Abstract
Erythromycin resistance rates were found to be increased, from 7.1 in 1993 to 32.8% in 1997, among community-acquired Streptococcus pneumoniae isolates from the Siena area of central Italy. Most of the erythromycin-resistant isolates carried ermAM determinants and were also resistant to josamycin and clindamycin, whereas a minority (5.8%) carried mefA determinants and remained susceptible to the latter drugs.
Macrolide and lincosamide antibiotics exhibit strong antimicrobial activity against streptococci and are among the drugs that can be used for chemotherapy of infections caused by Streptococcus pneumoniae. Macrolide resistance in pneumococci has been detected at variable rates in different epidemiological settings (see, for instance, references 1, 2, 4, 7, 15, 18, and 22), with a trend toward increasing resistance being reported by several investigators (4, 6, 9, 21). This is a matter of major concern, since macrolides are largely prescribed for the empiric chemotherapy of community-acquired respiratory tract infections and may be useful in case of intolerance to β-lactams as well as pneumococcal resistance to other antimicrobial agents.
In S. pneumoniae, macrolide resistance can be mediated by ribosomal modification (8) or active drug efflux (16). The former mechanism is associated with high-level resistance to macrolides, lincosamides, and streptogramin B (MLSB-type resistance pattern) (8), while the latter mechanism is associated with low-level resistance to 14- and 15-membered-ring macrolides only (M-type resistance pattern) (16, 19). Both resistance effectors are encoded by acquired determinants: the ermAM gene encoding the ribosome-modifying enzyme (8, 20) and the mefE gene for the efflux system (19). The contributions of these mechanisms to macrolide resistance in pneumococci appears to be variable in different epidemiological settings (1, 2, 7, 15, 18, 22), although a molecular analysis of the resistance determinants was carried out only in a minority of cases (7, 22).
In the work described here we determined the macrolide and lincosamide susceptibilities of 302 S. pneumoniae isolates isolated from an area of central Italy over a 5-year period (1993 to 1997) and investigated the resistance determinants carried by macrolide-resistant isolates.
Pneumococci were randomly selected from among those that were classified as community acquired and that were cultured at the Laboratory of Clinical Bacteriology of the Institute of Infectious Diseases, University of Siena, during the period 1 January 1993 to 31 December 1997 from samples from patients who were residents of the Siena area. Repeated isolates from the same individual were not included unless the isolations were separated by a period of at least 1 year. Identification of pneumococcal isolates was performed by standard procedures (12). In vitro susceptibility testing was performed by a broth microdilution method as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (10). Erythromycin and clindamycin were from Sigma Chemical Co. (St. Louis, Mo.); josamycin was from ICN (Costa Mesa, Calif.). The breakpoints for susceptibility classification were those specified by NCCLS (11). S. pneumoniae ATCC 49619 was used for quality control of susceptibility testing.
Of the 302 pneumococcal isolates, 233 (77.2%) were found to be susceptible to erythromycin (MICs, ≤0.015 to 0.06 μg/ml), josamycin (MICs, 0.06 to 0.25 μg/ml), and clindamycin (MICs, ≤0.06 to 0.25 μg/ml), while the remaining 69 (22.8%) were resistant to erythromycin. Of these, 65 (94.2%) exhibited high-level resistance to erythromycin (MICs, 128 to >1,024 μg/ml) and also to josamycin (MICs, 16 to >1,024 μg/ml) and clindamycin (MICs, 512 to >1,024 μg/ml), which is typical of a constitutive MLSB-type resistance pattern, while the remaining 4 (5.8%) exhibited low-level resistance (MICs, 1 to 8 μg/ml) to erythromycin only, which is typical of an M-type resistance pattern. A double-disk diffusion assay (8) confirmed that susceptibility to josamycin and clindamycin was not influenced by the presence of erythromycin in the isolates with M-type resistance.
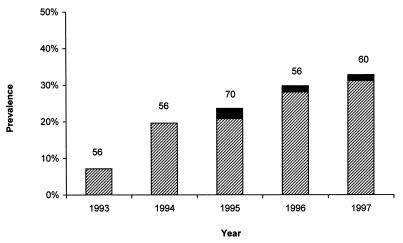
The prevalence of erythromycin-resistant isolates showed a steady increase during the study period, from 7.1% in 1993 to 32.8% in 1997 (Fig. 1).
FIG. 1.
Prevalences of MLSB-type resistance (▨) and M-type resistance (■) among S. pneumoniae isolates in different years. The numbers reported at the top of each column indicate the total number of isolates analyzed in that year.
The occurrence of ermAM- and mef-related genomic sequences was investigated in all the erythromycin-resistant isolates and in 54 (23.2%) randomly selected susceptible isolates by means of colony blot hybridization with 32P-labeled DNA probes. Colony blot hybridization was performed as described previously (13) after an initial exposure (twice for 10 min at room temperature) of pneumococcal colonies to 1% (wt/vol) Na-Sarkosyl–1% (wt/vol) sodium deoxycholate–1.5 M NaCl. With Streptococcus pyogenes and Streptococcus mitis, colony blotting was performed as described previously (5). The ermAM-specific probe was a 764-bp PCR amplicon that contains the entire ermAM-coding sequence (20). The mef-specific probes were 346-bp PCR amplicons which contained part of mefA or mefE, and the amplicons were obtained as described previously (17). S. pneumoniae 4C1 (ermAM+), S. pyogenes 1A77 (mefA+), and S. mitis 21A29 (mefE+) were included as hybridization controls.
The ermAM probe yielded a strong hybridization signal for all isolates with MLSB-type resistance, while it did not recognize any of the susceptible isolates or isolates with M-type resistance. The intensity of the hybridization signal was overall comparable to that obtained with a positive hybridization control for ermAM (data not shown). The presence of mef-related sequences was initially investigated with a mefE probe. This probe yielded a hybridization signal for the four isolates with M-type resistance, while it did not recognize any of the susceptible isolates or isolates with MLSB-type resistance. The hybridization signal, however, was weaker than that obtained with a positive hybridization control for mefE. By using a mefA probe, a stronger hybridization signal was obtained for the four isolates with M-type resistance, and the signal was comparable to that obtained with a positive hybridization control for mefA and stronger than that obtained with a positive hybridization control for mefE (data not shown).
To verify the identities of the mef alleles carried by the four isolates with M-type resistance, PCR was carried out with primers MEFA-up (5′-GACCAAAAGCCACATTGTGGA) and MEFA-dn (5′-CCTCCTGTCTATAATCGCATG), which were designed on the basis of sequences that flank the mefA gene from S. pyogenes 02C1064 (3), by using the following cycling conditions: 94°C for 30 s, 58°C for 60 s, and 70°C for 80 s, which were repeated for 40 cycles. In all cases an amplimer of the expected size (1,431 bp) was obtained. Restriction analysis of the amplimers with AvaII, BamHI, ClaI, HindIII, and NheI yielded in all cases a profile consistent with the sequence of mefA (with AvaII, products of 283, 419, and 729 bp; with BamHI, products of 223 and 1,208 bp; with ClaI, products of 620 and 811 bp; with HindIII, products of 437 and 994 bp; with NheI, a product of 1,431 bp) (3) but not with that of mefE (with AvaII, products of 283, 372, and 776 bp; with BamHI, a product of 1,431 bp; with ClaI, a product of 1,431 bp; with HindIII, a product of 1,431 bp; with NheI, products of 525 and 906 bp) (19).
Concluding remarks.
Similar to what has been observed elsewhere (4, 6, 9, 21), the prevalence of macrolide-resistant pneumococci has also recently undergone a substantial increase in the Siena area of central Italy, with macrolide-resistant pneumococci making up nearly one-third of the community-acquired isolates in 1997. A similar situation is probably consequent to an increased selective pressure generated by increased prescriptions of macrolides in community medicine, as has previously been demonstrated for S. pyogenes (14).
The results of this study revealed that, in our region, the most prevalent macrolide resistance phenotype among pneumococcal isolates is the MLSB type and that the relevant increase in the number of macrolide-resistant pneumococci observed during the study period was virtually completely a result of isolates that carry ermAM determinants. Isolates that have M-type resistance and that carry mef determinants have appeared only since 1995 and remain uncommon. A similar pattern of macrolide resistance determinants in pneumococci has also been observed in Spain (1), although it is different from those encountered in other countries where lower relative rates of isolates with MLSB-type resistance have been observed (7, 22). The relative prevalence of the two macrolide resistance mechanisms in pneumococci therefore exhibits remarkable geographical heterogeneity. This essentially results from large variations in the absolute rates of occurrence of isolates with MLSB-type resistance, whereas the prevalence of pneumococci with M-type resistance showed an overall low variability (0.45 to 1.6%) in different epidemiological settings (1, 7, 22; this study), suggesting that the erm determinants play a major role in the geographical and temporal variability in macrolide resistance rates among pneumococci. Molecular analysis also showed that all the pneumococci with M-type resistance encountered in this study apparently carried a mefA determinant, revealing that not only mefE but also mefA can be acquired by S. pneumoniae, resulting in an M-type resistance phenotype.
Acknowledgments
This work was partially supported by grant “Ricerca Scientifica-Quota 60%” from the University of Siena.
P.O., A.Z. and S.C. contributed equally to this work.
REFERENCES
- 1.Baquero F, García-Rodríguez J A, García de Lomas J, Aguilar L The Spanish Surveillance Group for Respiratory Pathogens. Antimicrobial resistance of 1,113 Streptococcus pneumoniae isolates from patients with respiratory tract infections in Spain: results of a 1-year (1996–1997) multicenter surveillance study. Antimicrob Agents Chemother. 1999;43:357–359. doi: 10.1128/aac.43.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breiman R F, Butler J C, Tenover F C, Eliott J A, Facklam R R. Emergence of drug-resistant pneumococcal infections in the United States. JAMA. 1994;271:1831–1835. [PubMed] [Google Scholar]
- 3.Clancy J, Petitpas J, Dib-Hajj F, Yaun W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 4.Geslin P, Buu-Hoi A, Frémaux A, Acar J F. Antimicrobial resistance in Streptococcus pneumoniae: an epidemiological survey in France, 1970–1990. Clin Infect Dis. 1992;15:95–98. doi: 10.1093/clinids/15.1.95. [DOI] [PubMed] [Google Scholar]
- 5.Huang T T, Malke H, Ferretti J J. Heterogeneity of the streptokinase gene in group A streptococci. Infect Immun. 1989;57:502–506. doi: 10.1128/iai.57.2.502-506.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson A P, Speller D C E, George R C, Warner M, Domingue G, Efstratiou A. Prevalence of antibiotic resistance and serotypes in pneumococci in England and Wales: results of observational surveys in 1990 and 1995. Br Med J. 1996;312:1454–1456. doi: 10.1136/bmj.312.7044.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston N J, De Azavedo J C, Kellner J D, Low D E. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2425–2426. doi: 10.1128/aac.42.9.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lonks J R, Medeiros A A. Emergence of erythromycin-resistant Streptococcus pneumoniae. Infect Med. 1994;11:415–418. [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 12.Rouff K L. Streptococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of Clinical Microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 299–307. [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Seppälä H, Klaukka T, Lehtonen R, Nenonen E, Huovinen P. Outpatient use of erythromycin: link to increased erythromycin resistance in group A streptococci. Clin Infect Dis. 1995;21:1378–1385. doi: 10.1093/clinids/21.6.1378. [DOI] [PubMed] [Google Scholar]
- 15.Sessegolo J F, Levin A S S, Levy C E, Asensi M, Facklam R R, Teixeira L M. Distribution of serotypes and antimicrobial resistance of Streptococcus pneumoniae strains isolated in Brazil from 1988 to 1992. J Clin Microbiol. 1994;32:906–911. doi: 10.1128/jcm.32.4.906-911.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. 1996;40:2562–2566. doi: 10.1128/aac.40.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syrogiannopoulos G A, Grivea I N, Beratis N G, Spiliopoulou A E, Fasola E L, Bajaksouzian S, Appelbaum P C, Jacobs M R. Resistance patterns of Streptococcus pneumoniae from carriers attending day-care centers in southwestern Greece. Clin Infect Dis. 1997;25:188–194. doi: 10.1086/514526. [DOI] [PubMed] [Google Scholar]
- 19.Tait-Kamradt A, Clancy A, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for erithromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trieu-Cout P, Poyart-Salmeron C, Carlier C, Courvalin P. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 1990;18:3660. doi: 10.1093/nar/18.12.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhaegen J, Glupczynski Y, Verbist L, Blogie M, Verbiest N, Vandeven J, Yourassowsky E. Capsular types and antibiotic susceptibility of pneumococci isolates from patients in Belgium with serious infections, 1980–1993. Clin Infect Dis. 1995;20:1339–1345. doi: 10.1093/clinids/20.5.1339. [DOI] [PubMed] [Google Scholar]
- 22.Widdowson C A, Klugman K P. Emergence of the M phenotype of erythromycin-resistant pneumococci in South Africa. Emerg Infect Dis. 1998;4:277–281. doi: 10.3201/eid0402.980216. [DOI] [PMC free article] [PubMed] [Google Scholar]