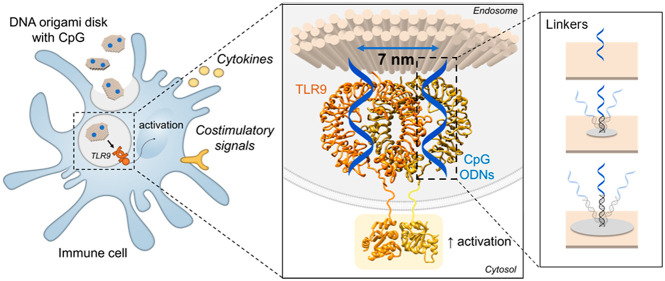
Abstract
First evidence of geometrical patterns and defined distances of biomolecules as fundamental parameters to regulate receptor binding and cell signaling have emerged recently. Here, we demonstrate the importance of controlled nanospacing of immunostimulatory agents for the activation of immune cells by exploiting DNA-based nanomaterials and pre-existing crystallography data. We created DNA origami nanoparticles that present CpG-motifs in rationally designed spatial patterns to activate Toll-like Receptor 9 in RAW 264.7 macrophages. We demonstrated that stronger immune activation is achieved when active molecules are positioned at the distance of 7 nm, matching the active dimer structure of the receptor. Moreover, we show how the introduction of linkers between particle and ligand can influence the spatial tolerance of binding. These findings are fundamental for a fine-tuned manipulation of the immune system, considering the importance of spatially controlled presentation of therapeutics to increase efficacy and specificity of immune-modulating nanomaterials where multivalent binding is involved.
Keywords: DNA, nanomaterials, CpG, TLR9, spacing, activation
Introduction
Ligand–receptor interactions are at the base of all biological processes, both in health and in disease.1 Their binding on the cell surface is defined not only by the type of molecules interacting but also by their interligand distances.2 This is in particular the case for immune cells, as the immune system developed to recognize the symmetric geometry of pathogen structures. Even for basic cell regulation, spatial patterns of membrane receptors have emerged as a fundamental feature for optimal signaling activation, as in the example of the immunological synapse.3
The advent of DNA nanotechnology has allowed the discovery of new insights on ligand spatial organization and its influence on immune activation.4 DNA nanomaterials offer an unprecedented control at the nanoscale level on the surface chemistry of nanoparticles (NPs) to create patterns of ligands with defined composition, number, and distance.5,6 As such, DNA origami structures have been employed to probe the influence of patterns and spacing on the minimal signaling unit for T-cell receptor activation7 and on the signaling cross-talk of its coreceptors8 and to investigate the impact of antigen valency in B-cell receptor activation.9 Recently, patterns on DNA nanomaterials have been shown to be transferable to cell surface receptors and as such to regulate apoptosis.10
Additionally, the expansion in crystallography data of biomolecules and proteins has paved the way for extremely fine-tuned material design. Extrapolating intra- and interligand distances from crystallography data narrows down the range of spatial patterns that need to be tested. In this way, a general screening strategy with multiple DNA structures with unlimited combinations of ligand spacing and valency can be substituted with a rationally designed restricted library to find the most potent binding patterns. Moreover, it allows the investigation of the spatial tolerance of binding for ligand–receptor systems at the nanoscale level. For example, Shaw et al. have exploited antibody X-ray crystallography data and DNA origami with patterns of antigens in a limited range of distances to investigate epitope–antibody bivalent binding.11 They have not only reported the optimal distance for binding, but they have also highlighted significant differences in spatial tolerance between IgM and IgG and between low- and high-affinity antibodies.
However, the relevance of spatial organization remains unexplored or understudied for many biological targets. DNA-based NPs have been used as carriers for immunostimulatory agents (e.g., adjuvants) for vaccination in preclinical models, but the focus has remained limited to general drug delivery.12−14 These previous studies have not taken full advantage of the unique DNA functionalization potential and knowledge of crystallography data on targeted receptors to understand the relation between spatially defined ligand presentation and cellular activation intensity.
The activation of Pattern Recognition Receptors (PRRs) by adjuvants on antigen-presenting cells (APCs) is the first key step in vaccination to potentiate the magnitude and quality of the immune response against the target antigen.15,16 The endosomal PRR Toll-like receptor 9 (TLR9) is triggered by oligodeoxynucleotides containing cytosine-phosphate-guanosine motifs (CpG-ODNs). However, it is thus far unclear whether spatial organization of CpG-ODNs has an impact on the activation of TLR9. Here, we demonstrate the importance of nanospacing of the immunostimulatory agents CpG-ODNs for the activation of TLR9 in an immune cell model. We exploit the recent characterization of the CpG-TLR9 interaction in published crystallography studies17−19 combined with the unique spatial control of ligands on DNA nanomaterials. We create DNA-based nanoparticles presenting CpG-ODNs at defined nanometer-scale distances to bind TLR9, as in the crystal structure of the active form. Additionally, we investigate the spatial tolerance of binding when linkers of different lengths are introduced.
Results and Discussion
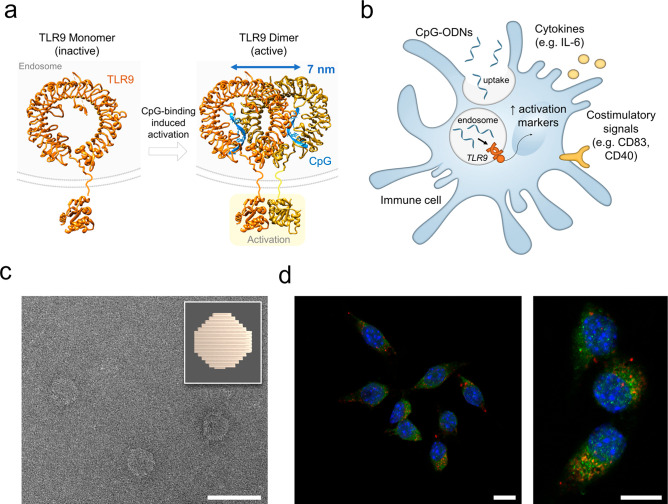
We start our study by extrapolating the interligand distance in the target receptor–ligand system using pre-existing protein crystallography data openly accessible in the protein database (PDB). In the inactive form, TLR9 is present as a monomer on the endosomal membrane. It dimerizes and activates upon binding of two CpG-motifs (stoichiometry TLR9:CpG 2:2).17,19 From the crystal structure of the dimeric form of the TLR9-CpG complex (PDB entry 5zln),18 we have measured the interligand distance of two CpG molecules bound in the active dimer, which we find to be 7 nm (Figure 1a). Therefore, we hypothesize that presenting two CpG molecules at this exact spacing may improve the triggering of TLR9 and immune cell activation.
Figure 1.
Toll-like receptor 9 (TLR9) activation by CpG and DNA-based disk for delivery of CpG-ODNs. (a) Representation of TLR9 activation with crystal structures of monomer of TLR9 (inactive form) and CpG-bound TLR9 dimer (active form) at binding stoichiometry TLR9:CpG 2:2. PBD codes: 3wpf (monomer), 5zln (dimer with CpG), 4om7 (cytosolic domain). (b) Representation of immune cell activation upon uptake of free CpG-ODNs. Surface markers analyzed in this study are reported. (c) Transmission electron microscopy image of a DNA origami disk and schematic representation (top right corner). Scale bar: 100 nm. (d) Confocal microscopy images, overview (left) and zoom-in (right), of the uptake of Cy5-labeled disks by RAW 264.7 cell line. Nuclei stained with DAPI (blue), late endosome marker (RAB7) in green and Cy5-disks in red. Cells were treated with DNase I after uptake to remove noninternalized DNA structures binding to the cell surface, as previously reported.22 Scale bars: 10 μm.
First, we have established the uptake of our DNA origami platform to reach the target, since the CpG-binding site of TLR9 is facing the endosomal environment.20 When used as adjuvant in vaccines, CpG-ODNs are promptly taken up by APCs, where they subsequently bind to TLR9 in the endosome triggering intracellular signaling cascades for immune cell activation (Figure 1b). For CpG-delivery in our study, we have employed DNA-based disk shaped NPs21 (Figure 1c): besides nanoscale control on ligand spacing, our DNA NP exhibits a compact shape and a reduced size (approximately 60 nm diameter), optimal for uptake by immune cells.22,23 For our study, we have chosen the RAW 264.7 macrophages as a cell model: these cells express high levels of TLR9 and are frequently selected in immune-engineering studies with CpG-functionalized nanomaterials.24 Confocal microscopy images of a Cy5-labeled disk incubated with RAW 264.7 indeed show that our DNA NP is rapidly taken up by these cells (<30 min) without the need of targeting molecules, and particles quickly colocalize in the late endosome, which is optimal for CpG delivery (Figures 1d and S1). Indeed, it has been previously reported that endosomal maturation via acidification is a prerequisite for CpG-ODNs signaling because of the pH-dependent nature of TLR9-CpG interaction.25 At physiological pH (pH 7.4) CpG motifs show weak binding affinity for TLR9, whereas stronger affinity is observed upon lowering the pH to acidic conditions as in the late endosome (pH 5.5).18,26 We have therefore tested and confirmed the stability of the DNA disk for the entire time scale of our assays both in the cell medium utilized for this study and at a low pH value as in the late endosome (pH 5.5) (Figure S2). The DNA platform maintains its stability throughout all conditions found in the biological assays; thus, it is suitable to deliver ligands in the endosome to the TLR9 target.
With endosomal delivery confirmed, we have focused our attention on the spatial presentation of CpG-ODNs to their target receptor. We exploit once again the programmability of DNA origami, where each strand can be site-specifically functionalized by design, to present molecules at a precise distance.6 Using DNA-PAINT super-resolution microscopy, we have previously demonstrated that our structure represents a versatile platform for multivalent ligand presentation with accurate matching between in silico design and experimental measures in functionalization patterns.21
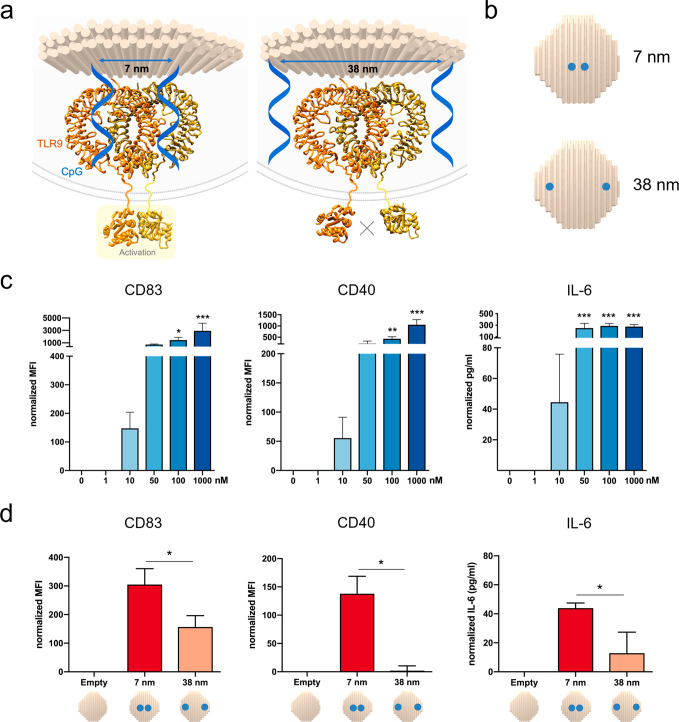
On the basis of the TLR9-CpG crystal structure analysis, we hypothesize that presenting CpG-ODNs spatially organized at an interligand spacing of 7 nm, matching the dimerization distance of TLR9, would be optimal for immune cell activation. Therefore, we have created CpG-presenting DNA disks displaying CpG at the distance of 7 nm to match the spacing of TLR9 dimers and as control at a larger distance (i.e., 38 nm) (Figure 2a,b). In the latter situation, we hypothesize that the dimer formation with two CpG molecules on their binding sites would be impaired and thus a significantly reduced cellular activation is expected. Dimers without CpG molecules are unable to induce the conformational changes in TLR9 cytosolic domains, which are necessary to trigger the signaling cascade via adaptor protein docking.27
Figure 2.
Spatially controlled activation of TLR9 by nanoscale controlled CpG-spacing on a DNA origami disk. (a) Representation of the spatially controlled presentation of two CpG molecules at 7 nm spacing (left) and larger spacing of 38 nm (right), with consequent activation effects. (b) Schematic of the disks representing the different locations and distances of the CpG molecules. (c) Activation assays of free CpG-ODNs, in the range 1 nM to 1 μM. Quantitative analysis of the surface markers CD83 and CD40 in RAW 264.7 assessed by flow cytometry (left and center). Median Fluorescence intensity (MFI) data are normalized subtracting the MFI of the cells without CpG activation (control). IL-6 production quantification assessed by ELISA (right). Data are normalized by subtracting the IL-6 production in pg/mL of unstimulated cells (control). (d) Activation assays of RAW 264.7 incubated with Cy5-labeled disk (0.5 nM) functionalized with two CpG molecules at different distances. Quantitative analysis of the surface markers CD83 and CD40 in the Cy5 gate in RAW 264.7 assessed by flow cytometry (left and center). Median Fluorescence Intensity (MFI) data are normalized subtracting the MFI of cells treated with a Cy5-labeled disk without CpG (empty disk) as a control. IL-6 production quantification assessed by ELISA (right). Data are normalized by subtracting the IL-6 production in pg/mL of cells treated with an empty disk. Data are represented as means of three biological replicates ± standard deviation (n = 3). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; one-way ANOVA compared to untreated cells for free CpG, t test 7 nm vs 38 nm for CpG disks.
As readout for the immune activation assays with RAW 264.7 cells, we have screened the release of the pro-inflammatory cytokine IL-6 in cell culture supernatants by ELISA and the increase in surface expression of the markers CD83 and CD40 by flow cytometry (Figure 2c,d). Cells are incubated with samples for 30 min; then DNase I treatment is applied to remove extra NPs in medium or externally membrane-bound as previously reported to avoid a false positive uptake signal.28 Subsequently, cells are incubated for four additional hours in fresh medium to allow cells to express surface markers before analysis.
Using this setup, we have first analyzed the effect of free CpG-ODNs in the RAW 264.7 cells at the nanomolar range and up to 1 μM, the standard dose for in vitro studies29 (Figure 2c). Subsequently, we have assessed the activation of RAW 264.7 incubated with disks without CpG (defined as “empty disk”) as a control and disks with two CpG-ODNs presented on their surface either at 7 nm or at 38 nm of interligand distance (Figure 2d). Disk samples are tested at 0.5 nM concentration, with a corresponding 1 nM concentration of CpG-ODNs (two molecules per structure) and are labeled with six integrated Cy5 dyes for quantification. To exclude artifacts, we have applied several control strategies described in the Supporting Information (Supplementary Text, Figures S3–S5).
We have observed statistically significant upregulation of both surface markers CD83 and CD40 and increased IL-6 release when two CpG molecules are present at 7 nm distance, matching the active dimer form, compared to a structure where they are conjugated at the larger, suboptimal spacing of 38 nm (Figure 2d). Simply changing the spatial organization of ligands presented on NPs using a rational design based on crystallography data can thus modulate cellular activation levels. While free CpG at 1 nM concentration does not induce cell activation (Figure 2c), the disk improves the intracellular delivery of CpG-ODNs and variances in activation due only to differential spacings can be observed in all readouts even at a concentration as low as 1 nM (Figure 2d). Thus, by combining delivery and correct spatial presentation, a significant reduction in therapeutic dose can be achieved.
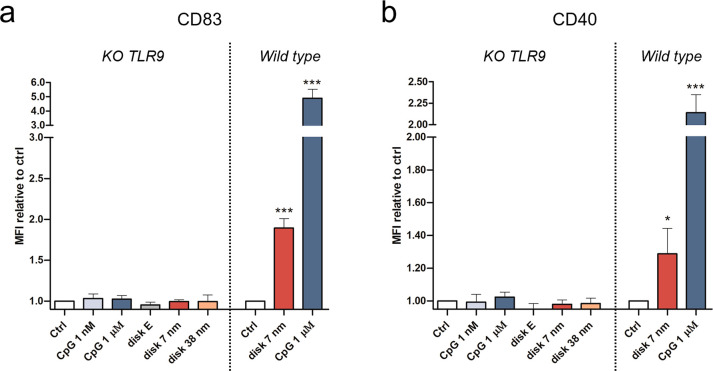
To confirm that the spatial activation effect of our CpG-disks is due to TLR9-specific stimulation and does not involve other immune pathways, we have generated a TLR9 knockout (KO) RAW 264.7 cell line (Figure S6). Free CpG-ODNs and our CpG-disk platform have been tested on the TLR9 KO cells. Disk uptakes between TLR9-KO RAW 264.7 and wild type are found to be comparable (Figure S7). Interestingly, no activation in any of the samples in the KO cell line has been observed (Figure 3), confirming our previously measured cellular activation exclusively results from the CpG-TLR9 pathway.
Figure 3.
Control of specific activation using standard and TLR9 knockout (KO) RAW 264.7. Quantitative analysis of the cell-surface expression of the markers CD83 (a) and CD40 (b) in wild-type and KO RAW 264.7, incubated with CpG free and disk-conjugated, assessed by flow cytometry. Median Fluorescence Intensity (MFI) data are reported relative to MFI of the corresponding untreated control cells (wild type or KO). Data are represented as means of three biological replicates ± standard deviation (n = 3). *p ≤ 0.05, ***p ≤ 0.001; one-way ANOVA compared to corresponding control.
Taken together, our results demonstrate how spatial presentation of ligands on nanomaterials, rationally designed on the basis of crystallographic data, can significantly affect the activation of immune cells. NP-mediated delivery and preassembly of ligands on the NP surface increase the local concentration of the ligand next to the receptors, benefiting from a controlled spatial multivalency effect compared to the case for free CpG at the same dose. While CpG-ODNs in free form require a high dose to stimulate the cells, strong activation even at very low concentrations of ligands can be achieved simply by proper spatial organization. This finding may have significant consequences for therapeutic ligands that are subject to dose-dependent toxicity.
Active molecules are often conjugated to nanomaterials via linkers of diverse chemical nature and length. These linkers are considered “functionally inert”, but their flexibility influences the spacing at which ligands are presented to cell surface receptors.30−32 Therefore, we have investigated the spatial tolerance of the TLR9 activation pathway when linkers of various length are introduced between the DNA platform and the CpG-ODN ligands.
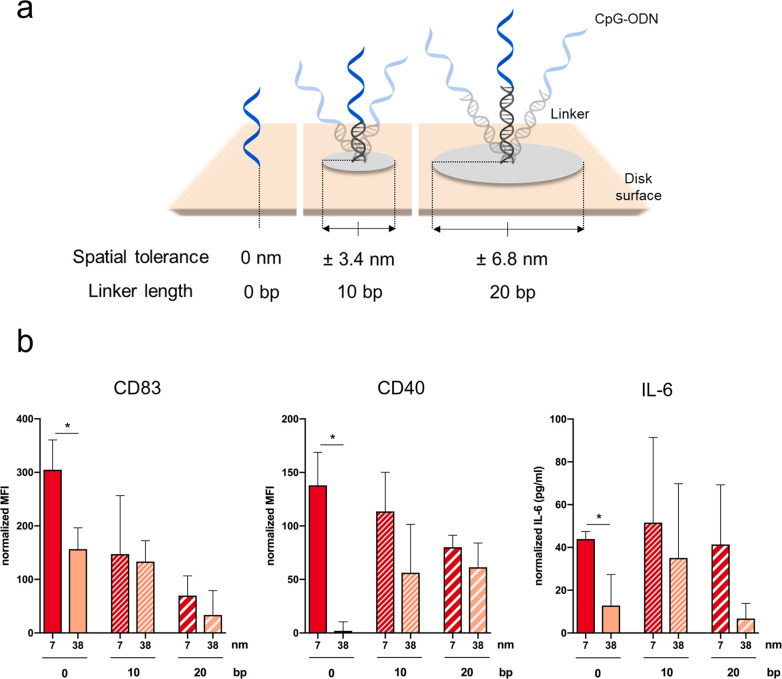
Exploiting once again the programmability of DNA, we have introduced a DNA linker of 10 or 20 base pairs (bp) between the core origami structure and the CpG-ODNs (Figures 4a and S8). Assuming an ideal case of maximal extension, the introduction of a 10 bp or 20 bp DNA double helix adds a space of ±3.4 or ±6.8 nm, respectively, to the 7 and 38 nm distances (calculated from the disk surface base).
Figure 4.
Spatial tolerance of CpG-ODNs binding with flexible DNA linkers. (a) Representation of the CpG-ODNs functionalization to the disk surface in the absence of linkers and in the presence of linkers of 10 or 20 base pairs (bp). (b) Activation assays of RAW 264.7 incubated with Cy5-labeled disk functionalized with two CpG molecules at different distances (7 and 38 nm) and with different linker lengths (0, 10, or 20 bp). Quantitative analysis of the cell-surface expression of the markers CD83 (left) and CD40 (center) in the Cy5 gate assessed by flow cytometry and IL-6 production quantification assessed by ELISA (right) in RAW 264.7. Data normalized compared to cells treated with empty disk. Data are represented as mean of three biological replicates ± standard deviations (n = 3). *p ≤ 0.05; t test 7 nm vs 38 nm CpG disks.
Interestingly, we have observed (1) the loss of significant difference in activation between 7 and 38 nm spacing in all the readouts when a linker is included, (2) a progressively lower activation correlating with the increase in linker length, and (3) higher standard deviation for several samples where a linker is introduced between the disk surface and CpG ligand (Figure 4b). While the first observation may suggest that ligands can perfectly fit receptors no matter what their spacing is, if a sufficiently long linker is included, the second point shows that this is not the case. Hence, we deduce that reduced flexibility in the presentation of ligands at the correct spacing is required to fit with the active form of the receptor. Ligand rigidity and matching of receptor distances combine in a phenomenon that we define as spatial tolerance of binding. It has been shown that reduced flexibility confers an advantage for the thermodynamics of ligand binding. A low spatial tolerance decreases the conformational and combinatorial entropy penalties.33 On the contrary, the higher conformational possibilities given by the linkers increases the entropic penalty of binding. The third effect may derive from a combination of the previous two, indicating that the observed average effect comes from a mixture of specific and nonspecific activation stimuli. Taken together, the flexibility introduced by the linkers negatively impacts the improved efficacy given by spatial presentation. However, spatial tolerance requires a minimum range for interactions: in other words, a balance between a rigid interaction and adjustability to a certain extent is required for binding. In this case, it is important to note that a CpG-ODN is a single strand DNA sequence itself with a length of 20 nucleotides. Therefore, the ligand has an intrinsic flexibility that allows ligand–receptor interactions with favorable binding enthalpy.34
In summary, the introduction of linkers between a nanoparticle surface and active molecules has a clear impact on ligand–receptor complexation and, consequently, cell activation. In the design of active nanomaterials, the static spatial organization as well as the dynamics of ligand–receptor binding are closely correlated and therefore demand special attention. Not only spatial presentation but also the influence of spatial tolerance need to be carefully investigated related to the target pathway. If properly executed, spatially controlled ligand presentation can be a breakthrough for the development of immune-modulating nanomaterials where multivalent binding is involved.
Conclusions
While multiple clinical trials have shown the efficacy of CpG-ODNs for activation of APCs through the TLR9 pathway,35 the current method of administration through subcutaneous injection of free CpG-ODNs requires high doses.36 This can lead to side effects such as systemic inflammation in the case of nonspecific off-target delivery and/or autoimmune diseases in the case of excessive immune reactions.37 Therefore, continuous research to improve safety though specificity and dose reduction, while maintaining efficacy, is a medical need.37
While intensive effort has been made in immune-engineering research to create nanomaterials for drug delivery and new therapeutic molecules, the importance of interligand distance in multivalent binding has been neglected.38,39 DNA nanotechnology, with its unprecedented control of spacing for biological ligands, has emerged as a powerful tool to investigate fundamental details of ligand–receptor interactions and to understand how to manipulate the activation of signaling pathways on the nano level.4 Early examples of delivery of CpG-loaded DNA nanostructures, such as a DNA nanotube and a wireframe DNA tetrahedron, have shown in vitro and in vivo increased immune activation compared to results for free CpG-ODNs.12−14 However, in these studies, the focus of the nanocarriers has been simply to maximize CpG delivery rather than to investigate the foundational ligand–receptor interaction in this pathway.
On the basis of crystallographic data, we here have demonstrated how nanoscale variations in ligand spacing produce significantly different immunological responses. Matching the CpG spacing with the distance of the binding sites of the TLR9 active dimer significantly enhances cellular activation in comparison with random ligand presentation or free form CpG at an equal dose. Our results demonstrate that dose reduction can be achieved not only by proper delivery systems but also via nanoscale spatial control of ligand presentation on nanomaterial surfaces. Additionally, we have shown the negative consequences of flexible linkers used to conjugate active molecules to nanomaterials, as they impact the spatial tolerance and consequently the efficacy of binding. Not only pattern design but also a certain rigidity in ligand presentation are required to efficiently orchestrate a multivalent interaction.
The spatial tolerance of a signaling pathway therefore is a means to control therapeutic intervention. At the same concentration, higher activation can be achieved simply by presenting ligands in the proper way to their receptors. Introducing uniformity and proper rigidity in ligand presentation, offering a full ligand dose to bioreceptors in the optimal way, reduces the amount of drug needed for immune activation and to lower variability in efficacy. Herein, DNA-based nanomaterials are starting to set the basis in the understanding of the structure–activity relationships behind multivalent ligand–receptor signaling pathways. Future vaccine design strategies with fine-tuned control of immune-modulating ligand, valency, pattern, rigidity and spacing may therefore ensure decreased systemic disorders by dose reduction, while maintaining efficacy.
Acknowledgments
We thank Dr. Jorieke Weiden for critical discussions on data analysis and on the writing of the manuscript. We thank Kaltrina Paloja, Vincenzo Caroprese, and Hugo J. Rodríguez-Franco for meaningful discussion on experiments and Christine Lavanchy for technical support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.2c00275.
Materials and methods, supplementary text on controls, agarose gels, confocal pictures, stability assays, flow cytometry gating strategy and uptake data, agarose gel electrophoresis, and tables with DNA sequences for the disk structure (PDF)
Author Contributions
A.C. contributed to the conception of the study, performed sample preparation, microscopy imaging, stability, and activation assays, analyzed data, and wrote the manuscript. M.M.K. contributed to activation assays, to critical discussions, and to the writing of the manuscript. S.N. created and characterized the TLR9 knockout (KO) RAW 264.7 cell line. I.A.P. contributed to critical discussions for the design of activation assays and TLR9 KO. M.M.C.B. conceived and supervised the study, interpreted data, and wrote the manuscript. All authors reviewed and approved the manuscript.
This work has been supported by the “Fondation Pierre Mercier pour la Science”, by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 948334 InActioN), and by the Human Frontier Science Program (HFSP) through the Young Investigator Grant RGY0065/2018.
The authors declare no competing financial interest.
Supplementary Material
References
- Li D.; Wu M. Pattern Recognition Receptors in Health and Diseases. Sig Transduct Target Ther 2021, 6 (1), 291. 10.1038/s41392-021-00687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz B. N.; Groves J. T. Spatial Organization and Signal Transduction at Intercellular Junctions. Nat. Rev. Mol. Cell Biol. 2010, 11 (5), 342–352. 10.1038/nrm2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P.; den Boer A. Th.; Gunzer M. Tuning Immune Responses: Diversity and Adaptation of the Immunological Synapse. Nat. Rev. Immunol 2005, 5 (7), 532–545. 10.1038/nri1647. [DOI] [PubMed] [Google Scholar]
- Tseng C. Y.; Wang W. X.; Douglas T. R.; Chou L. Y. T. Engineering DNA Nanostructures to Manipulate Immune Receptor Signaling and Immune Cell Fates. Adv. Healthcare Mater. 2022, 11, 2101844. 10.1002/adhm.202101844. [DOI] [PubMed] [Google Scholar]
- Seeman N. C.; Sleiman H. F. DNA Nanotechnology. Nat. Rev. Mater. 2018, 3 (1), 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Wickham S. F. J.; Auer A.; Min J.; Ponnuswamy N.; Woehrstein J. B.; Schueder F.; Strauss M. T.; Schnitzbauer J.; Nathwani B.; Zhao Z.; Perrault S. D.; Hahn J.; Lee S.; Bastings M. M.; Helmig S. W.; Kodal A. L.; Yin P.; Jungmann R.; Shih W. M. Complex Multicomponent Patterns Rendered on a 3D DNA-Barrel Pegboard. Nat. Commun. 2020, 11 (1), 5768. 10.1038/s41467-020-18910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmeier J.; Platzer R.; Eklund A. S.; Schlichthaerle T.; Karner A.; Motsch V.; Schneider M. C.; Kurz E.; Bamieh V.; Brameshuber M.; Preiner J.; Jungmann R.; Stockinger H.; Schütz G. J.; Huppa J. B.; Sevcsik E. DNA Origami Demonstrate the Unique Stimulatory Power of Single PMHCs as T Cell Antigens. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (4), e2016857118 10.1073/pnas.2016857118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang T.; Alvelid J.; Spratt J.; Ambrosetti E.; Testa I.; Teixeira A. I. Spatial Regulation of T-Cell Signaling by Programmed Death-Ligand 1 on Wireframe DNA Origami Flat Sheets. ACS Nano 2021, 15 (2), 3441–3452. 10.1021/acsnano.0c10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneziano R.; Moyer T. J.; Stone M. B.; Wamhoff E.-C.; Read B. J.; Mukherjee S.; Shepherd T. R.; Das J.; Schief W. R.; Irvine D. J.; Bathe M. Role of Nanoscale Antigen Organization on B-Cell Activation Probed Using DNA Origami. Nat. Nanotechnol. 2020, 15 (8), 716–723. 10.1038/s41565-020-0719-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Baars I.; Fördös F.; Högberg B. Clustering of Death Receptor for Apoptosis Using Nanoscale Patterns of Peptides. ACS Nano 2021, 15 (6), 9614–9626. 10.1021/acsnano.0c10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.; Hoffecker I. T.; Smyrlaki I.; Rosa J.; Grevys A.; Bratlie D.; Sandlie I.; Michaelsen T. E.; Andersen J. T.; Högberg B. Binding to Nanopatterned Antigens Is Dominated by the Spatial Tolerance of Antibodies. Nat. Nanotechnol. 2019, 14 (2), 184–190. 10.1038/s41565-018-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller V. J.; Heidegger S.; Sandholzer N.; Nickels P. C.; Suhartha N. A.; Endres S.; Bourquin C.; Liedl T. Cellular Immunostimulation by CpG-Sequence-Coated DNA Origami Structures. ACS Nano 2011, 5 (12), 9696–9702. 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- Li J.; Pei H.; Zhu B.; Liang L.; Wei M.; He Y.; Chen N.; Li D.; Huang Q.; Fan C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5 (11), 8783–8789. 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- Liu X.; Xu Y.; Yu T.; Clifford C.; Liu Y.; Yan H.; Chang Y. A DNA Nanostructure Platform for Directed Assembly of Synthetic Vaccines. Nano Lett. 2012, 12 (8), 4254–4259. 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S. H. E. The Contribution of Immunology to the Rational Design of Novel Antibacterial Vaccines. Nat. Rev. Microbiol 2007, 5 (7), 491–504. 10.1038/nrmicro1688. [DOI] [PubMed] [Google Scholar]
- Reed S. G.; Orr M. T.; Fox C. B. Key Roles of Adjuvants in Modern Vaccines. Nature Medicine 2013, 19 (12), 1597–1608. 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- Ohto U.; Shimizu T. Structural Aspects of Nucleic Acid-Sensing Toll-like Receptors. Biophys Rev. 2016, 8 (1), 33–43. 10.1007/s12551-015-0187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U.; Shibata T.; Tanji H.; Ishida H.; Krayukhina E.; Uchiyama S.; Miyake K.; Shimizu T. Structural Basis of CpG and Inhibitory DNA Recognition by Toll-like Receptor 9. Nature 2015, 520 (7549), 702–705. 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- Ohto U.; Ishida H.; Shibata T.; Sato R.; Miyake K.; Shimizu T. Toll-like Receptor 9 Contains Two DNA Binding Sites That Function Cooperatively to Promote Receptor Dimerization and Activation. Immunity 2018, 48 (4), 649–658.e4. 10.1016/j.immuni.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Desmet C. J.; Ishii K. J. Nucleic Acid Sensing at the Interface between Innate and Adaptive Immunity in Vaccination. Nat. Rev. Immunol 2012, 12 (7), 479–491. 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- Eklund A. S.; Comberlato A.; Parish I. A.; Jungmann R.; Bastings M. M. C. Quantification of Strand Accessibility in Biostable DNA Origami with Single-Staple Resolution. ACS Nano 2021, 15 (11), 17668–17677. 10.1021/acsnano.1c05540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastings M. M. C.; Anastassacos F. M.; Ponnuswamy N.; Leifer F. G.; Cuneo G.; Lin C.; Ingber D. E.; Ryu J. H.; Shih W. M. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018, 18 (6), 3557–3564. 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- Mitchell M. J.; Billingsley M. M.; Haley R. M.; Wechsler M. E.; Peppas N. A.; Langer R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Discov 2021, 20 (2), 101–124. 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist S. E. Variable Expression of Toll-like Receptor in Murine Innate and Adaptive Immune Cell Lines. Int. Immunol. 2002, 14 (9), 1065–1074. 10.1093/intimm/dxf069. [DOI] [PubMed] [Google Scholar]
- Rutz M.; Metzger J.; Gellert T.; Luppa P.; Lipford G. B.; Wagner H.; Bauer S. Toll-like Receptor 9 Binds Single-Stranded CpG-DNA in a Sequence- and PH-Dependent Manner. Eur. J. Immunol. 2004, 34 (9), 2541–2550. 10.1002/eji.200425218. [DOI] [PubMed] [Google Scholar]
- Casey J. R.; Grinstein S.; Orlowski J. Sensors and Regulators of Intracellular PH. Nat. Rev. Mol. Cell Biol. 2010, 11 (1), 50–61. 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- Gay N. J.; Symmons M. F.; Gangloff M.; Bryant C. E. Assembly and Localization of Toll-like Receptor Signalling Complexes. Nat. Rev. Immunol 2014, 14 (8), 546–558. 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Ponnuswamy N.; Bastings M. M. C.; Nathwani B.; Ryu J. H.; Chou L. Y. T.; Vinther M.; Li W. A.; Anastassacos F. M.; Mooney D. J.; Shih W. M. Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation. Nat. Commun. 2017, 8 (1), 15654. 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker H.; Vabulas R. M.; Takeuchi O.; Hoshino K.; Akira S.; Wagner H. Immune Cell Activation by Bacterial Cpg-DNA through Myeloid Differentiation Marker 88 and Tumor Necrosis Factor Receptor–Associated Factor (Traf)6. Journal of Experimental Medicine 2000, 192 (4), 595–600. 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia C. H.; Tian S.; Perry J. L.; Luft J. C.; DeSimone J. M. Role of Linker Length and Antigen Density in Nanoparticle Peptide Vaccine. ACS Omega 2019, 4 (3), 5547–5555. 10.1021/acsomega.8b03391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L. J.; Tacken P. J.; Fokkink R.; Figdor C. G. The Influence of PEG Chain Length and Targeting Moiety on Antibody-Mediated Delivery of Nanoparticle Vaccines to Human Dendritic Cells. Biomaterials 2011, 32 (28), 6791–6803. 10.1016/j.biomaterials.2011.04.082. [DOI] [PubMed] [Google Scholar]
- Maslanka Figueroa S.; Fleischmann D.; Beck S.; Goepferich A. The Effect of Ligand Mobility on the Cellular Interaction of Multivalent Nanoparticles. Macromol. Biosci. 2020, 20 (4), 1900427. 10.1002/mabi.201900427. [DOI] [PubMed] [Google Scholar]
- Morzy D. K.; Bastings M. Significance of Receptor Mobility in Multivalent Binding on Lipid Membranes. Angewandte Chemie Intl Edit 2022, 14167. 10.1002/anie.202114167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Veracoechea F. J.; Leunissen M. E. The Entropic Impact of Tethering, Multivalency and Dynamic Recruitment in Systems with Specific Binding Groups. Soft Matter 2013, 9 (12), 3213. 10.1039/c3sm27766f. [DOI] [Google Scholar]
- Cooper C.; Mackie D. Hepatitis B Surface Antigen-1018 ISS Adjuvant-Containing Vaccine: A Review of HEPLISAVTM Safety and Efficacy. Expert Review of Vaccines 2011, 10 (4), 417–427. 10.1586/erv.10.162. [DOI] [PubMed] [Google Scholar]
- Krieg A. M. Therapeutic Potential of Toll-like Receptor 9 Activation. Nat. Rev. Drug Discov 2006, 5 (6), 471–484. 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- Vanpouille-Box C.; Hoffmann J. A.; Galluzzi L. Pharmacological Modulation of Nucleic Acid Sensors — Therapeutic Potential and Persisting Obstacles. Nat. Rev. Drug Discov 2019, 18 (11), 845–867. 10.1038/s41573-019-0043-2. [DOI] [PubMed] [Google Scholar]
- Comberlato A.; Paloja K.; Bastings M. M. C. Nucleic Acids Presenting Polymer Nanomaterials as Vaccine Adjuvants. J. Mater. Chem. B 2019, 7, 6321. 10.1039/C9TB01222B. [DOI] [PubMed] [Google Scholar]
- Smith D. M.; Simon J. K.; Baker J. R. Jr Applications of Nanotechnology for Immunology. Nat. Rev. Immunol 2013, 13 (8), 592–605. 10.1038/nri3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.