology and clinical outcomes. Integration of genome-wide association study (GWAS) results with additional “omic” measures to evaluate AAO has not been reported and may reveal novel markers of susceptibility and/or resistance to major depressive disorder (MDD). To address this gap, we integrated genomics with metabolomics using data-driven network analysis to characterize and differentiate MDD based on AAO. This study first performed two GWAS for AAO as a continuous trait in (a) 486 adults from the Pharmacogenomic Research Network-Antidepressant Medication Pharmacogenomic Study (PGRN-AMPS), and (b) 295 adults from the Combining Medications to Enhance Depression Outcomes (CO-MED) study. Variants from top signals were integrated with 153 p180-assayed metabolites to establish multi-omics network characterizations of early (<age 18) and adult-onset depression. The most significant variant (p = 8.77 × 10−8) localized to an intron of SAMD3. In silico functional annotation of top signals (p < 1 × 10−5) demonstrated gene expression enrichment in the brain and during embryonic development. Network analysis identified differential associations between four variants (in/near INTU, FAT1, CNTN6, and TM9SF2) and plasma metabolites (phosphatidylcholines, carnitines, biogenic amines, and amino acids) in early- compared with adult-onset MDD. Multi-omics integration identified differential biosignatures of early- and adult-onset MDD. These biosignatures call for future studies to follow participants from childhood through adulthood and collect repeated -omics and neuroimaging measures to validate and deeply characterize the biomarkers of susceptibility and/or resistance to MDD development.
Keywords: genomics, metabolomics, major depressive disorder, age at onset, network analysis
1. Introduction
Major depressive disorder (MDD) etiology and prognosis vary by the age at depressive onset. Early onset is characterized by poorer quality of life, greater psychiatric and medical comorbidity, higher heritability, and increased suicidality [1,2,3,4,5]. This suggests that individuals with early-onset MDD may benefit from a tailored pharmacologic treatment approach [6,7,8,9,10]. However, psychotherapy and pharmacotherapy remain largely consistent across the age spectrum [11]. Before pharmacotherapy can be individualized according to the age at MDD onset, deeper characterizations of the biological differences between early and later-onset presentations are necessary.
Several genome-wide association studies (GWAS) and one exome-wide association study have been performed to understand the genomics of MDD specific to age at onset [8,12,13,14,15]. Collectively, and in conjunction with polygenic risk analyses, these studies suggest that earlier-onset MDD may share greater genetic overlap with schizophrenia, bipolar disorders, and attention deficit/hyperactivity disorder (ADHD) than later onset [8,12,16,17]. Two GWAS investigating the age at depressive onset (N = 2746 [14]) and (N = 9238 [13]) did not identify overlapping top signals (p < 1 × 10−5), highlighting phenotypic heterogeneity that may require additional biological measures to understand. Circulating metabolite concentrations are highly heritable (up to 62%) [18,19] and are differentially perturbed in MDD according to age [10], yet the genetic factors associated with these differences have not yet been investigated. To our knowledge, no integrative genomic and metabolomic approaches have been undertaken to identify multi-omics signatures which best characterize and differentiate patients with early- and late-onset depression. Such an analysis may yield insights into heritable (genomic) and downstream (metabolomic) differences between age groups.
Multi-omics strategies are amongst the approaches at the forefront of individualized medicine in psychiatry and are grounded in the understanding that complex traits (e.g., onset of depression) cannot be fully characterized by isolate biomeasures [20,21,22,23,24,25,26,27,28]. Rather, interactions between genes, transcripts, proteins, metabolites, and the environment determine the development and trajectory of complex diseases [29]. Multi-omics strategies can aide in biomarker selection for future experimental validation by identifying the features which best characterize conditions of interest (e.g., early- vs. adult-onset MDD) [30]. This is imperative in this context given the breadth of the metabolomic and genomic platforms. The objectives of this study were therefore to (1) identify risk and protective single nucleotide variants (SNVs) associated with age at onset of MDD using a GWAS approach, and (2) integrate genomic findings with plasma metabolites to identify multi-omics differences between individuals with early- (<age 18) versus adult-onset MDD. We hypothesized that this multi-omics approach would derive distinct molecular signatures of early- versus adult-onset MDD.
2. Materials and Methods
2.1. Data Sources
This was a cross-sectional, secondary analysis of Caucasian adults with MDD from the PGRN-AMPS (N = 486 with genomics; N = 245 with multiple omics) and CO-MED (N = 295 with genomics; N = 76 with multiple omics) studies (Table 1; see Supplementary Figures S1 and S2 for sample inclusion). The PGRN-AMPS (NCT00613470) and CO-MED (NCT00590863) trials enrolled outpatients with moderate to severe nonpsychotic MDD. These trials have been characterized in detail in prior publications [31,32]. The CO-MED study included patients who met DSM-IV-TR criteria for recurrent or chronic MDD (current episode lasting ≥ 2 years), while PGRN-AMPS included patients with DSM-IV diagnosed MDD, without the requirement of recurrent or chronic presentation. The CO-MED cohort also represents a more regionally diverse population—the trial was conducted across six primary and nine psychiatric care sites (compared to the single-site PGRN-AMPS trial). The inclusion and exclusion criteria were otherwise similar between studies. Both studies were conducted in accordance with the approval of their respective Institutional Review Boards. Informed consent was obtained from all participants.
Table 1.
Demographics and patient characteristics.
| Genomics Analyses | Multi-Omics Analyses | |||
|---|---|---|---|---|
| CO-MED | PGRN-AMPS | Early Onset (<18 Years of Age) | Adult Onset (≥18 Years of Age) | |
| Total (N) | 295 | 486 | 130 | 191 |
| PGRN-AMPS (N) | - | 486 | 100 | 145 |
| CO-MED (N) | 295 | - | 30 | 46 |
| Age [mean (SD)] | 43.2 (12.9) * | 39.9 (13.7) * | 37.3 (12.7) * | 43.8 (12.8) * |
| Ethnicity (% Hispanic) | 20% * | 2.1% †* | 4.8% † | 9.2% † |
| Sex (% Women) | 68% | 62% | 71% | 62% |
| Age at depressive onset (Median [min, max]) | 18 [0, 60] | 20 [4, 83] | 14 [4, 17] * | 33 [18, 83] * |
| Baseline depressive severity [mean (SD)] | 15.7 (3.4) | 15.1 (3.5) | 15.3 (3.6) | 14.8 (3.0) |
* Significantly different (p < 0.05) by Fisher’s Exact test or independent-samples t-test (calculated for all variables excluding study Ns). † Ethnicity characterizations calculated from a smaller subset of samples based upon data availability (117 individuals with early depressive onset; 177 individuals with adult depressive onset; 338 PGRN-AMPS individuals).
2.2. Measures and Outcomes
Age at depressive onset was self-reported with a baseline case report form in PGRN-AMPS and with the Mini-International Neuropsychiatric Interview (MINI) [7] in CO-MED. Baseline depression severity was assessed with the 16-item Quick Inventory of Depressive Symptomatology—Clinician-Rated (QIDS-C) [33]. Race and ethnicity were self-reported and validated through genomic techniques (described in Genotyping, Genome-Wide Imputation, and Quality Control).
2.3. Genotyping, Genome-Wide Imputation, and Quality Control
PGRN-AMPS: Genotyping was performed at the RIKEN Center for Genomic Medicine (Yokohama, Japan) using Illumina human 610-Quad BeadChips (Illumina, San Diego, CA, USA), with quality control procedures and imputation as previously described [26,31,34]. Following genotyping, quality control, and imputation, 7,017,931 variants meeting a minor allele frequency threshold of 0.01 and an imputation R2 > 0.3 in 486 Caucasian individuals were retained for statistical analysis.
CO-MED: Genotyping was performed using Illumina Quad, Human Omni 2.5 bead chips, as previously described [35]. During quality control, samples were removed if self-reported sex was incongruent with X-chromosome estimated sex, call rate was <98%, or heterozygosity ratio was outside the mean (≤0.7 on any chromosome). One individual from each pair determined to have kinship > 0.08 by the King-Robust test was removed. Concordance between intended duplicates was assessed. Variants were removed if unmapped, duplicated, achieved call rate < 95% or achieved Hardy Weinberg equilibrium p < 1 × 10−10. These criteria resulted in 2,356,856 variants from 464 individuals available for phasing and imputation. Imputation was performed using the Michigan Imputation Server [36] with EAGLE2 and the Haplotype Reference Consortium (HRC) reference panel (version r1.1 2016) for phasing [37]. Data were aligned to the human reference genome GRCh37. Following quality control and imputation, 8,286,413 variants meeting a minor allele frequency threshold of 0.01 and an imputation R2 > 0.3 in 295 Caucasian individuals were retained for statistical analysis. Race was self-reported and concordant with genetic clustering based upon multidimensional scaling [38].
2.4. Genome-Wide Association Study Statistical Analyses
GWAS for age at depressive onset as a continuous trait was performed with univariate linear regression under an additive genetic model [39]. GWAS was performed separately in the PGRN-AMPS and CO-MED cohorts to assess the replicability of signals across independent cohorts. Prior to analysis, age at depressive onset was log2 transformed to meet the assumption of normality. Analyses were adjusted for sex, and the first ten principal components of ancestry and filtered for minor allele frequency >5%. Quantile-quantile plots of expected vs. observed p-values were constructed to assess for statistical inflation (Supplemental Figure S3). Given that the purpose of GWAS was to identify candidates for subsequent multi-omics integration analyses, index variants associated with age at depressive onset p < 1 × 10−5 were retained. Index variants were defined as the variant with the lowest p-value amongst all associated variants (r2 > 0.8) in each signal. Sensitivity analyses were conducted excluding individuals with self-reported depressive onset prior to age three, as evidence suggests that clinically significant depression can arise as early as age three [40,41,42,43]. Analysis was performed in PLINK version 2.0 (Mountain View, CA, USA) [44], and Manhattan plots were created with LocusZoom.org [45].
2.5. Variant Annotation
All variants meeting suggestive significance were annotated with the nearest gene, location to nearest gene, and consequences (e.g., non-synonymous, missense, synonymous variant). Next, these variants and variants in linkage disequilibrium (r2 > 0.8) were assessed for known cis- and trans- expression quantitative trait loci (eQTL) gene labels. Gene annotation was done with the HaploReg database [46], implemented in R studio with haploR [47]. Following variant annotation, the nearest genes and eQTL genes for the 36 index SNVs were input into the Database for Annotation, Visualization and Integrated Discovery (DAVID) [48] v6.8 (Frederick, MD, USA) for tissue-specific expression analysis, tissue-associated protein enrichment, and pathway and disease over-representation.
2.6. Metabolomics
Plasma metabolites were assayed in a subset of PGRN-AMPS and CO-MED patients using the AbsoluteIDQ p180 platform [49]. This targeted assay utilizes triple quadrupole tandem mass spectrometry to detect metabolites from five analyte classes (acylcarnitines, amino acids, biogenic amines, glycerophospholipids and sphingolipids). This assay is not exhaustive of all potential metabolite correlates of early onset depression, and it does not measure several metabolites which are well-studied in psychiatry (i.e., dopamine, GABA). However, these analytes have enhanced our understanding of the biology of MDD [20,50,51,52], schizophrenia [53], and psychosis [54,55], warranting their investigation in the context of early-onset MDD. Metabolomic profiling and quality control were conducted separately for the PGRN-AMPS and CO-MED cohorts, as described in previous publications [51,52,56]. Metabolites with ≥10% missingness were excluded from the current analysis, leaving 153 metabolites available for analysis (Supplementary Table S1). All metabolites meeting quality control criteria were included in the current integration analysis to identify which may be candidates for future, targeted studies.
2.7. Multi-Omics Integration Network Analysis
The purpose of the current multi-omics integration network analysis is to (a) compare differences in individuals with early (<age 18) vs. adult onset MDD and (b) identify biomarkers capable of characterizing such phenotypes.
Inputs to the analysis included the 36 index SNVs from GWAS signals with suggestive association with age at depressive onset (p < 1 × 10−5 in either cohort) and 153 p180-assayed metabolites meeting quality control criteria. The threshold of suggestive significance was defined in accordance with several GWAS studies and the GWAS catalog [57]. These metabolites represent five classes (glycerophospholipids, amino acids, biogenic amines, acylcarnitines, and sphingolipids), and are synthesized and metabolized by enzymes encoded in the host genome. These metabolic classes are amongst those which are essential for growth, development, and many key physiological functions [58].
The analysis utilizes a sparse partial least squares discriminant approach [59] to identify significant correlations between SNVs and metabolites (p < 0.05; |r| > 0.1) in individuals with early and adult onset MDD. Furthermore, the analysis includes a multi-level community detection step [60] to identify communities (sub-networks) of SNVs and metabolites which are strongly associated within their community and less associated outside of the community. Community detection accounts for the correlation structure amongst metabolites when assessing metabolite-variant associations. The assumption underlying community detection is that communities are comprised of functionally related biomolecules [30].
The analysis was performed using xMWAS (Atlanta, GA, USA) [30] as implemented in R v4.0.3 (Vienna, Austria) [61] with RStudio version 1.3 (Vienna, Austria) [62]. Results were visualized using Cytoscape (Bethesda, MD, USA) [63].
3. Results
3.1. Sample Characteristics
Table 1 provides the demographic characteristics for the samples. Generally, the sample characteristics for the two studies were similar, although the CO-MED sample had a higher ratio of Hispanic participants than PGRN-AMPS. Mean age at baseline was younger in those with early onset than adult onset (37.3 ± 12.7 vs. 43.8 ± 12.8; p < 0.05); all other baseline characteristics were similar between early- and adult-onset groups.
3.2. Genome-Wide Association Study Statistical Analyses
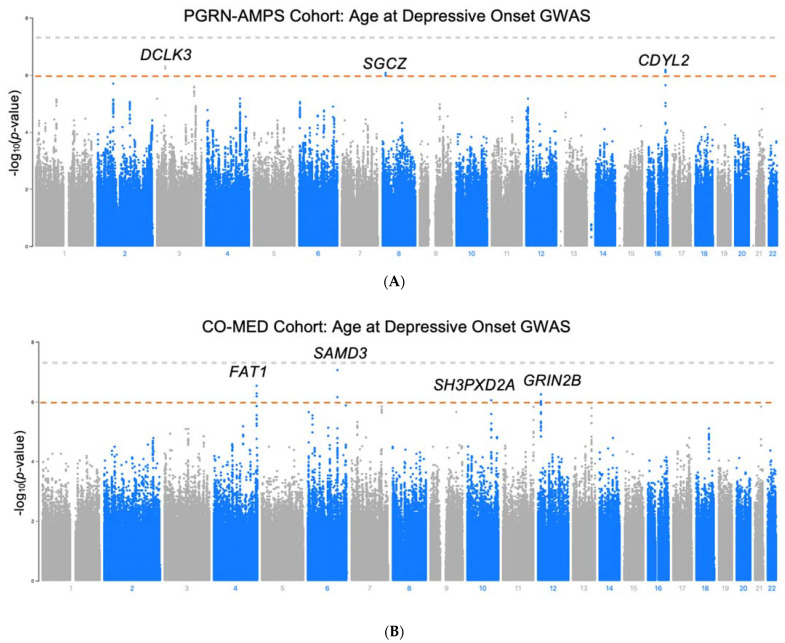
GWAS for age at depressive onset was performed in the PGRN-AMPS and in CO-MED cohorts (Manhattan plots illustrated in Figure 1). No SNVs met genome-wide statistical significance after accounting for multiple comparisons (p < 5 × 10−8) in either cohort. However, a signal of three SNVs in tight linkage-disequilibrium (LD) in an intron of the Sterile Alpha Motif Domain Containing 3 (SAMD3) gene (chromosome 6) achieved near statistical significance in the CO-MED cohort, with the tag SNV (rs870816) reaching p = 8.8 × 10−8 (t = −4.49). The negative direction of the t-statistic effect size for rs870816 signifies that the minor allele associates with earlier MDD onset. There was no overlap between cohorts in the signals which achieved suggestive significance (p < 1 × 10−5). Fifty-five SNVs met suggestive significance (p < 1 × 10−5) in PGRN, and 79 SNVs met suggestive significance in CO-MED (Supplementary Tables S2 and S3A). One individual in the CO-MED study reported depressive onset prior to three years of age (self-reported age = 0) and was excluded in a subsequent GWAS sensitivity analysis. The top signals found in the sensitivity analysis remain consistent with the analysis which includes this individual (Supplementary Table S3B). Of these total 134 SNVs with suggestive significance, minor alleles for 91 of them associated with earlier MDD onset (negative effect size), while 54 associated with later onset (positive effect size). The 36 index SNVs for these signals are shown in Table 2. Genomic control λ values were within 1 ± 0.05 for each analysis.
Figure 1.
Manhattan plots for GWAS for age at depressive onset in (A) PGRN-AMPS cohort (N = 487) and (B) CO-MED (N = 295). Grey line: genome-wide significance (5 × 10−8). Red line: suggestive significance (1 × 10−6).
Table 2.
Index variants by study.
| A. PGRN-AMPS GWAS Index Variants | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant ID | SNV | Minor Allele | Nearest Gene | Distance to Nearest Gene | Location | eQTL | Minor Allele Frequency | Beta | Standard Error | T-Statistic | p-Value |
| 1-90283515-A-G | rs7545243 | G | LRRC8D | 3056 | RP5-943J3.1 (subcutaneous adipose, whole blood, lung), LRRC8B, LRRC8C, LRRC8D (whole blood) | 0.29 | −0.25 | 0.06 | −4.54 | 7.16 × 10−6 | |
| 2-71442773-G-C | rs59432780 | C | PAIP2B | 0 | Intron | PAIP2B (skin) | 0.12 | −0.38 | 0.08 | −4.81 | 2.01 × 10−6 |
| 2-71456834-T-C | rs56796378 | C | PAIP2B | 0 | Intron | MPHOSPH10 (whole blood) | 0.10 | −0.40 | 0.09 | −4.53 | 7.48 × 10−6 |
| 2-144491308-G-A | rs10928197 | A | ARHGAP15 | 17,222 | 0.43 | −0.23 | 0.05 | −4.49 | 8.8 × 10−6 | ||
| 2-144477195-G-A | rs12465492 | A | ARHGAP15 | 3681 | 0.38 | −0.24 | 0.05 | −4.48 | 9.43 × 10−6 | ||
| 3-36755107-G-A | rs35721771 | A | DCLK3 | 610,866 | 0.35 | 0.28 | 0.06 | 5.09 | 5.16 × 10−7 | ||
| 3-1006912-A-G | rs10510204 | A | CNTN6 | 613,027 | 0.40 | 0.24 | 0.05 | 4.55 | 6.75 × 10−6 | ||
| 4-150429619-G-A | rs6853045 | G | DCLK2 | 569,805 | Intron | 0.46 | −0.24 | 0.05 | −4.56 | 6.67 × 10−6 | |
| 6-5389403-C-A | rs73350538 | A | FARS2 | 0 | Intron | 0.06 | 0.51 | 0.11 | 4.49 | 8.79 × 10−6 | |
| 8-14827133-A-C | rs76522180 | C | SGCZ | 0 | Intron | 0.06 | −0.57 | 0.11 | −4.99 | 8.43 × 10−7 | |
| 12-9058993-A-T | rs7299653 | T | PHC1 | 8321 | KLRG1 (subcutaneous and visceral omentum adipose, aorta and tibial artery, brain (cortex), mammary tissue, fibroblasts, esophagus, atrial appendage, tibial nerve, pancreas), PHC1 (esophagus), RP11-436I9.6 (lung), RP11-705C15.3 (skeletal muscle), LINC00987 (subcutaneous adipose, whole blood), RP11-118B22.4 (whole blood), M6PR (whole blood), A2MP1 (whole blood) | 0.48 | −0.24 | 0.05 | −4.55 | 6.76 × 10−6 | |
| 16-80882476-T-C | rs9926993 | C | CDYL2 | 44,300 | 0.07 | −0.51 | 0.10 | −5.04 | 6.72 × 10−7 | ||
| 16-80842302-A-T | rs62052150 | T | CDYL2 | 4126 | 0.08 | −0.46 | 0.10 | −4.48 | 9.57 × 10−6 | ||
| B. CO-MED GWAS Index Variants | |||||||||||
| 3-93956553-T-A | rs143801763 | A | NSUN3 | 110,922 | 0.08 | 0.63 | 0.14 | 4.54 | 8.27 × 10−6 | ||
| 3-104317729-G-A | rs4450851 | A | MIR548A3 | 371,623 | 0.44 | −0.34 | 0.07 | −4.55 | 8.10 × 10−6 | ||
| 4-187604429-T-C | rs162181 | C | FAT1 | 0 | Intron | 0.20 | −0.48 | 0.09 | −5.25 | 2.96 × 10−7 | |
| 4-187592223-T-A | rs11723473 | A | FAT1 | 0 | Intron | 0.16 | −0.45 | 0.09 | −4.78 | 2.83 × 10−6 | |
| 4-128523964-T-C | rs1399212 | C | INTU | 30,121 | 0.06 | −0.71 | 0.15 | −4.59 | 6.64 × 10−6 | ||
| 6-130472920-C-T | rs870816 | T | SAMD3 | 0 | Intron | L3MBTL3, SAMD3 (whole blood) | 0.34 | −0.40 | 0.07 | −5.49 | 8.77 × 10−8 |
| 6-130474597-T-G | rs1932106 | G | SAMD3 | 0 | Intron | SAMD3 (whole blood) | 0.45 | 0.38 | 0.07 | 5.08 | 6.94 × 10−7 |
| 6-166957334-T-C | rs6456092 | C | RPS6KA2 | 0 | Intron | 0.37 | 0.37 | 0.07 | 4.95 | 1.32 × 10−6 | |
| 6-5367273-G-A | rs797147 | A | FARS2 | 0 | Intron | 0.11 | −0.55 | 0.11 | −4.83 | 2.21 × 10−6 | |
| 6-21867547-A-G | rs6940645 | G | FLJ22536 | 0 | Intron | 0.38 | −0.36 | 0.08 | −4.78 | 2.86 × 10−6 | |
| 6-89987882-G-T | rs12213221 | T | GABRR2 | 0 | Intron | 0.47 | −0.34 | 0.07 | −4.57 | 7.45 × 10−6 | |
| 6-130499476-C-T | rs1034263 | T | SAMD3 | 0 | Intron | SAMD3 (whole blood) | 0.07 | −0.67 | 0.15 | −4.55 | 7.82 × 10−6 |
| 7-132382054-C-T | rs10233511 | T | PLXNA4 | 48,606 | Intron | 0.29 | −0.40 | 0.08 | −4.92 | 1.45 × 10−6 | |
| 7-26421269-T-G | rs74409431 | G | SNX10 | 7319 | AC004540.4, SNX10 (subcutaneous adipose) | 0.13 | 0.51 | 0.11 | 4.66 | 4.80 × 10−6 | |
| 9-114014832-T-C | rs112014566 | C | OR2K2 | 74,929 | 0.11 | −0.59 | 0.12 | −4.84 | 2.19 × 10−6 | ||
| 10-105498155-T-A | rs7085238 | A | SH3PXD2A | 0 | Intron | SH3PXD2A (whole blood) | 0.22 | −0.44 | 0.09 | −5.03 | 8.86 × 10−7 |
| 11-134403616-C-A | rs12287910 | A | LOC283177 | 28,060 | GLB1L2|B3GAT1 (whole blood) | 0.42 | 0.36 | 0.07 | 4.92 | 1.48 × 10−6 | |
| 12-13883111-C-T | rs11609779 | T | GRIN2B | 0 | Intron | 0.18 | 0.50 | 0.10 | 5.12 | 5.61 × 10−7 | |
| 12-13880168-G-C | rs10845837 | C | GRIN2B | 0 | Intron | 0.30 | 0.39 | 0.08 | 4.83 | 2.24 × 10−6 | |
| 13-100223130-G-A | rs4297561 | A | TM9SF2 | 7853 | 0.33 | 0.39 | 0.08 | 4.90 | 1.61 × 10−6 | ||
| 13-100224460-G-A | rs2793779 | A | TM9SF2 | 9183 | 0.33 | 0.38 | 0.08 | 4.77 | 2.97 × 10−6 | ||
| 18-55933912-C-T | rs62094545 | T | NEDD4L | 0 | Intron | 0.17 | 0.44 | 0.10 | 4.55 | 7.86 × 10−6 | |
| 21-40135684-T-A | rs8127960 | A | NCRNA00114 | 0 | Intron | LINC00114 (tibial artery) | 0.25 | 0.43 | 0.09 | 4.93 | 1.44 × 10−6 |
3.3. Variant Annotation
Twenty of the 36 index SNVs localized to introns; the remaining SNVs localized outside of the open reading frames. Given that no SNVs localized within coding regions, there were no non-synonymous or missense variant candidates. Eleven variants are known eQTLs for one or more genes in one or more tissues (Table 2). Functional annotation of the genes using DAVID [48] at a nominal p-value (p < 0.05) demonstrated that the tissue-associated protein enrichment for these genes was greatest in the brain (“UP_TISSUE” annotation). Furthermore, the tissue-specific expression analysis (“UNIGENE EST QUARTILE” annotation) most strongly highlighted tissues involved in embryonic development. Multiple psychiatric traits are also represented at p < 0.05, including tobacco use disorder and alcohol use disorder (Supplementary Table S4).
3.4. Multi-Omics Integration Network Analysis
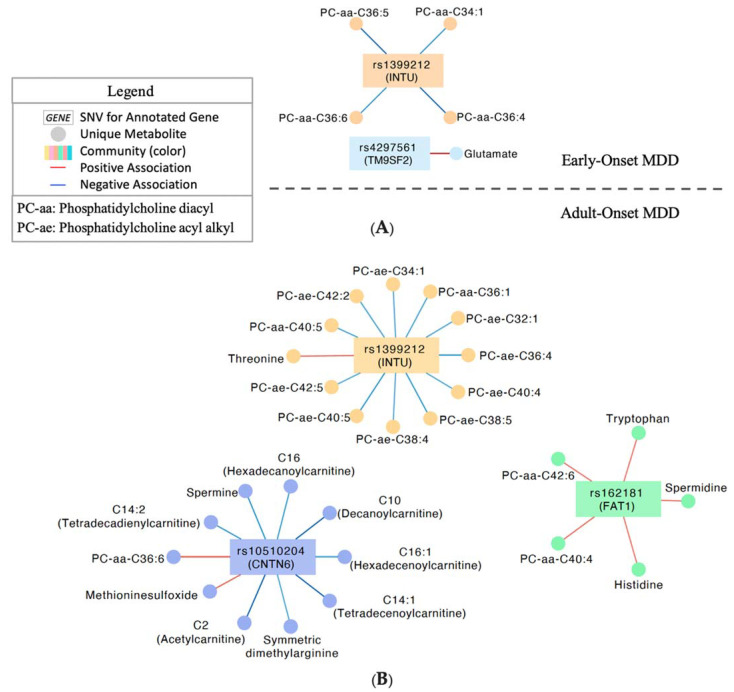
Integration analysis identified two communities of correlated (p < 0.05; |r| > 0.1) SNVs and metabolites in individuals with early (<age 18) onset MDD (Figure 2A), and three communities in individuals with adult onset MDD (Figure 2B). Four SNVs in total were represented (locus zoom plots, see Supplementary Figure S4). Notably, the SNV nearest INTU correlated with four phosphatidylcholine diacyl species (34:1, 36:4, 36:5, 36:6) in individuals with early-onset MDD, and 11 phosphatidylcholine species, (nine of which are acyl-alkyl species), in individuals with adult-onset (Figure 2). Early onset was also characterized by a strong positive correlation between glutamine and rs2793779 near TM9SF2, which is absent from the adult-onset network. Individuals with adult onset MDD demonstrate unique correlations between the (FAT1) intronic SNV and phosphatidylcholines, spermidine, histidine, tryptophan, and three lysophosphatidylcholine metabolites. Finally, several carnitines correlated with the SNV closest to CNTN6 in individuals with adult onset MDD but not in individuals with early-onset MDD (Figure 2). Pearson correlations for all SNVs and metabolites represented in these networks can be found in Supplementary Table S5.
Figure 2.
Multi-Omics integration network analysis. (A) Significant (p < 0.05; |r| > 0.1) associations of SNVs and metabolites in individuals with depressive onset prior to age 18. (B) Significant (p < 0.05; |r| > 0.1) associations in individuals with depressive onset at or after age 18. Legend applies to both (A) and (B).
4. Discussion
This work establishes multi-omics characterization of early- versus adult-onset MDD using data from the PGRN-AMPS and CO-MED studies. GWAS for age at depressive onset were performed for both PGRN-AMPS and CO-MED, and top variants were then integrated with plasma metabolomics to identify biological signatures which best differentiated early and adult-onset MDD. These multi-omics networks enabled biological characterization of MDD by age at onset and provide a basis for future functional experiments aiming to investigate the mechanisms underlying the development of MDD across the lifespan. Derived early- and adult-onset MDD biosignatures showed distinct associations between variants in/near INTU, FAT1, CNTN6, and TM9SF2 with plasma metabolites (phosphatidylcholines, carnitines, biogenic amines, and amino acids) for continued future investigation.
Expression of top variants identified through GWAS for age at depressive onset was enriched in the brain and during embryonic development according to the DAVID database, which provides functional annotations for lists of variants [48]. No variant achieved genome-wide significance, although this is not uncommon for GWAS for psychiatric traits given the phenotypic complexity [8,13,64,65,66]. Valuable mechanistic insights into biological drivers of neuropsychiatric diseases and drug response have been gained from GWAS signals of suggestive significance from the PGRN-AMPS cohort, despite its limited sample size [26,67,68]. Several variants meeting suggestive significance (mapping to SAMD3, GRIN2B) have previously been implicated in MDD [69,70], with our current study adding to this body of literature by revealing novel associations with age at MDD onset. In several instances, the minor allele of GWAS variants associated with a later onset of MDD, suggesting they might be protective against disease onset. Alternatively, these variants may confer MDD susceptibility by interacting with risk factors specific to later life, for example, functional/cognitive impairment or marital challenges [71]. In this context, continued investigation of the biological implications of top GWAS signals through multi-omics integration analysis was warranted.
The multi-omics integration analysis characterized the variant-metabolite associations that best differentiated early (<18 years) and adult-onset MDD. Onset of MDD was dichotomized at age 18 for this assessment for several reasons. For many, the transition at age 18 into legal adulthood includes major changes in determinants of health such as financial resources and social environments [72,73,74]. Clinically, age 18 also corresponds to a transition from adolescent to adult medical care. Furthermore, most research studies use age 18 to define early versus adult samples. Finally, age 18 was the median age at MDD onset in CO-MED and near the median (20 years) in PGRN-AMPS, so this threshold enabled a relatively balanced sample split.
The multi-omics networks demonstrated that rs1399212, nearest the “Inturned Planar Cell Polarity Protein (INTU)” gene, negatively associated with phosphatidylcholines of both networks, and had unique associations with the phosphatidylcholine acyl-alkyl (PC-ae) species in individuals with adult-onset MDD. The ether phosphatidylcholines (PC-ae species) play distinct roles from conventional phosphatidylcholines (PC-aa species) in cell differentiation, cell signaling, and reduction of oxidative stress [75]. These network associations suggest that INTU variation may impact concentrations of phosphatidylcholine metabolites, which is corroborated by associations of INTU and trans fatty acid levels in a sample of >9000 individuals with replication across diverse ancestries [76]. Furthermore, the differential INTU-phosphatidylcholine associations in early vs. adult-onset MDD, with adult-onset having many PC-ae associations, suggest that regulation of phosphatidylcholine biosynthesis or metabolism by INTU may be biologically distinct between groups, potentially conferring effects on cell differentiation, cell signaling, and reduction of oxidative stress.
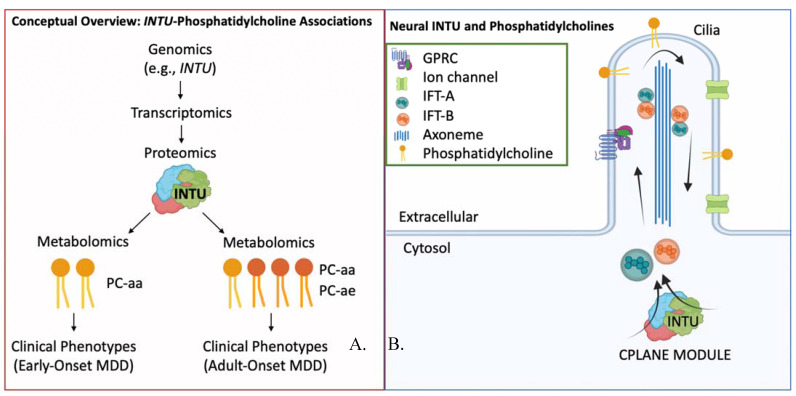
INTU protein is a central component of the CPLANE (ciliogenesis and planar polarity effector) module [77], which is critical for ciliogenesis and transport of ciliary proteins [77,78,79] (Figure 3). Cilia are highly conserved organelles which form specialized extensions of the cell membrane, and they may be motile or non-motile. In the brain, motile cilia are present on the ependymal cells of the ventricles and in the choroid plexus, whereas non-motile cilia are widely observed, including on astrocytes, neurons, and progenitors [80]. Cilia are critical for embryonic development [81,82,83,84]. Both during development and in adult homeostasis, cilia facilitate cellular signal transduction along major signaling pathways, including Hedgehog, Wnt, Notch, transforming growth factor B, G protein-coupled receptors, receptor tyrosine kinases, extracellular matrix receptors [85,86,87,88], pathways that are known to be perturbed in MDD [89,90]. Perturbations in cilia structure and/or function can lead to a spectrum of diseases (‘ciliopathies’) with broad developmental and adult phenotypes, including cognitive defects and neuropsychiatric phenotypes [91,92]. The cilia interactome, defined as interactions between ciliary proteins, also demonstrates extensive overlap with neuropsychiatric disease interactomes and genes differentially expressed in neuropsychiatric diseases, including MDD, schizophrenia, bipolar disorder, Alzheimer’s disease, ADHD, autism spectrum disorder, and Parkinson’s disease [93,94]. At the macroscopic level, INTU variants are amongst the polygenic signals associating with cortical thickness (5 × 10−9) in children of the Adolescent Brain Cognitive Development (ABCD) Study [95]. Cortical thickness, in turn, is dysregulated in MDD compared with controls [96,97,98], and a preliminary study suggests differences in cortical thickness between pediatric and adult onset MDD [99].
Figure 3.
Analysis conclusions and future aims. (A) Modulators (e.g., transcriptomics and proteomics) may mediate the differential associations between variation near INTU and phosphatidylcholine metabolites in early vs. adult-onset MDD. Future investigation into such modulators may enhance our understanding of the development of MDD at various stages across the lifespan. (B) INTU is a member of the CPLANE protein module which facilitates intraflagellar transport of proteins and lipids throughout the cilia via the intraflagellar transport A and B (IFT-A, IFT-B) complexes. Phosphatidylcholines are lipids widely present in the lipid membrane which facilitate receptor localization in the membrane and signaling via intracellular cascades.
The differential associations of specific phosphatidylcholine species and INTU variants according to age at depressive onset may be indicative of a mechanistic basis distinguishing early and adult-onset MDD. These associations may be mediated by additional biological and environmental factors (Figure 3), and they may have a functional relationship with the observed cortical thickness differences in patients with MDD across the lifespan. Given the role of INTU in neurodevelopment, whether these differences are specific to the neurodevelopmental period or whether they persist into adulthood remains a question for future investigation. Based on these networks, further characterization of the precise lipid content within ciliary membranes and their trafficking may help advance our biological understanding of early versus adult-onset MDD.
The multi-omics networks also reveal additional associations for future mechanistic studies. This includes the associations of variation near CNTN6 with plasma carnitines, which are specific to adult-onset MDD. Such an investigation will add to the current characterizations of subgroups of MDD by acylcarnitine metabolomic profiles [50]. This also includes the associations of variants in FAT1 with spermidine, given that spermidine has recently been proposed as an antidepressant drug [100,101]. Perhaps the most critical future direction arising from this work’s identification of differential variant-metabolite associations in early and adult-onset MDD will be for future studies to assay biomeasures across the lifespan to enhance our understanding of the development of MDD. The present data were sourced from adults. A characterization across the lifespan of biochemical (e.g., omics) and neuroimaging measures is needed, given the suggested interplay between INTU, plasma metabolomics, and cortical thickness in differentiating individuals by the age at MDD onset. Except in the case of rare mutation, an individual’s genome remains consistent throughout the lifespan. Meanwhile, the regulation of genomics via downstream biological and environmental factors creates a dynamic metabolome. Epigenetics may be particularly valuable in future studies for explaining the differential gene-metabolite associations in early versus adult-onset depression. Early life adversity can confer epigenetic modifications linked to development of depression in youth and adults [102]. Epigenomic alterations may drive neuropsychiatric disease through changes in gene expression and neural function, and individuals with adult versus later onset depression have been successfully discriminated by genome-wide DNA methylation markers [103,104]. Integrating biomeasures (e.g., -omics including epigenomics) in children and older adults alongside environmental (e.g., stress) information will further clarify mechanisms of MDD development at various stages of life and may enable strategies to mitigate risk.
While these findings are novel, this study acknowledges limitations. The top GWAS signals were not genome-wide significant and did not overlap in the PGRN-AMPS and CO-MED studies despite comparable age at depressive onset, sex, and baseline depression levels upon study enrollment. Differences may be attributed to ethnicity differences across cohorts (higher proportion of Hispanic individuals in CO-MED), enrollment criteria differences (PGRN-AMPS patients included all MDD, while CO-MED patients specifically had chronic or recurrent MDD), and/or limited sample sizes to investigate these hypotheses. Findings should therefore be validated in larger cohorts. Age at depressive onset was self-reported without independent verification. The multi-omics signatures of early and adult-onset MDD were derived from samples of adults with MDD. The extent to which these signatures replicate in the context of child and adolescent patient samples must be evaluated. Ideally, as MDD is not a static disorder but rather a disorder that may recur and change, longitudinal studies should exist which collect measures (e.g., metabolomics, neuroimaging) throughout various stages of health and disease to assess the trajectory of the illness. This may also enable identification of ‘hidden nodes’ underlying genomic-metabolomic associations to derive insights into their functional relationships. PGRN-AMPS and CO-MED did not uniformly collect data on environmental contributors to MDD or age-specific risk factors including puberty or menopause. Therefore, the extent of environmental stressors or hormonal influences were unaccounted for in the analyses. This study was performed in Caucasian individuals to maximize sample size and to avoid confounding of genomic findings from differences in minor allele frequency across ancestries. The characterized network signatures should be replicated in larger cohorts and across ancestries to assess the generalizability of these findings. These studies lacked uniform data on the duration of depressive illness and the number of prior depressive episodes at the time of metabolomic assay. Metabolites were not collected in a systematic manner (e.g., under fasting conditions, at a uniform time of day), which may confer noise to the analyses, although fasting status may not significantly impact laboratory variability for most metabolites [105]. The p180 platform does not assay several key psychiatric neurotransmitters, including dopamine and GABA, and the platform was unable to detect serotonin at the quality control threshold employed. Lipids assayed by current mass spectrometry technology may actually reflect sum signals of all isomeric/isobaric compounds having the same parent and daughter ions [49]. Therefore, future studies should validate the identified lipid metabolites with additional assays.
In conclusion, this study identified candidate variants through genome-wide analyses which associate with the development of MDD across the lifespan. Novel multi-omics integration analysis with top GWAS variants and plasma metabolomics enabled characterization of biosignatures of early and adult onset MDD. Such networks serve two purposes: First, they enable hypothesis generation for future mechanistic studies of the development of MDD throughout the lifespan. For example, this may include follow-up investigation of INTU-phosphatidylcholine functional relationships. Second, they demonstrate potential differential genomic regulation of the plasma metabolome by the age at MDD onset. The results here encourage future longitudinal studies to collect and integrate additional -omics (e.g., proteomics, exposomics) and neuroimaging at multiple timepoints to enable deeper biological characterization of MDD. This may ultimately help parse the heterogeneity of MDD and enable insights into biological drivers and protectors of the disease across the lifespan.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm12030412/s1. Figure S1: CO-MED sample inclusion for GWAS and Multi-Omics Integration Analysis; Figure S2: PGRN-AMPS sample inclusion for genomic replication and Multi-Omics Integration Analysis; Figure S3: GWAS QQ-Plot for PGRN-AMPS and CO-MED; Figure S4: GWAS locus zoom plots for variants retained following multi-omics integration analysis; Table S1: Metabolites common to PGRN-AMPS and CO-MED and included in multi-omics integration analysis; Table S2: GWAS associations of SNVs with age of depressive onset in the PGRN-AMPS cohort; Table S3: GWAS associations of SNVs with age of depressive onset in the CO-MED cohort; Table S4: Functional Annotation via DAVID; Table S5: Multi-Omics Integration Network Correlations and p-Values. Reference [106] is cited in the supplementary materials.
Author Contributions
Conceptualization, C.W.G., E.F.B., R.K., W.V.B., P.E.C., and A.P.A.; formal analysis, C.W.G. and A.P.A.; investigation, M.H.T., L.W., R.W., W.V.B., T.M., M.S., T.C., and J.B.; data curation, R.K.-D.; writing—original draft preparation, C.W.G., E.F.B., R.K., A.P.A., W.V.B., and P.E.C.; writing—reviewing and editing, C.W.G., E.F.B., A.P.A., R.W., W.V.B., P.E.C., and M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Harry C. and Debra A. Stonecipher Predoctoral Fellowship at the Mayo Clinic Graduate School of Biomedical Sciences, National Science Foundation (NSF) under grant 2041339; National Institutes of Health (NIH) under grants U19 GM61388, R01 GM028157, R01 AA027486, R01 MH108348, R24 GM078233, RC2 GM092729, U19 AG063744, N01 MH90003, R01 AG04617, U01 AG061359, RF1 AG051550, R01 MH113700 and R01 MH124655; K23AI143882; the Hersh Foundation, the Duke Psychiatry Pharmacometabolomics Center, and The Mayo Clinic Center for Individualized Medicine. The CO-MED study received medications at no cost from Forest Pharmaceuticals, GlaxoSmithKline, Organon, and Wyeth Pharmaceuticals. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the NSF or the NIH.
Institutional Review Board Statement
Both the PGRN-AMPS and CO-MED studies were conducted in accordance with the approval of their respective Institutional Review Boards.
Informed Consent Statement
Informed consent was obtained from all participants.
Data Availability Statement
All raw and analyzed data and related materials, including programming code, are available upon request to Mayo Clinic. PGRN-AMPS data have been deposited on dbGaP, study accession phs000670.v1.p.1.
Conflicts of Interest
Barreto consults for FAST Biomedical and Wolters Kluwer, both unrelated to the present work. L. Wang and R. M. Weinshilboum are co-founders of and stockholders in OneOme, LLC. W. V. Bobo’s research has been supported by the National Institute of Mental Health, the Agency for Healthcare Quality and Research, the National Science Foundation, the Myocarditis Foundation, and the Mayo Foundation for Medical Education and Research. He has contributed chapters to UpToDate concerning the pharmacological treatment of adults with bipolar major depression. P. E. Croarkin has received research grant support from Neuronetics, Inc., NeoSync, Inc., and Pfizer, Inc. He has received grant in-kind (equipment, supply, and genotyping support for research studies) from Assurex Health, Neuronetics, Inc., and MagVenture, Inc. P. E. Croarkin has served as a consultant for Engrail Therapeutics, Myriad Neuroscience, Procter & Gamble, and Sunovion. M. H. Trivedi has provided consulting services to Acadia Pharmaceuticals, Inc., Alkermes Inc, Alto Neuroscience Inc, Axsome Therapeutics, GH Research Limited, GreenLight VitalSign6 Inc, Janssen, Merck Sharp & Dohme Corp., Mind Medicine (MindMed) Inc., Neurocrine Biosciences Inc, Orexo US Inc, Otsuka, SAGE Therapeutics, Signant Health, Titan Pharmaceuticals, Inc. He has received grant/research funding from NIMH, NIDA, NCATS, American Foundation for Suicide Prevention, Patient-Centered Outcomes Research Institute (PCORI), and Blue Cross Blue Shield of Texas. Additionally, he has received editorial compensation from Oxford University Press. T. Carmody has provided consulting services to Alkermes, Inc. R. Kaddurah-Daouk in an inventor on a series of patents on use of metabolomics for the diagnosis and treatment of CNS diseases and holds equity in Metabolon Inc., Chymia LLC and PsyProtix. All other authors declared no competing interests for this work.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zisook S., Lesser I., Stewart J.W., Wisniewski S., Balasubramani G., Fava M., Gilmer W.S., Dresselhaus T.R., Thase M.E., Nierenberg A.A., et al. Effect of Age at Onset on the Course of Major Depressive Disorder. Am. J. Psychiatry. 2007;164:1539–1546. doi: 10.1176/appi.ajp.2007.06101757. [DOI] [PubMed] [Google Scholar]
- 2.Zisook S., Rush A.J., Albala A., Alpert J., Balasubramani G., Fava M., Husain M., Sackeim H., Trivedi M., Wisniewski S. Factors that differentiate early vs. later onset of major depression disorder. Psychiatry Res. 2004;129:127–140. doi: 10.1016/j.psychres.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Weissman M.M., Wickramaratne P., Merikangas K.R., Leckman J.F., Prusoff B.A., Caruso K.A., Kidd K.K., Gammon G.D. Onset of Major Depression in Early Adulthood. Arch. Gen. Psychiatry. 1984;41:1136–1143. doi: 10.1001/archpsyc.1984.01790230022003. [DOI] [PubMed] [Google Scholar]
- 4.Nierenberg A.A., Trivedi M.H., Fava M., Biggs M.M., Shores-Wilson K., Wisniewski S.R., Balasubramani G., Rush A.J. Family history of mood disorder and characteristics of major depressive disorder: A STAR∗D (sequenced treatment alternatives to relieve depression) study. J. Psychiatr. Res. 2007;41:214–221. doi: 10.1016/j.jpsychires.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tozzi F., Prokopenko I., Perry J.D., Kennedy J.L., McCarthy A.D., Holsboer F., Berrettini W., Middleton L.T., Chilcoat H.D., Muglia P. Family history of depression is associated with younger age of onset in patients with recurrent depression. Psychol. Med. 2008;38:641–649. doi: 10.1017/S0033291707002681. [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulos G.S., Young R., Abrams R.C., Meyers B., Shamoian A.C. Chronicity and relapse in geriatric depression. Biol. Psychiatry. 1989;26:551–564. doi: 10.1016/0006-3223(89)90080-2. [DOI] [PubMed] [Google Scholar]
- 7.Sheehan D.V., Lecrubier Y., Sheehan K.H., Amorim P., Janavs J., Weiller E., Hergueta T., Balker R., Dunbar G.C. The Mini-International Neuropsychiatric Interview (M.I.N.I.): The development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry. 1998;59((Suppl. S20)):22–33. [PubMed] [Google Scholar]
- 8.Shi J., Potash J.B., Knowles A.J., Weissman M., Coryell W., Scheftner A.W., Lawson W., DePaulo J.R., Gejman P.V., Sanders A.R., et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol. Psychiatry. 2010;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan P.F., Neale M.C., Kendler K.S. Genetic Epidemiology of Major Depression: Review and Meta-Analysis. Am. J. Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X., Liu L., Lan X., Cohen D., Zhang Y., Ravindran A.V., Yuan S., Zheng P., Coghill D., Yang L., et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol. Psychiatry. 2019;24:1478–1488. doi: 10.1038/s41380-018-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Psychological Association Clinical Practice Guideline for the Treatment of Depression Across Three Age Cohorts. [(accessed on 2 March 2022)]. Available online: https://www.apa.org/depression-guideline.
- 12.Power R.A., Tansey K.E., Buttenschøn H.N., Cohen-Woods S., Bigdeli T., Hall L.S., Kutalik Z., Lee S.H., Ripke S., Steinberg S., et al. Genome-wide Association for Major Depression Through Age at Onset Stratification: Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Biol. Psychiatry. 2017;81:325–335. doi: 10.1016/j.biopsych.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Power R., Keers R., Ng M.Y., Butler A.W., Uher R., Cohen-Woods S., Ising M., Craddock N., Owen M.J., Korszun A., et al. Dissecting the Genetic Heterogeneity of Depression Through Age at Onset. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2012;159:859–868. doi: 10.1002/ajmg.b.32093. [DOI] [PubMed] [Google Scholar]
- 15.Kang H.-J., Kim K.-T., Park Y., Yoo K.-H., Kim J.-W., Lee J.-Y., Kim S.-W., Shin I.-S., Kim J.-M. Genetic markers for depressive disorders with earlier age at onset. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2021;108:110176. doi: 10.1016/j.pnpbp.2020.110176. [DOI] [PubMed] [Google Scholar]
- 16.Rice F., Riglin L., Thapar A.K., Heron J., Anney R., O’Donovan M.C., Thapar A. Characterizing Developmental Trajectories and the Role of Neuropsychiatric Genetic Risk Variants in Early-Onset Depression. JAMA Psychiatry. 2019;76:306–313. doi: 10.1001/jamapsychiatry.2018.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong A.S.F., López J., Hammerton G., Manley D., Timpson N., Leckie G., Pearson R.M. Genetic and Environmental Risk Factors Associated with Trajectories of Depression Symptoms from Adolescence to Young Adulthood. JAMA Netw. Open. 2019;2:e196587. doi: 10.1001/jamanetworkopen.2019.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotta L.A., MacTel Consortium. Pietzner M., Stewart I.D., Wittemans L.B.L., Li C., Bonelli R., Raffler J., Biggs E.K., Oliver-Williams C., et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat. Genet. 2021;53:54–64. doi: 10.1038/s41588-020-00751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin S.-Y., The Multiple Tissue Human Expression Resource (MuTHER) Consortium. Fauman E., Petersen A.-K., Krumsiek J., Santos R., Huang J., Arnold M., Erte I., Forgetta V., et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyce J.B., Grant C.W., Liu D., MahmoudianDehkordi S., Kaddurah-Daouk R., Skime M., Biernacka J., Frye M.A., Mayes T., Carmody T., et al. Multi-omics driven predictions of response to acute phase combination antidepressant therapy: A machine learning approach with cross-trial replication. Transl. Psychiatry. 2021;11:1–11. doi: 10.1038/s41398-021-01632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen T.T.L., Liu D., Ho M.-F., Athreya A.P., Weinshilboum R. Selective Serotonin Reuptake Inhibitor Pharmaco-Omics: Mechanisms and Prediction. Front. Pharmacol. 2021;11:614048. doi: 10.3389/fphar.2020.614048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ressler K.J., Williams L.M. Big data in psychiatry: Multiomics, neuroimaging, computational modeling, and digital phenotyping. Neuropsychopharmacol. 2021;46:1–2. doi: 10.1038/s41386-020-00862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentea E., Depasquale E.A.K., O’Donovan S.M., Sullivan C.R., Simmons M., Meador-Woodruff J.H., Zhou Y., Xu C., Bai B., Peng J., et al. Kinase network dysregulation in a human induced pluripotent stem cell model of DISC1 schizophrenia. Mol. Omics. 2019;15:173–188. doi: 10.1039/C8MO00173A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Athreya A., Iyer R., Neavin D., Wang L., Weinshilboum R., Kaddurah-Daouk R., Rush J., Frye M., Bobo W. Augmentation of Physician Assessments with Multi-Omics Enhances Predictability of Drug Response: A Case Study of Major Depressive Disorder. IEEE Comput. Intell. Mag. 2018;13:20–31. doi: 10.1109/MCI.2018.2840660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Athreya A.P., Neavin D., Carrillo-Roa T., Skime M., Biernacka J., Frye M.A., Rush A.J., Wang L., Binder E.B., Iyer R.K., et al. Pharmacogenomics-Driven Prediction of Antidepressant Treatment Outcomes: A Machine-Learning Approach with Multi-trial Replication. Clin. Pharmacol. Ther. 2019;106:855–865. doi: 10.1002/cpt.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta M., Neavin D., Liu D., Biernacka J., Hall-Flavin D., Bobo W.V., Frye A.M., Skime M., Jenkins G.D., Batzler A., et al. TSPAN5, ERICH3 and selective serotonin reuptake inhibitors in major depressive disorder: Pharmacometabolomics-informed pharmacogenomics. Mol. Psychiatry. 2016;21:1717–1725. doi: 10.1038/mp.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neavin D., Kaddurah-Daouk R., Weinshilboum R. Pharmacometabolomics informs pharmacogenomics. Metabolomics. 2016;12:121. doi: 10.1007/s11306-016-1066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shukla R., Henkel N.D., Alganem K., Hamoud A.-R., Reigle J., Alnafisah R.S., Eby H.M., Imami A.S., Creeden J., Miruzzi S.A., et al. Signature-based approaches for informed drug repurposing: Targeting CNS disorders. Neuropsychopharmacology. 2021;46:116–130. doi: 10.1038/s41386-020-0752-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barabási A.-L., Gulbahce N., Loscalzo J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2010;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uppal K., Ma C., Go Y.-M., Jones D.P. xMWAS: A data-driven integration and differential network analysis tool. Bioinformatics. 2018;34:701–702. doi: 10.1093/bioinformatics/btx656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y., Biernacka J.M., Hebbring S., Chai Y., Jenkins G.D., Batzler A., Snyder K.A., Drews M.S., Desta Z., Flockhart D., et al. Pharmacogenomics of selective serotonin reuptake inhibitor treatment for major depressive disorder: Genome-wide associations and functional genomics. Pharmacogenomics J. 2012;13:456–463. doi: 10.1038/tpj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rush A.J., Trivedi M.H., Stewart J.W., Nierenberg A.A., Fava M., Kurian B.T., Warden D., Morris D.W., Luther J.F., Husain M.M., et al. Combining Medications to Enhance Depression Outcomes (CO-MED): Acute and Long-Term Outcomes of a Single-Blind Randomized Study. Am. J. Psychiatry. 2011;168:689–701. doi: 10.1176/appi.ajp.2011.10111645. [DOI] [PubMed] [Google Scholar]
- 33.Trivedi M.H., Rush A., Ibrahim H.M., Carmody T.J., Biggs M.M., Suppes T., Crismon M.L., Shores-Wilson K., Toprac M.G., Dennehy E.B., et al. The Inventory of Depressive Symptomatology, Clinician Rating (IDS-C) and Self-Report (IDS-SR), and the Quick Inventory of Depressive Symptomatology, Clinician Rating (QIDS-C) and Self-Report (QIDS-SR) in public sector patients with mood disorders: A psychometric evaluation. Psychol. Med. 2004;34:73–82. doi: 10.1017/s0033291703001107. [DOI] [PubMed] [Google Scholar]
- 34.Ji Y., Schaid D.J., Desta Z., Kubo M., Batzler A.J., Snyder K., Mushiroda T., Kamatani N., Ogburn E., Hall-Flavin D., et al. Citalopram and escitalopram plasma drug and metabolite concentrations: Genome-wide associations. Br. J. Clin. Pharmacol. 2014;78:373–383. doi: 10.1111/bcp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadad B., Raj P.D., Jha M.K., Carmody T., Dozmorov I., Mayes T.L., Wakeland E.K., Trivedi M.H. Association of Novel ALX4 Gene Polymorphisms with Antidepressant Treatment Response: Findings from the CO-MED Trial. Mol. Neuropsychiatry. 2018;4:7–19. doi: 10.1159/000487321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., Reshef Y.A., Finucane H.K., Schoenherr S., Forer L., McCarthy S., Abecasis C.F.G.R., et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miclaus K., Wolfinger R., Czika W. SNP selection and multidimensional scaling to quantify population structure. Genet. Epidemiol. 2009;33:488–496. doi: 10.1002/gepi.20401. [DOI] [PubMed] [Google Scholar]
- 39.Bush W.S., Moore J.H. Chapter 11: Genome-Wide Association Studies. PLOS Comput. Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luby J.L., Heffelfinger A.K., Mrakotsky C., Brown K.M., Hessler M.J., Wallis J.M., Spitznagel E.L. The Clinical Picture of Depression in Preschool Children. J. Am. Acad. Child Adolesc. Psychiatry. 2003;42:340–348. doi: 10.1097/00004583-200303000-00015. [DOI] [PubMed] [Google Scholar]
- 41.Luby J.L., Belden A., Pautsch J., Si X., Spitznagel E. The clinical significance of preschool depression: Impairment in functioning and clinical markers of the disorder. J. Affect. Disord. 2009;112:111–119. doi: 10.1016/j.jad.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egger H.L., Angold A. Common emotional and behavioral disorders in preschool children: Presentation, nosology, and epidemiology. J. Child Psychol. Psychiatry. 2006;47:313–337. doi: 10.1111/j.1469-7610.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- 43.Bufferd S.J., Dougherty L.R., Carlson G.A., Rose S., Klein D.N. Psychiatric Disorders in Preschoolers: Continuity from Ages 3 to 6. Am. J. Psychiatry. 2012;169:1157–1164. doi: 10.1176/appi.ajp.2012.12020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boughton A.P., Welch R.P., Flickinger M., VandeHaar P., Taliun D., Abecasis G.R., Boehnke M. LocusZoom.js: Interactive and embeddable visualization of genetic association study results. Bioinformatics. 2021;37:3017–3018. doi: 10.1093/bioinformatics/btab186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward L.D., Kellis M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhbannikov I.Y., Arbeev K., Ukraintseva S., Yashin A.I. haploR: An R package for querying web-based annotation tools. F1000Res. 2017;6:97. doi: 10.12688/f1000research.10742.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 49.Biocrates Life Sciences Absolute IDQ p180kit. Analytical Specifications p180 (AS-p180-2 for Research Use) Innsbruck: Biocrates Life Sciences. [(accessed on 2 March 2022)]. Available online: https://biocrates.com/absoluteidq-p180-kit/
- 50.Ahmed A.T., Mahmoudian Dehkordi S., Bhattacharyya S., Arnold M., Liu D., Neavin D., Moseley M.A., Thompson J.W., Williams L.S.J., Louie G., et al. Acylcarnitine metabolomic profiles inform clinically-defined major depressive phenotypes. J. Affect. Disord. 2020;264:90–97. doi: 10.1016/j.jad.2019.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharyya S., Dunlop B.W., Mahmoudiandehkordi S., Ahmed A.T., Louie G., Frye M.A., Weinshilboum R.M., Krishnan R.R., Rush A.J., Mayberg H.S., et al. Pilot Study of Metabolomic Clusters as State Markers of Major Depression and Outcomes to CBT Treatment. Front. Neurosci. 2019;13:926. doi: 10.3389/fnins.2019.00926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Czysz A.H., South C., Gadad B., Arning E., Soyombo A., Bottiglieri T., Trivedi M.H. Can targeted metabolomics predict depression recovery? Results from the CO-MED trial. Transl. Psychiatry. 2019;9:1–11. doi: 10.1038/s41398-018-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parksepp M., Leppik L., Koch K., Uppin K., Kangro R., Haring L., Vasar E., Zilmer M. Metabolomics approach revealed robust changes in amino acid and biogenic amine signatures in patients with schizophrenia in the early course of the disease. Sci. Rep. 2020;10:13983. doi: 10.1038/s41598-020-71014-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balõtšev R., Haring L., Koido K., Leping V., Kriisa K., Zilmer M., Vasar V., Piir A., Lang A., Vasar E. Antipsychotic treatment is associated with inflammatory and metabolic biomarkers alterations among first-episode psychosis patients: A 7-month follow-up study. Early Interv. Psychiatry. 2017;13:101–109. doi: 10.1111/eip.12457. [DOI] [PubMed] [Google Scholar]
- 55.Kriisa K., Leppik L., Balõtšev R., Ottas A., Soomets U., Koido K., Volke V., Innos J., Haring L., Vasar E., et al. Profiling of Acylcarnitines in First Episode Psychosis before and after Antipsychotic Treatment. J. Proteome Res. 2017;16:3558–3566. doi: 10.1021/acs.jproteome.7b00279. [DOI] [PubMed] [Google Scholar]
- 56.MahmoudianDehkordi S., The Mood Disorders Precision Medicine Consortium (MDPMC) Ahmed A.T., Bhattacharyya S., Han X., Baillie R.A., Arnold M., Skime M.K., John-Williams L.S., Moseley M.A., et al. Alterations in acylcarnitines, amines, and lipids inform about the mechanism of action of citalopram/escitalopram in major depression. Transl. Psychiatry. 2021;11:1–14. doi: 10.1038/s41398-020-01097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wishart D.S. Metabolomics for Investigating Physiological and Pathophysiological Processes. Physiol. Rev. 2019;99:1819–1875. doi: 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- 59.Lê Cao K.-A., Boitard S., Besse P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinform. 2011;12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008;2008:10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 61.R Code Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2020. [Google Scholar]
- 62.RStudio . RStudio: Integrated Development for R. RStudio, PBC; Boston, MA, USA: 2020. [Google Scholar]
- 63.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewis C.M., Ng M.Y., Butler A.W., Cohen-Woods S., Uher R., Pirlo K., Weale M., Schosser A., Paredes U.M., Rivera M., et al. Genome-Wide Association Study of Major Recurrent Depression in the U.K. Population. Am. J. Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- 65.Sullivan P.F., de Geus E.J.C., Willemsen G., James M.R., Smit J.H., Zandbelt T., Arolt V., Baune B.T., Blackwood D., Cichon S., et al. Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol. Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wray N.R., Pergadia M.L., Blackwood D., Penninx B.W.J.H., Gordon S.L., Nyholt D., Ripke S., MacIntyre D.J., McGhee A.K., MacLean A., et al. Genome-wide association study of major depressive disorder: New results, meta-analysis, and lessons learned. Mol. Psychiatry. 2010;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu D., Ray B., Neavin D.R., Zhang J., Athreya A.P., Biernacka J.M., Bobo W.V., Hall-Flavin D.K., Skime M.K., Zhu H., et al. Beta-defensin 1, aryl hydrocarbon receptor and plasma kynurenine in major depressive disorder: Metabolomics-informed genomics. Transl. Psychiatry. 2018;8:1–13. doi: 10.1038/s41398-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu D., Zhuang Y., Zhang L., Gao H., Neavin D., Carrillo-Roa T., Wang Y., Yu J., Qin S., Kim D.C., et al. ERICH3: Vesicular association and antidepressant treatment response. Mol. Psychiatry. 2021;26:2415–2428. doi: 10.1038/s41380-020-00940-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levey D.F., Stein M.B., Wendt F.R., Pathak G.A., Zhou H., Aslan M., Quaden R., Harrington K.M., Nuñez Y.Z., Overstreet C., et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci. 2021;24:954–963. doi: 10.1038/s41593-021-00860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.GENDEP Investigators. Uher R., Tansey K., Henigsberg N., Wolfgang M., Mors O., Hauser J., Placentino A., Souery D., Farmer A., et al. Common Genetic Variation and Antidepressant Efficacy in Major Depressive Disorder: A Meta-Analysis of Three Genome-Wide Pharmacogenetic Studies. Am. J. Psychiatry. 2013;170:207–217. doi: 10.1176/appi.ajp.2012.12020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cole M.G., Dendukuri N. Risk Factors for Depression Among Elderly Community Subjects: A Systematic Review and Meta-Analysis. Am. J. Psychiatry. 2003;160:1147–1156. doi: 10.1176/appi.ajp.160.6.1147. [DOI] [PubMed] [Google Scholar]
- 72.Arnett J.J. Emerging adulthood. A theory of development from the late teens through the twenties. Am. Psychol. 2000;55:469–480. doi: 10.1037/0003-066X.55.5.469. [DOI] [PubMed] [Google Scholar]
- 73.Larson N.I., Neumark-Sztainer D., Story M.T., Wall M.M., Harnack L.J., Eisenberg M.E. Fast Food Intake: Longitudinal Trends during the Transition to Young Adulthood and Correlates of Intake. J. Adolesc. Health. 2008;43:79–86. doi: 10.1016/j.jadohealth.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 74.Winpenny E.M., Winkler M.R., Stochl J., Van Sluijs E.M.F., Larson N., Neumark-Sztainer D. Associations of early adulthood life transitions with changes in fast food intake: A latent trajectory analysis. Int. J. Behav. Nutr. Phys. Act. 2020;17:1–11. doi: 10.1186/s12966-020-01024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dean J.M., Lodhi I.J. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mozaffarian D., Kabagambe E.K., Johnson O.C., Lemaitre R.N., Manichaikul A., Sun Q., Foy M., Wang L., Wiener H., Irvin M.R., et al. Genetic loci associated with circulating phospholipid trans fatty acids: A meta-analysis of genome-wide association studies from the CHARGE Consortium. Am. J. Clin. Nutr. 2015;101:398–406. doi: 10.3945/ajcn.114.094557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toriyama M., University of Washington Center for Mendelian Genomics. Lee C., Taylor S.P., Duran I., Cohn D.H., Bruel A.-L., Tabler J.M., Drew K., Kelly M., et al. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat. Genet. 2016;48:648–656. doi: 10.1038/ng.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adler P.N., Wallingford J.B. From Planar Cell Polarity to Ciliogenesis and Back: The Curious Tale of the PPE and CPLANE proteins. Trends Cell Biol. 2017;27:379–390. doi: 10.1016/j.tcb.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Butler M., Wallingford M.T.B.J.B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 2017;18:375–388. doi: 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guemez-Gamboa A., Coufal N., Gleeson J.G. Primary Cilia in the Developing and Mature Brain. Neuron. 2014;82:511–521. doi: 10.1016/j.neuron.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adler P.N. Planar Signaling and Morphogenesis in Drosophila. Dev. Cell. 2002;2:525–535. doi: 10.1016/S1534-5807(02)00176-4. [DOI] [PubMed] [Google Scholar]
- 82.Peng Y., Axelrod J.D. Asymmetric Protein Localization in Planar Cell Polarity. Planar Cell Polarity During Dev. 2012;101:33–53. doi: 10.1016/b978-0-12-394592-1.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simons M., Mlodzik M. Planar Cell Polarity Signaling: From Fly Development to Human Disease. Annu. Rev. Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wallingford J.B. Planar Cell Polarity and the Developmental Control of Cell Behavior in Vertebrate Embryos. Annu. Rev. Cell Dev. Biol. 2012;28:627–653. doi: 10.1146/annurev-cellbio-092910-154208. [DOI] [PubMed] [Google Scholar]
- 85.Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L.B., Christensen S.T. Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 2019;15:199–219. doi: 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guo J., Otis J.M., Higginbotham H., Monckton C., Cheng J., Asokan A., Mykytyn K., Caspary T., Stuber G.D., Anton E. Primary Cilia Signaling Shapes the Development of Interneuronal Connectivity. Dev. Cell. 2017;42:286–300. doi: 10.1016/j.devcel.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mykytyn K., Askwith C. G-Protein-Coupled Receptor Signaling in Cilia. Cold Spring Harb. Perspect. Biol. 2017;9:a028183. doi: 10.1101/cshperspect.a028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nechipurenko I.V. The Enigmatic Role of Lipids in Cilia Signaling. Front. Cell Dev. Biol. 2020;8:777. doi: 10.3389/fcell.2020.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tayyab M., Shahi M.H., Farheen S., Mariyath M.P., Khanam N., Castresana J.S., Hossain M.M. Sonic hedgehog, Wnt, and brain-derived neurotrophic factor cell signaling pathway crosstalk: Potential therapy for depression. J. Neurosci. Res. 2018;96:53–62. doi: 10.1002/jnr.24104. [DOI] [PubMed] [Google Scholar]
- 90.Duman R.S., Voleti B. Signaling pathways underlying the pathophysiology and treatment of depression: Novel mechanisms for rapid-acting agents. Trends Neurosci. 2012;35:47–56. doi: 10.1016/j.tins.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo J., Higginbotham H., Li J., Nichols J., Hirt J., Ghukasyan V., Anton E. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat. Commun. 2015;6:7857. doi: 10.1038/ncomms8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waters A.M., Beales P.L. Ciliopathies: An expanding disease spectrum. Pediatr. Nephrol. 2011;26:1039–1056. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karunakaran K.B., Chaparala S., Lo C.W., Ganapathiraju M.K. Cilia interactome with predicted protein–protein interactions reveals connections to Alzheimer’s disease, aging and other neuropsychiatric processes. Sci. Rep. 2020;10:1–16. doi: 10.1038/s41598-020-72024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karam C.S., Ballon J.S., Bivens N.M., Freyberg Z., Girgis R.R., Lizardi-Ortiz J.E., Markx S., Lieberman J.A., Javitch J.A. Signaling pathways in schizophrenia: Emerging targets and therapeutic strategies. Trends Pharmacol. Sci. 2010;31:381–390. doi: 10.1016/j.tips.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shadrin A.A., Kaufmann T., van der Meer D., Palmer C.E., Makowski C., Loughnan R., Jernigan T.L., Seibert T.M., Hagler D.J., Smeland O.B., et al. Vertex-wise multivariate genome-wide association study identifies 780 unique genetic loci associated with cortical morphology. NeuroImage. 2021;244:118603. doi: 10.1016/j.neuroimage.2021.118603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suh J.S., Schneider M.A., Minuzzi L., MacQueen G.M., Strother S.C., Kennedy S.H., Frey B.N. Cortical thickness in major depressive disorder: A systematic review and meta-analysis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2019;88:287–302. doi: 10.1016/j.pnpbp.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 97.Li Q., Zhao Y., Chen Z., Long J., Dai J., Huang X., Lui S., Radua J., Vieta E., Kemp G.J., et al. Meta-analysis of cortical thickness abnormalities in medication-free patients with major depressive disorder. Neuropsychopharmacology. 2020;45:703–712. doi: 10.1038/s41386-019-0563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou Z., Wang K., Tang J., Wei D., Song L., Peng Y., Fu Y., Qiu J. Cortical thickness distinguishes between major depression and schizophrenia in adolescents. BMC Psychiatry. 2021;21:1–9. doi: 10.1186/s12888-021-03373-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jaworska N., MacMaster F., Gaxiola I., Cortese F., Goodyear B., Ramasubbu R. A Preliminary Study of the Influence of Age of Onset and Childhood Trauma on Cortical Thickness in Major Depressive Disorder. BioMed Res. Int. 2014;2014:1–9. doi: 10.1155/2014/410472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Filfan M., Olaru A., Udristoiu I., Margaritescu C., Petcu E., Hermann D.M., Popa-Wagner A. Long-term treatment with spermidine increases health span of middle-aged Sprague-Dawley male rats. GeroScience. 2020;42:937–949. doi: 10.1007/s11357-020-00173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fiori L.M., Turecki G. Implication of the polyamine system in mental disorders. J. Psychiatry Neurosci. 2008;33:102–110. [PMC free article] [PubMed] [Google Scholar]
- 102.Soga T., Teo C.H., Parhar I. Genetic and Epigenetic Consequence of Early-Life Social Stress on Depression: Role of Serotonin-Associated Genes. Front. Genet. 2021;11:601868. doi: 10.3389/fgene.2020.601868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Archer T., Beninger R.J., Palomo T., Kostrzewa R.M. Epigenetics and Biomarkers in the Staging of Neuropsychiatric Disorders. Neurotox. Res. 2010;18:347–366. doi: 10.1007/s12640-010-9163-5. [DOI] [PubMed] [Google Scholar]
- 104.Yamagata H., Ogihara H., Matsuo K., Uchida S., Kobayashi A., Seki T., Kobayashi M., Harada K., Chen C., Miyata S., et al. Distinct epigenetic signatures between adult-onset and late-onset depression. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-81758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Townsend M.K., Clish C.B., Kraft P., Wu C., Souza A.L., Deik A.A., Tworoger S.S., Wolpin B.M. Reproducibility of Metabolomic Profiles among Men and Women in 2 Large Cohort Studies. Clin. Chem. 2013;59:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. Systematic Identification of Trans Eqtls as Putative Drivers of Known Disease Associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and analyzed data and related materials, including programming code, are available upon request to Mayo Clinic. PGRN-AMPS data have been deposited on dbGaP, study accession phs000670.v1.p.1.