Abstract
Nine ciprofloxacin-resistant viridans group streptococci isolated from asymptomatic carriers were analyzed. Identification to the species level by using three different commercial systems and a PCR-based approach was inconsistent. The nucleotide sequences of fragments of the parC, parE, gyrA, and gyrB genes showed considerable intra- and interspecies variations, and these variations mainly involved silent mutations. Three isolates had changes in Ser-79 of ParC (to Phe or Tyr). Phenotypic characterization indicated that eight of the nine isolates had a putative efflux mechanism that would confer low-level resistance to ciprofloxacin.
Although viridans group streptococci (VGS) form part of the normal flora of the human oral cavity, they cause infective endocarditis (9, 25, 27) and are a major cause of bacteremia in neutropenic cancer patients (4, 6, 8, 10). Increasing levels of resistance to penicillin and macrolide antibiotics have been observed in these bacteria (2, 3, 6, 8). Fluoroquinolone (Fq) resistance (Fqr) has been reported in VGS isolates from the blood of neutropenic cancer patients who received quinolone prophylaxis (14, 29). In general, there are two major mechanisms of Fqr including alterations in DNA topoisomerase IV (topo IV) and DNA gyrase (gyrase) and reduced levels of drug accumulation as a result of enhanced efflux. Efflux mechanisms as a cause of low-level Fqr have been described in Streptococcus pneumoniae (5, 7, 31) and other gram-positive bacteria (1, 16, 20–23, 30). In a recent study, the pmrA gene has been identified as a gene that codes for a pneumococcal Fq efflux pump (13). Genetic disruption of the transporter genes or inhibition of the transporter activity by the alkaloid reserpine decreased the MICs of Fqs and other toxins for those gram-positive bacteria (1, 13, 18, 21).
Fluoroquinolone susceptibility and classification of VGS isolates.
The VGS strains were isolated from asymptomatic patients during 1998 at the Hospital of Móstoles and were identified by standard methods (11, 26). The strains were first screened for decreased ciprofloxacin (Cp) susceptibility by disk diffusion (5 μg/disk), and those that yielded inhibition zone diameters of <12 mm were interpreted as resistant. By these criteria, a sample of 50 Cp-resistant (Cpr) VGS isolates was selected. These isolates were preliminarily classified at the species level by using three different standard phenotypic systems: API 20-Strep and ID 32-Strep (Biomerieux, Marcy L’Etoile, France) and BBL Crystal (Becton-Dickinson Europe, Meylan, France). The API 20-Strep and ID 32-Strep systems identified 48 and 49 of the 50 strains, respectively, while the BBL Crystal system identified only 34 of the 50 strains. The three methods allowed the coincidental identification of only 3 of the 50 isolates analyzed, while the two systems from the same manufacturer showed concordance for the identification of about half of the isolates (26 strains).
Further characterization of Fq susceptibility among the 50 Cpr VGS isolates was performed by E-test analysis on Mueller-Hinton agar plates (Difco) supplemented with 5% defibrinated sheep blood. This analysis revealed that nine strains had high-level Cpr (≥16 μg/ml) (Table 1). Their susceptibilities to Cp, sparfloxacin, and clinafloxacin were determined by the agar dilution method as described previously (14). Among the nine strains, for seven strains the Cp MICs by the agar dilution method were 4 μg/ml (a value fourfold lower than that observed by the E test), and the seven strains were then classified as having low-level Cpr. However, two strains had high-level Cpr by both the E-test and the agar dilution methods. Similar differences between the E-test and the agar dilution methods were observed when the susceptibilities to sparfloxacin were considered (Table 1).
TABLE 1.
Susceptibilities of strains to selected Fqs and mutations in the parC, parE, gyrA, and gyrB genesa
| Strain | MIC (μg/ml)
|
Amino acid change (codon change)b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cp
|
SPA
|
CLX, agar dilution | |||||||
| E test | Agar dilution | E test | Agar dilution | ParCc | ParE | GyrAd | GyrB | ||
| S. pneumoniae R6 | 0.5 | 0.5 | 0.25 | 0.25 | 0.125 | — | — | — | — |
| S. oralis NCTC11427 | 4 | 2 | 0.5 | 0.5 | 0.125 | — | — | — | ND |
| S. oralis ATCC10557 | 2 | 2 | 0.5 | 0.5 | 0.125 | — | — | — | ND |
| S. mitis NCTC12261 | 1 | 1 | 0.25 | 0.25 | 0.06 | — | — | ND | — |
| S. mitis V1e | 16 | 4 | 1 | 0.25 | 0.125 | — | — | ND | ND |
| S. mitis V2 | 16 | 4 | 1 | 0.5 | 0.125 | — | — | ND | ND |
| S. mitis V3 | 16 | 4 | 1 | 0.25 | 0.125 | — | — | ND | ND |
| S. mitis V4 | 16 | 4 | 1 | 0.5 | 0.125 | — | — | ND | ND |
| S. sanguis V5 | 16 | 4 | 1 | 0.5 | 0.125 | — | — | ND | 425A→G (GCT→GGT) |
| S. mitis V6 | 16 | 4 | 2 | 0.5 | 0.125 | — | — | — | 425A→G (GCT→GGT) |
| S. mitis V10 | 16 | 4 | 1 | 0.5 | 0.125 | 79S→F (TCT→TTT) | — | — | ND |
| S. mitis V8 | >32 | 16 | 2 | 1 | 0.25 | 79S→F (TCT→TTT) | — | — | — |
| S. oralis V9 | >32 | 32 | 2 | 1 | 0.5 | 79S→Y (TCT→TAT) | — | — | — |
SPA, sparfloxacin; CLX, clinafloxacin; ND, not determined.
Positions of substitutions are according to the coordinates for S. pneumoniae R6. —, no change.
The sequenced strains had an additional 91N→D (AAC→GAC) change compared to the sequence of S. pneumoniae R6.
The sequenced strains had an additional 114S→G (AGT→GGT) change compared to the sequence of S. pneumoniae R6.
Classification of the VGS clinical isolates to the species level was done with the API 20-Strep and ID 32-Strep systems.
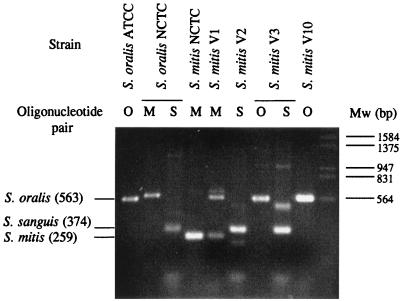
Although these nine isolates were coincidentally identified by both the API 20-Strep and the ID 32-Strep systems (Table 1), they were also subjected to an additional identification by the PCR method described by Garnier et al. (12). Three pairs of specific oligonucleotides, each one being specific for one species, were used in PCRs with DNA from each isolate obtained as described previously (14). This method allowed the identification of Streptococcus mitis NCTC 12261 and Streptococcus oralis ATCC 10557. However, S. oralis NCTC 11427 was amplified with the pairs of primers specific for both S. mitis and Streptococcus sanguis, with the size of the PCR fragment obtained with the S. mitis pair being larger than expected (Fig. 1). Among the nine clinical isolates, only four, phenotypically identified as S. mitis, were amplified with any pair of oligonucleotides. By this PCR method, strains V2 and V10 were identified as S. sanguis and S. oralis, respectively. Isolate V3 was amplified with the S. oralis-specific primers, but it also showed two bands with the S. sanguis-specific pair of primers. Likewise, isolate V1 also showed two PCR products with the S. mitis-specific pair of primers; one of the products corresponded to S. mitis and the other corresponded to S. oralis.
FIG. 1.
Identification of Cpr VGS isolates by the PCR method described by Garnier et al. (12). The DNAs of S. oralis ATCC 10557, S. oralis NCTC 11427, S. mitis NCTC 12261, and clinical isolates (S. mitis V1, V2, V3, and V10) were amplified with the oligonucleotide pairs indicated. O, M, and S, the S. oralis-, S. mitis-, and S. sanguis-specific pairs, respectively; Mw, bacteriophage λ DNA digested with EcoRI and HindIII. PCR products were resolved by electrophoresis on a 1.5% agarose-Tris-acetate-EDTA gel that was stained with a 0.5-μg/ml ethidium bromide solution. The sizes (in base pairs) of the PCR products expected for the different species are indicated on the left.
Sequencing of the parC, parE, gyrA, and gyrB QRDRs.
Because topo IV is a primary target for Cp in VGS, the quinolone resistance-determining regions (QRDRs) of the parC and parE genes were amplified with pneumococcus-specific oligonucleotides and the sequences of both DNA strands were determined as described previously (14). The differences in the 185-nucleotide (nt) parC sequences among the VGS (excluding those mutations involved in Cpr) were 1.1 to 13.0%, and those in the 210-nt parE sequences were 1.1 to 15.7%. Likewise, the differences in the VGS nt sequences compared with the sequence of S. pneumoniae R6 were 4.9 to 10.8% for parC and 4.3 to 14.6% for parE. Comparisons of the amino acid sequences of the VGS with that of S. pneumoniae R6 showed a single change in residue 91 of ParC: Asn in R6 and Asp in the VGS. Point mutations that affect Ser-79 of parC (change to Tyr or Phe) were found in the two high-level-Cpr strains (strains V8 and V9) and also in the low-level-Cpr strain (strain V10) (Table 1). This residue position has been found to be involved in Cpr in pneumococci (19) as well as in VGS (14).
The sequences of the gyrA and gyrB QRDRs were also determined for the two high-level-Cpr isolates and a low-level-Cpr isolate (isolate V6). Additionally, the gyrB QRDR sequence of isolate V5 was also determined. The differences in a 280-nt fragment of gyrA were 2.1 to 12.6% (among the VGS) and 4.6 to 12.5% (compared with the sequence of S. pneumoniae R6). Similar differences were found for a 311-nt fragment of gyrB: 1.6 to 11.2% (among the VGS) and 2.2 to 7.1% (compared with the sequence of S. pneumoniae R6). A single change at residue 114 of GyrA was found: Ser in R6 and Gly in the VGS. The change observed in GyrB was Ala-425-Gly in isolates V5 and V6. The significance of this change will be discussed below.
The high rate of variation observed between the nt sequences of identical genes from VGS (type strains and clinical isolates) is an indication of the poor classification of the group, as suggested by several investigators (17, 24, 28), and possibly reflects an interchange of genetic material between these bacteria. Thus, sequence comparisons cannot be used to classify VGS to the species level.
Characterization of efflux phenotype of Fq resistance.
The susceptibilities of the strains to two hydrophilic fluoroquinolones in the presence or absence of reserpine and known efflux pump substrates were determined by the validated agar dilution method of Brenwald et al. (7). The results are shown in Table 2. For these studies, S. pneumoniae R6 was used as a control strain since no differences in the Cp or norfloxacin MICs in the presence or absence of reserpine were found. However, for the VGS type strains, twofold reductions in the Cp MICs and fourfold reductions in the norfloxacin MICs were found in the presence of reserpine. For eight of the nine Cpr VGS isolates, fourfold or greater reductions in the Cp MICs and eightfold or greater reductions in the norfloxacin MICs were found in the presence of reserpine. In contrast, no change in the Cp MIC and a twofold reduction in the norfloxacin MIC were found for isolate V10 in the presence of reserpine.
TABLE 2.
Susceptibilities of strains to Fqs and efflux pump substrates
| Strain | MIC (μg/ml)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cp | Cp + RS | CP/CIP + RS | NOR | NOR + RS | NOR/NOR + RS | EBR | EBR + RS | EBR/EBR + RS | ACR | ACR + RS | ACR/ACR + RS | |
| S. pneumoniae R6 | 0.5 | 0.5 | 1 | 4 | 4 | 1 | 2 | 0.5 | 4 | 4 | 2 | 2 |
| S. oralis NCTC 11427 | 2 | 1 | 2 | 16 | 4 | 4 | 16 | 1 | 16 | 8 | 1 | 8 |
| S. oralis ATCC 10557 | 2 | 1 | 2 | 16 | 4 | 4 | 16 | 1 | 16 | 8 | 2 | 4 |
| S. mitis NCTC 12261 | 1 | 0.5 | 2 | 8 | 2 | 4 | 16 | 1 | 16 | 16 | 2 | 8 |
| S. mitis V1b | 4 | 1 | 4 | 16 | 2 | 8 | 32 | 0.25 | 128 | 8 | 0.5 | 16 |
| S. mitis V2 | 4 | 1 | 4 | 32 | 4 | 8 | 32 | 1 | 32 | 8 | 1 | 8 |
| S. mitis V3 | 4 | 1 | 4 | 32 | 4 | 8 | 32 | 1 | 32 | 16 | 2 | 8 |
| S. mitis V4 | 4 | 1 | 4 | 32 | 4 | 8 | 32 | 1 | 32 | 16 | 0.5 | 32 |
| S. sanguis V5 | 4 | 1 | 4 | 32 | 4 | 8 | 32 | 0.5 | 64 | 8 | 0.5 | 16 |
| S. mitis V6 | 4 | 1 | 4 | 32 | 4 | 8 | 32 | 0.5 | 64 | 16 | 0.5 | 32 |
| S. mitis V10 | 4 | 4 | 1 | 32 | 16 | 2 | 8 | 2 | 4 | 4 | 2 | 2 |
| S. mitis V8 | 16 | 4 | 4 | 128 | 16 | 8 | 32 | 1 | 32 | 16 | 1 | 16 |
| S. oralis V9 | 32 | 4 | 8 | 256 | 16 | 16 | 64 | 1 | 64 | 16 | 1 | 16 |
RS, reserpine; NOR, norfloxacin; EBR, ethidium bromide; ACR, acriflavine. MICs are the averages of at least two determinations, which did not differ by more than a twofold dilution.
Classification of the VGS clinical isolates to the species level was done with the API 20-Strep and ID 32-Strep systems.
From these results, we could assume that no efflux mechanism is involved in the Cp resistance of isolate V10, probably due to some mutation that abolishes the functionality of the pump, as has been observed in the Staphylococcus aureus Smr protein (15). We consider that if an isolate has an efflux mechanism, its (drug MIC)/(drug MIC in the presence of reserpine) ratio should be at least fourfold greater that those for S. pneumoniae R6 and isolate V10. Given this assumption, it can be presumed that all the VGS clinical isolates studied (with exception of isolate V10) have an Fq efflux mechanism (Table 2). These included isolates with low-level Cpr (MICs, 4 μg/ml) in which the only resistance mechanism was the efflux pump, such as isolates V1, V2, V3, V4, V5, and V6. Since isolates V5 and V6 have low-level Cpr and an efflux mechanism and gyrase is a secondary target for Cp in VGS (14), the mutations in their gyrB genes (Table 1) would not be involved in resistance. Isolates with high-level Cpr (MICs, ≥16 μg/ml), isolates V8 and V9, had both active efflux mechanisms and mutations in parC. Isolate V10 had a parC mutation and no efflux mechanism and the Cp MIC for the isolate was 4 μg/ml. These results suggest that the Cp efflux mechanism would confer a fourfold increase in Cpr, while a mutation in parC would also confer a fourfold increase. The Cp MIC for strain V9 was 32 μg/ml. This represents a 16-fold increase (compared with the MICs for the type strains), suggesting a synergistic effect of the putative efflux mechanism (which confers a 4-fold increase in the MIC) and the parC mutation (which confers a 4-fold increase in the MIC).
When the ethidium bromide and acriflavine susceptibilities are considered, 32- to 128-fold and 8- to 32-fold reductions in the ethidium bromide and acriflavine MICs, respectively, in the presence of reserpine were found for the eighth Cpr VGS with a putative efflux mechanism. A 16-fold reduction in ethidium bromide MICs and an 8-fold reduction in acriflavine MICs (with the exception of that for S. oralis ATCC 10557) were also observed for the VGS type strains in the presence of reserpine. In the presence or absence of reserpine small differences (two- to fourfold) were observed for S. pneumoniae R6 and isolate V10 with ethidium bromide and acriflavine. These results suggest the activity of an efflux pump(s) for these drugs in the VGS type strains studied (with the exception of S. oralis ATCC 10557 and acriflavine) and clinical isolates (with the exception of strain V10). The differences observed with Fqs, ethidium bromide, and acriflavine in the strains studied suggest that the efflux pump(s) in the strains studied would differ in their substrate specificities.
Nucleotide sequence accession number.
The new DNA sequences reported in this paper have been assigned the following GenBank accession numbers: AF144766 to AF144774 (parC regions), AF144784 to AF144787 (gyrA regions), AF144775 to AF144783 (parE regions), and AF144788 to AF144791 (gyrB regions).
Acknowledgments
We thank P. A. Lazo for allowing us to use the PCGENE program on his computer and for critical reading of the manuscript. The technical assistance of A. Rodriguez-Bernabé is acknowledged.
M.J.F. has a postdoctoral fellowship from Comunidad Autónoma de Madrid. This work was supported by grants 97/2026 from Fondo de Investigación Sanitaria and 08.2/0007/1997 from the Comunidad Autónoma de Madrid.
REFERENCES
- 1.Ahmed M, Borsch C M, Neyfakh A A, Schuldiner S. Mutants of the Bacillus subtilis multidrug transporter Bmr with altered sensitivity to the antihypertensive alkaloid reserpine. J Biol Chem. 1993;268:11086–11089. [PubMed] [Google Scholar]
- 2.Alcaide F, Carratala J, Liñares J, Gudiol F, Martín R. In vitro activities of eight macrolide antibiotics and RP-59500 (quinupristin-dalfopristin) against viridans group streptococci isolated from blood of neutropenic cancer patients. Antimicrob Agents Chemother. 1996;40:2117–2120. doi: 10.1128/aac.40.9.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcaide F, Liñares J, Pallarés R, Carratala J, Benítez M A, Gudiol F, Martín R. In vitro activity of 22 β-lactam antibiotics against penicillin-resistant and penicillin-susceptible viridans group streptococci isolated from blood. Antimicrob Agents Chemother. 1995;39:2243–2247. doi: 10.1128/aac.39.10.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awada A P, Van der Auwera P, Meunier P, Daneau D, Klastersky J. Streptococcal and enterococcal bacteremia in patients with cancer. Clin Infect Dis. 1992;15:33–48. doi: 10.1093/clinids/15.1.33. [DOI] [PubMed] [Google Scholar]
- 5.Baranova N N, Neyfakh A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochud P Y, Egglman P H, Calandra T H, Van Melle G, Saghafi L, Francioli P. Bacteremia due to viridans streptococcus in neutropenic patients with cancer: clinical spectrum and risk factors. Clin Infect Dis. 1994;18:25–31. doi: 10.1093/clinids/18.1.25. [DOI] [PubMed] [Google Scholar]
- 7.Brenwald N P, Gill M J, Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carratala J, Alcaide F, Fernandez-Sevilla A, Corbell X, Liñares J, Gudiol F. Bacteremia due to viridans streptococci that are highly resistant to penicillin: increase among neutropenic patients with cancer. Clin Infect Dis. 1995;20:1169–1173. doi: 10.1093/clinids/20.5.1169. [DOI] [PubMed] [Google Scholar]
- 9.Douglas C W I, Heath J, Hampton K K, Preston F E. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 10.Elting L S, Bodey G P, Keefe B H. Septicemia and shock syndrome due to viridans streptococci: a case-control predisposing factors. Clin Infect Dis. 1992;14:1201–1207. doi: 10.1093/clinids/14.6.1201. [DOI] [PubMed] [Google Scholar]
- 11.Facklam R R, Washington J A., II . Streptococcus and related catalase-negative gram-positive cocci. In: Balows W J, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 238–257. [Google Scholar]
- 12.Garnier F, Gerbaud G, Courvalin P, Galimand M. Identification of clinically relevant viridans group streptococci to the species level by PCR. Antimicrob Agents Chemother. 1997;35:2337–2341. doi: 10.1128/jcm.35.9.2337-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill M J, Brenwald N P, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:187–189. doi: 10.1128/aac.43.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González I, Georgiou M, Alcaide F, Balas D, Liñares J, de la Campa A G. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob Agents Chemother. 1998;42:2792–2798. doi: 10.1128/aac.42.11.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinius L L, Goldberg E B. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J Biol Chem. 1994;269:29998–30004. [PubMed] [Google Scholar]
- 16.Kaatz G W, Seo S M, Ruble C A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi K, Enari T, Totsuka K, Shimizu K. Comparison of phenotypic characteristics, DNA-DNA hybridization results, and results with a commercial rapid biochemical and enzymatic reaction system for identification of viridans group streptococci. J Clin Microbiol. 1995;33:1215–1222. doi: 10.1128/jcm.33.5.1215-1222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markham P N, Neyfakh A A. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:2673–2674. doi: 10.1128/aac.40.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz R, de la Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neyfakh A A. The multidrug efflux transporter of Bacillus subtilis is a structural and functional homolog of the Staphylococcus NorA protein. Antimicrob Agents Chemother. 1992;36:484–485. doi: 10.1128/aac.36.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neyfakh A A, Bidnenko V, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neyfakh A A, Borsch C M, W. K G. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng E Y W, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiological characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulsen K, Kilian M. Program and abstracts of the American Society for Microbiology Conference on Streptococcal Genetics, abstr. 2D-05. Washington, D.C: American Society for Microbiology; 1998. [Google Scholar]
- 25.Roberts R B, Krieger A G, Schiller N L, Gross K C. Viridans streptococcal endocarditis: the role of various species, including pyridoxal-dependent streptococci. Rev Infect Dis. 1979;1:955–965. doi: 10.1093/clinids/1.6.955. [DOI] [PubMed] [Google Scholar]
- 26.Ruoff K L. Streptococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 299–307. [Google Scholar]
- 27.Sussman J I, Baron J, Tenenbaum M J, Kaplan M H, Greenspan R R, Facklam R R, Tyburski M B, Goldman M A, Kanzer B F, Pizzarello R A. Viridans streptococcal endocarditis: clinical, microbiological, and echocardiographic correlations. J Infect Dis. 1986;154:597–603. doi: 10.1093/infdis/154.4.597. [DOI] [PubMed] [Google Scholar]
- 28.Vandamme P, Torck U, Falsen E, Pot B, Goossens H, Hersters K. Whole-cell protein electrophoretic analysis of viridans streptococci: evidence for heterogeneity among Streptococcus mitis biovars. Int J Syst Bacteriol. 1998;48:117–125. doi: 10.1099/00207713-48-1-117. [DOI] [PubMed] [Google Scholar]
- 29.Venditti M, Baiocchi P, Barandimarte C, Serra P, Gentile G, Girmenia C, Martino P. Antimicrobial susceptibilities of Streptococcus species that cause septicemia in neutropenic patients. Antimicrob Agents Chemother. 1989;33:580–582. doi: 10.1128/aac.33.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeller V, Janoir C, Kitzis M-D, Gutmann L, Moreau N J. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1973–1978. doi: 10.1128/aac.41.9.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]