Abstract
AG7088 is a potent, irreversible inhibitor of human rhinovirus (HRV) 3C protease {inactivation rate constant (kobs/[I]} = 1,470,000 ± 440,000 M−1 s−1 for HRV 14) that was discovered by protein structure-based drug design methodologies. In H1-HeLa and MRC-5 cell protection assays, AG7088 inhibited the replication of all HRV serotypes (48 of 48) tested with a mean 50% effective concentration (EC50) of 0.023 μM (range, 0.003 to 0.081 μM) and a mean EC90 of 0.082 μM (range, 0.018 to 0.261 μM) as well as that of related picornaviruses including coxsackieviruses A21 and B3, enterovirus 70, and echovirus 11. No significant reductions in the antiviral activity of AG7088 were observed when assays were performed in the presence of α1-acid glycoprotein or mucin, proteins present in nasal secretions. The 50% cytotoxic concentration of AG7088 was >1,000 μM, yielding a therapeutic index of >12,346 to >333,333. In a single-cycle, time-of-addition assay, AG7088 demonstrated antiviral activity when added up to 6 h after infection. In contrast, a compound targeting viral attachment and/or uncoating was effective only when added at the initiation of virus infection. Direct inhibition of 3C proteolytic activity in infected cells treated with AG7088 was demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of radiolabeled proteins, which showed a dose-dependent accumulation of viral precursor polyproteins and reduction of processed protein products. The broad spectrum of antiviral activity of AG7088, combined with its efficacy even when added late in the virus life cycle, highlights the advantages of 3C protease as a target and suggests that AG7088 will be a promising clinical candidate.
The Picornaviridae family consists of more than 200 different viruses, which are associated with a wide variety of medically important diseases including the common cold, aseptic meningitis, conjunctivitis, encephalitis, and respiratory disease (reviewed in references 7, 39, and 44). Human rhinoviruses (HRV), which include over 100 different virus serotypes, are the most important etiological agents of the common cold. Although HRV-induced upper respiratory illness is often mild and self-limiting, HRV infection may ultimately result in sinusitis, otitis media, and lower respiratory tract illnesses including exacerbations of asthma, cystic fibrosis, and bronchitis in individuals with underlying respiratory disorders (3). Although no effective antiviral therapies for either the prevention or treatment of diseases caused by HRV infection are currently available, considerable progress in the discovery and development of new antirhinoviral drugs directed towards a novel target, the HRV 3C protease, has recently been made (11–15, 19, 22, 26, 36, 41, 47, 51–53).
The HRV 3C protease is responsible for the cleavage of viral precursor polyproteins into structural and enzymatic proteins which are essential for viral replication. DNA sequence comparisons among HRV serotypes, and even among several related picornaviruses, have identified a significant degree of homology within the 3C coding region including the strict conservation of the active-site residues, thus providing an additional rationale for targeting drug discovery efforts (8, 16, 20, 21, 29, 31, 38, 45, 48, 49, 55). AG7088 is a potent, irreversible inhibitor of HRV 3C protease that was discovered by protein structure-based drug design methodologies (12, 35, 36) and is currently undergoing evaluation in phase I clinical studies with humans. In this study, we describe the in vitro activity of AG7088 against a variety of different HRV serotypes as well as related picornaviruses in different cell-based systems, as well as its cytotoxicity in these systems.
MATERIALS AND METHODS
Compounds.
AG7088 and pleconaril (17) were synthesized at Agouron Pharmaceuticals, Inc. Pirodavir (1) was kindly provided by Janssen Research Foundation (Beerse, Belgium), and WIN 51711 (40) was kindly provided by Sterling Winthrop Research Institute (Collegeville, Pa.). Ganciclovir (Syntex Corp., Palo Alto, Calif.) was obtained from a local pharmacy, and acyclovir was purchased from Sigma (St. Louis, Mo.).
Cells and virus strains.
All numbered HRV serotypes, echovirus type 11 (EV 11), enterovirus type 70 (ETV 70), coxsackievirus types A21 (CAV 21) and B3 strain Nancy (CVB 3), human cytomegalovirus (HCMV) strain AD169, and herpes simplex virus type 1 (HSV-1) strain McIntyre were purchased from the American Type Culture Collection (ATCC; Manassas, Va.). HRV Hanks and a nasal lavage from a patient challenged with HRV Hanks were kindly provided by Ronald Turner from the Medical University of South Carolina, Charleston, S.C. HRV and coxsackievirus stocks were propagated, and antiviral assays were performed, in H1-HeLa cells (ATCC) incubated at 34 and 37°C, respectively. ETV 70, EV 11, and HCMV stocks were propagated, and antiviral assays were performed, in MRC-5 (ATCC) cells at 37°C. HSV-1 stocks were propagated, and antiviral assays were performed, in Vero (ATCC) cells incubated at 37°C. Vero cells were grown in minimal essential medium (Life Technologies, Gaithersburg, Md.) supplemented with 5% fetal bovine serum (Hyclone, Logan, Utah). H1-HeLa cells and MRC-5 cells were grown in minimal essential medium supplemented with 10% fetal bovine serum.
Enzyme assays.
The proteolytic activity of HRV 14 3C protease was measured by a continuous fluorescence resonance energy transfer assay as described previously (11–15). In brief, cleavage of the substrate peptide was monitored by the appearance of fluorescent emission at 490 nm (following excitation at 336 nm) in a Perkin-Elmer LS50-B spectrophotometer. Data were analyzed with the nonlinear regression analysis program ENZFITTER, which calculates a first-order rate constant for the inactivation of HRV 14 3C protease. Protease selectivity assays were performed with commercially available proteases (at approximately 10 nM concentrations) essentially as described by the supplier. Human liver cathepsin B, porcine erythrocyte calpain I, and human neutrophil elastase were purchased from Calbiochem (San Diego, Calif.), bovine chymotrypsin and human thrombin were purchased from Boehringer Mannheim (Indianapolis, Ind.), and bovine trypsin was purchased from Sigma.
Cell protection assay.
The ability of compounds to protect cells against infection was measured by a dye reduction method (54). Briefly, H1-HeLa and MRC-5 cells were resuspended at 2 × 105 and 5 × 104 cells per ml, respectively, in medium containing appropriate concentrations of compound or medium only. In some experiments, assays were performed in the presence or absence of either human α1-acid glycoprotein (AAG) or type 1-S bovine submaxillary gland bovine mucin (Sigma), both at a final concentration of 1 mg/ml in medium containing 10% fetal bovine serum. Cells were infected with HRV, CAV 21, and CVB 3 at a multiplicity of infection (MOI) of 0.004 to 0.5, 0.03, and 0.08, respectively, or mock-infected with medium only. An MOI of 2.0 to 6.0 was used in assays utilizing HRV 25. MRC-5 cells were infected with EV 11 or ETV 70 at an MOI of 0.003 or 0.004, respectively, or mock-infected with medium only. Vero cells were resuspended at 1.5 × 105 cells per ml and infected with HSV-1 at an MOI of 0.05 or mock-infected with medium only. One to five days later, 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT; Sigma) with phenazine methosulfate (Sigma) was added to the test plates and the amount of formazan produced was quantified spectrophotometrically at a test reference of 450 nm and a reference wavelength of 650 nm. Data were expressed as the percentage of formazan produced in compound-treated cells compared to formazan produced in wells of uninfected, compound-free cells. The 50% effective concentration (EC50) was calculated as the concentration of compound that increased the percentage of formazan production in infected, compound-treated cells to 50% of that produced by uninfected, compound-free cells. The 50% cytotoxicity concentration (CC50) was calculated as the concentration of compound that decreased the percentage of formazan produced in uninfected, compound-treated cells to 50% of that produced in uninfected, compound-free cells. The therapeutic index was calculated by dividing the cytotoxicity (CC50) by the antiviral activity (EC50).
Time-of-addition assay.
Subconfluent monolayers of H1-HeLa cells in six-well plates were infected with HRV 14 at an MOI of 15. After 1 h of adsorption, cell monolayers were washed three times with phosphate-buffered saline (PBS) and replenished with medium. AG7088 (0.5 μM) or WIN 51711 (3.0 μM) was added at concentrations 20-fold above the EC50 (as determined by the cell protection assay) at the time of infection and at various times thereafter. Eight hours after infection, samples were processed by three freeze-thaw cycles followed by sonication for 15 s and clarification by centrifugation (5 min at 15,000 × g at 4°C). Clarified cell and supernatant lysates were stored at −70°C for subsequent analysis for infectious virus.
Virus yield assay.
Infectious virus titers were determined by a virus plaque assay. Briefly, 0.2 ml of serial 10-fold dilutions of virus were allowed to adsorb onto monolayers of H1-HeLa cells. After 1 h of adsorption, the cell monolayers were washed twice with PBS and overlayed with medium containing 0.5% SeaPlaque agarose (FMC Bioproducts, Rockland, Maine). After 3 days of incubation at 34°C, the cell monolayers were fixed with EAF (65% ethanol, 22% acetic acid, and 4% formaldehyde) and stained with 1% crystal violet, and virus plaques were enumerated. Data were expressed as PFU per milliliter.
Analysis of proteolytic processing.
The ability of AG7088 to inhibit HRV 14 3C-mediated proteolytic processing was assessed by polyacrylamide gel electrophoresis (PAGE) of radiolabeled sodium dodecyl sulfate (SDS)-solubilized lysates of HRV 14-infected cells. Initially, H1-HeLa cells were infected with HRV 14 at an MOI of 10. Eight and one-half hours after infection, the cells were washed with PBS and the medium was replaced with methionine- and cysteine-deficient medium (Life Technologies). At 9 h after infection, appropriate concentrations of compounds were added. After a 30-min exposure to compounds, 50 μCi of [35S]Met-[35S]Cys (Expre35S 35S protein label; New England Nuclear, Boston, Mass.) was added. One hour later, the monolayers were washed twice with cold PBS and lysed in 250 μl of radioimmunoprecipitation assay buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS), sonicated, and stored at −70°C for subsequent analysis. Proteins present in the solubilized cell lysates were resolved by 12% PAGE. Following electrophoresis, gels were stained with Coomassie brilliant blue, destained, and treated with Amplify (Amersham, Arlington Heights, Ill.). Gels were air dried overnight in cellulose sheets and exposed to film at −80°C.
HCMV antiviral assay.
The antiviral activity of AG7088 against HCMV AD169 replication in MRC-5 cells was determined by an enzyme-linked immunosorbent assay (ELISA) using a monoclonal antibody MAb directed against the HCMV major immediate-early gene product (MAb 810; Chemicon, Temecula, Calif.). Briefly, following a 2-h virus adsorption, the inoculum was removed and medium containing the appropriate concentrations of compound was added. Five days after infection, the MRC-5 monolayers were incubated with MAb 810, followed by goat anti-mouse antibody conjugated with horseradish peroxidase (Bio-Rad, Hercules, Calif.), and viral antigen was then detected spectrophotometrically at 650 nm with the tetramethylbenzidine liquid substrate system (Sigma). The EC50 was calculated as the concentration of compound that reduced the optical density to 50% of that of the virus control. The CC50 was measured by the XTT reduction method as described above.
Statistical analyses.
Determination of statistical significance was made by using Student’s unpaired t test with the Statview (SAS Institute, Inc., Cary, N.C.) software program.
RESULTS
Activity against HRV 3C protease.
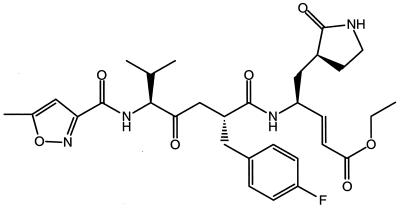
AG7088 is a ketomethylene-containing peptidomimetic compound which incorporates an unsaturated ethyl ester Michael acceptor (Fig. 1). AG7088 has demonstrated potent and irreversible inhibition of HRV 3C protease with an inactivation rate constant (kobs/[I]) of 1,470,000 ± 440,000 M−1 s−1 for HRV 14. Enzyme inhibition was shown to be specific for the viral 3C protease since 10 μM AG7088 produced no significant inhibition against a variety of serine or cysteine proteases, e.g., human elastase, human thrombin, bovine trypsin, bovine chymotrypsin, human and bovine cathepsin B, and porcine calpain. In addition, lack of reactivity with nonenzymatic thiols was demonstrated by AG7088’s stability when incubated in the presence of dithiothreitol (5 mM) (12).
FIG. 1.
Chemical structure of AG7088.
Activity against HRV serotypes and cytotoxicity of AG7088.
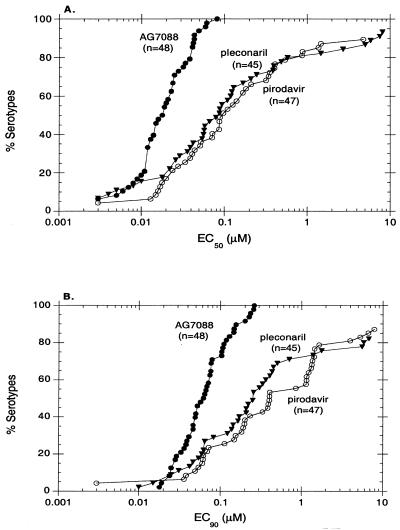
The efficacy of AG7088 against a panel of 48 different HRV serotypes was evaluated in a cell protection assay utilizing H1-HeLa cells (Fig. 2; Table 1). These included representative virus strains derived from minor and major receptor groups (50) as well as from two antiviral groups (A and B) previously defined based on differing susceptibilities to capsid-binding molecules (2). Pirodavir and pleconaril, compounds which inhibit virus capsid attachment and/or uncoating (1, 17), were included for comparison. Results indicate that AG7088 was active against all HRV serotypes (48 of 48) tested with a mean EC50 of 0.023 μM (range, 0.003 to 0.081 μM) and a mean EC90 of 0.082 μM (range, 0.018 to 0.261 μM). These values were comparable to or significantly better than those obtained with both pirodavir and pleconaril; pirodavir inhibited the replication of 42 of 47 (89%) HRV serotypes tested with a mean EC50 of 0.329 μM (range, 0.003 to 4.770 μM), and pleconaril inhibited the replication of 42 of 45 (93%) HRV serotypes tested with a mean EC50 of 0.822 μM (range, 0.003 to 8.122 μM). Furthermore, although AG7088 was able to inhibit approximately 80% of the HRV serotypes tested with EC50 and EC90 of less than or equal to 0.038 and 0.110 μM, respectively, EC50 of less than or equal to 0.579 and 0.862 μM and EC90 of less than or equal to 5.84 and 3.96 μM were necessary for pleconaril and pirodavir, respectively, to inhibit this same percentage of HRV serotypes (Fig. 2). Consistent with results obtained with pirodavir, AG7088 demonstrated comparable levels of activity against HRV serotypes derived from either major or minor receptor groups as well as from antiviral groups A or B (Table 1). In contrast, significant reductions in activity against HRV serotypes classified in antiviral group A were observed for pleconaril.
FIG. 2.
In vitro activity of AG7088 against HRV serotypes. EC50 (A) and EC90 (B) of AG7088, pleconaril, and pirodovir for 48, 45, and 47 HRV serotypes, respectively, were determined by measuring XTT dye reduction following 2 to 5 days of infection of H1-HeLa cells as described in Materials and Methods.
TABLE 1.
In vitro activity of AG7088 against HRV serotypesa
| HRV serotype | EC50 (μM) of:
|
EC90 (μM) of:
|
||||
|---|---|---|---|---|---|---|
| AG7088 | Pirodavir | Pleconaril | AG7088 | Pirodavir | Pleconaril | |
| 1ace | 0.041 ± 0.025 | 0.353 ± 0.311 | 0.388 ± 0.299 | 0.256 | 0.375, 2.497 | 0.653, 2.894 |
| 2ce | 0.012 ± 0.005 | 0.013 ± 0.009 | <0.003 | 0.030, 0.027 | 0.060 ± 0.050 | 0.054 |
| 3bd | 0.017, 0.019 | 0.015, 0.052 | 0.056, 0.055 | 0.027, 0.048 | 0.021, 0.087 | 0.178, 0.097 |
| 8bd | 0.023 ± 0.019 | >10 ± 0 | 5.016 ± 0.944 | 0.049 ± 0.037 | >10 ± 0 | >10 ± 0 |
| 9cd | 0.017, 0.006 | 0.004, 0.026 | <0.003, 0.007 | 0.083, 0.031 | 0.027, 0.439 | 0.100, 0.562 |
| 10cd | 0.024 ± 0.007 | 0.041 ± 0.016 | 0.123 | 0.147 ± 0.091 | 0.112 ± 0.037 | 0.302 |
| 11cd | 0.081 ± 0.133 | 0.141 ± 0.204 | 1.465 ± 1.586 | 0.021, 0.289 | 1.644 ± 2.036 | 6.710 ± 5.699 |
| 12cd | 0.041 | 0.018 | 0.109 | 0.110 | 0.062 | 0.256 |
| 13bd | 0.012 | 0.183 | 0.579 | 0.079 | 0.405 | 5.600 |
| 14bd | 0.013 ± 0.012 | 0.044 ± 0.045 | 0.058 ± 0.043 | 0.050 ± 0.048 | 0.154 ± 0.192 | 0.208 ± 0.169 |
| 15cd | 0.020 ± 0.022 | 0.517, 0.166 | 0.495, 0.478 | 0.103 ± 0.107 | 0.906, 1.995 | 1.200, 1.585 |
| 16cd | 0.015 ± 0.003 | 0.051 ± 0.016 | ND | 0.077 ± 0.021 | 0.191 ± 0.073 | ND |
| 17bd | 0.014, 0.013 | 0.148, 0.654 | >10, >10 | 0.027, 0.029 | 0.275, 2.626 | >10, >10 |
| 19cd | 0.013, 0.007 | 0.211 ± 0.295 | 0.161, 0.076 | 0.029, 0.042 | 1.226 ± 1.338 | 0.487, 0.383 |
| 21cd | 0.011 ± 0.005 | 0.008, 0.023 | 0.012, 0.045 | 0.076 ± 0.024 | 0.021, 0.054 | 0.028, 0.063 |
| 22cd | 0.034 ± 0.016 | 0.042, 0.005 | 0.010, 0.010 | 0.066 ± 0.025 | 0.084, 0.014 | 0.034, 0.027 |
| 23cd | 0.038 ± 0.043 | 0.043, 0.150 | 0.018 ± 0.023 | 0.225 ± 0.313 | 0.062, 0.739 | 0.042 ± 0.030 |
| 24cd | 0.049 ± 0.035 | 0.072, 0.261 | 0.048, 0.370 | 0.074 | 0.390 | 0.144 |
| 25cd | 0.061 ± 0.047 | >10 ± 0 | 0.054 ± 0.014 | 0.201 ± 0.147 | >10 ± 0 | 0.250 ± 0.237 |
| 28cd | 0.043 ± 0.026 | 0.046, 0.131 | 0.014, 0.057 | 0.118 ± 0.067 | 0.119, 0.246 | 0.031, 0.096 |
| 29ce | 0.025 ± 0.018 | 0.018, 0.016 | 0.050, 0.027 | 0.062 ± 0.039 | 0.044, 0.028 | 0.095, 0.068 |
| 30ce | 0.036, 0.013 | 0.025, 0.119 | 0.089, 0.084 | 0.071, 0.033 | 0.079, 0.316 | 0.338, 0.562 |
| 31ce | 0.021 ± 0.014 | 4.621, 4.919 | 0.019, 0.025 | 0.039 ± 0.024 | 7.943, 7.833 | 0.053, 0.071 |
| 32bd | 0.015 ± 0.007 | 0.418, 0.400 | 0.118, 0.140 | 0.052 ± 0.022 | 0.840, 1.896 | 0.536, 0.876 |
| 36cd | 0.013 ± 0.007 | 0.087 ± 0.067 | 0.018 ± 0.013 | 0.064 ± 0.080 | 1.323 ± 0.572 | 0.062 ± 0.023 |
| 38cd | 0.069, 0.048 | 0.030, 0.222 | 0.019, 0.025 | 0.208, 0.082 | 0.112, 0.700 | 0.261, 0.080 |
| 39cd | 0.051, 0.012 | 0.053, <0.003 | ND | 0.095, 0.052 | 0.022, 0.100 | ND |
| 40cd | 0.043 ± 0.043 | 0.020 | 0.026 | 0.129 ± 0.110 | 0.069 | 0.059 |
| 45bd | 0.023 ± 0.007 | >10, >10 | >10, >10 | 0.076 ± 0.021 | >10, >10 | >10, >10 |
| 49ce | 0.012, 0.018 | ND | ND | 0.062, 0.408 | ND | ND |
| 52bd | 0.016 ± 0.003 | 0.530, 0.547 | 6.244, >10 | 0.046 ± 0.037 | 0.856, 0.848 | >10, >10 |
| 53cd | 0.006 ± 0.004 | 0.093, 0.124 | 0.061, 0.114 | 0.025 ± 0.018 | 2.371, 0.316 | 0.197, 0.243 |
| 54bd | 0.012 | 0.880 | 2.721 | 0.261 | >10 | >10 |
| 56cd | <0.003 | <0.003 | 0.093 | 0.033 | <0.003 | 0.407 |
| 59cd | <0.003 | 0.071 | 0.394 | 0.018 | 1.136 | 5.843 |
| 63cd | 0.024 ± 0.019 | 0.039 ± 0.013 | 0.168 ± 0.016 | 0.071 ± 0.068 | 0.201 ± 0.033 | 0.362 ± 0.100 |
| 67cd | 0.019 ± 0.016 | 0.052 ± 0.012 | 0.066 ± 0.026 | 0.046 ± 0.027 | 0.148 ± 0.101 | 0.165 ± 0.064 |
| 68cd | 0.042 ± 0.043 | 1.470 ± 0.079 | 0.004 ± 0.002 | 0.107 ± 0.082 | 5.290 ± 4.079 | 0.022, 0.008 |
| 73cd | 0.019 ± 0.011 | 0.087 ± 0.051 | 0.057 ± 0.013 | 0.046 ± 0.027 | 0.268, 0.495 | 0.209 ± 0.023 |
| 75cd | 0.011, 0.003 | 1.624, 0.100 | 0.109, 0.049 | 0.024, 0.023 | 7.017, 0.909 | 0.270, 0.241 |
| 78cd | 0.005 | 1.379 | <0.003 | 0.019 | 6.629 | 0.010 |
| 81cd | 0.012, 0.004 | 0.617, 0.019 | 0.357, 0.133 | 0.025, 0.013 | 2.248, 0.060 | 0.718, 0.282 |
| 84bd | 0.020, <0.003 | 0.050, 0.231 | 7.504 ± 4.323 | 0.071, 0.027 | 1.413, 0.903 | >10 ± 0 |
| 86bd | 0.012, 0.005 | 0.045, 0.127 | 0.048, 0.036 | 0.029, 0.052 | 0.183, 0.479 | 0.178, 0.066 |
| 87bf | 0.060, 0.021 | >10, >10 | >10, >10 | 0.121, 0.087 | >10, >10 | >10, >10 |
| 89cd | 0.015, 0.009 | 0.041, 0.007 | <0.003 | 0.026, 0.024 | 0.112, 0.031 | 0.025 |
| 95bd | 0.028 ± 0.017 | >10, >10 | 4.977, 6.651 | 0.068 ± 0.053 | >10, >10 | >10, >10 |
| Hankscd | <0.003, <0.003 | <0.003 | 0.003, 0.013 | 0.026, 0.021 | <0.003 | 0.023, 0.027 |
Antiviral activity was determined by measuring XTT dye reduction following 2 to 5 days of infection of H1-HeLa cells as described in Materials and Methods. Results represent the means ± standard deviations (3 to 33 experiments) or individual values (1 or 2 experiments). ND, not determined.
Antiviral group A (2).
Antiviral group B (2).
Major receptor group (50).
Minor receptor group (50).
Neither major nor minor receptor group (50).
AG7088 also demonstrated comparable activity against a low-passage clinical isolate, with an EC50 and EC90 of 0.006 and 0.030 μM, respectively, as well as against HRV 14 replication in a different host cell type, i.e., MRC-5 cells, with an EC50 and EC90 of 0.004 and 0.009 μM, respectively. The specificity of AG7088 for HRV was also demonstrated by the absence of activity (EC50 > 100 μM) against other heterologous viruses, e.g., HCMV strain AD169 and HSV-1 strain McIntyre (data not shown).
Cytotoxicity in H1-HeLa cells was measured in parallel with the determination of antiviral activity. Results indicated that the EC50 of AG7088 required for antiviral activity (range, 0.003 to 0.081 μM) was significantly less than the CC50 of >1,000 μM, yielding a therapeutic index of >12,346 to >333,333. CC50 of 150 and 77 μM were observed for pirodavir and pleconaril, respectively, yielding therapeutic indices of <15 to 49,967 and <8 to 25,667, respectively.
Antiviral activity of AG7088 after HRV infection.
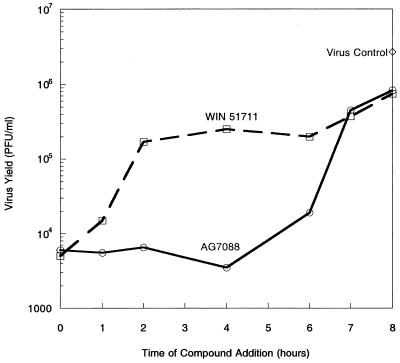
A time-of-addition assay was performed to determine the efficacy of AG7088 when added at various times after virus infection. For this purpose, H1-HeLa cells were infected with HRV 14 at a high MOI to achieve a single cycle of virus replication and levels of infectious virus were determined 8 h later. Results indicated that the addition of AG7088 to infected cells could be delayed up to 6 h after infection without a significant loss of in antiviral activity (Fig. 3). In contrast, a compound (WIN 51711) which binds to virus capsids and acts by inhibiting virus uncoating (40) was inhibitory only when provided at the initiation of the virus life cycle; a loss of virus suppression was observed when addition of the compound was delayed until 2 h after infection.
FIG. 3.
Antiviral activity of AG7088 when added at various times after virus infection. H1-HeLa cells were infected with HRV 14 at an MOI of 15. AG7088 (0.5 μM) and WIN 51711 (3.0 μM) were added at various times after infection. Virus control represents infected cells incubated with medium only. Eight hours after infection, cell lysates and supernatants were collected and the level of infectious virus was determined as described in Materials and Methods.
Inhibition of HRV 3C proteolytic processing.
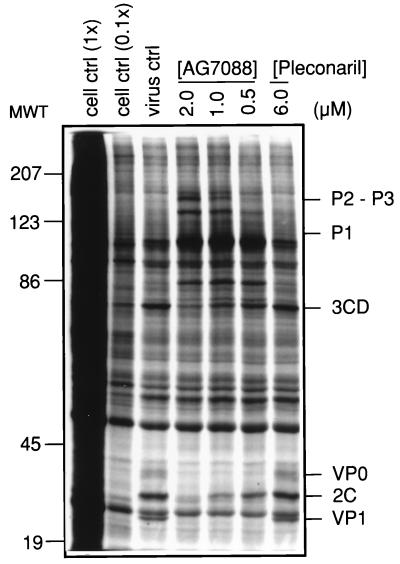
To confirm that the in vitro antiviral activity of AG7088 was derived from a direct inhibition of HRV 3C-mediated proteolytic processing, HRV14-infected H1-HeLa cells were treated with AG7088 (2.0 to 0.5 μM) and radiolabeled polyproteins were resolved by SDS-PAGE. Pleconaril was included as a negative control. SDS-PAGE analysis (Fig. 4) indicated a dose-dependent accumulation of large HRV 14 precursor polyproteins with a concomitant reduction of low-molecular-weight cleavage products in cells treated with AG7088 but not pleconaril. Polyprotein cleavage products predicted to accumulate following inhibition of 3C-mediated proteolytic processing, e.g., P1 (97 kDa) and P2-P3 (146 kDa) were observed. Likewise, a predicted reduction in the polyprotein cleavage products, e.g., VP1 (33 kDa), VP0 (37 kDa), 2C (38 kDa), and 3CD (72 kDa), was also observed.
FIG. 4.
Inhibition of HRV 14 3C-mediated proteolytic processing by AG7088. SDS-solubilized lysates were prepared from uninfected cells (cell ctrl; 0.1× indicates 1/10 the amount of uninfected cell lysate analyzed in lane designated 1×) or infected cells treated with AG7088, pleconaril, or medium only (virus ctrl), and equal amounts of protein were analyzed by PAGE as described in Materials and Methods. P2-P3, P1, 3CD, VP0, 2C, and VP1 designate HRV-specific polypeptides. MWT, molecular weight (in thousands).
Effect of protein on the antiviral activity of AG7088.
Since evaluation of AG7088 in human clinical studies involves delivery by the intranasal route, it was of interest to evaluate the potential effects of proteins present in nasal secretions, AAG and mucin, on the in vitro antiviral activity of AG7088 in a H1-HeLa cell protection assay. Although the physiological concentration of AAG or mucin in nasal fluid is not known with certainty, AAG at a concentration of 1 mg/ml, which represents a physiologically relevant concentration in plasma (30), and mucin at a concentration of 1 mg/ml, which represents the maximum soluble concentration, were resuspended in medium containing 10% fetal bovine serum for these experiments. Under these conditions, no statistically significant differences between the antiviral activity of AG7088 in the presence of either AAG or mucin (P > 0.05; Table 2) and the antiviral activity of AG7088 in the absence of AAG or mucin were observed. The mean EC50 and EC90 for AG7088 against HRV 14 in the absence of AAG were 0.021 and 0.040 μM, respectively, while in the presence of AAG, the mean EC50 and EC90 were 0.043 and 0.108 μM, respectively. Similarly, the EC50 and EC90 in the absence of mucin were 0.059 and 0.160 μM, respectively, while in the presence of mucin the EC50 and EC90 were 0.030 μM and 0.074 μM, respectively. No cytotoxicity was observed up to concentrations of 1 and 10 μM AG7088 in the presence of either AAG or mucin, respectively.
TABLE 2.
In vitro antiviral efficacy of AG7088 in the presence of AAG or mucina
| Treatmentb | EC50 (μM)c | EC90 (μM)c | CC50 (μM) | Therapeutic indexd |
|---|---|---|---|---|
| No AAG | 0.009, 0.033 | 0.031, 0.048 | >1 | >48 |
| AAG | 0.043 ± 0.020 | 0.108 ± 0.038 | >1 | >23 |
| No mucin | 0.059 ± 0.019 | 0.086, 0.234 | >10 | >169 |
| Mucin | 0.030 ± 0.019 | 0.074 ± 0.033 | >10 | >333 |
Antiviral efficacy was determined by measuring XTT dye reduction following 2 or 3 days of infection of H1-HeLa cells with HRV 14 as described in Materials and Methods.
Experiments were conducted in the presence or absence of AAG or mucin (1 mg/ml), both resuspended in medium containing 10% fetal bovine serum.
Results represent the means ± standard deviations (three or four experiments) or individual values (one or two experiments).
Therapeutic index = CC50/EC50.
Activity of AG7088 against related picornaviruses.
The efficacy of AG7088 against four related picornaviruses was also examined. Pirodavir and pleconaril were included for comparison. In H1-HeLa or MRC-5 cell protection assays, AG7088 was active against all four picornaviruses tested, with EC50 ranging from 0.007 to 0.183 μM and EC90 ranging from 0.033 to 0.340 μM (Table 3). These values were comparable to or significantly better than those obtained with both pirodavir and pleconaril; pirodavir inhibited the replication of two of the four (50%) picornaviruses tested with EC50 of 4.833 and 0.443 μM, and pleconaril inhibited the replication of three of the four (75%) picornaviruses tested with EC50 ranging from 0.037 to 1.012 μM. SDS-PAGE analysis of CAV 21-infected cells treated with AG7088 indicated a dose-dependent accumulation of large viral precursor polyproteins with a concomitant reduction of low-molecular-weight cleavage products, confirming that the in vitro antiviral efficacy of AG7088 is due to a direct inhibition of picornavirus 3C protease (data not shown).
TABLE 3.
In vitro activity of AG7088 against picornavirus infectiona
| Virus strain | AG7088
|
Pirodavir
|
Pleconaril
|
|||
|---|---|---|---|---|---|---|
| EC50 (μM) | EC90 (μM) | EC50 (μM) | EC90 (μM) | EC50 (μM) | EC90 (μM) | |
| CAV 21 | 0.147 ± 0.024 | 0.314 ± 0.073 | 4.833 | >10 | 0.037 ± 0.048 | 0.148 ± 0.150 |
| CVB 3 | 0.183 ± 0.161 | 0.340 ± 0.321 | >10 | >10 | >10 | >10 |
| EV 11 | 0.014 ± 0.003 | 0.033 ± 0.015 | >10 | >10 | 0.165 | 0.275 |
| ETV 70 | 0.007 ± 0.006 | 0.066 ± 0.119 | 0.443 | 0.899 | 1.297, 0.726 | 2.726, 2.492 |
Antiviral efficacy was determined by measuring XTT dye reduction following 1 to 5 days of infection as described in Materials and Methods. Results represent the means ± standard deviations (three to six experiments) or individual values (one or two experiments).
DISCUSSION
As members of one of the largest families of medically important human pathogens, HRVs are the single major cause of the common cold. HRV-induced upper respiratory illness has also been associated with serious medical complications in individuals with underlying respiratory disorders (3). Although potent in vitro antirhinoviral activity has been described for numerous compounds to date (reviewed in references 5, 9, and 37), in only a few instances have reductions in clinical symptoms and/or virus infection been achieved in clinical trials (3, 9). The vast majority of these compounds act by binding to virus capsids and inhibiting either virus attachment or subsequent uncoating. However, recent reports have described novel inhibitors of 3C protease. The latter include peptide aldehydes (19, 22, 47, 53), isatins (52), and homophthalimides (51). Recently a class of irreversible inhibitors incorporating Michael acceptors, which exhibits potent cell-based antiviral activity with little to no cellular cytotoxicity, has been described (11–15, 26). In this study, we describe the antiviral activity and cytotoxicity of AG7088, a potent, peptidomimetic inhibitor of HRV 3C protease that has recently begun to be evaluated in human clinical studies.
In cell-based assays, AG7088 demonstrated comparable antiviral potencies (27-fold range in EC50) against all 48 HRV serotypes tested. These results were in contrast to those observed with the capsid-binding compounds tested. The latter compounds demonstrated a significantly wider range in potency (1,590- to 2,707-fold range in EC50) and activity against most but not all HRV serotypes tested. The finding of comparable ranges in the activity of AG7088 against numerous HRV serotypes is consistent with DNA sequence analyses performed on 3C protease-coding gene regions, which demonstrate a significant level of homology in substrate/inhibitor binding regions (8, 16, 20, 21, 29, 31, 36, 38, 45, 48, 49, 55). This same level of homology, reflected in the 3C protease-coding regions derived from other picornaviruses, is also consistent with levels of activity demonstrated against four related picornaviruses.
Nasal secretions are biochemically complex and contain many serum proteins and mucous glycoproteins (33, 43, 46) including acid glycoproteins (34, 42) (unpublished observations). Since human clinical studies of AG7088 involve delivery by the intranasal route, it was of interest to determine the potential effects of mucin and AAG, proteins that are present in nasal washings. AAG is a major serum glycoprotein that has been shown to reduce the in vitro antiviral activity of several human immunodeficiency virus protease inhibitors (4, 28, 32). AAG has also been detected in various concentrations in nasal lavages (unpublished data). No significant reductions, however, in the antiviral efficacy of AG7088 in the presence of either AAG or mucin were observed.
HRVs, as members of the picornavirus family, encode a single large polyprotein precursor (reviewed in references 7, 10, 23, 27, 39, and 44) that depends on the virally encoded 3C protease for all posttranslational cleavages with the exception of an initial autocatalytic cleavage by the 2A protease to release P1, the precursor to the viral capsid proteins, an alternative cleavage by 2A to generate 3C′ and 3D′ products, and a late autocatalytic cleavage of VP0, which occurs during final viral assembly. The necessity of the 3C protease throughout the virus life cycle was confirmed in experiments that demonstrated that the ability of AG7088 to suppress virus replication when added throughout a single cycle of virus replication in a time-of-addition assay. That the antiviral activity of AG7088 was due directly to its inhibition of 3C protease was demonstrated by SDS-PAGE analysis of infected cell lysates, which indicated a dose-dependent accumulation of large HRV 14 precursor polyproteins with a concomitant reduction of low-molecular-weight cleavage products. Profiles of polyproteins predicted to accumulate or be reduced were consistent with the cleavage profile observed when certain amino acid substitutions are introduced into the 3C protease catalytic site (6, 18, 24, 25).
These studies describe the antiviral activity and cytotoxicity of a novel inhibitor of HRV 3C protease. As an irreversible inhibitor, AG7088 may be capable of forming covalent interactions with other proteins and inducing possible toxicities in vivo. The potential for these types of interactions have, however, been minimized by selection of a Michael acceptor with only mild chemical reactivity (12) and certainly can be explored in appropriate clinical trial studies. In summary, the potent activity against all HRV serotypes tested combined with the ability to inhibit virus replication throughout a single cycle of virus replication indicates that AG7088 is a promising new clinical candidate.
ACKNOWLEDGMENT
We thank Jules Beardsley for help in preparation of the manuscript.
REFERENCES
- 1.Andries K, Dewindt B, Snoeks J, Willebrords R, Van Eemeren K, Stokbroekx R, Janssen P A J. In vitro activity of pirodavir ( R77975), a substituted phenoxy-pyridazinamine with broad-spectrum antipicornaviral activity. Antimicrob Agents Chemother. 1992;36:100–107. doi: 10.1128/aac.36.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andries K, Dewindt B, Snoeks J, Wouters L, Moereels H, Lewi P J, Janssen P A J. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J Virol. 1990;64:1117–1123. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arruda E, Hayden F G. Clinical studies of antiviral agents for picornaviral infections. In: Jeffries D J, De Clerq E, editors. Antiviral chemotherapy. Chichester, N.Y: John Wiley & Sons, Ltd.; 1995. pp. 321–355. [Google Scholar]
- 4.Bilello J A, Bilello P A, Prichard M, Robins T, Drusano G L. Reduction of the in vitro activity of A77003, an inhibitor of human immunodeficiency virus protease, by human serum alpha-1 acid glycoprotein. J Infect Dis. 1995;171:546–551. doi: 10.1093/infdis/171.3.546. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco L. Picornavirus inhibitors. Pharmacol Ther. 1994;64:119–128. doi: 10.1016/0163-7258(94)90040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheah K-C, Leong L E-C, Porter A G. Site-directed mutagenesis suggests close functional relationship between a human rhinovirus 3C cysteine protease and cellular trypsin-like serine proteases. J Biol Chem. 1990;265:7180–7187. [PubMed] [Google Scholar]
- 7.Couch R B. Rhinoviruses. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 607–629. [Google Scholar]
- 8.Dahllund L, Nissinen L, Pulli T, Hyttinen V P, Stanway G, Hyypia T. The genome of echovirus 11. Virus Res. 1995;35:215–222. doi: 10.1016/0168-1702(94)00104-k. [DOI] [PubMed] [Google Scholar]
- 9.Diana G D, Pevear D C. Antipicornavirus drugs: current status. Antiviral Chem Chemother. 1997;8:401–408. [Google Scholar]
- 10.Dougherty W G, Semler B L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragovich P S, Prins T J, Zhou R, Fuhrman S A, Patick A K, Matthews D A, Ford C E, Meador III J W, Ferre R A, Worland S T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 3. Structure-activity studies of ketomethylene-containing peptidomimetics. J Med Chem. 1999;42:1203–1212. doi: 10.1021/jm980537b. [DOI] [PubMed] [Google Scholar]
- 12.Dragovich P S, Prins T J, Zhou R, Webber S E, Marakovits J T, Fuhrman S A, Patick A K, Matthews D A, Lee C A, Ford C E, Burke B J, Rejto P A, Hendrickson T F, Tuntland T, Brown E L, Meador III J W, Ferre R A, Harr J E V, Kosa M B, Worland S T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as l-glutamine replacements. J Med Chem. 1999;42:1213–1224. doi: 10.1021/jm9805384. [DOI] [PubMed] [Google Scholar]
- 13.Dragovich P S, Webber S E, Babine R E, Fuhrman S A, Patick A K, Matthews D A, Lee C A, Reich S H, Prins T J, Marakovits J T, Littlefield E S, Zhou R, Tikhe J, Ford C E, Wallace M, Meador III J W, Ferre R A, Brown E L, Binford S L, Harr J E V, DeLisle D M, Worland S T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure-activity studies. J Med Chem. 1998;41:2806–2818. doi: 10.1021/jm980068d. [DOI] [PubMed] [Google Scholar]
- 14.Dragovich P S, Webber S E, Babine R E, Fuhrman S A, Patick A K, Matthews D A, Reich S H, Marakovits J T, Prins T J, Zhou R, Tikhe J, Littlefield E S, Bleckman T M, Wallace M, Little T, Ford C E, Meador III J W, Ferre R A, Brown E L, Binford S L, DeLisle D M, Worland S T. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure-activity studies. J Med Chem. 1998;41:2819–2834. doi: 10.1021/jm9800696. [DOI] [PubMed] [Google Scholar]
- 15.Dragovich P S, Zhou R, Skalitzky D J, Fuhrman S A, Patick A K, Ford C E, Meador III J W, Worland S T. Solid-phase synthesis of irreversible human rhinovirus 3C protease inhibitors. 1. Optimization of tripeptides incorporating N-terminal amides. Bioorg Med Chem Lett. 1999;7:589–598. doi: 10.1016/s0968-0896(99)00005-x. [DOI] [PubMed] [Google Scholar]
- 16.Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, Blaas D, Kuechler E. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2, and 14. Proc Natl Acad Sci USA. 1987;84:2605–2609. doi: 10.1073/pnas.84.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromtling R A, Castañer J. VP-63843 pleconaril WIN-63843. Drugs Future. 1997;22:40–44. [Google Scholar]
- 18.Hammerle T, Hellen C U T, Wimmer E. Site-directed mutagenesis of the putative catalytic triad of poliovirus 3C proteinase. J Biol Chem. 1991;266:5412–5416. [PubMed] [Google Scholar]
- 19.Heinz B A, Tang J, Labus J M, Chadwell F W, Kaldor S W, Hammond M. Simple in vitro translation assay to analyze inhibitors of rhinovirus proteases. Antimicrob Agents Chemother. 1996;40:267–270. doi: 10.1128/aac.40.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes P J, North C, Jellis C H, Minor P D, Stanway G. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J Gen Virol. 1988;69:49–50. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- 21.Hughes P J, North C, Minor P D, Stanway G. The complete nucleotide sequence of coxsackievirus A21. J Gen Virol. 1989;70:2943–2952. doi: 10.1099/0022-1317-70-11-2943. [DOI] [PubMed] [Google Scholar]
- 22.Kaldor S W, Hammond M, Dressman B A, Labus J M, Chadwell F W, Kline A D, Heinz B A. Glutamine-derived aldehydes for the inhibition of human rhinovirus 3C protease. Bioorg Med Chem Lett. 1995;5:2021–2026. [Google Scholar]
- 23.Kay J, Dunn B M. Viral proteinases: weakness in strength. Biochim Biophys Acta. 1990;1048:1–18. doi: 10.1016/0167-4781(90)90015-t. [DOI] [PubMed] [Google Scholar]
- 24.Kean K M, Howell M T, Grünert S, Girard M, Jackson R J. Substitution mutations at the putative catalytic triad of the poliovirus 3C protease have differential effects on cleavage at different sites. Virology. 1993;194:360–364. doi: 10.1006/viro.1993.1268. [DOI] [PubMed] [Google Scholar]
- 25.Kean K M, Teterina N L, Marc D, Girard M. Analysis of putative active site residues of the poliovirus 3C protease. Virology. 1991;181:609–619. doi: 10.1016/0042-6822(91)90894-h. [DOI] [PubMed] [Google Scholar]
- 26.Kong J-S, Venkatraman S, Furness K, Nimkar S, Shepherd T A, Wang Q M, Aube J, Hanzlik R P J. Synthesis and evaluation of peptidyl Michael acceptors that inactivate human rhinovirus 3C protease and inhibit virus replication. J Med Chem. 1998;41:2579–2587. doi: 10.1021/jm980114+. [DOI] [PubMed] [Google Scholar]
- 27.Kräusslich H-G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 28.Lazdins J K, Mestan J, Goutte G, Walker M R, Bold G, Capraro H G, Klimkait T. In vitro effect of alpha-acid glycoprotein on the anti-human immunodeficiency virus (HIV) activity of the protease inhibitor CGP 61755: a comparative study with other relevant HIV protease inhibitors. J Infect Dis. 1997;175:1063–1070. doi: 10.1086/520352. [DOI] [PubMed] [Google Scholar]
- 29.Lee W M, Wang W, Rueckert R R. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes. 1995;9:177–181. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- 30.Lentner C, editor. Geigy scientific tables. Basel, Switzerland: Ciba-Geigy Ltd.; 1984. p. 142. [Google Scholar]
- 31.Lindberg A M, Stalhandske P O, Pettersson U. Genome of coxsackievirus B3. Virology. 1987;156:50–63. doi: 10.1016/0042-6822(87)90435-1. [DOI] [PubMed] [Google Scholar]
- 32.Livingston D J, Pazhanisamy S, Porter D J T, Partaledis J A, Tung R D, Painter G R. Weak binding of VX-478 to human plasma proteins and implications for anti-human immunodeficiency virus therapy. J Infect Dis. 1995;172:1238–1245. doi: 10.1093/infdis/172.5.1238. [DOI] [PubMed] [Google Scholar]
- 33.Lorin M I, Gaerlin P F, Mandel I D. Quantitative composition of nasal secretions in normal subjects. J Clin Med. 1972;80:275–281. [PubMed] [Google Scholar]
- 34.Marom Z, Shelhamer J, Kaliner M. Nasal mucus secretion. Ear Nose Throat J. 1984;63:36–37. [PubMed] [Google Scholar]
- 35.Matthews, D. A., P. S. Dragovich, S. E. Webber, S. A. Fuhrman, A. K. Patick, L. S. Zalman, T. Hendrickson, T. J. Prins, J. T. Marakovits, R. Zhou, J. Tikhe, C. E. Ford, J. W. Meador, R. A. Ferre, E. L. Brown, S. L. Binford, M. A. Brothers, D. M. DeLisle, and S. T. Worland. Structure-assisted design of mechanism based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 36.Matthews D A, Smith W W, Ferre R A, Condon B, Budahazi G, Sisson W, Villafranca J E, Janson C A, McElroy H E, Gribskov C L, Worland S. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 37.McKinlay M A, Pevear D C, Rossmann M G. Treatment of the picornavirus common cold by inhibitors of viral uncoating and attachment. Annu Rev Microbiol. 1992;46:635–654. doi: 10.1146/annurev.mi.46.100192.003223. [DOI] [PubMed] [Google Scholar]
- 38.Meador J W, III, Ngo H, Ford C E, Patick A K, Ferre R A, Matthews D A, Worland S T. PCR amplification and determination of the RNA sequences for the P3 coding region of human rhinoviral serotypes. Antivir Res. 1998;37:A72. [Google Scholar]
- 39.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 549–605. [Google Scholar]
- 40.Otto M J, Fox M P, Fancher M J, Kuhrt M F, Diana G D, McKinlay M A. In vitro activity of WIN 51711, a new broad-spectrum antipicornavirus drug. Antimicrob Agents Chemother. 1985;27:883–886. doi: 10.1128/aac.27.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patick A, Potts K. Protease inhibitors as antiviral agents. Clin Microbiol Rev. 1998;11:614–627. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patow C A, Shelhamer J, Marom Z, Logun C, Kaliner M. Analysis of human nasal mucous glycoproteins. Am J Otolaryngol. 1984;5:334–343. doi: 10.1016/s0196-0709(84)80003-4. [DOI] [PubMed] [Google Scholar]
- 43.Platts-Mills T A. Local production of IgG, IgA, and IgE antibodies in grass pollen hay fever. J Immunol. 1979;12:2218–2225. [PubMed] [Google Scholar]
- 44.Rueckert R R. Picornaviridae and their replication. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 507–548. [Google Scholar]
- 45.Ryan M D, Jenkins O, Hughes P J, Brown A, Knowles N J, Booth D, Minor P D, Almond J W. The complete nucleotide sequence of enterovirus type 70: relationships with other members of the picornaviridae. J Gen Virol. 1990;71:2291–2299. doi: 10.1099/0022-1317-71-10-2291. [DOI] [PubMed] [Google Scholar]
- 46.Schorn D, Hochestrasser M. Biochemical investigations of nasal secretions. Acta Otorhinolaryngol Belgica. 1979;33:603–606. [PubMed] [Google Scholar]
- 47.Shepherd T A, Cox G A, McKinney E, Tang J, Wakulchik M, Zimmerman R E, Villarreal E C. Small peptidic aldehyde inhibitors of human rhinovirus 3C protease. Bioorg Med Chem Lett. 1996;6:2893–2896. [Google Scholar]
- 48.Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C, Fogy I, Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13:2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanway G, Hughes P J, Mountford R C, Minor P D, Almond J W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uncapher C R, DeWitt C M, Colonno R J. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q M, Johnson R B, Hungheim L N, Cohen J D, Villarreal E C. Dual inhibition of human rhinovirus 2A and 3C proteases by homophthalimides. Antimicrob Agents Chemother. 1998;42:916–920. doi: 10.1128/aac.42.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webber S, Tikhe J, Worland S T, Fuhrman S A, Hendrickson T F, Matthews D A, Love R A, Patick A K, Meador J W, Ferre R A, Brown E L, DeLisle D M, Ford C E, Binford S L. Design, synthesis, and evaluation of nonpeptidic inhibitors of human rhinovirus 3C protease. J Med Chem. 1996;39:5072–5082. doi: 10.1021/jm960603e. [DOI] [PubMed] [Google Scholar]
- 53.Webber S E, Okano K, Little T L, Reich S, Xin Y, Worland S T, Fuhrman S A, Matthews D A, Hendrickson T F, Love R A, Patick A K, Meador III J W, Ferre R A, Brown E L, Ford C E, Binford S L. Tripeptide aldehyde inhibitors of human rhinovirus 3C protease: design, synthesis, biological evaluation, and cocrystal structure solution of P1 glutamine isosteric replacements. J Med Chem. 1998;41:2786–2805. doi: 10.1021/jm980071x. [DOI] [PubMed] [Google Scholar]
- 54.Weislow O S, Kiser R, Fine D L, Bader J, Shoemaker R H, Boyd M R. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J Natl Cancer Inst. 1989;81:577–586. doi: 10.1093/jnci/81.8.577. [DOI] [PubMed] [Google Scholar]
- 55.Werner G, Rosenwirth B, Bauer E, Seifert J M, Werner F J, Besemer J. Molecular cloning and sequence determination of the genomic regions encoding protease and genome-linked protein of three picornaviruses. J Virol. 1986;57:1084–1093. doi: 10.1128/jvi.57.3.1084-1093.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]