Abstract
Chitooligosaccharides (COSs) have been widely used in agriculture, medicine, cosmetics, and foods, which are commonly prepared from chitin with chitinases. So far, while most COSs are prepared from colloidal chitin, chitinases used in preparing COSs directly from natural crystalline chitin are less reported. Here, we characterize three chitinases, which were identified from the marine bacterium Pseudoalteromonas flavipulchra DSM 14401T, with an ability to degrade crystalline chitin into (GlcNAc)2 (N,N’-diacetylchitobiose). Strain DSM 14401 can degrade the crystalline α-chitin in the medium to provide nutrients for growth. Genome and secretome analyses indicate that this strain secretes six chitinolytic enzymes, among which chitinases Chia4287, Chib0431, and Chib0434 have higher abundance than the others, suggesting their importance in crystalline α-chitin degradation. These three chitinases were heterologously expressed, purified, and characterized. They are all active on crystalline α-chitin, with temperature optima of 45–50 °C and pH optima of 7.0–7.5. They are all stable at 40 °C and in the pH range of 5.0–11.0. Moreover, they all have excellent salt tolerance, retaining more than 92% activity after incubation in 5 M NaCl for 10 h at 4 °C. When acting on crystalline α-chitin, the main products of the three chitinases are all (GlcNAc)2, which suggests that chitinases Chia4287, Chib0431, and Chib0434 likely have potential in direct conversion of crystalline chitin into (GlcNAc)2.
Keywords: chitinases; crystalline chitin; chitooligosaccharides; N,N′-diacetylchitobiose; Pseudoalteromonas
1. Introduction
Chitin is a polymer of N-acetyl-d-glucosamine (GlcNAc) and is the second most abundant polysaccharide after cellulose in nature. Chitin is mainly present in arthropod exoskeletons, fungal cell walls, and insect cuticles in a crystalline form, which is intractable, highly hydrophobic, and insoluble in water [1]. Chitin has three polymorphic isomers, including α-chitin, β-chitin, and γ-chitin. Among them, α-chitin is the most common form found in fungi, insect exoskeletons, and shells of crustaceans. α-chitin is harder to degrade than β-chitin and γ-chitin as it has a higher degree of recalcitrance, which decreases the accessibility of the individual polymer chains [2]. Colloidal chitin is normally prepared by treating natural chitin with strong acids to break the crystal structure and increase the accessibility of the substrate to enzymes. Therefore, colloidal chitin is usually used as the substrate for chitinase characterization.
The annual production of chitin in the ocean exceeds billions of tons [3,4], which is a good source for the production of chitooligosaccharides (COSs) and GlcNAc. Due to their various bioactive activities, COSs and GlcNAc have been widely applied in agriculture, medicine, cosmetics, and foods. For example, COSs have protective effects against infections and enhanced antitumor properties [5,6]. GlcNAc and (GlcNAc)2 (N,N′-diacetylchitobiose) can serve as cosmetic ingredients, dietary supplements, and osteoarthritis therapeutics [7,8,9].
The chitinolytic enzymes contain chitinases (EC 3.2.1.14), mainly from the GH18 and GH19 families, and β-N-acetylglucosaminidases (EC 3.2.1.52), mainly from the GH20 and GH3 families. While several β-N-acetylglucosaminidases have been reported to be active on chitin [10,11,12], the hydrolysis of chitin into COSs and/or GlcNAc is predominantly catalyzed by chitinases [13]. Chitinases include endochitinases and exochitinases, which are widely produced by bacteria [14], fungi [15], and plants [16], playing key roles in natural chitin degradation and recycling. Many bacteria-derived chitinases have been characterized, predominantly with colloidal chitin or chitooligosaccharides (or synthetic chitooligosaccharide analogs) as the substrate. Reported bacterial chitinases are mostly mesophilic enzymes with optimal temperatures at 40–60 °C [17,18,19,20,21,22,23,24,25]; only a few have been found to be cold-active enzymes, such as CHI II of Glaciozyma antarctica PI12 (15 °C) [26] and ChiA of Pseudoalteromonas sp. DL-6 (20 °C) [27]. The pH optima of bacterial chitinases are over a wide range. For example, chitinases from Streptomyces chilikensis RC1830 [24], Pseudoalteromonas tunicata CCUG 44952T [25], and Bacillus sp. R2 [21] showed their highest activity at neutral pHs (7.0–7.5), those from Micrococcus sp. AG84 [22], Pseudoalteromonas sp. DC14 [23], and Citrobacter freundii haritD11 [28] at basic pHs (8.0–9.0), and those from Moritella marina ATCC 15381 [29] and Paenicibacillus barengoltzii CAU904 [17] at acidic pHs (3.5 and 5.0, respectively). Chitinases are good tools to prepare COSs and GlcNAc from chitin. Because the natural source of chitin is crystalline chitin, chitinases that can efficiently hydrolyze crystalline chitin have better application potential in preparing COSs and GlcNAc from natural chitin sources than those only active on colloidal chitin. However, so far, only a few crude enzymes produced by wild strains and recombinant chitinases have been reported to be used in preparing COSs and GlcNAc from crystalline chitin [27,30,31,32,33]. Thus, it is necessary to identify and characterize more chitinases that can efficiently hydrolyze crystalline chitin for preparing COSs and GlcNAc from natural chitin sources.
Bacteria of the genus Pseudoalteromonas are widely distributed in the ocean, accounting for 2–3% of total bacterial abundance in upper ocean waters [34,35]. Many strains in this genus contain multiple chitinase-encoding genes [36], and some have been reported to secrete chitinases [18,23,25,27,37,38]. Furthermore, some chitinases from Pseudoalteromonas have been characterized. The GH18 chitinase Chi23, from Pseudoalteromonas aurantia DSM6057, is a thermostable enzyme with activity towards crystalline chitin in acidic conditions (pH 3.0–6.0) [18]. The GH18 chitinases ChiA and ChiC from Pseudoalteromonas sp. DL-6 [27,37] and ChiB from Pseudoalteromonas sp. O-7 [38] are cold-active enzymes with temperature optima at 20–30 °C. The GH19 chitinase Ptchi19 from Pseudoalteromonas tunicata CCUG 44952T was active at 20–50 °C and pH 6.0–9.5 [25]. The chitinase purified from the fermentation broth of Pseudoalteromonas sp. DC14 exhibited halo-alkali and thermo-stable properties [23]. Despite these studies, Pseudoalteromonas chitinases with potential in preparing COSs/GlcNAc from natural crystalline chitin have rarely been reported. The aim of this study is to identify and characterize chitinases with activity on crystalline chitin from marine Pseudoalteromonas bacteria and to evaluate their potential in preparing COSs/GlcNAc from natural crystalline chitin. In this study, the ability of 26 Pseudoalteromonas type strains to use crystalline chitin as a carbon source for growth was investigated, and Pseudoalteromonas flavipulchra DSM 14401T (hereafter strain DSM 14401), which was isolated from surface seawater [39], was found to have the highest degradation rate on crystalline α-chitin. The extracellular chitinases secreted by strain DSM 14401 were further identified by genome and secretome analyses. Three chitinases with high abundance in the secretome were heterologously expressed in Escherichia coli BL21 (DE3) and biochemically characterized. The hydrolytic products released from crystalline chitin by these chitinases were further investigated. The results suggest that these chitinases likely have potential in the preparation of (GlcNAc)2 from natural crystalline chitin.
2. Results and Discussion
2.1. The Ability of Strain DSM 14401 to Utilize Crystalline Chitin
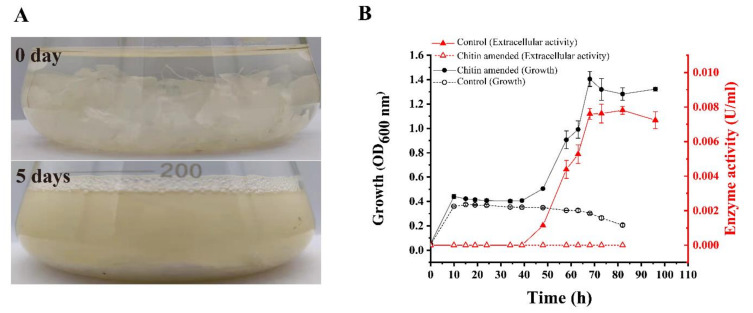
To obtain Pseudoalteromonas strains that can secrete chitinases to efficiently degrade crystalline chitin, 26 type Pseudoalteromonas strains (Table S1) were cultured in a liquid medium containing chitin flakes (crystalline α-chitin) as carbon source, and their growth and the degree of degradation of chitin flakes were observed. Strain DSM 14401 showed the greatest degradation rate of chitin flakes. This strain was able to degrade most of the chitin flakes in the medium in 5 days (Figure 1A). The growth curve and the extracellular chitinase activity of strain DSM 14401 during cultivation were also investigated (Figure 1B). The strain was cultured in a medium containing 0.05% peptone, 0.01% yeast powder, and 3% chitin flakes; the same medium without chitin flakes was used as a control. Strain DSM 14401 grew rapidly in the first 10 h in both media, with or without chitin flakes. After 10 h, the growth stagnated in both media, likely due to the depletion of the absorbable nutrients, such as peptone and yeast powder. After 40 h, while the cell number in the control medium began to decrease slowly and no extracellular chitinase activity was detected during the cultivation, both the cell number and the extracellular chitinase activity in the medium containing chitin flakes began to continuously increase until 68 h (Figure 1B). Based on this result, it can be speculated that, after absorbable nutrients were depleted, strain DSM 14401 began to secrete chitinases to degrade the chitin flakes in the medium into COSs/GlcNAc, which were absorbed by the strain to support its growth.
Figure 1.
Growth and extracellular chitinase activity of P. flavipulchra DSM 14401T cultured on crystalline chitin. (A) Cultures of strain DSM 14401 at 25 °C for 0 and 5 days. (B) The growth curve (black line) and the extracellular chitinase activity (red line) of strain DSM 14401. Strain DSM 14401 was cultured in a minimal medium containing 0.05% peptone, 0.01% yeast powder, and 3% (w/v) chitin flakes at 25 °C and 180 rpm. The extracellular chitinase activity was measured with chitin powder as the substrate at 50 °C. Strain DSM 14401 cultured in the same medium without chitin flakes and under the same conditions was used as the control.
Paulsen et al. reported that 27 Pseudoalteromonas strains have the ability to degrade crystalline chitin [36]. Strain Pseudoalteromonas sp. DC14 was also reported to be able to degrade crystalline chitin [23]. In addition, 5 chitinases from Pseudoalteromonas strains have been expressed and characterized, including ChiA and ChiC from Pseudoalteromonas sp. DL-6 [27,37], ChiB from Pseudoalteromonas sp. O-7 [38], PtChi19p from P. tunicata CCUG 44952T [25] and Chi23 from P. aurantia DSM6057 [18]. Among them, chitinases ChiA, PtChi19p, and Chi23 have activity on crystalline chitin based on substrate specificity analysis [18,25,27]. These reports indicate that many Pseudoalteromonas strains can produce chitinases with activity on crystalline chitin. Consistently, strain DSM 14401 was most likely to secrete chitinases with activity on crystalline chitin due to its high degradation rate on crystalline α-chitin.
2.2. Identification of the Chitinases Secreted by Strain DSM 14401
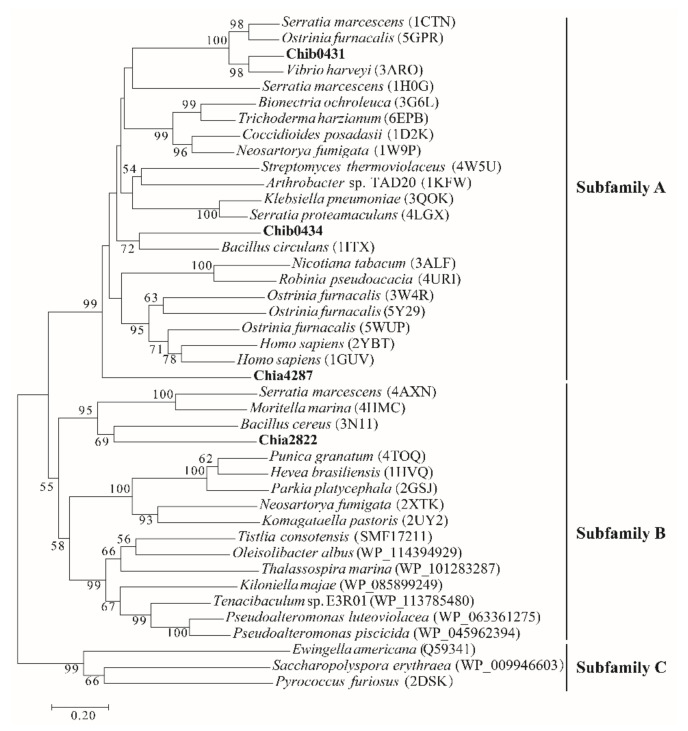
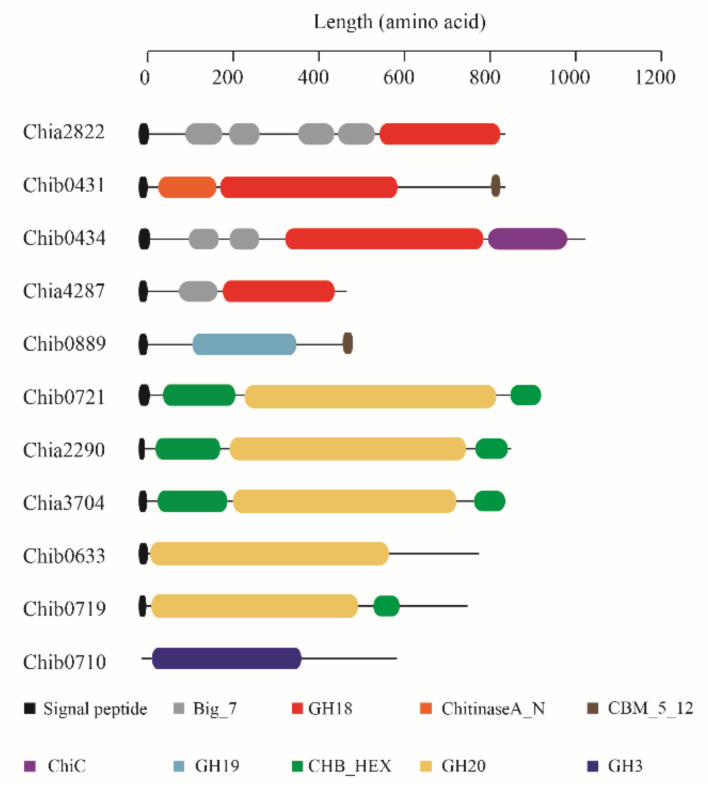
To ascertain the chitinolytic enzymes secreted by strain DSM 14401, genomic analysis was carried out to find putative chitinolytic enzyme-encoding genes in strain DSM 14401. There are 11 genes encoding putative chitinolytic enzymes in strain DSM 14401, which were named Chia2822, Chib0431, Chib0434, Chia4287, Chib0889, Chib0721, Chia2290, Chia3704, Chib0633, Chib0719 and Chib0710. Chia2822, Chib0431, Chib0434, and Chia4287 are potential chitinases belonging to the GH18 family (Figure 2). Of these, Chib0431, Chib0434, and Chia4287 belong to the GH18A subfamily that mainly contains processive exochitinases [40,41,42], and Chia2822 belongs to the GH18B subfamily that mainly contains non-processive endochitinases [43,44]. Multiple sequence alignments suggest that all these GH18 chitinases of strain DSM 14401 contain a DxDxE catalytic motif (Figure S1), which is conserved in the GH18 chitinases [45]. Chitinase Chib0889 belongs to the GH19 family that mainly contains chitinases found in plants [46]. Two GH19 chitinases, LYS177 and LYS188, from Pseudomonas Ef1 have been reported to have lysozyme activity and they are clustered with phage/prophage endolysins based on the phylogenetic analysis [47]. However, the GH19 chitinase, Chib0889, of strain DSM 14401 was nested in the cluster of chitinases from Proteobacteria (Figure S2), implying that Chib0889 may function as a chitinase rather than a lysozyme. Chib0721, Chia2290, Chia3704, Chib0633, and Chib0719 from the GH20 family, and Chib0710 from the GH3 family are potential β-N-acetylglucosaminidases. The predicted domain architectures of these chitinolytic enzymes are shown in Figure 3. Except for Chib0710, the other chitinolytic enzymes all have a signal peptide predicted by SignalP 5.0, implying that they are likely secreted enzymes. Among these enzymes, Chib0633 and Chib0710 are single-domain enzymes, while the others are all multi-domain enzymes containing one or more carbohydrate-binding domains (Big_7, CBM_5_12, and CHB_HEX) in addition to their catalytic domains. The CBMs (carbohydrate-binding modules) in chitinases were reported to facilitate enzyme movement along a chitin chain during processive action and to stimulate the substrate to decrystallize [48,49,50,51].
Figure 2.
Phylogenetic analysis of chitinases Chib0431, Chib0434, Chia4287, and Chia2822 with other GH18 chitinases. The phylogenetic tree was constructed by the Neighbor-Joining method. Bootstrap analysis of 1000 replicates was conducted.
Figure 3.
Domain architecture of the 11 chitinolytic enzymes of P. flavipulchra DSM 14401T. Protein sequences were analyzed on the HMMER website, and domains were illustrated by different colors based on their functional annotations. The Pfam IDs corresponding to the function annotations are as follows: Big_7, bacterial Ig domain (PF17957); GH18, glycosyl hydrolases family 18 (PF00704); ChitinaseA_N, ChitinaseA_N-terminal domain (PF08329); CBM_5_12, carbohydrate-binding module (PF02839), ChiC, Chitinase C (PF06483); GH19, glycoside hydrolase family 19 (PF00182), CHB_HEX, putative carbohydrate-binding domain (PF03173); GH20, glycosyl hydrolase family 20 (PF00728); GH3, glycosyl hydrolase family 3 (PF00933).
Secretome analysis was further performed to identify the chitinolytic enzymes secreted by strain DSM 14401 cultured in the medium containing 3% chitin flakes as the sole carbon source. The extracellular proteins tightly absorbed on the chitin flakes were collected for secretome analysis when approximately half of the chitin flakes in the medium were degraded after 85 h. Finally, 6 of the putative chitinolytic enzymes were detected in the secretome. Of these, the 4 GH18 chitinases accounted for 97.50% of the abundance, and the GH19 and GH20 chitinolytic enzymes each accounted for 1.25% (Table 1), which suggests the importance of the GH18 chitinases in the degradation of crystalline chitin. Of the GH18 chitinases, Chia4287 was the most abundant (48.75%), followed by Chib0431 (25.00%), Chib0434 (15.00%) and Chia2822 (8.75%). The five putative β-N-acetylglucosaminidases with a predicted signal peptide were not found in the secretome, which may be secreted to the periplasm.
Table 1.
The extracellular chitinolytic enzymes secreted by strain DSM 14401 identified by secretome analysis.
| Chitinolytic Enzyme | Accession Number |
Family | Length (aa) | Molecular Weight (kDa) | PSMs a | Abundance b |
|---|---|---|---|---|---|---|
| Chia4287 | WP_039494805 | GH18 | 479 | 50.86 | 39 | 48.75% |
| Chib0431 | WP_039495329 | GH18 | 822 | 87.51 | 20 | 25.00% |
| Chib0434 | WP_039495331 | GH18 | 1037 | 112.17 | 12 | 15.00% |
| Chia2822 | WP_039492151 | GH18 | 850 | 90.42 | 7 | 8.75% |
| Chib0889 | WP_084204324 | GH19 | 470 | 53.05 | 1 | 1.25% |
| Chib0721 | WP_039496328 | GH20 | 915 | 101.45 | 1 | 1.25% |
a Peptide-Spectrum Matches. b Abundance was calculated based on the proportion of the PSMs of a chitinolytic enzyme in the sum of PSMs of all chitinolytic enzymes in the secretome.
It has been reported that chitinolytic strains belonging to the genus Pseudoalteromonas usually have two GH18 chitinase genes in their chitin degradation clusters [36]. In addition, many Pseudoalteromonas species also contain one or more GH19 chitinase genes [36]. However, the removal of the GH19 chitinase gene from strain Pseudoalteromonas rubra S4059 had no significant influence on the growth of the strain on crystalline α-chitin [52], suggesting that the GH19 chitinase is likely unimportant in the utilization of crystalline chitin. In contrast, the removal of the GH18 chitinase gene chiD from strain Cellvibrio japonicus Ueda107 made it unable to grow on crystalline α-chitin [53], indicating that the GH18 chitinase plays an important role in the crystalline chitin degradation of this strain. Moreover, it has been reported that (GlcNAc)2 and larger chitooligosaccharides can induce the expression of chitinases in Vibrio furnissii 7225 and Vibrio cholerae O1 [54]. For strain DSM 14401, although its genome contains a GH19 chitinase gene, a GH3 β-N-acetylglucosaminidase gene, and 5 GH20 β-N-acetylglucosaminidase genes in addition to 4 GH18 chitinase genes, secretome analysis showed that it mainly secreted the GH18 chitinases when crystalline α-chitin was present, which suggests that the GH18 chitinases likely play a main role in the degradation of crystalline α-chitin in this strain.
2.3. Characterization of the GH18 Chitinases with Activity on Crystalline Chitin
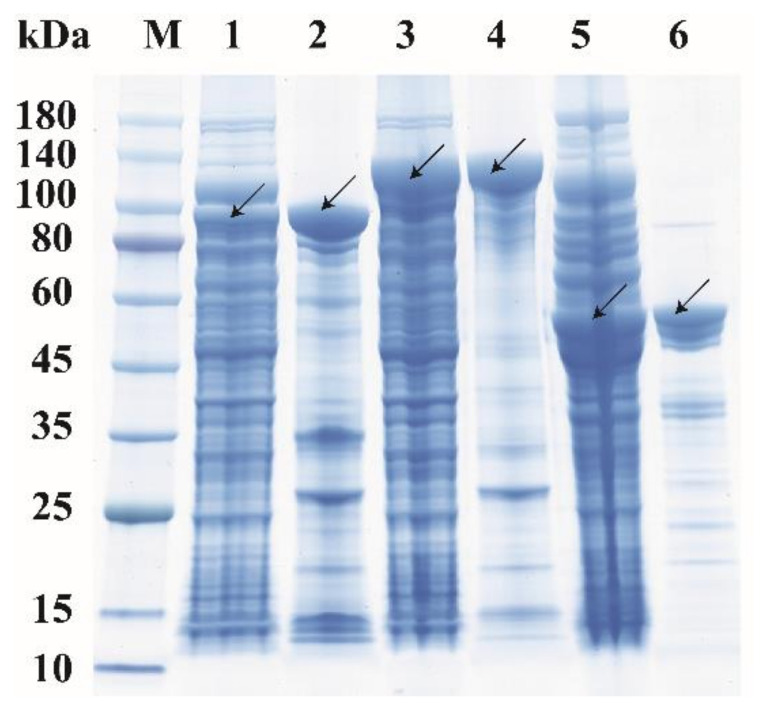
The high abundance of the GH18 chitinases in the secretome of strain DSM 14401 implies that they are likely to be the chitinases with activity on crystalline chitin. Thus, 3 GH18 chitinases, Chia4287, Chib0431, and Chib0434, with high abundance in the secretome, were selected to be expressed and characterized. Genes encoding Chia4287, Chib0431, and Chib0434 were heterologously expressed in E. coli BL21 (DE3), and the recombinant proteins were purified by NTA-Ni Sepharose affinity chromatography (Figure 4). The purification folds for Chib0431, Chib0434 and Chia4287 were 6.75, 5.33, and 7.30, respectively (Table S2). As shown in Figure 4, the 3 purified recombinant proteins have apparent molecular weights of approximately 88 kDa (Chib0431), 112 kDa (Chib0434), and 51 kDa (Chia4287), consistent with their theoretical molecular weights (Table 1).
Figure 4.
The SDS-PAGE analysis of recombinant proteins Chib0431, Chib0434, and Chia4287. Lane M, protein molecular mass marker; Lane 1, the cell lysate of E. coli containing recombinant protein Chib0431; Lane 2, the purified recombinant protein Chib0431; Lane 3, the cell lysate of E. coli containing recombinant protein Chib0434; Lane 4, the purified recombinant protein Chib0434; Lane 5, the cell lysate of E. coli containing recombinant protein Chia4287; Lane 6, the purified recombinant protein Chia4287. The enzyme bands are indicated by arrows.
To investigate the substrate specificity of these 3 chitinases, the enzyme activities of Chib0431, Chib0434, and Chia4287 toward colloidal chitin, chitin powder, chitosan, microcrystalline cellulose, 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide (MUF-GlcNAc) [55], 4-Methylumbelliferyl-β-D-N,N′-diacetylchitobioside hydrate (MUF-(GlcNAc)2) [56], and 4-Methylumbelliferyl-β-D-N,N′,N″-triacetylchitotrioside (MUF-(GlcNAc)3) [57] were determined. As shown in Table 2, all the three chitinases had activity toward colloidal chitin, crystalline chitin, MUF-(GlcNAc)2, and MUF-(GlcNAc)3, but neither had activity toward chitosan, microcrystalline cellulose, or MUF-GlcNAc. Among them, Chia4287 had the highest activity towards chitin powder, followed by Chib0431 and Chib0434, which is consistent with their amount in the secretome. Chitinases Chia4287 and Chib0431 exhibited higher activities toward MUF-(GlcNAc)3 than MUF-(GlcNAc)2, suggesting that both enzymes likely function as endochitinases. In contrast, Chib0434 showed approximately 10-fold higher activity toward MUF-(GlcNAc)2 than MUF-(GlcNAc)3, suggesting that Chib0434 tends to act as an exochitinase.
Table 2.
The substrate specificity of the three chitinases of strain DSM 14401 a.
| Substrate | Specific Activity (U/mg) | ||
|---|---|---|---|
| Chia4287 | Chib0431 | Chib0434 | |
| Colloidal chitin | 0.53 ± 0.05 | 0.15 ± 0.04 | 0.09 ± 0.02 |
| Chitin powder | 0.17 ± 0.005 | 0.04 ± 0.002 | 0.01 ± 0.001 |
| Chitosan | ND b | ND | ND |
| Microcrystalline cellulose | ND | ND | ND |
| MUF-GlcNAc | ND | ND | ND |
| MUF-(GlcNAc)2 | 130.32 ± 3.29 | 19.69 ± 1.30 | 221.68 ± 12.15 |
| MUF-(GlcNAc)3 | 139.33 ± 26.96 | 423.12 ± 9.82 | 23.47 ± 3.57 |
a The data in the table are from three experiment repeats (mean ± SD). b ND means that the enzyme activity was not detectable.
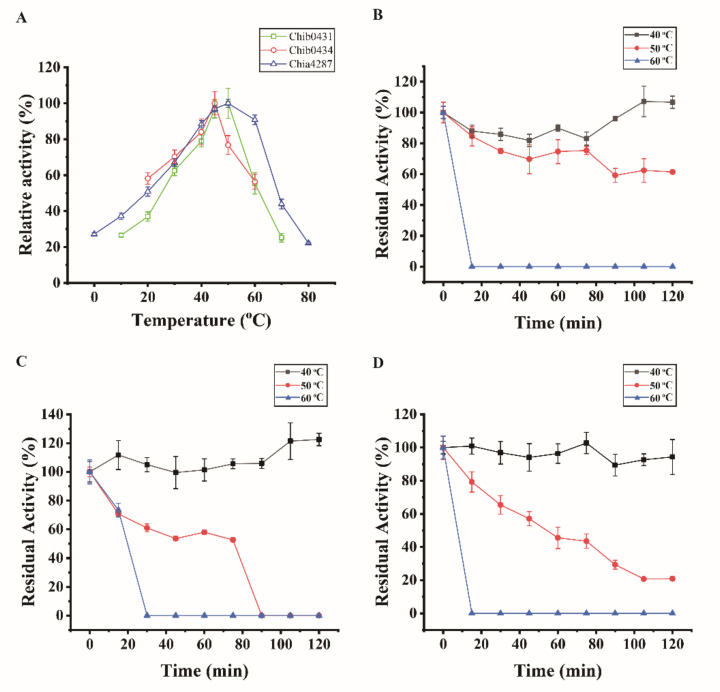
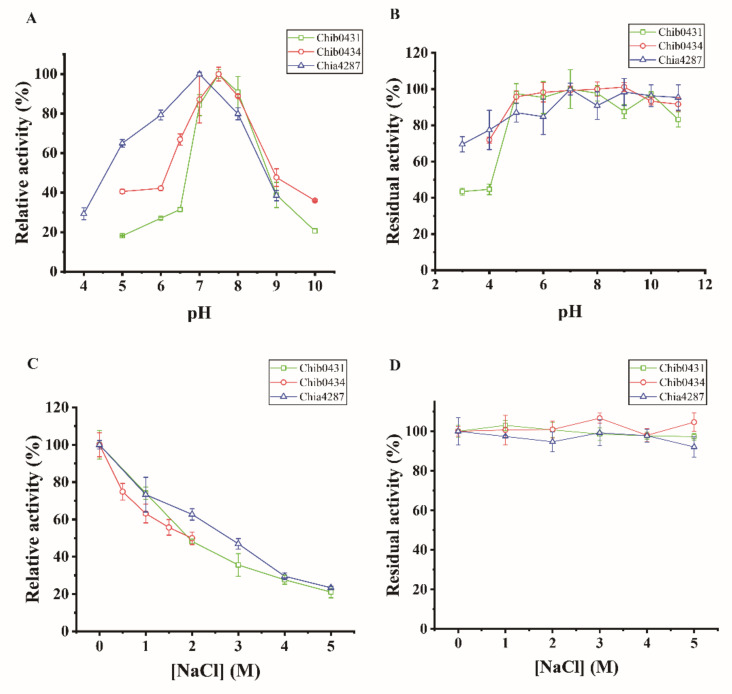
With chitin powder as the substrate, the three chitinases were biochemically characterized. Both chitinases Chib0431 and Chia4287 showed optimum temperatures at 50 °C, and Chib0434 at 45 °C (Figure 5A). For their thermal stability, Chib0431 retained approximately 100% activity at 40 °C and more than 61% at 50 °C after 120 min incubation but lost all its activity at 60 °C in 15 min (Figure 5B). Chib0434 retained 100% activity at 40 °C after 120 min incubation but lost all its activity at 50 °C in 90 min and at 60 °C in 30 min (Figure 5C). Chitinase Chia4287 retained high activity (≥89%) when incubated at 40 °C for 120 min (Figure 5D). Chitinases Chib0434 and Chia4287 both showed highest activity at pH 7.5 and Chib0431 at pH 7.0 (Figure 6A). For their pH stability, the 3 chitinases all exhibited high stability (retaining ≥80% activity) from pH 5.0 to 11.0 in the Britton–Robinson buffer after 10 h incubation at 4 °C (Figure 6B). They all showed highest activity at 0 M NaCl (Figure 6C) but maintained high activity (≥92%) in 1–5 M NaCl after 10 h incubation at 4 °C (Figure 6D). Therefore, the 3 chitinases have temperature optima of 45–50 °C and pH optima of 7.0–7.5, indicating that they are all neutral and mesophilic enzymes. They are all stable at 40 °C and in the pH range of 5.0–11.0, and all have excellent salt tolerance.
Figure 5.
Effect of temperature on the activities and stabilities of chitinases Chib0431, Chib0434, and Chia4287. (A) Effect of temperature on the activities of Chib0431, Chib0434, and Chia4287. The activities of each enzyme were measured at its optimal pH with chitin powder as the substrate. The highest activity of each enzyme was defined as 100%. (B) Effect of temperature on the stability of Chib0431. (C) Effect of temperature on the stability of Chib0434. (D) Effect of temperature on the stability of Chia4287. In B, C, and D, the residual activities of each enzyme were measured at its optimal temperature and pH with chitin powder as the substrate, and the activity of each enzyme without incubation was defined as 100%. The graphs show data from triplicate experiments (mean ± SD).
Figure 6.
Effects of pH and NaCl on the activities and stabilities of chitinases Chib0431, Chib0434, and Chia4287. (A) Effect of pH on the activities of Chib0431, Chib0434, and Chia4287. The activities of each enzyme were measured at its optimal temperature with chitin powder as the substrate. The highest activity of each enzyme was defined as 100%. (B) Effect of pH on the stabilities of Chib0431, Chib0434, and Chia4287. The residual activities of each enzyme were measured at its optimal temperature and pH with chitin powder as the substrate. (C) Effect of NaCl concentration on the activities of Chib0431, Chib0434, and Chia4287. The activities of each enzyme were measured at its optimal temperature and pH with chitin powder as the substrate. The activity of each enzyme in 0 M NaCl was defined as 100%. (D) Effect of NaCl concentration on the stabilities of Chib0431, Chib0434, and Chia4287. The residual activities of each enzyme were measured at its optimal temperature and pH with chitin powder as the substrate. The highest activity of each enzyme was defined as 100%. The graphs show data from triplicate experiments (mean ± SD).
Many chitinases have been heterologously expressed and characterized with colloidal chitin or synthetic chitooligosaccharide analogs. As shown in Table 3, the temperature and pH optima of the reported chitinases and their thermostability are quite diverse. So far, several Pseudoalteromonas GH18 chitinases have been characterized (Table 3). The chitinase Chi23 from P. aurantia DSM6057 was reported to be thermostable but active toward crystalline chitin only in acidic conditions (pH of 3.0–6.0) [18]. Chitinases ChiA and ChiC from Pseudoalteromonas sp. DL-6 [27,37] and ChiB from Pseudoalteromonas sp. O-7 [38] are all cold-active enzymes with optimal activities at 20–30 °C and low thermostability. The 3 mesophilic chitinases, Chib0431, Chib0434, and Chia4287, characterized in this study are active toward crystalline chitin at neutral pH conditions (pH 7.0–7.5) and have good thermostability and pH- and salt-tolerance, which, therefore, may be good candidates for industrial application.
Table 3.
Characteristics of bacterial chitinases.
| Enzyme | Family | Molecular Weight (kDa) |
pH Optimum |
Temperature Optimum (°C) |
NaCl Optimum (M) |
Thermostability (Half-Life) |
Substrate (Specific Activity) |
Hydrolytic Products (Substrate) |
References |
|---|---|---|---|---|---|---|---|---|---|
| Chib0431 from Pseudoalteromonas flavipulchra DSM 14401T | GH18 | 87.51 | 7.5 | 50 | 0 | >2 h at 50 °C | α-chitin (0.04 ± 0.002 U/mg) |
GlcNAc and (GlcNAc)2 (α-chitin) |
This study |
| Chib0434 from Pseudoalteromonas flavipulchra DSM 14401T | GH18 | 112.17 | 7.5 | 45 | 0 | ~80 min at 50 °C | α-chitin (0.01 ± 0.001 U/mg) |
GlcNAc and (GlcNAc)2 (α-chitin) |
This study |
| Chia4287 from Pseudoalteromonas flavipulchra DSM 14401T | GH18 | 50.86 | 7.0 | 50 | 0 | <60 min at 50 °C | α-chitin (0.17 ± 0.005 U/mg) |
GlcNAc and (GlcNAc)2 (α-chitin) | This study |
| CHI II from Glaciozyma antarctica PI12 | GH18 | 39 and 50 | 4.0 | 15 | - | <30 min at 30 °C | Colloidal chitin (-) |
- | [26] |
| MmChi60 from Moritella marina | GH18 | 60.8 | 5.0 | 28 | - | ~5 h at 50 °C | Colloidal chitin (0.016 U/mg) |
- | [29] |
| ChiA from Pseudoalteromonas sp. DL-6 | GH18 | 113.5 | 8.0 | 20 | - | ~1 h at 40 °C | α-chitin (0.128 ± 0.001 U/mL) |
(GlcNAc)2 (α-chitin) |
[27] |
| ChiC from Pseudoalteromonas sp. DL-6 | GH18 | 91 | 9.0 | 30 | 2 | ~1 h at 50 °C | α-chitin (4.8 ± 0.2 U/mg) |
(GlcNAc)2 (colloidal chitin) |
[37] |
| Chi23 from Pseudoalteromonas aurantia DSM6057 | GH18 | 30.4 | 5.0 | 60 | 3 | ~40 min at 70 °C | Crystalline Chitin (0.1 ± 0.01 U/mg) |
(GlcNAc)2 and GlcNAc)3 (α-chitin) |
[18] |
| ChiB from Pseudoalteromonas sp. O-7 | GH18 | 90.2 | 6.0 | 30 | - | - | pNP-(GlcNAc)2 (30.8 U/mg) |
- | [38] |
| ScChiC from Streptomyces coelicolor A3(2) | GH18 | - | 5 | 55 | - | ~1 h at 60 °C | (GlcNAc)6 (4120 ± 80 U/mg) |
(GlcNAc)2 (crab shell chitin) |
[19] |
| StmChiA from Stenotrophomonas maltophilia | GH18 | 70.5 | 5.0 | 40 | - | >90% of initial activity at 30–50 °C (up to 1 h) | (GlcNAc)6 (-) |
GlcNAc and (GlcNAc)2 (α-chitin) |
[20] |
| StmChiB from Stenotrophomonas maltophilia | GH18 | 41.6 | 7.0 | 40 | - | >90% of initial activity at 30–50 °C (up to 1 h) | (GlcNAc)6 (-) |
- | [20] |
| PbChi67 from Paenicibacillus barengoltzii CAU904 | - | 67.9 | 3.5 | 60 | - | 43 min at 65 °C | α-chitin (0.3 ± 0.04 U/mg) |
(GlcNAc)2, (GlcNAc)3 and(GlcNAc)4 (colloidal chitin) |
[17] |
| A chitinase from Bacillus sp. R2 | - | 41.69 | 7.5 | 40 | - | >30 min at 50 °C | Colloidal chitin (-) |
- | [21] |
| A chitinase from Citrobacter freundii haritD11 | - | 64 | 8.0 | 35 | - | ~1 h at 60 °C | Colloidal chitin (140.55 U/mg) | - | [28] |
| A chitinase from Micrococcus sp. AG84 | - | 33 | 8.0 | 40 | - | >1 h at 80 °C | Colloidal chitin (93.02 U/mg) |
- | [22] |
| A chitinase from Pseudoalteromonas sp. DC14 | - | 65 | 9.0 | 40 | 10% (w/v) | >30 min at 60 °C | Colloidal chitin (5.6 U/mg) |
- | [23] |
| A chitinase from Streptomyces chilikensis RC1830 | - | 10.5 | 7.0 | 60 | - | - | Colloidal chitin (60.53 U/mg) | - | [24] |
| PtChi19 from Pseudoalteromonas tunicata CCUG 44952T | GH19 | 53.5 | 7.5 | 45 | 2 | >40 min at 50 °C | Crystalline Chitin (16.4 mU/ mg) | - | [25] |
- Not available.
2.4. Analysis of the Products of the Chitinases on Crystalline Chitin
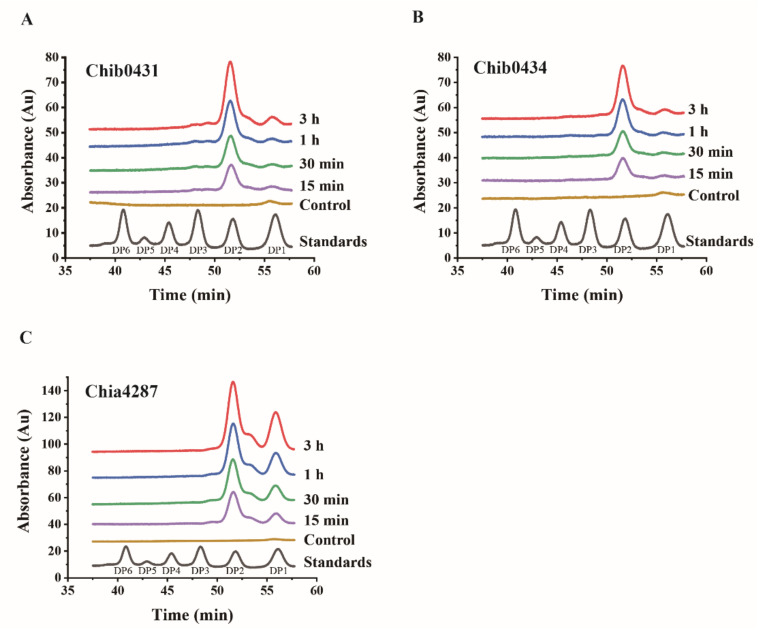
In order to investigate the application potential of the three chitinases in preparing COSs/GlcNAc from natural chitin, we analyzed the degradation products of Chia4287, Chib0431, and Chib0434 towards crystalline chitin. The reaction mixtures, containing chitin powder and chitinases, were incubated at their respective optimal temperatures for different time periods (15 min, 30 min, 1 h, and 3 h). The COSs/GlcNAc released from chitin in the supernatants of the mixtures were analyzed by gel filtration chromatography on a Superdex Peptide 10/300 GL column. For Chib0431 and Chib0434, during the 3 h degradation of crystalline chitin, (GlcNAc)2 was always the predominant product, with only a slight amount of GlcNAc (Figure 7A,B). However, in the hydrolytic products of Chia4287 on crystalline chitin, although (GlcNAc)2 was also the main product, the proportion of GlcNAc was much higher compared to that in the hydrolytic products of Chib0431 and Chib0434 (Figure 7C). Together, these results indicate that Chia4287, Chib0431, and Chib0434 can degrade crystalline chitin into (GlcNAc)2 and GlcNAc, with (GlcNAc)2 as the main product. These results imply that they may have potential in the preparation of (GlcNAc)2 from natural crystalline chitin.
Figure 7.
Analysis of the degradation products of the three chitinases on crystalline chitin. (A) The degradation product of Chib0431. (B) The degradation product of Chib0434. (C) The degradation product of Chia4287. Chitin powder was degraded by the chitinases at their respective optimal temperatures for different times (15 min, 30 min, 1 h, and 3 h). The reaction system with enzyme inactivated at 100 °C for 10 min was used as the control. The reaction was terminated by boiling at 100 °C for 10 min, and then the reaction mixtures were centrifuged at 17,949× g for 10 min. The products in the supernatants were analyzed by gel filtration chromatography on a Superdex Peptide10/300 GL column (GE Healthcare, Sweden), which were monitored at a wavelength of 210 nm. The injected volume was 10 μL. DP1-DP6 are chitooligosaccharide markers. DP1, GlcNAc; DP2, (GlcNAc)2; DP3, (GlcNAc)3; DP4, (GlcNAc)4; DP5, (GlcNAc)5; DP6, (GlcNAc)6.
COSs/GlcNAc have been widely prepared with a variety of crude enzymes from wild strains and purified recombinant chitinases, most of which were prepared with colloidal chitin [17,58,59,60,61]. So far, however, there have been only a few chitinases used to prepare COSs/GlcNAc from natural crystalline chitin. The enzyme cocktail of strain Paenibacillus sp. LS1 can produce GlcNAc and (GlcNAc)2 with minor (GlcNAc)3 from crystalline α-chitin [30]. The crude enzyme of Aeromonas hydrophila H-2330 mainly produces GlcNAc from crystalline α-chitin [31]. The chitinase ChiA of strain Pseudoalteromonas sp. DL-6 is an endochitinase, and its products on crystalline α-chitin are a mixture of chitin COSs (DP 2–6), with (GlcNAc)2 as the major product [27]. The mixture of purified chitinases SaChiB and SaHEX of strain Streptomyces alfalfa ACCC40021 can enhance the conversion of crystalline α-chitin to GlcNAc [62]. The chitinase of strain Chitinibacter sp. GC72 can degrade practical-grade chitin into GlcNAc [33]. The three chitinases characterized in this study can degrade crystalline α-chitin into (GlcNAc)2, suggesting their potential in direct conversion of natural crystalline chitin into (GlcNAc)2.
3. Materials and Methods
3.1. Bacterial Strains and Experimental Materials
The 26 type strains of genus Pseudoalteromonas were purchased from Deutsche Sammlung von Mikroorganismen and Zelkulturen (DSMZ) or Japan Collection of Microorganisms (JCM). Chitin powder (crystalline α-chitin), MUF-GlcNAc, MUF-(GlcNAc)2, and MUF-(GlcNAc)3 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Chitin flakes, purchased from Yuan Cheng Group (Wuhan, China), are crystalline α-chitin. Colloidal chitin was prepared as previously described [18]. GlcNAc, (GlcNAc)2, (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6 were purchased from BZ Oligo Biotech Co., LTD (Qingdao, China). Chitosan was purchased from Sangon Biotech (Shanghai, China). BCA protein assay kit was purchased from Thermo Scientific (Boston, MA, USA). Other chemicals were of analytical grade and commercially available.
3.2. Screening of Strain DSM 14401
The 26 type strains of genus Pseudoalteromonas (Table S1) were cultivated at 25 °C and 180 rpm in the TYS medium composed of 0.5% (w/v) peptone, 0.1% (w/v) yeast powder, and artificial seawater (pH 7.8). When the OD600 of the culture was approximately 1.0, 2 mL cell suspension was collected and the cells were washed with the minimal medium (30 g/L NaCl, 0.5 g/L NH4Cl, 3 g/L MgCl2·6H2O, 2 g/L K2SO4, 0.2 g/L K2HPO4, 0.01 g/L CaCl2, 0.006 g/L FeCl3·6H2O, 0.005 g/L Na2MoO4·7H2O, 0.004 g/L CuCl2·2H2O, 6 g/L Tris, pH 7.6) three times. Then, the washed cells were inoculated into the minimal medium supplemented with 0.05% (w/v) peptone, 0.01% (w/v) yeast powder, and 3% (w/v) chitin flakes and cultivated at 25 °C and 180 rpm for 5 days. Their growth and the degree of degradation of the chitin flakes were observed every day. Among them, strain DSM 14401 showed the highest degradation rate on crystalline α-chitin, which was then chosen for further study. The OD600 of the culture of this strain in the medium was measured at different time intervals, as indicated in Figure 1, to produce its growth curve. The washed cells were cultured in the same medium without chitin flakes and in the same conditions as the control.
3.3. Extracellular Chitinase Activity Assay of Strain DSM 14401
During the cultivation of strain DSM 14401 in the above liquid medium with or without chitin flakes, 1 mL of culture was taken out at different intervals, as indicated in Figure 1. The cultures were filtered with a 0.22 μm filter to remove the bacterial cells, and the filtrate was used for the extracellular chitinase activity assay. A 200 μL mixture consisting of 50 mM Tris-HCl (pH 7.0), 3% chitin powder, and 50 μL of filtrate was incubated at 50 °C for 2 h. The mixture was then centrifuged at 17,949× g for 2 min at 4 °C and the supernatant obtained was used for the reducing-sugar assay by the DNS method [63]. The control mixture contained a pre-boiled filtrate instead of the filtrate. Subsequently, the optical density at 550 nm was measured to quantify the released reducing sugar. The amount of reducing sugar generated was calculated using GlcNAc as a standard. One unit of enzyme activity was defined as the amount of enzyme that liberated 1 μmol of reducing sugar per minute.
3.4. Bioinformatics Analysis
The genome DNA of strain DSM 14401 was sequenced by our lab [64]. The putative chitinases of this strain were determined according to dbCAN [65] analyses. Signal peptides of the chitinases were predicted by SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/ (accessed on 12 January 2022)) [66]. The domain architectures of the chitinases were predicted on the HMMER website (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan (accessed on 12 January 2022)) [67]. The phylogenetic tree was constructed based on the Neighbor-Joining method and using the Poisson model with MEGA X after multiple alignments of the sequences by MUCLE [68]. Sequences alignment results were visualized using the ESPript 3.0 server [69]. The molecular weights of the chitinases were predicted by the ExPASy Server (https://web.expasy.org/compute_pi/ (accessed on 12 January 2022)) [70].
3.5. Secretome Analysis
Strain DSM 14401 was cultured at 25 °C and 180 rpm in a medium containing the minimal medium and 3% chitin flakes. When approximately half of the chitin flakes were degraded, the culture was centrifuged at 8228× g at 4 °C for 6 min. The precipitates were resuspended using 20 mM Tris-HCl (pH 8.0) containing 1 M NaCl, and then centrifuged at 1157× g at 4 °C for 3 min. This step was repeated three times. The resultant precipitates were resuspended using 50 mM Tris-HCl (pH 8.0) containing 6 M Guanadine-HCl, and then centrifuged at 15,557× g at 4 °C for 10 min. The supernatant was moved into an ultrafiltration tube (15 mL, 3 kDa). The Guanadine-HCl in the supernatant was removed by adding 50 mM Tris-HCl to the ultrafiltration tube (molecular weight cut-off, 3 kDa) and centrifugation (4629× g for 10 min at 4 °C) for three times. Then, the proteins in the supernatant were precipitated by 50 mL acetone containing 10% trichloroacetic acid and 0.1% dithiothreitol overnight at −20 °C. The precipitates were harvested and washed by 80% acetone and 100% acetone successively, and then lyophilized. The lyophilized sample was successively denatured, reduced, and alkylated by denaturation buffer (0.5 M Tris-HCl, 2.75 mM EDTA, 6 M Guanadine-HCl), dithiothreitol (1 M), and iodoacetamide (1 M), respectively. The sample solution was further replaced with 25 mM NH4HCO3 solution by centrifugation ultrafiltration (15,294× g for 15 min at 4 °C) in an ultrafiltration tube (1 mL, 3 kDa). The sample was digested using trypsin at 37 °C for 12 h, and the resultant peptides were desalted on a C18 column (ZipTip C18, Millipore, Billerica, MA, USA). The desalted peptides were analyzed using the mass spectrometer Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany) coupled with Easy-nLC 1000 (Thermo Fisher Scientific, Bremen, Germany). Finally, the raw data was analyzed against the genome of strain DSM 14401 using Thermo Scientific Proteome DiscovererTM 1.4. The mass spectrometry proteomics data have been deposited to the ProteomeXchange [71] Consortium via the PRIDE [72] partner repository with the dataset identifier PXD030600. The reviewer account details: Username: reviewer_pxd030600@ebi.ac.uk; Password: 1QCP2jqI.
3.6. Expression and Purification of Chitinases Chib0431, Chib0434, Chia4287
The gene sequences of Chib0431, Chib0434, and Chia4287 without the signal peptide were cloned from the genomic DNA of strain DSM 14401 and inserted into the NdeI and XhoI sites of the expression vector pET-22b(+). The constructed recombinant plasmids were then transformed into E. coli BL21(DE3) for protein expression. The constructed recombinant E. coli BL21(DE3) strains were cultured at 37 °C in liquid LB medium containing 100 μg/mL ampicillin. When the OD600 of the cultures reached 0.6–1.0, 0.45 mM isopropyl thio-β-D-galactoside (IPTG), used as an inducer, was added into the cultures, and the cultures were incubated at 18 °C for 16 h. Then, the recombinant E. coli cells in the cultures were collected via centrifugation and crushed by sonication in the lysis buffer (100 mM NaCl, 5 mM imidazole, 50 mM Tris-HCl pH 8.0). The recombinant proteins of Chib0431, Chib0434, and Chia4287 in the cell extracts were further purified by affinity chromatography with Ni-NTA agarose resins (Qiagen, Santa Clarita, CA, USA), followed by desalination on PD-10 Desalting Columns (GE Healthcare, Piscataway, NJ, USA), using 10 mM Tris-HCl containing 100 mM NaCl (pH 8.0) as the running buffer. The purified proteins were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [73]. The protein concentrations were determined using a BCA protein assay kit with bovine serum albumin (BSA) as the standard.
3.7. Enzyme Assays
The activities of the three purified chitinases towards chitin powder, colloidal chitin, chitosan, microcrystalline cellulose, MUF-GlcNAc, MUF-(GlcNAc)2 and MUF-(GlcNAc)3 were assayed in 50 mM Tris-HCl at their respective optimal temperatures and pHs (50 °C and pH 7.0 for Chia4287, 50 °C and pH 7.5 for Chib0431, 45 °C and pH 7.5 for Chib0434). When the insoluble chitin powder, chitosan, or microcrystalline cellulose was used as the substrate, the reaction mixture contained 190 µL 50 mM Tris-HCl, 3% (w/v) substrate and 10 µL enzyme, which was incubated for 1 h for Chia4287 or 2 h for Chib0431 and Chib0434. When colloidal chitin was used as the substrate, the reaction mixture contained 190 µL 0.75% (w/v) colloidal chitin in 50 mM Tris-HCl and 10 µL enzyme, which was incubated for 40 min. After incubation, the activities of the chitinases towards these substrates were determined using the DNS method [63]. The enzyme activity (U) was defined as the amount of enzyme that required to release 1 μmol GlcNAc equivalent reducing sugar from the substrate per minute. When MUF- GlcNAc, MUF-(GlcNAc)2, or MUF-(GlcNAc)3 was used as the substrate, the enzyme activity was assayed for 15 min with the reaction mixture contained 790 µL 1 mM substrate in 50 mM Tris-HCl and 10 µL enzyme, which was incubated for 15 min and then terminated by an addition of 0.4 M NaCO3. The enzyme activity (U) was defined as the amount of enzyme that required to release 1 μmol MUF from the substrate per minute.
3.8. Characterization of the Chitinases
The purified Chib0431, Chib0434, and Chia4287 were characterized with chitin powder as substrate. The effect of temperature on the enzyme activity was measured by assaying the enzyme activity at different temperatures (0–80 °C for Chia4287; 10–70 °C for Chib0431 and 20–60 °C for Chib0434) and their respective optimal pHs. The effect of pH on the enzyme activity was measured by assaying the enzyme activity in the Britton-Robinson buffer at different pHs (pH 4.0–9.0 for Chia4287; pH 5.0–10.0 for Chib0431 and Chib0434) and their respective optimal temperatures. Effect of salinity on the enzyme activity was assayed by assaying the enzyme activity in 50 mM Tris-HCl containing different concentrations of NaCl (0–5 M for Chib0431 and Chia4287; 0–2 M for Chib0434) at their respective optimal temperatures and pHs.
For the thermal stability assay, the purified chitinases were incubated at 40 °C, 50 °C, or 60 °C for 0–120 min, and the residual activities towards chitin powder were measured at an interval of 15 min under their respective optimal temperatures and pHs. For the pH stability assay, the purified chitinases were incubated in the Britton-Robinson buffers ranging from pH 3.0 to pH 11.0 at 4 °C for 10 h, and the residual activities towards chitin powder were measured at their respective optimal temperatures and pHs. For the halotolerance assay, the purified chitinases were incubated in 50 mM Tris-HCl containing different concentrations of NaCl (0–5 M) at 4 °C for 10 h, and the residual enzyme activities towards chitin powder were measured at their respective optimal temperatures and pHs.
3.9. Analysis of the Products Released from Crystalline Chitin by the Chitinases
The purified Chib0431, Chib0434, and Chia4287 (10 μL) were incubated with 3.0% chitin powder in 190 μL of 50 mM Tris-HCl (pH 7.0) for different times (15 min, 30 min, 1 h, and 3 h) at their respective optimal temperatures. The reaction was terminated by boiling at 100 °C for 10 min, and the reaction mixtures were centrifuged at 17,949× g for 10 min. Then, the products in the supernatants were analyzed by gel filtration chromatography on a Superdex Peptide 10/300 GL column (GE Healthcare, Uppsala, Sweden), which were monitored at 210 nm using a UV detector. The injected volume was 10 μL. The products were eluted with 0.2 M ammonium hydrogen carbonate for 90 min with a flow rate of 0.3 mL/min. The reaction system containing 10 μL enzyme pre-heated at 100 °C for 10 min was used as the control. A mixture of GlcNAc, (GlcNAc)2, (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6 was used as the marker.
4. Conclusions
COSs have wide application in agriculture, medicine, cosmetics, and foods. While most COSs are now prepared with colloidal chitin, there are only a few reports of chitinases with potential in the preparation of COSs from natural crystalline chitin. In this study, three chitinases with activity on crystalline chitin were identified from a marine Pseudoalteromonas strain and characterized. These chitinases are all neutral mesophilic enzymes, which are most active at 45–50 °C and pH 7.0–7.5, and have high stability at 40 °C, pH 5.0–11.0, and in 5 M NaCl. The main products of the three chitinases on crystalline chitin are all (GlcNAc)2, suggesting that these chitinases have potential in preparing (GlcNAc)2 via direct degradation of natural crystalline chitin. Further studies such as improving the expression amount of these chitinases and their degradation efficiency on crystalline chitin are underway.
Acknowledgments
We would like to thank Andrew McMinn from the University of Tasmania, Australia, for editing this paper. We would like to thank Cai-Yun Sun and Rui Wang from State Key Laboratory of Microbial Technology of Shandong University for help and guidance in Bioscreen C Microbiology reader.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md20030165/s1, Figure S1: Multiple sequence alignments of Chib0431, Chib0434, Chia4287, and Chia2822 with known GH18 chitinases.; Figure S2: Phylogenetic analysis of Chib0889 with other GH19 chitinases.; Table S1: General information of 26 type strains of Pseudoalteromonas.; Table S2: Purification of the recombinant enzymes Chib0431, Chib0434, and Chia4287. References [39,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96] are cited in the supplementary materials.
Author Contributions
Conceptualization, X.-L.C. and Y.-Z.Z.; Investigation, Y.-J.W.; Methodology, X.-B.R., S.-S.L. and Y.-J.W.; Project administration, X.-L.C. and P.-Y.L.; Resources, X.-L.C. and Y.-Z.Z.; Software, Y.-R.D. and K.-X.H.; Supervision, Q.-L.Q., X.-L.C. and P.-Y.L.; Writing—original draft, X.-B.R.; Writing—review & editing, X.-L.C. and P.-Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Science Foundation of China (grants 42176229, 31870101, U2006205, 31870052 and awarded to P.-Y.L., Q.-L.Q., X.-L.C. and X.-L.C., respectively), the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817 awarded to Y.-Z.Z.), Taishan Scholars Program of Shandong Province (tspd20181203 awarded to Y.-Z.Z.).
Institutional Review Board Statement
This article does not contain any studies involving human participants or animals performed by any of the authors.
Data Availability Statement
Proteomic data are available via ProteomeXchange with identifier PXD030600.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rinaudo M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006;31:603–632. doi: 10.1016/j.progpolymsci.2006.06.001. [DOI] [Google Scholar]
- 2.Jang M.K., Kong B.G., Jeong Y.I., Lee C.H., Nah J.W. Physicochemical characterization of α-chitin, β-chitin, and γ-chitin separated from natural resources. J. Polym. Sci. Part A Polym. Chem. 2004;42:3423–3432. doi: 10.1002/pola.20176. [DOI] [Google Scholar]
- 3.Johnstone J. Conditions of Life in the Sea. Cambridge University Press; Cambridge, UK: 1908. p. 332. [Google Scholar]
- 4.Poulicek M., Jeauniaux C. Chitin Chitosan. Elsevier; London, UK: 1988. Chitin biomass in marine sediments; pp. 152–160. [Google Scholar]
- 5.Tokoro A., Kobayashi M., Tatewaki N., Suzuki K., Okawa Y., Mikami T., Suzuki S., Suzuki M. Protective effect of N-acetyl chitohexaose on Listeria monocytogenes infection in mice. Microbiol. Immunol. 1989;33:357–367. doi: 10.1111/j.1348-0421.1989.tb01983.x. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura K., Nishimura S., Nishi N., Saiki I., Tokura S., Azuma I. Immunological activity of chitin and its derivatives. Vaccine. 1984;2:93–99. doi: 10.1016/S0264-410X(98)90039-1. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.K., Ravichandran Y.D., Khan S.B., Kim Y.T. Prospective of the cosmeceuticals derived from marine organisms. Biotechnol. Bioprocess Eng. 2008;13:511–523. doi: 10.1007/s12257-008-0113-5. [DOI] [Google Scholar]
- 8.Bak Y.K., Lampe J.W., Sung M.K. Effects of dietary supplementation of glucosamine sulfate on intestinal inflammation in a mouse model of experimental colitis. J. Gastroenterol. Hepatol. 2014;29:957–963. doi: 10.1111/jgh.12485. [DOI] [PubMed] [Google Scholar]
- 9.Tamai Y., Miyatake K., Okamoto Y., Takamori Y., Sakamoto K., Minami S. Enhanced healing of cartilaginous injuries by N-acetyl-D-glucosamine and glucuronic acid. Carbohydr. Polym. 2003;54:251–262. doi: 10.1016/S0144-8617(03)00170-X. [DOI] [Google Scholar]
- 10.Wang Y.C., Lien T.S., Chen N.Y., Hsu T.H. Purification and characterization of β-N-acetylglucos-aminidase from Grifola frondosa. BioResources. 2021;16:7234–7248. doi: 10.15376/biores.16.4.7234-7248. [DOI] [Google Scholar]
- 11.Jiang S., Jiang H., Zhou Y., Jiang S., Zhang G. High-level expression of β-N-Acetylglucosaminidase BsNagZ in Pichia pastoris to obtain GlcNAc. Bioprocess Biosyst. Eng. 2019;42:611–619. doi: 10.1007/s00449-018-02067-5. [DOI] [PubMed] [Google Scholar]
- 12.Suginta W., Chuenark D., Mizuhara M., Fukamizo T. Novel β-N-acetylglucosaminidases from Vibrio harveyi 650: Cloning, expression, enzymatic properties, and subsite identification. BMC Biochem. 2010;11:40. doi: 10.1186/1471-2091-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar M., Brar A., Vivekanand V., Pareek N. Bioconversion of Chitin to Bioactive Chitooligosaccharides: Amelioration and Coastal Pollution Reduction by Microbial Resources. Mar. Biotechnol. 2018;20:269–281. doi: 10.1007/s10126-018-9812-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu C., Shen N., Wu J., Jiang M., Shi S., Wang J., Wei Y., Yang L. Cloning, expression and characterization of a chitinase from Paenibacillus chitinolyticus strain UMBR 0002. PeerJ. 2020;8:e8964. doi: 10.7717/peerj.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung D., Baek K., Bae S.S., Jung J. Identification and characterization of a marine-derived chitinolytic fungus, Acremonium sp. YS2-2. J. Microbiol. 2019;57:372–380. doi: 10.1007/s12275-019-8469-0. [DOI] [PubMed] [Google Scholar]
- 16.Song Y.S., Lee S.H., Cho J.A., Moon C., Seo D.J., Jung W.J. Expression and degradation patterns of chitinase purified from Xuehuali (Pyrus bretschneiderilia) pollen. Int. J. Biol. Macromol. 2018;107:446–452. doi: 10.1016/j.ijbiomac.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Fu X., Yan Q., Wang J., Yang S., Jiang Z. Purification and biochemical characterization of novel acidic chitinase from Paenicibacillus barengoltzii. Int. J. Biol. Macromol. 2016;91:973–979. doi: 10.1016/j.ijbiomac.2016.06.050. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y.J., Jiang W.X., Zhang Y.S., Cao H.Y., Zhang Y., Chen X.L., Li C.Y., Wang P., Zhang Y.Z., Song X.Y., et al. Structural Insight into Chitin Degradation and Thermostability of a Novel Endochitinase from the Glycoside Hydrolase Family 18. Front. Microbiol. 2019;10:2457. doi: 10.3389/fmicb.2019.02457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen-Thi N., Doucet N. Combining chitinase C and N-acetylhexosaminidase from Streptomyces coelicolor A3(2) provides an efficient way to synthesize N-acetylglucosamine from crystalline chitin. J. Biotechnol. 2016;220:25–32. doi: 10.1016/j.jbiotec.2015.12.038. [DOI] [PubMed] [Google Scholar]
- 20.Suma K., Podile A.R. Chitinase A from Stenotrophomonas maltophilia shows transglycosylation and antifungal activities. Bioresour. Technol. 2013;133:213–220. doi: 10.1016/j.biortech.2013.01.103. [DOI] [PubMed] [Google Scholar]
- 21.Cheba B.A., Zaghloul T.I., EL-Mahdy A.R., EL-Massry M.H. Effect of pH and Temperature on Bacillus sp. R2 Chitinase A activity and Stability. Procedia Technol. 2016;22:471–477. doi: 10.1016/j.protcy.2016.01.092. [DOI] [Google Scholar]
- 22.Neelamegam A., Sadhasivam G., Muthuvel A., Thangavel B. Purification and characterization of chitinase from micrococcus sp.AG84 isolated from marine environment. Afr. J. Microbiol. Res. 2011;4:2822–2827. [Google Scholar]
- 23.Makhdoumi A., Dehghani-Joybari Z., Mashreghi M., Jamialahmadi K., Asoodeh A. A novel halo-alkali-tolerant and thermo-tolerant chitinase from Pseudoalteromonas sp. DC14 isolated from the Caspian Sea. Int. J. Environ. Sci. Technol. 2015;12:3895–3904. doi: 10.1007/s13762-015-0848-4. [DOI] [Google Scholar]
- 24.Ray L., Panda A.N., Mishra S.R., Pattanaik A.K., Adhya T.K., Suar M., Raina V. Purification and characterization of an extracellular thermo-alkali stable, metal tolerant chitinase from Streptomyces chilikensis RC1830 isolated from a brackish water lake sediment. Biotechnol. Rep. 2019;21:e00311. doi: 10.1016/j.btre.2019.e00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsujibo H., Orikoshi H., Shiotani K., Hayashi M., Umeda J., Miyamoto K., Imada C., Okami Y., Inamori Y. A novel family 19 chitinase from the marine-derived Pseudoalteromonas tunicata CCUG 44952T: Heterologous expression, characterization and antifungal activity. Biochem. Eng. J. 2015;93:84–93. [Google Scholar]
- 26.Ramli A.N., Mahadi N.M., Rabu A., Murad A.M., Bakar F.D., Illias R.M. Molecular cloning, expression and biochemical characterisation of a cold-adapted novel recombinant chitinase from Glaciozyma antarctica PI12. Microb. Cell Fact. 2011;10:94. doi: 10.1186/1475-2859-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X.H., Zhao Y., Tan H.D., Chi N.Y., Zhang Q.F., Du Y.G., Yin H. Characterisation of a chitinase from Pseudoalteromonas sp. DL-6, a marine psychrophilic bacterium. Int. J. Biol. Macromol. 2014;70:455–462. doi: 10.1016/j.ijbiomac.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 28.Meruvu H., Donthireddy S.R. Purification and characterization of an antifungal chitinase from Citrobacter freundii str. nov. haritD11. Appl. Biochem. Biotechnol. 2014;172:196–205. doi: 10.1007/s12010-013-0540-4. [DOI] [PubMed] [Google Scholar]
- 29.Stefanidi E., Vorgias C.E. Molecular analysis of the gene encoding a new chitinase from the marine psychrophilic bacterium Moritella marina and biochemical characterization of the recombinant enzyme. Extremophiles. 2008;12:541–552. doi: 10.1007/s00792-008-0155-9. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee S., Behera P.K., Madhuprakash J. Efficient conversion of crystalline chitin to N-acetylglucosamine and N,N′-diacetylchitobiose by the enzyme cocktail produced by Paenibacillus sp. LS1. Carbohydr. Polym. 2020;250:116889. doi: 10.1016/j.carbpol.2020.116889. [DOI] [PubMed] [Google Scholar]
- 31.Sashiwa H., Fujishima S., Yamano N., Kawasaki N., Nakayama A., Muraki E., Hiraga K., Oda K., Aiba S. Production of N-acetyl-D-glucosamine from alpha-chitin by crude enzymes from Aeromonas hydrophila H-2330. Carbohydr. Res. 2002;337:761–763. doi: 10.1016/S0008-6215(02)00034-4. [DOI] [PubMed] [Google Scholar]
- 32.Lv C.Y., Gu T.Y., Ma R., Yao W., Huang Y.Y., Gu J.G., Zhao G.G. Biochemical characterization of a GH19 chitinase from Streptomyces alfalfae and its applications in crystalline chitin conversion and biocontrol. Int. J. Biol. Macromol. 2021;167:193–201. doi: 10.1016/j.ijbiomac.2020.11.178. [DOI] [PubMed] [Google Scholar]
- 33.Gao C., Zhang A., Chen K., Hao Z., Tong J., Ouyang P. Characterization of extracellular chitinase from Chitinibacter sp. GC72 and its application in GlcNAc production from crayfish shell enzymatic degradation. Biochem. Eng. J. 2015;97:59–64. doi: 10.1016/j.bej.2015.02.010. [DOI] [Google Scholar]
- 34.Chen X.L., Wang Y., Wang P., Zhang Y.Z. Proteases from the marine bacteria in the genus Pseudoalteromonas: Diversity, characteristics, ecological roles, and application potentials. Mar. Life Sci. Technol. 2020;2:309–323. doi: 10.1007/s42995-020-00058-8. [DOI] [Google Scholar]
- 35.Wietz M., Gram L., Jørgensen B., Schramm A. Latitudinal Patterns in the Abundance of Major Marine Bacterioplankton Groups. Aquat. Microb. Ecol. 2010;61:179–189. doi: 10.3354/ame01443. [DOI] [Google Scholar]
- 36.Paulsen S.S., Strube M.L., Bech P.K., Gram L., Sonnenschein E.C. Marine Chitinolytic Pseudoalteromonas Represents an Untapped Reservoir of Bioactive Potential. mSystems. 2019;4:e00060-19. doi: 10.1128/mSystems.00060-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Chi N., Bai F., Du Y., Zhao Y., Yin H. Characterization of a cold-adapted and salt-tolerant exo-chitinase (ChiC) from Pseudoalteromonas sp. DL-6. Extremophiles. 2016;20:167–176. doi: 10.1007/s00792-016-0810-5. [DOI] [PubMed] [Google Scholar]
- 38.Orikoshi H., Baba N., Nakayama S., Kashu H., Miyamoto K., Yasuda M., Inamori Y., Tsujibo H. Molecular analysis of the gene encoding a novel cold-adapted chitinase (ChiB) from a marine bacterium, Alteromonas sp. strain O-7. J. Bacteriol. 2003;185:1153–1160. doi: 10.1128/JB.185.4.1153-1160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanova E.P., Shevchenko L.S., Sawabe T., Lysenko A.M., Svetashev V.I., Gorshkova N.M., Satomi M., Christen R., Mikhailov V.V. Pseudoalteromonas maricaloris sp. nov., isolated from an Australian sponge, and reclassification of [Pseudoalteromonas aurantia] NCIMB 2033 as Pseudoalteromonas flavipulchra sp. nov. Int. J. Syst. Evol. Microbiol. 2002;52:263–271. doi: 10.1099/00207713-52-1-263. [DOI] [PubMed] [Google Scholar]
- 40.Chen W., Qu M., Zhou Y., Yang Q. Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects. J. Biol. Chem. 2018;29:2652–2660. doi: 10.1074/jbc.RA117.000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pantoom S., Vetter I.R., Prinz H., Suginta W. Potent family-18 chitinase inhibitors: X-ray structures, affinities, and binding mechanisms. J. Biol. Chem. 2011;286:24312–24323. doi: 10.1074/jbc.M110.183376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houston D.R., Shiomi K., Arai N., Omura S., Peter M.G., Turberg A., Synstad B., Eijsink V.G., van Aalten D.M. High-resolution structures of a chitinase complexed with natural product cyclopentapeptide inhibitors: Mimicry of carbohydrate substrate. Proc. Natl. Acad. Sci. USA. 2002;99:9127–9132. doi: 10.1073/pnas.132060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun X., Li Y., Tian Z., Qian Y., Zhang H., Wang L. A novel thermostable chitinolytic machinery of Streptomyces sp. F-3 consisting of chitinases with different action modes. Biotechnol. Biofuels. 2019;12:136. doi: 10.1186/s13068-019-1472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki K., Taiyoji M., Sugawara N., Nikaidou N., Henrissat B., Watanabe T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 1999;343:587–596. doi: 10.1042/bj3430587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu T., Zhu W., Wang J., Zhou Y., Duan Y., Qu M., Yang Q. The deduced role of a chitinase containing two nonsynergistic catalytic domains. Acta Crystallogr. D Struct. Biol. 2018;74:30–40. doi: 10.1107/S2059798317018289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai Y., Eijsink V.G., Kielak A.M., van Veen J.A., de Boer W. Genomic comparison of chitinolytic enzyme systems from terrestrial and aquatic bacteria. Environ. Microbiol. 2016;18:38–49. doi: 10.1111/1462-2920.12545. [DOI] [PubMed] [Google Scholar]
- 47.Orlando M., Pucciarelli S., Lotti M. Endolysins from Antarctic Pseudomonas Display Lysozyme Activity at Low Temperature. Mar. Drugs. 2020;18:579. doi: 10.3390/md18110579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe T., Ito Y., Yamada T., Hashimoto M., Sekine S., Tanaka H. The roles of the C-terminal domain and type-Iii domains of chitinase A1 from Bacillus Circulans Wl-12 in chitin degradation. J. Bacteriol. 1994;176:4465–4472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svitil A.L., Kirchman D.L. A chitin-binding domain in a marine bacterial chitinase and other microbial chitinases: Implications for the ecology and evolution of 1,4-beta-glycanases. Microbiology. 1998;144:1299–1308. doi: 10.1099/00221287-144-5-1299. [DOI] [PubMed] [Google Scholar]
- 50.Eijsink V.G.H., Vaaje-Kolstad G., Varum K.M., Horn S.J. Towards new enzymes for biofuels: Lessons from chitinase research. Trends. Biotechnol. 2008;26:228–235. doi: 10.1016/j.tibtech.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Nimlos M.R., Beckham G.T., Matthews J.F., Bu L.T., Himmel M.E., Crowley M.F. Binding preferences, surface attachment, diffusivity, and orientation of a family 1 carbohydrate-binding module on cellulose. J. Biol. Chem. 2012;287:20603–20612. doi: 10.1074/jbc.M112.358184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X., Isbrandt T., Strube M.L., Paulsen S.S., Nielsen M.W., Buijs Y., Schoof E.M., Larsen T.O., Gram L., Zhang S.D. Chitin Degradation Machinery and Secondary Metabolite Profiles in the Marine Bacterium Pseudoalteromonas rubra S4059. Mar. Drugs. 2021;19:108. doi: 10.3390/md19020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monge E.C., Tuveng T.R., Vaaje-Kolstad G., Eijsink V.G.H., Gardner J.G. Systems analysis of the glycoside hydrolase family 18 enzymes from Cellvibrio japonicus characterizes essential chitin degradation functions. J. Biol. Chem. 2018;293:3849–3859. doi: 10.1074/jbc.RA117.000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meibom K.L., Li X.B., Nielsen A.T., Wu C.Y., Roseman S., Schoolnik G.K. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Brien M., Colwell R.R. A rapid test for chitinase activity that uses 4-methylumbelliferyl-N-acetyl-beta-D-glucosaminide. Appl. Environ. Microbiol. 1987;53:1718–1720. doi: 10.1128/aem.53.7.1718-1720.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howard M.B., Ekborg. N.A., Taylor L.E., Weiner R.M., Hutcheson S.W. Genomic analysis and initial characterization of the chitinolytic system of Microbulbifer degradans strain 2-40. J. Bacteriol. 2003;185:3352–3360. doi: 10.1128/JB.185.11.3352-3360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reindl M., Stock J., Hussnaetter K.P., Genc A., Brachmann A., Schipper K. A Novel Factor Essential for Unconventional Secretion of Chitinase Cts1. Front. Microbiol. 2020;11:1529. doi: 10.3389/fmicb.2020.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H.J., Lee Y.S., Choi Y.L. Cloning, purification, and characterization of an organic solvent-tolerant chitinase, MtCh509, from Microbulbifer thermotolerans DAU221. Biotechnol. Biofuels. 2018;11:303. doi: 10.1186/s13068-018-1299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan M., Li J., Lv X., Du G., Liu L. Molecular engineering of chitinase from Bacillus sp. DAU101 for enzymatic production of chitooligosaccharides. Enzyme. Microb. Technol. 2019;124:54–62. doi: 10.1016/j.enzmictec.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 60.Le B., Yang S.H. Characterization of a chitinase from Salinivibrio sp. BAO-1801 as an antifungal activity and a biocatalyst for producing chitobiose. J. Basic Microbiol. 2018;58:848–856. doi: 10.1002/jobm.201800256. [DOI] [PubMed] [Google Scholar]
- 61.Hosny A., El-Shayeb N.A., Abood A., Abdel-Fattah A.M. A Potent Chitinolytic Activity of Marine Actinomycete sp. and Enzymatic Production of Chitooligosaccharides. Aust. J. Basic Appl. Sci. 2010;4:615–623. [Google Scholar]
- 62.Lv C., Gu T., Xu K., Gu J., Li L., Liu X., Zhang A., Gao S., Li W., Zhao G. Biochemical characterization of a β-N-acetylhexosaminidase from Streptomyces alfalfae and its application in the production of N-acetyl-D-glucosamine. J. Biosci. Bioeng. 2019;128:135–141. doi: 10.1016/j.jbiosc.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 63.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 64.Xie B.B., Rong J.C., Tang B.L., Wang S., Liu G., Qin Q.L., Zhang X.Y., Zhang W., She Q., Chen Y., et al. Evolutionary Trajectory of the Replication Mode of Bacterial Replicons. mBio. 2021;26:e02745-20. doi: 10.1128/mBio.02745-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 67.Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 71.Deutsch E.W., Bandeira N., Sharma V., Perez-Riverol Y., Carver J.J., Kundu D.J., García-Seisdedos D., Jarnuczak A.F., Hewapathirana S., Pullman B.S., et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020;48:D1145–D1152. doi: 10.1093/nar/gkz984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 74.Romanenko L.A., Zhukova N.V., Rohde M., Lysenko A.M., Mikhailov V.V., Stackebrandt E. Pseudoalteromonas agarivorans sp. nov., a novel marine agarolytic bacterium. Int. J. Syst. Evol. Microbiol. 2003;53:125–131. doi: 10.1099/ijs.0.02234-0. [DOI] [PubMed] [Google Scholar]
- 75.Ivanova E.P., Gorshkova N.M., Zhukova N.V., Lysenko A.M., Zelepuga E.A., Prokof’eva N.G., Mikhailov V.V., Nicolau D.V., Christen R. Characterization of Pseudoalteromonas distincta-like sea-water isolates and description of Pseudoalteromonas aliena sp. nov. Int. J. Syst. Evol. Microbiol. 2004;54:1431–1437. doi: 10.1099/ijs.0.03053-0. [DOI] [PubMed] [Google Scholar]
- 76.Al Khudary R., Stosser N.I., Qoura F., Antranikian G. Pseudoalteromonas arctica sp. nov., an aerobic, psychrotolerant,marine bacterium isolated from Spitzbergen. Int. J. Syst. Evol. Microbiol. 2008;58:2018–2024. doi: 10.1099/ijs.0.64963-0. [DOI] [PubMed] [Google Scholar]
- 77.Gauthier M., Breittmayer V.A. A new antibiotic-producing bacterium from seawater: Alteromonas aurantia sp. nov. Int. J. Syst. Evol. Microbiol. 1979;29:366–372. doi: 10.1099/00207713-29-4-366. [DOI] [Google Scholar]
- 78.Akagawa-Matsushita M., Matsuo M., Koga Y., Yamasato K. Alteromonas atlantica sp. nov. and Alteromonas carrageenovora sp. nov., bacteria that decompose algal polysaccharides. Int. J. Syst. Evol. Microbiol. 1993;42:621–627. doi: 10.1099/00207713-43-2-400. [DOI] [Google Scholar]
- 79.Gauthier M. Alteromonas citrea, a new Gram-negative, yellow-pigmented species from seawater. Int. J. Syst. Evol. Microbiol. 1977;27:349–354. doi: 10.1099/00207713-27-4-349. [DOI] [Google Scholar]
- 80.Chan K., Baumann L., Garza M., Baumann P. Two new species of Alteromonas: Alteromonas espejiana and Alteromonas undina. Int. J. Syst. Evol. Microbiol. 1978;28:217–222. doi: 10.1099/00207713-28-2-217. [DOI] [Google Scholar]
- 81.Ivanova E.P., Sawabe T., Alexeeva Y.V., Lysenko A.M., Gorshkova N.M., Hayashi K., Zukova N.V., Christen R., Mikhailov V.V. Pseudoalteromonas issachenkonii sp. nov., a bacterium that degrades the thallus of the brown alga Fucus evanescens. Int. J. Syst. Evol. Microbiol. 2002;52:229–234. doi: 10.1099/00207713-52-1-229. [DOI] [PubMed] [Google Scholar]
- 82.Xu X.W., Wu Y.H., Wang C.S., Gao X.H., Wang X.G., Wu M. Pseudoalteromonas lipolytica sp. nov., isolated from the Yangtze River estuary. Int. J. Syst. Evol. Microbiol. 2009;60:2176–2181. doi: 10.1099/ijs.0.017673-0. [DOI] [PubMed] [Google Scholar]
- 83.Gauthier M. Validation of the name Alteromonas luteoviolacea. Int. J. Syst. Evol. Microbiol. 1982;32:82–86. doi: 10.1099/00207713-32-1-82. [DOI] [Google Scholar]
- 84.Yoon J.H., Kim I.G., Kang K.H., Oh T.K., Park Y.H. Alteromonas marina sp. nov., isolated from sea water of the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 2003;53:1625–1630. doi: 10.1099/ijs.0.02536-0. [DOI] [PubMed] [Google Scholar]
- 85.Romanenko L.A., Zhukova N.V., Lysenko A.M., Mikhailov V.V., Stackebrandt E. Assignment of ‘Alteromonas marinoglutinosa’NCIMB 1770 to Pseudoalteromonas mariniglutinosa sp. nov., nom. rev., comb. nov. Int. J. Syst. Evol. Microbiol. 2003;53:1105–1109. doi: 10.1099/ijs.0.02564-0. [DOI] [PubMed] [Google Scholar]
- 86.Ivanova E.P., Kiprianova E.A., Mikhailov V.V., Levanova G.F., Garagulya A.D., Gorshkova N.M., Yumoto N. Characterization and identification of marine Alteromonas nigrifaciens strains and emendation of the description. Int. J. Syst. Evol. Microbiol. 1996;46:223–228. doi: 10.1099/00207713-46-1-223. [DOI] [Google Scholar]
- 87.Ivanova E.P., Sawabe T., Lysenko A.M., Gorshkova N.M., Hayashi K., Zhukova N.V., Nicolau D.V., Christen R., Mikhailov V.V. Pseudoalteromonas translucida sp. nov. and Pseudoalteromonas paragorgicola sp. nov., and emended description of the genus. Int. J. Syst. Evol. Microbiol. 2002;52:1759–1766. doi: 10.1099/00207713-52-5-1759. [DOI] [PubMed] [Google Scholar]
- 88.Venkateswaran K., Dohmoto N. Pseudoalteromonas peptidolytica sp. nov., a novel marine mussel-thread-degrading bacterium isolated from the Sea of Japan. Int. J. Syst. Evol. Microbiol. 2000;50:565–574. doi: 10.1099/00207713-50-2-565. [DOI] [PubMed] [Google Scholar]
- 89.Isnansetyo A., Kamei Y. Pseudoalteromonas phenolica sp. nov., a novel marine bacterium that produces phenolic anti-methicillin-resistant Staphylococcus aureus substances. Int. J. Syst. Evol. Microbiol. 2003;53:583–588. doi: 10.1099/ijs.0.02431-0. [DOI] [PubMed] [Google Scholar]
- 90.Hansen A., Weeks O., Colwell R. Taxonomy of Pseudomonas piscicida (Bein) Buck, Meyers, and Leifson. J. Bacteriol. 1965;89:752–761. doi: 10.1128/jb.89.3.752-761.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bowman J.P. Pseudoalteromonas prydzensis sp. nov., a psychrotrophic, halotolerant bacterium from Antarctic sea ice. Int. J. Syst. Evol. Microbiol. 1998;48:1037–1041. doi: 10.1099/00207713-48-3-1037. [DOI] [PubMed] [Google Scholar]
- 92.Gauthier M. Alteromonas rubra sp. nov., a new marine antibiotic-producing bacterium. Int. J. Syst. Evol. Microbiol. 1976;26:459–466. doi: 10.1099/00207713-26-4-459. [DOI] [Google Scholar]
- 93.Lau S.C., Tsoi M.M., Li X., Dobretsov S., Plakhotnikova Y., Wong P.-K., Qian P.Y. Pseudoalteromonas spongiae sp. nov., a novel member of the γ-Proteobacteria isolated from the sponge Mycale adhaerens in Hong Kong waters. Int. J. Syst. Evol. Microbiol. 2005;55:1593–1596. doi: 10.1099/ijs.0.63638-0. [DOI] [PubMed] [Google Scholar]
- 94.Ivanova E.P., Romanenko L.A., Matte M.H., Matte G.R., Lysenko A.M., Simidu U., Kita-Tsukamoto K., Sawabe T., Vysotskii M.V., Frolova G.M., et al. Retrieval of the species Alteromonas tetraodonis Simidu et al. 1990 as Pseudoalteromonas tetraodonis comb. nov. and emendation of description. Int. J. Syst. Evol. Microbiol. 2001;51:1071–1078. doi: 10.1099/00207713-51-3-1071. [DOI] [PubMed] [Google Scholar]
- 95.Holmström C., James S., Neilan B.A., White D.C., Kjelleberg S. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Evol. Microbiol. 1998;48:1205–1212. doi: 10.1099/00207713-48-4-1205. [DOI] [PubMed] [Google Scholar]
- 96.Egan S., Holmström C., Kjelleberg S. Pseudoalteromonas ulvae sp. nov., a bacterium with antifouling activities isolated from the surface of a marine alga. Int. J. Syst. Evol. Microbiol. 2001;51:1499–1504. doi: 10.1099/00207713-51-4-1499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data are available via ProteomeXchange with identifier PXD030600.