Abstract
Varicella-zoster virus (VZV) is a common herpesvirus responsible for disseminated or chronic infections in immunocompromised patients. Effective drugs such as acyclovir (ACV), famciclovir (prodrug of penciclovir), and foscarnet are available to treat these infections. Here we report the phenotypic and genetic characterization of four ACV-resistant VZV strains isolated from AIDS patients and transplant recipients. Sensitivity to six antiviral drugs was determined by an enzyme-linked immunosorbent assay, viral thymidine kinase (TK) activity was measured by comparing [3H]thymidine and 1-β-d-arabinofuranosyl-[3H]thymine as substrates, and the TK gene open reading frame was sequenced. Three strains were found to be TK deficient, and the fourth was a mixed population composed of TK-positive and TK-deficient viruses. Each strain presented a unique TK gene mutation that could account for ACV resistance. In one strain, the deletion of two nucleotides at codon 215 induced a premature stop signal at codon 217. In another strain, a single nucleotide addition at codon 167 resulted in a premature stop signal at codon 206. In both other strains, we identified amino acid substitutions already described in other ACV-resistant VZV strains: either Glu→Gly at residue 48 or Arg→Gly at residue 143. According to our work and data previously reported on resistant VZV strains, there are three areas in the TK gene where 71% of the mutations described to date are located. These areas are putative candidates for a genotypic diagnosis of ACV resistance.
Varicella-zoster virus (VZV) is a widespread herpesvirus which is responsible for both a primary disease (varicella, or chickenpox) and a recurrent one (zoster, or shingles) following reactivation of the virus. VZV infections are associated with significant morbidity and mortality among immunocompromised patients because of either disseminated infections or chronic reactivations (17, 24). It is now well established that zoster can be an early sign of AIDS. The prognosis of these infections has completely changed since the introduction of antiviral treatments.
Among antiviral drugs available to treat these infections, acyclovir (ACV) remains the drug of preference (1). Penciclovir (PCV) and sorivudine (BVaraU) are two other nucleoside analogues which are effective in vitro on VZV; however, BVaraU toxicity has impaired its clinical use. These drugs act as triphosphates by competitive inhibition of viral DNA polymerase; in addition, ACV triphosphate is a DNA chain terminator. Their phosphorylation is dependent on VZV deoxypyrimidine kinase which possesses both thymidine and thymidylate phosphorylating activities. The viral thymidine kinase (TK) activity is involved in the first phosphorylation step of most nucleoside analogues such as ACV, PCV, and BVaraU, whereas the viral thymidylate kinase activity is required for the second phosphorylation step in only a few molecules such as BVaraU and bromovinyldeoxyuridine (BVDU). The next phosphorylations are achieved by cellular kinases. Foscarnet (phosphonoformic acid) is an antiviral drug that acts directly on viral DNA polymerase by impeding pyrophosphate release from deoxynucleotides during DNA synthesis (3). In recent years, a new class of compounds including cidofovir, (S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine, has been shown to exhibit antiviral activity on a broad range of DNA viruses including VZV. These compounds are acyclic nucleoside phosphonates which are not phosphorylated by the virus-encoded TK but by cellular kinases (6).
ACV-resistant VZV clinical strains have been reported, albeit rarely. They have mostly been described in AIDS patients (2, 10, 15, 19, 21, 24, 26). As for herpes simplex virus, VZV resistance has been detected in strains with deficient TK activity (TKD) or, less often, with TK having an altered substrate specificity (TKalt) (23). TK-deficient ACV-resistant mutants are usually cross resistant to other drugs dependent on viral TK activity. Resistance to ACV associated with an altered DNA polymerase has also been described (19).
Our aim was to fully characterize four ACV-resistant VZV clinical strains and compare their phenotypic and genetic features with those of previously described strains. This comparison has shown that genetic changes (deletions or substitutions) occur in putative hot spots that may become specific targets for the molecular screening of ACV-resistant VZV strains.
(Results of this study were presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, September 1998, in San Diego, Calif. [17a].)
MATERIALS AND METHODS
VZV isolates and strains.
The three ACV-sensitive reference strains were Ellen (ATCC VR-586), Oka (ATCC VR-795), and D9507 (our laboratory reference strain, given by Ulrich Krech, St. Gallen, Switzerland).
Four ACV-resistant VZV isolates were characterized. Two strains, WStr and WMad, were kindly supplied by P. Collins (Wellcome Laboratory, Beckenham, Great Britain); they were isolated from a transplant recipient and a human immunodeficiency virus-positive patient, respectively, both affected with zoster and previously treated with ACV. W. Wunderli (Geneva, Switzerland) kindly provided us with the SW689 isolate, which was isolated from the cerebrospinal fluid of an AIDS patient in 1994 presenting with disseminated skin lesions associated with outer retinal necrosis (28). LYON6625 was isolated in 1993 in our laboratory from the vesicular fluid of an AIDS patient suffering from disseminated zoster and repeatedly treated with ACV.
All isolates were cultivated in human diploid embryonic fibroblasts (MRC-5), by using cell-associated virus as an inoculum. When specified, virus isolates were plaque purified in MRC-5 cells with limited dilutions of cell-free virus.
Antiviral susceptibility studies.
We evaluated the sensitivities of the isolates to six antiviral agents: ACV (Glaxo Wellcome), PCV (SmithKline Beecham), ganciclovir (GCV; Roche), BVDU (Sigma), arabinofuranosylthymine (AraT; Sigma), and foscarnet (phosphonoformic acid; Astra).
Assays were performed in duplicate on MRC-5 cells seeded in 96-well microplates. We used a chessboard titration of each virus strain, allowing simultaneous titration of the virus both with various concentrations of antiviral drug and without antiviral drug. Dilutions of virus and drugs were prepared in Eagle’s minimum essential medium supplemented with 2% fetal calf serum. Cells were infected with five 10-fold dilutions of cell-associated virus. Each dilution was incubated with a series of six concentrations of each antiviral drug (from 1.2 to 300 μM in threefold dilutions for ACV, PCV, and GCV; from 0.49 to 500 μM in fourfold dilutions for cidofovir; from 0.16 to 40 μM in threefold dilutions for AraT; from 0.008 to 2 μM in threefold dilutions for BVDU; and from 15.6 to 500 μM in twofold dilutions for foscarnet).
Cycloheximide was used at a concentration of 2 μg/ml to completely inhibit viral replication, and a control of virus titer with no antiviral drug was also included. After 3 days at 36°C with 5% CO2, viral multiplication was checked by an enzyme-linked immunosorbent assay. Briefly, cells were fixed with 0.1% glutaraldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature. Cells were washed three times in PBS. The detection antibodies used were from a pool of human sera from patients presenting with zoster and were previously tested to determine the optimal dilution (usually 1/4,000) prepared in PBS containing 10% fetal calf serum. One hundred microliters was added to each well for 1 h at 37°C. After three washes with PBS, a peroxidase-conjugated goat anti-human immunoglobulin (Argène-Biosoft) was added (diluted 1/4,000 in PBS–10% fetal calf serum; 100 μl per well) for 30 min at 37°C. Microplates were then washed three times in PBS. The substrate, 2,2′-azinobis-3-ethylbenzthiazolinesulfonic acid (ABTS) diluted in ABTS buffer (Boehringer, Mannheim, Germany), was added to each well (100 μl). After 30 min at room temperature, the plates were briefly agitated and optical densities were read at 405 nm with a multichannel spectrophotometer (Titertek-Multiskan). Optical density values were analyzed with the Biolise program (Life Sciences International) to calculate the concentration of the drug causing a 50% inhibition of viral replication (IC50) by logistic regression analysis as described previously (13).
TK assays.
Viral stocks were prepared on TK-deficient HeLa cells (resistant to 100 μM bromodeoxyuridine; ATCC CCL2). These cells were infected with cell-associated virus. After 3 days of incubation at 36°C, the cells were trypsinized and centrifuged at 1,100 × g for 10 min at 4°C. The viral titer of each stock was determined on MRC-5 cells. Cell pellets were stored at −80°C until extraction.
Enzyme extraction was carried out at 4°C. A calibrated suspension of infected cells (20 × 106 cells/500 μl) was prepared in an appropriate extraction buffer (50 mM Tris-HCl [pH 8], 50 mM KCl, 1 mM MgCl2, 1 mM ATP, 1 mM dithiothreitol, and 5% glycerol). Cells were subsequently sonicated for three periods of 30 s at 50 mV. The suspensions were then centrifuged at 120,000 × g for 60 min at 4°C. The protein concentration of each enzyme extract was determined by the Lowry method (Bio-Rad protein assay). Crude extracts were aliquoted and stored at −80°C.
TK enzymatic activity was evaluated with two substrates: [3H]thymidine (33 Ci/mmol and 1 mCi/ml; ICN Pharmaceuticals, Inc.) and 1-β-d-arabinofuranosyl-[3H]thymine ([3H]araT) (3.1 Ci/mmol and 2 mCi/ml; Amersham Life Science), as previously described (7). Briefly, 150 μl of crude extracts was incubated at 37°C with 150 μl of substrate medium (2×) containing 150 mM phosphate buffer (pH 7.6), 20 mM ATP, 20 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol, and 10 mM NaF. The mixture was incubated with either 50 μM thymidine plus [3H]thymidine (2.5 μCi per 50 μl of buffer) or [3H]araT (2.5 μCi per 50 μl of buffer). The phosphorylation levels of [3H]thymidine or [3H]araT were determined in duplicate at three time points (15, 30, and 60 min) by spotting 40 μl of the reaction mixture on DEAE paper disks (DE81; Whatman). DEAE papers were subsequently washed in ammonium formate and ethanol. Liquid scintillation counting (UltimaGold MV; Packard) was done on dried DEAE papers.
TK gene sequencing.
VZV strains were cultivated on MRC-5 cells until a 70 to 100% cytopathic effect was observed. The infected cells were then harvested by trypsinization and rinsed once with PBS. After centrifugation, cell pellets were immediately used or stored at −80°C until DNA extraction. Cells were thawed and resuspended in Tris-EDTA buffer (10 mM Tris base and 1 mM EDTA). Cells were lysed by adding an equal volume of TENS lysis buffer (40 mM Tris, 40 mM EDTA, 300 mM NaCl, 2% Sarkosyl). DNA was extracted after 1 h at 55°C with proteinase K (final concentration of 200 μg/ml). DNA was purified with a standard phenol and chloroform-isoamylic alcohol protocol and precipitated with ethanol; the DNA pellet was dissolved in water and stored at −20°C.
The whole TK gene was amplified with two PCRs which amplified two overlapping fragments. The first one started 43 nucleotides before the ATG initiation codon and ended at nucleotide 521, with the primers TKA (5′-GGGGATCCCCGTCCCAGAAGATAAC) and TKE (3′-CAAAGTGAGGGGTCAGTCCTAGGGG). The second region started at nucleotide 471 and ended 67 nucleotides after the stop codon, with the primers TKB (5′-GGGGATCCCGGCGCTTCCTGGGTTA) and TKF (3′-CAGTATGAGCGCACATGCCTAGGGG).
PCR was carried out with a DNA thermal cycler (Hybaid) under the following conditions: five cycles at 94°C for 20 s, 55°C for 30 s, and 75°C for 5 min, followed by 30 cycles at 94°C for 20 s, 55°C for 30 s, and 75°C for 1 min, with a final extension step at 75°C for 5 min.
The size of the amplified DNA was checked under UV light after electrophoresis in a low-melting-point agarose gel. The expected band was excised and put in 100 μl of water. After dissolving the agarose at 65°C for 15 min, the DNA was purified with a phenol and chloroform-isoamylic alcohol protocol and precipitated with ethanol; the DNA pellet was dissolved in water and stored at −20°C. The coding strand of the amplified DNA was sequenced with an automated sequencer (ESGS Company, Evry, France). Results were corroborated by manual sequencing (Sequenase sequencing kit; Amersham Life Science) on DNA prepared from another PCR. The TK gene sequence of each strain was compared to that of the ACV-sensitive reference strain Dumas (5).
Nucleotide sequence accession numbers.
Sequences determined in this study have been submitted to GenBank under accession no. AF162437 through AF162440.
RESULTS
Antiviral susceptibility profiles.
Susceptibility profiles of three ACV-sensitive VZV strains and four ACV-resistant clinical strains are presented in Table 1. The IC50s for resistant strains are compared with the mean value for ACV-sensitive reference strains. An ACV IC50 15 times higher was observed for clinical strains WStr, LYON6625, and SW689. A similar increase was observed for PCV. The GCV IC50 was threefold higher for LYON6625 and SW689 and eightfold higher for WStr. Susceptibility to araT was lower for WStr and LYON6625 (IC50, ×20 to ×40) than for SW689 (IC50, ×5). The BVDU IC50 was higher for WStr than for LYON6625 and SW689 (×60 and ×10, respectively). Briefly, WStr presented a high level of resistance to all the TK-dependent drugs tested; LYON6625 and SW689 showed a high level of resistance to ACV and PCV and a low level of resistance to GCV and BVDU; however, these two strains differed in their resistance to araT, which was high for LYON6625 and intermediate for SW689. These three ACV-resistant strains were plaque purified in MRC-5 cells three times. Two purified viruses from each strain were tested for their susceptibility to ACV. All purified virus ACV IC50s were similar to that of the corresponding original strain.
TABLE 1.
Susceptibility profiles of VZV strains
| VZV strain | IC50 (μM)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACV
|
PCV
|
GCV
|
araT
|
BVDU
|
Foscarnet
|
|||||||
| Mean ± SD | Median (range) | Mean ± SD | Median (range) | Mean ± SD | Median (range) | Mean ± SD | Median (range) | Mean ± SD | Median (range) | Mean ± SD | Median (range) | |
| ACV-sensitive strains | ||||||||||||
| Ellen | 6.8 ± 4.9 | 5.7 (2.5–11) | 1.3 ± 0.1 | 1.2 (1.2–1.4) | 3.6 ± 3.4 | 3.8 (1.2–6.0) | 0.6 ± 0.4 | 0.55 (0.35–0.85) | 0.016 ± 0.004 | 0.015 (0.013–0.019) | 140 ± 90 | 145 (60–210) |
| Oka | 13 ± 3 | 11 (10–16) | 8.0 ± 2.6 | 8.4 (5.2–10) | 16 ± 9 | 17 (9.5–22) | 1.1 ± 0.1 | 1.0 (1.0–1.2) | 0.035 ± 0.007 | 0.035 (0.030–0.040) | 96 ± 21 | 90 (75–120) |
| D9507 | 12 ± 3 | 12 (9.0–15) | 8.3 ± 2.5 | 9.1 (6.0–10) | 7.5 ± 4.6 | 6.0 (2.7–14) | 0.4 ± 0.3 | 0.5 (0.16–0.54) | 0.025 ± 0.007 | 0.022 (0.020–0.033) | 133 ± 22 | 118 (76–203) |
| Mean | 11 | 10 | 6 | 6 | 9 | 9 | 0.7 | 0.7 | 0.025 | 0.025 | 123 | 118 |
| WMad | 51 ± 28 | 56 (13–83) | 32 ± 22 | 32 (11–54) | 27 ± 13 | 33 (11–36) | 3.2 ± 2.1 | 3.4 (1.0–6.0) | 1.5 ± 0.6 | 1.7 (0.7–2.0) | 148 ± 29 | 144 (125–189) |
| WStr | 342 ± 110 | 353 (246–423) | 486 ± 262 | 396 (254–500) | 76 ± 41 | 65 (33–130) | 26 ± 11 | 23 (18–39) | 1.5 ± 0.8 | 1.4 (0.9–2.0) | 116 ± 14 | 112 (94–142) |
| LYON6625 | 188 ± 102 | 147 (94–273) | 129 ± 94 | 82 (65–170) | 29 ± 18 | 26 (15–38) | 17 ± 5 | 16 (13–24) | 0.27 ± 0.10 | 0.29 (0.18–0.34) | 80 ± 15 | 87 (70–92) |
| SW689 | 221 ± 116 | 245 (130–292) | 97 ± 86 | 145 (33–162) | 29 ± 24 | 17 (8.9–47) | 3.7 ± 1.7 | 3.1 (2.3–5.6) | 0.32 ± 0.08 | 0.32 (0.26–0.37) | 107 ± 36 | 80 (75–160) |
All data were calculated from at least three independent assays performed in duplicate.
The WMad strain presented a low level of resistance to all the drugs tested except BVDU (IC50, ×5 for ACV, PCV, GCV, and araT and ×60 for BVDU). In order to check the heterogeneity of WMad viral populations, ACV susceptibility was tested on seven purified viruses obtained through one cycle of plaque purification: two were ACV sensitive (IC50, 12 and 5 μM) and five were resistant to ACV with a mean IC50 of 206 μM.
All the strains studied were sensitive to cidofovir and foscarnet.
TK activity.
TK activities were determined for the initial strains. Preliminary tests with extracts from cells infected with reference strains showed that enzymatic kinetics checked at 15, 30, and 60 min of incubation were linear, with either thymidine or araT substrates (data not shown). The viral TK levels detected at 60 min with different VZV strains are presented in Table 2. Compared to the activities induced by ACV-sensitive TK+ strains (Ellen, Oka, and D9507), WStr, LYON6625, and SW689 showed greatly reduced TK activities. Phosphorylation levels obtained with either thymidine or araT were comparable to those measured in noninfected cells. These results suggest that WStr, LYON6625, and SW689 have a deficient TK (TKD).
TABLE 2.
TK activity of VZV strains
| Strain | Activity (mean ± SD) (pmol/mg of protein/h)a of:
|
|
|---|---|---|
| [3H]thymidineb | [3H]araTc | |
| Noninfected HeLa cells | 217 ± 57 | 0 |
| VZV strains | ||
| Ellen | 13,472 ± 478 | 4,208 ± 643 |
| Oka | 12,868 ± 926 | 2,210 ± 89 |
| D9507 | 12,619 ± 1,642 | 2,737 ± 1,450 |
| WMad | 3,182 ± 619 | 485 ± 44 |
| WStr | 303 ± 169 | 0 |
| LYON6625 | 156 ± 49 | 0 |
| SW689 | 103 ± 41 | 0 |
Data were calculated from at least three independent assays as picomoles of the nucleoside analogue phosphorylated in 1 h per mg of protein at 37°C.
Final concentration of thymidine, 25 μM. Resulting specific activity in the reaction mixture, 1 Ci/mmol.
Final concentration of araT, 8.1 μM. Resulting specific activity in the reaction mixture, 3 Ci/mmol.
Concerning the WMad strain, the TK levels measured with thymidine and araT corresponded to 25 and 12 to 22% of the mean value obtained from the sensitive strains, respectively. These results are consistent with the presence of heterogeneous viral populations in WMad as detected with ACV susceptibility determination of plaque-purified virus.
Genetic analysis.
The TK genes of the three ACV-sensitive strains and the four ACV-resistant strains were sequenced and compared to that of the ACV-sensitive reference strain Dumas (5) (Table 3). The Ellen, Oka, and D9507 reference strains and the ACV-resistant isolates showed amino acid substitution Ser→Leu at position 288, which has already been observed in all non-Dumas VZV strains. Reference strain D9507 presented an additional amino acid substitution of Ser→Asn at residue 179.
TABLE 3.
TK gene sequence modificationsa of VZV strains
| Virus | Nucleotide position (change) | Amino acid position (change) |
|---|---|---|
| ACV-sensitive strains | ||
| Ellen | ||
| Oka | ||
| D9507 | 536 (AGT→AAT) | 179 (Ser→Asn) |
| ACV-resistant strains | ||
| WMad | 497 (addition C) | 167 (frameshift→206 stop codon) |
| WStr | 143 (GAG→GGG) | 48 (Glu→Gly) |
| LYON6625 | 514 (GTT→CTT) | 172 (Val→Cys) |
| 536 (AGT→ATT) | 179 (Ser→Ile) | |
| 641–642 (deletion T-A) | 215 (frameshift→217 stop codon) | |
| SW689 | 428 (AGA→GGA) | 143 (Arg→Gly) |
Compared to the sequence of the ACV-sensitive reference strain Dumas (5). All strains have an amino acid change at position 288 (Ser→Leu) due to a nucleotide change at position 863 (C→T).
The resistant strains had additional mutations. A nucleotide substitution at position 143 (A→G) was observed in WStr, responsible for an amino acid change (Glu→Gly) at codon 48. In strain SW689, a nucleotide substitution (A→G) was detected at position 428 causing an Arg→Gly switch at codon 143. The LYON6625 strain had two nucleotide substitutions (514 and 536), giving rise to changes in amino acids 172 and 179, respectively, and a deletion of two nucleotides, 641 and 642 (T-A), resulting in a premature stop codon at 217 and the synthesis of a truncated protein. For WStr, SW689, and LYON6625, the same mutations were identified in the corresponding plaque-purified virus.
Among purified viruses obtained through one cycle of plaque purification of parental WMad, all ACV-resistant viruses presented a single nucleotide addition (C) at position 497. It induced a frameshift at codon 167, resulting in a premature termination signal at codon 206. ACV-sensitive virus purified from WMad did not show that frameshift mutation.
DISCUSSION
From 1990 to 1998, 111 VZV clinical strains have been isolated in our laboratory from 105 patients, mainly during zoster episodes. Eighty percent of these strains were recovered from immunocompromised patients, mostly patients with AIDS. The sensitivity to ACV has been evaluated for all these strains. Only one virus resistant to ACV was isolated in 1993. The three other resistant strains characterized in this study were also recovered from severely immunocompromised patients affected with zoster and previously treated with ACV. Similar clinical presentations associated with the isolation of a resistant virus have already been described in human immunodeficiency virus-infected patients (2, 10, 14–16, 19).
The ACV-resistant clinical strain susceptibility profiles showed a cross-resistance towards TK-dependent drugs, which suggests viral TK involvement in ACV resistance that was assessed by TK activity. A possible alteration of viral DNA polymerase activity has not yet been checked and consequently cannot be excluded. But, according to foscarnet and cidofovir susceptibilities, viral DNA polymerase does not appear to be involved in ACV resistance of the strains characterized in this study.
Three strains were TK deficient (WStr, LYON6625, and SW689). The WMad strain presented low resistance levels to TK-dependent antiviral drugs, except for a high resistance level to BVDU, and a TK activity consistent with an altered or low-level TK phenotype. In fact, this strain was found to contain a mixture of virus either susceptible or highly resistant to ACV. Heterogeneous viral populations have also been reported by Talarico et al. in three ACV-resistant clinical strains (26).
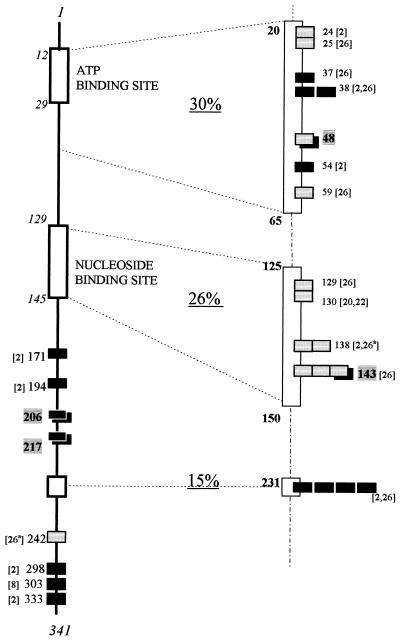
The molecular basis of resistance was determined by TK gene sequencing. The VZV TK open reading frame consists of 1,023 bp coding for a protein of 341 amino acids. By similarity to the ATP- and nucleoside-binding sites of herpes simplex virus TK (4, 11), VZV TK was presumed to have an ATP-binding site at amino acids 12 to 29 and a nucleoside binding site at amino acids 129 to 145 (22). Among all the ACV-resistant clinical strains of VZV described so far, 23 had their TK gene sequenced and 22 were found to bear a mutation in the coding part of the TK gene. In 95% of cases, only one site was modified, resulting in a single amino acid substitution or in a premature stop codon. These mutations were located in all areas of the TK gene, but careful analysis revealed three specific regions (Fig. 1): RI, amino acids 20 to 65, including the ATP-binding site and the next region; RII, amino acids 125 to 150, corresponding to the nucleoside binding site; and RIII, amino acid 231, which is the most frequently mutated site described to date.
FIG. 1.
Localization of TK gene mutations described in ACV-resistant VZV clinical strains. Numbers correspond to amino acid positions. Mutations detected in the same strain are indicated (a). Premature stop codons (black boxes) and amino acid substitutions (gray boxes) are indicated, determined either in references shown in square brackets (22 strains) or in this study (4 strains) (boxes on black box background).
Among the VZV clinical strains analyzed in our study, one (WStr) had a substitution at amino acid 48 (glutamic acid→glycine), which is located downstream of the VZV TK ATP-binding site (in region RI). Suzutani et al. (25) described a VZV mutant, obtained by random mutagenesis, which had the substitution glutamic acid→lysine at the same amino acid position, as well as a substitution at residue 185. This mutant had lost both TK and viral thymidylate kinase activities, but the exact role of each substitution has not been established. For the WStr strain, phenotypic data suggest that the nonconservative substitution at residue 48 is sufficient to reduce TK activity. This residue is conserved among many herpesviruses; it is equivalent to glutamic acid 83 of herpes simplex virus type 1 TK, which is the putative base in ester formation (27).
Strain SW689 had amino acid substitution Arg→Gly at residue 143, located in the nucleoside binding site (region RII) where many published mutations are clustered. Talarico et al. have described two strains with substitutions at residue 143 (26). The first strain (8812) had exactly the same amino acid substitution as strain SW689 (Arg→Gly), which is a nonconservative substitution. At the same residue, residue 143, the second strain (8919) reported by Talarico et al. had a distinct conservative amino acid substitution: Arg→Lys. This mutation can be correlated with the altered TK phenotype reported for strain 8919, as its TK was unable to bind ACV but could phosphorylate some nucleoside analogues such as PCV and BVaraU.
Homology studies of VZV and herpes simplex virus TK genes revealed that arginine 143 of VZV TK is equivalent to arginine 176 and 177 of the TK from herpes simplex virus type 1 and type 2, respectively. Mutations of these amino acids have also been found in ACV-resistant herpes simplex virus (4, 12, 18).
The TK gene of the ACV-resistant strain LYON6625 presented a two-nucleotide deletion at amino acid 215, resulting in a frameshift and a premature stop codon at amino acid 217. WMad had a single nucleotide addition at codon 167, resulting in a premature stop codon at amino acid 206. This addition occurs in a homopolymer run of G and C. Two TK-deficient strains with an addition or deletion of a cytosine in the same homopolymer have been previously reported (2). G or C repeats appear to be critical regions for the occurrence of mutations within the VZV TK gene, as has been pointed out for herpes simplex virus TK (9). The two strains LYON6625 and WMad must synthesize a truncated and possibly nonfunctional TK. Premature stop codons are responsible for nucleoside analogue resistance in about half of the VZV resistant strains sequenced to date.
In conclusion, among the four ACV-resistant VZV strains characterized in this study, two had a single nucleotide substitution and two others had a nucleotide insertion or deletion in the TK gene open reading frame. These mutations could account for their resistance to ACV. Genetic analysis of a larger number of ACV-resistant viruses, associated with site-directed mutagenesis studies, is required prior to the development of a genotypic diagnosis of resistance. Such a technique could be directly performed on biological samples. It would be useful to establish sensitivity survey networks for VZV clinical isolates with regard not only to ACV but also to new antiviral drugs.
REFERENCES
- 1.Bean B, Braun C, Balfour H H. Acyclovir therapy for acute herpes zoster. Lancet. 1982;ii:118–121. doi: 10.1016/s0140-6736(82)91090-x. [DOI] [PubMed] [Google Scholar]
- 2.Boivin G, Edelman C K, Pedneault L, Talarico C L, Biron K K, Balfour H H. Phenotypic and genotypic characterization of ACV-resistant varicella-zoster viruses isolated from persons with AIDS. J Infect Dis. 1994;170:68–75. doi: 10.1093/infdis/170.1.68. [DOI] [PubMed] [Google Scholar]
- 3.Chrips P, Clissold S P. Foscarnet. A review of its antiviral activity, pharmacokinetic properties and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs. 1991;41:104–129. doi: 10.2165/00003495-199141010-00009. [DOI] [PubMed] [Google Scholar]
- 4.Darby G, Larder B A, Inglis M M. Evidence that the “active centre” of the herpes simplex virus thymidine kinase involves an interaction between three distinct regions of the polypeptide. J Gen Virol. 1986;67:753–758. doi: 10.1099/0022-1317-67-4-753. [DOI] [PubMed] [Google Scholar]
- 5.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 6.De Clercq E, Holy A, Rosenberg I, Sakuma T, Balzarini J, Maugdal P C. A novel selective broad spectrum anti-DNA virus agent. Nature. 1986;323:464–467. doi: 10.1038/323464a0. [DOI] [PubMed] [Google Scholar]
- 7.de Turenne-Tessier M, Ooka T, de The G, Daillie J. Characterization of an Epstein-Barr virus-induced thymidine kinase. J Virol. 1986;57:1105–1112. doi: 10.1128/jvi.57.3.1105-1112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fillet A M, Dumont B, Caumes E, Visse B, Agut H, Bricaire F, Huraux J M. Acyclovir-resistant varicella-zoster virus: phenotypic and genetic characterization. J Med Virol. 1998;55:250–254. [PubMed] [Google Scholar]
- 9.Gaudreau A, Hill E, Balfour H H, Erice A, Boivin G. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J Infect Dis. 1998;178:297–303. doi: 10.1086/515626. [DOI] [PubMed] [Google Scholar]
- 10.Jacobson M A, Berger T G, Fikrig S, Bechere P, Moohr J W, Stanat S C, Biron K K. ACV-resistant varicella zoster virus infection after chronic oral ACV therapy in patients with the acquired immunodeficiency syndrome. Ann Intern Med. 1990;112:187–191. doi: 10.7326/0003-4819-112-3-187. [DOI] [PubMed] [Google Scholar]
- 11.Kit S, Sheppard M, Ichimura H, Nusinoff-Lehrman S, Ellis M N, Fyfe J A, Otsuka H. Nucleotide sequence changes in thymidine kinase gene of herpes simplex virus type 2 clones from an isolate of a patient treated with acyclovir. Antimicrob Agents Chemother. 1987;31:1483–1490. doi: 10.1128/aac.31.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kost R G, Hill E L, Tigges M, Straus S E. Brief report: recurrent acyclovir-resistant genital herpes in an immunocompetent patient. N Engl J Med. 1993;329:1777–1782. doi: 10.1056/NEJM199312093292405. [DOI] [PubMed] [Google Scholar]
- 13.Langlois M, Allard J P, Nugier F, Aymard M. A rapid and automated colorimetric assay for evaluating the sensitivity of herpes simplex strains to antiviral drugs. J Biol Stand. 1986;14:201–211. doi: 10.1016/0092-1157(86)90004-1. [DOI] [PubMed] [Google Scholar]
- 14.Leibovitz E, Kaul A, Rigaud M, Bebenroth D, Krasinski K, Borkowsky W. Chronic varicella zoster in a child infected with human immunodeficiency virus: case report and review of the literature. Cutis. 1992;49:27–31. [PubMed] [Google Scholar]
- 15.Linnemann C C, Biron K K, Hoppenjans W G, Solinger A M. Emergence of acyclovir-resistant varicella zoster virus in an AIDS patient on prolonged acyclovir therapy. AIDS. 1990;4:577–579. doi: 10.1097/00002030-199006000-00014. [DOI] [PubMed] [Google Scholar]
- 16.Lyall E G, Ogilvie M M, Smith N M, Burns S. Acyclovir resistant varicella zoster and HIV infection. Arch Dis Child. 1994;70:133–135. doi: 10.1136/adc.70.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masaoka T, Hiraoka A, Teshima H, Tominaga N. Varicella-zoster virus infection in immunocompromised patients. J Med Virol. 1993;41(Suppl. 1):82–84. doi: 10.1002/jmv.1890410515. [DOI] [PubMed] [Google Scholar]
- 17a.Morfin F, Thouvenot D, Aymard M, Lina B. Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Genetic and phenotypic characterisation of clinical strains of varicella-zoster virus resistant to acyclovir, abstr. H-20a; p. 320. [Google Scholar]
- 18.Nugier F, Colin J N, Aymard M, Langlois M. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J Med Virol. 1992;36:1–12. doi: 10.1002/jmv.1890360102. [DOI] [PubMed] [Google Scholar]
- 19.Pahwa S, Biron K, Lim W, Swenson P, Kaplan M H, Sadick N, Pahwa R. Continuous varicella-zoster infection associated with acyclovir resistance in a child with AIDS. J Virol. 1988;260:2879–2882. [PubMed] [Google Scholar]
- 20.Roberts G B, Fyfe J A, Gaillard R K, Short S A. Mutant varicella-zoster virus thymidine kinase: correlation of clinical resistance and enzyme impairment. J Virol. 1991;65:6407–6413. doi: 10.1128/jvi.65.12.6407-6413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Safrin S, Berger T G, Gilson I, Wolfe P R, Wofsy C B, Mills J, Biron K K. Foscarnet therapy in five patients with AIDS and acyclovir-resistant varicella-zoster virus infection. Ann Intern Med. 1991;115:19–21. doi: 10.7326/0003-4819-115-1-19. [DOI] [PubMed] [Google Scholar]
- 22.Sawyer M, Inchauspe G, Biron K K, Waters D J, Straus S E, Ostrove J M. Molecular analysis of the pyrimidine deoxyribonucleoside kinase gene of wild-type and acyclovir-resistant strains of varicella-zoster virus. J Gen Virol. 1988;69:2585–2593. doi: 10.1099/0022-1317-69-10-2585. [DOI] [PubMed] [Google Scholar]
- 23.Shiraki K, Ogino T, Yamanishi K, Takahashi M. Thymidine kinase with altered substrate specificity of acyclovir resistant varicella-zoster virus. Biken J. 1986;29:7–10. [PubMed] [Google Scholar]
- 24.Snoeck R, Gerard M, Sadzot-Delvaux C, Andrei G, Balzarini J, Reymen D, Ahadi N, DeBruyn J M, Piette J, Rentier B, Clumeck N, DeClercq E. Meningoradiculoneuritis due to acyclovir-resistant varicella-zoster virus in a patient with AIDS. J Med Virol. 1994;42:338–347. doi: 10.1002/jmv.1890420404. [DOI] [PubMed] [Google Scholar]
- 25.Suzutani T, Lacey S F, Powell K L, Purifoy D J M, Honess R W. Random mutagenesis of the thymidine kinase gene of varicella-zoster virus. J Virol. 1992;66:2118–2124. doi: 10.1128/jvi.66.4.2118-2124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talarico C L, Phelps W C, Biron K K. Analysis of thymidine kinase genes from acyclovir-resistant mutants of varicella-zoster virus isolates from patients with AIDS. J Virol. 1993;67:1024–1033. doi: 10.1128/jvi.67.2.1024-1033.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wild K, Bohner T, Folkers G, Schulz G E. The structures of thymidine kinase from herpes simplex virus type 1 in complex with substrates and a substrate analogue. Protein Sci. 1997;6:2097–2106. doi: 10.1002/pro.5560061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wunderli W, Miner R, Wintsch J, Von Gunten S, Hirsch H H, Hirschel B. Outer retinal necrosis due to a strain of varicella-zoster virus resistant to acyclovir, ganciclovir and sorivudine. Clin Infect Dis. 1996;22:864–865. doi: 10.1093/clinids/22.5.864. [DOI] [PubMed] [Google Scholar]