Abstract
Two methylenecyclopropane nucleoside analogues with a phenylphosphoralaninate moiety, QYL-685 and QYL-609, exert potent and specific activities against human immunodeficiency virus type 1 strain LAI (HIV-1LAI) and HIV-2 in vitro. In this study, we induced HIV-1 variants resistant to QYL-685 by exposing HIV-1LAI to increasing concentrations of QYL-685. After 16 passages, the virus (HIV-1P16) was less sensitive to QYL-685 (104-fold), QYL-609 (>41-fold), and (−)-β-2′,3′-dideoxy-3′-thiacytidine (3TC) (>1,100-fold) than was HIV-1LAI and contained an M184I mutation. Two infectious clones, HIV-1M184I and HIV-1M184V, were resistant to QYL-685, QYL-609, and 3TC, confirming that the M184I mutation was responsible for the observed resistance. Viral-fitness analyses (competitive HIV-1 replication assays) revealed that in the absence of drugs, M184I and M184V conferred a replication disadvantage on the virus compared to the replication efficiency of the wild-type infectious clone (HIV-1wt). However, in the presence of QYL-685 (4 μM), HIV-1M184I and HIV-1M184V showed greater fitness than HIV-1wt. These data may provide structural and virological relevance with regard to the emergence of M184I and M184V substitutions in HIV-1.
To date, a number of nucleoside reverse transcriptase (RT) inhibitors have been developed for the treatment of patients with human immunodeficiency virus type 1 (HIV-1) infection. However, the accumulation of data suggests that the development of HIV-1 variants with reduced sensitivities to RT inhibitors is related to the clinical deterioration of patients receiving such agents (2, 4, 9, 12, 13, 18, 22).
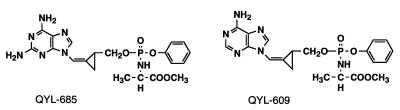
Recently, the replacement of the ribofuranose backbone of a nucleoside with a Z-methylenecyclopropane moiety (designated QYL compounds) was shown to confer potent antiviral properties on certain nucleoside analogues (15, 16, 24). Both QYL-685 and -609 have significant activities against human cytomegalovirus, herpes simplex virus 1 and 2, human hepatitis virus, Epstein-Barr virus, and varicella-zoster virus (14). In our previous study of structure-activity relationships, we found that adenine and 2,6-diaminopurine Z-methylenecyclopropanes (QYL-546 and QYL-284A, respectively) were moderately active against HIV-1 in vitro (24). The addition of a phenylphosphoralaninate (PPA) moiety to QYL-546 and QYL-284A, producing QYL-685 and QYL-609, respectively (Fig. 1), however, significantly increased their activities against HIV-1 in vitro (24). It is presumed that the agents possessing the highly lipophilic PPA moiety enter cells relatively quickly. It should also be noted that methylenecyclopropanes with the PPA moiety are monophosphorylated with the phosphate group within the PPA moiety and thus serve as monophosphate prodrugs. QYL-685 and QYL-609 were also active against HIV-2 and HIV-1 variants, including a zidovudine (AZT)-resistant infectious clone, HIV-1215, a didanosine-resistant clone, HIV-174, and a multidideoxynucleoside-resistant clone, HIV-162/75/77/116/151, in vitro (24).
FIG. 1.
Structures of QYL-685 and QYL-609.
In the present study, we induced HIV-1 variants resistant to QYL-685 in vitro by continuously exposing the virus to increasing concentrations of QYL-685 and defined the virological properties of infectious HIV-1 clones carrying the QYL-685 resistance-associated pol gene mutation M184I or M184V. The viral fitness of those clones was also examined in the presence or absence of QYL-685.
MATERIALS AND METHODS
Cells and viruses.
MT-2 cells were grown in an RPMI 1640-based culture medium, and Cos-7 cells were grown in Dulbecco’s modified Eagle medium. These media were supplemented with 15% fetal calf serum (HyClone Laboratories, Logan, Utah), 50 U of penicillin per ml, and 50 μg of streptomycin per ml. A laboratory HIV-1 strain, HIV-1LAI (26), was used as the source of wild-type infectious virions for induction of variants with reduced sensitivity to QYL-685.
Antiviral agents.
QYL-609 and -685 were synthesized as previously described (14). AZT and 2′,3′-dideoxyinosine (ddI or didanosine) were purchased from Sigma (St. Louis, Mo.) and Calbiochem (La Jolla, Calif.), respectively. (−)-β-2′,3′-Dideoxy-3′-thiacytidine (3TC; also called lamivudine) was a kind gift from R. F. Schinazi (Atlanta, Ga.).
Generation of QYL-685-resistant HIV-1 in vitro.
MT-2 cells (5 × 105) were infected with HIV-1LAI at 500 50% tissue culture infectious doses (TCID50) and cultured in the presence of QYL-685 at an initial concentration of 0.5 μM. Viral replication was monitored by confirming the HIV-1LAI-induced cytopathic effect in MT-2 cells. The culture supernatant was harvested on day 7 and used to infect fresh MT-2 cells for the next round of culture in the presence of increased concentrations of QYL-685. As soon as the virus started to propagate in the presence of the drug, the QYL-685 concentration was further increased. The selection was carried out for a total of 16 passages, with QYL-685 drug concentrations ranging from 0.5 to 8 μM. To determine infectivity titers, MT-2 cells (2,000 cells/well) in 96-well flat-bottomed microtiter culture plates (Costar, Cambridge, Mass.) were exposed to passage 16 HIV-1 (HIV-1P16). MT-2 cultures were examined for cytopathic effects on day 7 of culture, and the TCID50 was determined by the method of Reed and Muench (17). Six replications of all titration assays were performed. The profiles of cross-resistance of the selected viral stocks to several RT inhibitors were examined by the 3-(4,5-dimetylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay as described below. Proviral DNA derived from the HIV-1P16-infected-cell lysates was cloned and sequenced as described below.
Generation of infectious HIV-1 clones.
Infectious molecular HIV-1 clones with mutations of interest were constructed with the pHXB2RIP7-based plasmid pSUM9, as previously described (11, 20). The HIV-1 clone carrying the wild-type RT was designated HIV-1wt. The HIV-1 clones carrying an amino acid substitution, M184V or M184I, in the RT-encoding region were designated HIV-1M184V or HIV-1M184I, respectively. Determination of the nucleotide sequences of infectious clones confirmed that each contained the intended mutation. The infectivity titers of these clones were determined as described above. All titration assays were performed with six replicates.
Drug sensitivity assays.
The sensitivities of the HIV-1P16 and infectious molecular HIV-1 clones to various drugs were determined as previously described with minor modifications (7, 20, 23). Briefly, MT-2 cells (2 × 104/ml) were exposed to 100 TCID50 of the HIV-1P16 in the presence of various concentrations of drugs in 96-well microculture plates and incubated at 37°C for 7 days. After 100 μl of the medium was removed from each well, 10 μl of MTT solution (7.5 mg/ml) in phosphate-buffered saline was added to each well in the plate. The plate was then incubated at 37°C for 2 h. After incubation, to dissolve the formazan crystals, 100 μl of acidified isopropanol containing 4% (vol/vol) Triton X-100 was added to each well. The optical density (wavelength, 570 nm) was measured in a microplate reader (model 3550; Bio-Rad Laboratories, Hercules, Calif.). All assays were performed in triplicate.
Assays for HIV-1 p24 Gag protein production were performed with MT-2 cells as previously described (23). Briefly, MT-2 cells (2 × 103) cultured in 96-well microtiter culture plates in complete medium were treated with different compounds at various concentrations and exposed to 100 TCID50 of infectious molecular HIV-1 clones. On day 7 in culture, the supernatant was harvested and the amount of p24 Gag protein was determined by radioimmunoassays (Du Pont, NEN Research Products, Boston, Mass.). All assays were performed in triplicate. Drug concentrations which inhibited viral replication by 50% (50% effective concentrations [EC50]) were determined by comparison of the p24 production levels with the p24 production levels in drug-free control cell cultures as previously described (20, 23).
Molecular cloning and determination of nucleotide sequences.
High-molecular-weight DNA was extracted from HIV-1P16-infected MT-2 cells with InstaGene Matrix (Bio-Rad Laboratories) and was subjected to molecular cloning followed by sequence determination as previously described (21). The primers used for the first PCR of the RT region were SA009 (5′-TTT AAA TTT TCC CAT TAG CCC TAT-3′) and SA015 (5′-ACT CCA TGT ACT GGT TCT TTT AGA-3′). The first PCR mixture consisted of 5 μl of the proviral DNA solution, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.0 mM MgCl2, 0.01% gelatin, 0.2 mM deoxynucleoside triphosphates (dNTPs), 2.0 U of Taq DNA polymerase (Perkin-Elmer, Foster City, Calif.), and 12.5 pmol of each of the first PCR primers in a total volume of 50 μl. The PCR process consisted of an initial denaturation for 2 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, with a final 8-min incubation at 72°C. The first-round PCR product (1 μl) of the RT region was used directly in the second round of PCR with primers 881NMF (5′-ATG GAT GGC CCA AAA GTT AAA CAA-3′) and 891NMR (5′-CTG GCT AGC CCA ATT CAA TTT TCC CAC-3′). The PCR process involved an initial 2-min denaturation at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, with a final 8-min extension at 72°C. The second-round PCR products were purified with spin columns (PCR select III; 5 Prime 3 Prime, Boulder, Colo.) and cloned directly (PCR-Script Amp Cloning Kit; Stratagene, La Jolla, Calif.). Clones were sequenced with both M13 forward and reverse dye-labeled primers on an ABI model 373 automated DNA sequencer.
Competitive HIV-1 replication assay.
A pair of infectious clones to be compared for replicative ability was added to freshly prepared H9 cells (3 × 105) and cultured in the presence or absence of QYL-685 (8). To ensure that the infectivities of the two infectious clones were approximately equal, a fixed amount (200 TCID50) of one infectious clone was combined with three different amounts (100, 200, and 300 TCID50) of the other infectious clone. On day 1 of culture, one-third of the infected H9 cells were harvested and washed twice with phosphate-buffered saline and cellular DNA was purified with InstaGene Matrix. Purified DNA was subjected to nested PCR and sequencing as described above. The HIV-1 coculture which best approximated a 50:50 mixture on day 1 was further propagated, and the remaining cultures were discarded. Every seven days, the supernatant of the virus coculture was transmitted to new uninfected H9 cells, the cells harvested at each passage were subjected to direct DNA sequencing, and a viral population change was determined. The persistence of the original amino acid substitution(s) was confirmed for all infectious clones used in this study.
RESULTS
Selection of QYL-685-resistant variants.
In order to select HIV-1 variants resistant to QYL-685 in vitro, we exposed MT-2 cells to HIV-1LAI and the virus was serially passaged in the presence of increasing concentrations of QYL-685. The virus was initially propagated in the presence of 0.5 μM QYL-685, and during the course of the selection procedure, the drug concentration was increased to 8 μM. At passage 16, the supernatant containing the virus (HIV-1P16) was harvested and titrated for its infectivity and the sensitivities of the virus to AZT, ddI, 3TC, QYL-609, and QYL-685 were determined by the MTT assay (Table 1). The EC50 of QYL-685 against HIV-1LAI and HIV-1P16 were 0.047 and 4.9 μM, respectively. The fold difference in the EC50 was 104. The EC50 of QYL-609 against HIV-1P16 was >41-fold higher than that against HIV-1LAI (>0.49 versus 0.012 μM). HIV-1P16 was also examined for cross-resistance to AZT, ddI, and 3TC (Table 1). HIV-1P16 was found to be highly resistant to 3TC (with an EC50 >1,100-fold greater than that of HIV-1LAI) but remained sensitive to AZT and ddI. Highly 3TC-resistant mutants (HIV-1M184I and HIV-1M184V) were reportedly selected by passaging them two or three times in phytohemagglutinin-activated peripheral blood mononuclear cells (18). For the selection of QYL-685-resistant HIV-1 (carrying 184I), at least seven passages were required in vitro. These results suggest that the emergence of QYL-685-resistant variants possibly requires more time than that of 3TC-resistant HIV-1 in vitro.
TABLE 1.
Sensitivities to AZT, ddI, 3TC, QYL-609, and QYL-685 of HIV-1LAI and HIV-1P16a
| Drug | EC50b (μM) against:
|
CC50 (μM) | |
|---|---|---|---|
| HIV-1LAI | HIV-1P16 | ||
| AZT | 0.028 ± 0.005 | 0.02 ± 0.007 (1×) | >100 |
| ddI | 3.9 ± 1.0 | 5.7 ± 1.3 (2×) | 59 ± 4.3 |
| 3TC | 0.91 ± 0.34 | >1,000 (>1,100×) | |
| QYL-685 | 0.047 ± 0.014 | 4.9 ± 1.3 (104×) | 28 ± 1.4 |
| QYL-609 | 0.012 ± 0.004 | >0.49 (>41×) | 0.49 ± 0.01 |
Data shown are the mean values (with standard deviations) derived from the results of more than three independent experiments conducted in triplicate.
MT-2 cells (2 × 103) were exposed to 100 TCID50 of HIV-1LAI or HIV-1P16 and cultured in the presence of various concentrations of AZT, ddI, QYL-609, or QYL-685, and the EC50 were determined by MTT assay on day 7 of culture. The numbers in parentheses indicate fold differences from EC50 against HIV-1LAI.
Amino acid substitution associated with QYL-685 exposure.
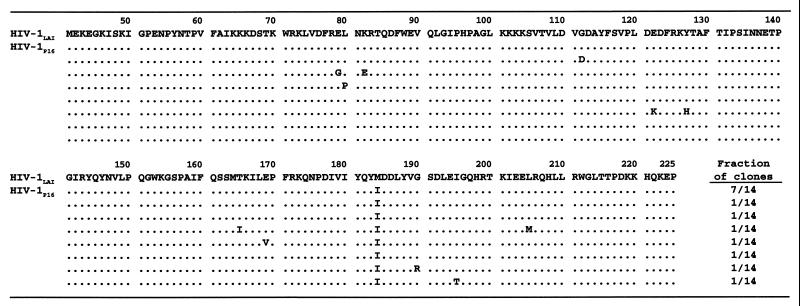
We then determined the amino acid sequence of the RT-encoding region of HIV-1P16. Proviral DNA obtained from HIV-1P16-infected cells was subjected to PCR amplification, followed by cloning and sequencing. At passage 7 (in the presence of 4 μM QYL-685), a mutation at codon 184 from Met to Ile was first identified. The cell-free transmission of HIV-1 in the presence of 8 μM QYL-685 was continued for an additional nine passages. However, Ile at codon 184 persisted and did not change into Val, in contrast to results in previous publications describing the development of the M184I mutation in RT of HIV-1 when the virus was exposed to 3TC in vitro (1, 3, 18). When HIV-1P16 was cloned and the sequence of the pol region was determined, the M184I mutation was seen in 14 of 14 clones examined, although several substitutions were sporadically observed at other residues (Fig. 2).
FIG. 2.
Sequence analysis of the RT-encoding regions (codons 41 through 225) of molecular clones derived from HIV-1P16. The fractions of clones containing each unique RT sequence are indicated at the lower right. The HIV-1LAI sequence as a consensus sequence is shown at the top. Identity with the consensus sequence at individual amino acid positions is indicated by dots.
Drug susceptibility of HIV-1 containing a QYL-685-associated substitution.
In order to confirm that the observed M184I mutation was associated with the observed reduced sensitivity of HIV-1P16 to QYL-685, two recombinant infectious clones containing M184V (HIV-1M184V) and M184I (HIV-1M184I) were constructed. The EC50 of QYL-685 against HIV-1M184V and HIV-1M184I were 11- and 9-fold higher, respectively, than that against HIV-1wt. The EC50 of QYL-609 against HIV-1M184V and HIV-1M184I were 12- and 9-fold higher, respectively, than that against HIV-1wt (Table 2). We also examined these clones with respect to 3TC, AZT, and ddI. In agreement with previous reports (10, 19), HIV-1M184V and HIV-1M184I were highly resistant to 3TC (showing an IC50 >590-fold higher than that of HIV-1wt); however, both were sensitive to AZT and ddI.
TABLE 2.
Sensitivities to AZT, ddI, 3TC, QYL-609 and -685 of infectious HIV-1 clonesa
| Drug | EC50b (μM) against:
|
CC50 (μM) | ||
|---|---|---|---|---|
| HIV-1wt | HIV-1M184I | HIV-1M184V | ||
| AZT | 0.04 ± 0.001 | 0.008 ± 0.002 (0.4×) | 0.006 ± 0.001 (0.2×) | >100 |
| ddI | 3.0 ± 0.6 | 2.3 ± 0.5 (0.8×) | 4.5 ± 1.1 (2×) | 61 ± 19 |
| 3TC | 1.7 ± 0.2 | >1,000 (>590×) | >1,000 (>590) | >1,000 |
| QYL-685 | 0.29 ± 0.03 | 2.6 ± 0.5 (9×) | 3.1 ± 0.5 (11×) | 23 ± 1.3 |
| QYL-609 | 0.026 ± 0.008 | 0.24 ± 0.05 (9×) | 0.3 ± 0.06 (12×) | 0.52 ± 0.04 |
Data shown are mean values (with standard deviations) derived from the results of three independent experiments conducted in triplicate.
The EC50 were determined by employing MT-2 cells (2 × 109) exposed to infectious HIV-1 clones (100 TCID50) and by using the inhibition of p24 Gag protein production as an endpoint on day 7 of culture. The numbers in parentheses indicate fold differences from EC50 against HIV-1wt.
Viral fitness of HIV-1wt, HIV-1M184I, and HIV-1M184V.
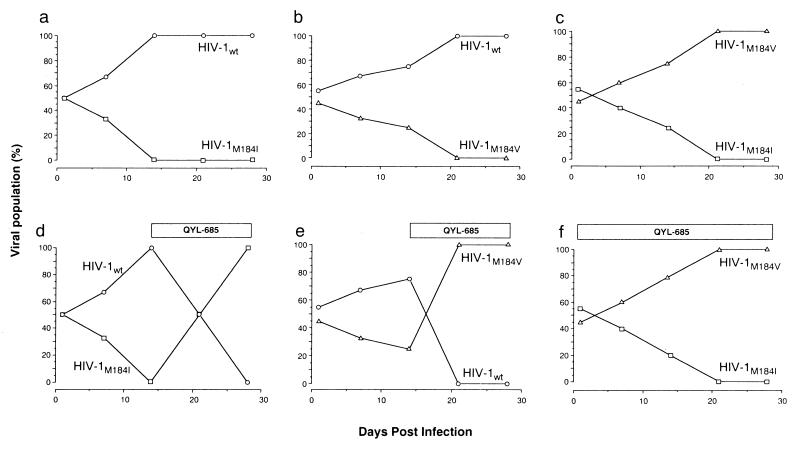
To further define the significance of the mutations M184I and M184V, the levels of viral fitness were compared among HIV-1wt, HIV-1M184V, and HIV-1M184I in the presence or absence of 4 μM QYL-685 by a competitive HIV-1 replication assay (CHRA) (8) (Fig. 3). Two infectious clones were mixed ∼50:50 and were used as a source of infectious virions. Every seven days, the cell-free supernatant of virus coculture was transmitted to new uninfected H9 cells. High-molecular-weight DNA extracted from the cells harvested at the end of each passage was subjected to direct DNA sequencing, and shifts in the viral population were assessed. In the absence of drugs, HIV-1wt outgrew both HIV-1M184I and HIV-1M184V (Fig. 3a and b). However, after QYL-685 was added at passage 3, the predominating HIV-1wt was readily outgrown by these two mutant HIV-1 clones (Fig. 3d and e), indicating that these two mutant clones have a replication advantage over HIV-1wt in the presence of QYL-685. When we compared the replication rates of HIV-1M184I and HIV-1M184V, regardless of the presence of QYL-685, HIV-1M184V predominated over HIV-1M184I, demonstrating that the former has a replication advantage over the latter (Fig. 3c and f).
FIG. 3.
Replication profiles of HIV-1wt, HIV-1M184I, and HIV-1M184V in CHRA. Three infectious clones to be compared for their fitness were mixed 50:50 and used to infect H9 cells in the absence (a through c) or presence (d through f) of QYL-685 (4 μM). The cell-free supernatant was transferred to fresh H9 cells every seven days. High-molecular-weight DNAs extracted from infected cells at the end of each passage were subjected to nucleotide sequencing, and the proportions of Met, Val, and Ile at position 184 were determined.
DISCUSSION
In the present study, we selected HIV-1 variants resistant to a novel methylenecyclopropane nucleoside RT inhibitor, QYL-685, by exposing HIV-1LAI to increasing concentrations of the drug and defined the properties of the variants obtained. HIV-1, which propagated in the presence of 8 μM QYL-685 (HIV-1P16), proved to be highly resistant to QYL-685 (Table 1). We analyzed the nucleotide sequence of the RT-encoding region at each passage beyond passage 3 and first identified the M184I mutation at passage 7. The M184I mutation was confirmed to be responsible for the observed viral resistance to QYL-685 (Table 2).
We and others have reported that HIV-1M184I emerges first, and shortly thereafter HIV-1M184V becomes dominant in patients receiving 3TC (6, 18, 25). Boucher and his colleagues also reported that HIV-1M184I develops first, followed by the appearance of HIV-1M184V when the virus is selected against 3TC in vitro (1). Therefore, expecting to detect the M184V mutation, we continued to select HIV-1LAI for an additional nine passages in the presence of up to 8 μM QYL-685; however, M184V failed to emerge even at passage 16, and the absence of M184V was confirmed by cloning HIV-1p16 (data not shown). We also increased the QYL-685 concentration to 10 μM to provide the virus with more stringent selection pressure; however, this concentration was rather toxic to the cells and we could not continue the selection process. It is, therefore, of note that there may be a limitation in our finding that HIV-1M184V did not emerge with QYL-685 in the present study.
Although the diaminopurine moiety of QYL-685 may resemble the amino-oxopyrimidine (cytosine) portion of 3TC, their structures are fundamentally different in other aspects (methylenecyclopropane versus oxathiolane ring), and the selection pressure of QYL-685 on HIV-1 can substantially differ from that of 3TC. Indeed, the fold difference in the EC50 of 3TC determined for HIV-1wt and QYL-685-resistant HIV-1 was much greater than that of QYL-685 (>1,100 versus 104 in Table 1 and >590 versus 11 in Table 2). The EC50 of QYL-685 against HIV-1M184I and HIV-1M184V were 2.6 and 3.1 μM, respectively. These EC50 were far from QYL-685’s 50% cytotoxic concentration (CC50), 23 μM. The data indicate that HIV-1M184I and HIV-1M184V are far more resistant to 3TC than to QYL-685. In a recent study by Huang et al. analyzing the structure of a covalently trapped catalytic complex of HIV-1 RT, the M184V mutation is shown to influence both the incoming dNTP and the primer’s 3′ terminus (5). In the catalytic site of RT, the codon 151 methionine side chain contacts the sugar and base of the 3′ nucleotide in the primer, although introduction of Ile or Val also reportedly creates a contact with the incoming dNTP sugar ring (5). Thus, the interaction of the Ile or Val side chain with the incoming triphosphate form of QYL-685 should differ from that with the triphosphate form of 3TC, likely producing the difference in EC50 of QYL-685 and 3TC against HIV-1M184I as well as HIV-1M184V. Furthermore, modeling of 3TC in the covalently trapped catalytic complex has shown an enhanced interference with Ile or Val with respect to the incoming dNTP by the oxathiolane ring configuration of 3TC (5), also suggesting that the interference by the Z-methylenecyclopropane group of QYL-685 can substantially differ from that by 3TC.
The viral-fitness assay or CHRA allows a determination as to whether the presence of QYL-685 has a significant impact on the replication rate of HIV-1M184I, which was selected with QYL-685 in vitro, as well as whether QYL-685 affects HIV-1M184V replication, which was not selected for during a total of 16 passages. CHRA revealed that in the absence of drugs, the M184I and M184V mutations conferred a replication disadvantage on the virus compared to HIV-1wt. However, in the presence of QYL-685 (4 μM), HIV-1M184I and HIV-1M184V showed greater fitness than HIV-1wt.
It is worth noting that QYL-685 is derived from a racemic form of the parent methylenecyclopropane analogue and, as such, consists of four diastereoisomers. Prodrugs of the corresponding R- and S-enantiomers (two diastereoisomeric pairs) were prepared, and their in vitro anti-HIV-1 activities have been confirmed (unpublished data). Compounds derived from the R-enantiomer have proven to be more active against HIV-1 than those from the S-enantiomer (unpublished data). The intriguing question of whether both diastereoisomeric pairs contribute to the resistance profile observed for QYL-685 should be answered.
Taken together, the present data may be structurally and virologically relevant with regard to the emergence of M184I and M184V in the therapy of HIV-1 infection. These data may also lead to useful tactics for potentially developing Z-methylenecyclopropane nucleoside RT inhibitors, including QYL-685, for treatment of HIV-1 infection.
ACKNOWLEDGMENTS
We thank Robert E. Wittes, Carmen Allegra, and Edison Liu for advice and Pope Kosalaraksa for helpful discussion.
H.M. is a recipient of grants from the Ministry of Health and Welfare of Japan (Promotion of AIDS Research) and the Japan Society for the Promotion of Science.
REFERENCES
- 1.Boucher C A B, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzgibbon J E, Howell R M, Harberzetti C A, Sperber S J, Gocke D J, Dubin D T. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1992;36:153–157. doi: 10.1128/aac.36.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu Z, Gao Q, Fang H, Salomon H, Parniak M A, Goldberg E, Cameron J, Wainberg M A. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 6.Kavlick M F, Shirasaka T, Kojima E, Pluda J M, Hui F, Jr, Yarchoan R, Mitsuya H. Genotypic and phenotypic characterization of HIV-1 isolated from patients receiving (−)-2′,3′-dideoxy-3′-thiacytidine. Antivir Res. 1995;28:133–146. doi: 10.1016/0166-3542(95)00044-m. [DOI] [PubMed] [Google Scholar]
- 7.Kodama E, Shigeta S, Suzuki T, De Clercq E. Application of a gastric cancer cell line (MKN-28) for anti-adenovirus screening using the MTT method. Antivir Res. 1996;31:159–164. doi: 10.1016/0166-3542(96)06966-5. [DOI] [PubMed] [Google Scholar]
- 8.Kosalaraksa P, Kavlick M F, Maroun V, Le R, Mitsuya H. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J Virol. 1999;73:5356–5363. doi: 10.1128/jvi.73.7.5356-5363.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larder B A. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J Gen Virol. 1994;75:951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- 10.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 11.Maeda Y, Venzon D J, Mitsuya H. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J Infect Dis. 1998;177:1207–1213. doi: 10.1086/515282. [DOI] [PubMed] [Google Scholar]
- 12.Mitsuya H, Erickson J. Discovery and development of antiretroviral therapeutics for HIV infection. In: Merigan T C, Bartlet J G, Bolognesi D, editors. Textbook of AIDS medicine. Baltimore, Md: Williams & Wilkins; 1999. pp. 751–780. [Google Scholar]
- 13.Nunberg J H, Schleif W A, Boots E J, O’Brien J A, Quintero J C, Hoffman J M, Emini E A, Goldman M E. Viral resistance to human immunodeficiency virus type 1-specific pyridinone reverse transcriptase inhibitors. J Virol. 1991;65:4887–4892. doi: 10.1128/jvi.65.9.4887-4892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu, Y.-L., R. G. Ptak, J. M. Breienbach, J.-S. Lin, Y.-C. Cheng, J. C. Drach, E. R. Kern, and J. Zemlicka. Synthesis and antiviral activity of phosphoralaninate prodrugs of methylenecyclopropane analogues of nucleosides. Antivir. Res., in press. [DOI] [PubMed]
- 15.Qiu Y L, Ksebati M B, Ptak R G, Fan B Y, Breitenbach J M, Lin J S, Cheng Y C, Kern E R, Drach J C, Zemlicka J. (Z)- and (E)-2-((hydroxymethyl)cyclopropylidene)methyladenine and -guanine. New nucleoside analogues with a broad-spectrum antiviral activity. J Med Chem. 1998;41:10–23. doi: 10.1021/jm9705723. [DOI] [PubMed] [Google Scholar]
- 16.Qiu Y L, Ptak R G, Breitenbach J M, Lin J S, Cheng Y C, Kern E R, Drach J C, Zemlicka J. (Z)- and (E)-2-(hydroxymethylcyclopropylidene)-methylpurines and pyrimidines as antiviral agents. Antivir Chem Chemother. 1998;9:341–352. [PubMed] [Google Scholar]
- 17.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 18.Schinazi R F, Lloyd R M, Jr, Nguyen M-H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 20.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type-1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirasaka T, Yarchoan R, O’Brien M C, Husson R N, Anderson B D, Kojima E, Shimada T, Broder S, Mitsuya H. Changes in drug sensitivity of human immunodeficiency virus type 1 during therapy with azidothymidine, dideoxycytidine, and dideoxyinosine: an in vitro comparative study. Proc Natl Acad Sci USA. 1993;90:562–566. doi: 10.1073/pnas.90.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka M, Srinivas R V, Ueno T, Kavlick M F, Hui F K, Fridland A, Driscoll J S, Mitsuya H. In vitro induction of human immunodeficiency virus type 1 variants resistant to 2′-beta-fluoro-2′,3′-dideoxyadenosine. Antimicrob Agents Chemother. 1997;41:1313–1318. doi: 10.1128/aac.41.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchida H, Kodama N E, Yoshimura K, Maeda Y, Kasalaraksa P, Maroun V, Qiu Y L, Zemlicka J, Mitsuya H. In vitro anti-HIV activity of Z- and E-methylenecyclopropane nucleoside analogues and their phosphoro-l-alaninate diesters. Antimicrob Agents Chemother. 1999;43:1487–1490. doi: 10.1128/aac.43.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wainberg M A, Salomon H, Gu Z, Montaner J S G, Cooley T P, McCaffrey R, Ruedy J, Hilary M H, Cammack N, Cameron J, Nicholson W. Development of HIV-1 resistance to (−)2′-deoxy-3′-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS. 1995;9:351–357. [PubMed] [Google Scholar]
- 26.Wain-Hobson S, Vartanian J-P, Henry M, Chenciner N, Cheynier R, Delassus S, Martins L, Sala M, Nugeyre M-T, Guetard D, Klatzman D, Gluckman J-C, Rozenbaum W, Barre-Sinoussi F, Montagnier L. LAV revisited: origins of the early HIV-1 isolates from Institut Pasteur. Science. 1991;252:961–965. doi: 10.1126/science.2035026. [DOI] [PubMed] [Google Scholar]