SUMMARY
Dicer controls the biogenesis of microRNAs (miRNAs) and is essential for neurogenesis. Recent reports show that the levels and substrate selectivity of dicer result in the preferential biogenesis of specific miRNAs in vitro. However, how dicer expression levels and miRNA biogenesis are regulated in vivo and how this affects neurogenesis is incompletely understood. Here we show that during zebrafish hindbrain development dicer expression levels are controlled by miR-107 to tune the biogenesis of specific miRNAs, such as miR-9, whose levels regulate neurogenesis. Loss of miR-107 function stabilizes dicer levels and miR-9 biogenesis across the ventricular hindbrain zone resulting in an increase of both proliferating progenitors and post-mitotic neurons. miR-9 ectopic accumulation in differentiating neuronal cells recapitulated the excessive neurogenesis phenotype. We propose that miR-107 modulation of dicer levels in differentiating neuronal cells is required to maintain the homeostatic levels of specific miRNAs, whose precise accumulation is essential for neurogenesis.
Keywords: zebrafish, neurogenesis, dicer, miR-107, miR-9
INTRODUCTION
miRNAs are highly conserved non-coding small RNAs that bind to the 3′-untranslated region (3′UTR) of target messenger-RNAs (mRNAs) and post-transcriptionally regulate the expression of genes by repressing translation and/or decreasing transcript stability (Bazzini et al., 2012; Guo et al., 2010). miRNAs are transcribed in a tissue- and time-specific manner as large primary miRNA transcripts (pri-miRNAs), and subsequently cleaved by the RNAse enzyme DROSHA and the cofactor DGCR8 to form stem-loop hairpins (pre-miRNAs). Pre-miRNAs are bound by dicer which generates the mature miRNAs (Denli et al., 2004). The levels of miRNAs within a single cell type are important to determine which mRNAs are regulated (Mukherji et al., 2011). Accordingly, miRNA accumulation varies greatly between tissues (Ason et al., 2006; Lagos-Quintana et al., 2002) and regulates a large variety of cellular processes including oncogenesis (He et al., 2005), cardiogenesis (Zhao et al., 2007), muscle development (Mishima et al., 2009), and brain morphogenesis (Giraldez et al., 2005).
Neurogenesis is a highly conserved process among vertebrates (Gage and Temple, 2013). During the development of the central nervous system the proper balance between the proliferation and differentiation of neural progenitors allows the temporal production of post-mitotic neural cells. This process is modulated by miRNAs as suggested by their differential expression during differentiation (Gangaraju and Lin, 2009). Mice, homozygous for dicer deletion, die at embryonic day 7.5, while the tissue specific deletion of dicer at later time points during mouse neural development results in several cerebral cortex defects, including apoptosis, neurons with increased spine and dendritic length, and premature neural stem cell differentation and migration (Cuellar et al., 2008; Volvert et al., 2012) (Kawase-Koga et al., 2009; Saurat et al., 2013). While these studies support a requirement for dicer in neural development, often the reduction of only a few miRNAs (for example miR-124 and miR-9) are reported to be the primary cause of the neurogenesis defects in the dicer-null embryos (Huang et al., 2010). Similary, in zebrafish, removal of dicer leads to profound brain morphogenesis defects that can be recapitulated solely by the absence of miR-430 biogenesis (Giraldez et al., 2005). The importance of dicer for neural cell differentiation led us to investigate the mechanisms that regulate dicer levels necessary for the biogenesis of key miRNAs during neurogenesis.
miR-107 belongs to the higly conserved miR-103/107 family (Polster et al., 2010) and is associated with several pathological conditions, including defects in glucose homeostasis, cancer and neurodegenerative diseases (Martello et al., 2010; Scarr et al., 2013; Trajkovski et al., 2011). In breast cancer cells, miR-103/107 downregulates the expression level of dicer to globally reduce miRNA biogenesis in the long term, leading to metastasis and a poor outcome in patients (Martello et al., 2010). Expression profiles of miR-103/107 family members show that miR-103 is physiologically expressed at similar levels across various human tissues including the cerebral and frontal cortex, heart, lung, and liver, whereas miR-107 is specifically enriched in brain samples (Wang et al., 2014). The regulatory role of miR-107 on dicer function in cancer cells raises questions about the role of miR-107 in dicer-dependent physiological processes such as neurogenesis.
We investigated whether the regulation of dicer activity by miR-107 is a canonical homeostatic mechanism controlling miRNA levels during neuronal differentiation. Using zebrafish as a model we demonstrated that miR-107 is enriched from neural/neuronal progenitor cells to newborn neurons detaching from the ventricular zone of the developing hindbrain. miR-107 interacts with dicer at the post-transcriptional level, negatively tuning its levels and function in neuronal differentiating cells. Gain of miR-107 function causes a profound loss of brain morphogenesis consequential to a loss of dicer and miR-430 biogenesis. On the contrary, loss of endogenous miR-107 increases dicer mRNA stability during late hindbrain neurogenesis thus increasing the number of cycling progenitors generating neurons. Similarly, using adult tectum-derived neurospheres as an in vitro model of neurogenesis, we observed that the silencing of miR-107 results in the stabilization of dicer levels that predominantly promotes miR-9 biogenesis. Loss of miR-107 or the ectopic expression of miR-9 promotes neurosphere neurogenesis in vitro. Congruently, in the embryos lacking miR-107, miR-9 biogenesis increases across the hindbrain ventricular zone as a consequence of dicer stabilization. Thus, the gain of miR-9 function recapitulated the neurogenic defect observed in the absence of miR-107. In conclusion, miR-107 controlled dicer levels along the hindbrain ventricular zone, to tune the biogenesis of specific miRNAs, such as miR-9, whose accumulation in neural and neuronal progenitor cells is crucial to generating neurons. Our data proposes a model in which a tissue specific miRNA-dicer interaction regulates the biogenesis of key miRNAs whose function during cell development is dependent on their levels.
RESULTS
miR-107 expression pattern during hindbrain development
Employing the zebrafish as an experimental model, we first analyzed the spatiotemporal pattern of miR-107 expression in the developing brain.
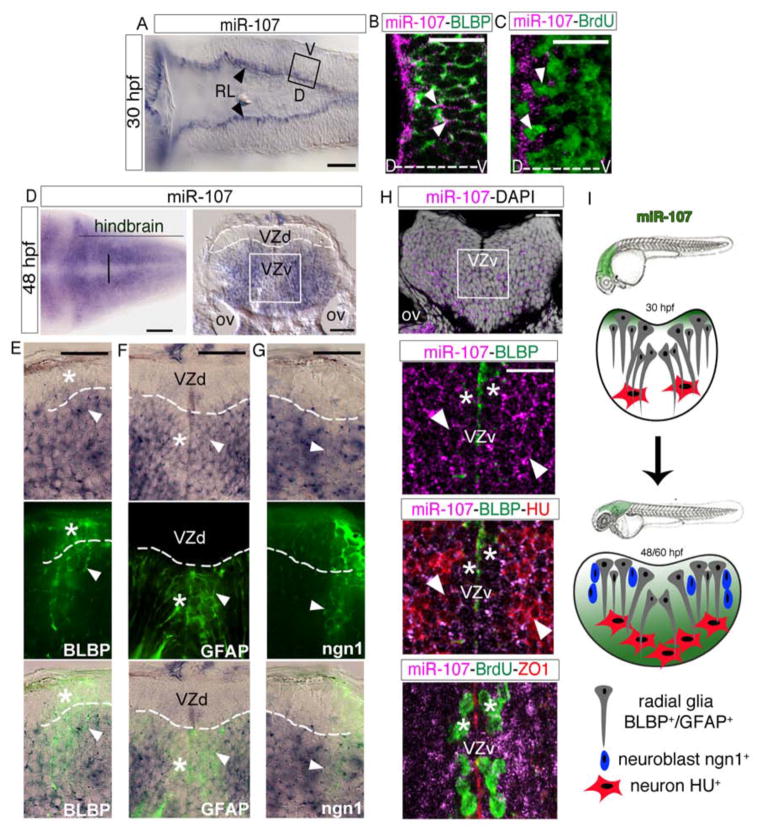
We used a labeled Locked Nucleic Acid (LNA) RNA probe to detect the expression pattern of the mature miR-107 sequence. At 30 hours post-fertilization (hpf), miR-107 started to be expressed by a few proliferating radial glial progenitor cells [incorporating BrdU and expressing the radial glia-marker BLBP(Diotel et al., 2010)] localized in the rhombic lip (RL) of the hindbrain ventricular zone (Figure 1A–C, arrowheads). Interestingly, at later neurogenesis stages (48 and 60 hpf) (Schmidt et al., 2013), miR-107 expression was excluded from the dorsal hindbrain ventricular zone and detected in both the periventricular and post-mitotic mantle zone (Figure 1D–H). To determine the identity, commitment, and fate of the miR-107 positive cells we performed a series of double staining with the miR-107 LNA probe and several protein markers of neural/neuronal progenitors in post-mitotic neurons. We detected that miR-107 was expressed in neural progenitor cells that were BLBP and GFAP positive detaching from the dorsal and ventral ventricular zone, respectively (Figure 1E, F arrowheads). On the contrary, the most ventricular row of BLBP and GFAP positive cells had little to no miR-107 expression, suggesting that miR-107 is excluded in neural cells in the progenitor state at these developmental stages (Figure 1E, F, stars). Based on the strong distribution of miR-107 in the periventricular area, we hypothesized that miR-107 expression would demark a later commitment state of neural cells that exit the ventricular area. Thus, we investigated the expression of miR-107 in neuronal progenitor cells using the Tg(-8.4neurog1:GFP) zebrafish line (Blader et al., 2004). As expected, only the neurogenin (neurog1, hereafter referred as ngn1) GFP positive cells invading the mantle zone co-expressed miR-107 (Figure 1G, arrowheads). This expression pattern further confirms that miR-107 highlights the commitment of neural/neuronal progenitors exiting the ventricular area to generate newborn neurons. Congruently, we detected miR-107 in post-mitotic neurons labeled with the HU marker (Kim et al., 1996) exiting the periventricular area, demarked by the position of BrdU+ proliferating progenitors, and migrating into the mantle zone (Figure 1H, stars and arrowheads respectively). Moreover, miR-107 showed a similar expression pattern in the hindbrain of 60 hpf embryos (Figure S1A–D). Altogether, these data indicate that the expression of miR-107 in the developing hindbrain followed the maturation of neuronal progenitor cells leaving the ventricular area and differentiating into newborn neurons (Figure 1I).
Figure 1. miR-107 is expressed in newborn neurons and in neural/neuronal progenitors across the hindbrain periventricular zone.
(A) Expression of miR-107 at 30hpf is restricted to the rhombic lip (RL) (arrowheads). Dorsal is up and anterior to the left. (B–C) Whole-mount confocal images of the boxed area in (A), stained with miR-107 LNA probe and anti-BLBP antibody in (B) or anti-BrdU antibody in (C). Brain orientation is indicated as D (dorsal) and V (ventral). Arrowheads indicate miR-107 expression. (D–H) Expression of miR-107 across the hindbrain of 48 hpf embryos. (D) Bright-field images of whole mount embryos (left panel) and hindbrain transverse section (right panel). VZd indicates the dorsal ventricular hindbrain zone (white dotted line), VZv indicates ventral ventricular hindbrain zone (white solid square). (E–G) High magnification images (60X) of the VZd zone representative of the region highlighted in D (left panel). miR-107 and protein co-localization is indicated with arrowheads, while miR-107 negative or low expressing cells are indicated with stars. (H) Top panel is a confocal image of a transverse hindbrain section at the level of the otic vesicle (OV). miR-107 is shown in purple and DAPI nuclei are in white. Stars indicate miR-107 expression in the ventral ventricular zone (VZv). Bottom panels are coronal confocal sections at the VZv level highlighted by the white box in the top panels and stained for the indicated proteins. ZO1 staining indicates the apical region of the VZv where BrdU positive cells are nested (white stars). (I) Schematic representation of the dynamic expression of miR-107 (green) in the developing hindbrain from 30 to 48/60 hpf. Scale bars= 50μm.
miR-107 targets dicer during neural development
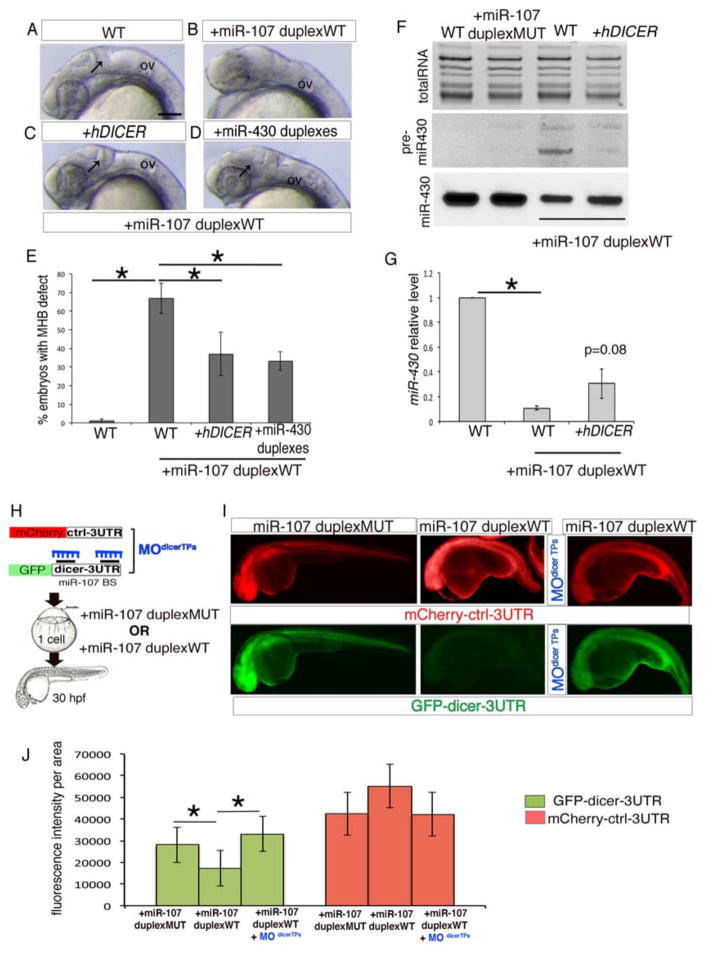
To address whether miR-107 expression was able to post-transcriptionally regulate dicer activity during neurogenesis, we analyzed miRNA biogenesis in miR-107-gain of function embryos obtained by injecting a miR-107 oligonucleotide duplex that mimics the mature sequence of miR-107 (miR-107-duplex-WT). At 24hpf, the majority of the embryos injected with 100 μM miR-107-duplex-WT (around 70%) displayed a loss of the midbrain-hindbrain boundary (MHB) (Figure 2A, B and E), similar to maternal zygotic dicer mutants (MZ dicer) that present a loss of MHB and embryonic miRNAs (Giraldez et al., 2005). Indeed, we demonstrated that this defect predominantly resulted from the repression of dicer transcripts, since the MHB was significantly rescued in embryos injected with both a miR-107 duplex and an in vitro transcribed human DICER mRNA lacking the 3′UTR (hDICER) (Figure 2C, arrow, and E). Moreover, the co-injection of 10 μM miR-430 duplexes was sufficient to rescue the MHB in miR-107-duplex-WT embryos, suggesting that the neural defect in the miR-107-gain of function embryos was consequential to an impaired miR-430 biogenesis (Figure 2D, arrow, and E). To test this hypothesis we analyzed the levels of miR-430 in miR-107-duplex embryos by northern blot and qRT-PCR. As expected, the gain of miR-107 function showed a loss of miR-430 mature sequence and an accumulation of its pre-miRNA indicating loss of dicer activity, similar to the MZ dicer mutants (Giraldez et al., 2005) (Figure 2F and G). Accordingly, the miR-430 biogenesis was partially restored after hDICER co-injection with the miR-107-duplex-WT (Figure 2F and G). In contrast, no accumulation of pre-miR-430 or a significant decrease of mature miR-430 was observed in embryos injected either with 100 μM miR-107 duplex with 8 mutated nucleotides in the seed region (miR-107-duplex-MUT) or with 100 μM miR-221 duplex (Nicoli et al., 2012) or 100 μM miR-1 duplex (Mishima et al., 2009) (Figure 2F and Figure S1E). Thus, excluding the possibility that the injection of the miR-107-duplex-WT at the dose of 100μM may generally compete with the endogenous pool of miRNAs for the intracellular small RNA machinery (Khan et al., 2009). Therefore, all together, our data demonstrate that the ectopic miR-107 expression affects embryonic brain morphogenesis by specifically repressing dicer activity and miRNAs biogenesis.
Figure 2. miR-107 post-transcriptionally regulates dicer during neural development.
(A–D) Lateral view of 24 hpf zebrafish embryos treated as indicated. Anterior is to the left. Black arrows indicate MHB in (A), (C) and (D). (E) Percentage of embryos without MHB observed in (A–D). Data are presented as mean ± SEM of five independent experiments (n~20 embryos each). *p<0.05. (F) Northern Blot showing miR-430 biogenesis in 24 hpf embryos treated as indicated. WT (un-injected embryos) and miR-107-duplex-MUT were used as controls. The image is representative of two independent experiments. (G) qRT-PCR of miR-430 levels in 24 hpf embryos treated as indicated. Levels are relative to the WT control. Data are mean ± SEM (n=4). *p<0.05. (H) Schematic representation of the miR-107 sensor assay shown in (I–J). (I) Fluorescent images of whole-mount zebrafish embryos at 24 hpf treated as indicated. Anterior is to the left. (J) Quantification of GFP and mCherry fluorescence levels in each experimental condition measured as fluorescence intensity per area. Data are presented as mean ± SD of 3 independent experiments (n=5 to 10 embryos each). *p<0.05. Scale bars= 50μm. CNS: Central Nervous System; OV: otic vesicle.
To further assess if miR-107 directly regulates dicer mRNA translation, we used a previously validated miRNA sensor strategy (Mishima et al., 2009) (Figure 2H). Embryos were co-injected with a green fluorescent protein (GFP) fused with a zebrafish dicer-3′UTR sequence containing two conserved miR-107 binding sites (GFP-dicer-3′UTR) (Figure S1F), and the miR-107-duplex-WT or the miR-107 duplex-MUT. Co-injection of the GFP-dicer-3′UTR construct and miR-107-duplex-WT inhibited GFP expression compared with the embryos injected with the miR-107-duplex-MUT. In contrast, both miR-107 duplexes did not change the expression of an mCherry mRNA fused with a control-3′UTR. Therefore, miR-107 repressed GFP expression by specifically binding to the dicer-3′UTR sequence (Figure 2I and J). To further validate the specificity of the miR-107 binding sites in the dicer-3′UTR we designed two morpholino target protectors (MOdicer TPs) that bind to the dicer-3′UTR to prevent miR-107 repression (Figure S1F). In agreement with our prediction, the injection of the MOdicer TPs was sufficient to prevent the binding of the miR-107 duplex to the GFP-dicer-3′UTR mRNA as demonstrated by the recovery of the GFP expression in the miR-107-duplex-WT co-injected embryos (Figure 2I and J). Altogether, these results provide evidence that miR-107 can post-transcriptionally regulate dicer activity by specific binding to the dicer-3′UTR.
Dicer and miR-107 have inverse spatial and temporal expression patterns in the developing hindbrain
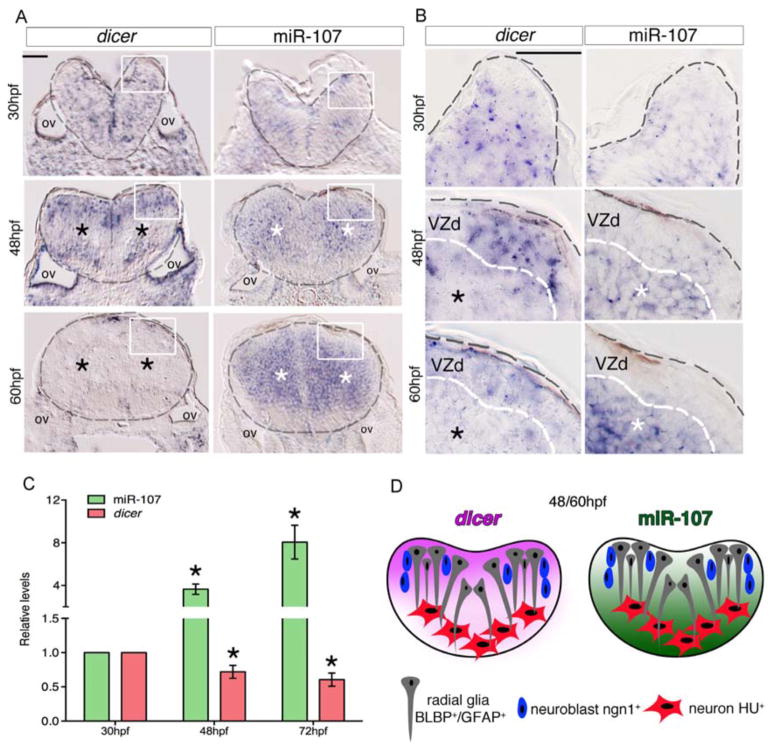
We hypothesized that if miR-107 expression in the developing hindbrain is necessary to repress dicer, miR-107 and dicer should localize temporally and spatially in an opposing manner. To test this prediction we first analyzed dicer and miR-107 expression by in situ hybridization, using an antisense dicer riboprobe and miR-107 LNA probe. Transversal hindbrain sections of 30 hpf embryos showed that miR-107 was scarcely expressed whereas dicer mRNA was broadly detected in every cell, accordingly with its important role in miRNA processing during early brain morphogenesis (Figure 3A and B). Interestingly, dicer and miR-107 expression at 48 and 60 hpf showed an opposite spatial distribution particularly evident in the periventricular and mantle hindbrain zone (Figure 3A, black and white stars respectively). High magnification pictures of these regions showed that, while dicer was restricted to the most dorsal ventricular area, miR-107 was diffusely expressed in the periventricular and post-mitotic hindbrain zone (Figure 3B, white and black stars, respectively). The levels of miR-107 and dicer were quantified by qRT-PCR in whole embryonic brain tissue confirming the progressive decline of dicer transcripts from 30hpf to 60hpf where miR-107 was significantly increased (Figure 3C). Altogether, these data showed that dicer mRNA levels progressively decreased in the miR-107 positive cells previously identified as neuronal differentiating cells and newborn neurons (Figure 3D and Figure 1E–H).
Figure 3. miR-107 and dicer show an opposite expression pattern along the hindbrain ventricular zone.
(A–B) Expression of dicer and miR-107 in 15 μm hindbrain cross-sections at 30, 48 and 60 hpf. Gray dashed line indicates the hindbrain area; black stars indicate the area with low dicer expression while white stars show the region of high miR-107 expression. (B) High magnification pictures (60X) of the region boxed in (A). (C) qRT-PCR of dicer and miR-107 levels at 30, 48 and 72 hpf and relative to the expression levels at 30 hpf. Data are presented as mean ± SEM of three independent experiments (n~20 embryos each). * p<0.05. (D) Graphic representation of the opposite spatial expression of dicer (purple) and miR-107 (green) in the hindbrain from 48 to 60 hpf. Scale bars= 50μm.
Loss of miR-107 function promotes hindbrain neurogenesis
The miR-107 pattern of expression in the neuronal cells of the hindbrain suggests its possible function during neurogenesis. To test this hypothesis, we performed loss of function experiments to deplete the embryos of miR-107a, the miR-107 mammalian homolog in the zebrafish. We generated a zebrafish miR-107a mutant using a transcription activator-like effector nuclease strategy (TALENs107a) (Bedell et al., 2012) (Figure S2). Injection of TALENs107a introduced specific embryonic mutations within the miR-107a genomic region and blocked miR-107 expression in the brain similarly to a previously validated morpholino-antisense oligonucleotide (MO107a) (Figure S2A) (Nicoli et al., 2012).
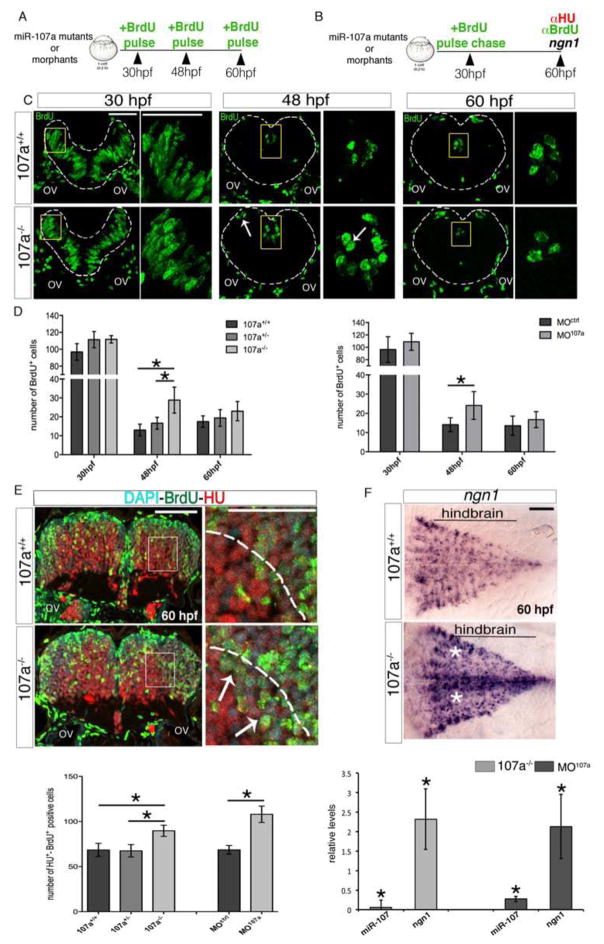
We next investigated the effect of miR-107a loss of function on cycling progenitors and neuron production in the developing hindbrain (Figure 4A and B). To test the number of cycling progenitors, F2 miR-107a mutant embryos as well as MO107a and MOctrl injected embryos were briefly pulsed at 30, 48 and 60 hpf with BrdU (Figure 4A, C and Figure S2C, D). At 30 hpf, when miR-107 was expressed in a few proliferating radial glia progenitors (Figure 1B–C), the number of BrdU+ cells was only modestly increased in miR-107a morphant and miR-107a−/− mutant embryos compared with the respective controls (Figure 4C and D). At 48 hpf, both miR-107a mutants and morphants showed the most robust increase of BrdU+ cells counted in the ventricular area while at 60 hpf their number was comparable between embryos with and without miR-107a (Figure 4C, arrows, and D). Thus, suggesting that the lack of miR-107a promotes cycling progenitors between 30 and 48 hpf. This transient increase of BrdU+ cells could be caused either by a block in cell-cycle exit or by an increase in the number of cycling progenitors. To discriminate between these two possibilities we labeled dividing progenitors with a brief BrdU pulse at 30 hpf and followed their fate at 60 hpf, the time point at which most of the neural progenitors exit the cell cycle in both control and miR-107a mutants or morphants. Neuronal fate was determined by the expression of an early marker of neuronal differentiation, HU (Figure 4B). In the hindbrain of the miR-107a−/− and MO107a injected embryos we observed an increase of progenitors that differentiated into neurons, as counted by the HU-BrdU double-positive cells, migrating in the mantle zone (Figure 4E, arrows). Thus, the observed supernumerary BrdU+ neural progenitors from 30 to 48 hpf can exit the cell cycle and differentiate into neurons by 60 hpf. These data suggest that the absence of miR-107a led to an increased number of cycling progenitors generating neurons. To further corroborate this hypothesis we detected and quantified the expression of ngn1 as a specific gene marker of neuronal progenitors (Korzh et al., 1998). As expected, embryos lacking miR-107a compared with the controls showed an expansion of the ngn1 expressing cells at 60 hpf confirming that the supernumerary number of cycling progenitors detected early on followed the neuronal fate (Figure 4F, white stars).
Figure 4. Loss of miR-107 function promotes embryonic hindbrain neurogenesis.
(A and B) Schematic representation of the experiments in (C–D) and (E), respectively. (C) Hindbrain cross-sections showing brief BrdU pulse labeling of miR-107a+/+ and miR-107a−/−mutants at 30, 48 and 60 hpf. Panels in the right are high magnifications of the area boxed in the left panels. White arrows indicate supernumerary BrdU+ cells. Gray dashed line indicates the hindbrain area. (D) Number of BrdU+ cells per section in (C) and in the MO107a and MOctrl injected embryos treated in the same conditions. Data are shown as mean ± SEM of two independent experiments for 107a mutants and three for the morphants (n= 5 to 10 embryos each). *p<0.05. (E) Top panel shows hindbrain cross-sections of 60 hpf miR-107a+/+ and miR-107a−/− mutants stained for BrdU and HU. Panels in the right are high magnifications of the boxed area in the left panels. Arrows indicate the increase of BrdU-HU double-positive cells migrating in the mantle area (dashed line). The graph at the bottom shows the quantification of the number of BrdU-HU double positive cells in the indicated samples. Data are shown as mean ± SEM of two independent experiments for 107a mutants and three for morpholino (n= 5 to 10 embryos each). *p<0.05. (F) Expression of ngn1 in 107a+/+ and 107a−/− mutants at 60hpf. Dorsal is up, anterior is to the left. White stars indicate increased ngn1 staining. The graph shows the quantification by qRT-PCR of miR-107 and ngn1 expression levels in 107a mutants and in MO107a injected embryos. Levels are relative to the respective controls (107a−/− and MOctrl respectively). Data are shown as mean ± SEM. *p<0.05. Scale bars= 50μm
Taken together, these data suggest that the absence of miR-107a during hindbrain development led to an increased number of cycling progenitors and promoted their differentiation into neuronal cells.
Loss of miR-107 stabilizes dicer and promotes miR-9 biogenesis and neurogenesis in zebrafish brain-derived neurospheres
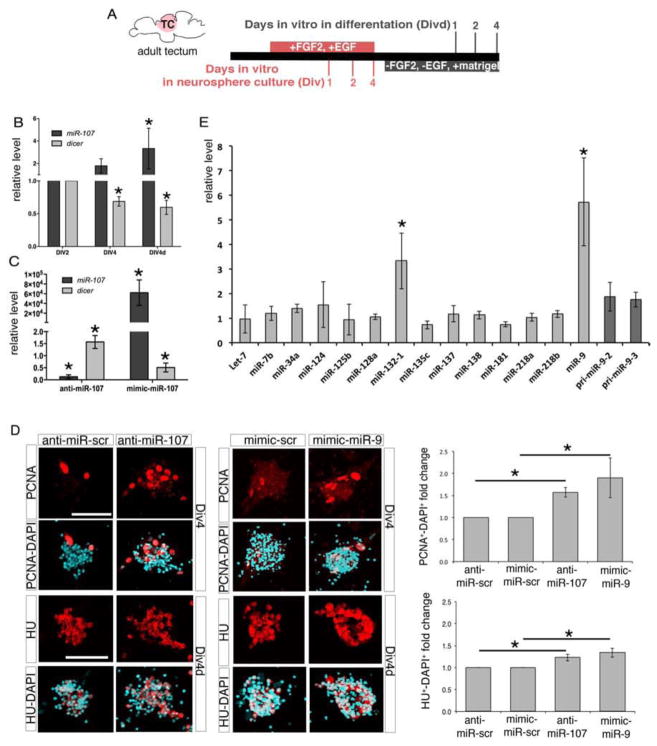
Based on the expression and function of miR-107 in the developing hindbrain, we hypothesized that miR-107 acts in a cell autonomous manner within neural cells by repressing dicer and driving specific inhibitory signaling to properly control neuron production. Therefore, we tested miR-107 funtion in an in vitro model of neurogenesis using zebrafish brain-derived neurospheres. The neurosphere assay is a valuable tool for isolating and understanding the signaling required for both stem and progenitor cells to differentiate into neurons, independently from their environment in vivo (Campos, 2004). In this study, we established for the first time a zebrafish neurosphere assay using adult neural progenitor cells (NPCs) cultured in the presence of a high concentration (10 ng/ml) of bFGF2 and EGF growth factors. Adult NPCs were isolated from different adult brain regions such as the telencephalon, tectum, and cerebellum (Figure S3A and B). Interestingly, zebrafish NPCs isolated from the tectum, generated after 4 days in vitro (Div4), had the highest number of neurospheres compared with other brain regions (Figure S3C and D). Although we cannot exclude contamination from the adjacent diencephalic area our data suggested that the tectum region is enriched in NPCs growing in our culture conditions. Thus, we only used adult tectal material for performing neurosphere assays. Neurospheres formed mainly from the proliferation of GFAP+ radial glia cells that served as neural progenitors able to self-renew and differentiate after withdrawal of growth factors for three consecutive culture passages (Figure S3E arrowhead and data not shown). After four days in vitro in the absence of growth factors (Div4d), zebrafish neurospheres differentiated into all neural cell types:. astrocyte-like cells (GFAP+) (Nielsen and Jorgensen, 2003), oligodendrocytes (Sox-10+) (Takada and Appel, 2010) and mature neurons (α-tubulin+) (Ulrich et al., 2011) and expressed gene markers of several post-mitotic neural cells (Figure S3F, arrowheads, and G). qRT-PCR analysis indicated that, during the course of neurosphere differentiation, dicer mRNA levels progressively declined while miR-107 expression increased (Figure 5A and B). Together with our in vivo data, this result confirms that miR-107 and dicer are expressed in an opposite time-course manner from the stage of progenitor to the stage of mature post-mitotic neural cells.
Figure 5. Loss of the miR-107-dicer interaction promotes neurogenesis and correlates with an increase in miR-9 biogenesis and activity in zebrafish brain-derived neurospheres.
(A) Schematic representation of the neurosphere assay. (B) qRT-PCR of dicer and miR-107 levels in neurospheres at the stages indicated. Expression levels are relative to Div2. Data are represented as mean ± SEM (n=3). *p<0.05. (C) qRT-PCR of dicer and miR-107 levels in neurospheres at Div4d treated as indicated at Div4. Levels are relative to cells transfected with anti-miR-scramble for anti-miR-107 samples and mimic-scrambled for mimic-miR-107 treatments. Values are presented as mean ± SEM (n=3 to 5). *p<0.05. (D) Left panels are confocal images showing neurospheres at Div4 and Div4d stages treated at Div2 and Div4 respectively with the indicated oligonucleotides. Neurospheres at Div4 are stained with the anti-PCNA antibody and neurospheres at Div4d are stained with an anti-HU antibody. Right charts show the quantification of the total number of PCNA and HU positive cells normalized to the total number of DAPI cells per neurospheres. Values are represented as fold change of PCNA/DAPI or HU/DAPI double positive cells in treated neurospheres compared to the respective controls. Data represent the mean ± SEM (n=4 for the PCNA and n=5 for HU immunodetection). *p<0.05. (E) qRT-PCR of indicated miRNAs expressed in neurospheres at Div4d treated at Div4 with anti-miR-107. Levels are relative to the control (anti-miR-scrambled). Data represent the mean ± SEM (n=4). *p<0.05. Scale bars= 50μm.
We then tested whether miR-107 directly controls the dicer mRNA expression level. Div2 neurospheres were treated with an LNA-anti-miR-107 (Lennox et al., 2013) or with miR-107-mimic oligonucleotides, to determine the effect on dicer expression of either the loss- or gain- of miR-107 function, respectively. miR-107-deficient derived neurospheres showed a stabilized level of dicer transcripts, while dicer mRNA expression was inhibited in gain-of-miR-107 function neurospheres (Figure 5C). These data suggested that during neurosphere neurogenesis dicer mRNA levels are regulated by miR-107 expression. To further investigate the neurogenic effect of miR-107-dicer crosstalk in NPCs, we analyzed the number of proliferating cell nuclear antigen (PCNA) positive proliferating progenitors and HU+ neurons in neurospheres cultured in the presence or absence of miR-107 (Figure 5D). The increased number of PCNA+ dividing progenitor cells and HU+ post-mitotic neurons in miR-107-deficient cultures indicates that the progressive destabilization of dicer by miR-107 normally represses neurogenesis in vitro (Figure 5D).
Next, we investigated the molecular mechanisms linking miR-107 expression, dicer destabilization, and neurogenesis progression. We first analyzed the biogenesis of miRNAs highly enriched in adult neural cells (Kapsimali et al., 2007) in differentiating neurospheres with or without miR-107. Among the 14 miRNAs analyzed, neurospheres differentiated after anti-miR-107 treatment showed a striking accumulation of miR-9 mature sequence (Figure 5E). Importantly, the expression of miR-9 precursors (pri-miR-9-2 and pre-miR-9-2) transcribed in neural cells (Bonev et al., 2012), were not increased in the absence of miR-107 (Figure 5E). Thus, the accumulation of miR-9 resulted from an excess of its biogenesis rather than from an increase of its transcription. miR-9 is a highly conserved miRNA essential for NPC differentation, i.e., the specific threshold at which miR-9 is accumulated in NPCs influences the timing of their proliferation and differentiation (Bonev et al., 2012). We found that increased miR-9 expression levels in floating undifferentiating neurospheres (Div4) via transfection with a miR-9-mimic miRNA, caused an increased number of PCNA+ cells, in comparison to control neurospheres. A similar miR-9-mimic treatment on differentiating neurospheres (Div4d) resulted in an increased number of HU+ post-mitotic neurons. Therefore, the miR-107-loss-of-function or the excess of miR-9 expression similarly promoted neurogenesis (Figure 5D).
Altogether, our in vitro data indicate that the absence of miR-107 increases dicer mRNA levels promoting the specific biogenesis of miR-9 and neurosphere neurogenesis.
miR-107 modulates miR-9 biogenesis across the hindbrain ventricular zone by controlling the spatial and temporal expression of dicer levels
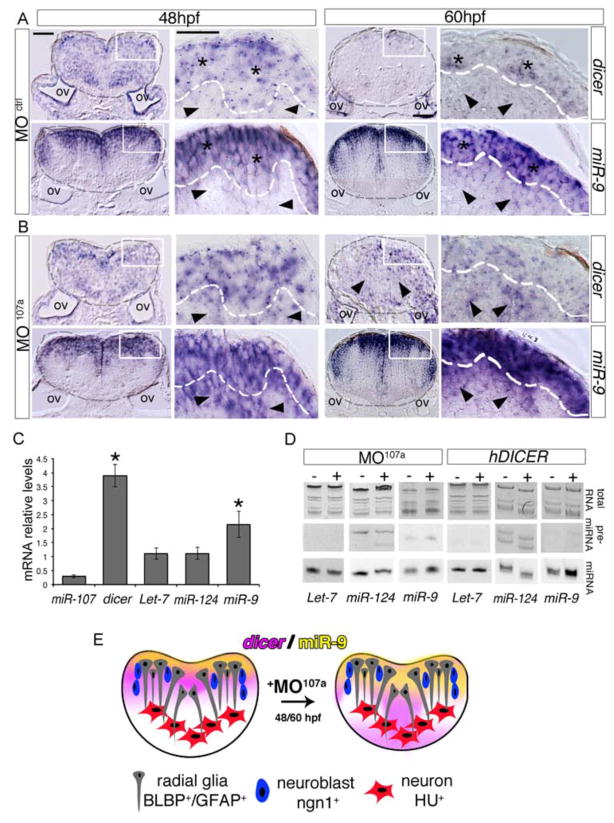
To assess if miR-107 controls dicer to predominantly modulate the level of miR-9 during hindbrain development, we analyzed the temporal and spatial expression of dicer transcripts and the miR-9 mature sequence in embryos injected with a miR-107a morpholino (MO107a), which phenocopied the hindbrain neurogenesis defects of the miR107a−/− mutants that lack miR-107a function and showed significant increases in dicer expression (Figure 4 and Figure S4A). In the hindbrain section of MO107a injected embryos at 30 hpf, dicer and miR-9 expression was unaffected compared with respective controls (Figure S4B). This was congruent with restricted miR-107 expression in a few proliferating radial glia progenitor cells at 30 hpf and the mild phenotype on the progenitor numbers in the miR107a morphants at this developmental stage.
At 48 and 60 hpf, in the control embryos dicer mRNA was compartmentalized toward the most dorsal ventricular row of cells where miR-9 was also highly expressed (Figure 6A, stars). The miR-9 expression in the ventricular area corresponds to neural and neuronal progenitor cells differentiating into newborn neurons whereas most of the miR-9 mature sequence was excluded from the region outside the ventricular area (Figure 6A, arrowheads) (Coolen et al., 2012). In contrast, in the hindbrain section of the MO107a embryos at the same developmental stages dicer mRNA levels were ectopically expressed outside the ventricular area towards the periventricular and post-mitotic region, where miR-107 expression is normally localized (Figure 6B, arrowheads, and Figure 1 and 3). Notably, several cell layers at the periventricular area showed a concomitant accumulation of miR-9 undetectable in the control embryos (Figure 6B, arrowheads). We did not detect ectopic expression of miR-9 in post-mitotic neurons of the most deep mantle hindbrain region suggesting that the miR-107-dicer interaction modulates miR-9 biogenesis predominantly in differentiating neuronal cells (Figure 6A and B).
Figure 6. miR-107 is required to control dicer and miR-9 expression across the hindbrain ventricular zone.
(A–B) Expression of dicer and miR-9 in hindbrain cross-sections of MOctrl and MO107a injected embryos at 48 and 60 hpf. Area inside of the gray dashed line indicates the hindbrain area; black stars in (A) show dicer and miR-9 positive areas while black arrowheads show dicer and miR-9 low expression. Black arrowheads in (B) show dicer and miR-9 ectopic expression. Dorsal is up. (C) qRT-PCR of dicer mRNA and the indicated miRNAs in MO107a embryos at 60 hpf. Levels are relative to MOctrl embryos. Error bars represent the ± SD between three independent experiments (n~20 embryos each). *p<0.05. (D) Northern blot analysis of total RNA extracted from 60 hpf embryos injected with MO107a, MOctrl or human DICER mRNA The images are representative of two independent experiments. (E) Schematic representation of the effect of MO107a on dicer (purple) and miR-9 (yellow) pattern expression at 48 and 60 hpf embryos. Scale bars= 50μm.
We further quantified the increase of dicer and miR-9 expression by qRT-PCR in 60 hpf embryos injected with MO107a or MOctrl (Figure 6C). We confirmed that, upon miR-107 removal, dicer mRNA levels increased significantly as well as the miR-9 mature sequences. Additionally, the expression of Let-7 and miR-124, two of the most abundant neural specific miRNAs in embryonic neural development (Kapsimali et al., 2007), was unchanged suggesting that the stabilization of dicer mRNA preferentially promotes the biogenesis of miR-9 in vivo (Figure 6C). Thus, we analyzed by northern blot the biogenesis of miR-9, Let-7 and miR-124 at 60 hpf embryos injected with MO107a or with a hDICER mRNA. We detected upon the respective treatments a rapid and specific accumulation of miR-9 mature sequence rather than miR-9 precursor, confirming that the accumulation of miR-9 is preferentially promoted by the increased dicer activity, induced by either exogenous hDICER mRNA injection or endogenous lack of miR-107a (Figure 6D). In summary, our data showed that at late hindbrain neurogenesis, dicer expression is destabilized by miR-107 in the periventricular and post-mitotic zone allowing the modulation of miR-9 in these regions. Upon removal of miR-107a, the ectopic levels of dicer lead to a preferential biogenesis of miR-9 leading to excess miR-9 accumulation in cells along the post-mitotic front normally expressing miR-107 (Figure 6E).
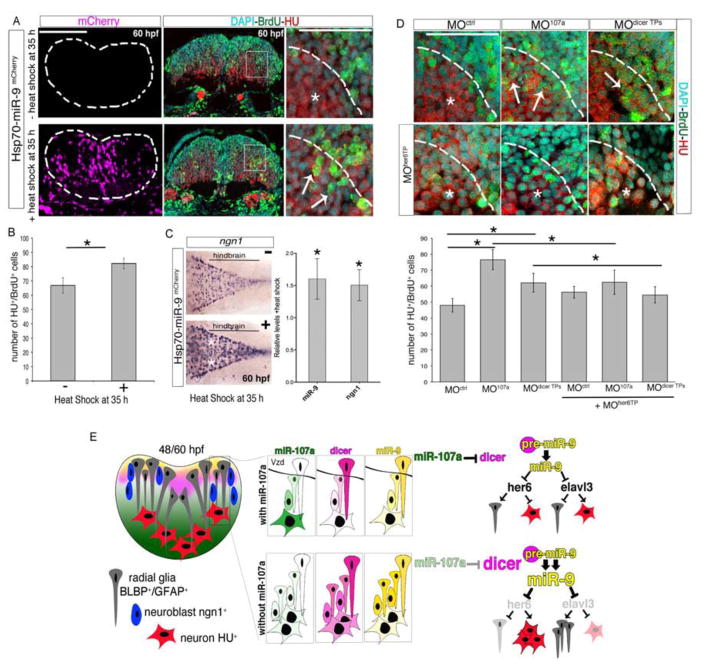
Increased miR-9 levels during late hindbrain neurogenesis promote the production of neurons in the absence of miR-107-dicer interactions
During hindbrain development, miR-9 regulates several antagonistic factors in both neural and neuronal progenitor cells, balancing the appropriate production of neurons over time. The downregulation of miR-9 increases the expression of progenitor- and cell-cycle exit-promoting genes to drive certain progenitors to become neurons (Bonev et al., 2011; Coolen et al., 2012). Interestingly, our data indicated that in the mutant or morphant embryos lacking miR-107a, hindbrain neurogenesis increased in correlation with dicer and miR-9 augmentation between 30, 48 and 60 hpf. We hypothesized that either the up- or downregulation of miR-9 at this developmental time causes an imbalance of pro- and anti-neurogenic signaling promoting neurons. To test this, we first assessed the hindbrain neurogenesis progression in embryos where miR-9 was ectopically induced between 30 to 60 hpf by the injection of a temperature inducible promoter driving mCherry fused to the miR-9 precursor (Hsp70-miR-9mCherry). Neurogenesis was evaluated by determining the number of neurons differentiating over the course of the heat-shock treatment as well as the expression of the neuroblast specific marker ngn1. Hsp70-miR-9mCherry injected embryos were briefly pulsed with BrdU and heat-shocked at 30 hpf. By 48 hpf mCherry was detected throughout the brain and trunk regions and the ectopic expression of miR-9 was confirmed by northern blot and qRT-PCR analysis (Figure 7A–B and Figure S5A–C). We found that in the 60 hpf embryos in which mCherry was abundantly expressed in the post-mitotic hindbrain area, a larger proportion of BrdU-positive cells pulsed at 30 hpf differentiated into HU positive neurons (Figure 7A, arrows). Accordingly, in the same embryos we detected an expansion of ngn1 expressing cells in the hindbrain (Figure 7C). Thus, the overexpression of miR-9, as well as the downregulation of miR-107a, promoted the number of progenitors committed to becoming neurons. Moreover, these data indicate that the increased neurogenesis in the absence of the miR-107-dicer interaction may be consequential to miR-9 accumulation.
Figure 7. The increase of miR-9 accumulation promotes hindbrain neurogenesis in the absence of miR-107 and dicer post-transcriptional regulation.
(A) Comparison of the number of BrdU/HU double positive cells in embryos injected with the Hsp70-miR-9mCherry plasmid with or without heat-shock. mCherry positive cells (purple) indicate the cells overexpressing miR-9. HU, BrdU and DAPI triple staining was performed in the same hindbrain cross-section at 60 hpf. White arrows in the high magnification (60X) pictures indicate increased number of BrdU/HU double positive cells migrating in the mantle area (dashed line). (B) Number of BrdU and HU double positive cells. Data are presented as mean ± SEM (n= 5 embryos each group). *p<0.05. (C) Expression of ngn1 in 60 hpf embryos treated as described above. Dorsal is up and anterior is left. White stars indicate ngn1 expression observed in 30% ± 5 of the heat-shocked embryos (n=15) compared with the ngn1 expression in non heat-shocked embryos (n=20). The chart shows the quantification by qRT-PCR of miR-9 and ngn1 expression level in the brain of 60 hpf embryos treated as above and following heat shock. Levels are relative to the control without heat shock. Data are presented as mean ± SEM (n=3). p<0.05. (D) High magnifications (60X) of hindbrain cross-sections showing BrdU/HU double positive cells in embryos at 60 hpf injected with MOctrl, MO107a, or MOdicer TPs with or without MOher6TP. Whole cross sections are shown in Figure S5D. Arrows indicate the BrdU/HU double positive cells. Stars indicate DAPI nuclei of BrdU negative and HU positive cells in the same area. The chart shows the number of HU and BrdU double positive cells in the indicated treatment. Data represent the mean ± SEM (n= 6 to 9 embryos for each experimental condition). *p<0.05. (E) Schematic representation of the miR-107 (green) - dicer (pink) signaling pathway controlling miR-9 (yellow) biogenesis during hindbrain neurogenesis. Scale bars= 50μm.
To test this hypothesis we reasoned that ectopic miR-9 expression could cause excessive neuronal differentiation by repressing her6, which is the Hes-1 mammalian homolog in zebrafish and the miR-9 progenitor-promoting target gene in the developing hindbrain (Bonev et al., 2012; Coolen et al., 2012). Thus, we assessed the production of neurons in embryos lacking the miR-107a-dicer interaction and miR-9 repression to her6, using a target protector morpholino previously used to prevent the binding of miR-9 to the her6 3′UTR (MOher6 TP)(Coolen et al., 2012). We found that embryos co-injected with MOher6 TP and MO107a showed a normal number of BrdU+ cells pulsed at 30 hpf and differentiated into HU neurons at 60 hpf when compared with the BrdU-HU double positive cells counted in the MO107a injected embryos alone (Figure 7D and Figure S5D). Furthermore, the inhibition of miR-9-her6 binding was able to rescue the excessive neuronal differentiation detected in embryos injected with the MOdicer TPs to the control level (Figure 7D). Accordingly, in these embryos the protection of dicer-3′UTR to miR-107 binding sites was also able to stabilize dicer, miR-9 and ngn1 hindbrain expression similarly to the 107a morphants (Figure S5E).
Altogether, these data prove that the increased levels of miR-9 are responsible for the excessive number of hindbrain neurons in the embryos lacking the miR-107a specific repression of dicer-3′UTR.
DISCUSSION
miRNAs act at many different levels of the neurogenesis process and neuronal identity. Indeed, specific subsets of miRNAs are differentially regulated between neural stem cell proliferation and differentiation (neurogenesis-related miRNAs) and neuronal identity (neuronal-specification related miRNAs) (Li and Jin, 2010). However, the mechanisms that control their differential accumulation during development are poorly understood. Here we propose that the brain-enriched miRNA, miR-107, negatively regulates dicer at the post-transcriptional level tuning its function to control the precise accumulation of specific miRNAs decisive for neurogenesis progression. This finding may appear paradoxical at first, considering the well-established notion that miRNAs are necessary for both development and maintenance of the brain’s functions (Volvert et al., 2012). However, we observed that the reduction of dicer levels did not correlate with a rapid and global loss of neurogenesis-related miRNAs. Indeed, we found that miR-107 itself is accumulating over the time course of dicer reduction in both in vivo and in vitro neurogenesis models. miRNAs are well known to be stable molecules and global miRNA analysis upon transcription shutoff showed that the majority of miRNAs do not decrease during the course of the first eight hours post-treatment and some miRNAs can have a half-life of days (Bail et al., 2010). Therefore, during the time course of dicer depletion, miRNAs (such as miR-107) are sufficiently stable and accumulate.
We showed that during neurogenesis, the lack of miR-107 increases the functional levels of dicer to preferentially promote the biogenesis of miR-9. Previous in vitro studies showed that human DROSHA and DICER have preferential cleavage efficiencies on pri- and pre-miRNAs respectively (Feng et al., 2012; Feng et al., 2011). In these studies, DROSHA has an average cleavage efficiency on pri-miR-9, while the cleavage efficiency of DICER for pre-miR-9 is one of the highest of the ~160 pre-miRNAs analyzed (Feng et al., 2012). These biochemical data may explain why excessive levels of dicer, in the absence of miR-107, rapidly increased miR-9 biogenesis during neuronal differentiation. Accordingly, previous data indicates that the pre-miR-9-2 is rapidly cleaved into the mature miR-9 in vitro in NPCs. The high stability of the miR-9 mature sequence allows its accumulation over time and establishes the timing of neural progenitor proliferation and differentiation (Bonev et al., 2012). In our model we believe that the decrease of dicer during neurogenesis is a mechanism that “curbs” and tunes the accumulation of specific neurogenesis-related miRNAs, such as miR-9, whose precursors are highly unstable (rapidly processed) and whose mature sequence is highly stable (Bonev et al., 2012). This mechanism is particularly relevant during hindbrain development where, to properly allow the neurogenesis progression, miR-9 expression is temporally required in several cell types of the ventricular area and decreases in post-mitotic neuronal cells. Our data indicate that miR-107 expression is critical to post-transcriptionally destabilize dicer in cells exiting the ventricular area, to facilitate the gradual decay of miR-9 mature sequence in neuronal differentiating cells (Figure 7E).
We showed that loss of the miR-107-dicer interaction promotes hindbrain neurogenesis at late stages of development. Even though our results strongly suggest that this neurogenesis defect correlates predominantly with an increase of miR-9 biogenesis, we cannot exclude that other miRNAs may contribute to this phenotype. Investigating 14 out of ~40 miRNAs known to be expressed in brain tissue (Bak et al., 2008; Kapsimali et al., 2007), we found that miR-132 and miR-34 also showed increased expression levels upon loss of miR-107. However, miR-132 is involved in the control of neuronal identity rather than neurogenesis while miR-34, even if involved in neurogenesis, did not have significantly increased levels to justify the phenotype observed in the absence of miR-107 (Lopez-Ramirez and Nicoli, 2013).
miR-9 function in neural progenitor proliferation is debated and controversial, however, consistent with our findings, the increased levels of miR-9 in neural progenitor cells exhibits pro-differentiation function in numerous organisms (Bonev et al., 2011; Delaloy et al., 2010; Zhao et al., 2009). Interestingly, Coolen et al., showed that during zebrafish hindbrain development the loss of miR-9 leads to both an increased number of cycling progenitors and neurons at late neurogenesis stages. In this model, miR-9 controls the ambivalent state of neural cells committed to become neurons by blocking antagonistic genes like her6, which promotes progenitor proliferation and inhibits their differentiation, and elavl3, which promotes neuronal differentiation and inhibits progenitors proliferation (Coolen et al., 2012) (Figure 7E, top panel). Therefore, the depletion of miR-9 increases both the number of cycling progenitors (via augmentation of her6) and neuronal differentiation (via augmentation of elavl3). On the contary, in our model we show that the augmentation of miR-9 across the hindbrain ventricular zone promotes progenitor proliferation (possibly via repression of elavl3) and an excessive production of neurons (via repression of her6) (Figure 7E, bottom panel). Indeed, the protection assay of her6 alone was sufficient to rescue the supernumerary number of neurons produced in the absence of miR-107-dicer interaction. Therefore, both the excessive inhibition and de-repression of antagonist genes operated by miR-9, shift the ambivalent state of neural progenitors to generate neurons. Hence, our data strongly highlights the necessity of the miR-107-dicer interaction to modulate miR-9 whose functions depend upon its specific level of accumulation during hindbrain neurogenesis.
Interestingly, during the in vitro neurogenesis of NPCs the over-expression and knockdown of miR-9 function to dampen Hes-1 oscillation and promote neuron differentiation (Bonev et al., 2012). Our data indicate that the regulation of miR-9 by the miR-107-dicer interaction also functions during in vitro neurogenesis of NPCs derived from zebrafish adult tectum; loss of miR-107 increased dicer mRNA as well as miR-9 biogenesis whose augmentation promoted neurosphere neurogenesis. The effects of miR-9 in the differentiation of the tectum derived NPCs are consistent with its pro-neurogenic function previously documented in the NPCs of the midbrain-hindbrain region (Leucht et al., 2008). Additionally, our in vitro data unambiguously confirmed that the miR-107-dicer interaction controls miR-9 in neurogenesis excluding any possible non-cell-autonomous contribution from other cell types expressing miR-107 in vivo (Nicoli et al., 2012).
Our findings establish that in physiological conditions dicer levels are subjected to tissue-specific post-transcriptional regulation to maintain the homeostasis of neural development. The dicer 3′UTR has several dozen different miRNA binding sites suggesting the existence of alternative miRNAs-mRNAs regulatory pathways (Guo et al., 2010). Our data launch the fascinating hypothesis that different cell types may use specific miRNAs to set their steady state level of dicer and therefore tune its function in physiological and tissue specific contexts.
Finally, the regulatory activity of miR-107 during neurogenesis can open novel perspectives for the use of miR-107 manipulation in the treatment of neurological disorders such as Alzheimer’s disease and schizophrenia, where altered expression levels of miR-107 have been observed and whose root causes remains to be elucidated (Scarr et al., 2013; Wang et al., 2008).
EXPERIMENTAL PROCEDURES
Zebrafish husbandry and strains
Zebrafish were grown and maintained according to protocols approved by the Yale University Animal Care. The Tg(sox10(7.2):mrfp)sn1 (Kirby et al., 2006) and Tg(-8.4neurog1:GFP) (Blader et al., 2004) were used in addition to wild-type strain.
Whole-mount in situ hybridization and immunohistochemistry
Zebrafish embryos were fixed and processed for whole-mount in situ hybridization as described previously (Nicoli et al., 2012). Detailed in situ hybridization and immunohistochemistry procedures described in Supplemental Experimental Procedures.
mRNAs, miRNA duplexes and morpholino injections
Injections were performed using 250 pg of hDICER, 75 pg of eGFP-dicer-3′UTR and mCherry control. Duplex mature miRNAs (miR-107-duplex-WT and miR-107-duplex-MUT) were injected at the one-cell stage at a concentration of 100 μM per embryo. miR-430s duplexes were injected at 10 μM (Giraldez et al., 2005). Antisense morpholinos were purchased from GeneTools. The miR-107a and control morpholinos (Nicoli et al., 2012) were injected at a concentration of 7.5 ng per embryos. The dicer-target protector morpholinos against two specific binding sites (TP1 and TP2) for miR-107a seed region were designed (Figure S1F, Table in Supplemental Experimental Procedures) and injected in one-cell stage embryos at a concentration of 5 ng per embryo. The target protector morpholino against the her6–miR-9 binding site was injected at a concentration of 0.1 ng per embryo (Coolen et al., 2012).
Inducible miR-9 gain of function
We generated a Hsp70-miR-9mCherry construct containing transposable element regions flanking a temperature inducible promoter (hsp70) and an mCherry coding sequence fused to an intronic region in which the precursor of miR-9 was cloned as previously described (Nicoli et al., 2010). Following heat-shock, subsequent splicing and processing yield separate mature miR-9 and mCherry mRNAs. Embryos were injected with 50 pg per embryo of Hsp70-miR-9mCherry and 50 pg of transposase at the one-cell stage. Heat shock was performed at 42°C for 15 min. Embryos were fixed at 60 hpf and processed for whole-mount in situ hybridization or cryosectioning prior to staining.
BrdU incorporation and immunohistochemistry in zebrafish embryos
The BrdU pulse experiment on zebrafish embryos at 30, 48, and 60 hpf was performed as previously described (Coolen et al., 2012). Briefly, embryos were pulsed in a 10 mM BrdU solution (Sigma) for 20 min at 4°C. After cryoprotection in sucrose-gelatin, samples were cross-sectioned at 15 μm thickness and mounted onto SuperFrost Plus slides (VWR International). Slides were incubated with an anti-HuC/HuD mouse monoclonal antibody (1:250, Life Technologies) and a rat anti-BrdU (1:200, Accurate Chemical & Scientific Corp). Goat anti-mouse and anti-rat antibodies conjugated to Alexa Fluor 546 or Alexa Fluor 642 (1:400, Life Technologies) were used as secondary antibodies. Cell nuclei were stained with DAPI using mounting media obtained from SouthernBiotech. BrdU-HU double positive cells were counted in the same area of the hindbrain by using ImageJ (multi point selection tool).
Zebrafish neurospheres
Primary neurospheres were isolated from Zebrafish adult brains. Culture conditions and treatments are described in the Supplemental Information.
Statistical analysis
Data are represented as mean ± standard error of the mean (SEM) unless otherwise indicated. For all experiments, the number of independent experiments, n, is indicated. For all other experiments, statistical significance was considered if P was lower than 0.05 as determined by paired, 1-tailed Student’s t-test.
Supplementary Material
Acknowledgments
The authors are grateful to Laure Bally-Cuif for providing her6 morpholino target protector and ISH probes; Antonio Giraldez for providing miR-430 duplexes and hDICER plasmid; and Anne Eichmann, Suk-Won Jin, and Jeffrey Bender for critical reading and comments on the manuscript. This work was supported by grants from National Institute of Health (5R00HL105791 to S.N.) and from the Alzheimer’s Association (NIRP 12-259162 to S.N.).
Footnotes
Authors Contributions
S.N. conceived the project; E.M. and S.N. designed and carried out all the experiments; M.A.L.R. designed and carry out the experiments in Figure 5, Figure S3 and contribute to the Figure 2A–D, E and G; A.N. generated the miR-107a mutants; G.H.T. performed miR-107 in situ hybridization in Figure S2A; A.M.M. generated Figure S1F; C.F.C. and J.L.T. helped with the generation of the zebrafish neurospheres assay. S.N. wrote the manuscript. E.M., M.A.L.R. and J.L.T. contributed to write the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. Rna. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. Rna. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blader P, Lam CS, Rastegar S, Scardigli R, Nicod JC, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, et al. Conserved and acquired features of neurogenin1 regulation. Development. 2004;131:5627–5637. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Developmental cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Stanley P, Papalopulu N. MicroRNA-9 Modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos LS. Neurospheres: insights into neural stem cell biology. J Neurosci Res. 2004;78:761–769. doi: 10.1002/jnr.20333. [DOI] [PubMed] [Google Scholar]
- Coolen M, Thieffry D, Drivenes O, Becker TS, Bally-Cuif L. miR-9 controls the timing of neurogenesis through the direct inhibition of antagonistic factors. Developmental cell. 2012;22:1052–1064. doi: 10.1016/j.devcel.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell stem cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Diotel N, Vaillant C, Gueguen MM, Mironov S, Anglade I, Servili A, Pellegrini E, Kah O. Cxcr4 and Cxcl12 expression in radial glial cells of the brain of adult zebrafish. The Journal of comparative neurology. 2010;518:4855–4876. doi: 10.1002/cne.22492. [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhang X, Graves P, Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. Rna. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang X, Song Q, Li T, Zeng Y. Drosha processing controls the specificity and efficiency of global microRNA expression. Biochimica et biophysica acta. 2011;1809:700–707. doi: 10.1016/j.bbagrm.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nature reviews Molecular cell biology. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nature biotechnology. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Taira E, Kuo CH, Li BS, Okamoto H, Nakahira K, Ikenaka K, Higuchi H, Miki N. Neuron-specific expression of a chicken gicerin cDNA in transient transgenic zebrafish. Neurochemical research. 1996;21:231–237. doi: 10.1007/BF02529139. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Korzh V, Sleptsova I, Liao J, He J, Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Developmental dynamics: an official publication of the American Association of Anatomists. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology: CB. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lennox KA, Owczarzy R, Thomas DM, Walder JA, Behlke MA. Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Molecular therapy Nucleic acids. 2013;2:e117. doi: 10.1038/mtna.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- Lopez-Ramirez MA, Nicoli S. Role of miRNAs and epigenetics in neural stem cell fate determination. Epigenetics: official journal of the DNA Methylation Society. 2013;9 doi: 10.4161/epi.27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nature genetics. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Knyphausen CP, Zhu LJ, Lakshmanan A, Lawson ND. miR-221 is required for endothelial tip cell behaviors during vascular development. Dev Cell. 2012;22:418–429. doi: 10.1016/j.devcel.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Jorgensen AL. Structural and functional characterization of the zebrafish gene for glial fibrillary acidic protein, GFAP. Gene. 2003;310:123–132. doi: 10.1016/s0378-1119(03)00526-2. [DOI] [PubMed] [Google Scholar]
- Polster BJ, Westaway SK, Nguyen TM, Yoon MY, Hayflick SJ. Discordant expression of miR-103/7 and pantothenate kinase host genes in mouse. Molecular genetics and metabolism. 2010;101:292–295. doi: 10.1016/j.ymgme.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurat N, Andersson T, Vasistha NA, Molnar Z, Livesey FJ. Dicer is required for neural stem cell multipotency and lineage progression during cerebral cortex development. Neural development. 2013;8:14. doi: 10.1186/1749-8104-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr E, Craig JM, Cairns MJ, Seo MS, Galati JC, Beveridge NJ, Gibbons A, Juzva S, Weinrich B, Parkinson-Bates M, et al. Decreased cortical muscarinic M1 receptors in schizophrenia are associated with changes in gene promoter methylation, mRNA and gene targeting microRNA. Transl Psychiatry. 2013;3:e230. doi: 10.1038/tp.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Strahle U, Scholpp S. Neurogenesis in zebrafish - from embryo to adult. Neural development. 2013;8:3. doi: 10.1186/1749-8104-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Appel B. Identification of genes expressed by zebrafish oligodendrocytes using a differential microarray screen. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239:2041–2047. doi: 10.1002/dvdy.22338. [DOI] [PubMed] [Google Scholar]
- Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- Ulrich F, Ma LH, Baker RG, Torres-Vazquez J. Neurovascular development in the embryonic zebrafish hindbrain. Developmental biology. 2011;357:134–151. doi: 10.1016/j.ydbio.2011.06.037. [DOI] [PubMed] [Google Scholar]
- Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell death and differentiation. 2012;19:1573–1581. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Danaher RJ, Miller CS, Berger JR, Nubia VG, Wilfred BS, Neltner JH, Norris CM, Nelson PT. Expression of miR-15/107 Family MicroRNAs in Human Tissues and Cultured Rat Brain Cells. Genomics, proteomics & bioinformatics. 2014 doi: 10.1016/j.gpb.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature structural & molecular biology. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.