Abstract
As the site of insertion of the aadB gene cassette on pRAY, from a clinical isolate of Acinetobacter, is almost identical to the preferred site on integrons, the composite 59-base element (59-BE) associated with this cassette is potentially recombinationally active. By using a conduction assay to quantitate site activity, the 59-BE was recognized by integrase with high frequency, indicating that the composite site is recombinationally active.
The acquisition of antibiotic resistance gene cassettes by integrons and the role of integrons in the mobilization of antibiotic resistance genes to and from members of the family Enterobacteriaceae have been reported extensively (17). There is much less information on these elements in species of Acinetobacter, and only recently have gene cassettes and integrons been reported in this genus (1, 9, 20, 22).
Integrons encode a site-specific recombinase, integrase (IntI1), that catalyzes the mobility of gene cassettes (10, 14, 15, 19). Although IntI1 preferentially recognizes the core sites (GTTRRRY) of attI1 and of 59-base element (59-BE) flanking gene cassettes, it also recognizes secondary sites, conforming to the consensus GWTMW (8).
A gene cassette encoding a 2"-adenylyltransferase, which has activity against aminoglycoside antibiotics, including gentamicin, tobramycin, and kanamycin, was identified at a secondary site on plasmid pRAY in a clinical isolate of Acinetobacter (20). The aadB gene is regulated by a promoter on pRAY (21). The boundaries of the cassette suggest that insertion of the cassette was catalyzed by IntI1 (20). Furthermore, the putative insertion site (GTTAGGA) on pRAY is similar to the preferred site on integrons, and it may be that the cassette is flanked by two putatively active recombination sites (20), which is unusual for a cassette at a secondary site (18).
Plasmids and bacterial strains.
The plasmids and bacterial strains used in this study are listed in Table 1. A 1.7-kb BamHI-HindIII fragment, containing the aadB gene cassette from pRAY, was restricted from pHS100 (20) and ligated to pSU18 to generate pSU1817. Escherichia coli DH5α was used as a host for plasmid construction and in gene cassette excision experiments. E. coli UB5201 (F− pro met recA56 gyrA Nalr) and UB1637 (his lys trp recA56 rpsL) were used as recipient and donor strains, respectively, in mating experiments.
TABLE 1.
Plasmids and bacterial strains
Conduction assays.
Gene cassettes at secondary sites are thought to be stably integrated because the composite 59-BE formed on insertion is recombinationally inactive (18). Following the insertion of an aadB cassette at a secondary site on RSF1010, generating pIE723, a 59-BE core sequence (GATCAAA) which differs from the consensus core sequence site (GTTRRRY) at one conserved position and two consensus positions was formed. Studies have shown that a T→A transversion in the second position of the core sequence reduced the frequency of recombination 1,000-fold (23). Changes in the first and third positions of the GTT triplet and a transition at the second position reduced recombinational activity 40- to 170-fold, showing that the central T is critical for site activity (23). That the aadB 59-BE from pIE723 was essentially recombinationally inactive is consistent with these data, and it was concluded that the cassette is stably integrated in pIE723 (18).
As the insertion site on pRAY (GTTAGGA) is almost identical to the consensus core and, importantly, contains the critical GTT, the core site of the composite 59-BE of the cassette on pRAY is potentially recombinationally active (20). To test whether the putative recombination sites could be recognized by integrase, the frequency of conduction of pSU1817 by R388 was determined. The integron In3 on R388 contains two integrated gene cassettes, dfrB2 and orfA, and consequently three recombination sites, the two cassette-associated 59-BEs and an attI1 site. Integrase was supplied in trans by including pSU2056, which expresses high levels of integrase (15). Conduction experiments were carried out as described previously (8). E. coli UB1637(pSU2056)(R388) was transformed with pSU1817 and conjugated with UB5201. Donor and recipient cells were mixed on a 0.22-μm-pore-size Millipore filter on Luria-Bertani (LB) agar and were left undisturbed at 37°C for 3 to 4 h. Filters were washed with 1 ml of physiological saline, and 10-fold dilutions of the suspension were plated onto M9 minimal medium (16) containing trimethoprim (20 μg/ml) and LB agar containing chloramphenicol (25 μg/ml) and nalidixic acid (20 μg/ml). Trimethoprim (20 μg/ml) was incorporated into the M9 minimal medium to select for UB5201 transconjugants containing R388. LB agar containing chloramphenicol (25 μg/ml) and nalidixic acid (20 μg/ml) was selective for UB5201 containing pSU1817/R388 cointegrates. The conduction frequency in each assay was calculated as the ratio of Cmr to Tpr transconjugants. The average frequency of conduction of pSU1817 (2.3 × 10−1) was more than 5 orders of magnitude higher than that of the vector pSU18, which does not contain any specific sites, and similar to that of pSU18R2 (1.3 × 10−2), which contains a 59-BE (Table 2).
TABLE 2.
Conduction frequencies of cointegrates
| Plasmida | Conduction frequencyb in expt:
|
Avg | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| pSU1817 | 5.8 × 10−2 | 5.4 × 10−1 | 7.8 × 10−2c | 2.3 × 10−1 |
| pSU18 | 1.3 × 10−7 | 3.9 × 10−8 | 1.3 × 10−6 | 4.9 × 10−7 |
| pSU18R2 | 3.2 × 10−3 | 1.7 × 10−2 | 2.0 × 10−2 | 1.3 × 10−2 |
The donor strain also contained plasmids R388 and pSU2056.
Averages of duplicate platings were scored in each experiment.
Average of four independent mating experiments.
Mapping of the recombination sites.
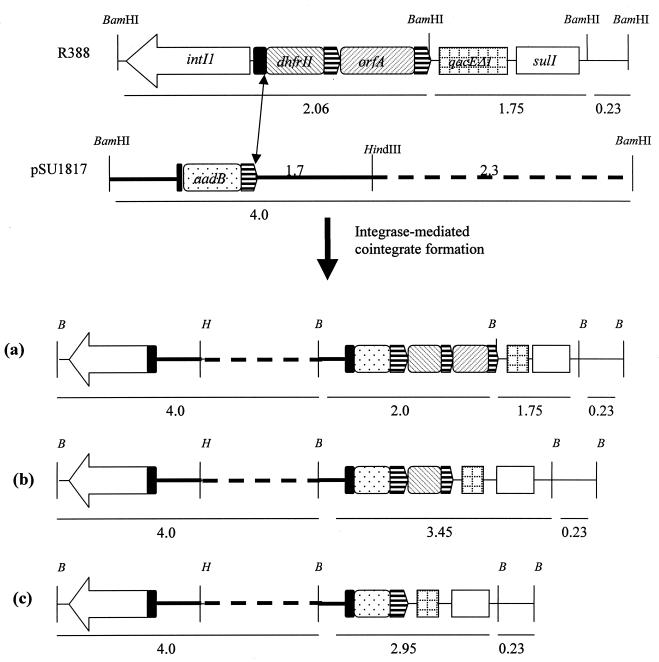
To determine the site of insertion of pSU1817 into R388, plasmid DNA from 24 transconjugants from two independent crosses was digested with BamHI, and the fragments were separated by agarose gel electrophoresis. Plasmid R388 contains four BamHI fragments of ∼29, 2.06, 1.75, and 0.23 kb (2). The potential site-specific sites of recombination of In3 on R388 are located in the 2.06-kb (attI1 and 59-BE of dfrB2) and 1.75-kb (59-BE of orfA) fragments. Recombination involving any of the sites in the 2.06- or 1.75-kb fragments would alter the sizes of these fragments. BamHI digestion of plasmid DNA from 24 Cmr transconjugants generated three profiles. Seventy percent (17 of 24) of the cointegrates had a profile consistent with a recombination event involving attI1 on R388 and the 59-BE of aadB on pSU1817 (Fig. 1a). The second profile (4 of 24) resulted from a similar recombination event between attI1 on R388 and the 59-BE on pSU1817 and excision of the orfA gene cassette (Fig. 1b). A recombination event between R388 attI1 and the 59-BE on pSU1817, combined with the excision of the orfA and dfrB2 cassettes, resulted in a third profile (Fig. 1c) in 3 of the 24 cointegrates analyzed. Antibiotic susceptibility testing confirmed that the Tpr gene cassette had been excised.
FIG. 1.
Cointegrate formation involving R388 attI1 site ( ) and aadB 59-BE (
) and aadB 59-BE ( ) on pSU1817. Note that the attI1 site flanking aadB on pSU1817 is incomplete. The double-headed arrow indicates sites involved in the formation of cointegrates. BamHI (B) and HindIII (H) sites are shown. (a) Recombination between R388 attI1 site and 3′ core site of 59-BE on pSU1817; (b) recombination between R388 attI1 site and 3′ core site of 59-BE on pSU1817 and excision of orfA gene cassette; (c) recombination between R388 attI1 site and 3′ core site of 59-BE on pSU1817 and excision of orfA and dfrB2 gene cassettes. Note that the same result would also be produced by recombination between the 59-BE of orfA of R3888 and aadB of pSU1817, combined with the excision of the orfA and dfrB2 cassettes. pSU18 is indicated by a broken line. Sizes below maps are in kilobases.
) on pSU1817. Note that the attI1 site flanking aadB on pSU1817 is incomplete. The double-headed arrow indicates sites involved in the formation of cointegrates. BamHI (B) and HindIII (H) sites are shown. (a) Recombination between R388 attI1 site and 3′ core site of 59-BE on pSU1817; (b) recombination between R388 attI1 site and 3′ core site of 59-BE on pSU1817 and excision of orfA gene cassette; (c) recombination between R388 attI1 site and 3′ core site of 59-BE on pSU1817 and excision of orfA and dfrB2 gene cassettes. Note that the same result would also be produced by recombination between the 59-BE of orfA of R3888 and aadB of pSU1817, combined with the excision of the orfA and dfrB2 cassettes. pSU18 is indicated by a broken line. Sizes below maps are in kilobases.
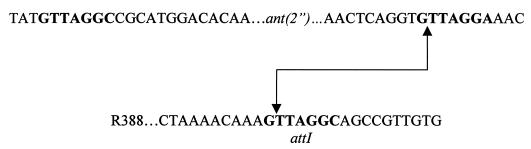
A primer from the 5′ side of attI1 in R388 was used to determine the nucleotide sequence of the sites involved in cointegrate formation. The oligonucleotide, 5′-GGGAATTCAGCAACGATGTTACGCA-3′, was labelled with [γ-32P]ATP and included in a sequencing reaction as described previously (8). DNA sequencing of one junction from a cointegrate of R388 and pSU1817 of each profile described above confirmed that recombination was site specific and involved attI1 on R388 and the 59-BE on pSU1817 (Fig. 2).
FIG. 2.
Sequence of boundary of the aadB gene cassette on pSU1817 and the attI1 insertion site on R388 involved in cointegrate formation. The secondary site and 59-BE core site flanking the aadB gene cassette and the attI1 site on R388 are indicated in boldface type. The arrows indicate the sites involved in the formation of the recombinant junction.
These data show that unlike the 59-BE from pIE723, the 59-BE from pRAY is highly active. Cointegrations involving the 59-BE on pSU1817 and attI1 on R388 occurred at frequencies comparable to those of recombination events involving two 59-BEs (i.e., 3.7 × 10−2) (4, 5), whereas the 59-BE from pIE723 was conducted at a frequency only slightly above background (18).
Cassette excision analysis.
The transfer of gene cassettes may proceed through a route that does not involve a cointegration event. Gene cassettes are excised from integrons as free circles. Following excision, a cassette can be inserted by a single IntI1-mediated event between the 59-BE of a circular cassette and either a specific site on an integron or a secondary site (4–6, 8, 10, 11, 13, 19). There is a strong requirement for two primary recombination sites in order for gene cassettes to be excised (4, 5). As the aadB gene in this study is flanked by a 59-BE and an incomplete attI1 site, excision of the cassette was anticipated to be a rare event. Two separate methods were used to detect IntI1-mediated excision of the aadB gene cassette from pSU1817. In the first methods, the plasmid was propagated in the presence of high levels of integrase encoded by pSU2056 (15). Plasmid DNA was isolated from DH5α cells containing pSU2056 and pSU1817 and reintroduced into DH5α by transformation. Transformants were selected on medium containing chloramphenicol (25 μg/ml). To screen for the loss of the aadB gene cassette from pSU1817, transformants were subsequently replica plated onto LB medium containing either chloramphenicol (25 μg/ml) or kanamycin (25 μg/ml). Cmr transformants that were Kms were indicative of loss of the aadB gene cassette from pSU1817. A total of 300 and, in a separate experiment, 700 Cmr E. coli DH5α (pSU2056)(pSU1817) transformants were replica plated onto medium containing either chloramphenicol (25 μg/ml) or kanamycin (25 μg/ml). No Kms transformants were isolated, indicating either that the aadB gene cassette could not be excised from pSU1817 or that excision occurred at a low frequency (<1.4 × 10−3).
In the second method, a PCR assay was used to check for cassette excision. Plasmid pSU1817 was completely digested with a restriction endonuclease that cleaves this plasmid at a single site within the aadB cassette. The cleaved DNA was used as a template in a PCR assay with two primers (CAGGAAACAGCTATGAC and GTAAAACGACGGCCAGTG) that correspond to the vector-located forward and reverse primer sites flanking the aadB cassette in pSU1817. After 30 cycles of sequential 30-s incubations at 94, 50, and 72°C in a Perkin-Elmer 2400 apparatus, the products were analyzed by electrophoresis. We were unable to obtain any amplification product, indicating again that cassette deletion was either a rare event or a nonexistent one.
We can conclude that upon integration of a gene cassette at a secondary site, the resistance gene in the cassette becomes flanked by a secondary site and a hybrid 59-BE. As shown here, the hybrid 59-BE may be fully recombination proficient. This quality in itself confers mobility on the resistance gene cassette, since IntI1-mediated replicon cointegration may occur at a high frequency with just the hybrid 59-BE present in the mobilized replicon and a functional integron in the driver. However, cassette excision is a reaction that requires further specificity and will probably not occur at all in the case of the cassettes integrated at secondary sites.
Acknowledgments
This work was supported by a UNESCO-MCBN short-term fellowship to H.S. and by grants from the University of Cape Town to B.G.E.
REFERENCES
- 1.Amyes S G B, Young H-K. Mechanisms of antibiotic resistance in Acinetobacter spp.—genetics of resistance. In: Bergogne-Bérézin E, Joly-Guillou M-L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 185–223. [Google Scholar]
- 2.Avila P, de la Cruz F. Physical and genetic characterization of the Inc W plasmid R388. Plasmid. 1988;20:155–157. doi: 10.1016/0147-619x(88)90019-4. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomé B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 4.Collis C M, Grammaticopoulos G, Briton J, Stokes H W, Hall R M. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 5.Collis C M, Hall R M. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 6.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De la Cruz F, Grinstead J. Genetic and molecular characterization of Tn21, a multiple resistance transposon of R100.1. J Bacteriol. 1982;151:222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francia M V, de la Cruz F, García Lobo J M. Secondary sites for integration mediated by the Tn21 integrase. Mol Microbiol. 1993;10:823–828. doi: 10.1111/j.1365-2958.1993.tb00952.x. [DOI] [PubMed] [Google Scholar]
- 8a.Francia, M. V., and J. M. García Lobo. Unpublished data.
- 9.Gonzalez G, Sossa K, Bello H, Dominguez M, Mella S, Zemelman R. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol Lett. 1998;161:125–128. doi: 10.1111/j.1574-6968.1998.tb12937.x. [DOI] [PubMed] [Google Scholar]
- 10.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 11.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Hansson K, Sköld O, Sundström L. Non-palindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol Microbiol. 1997;26:441–453. doi: 10.1046/j.1365-2958.1997.5401964.x. [DOI] [PubMed] [Google Scholar]
- 14.Martinez E, de la Cruz F. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol Gen Genet. 1988;211:320–325. doi: 10.1007/BF00330610. [DOI] [PubMed] [Google Scholar]
- 15.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 17.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 18.Recchia G D, Hall R M. Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the IncQ plasmid RSF1010. Mol Microbiol. 1995;15:179–187. doi: 10.1111/j.1365-2958.1995.tb02232.x. [DOI] [PubMed] [Google Scholar]
- 19.Recchia G D, Stokes H W, Hall R M. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 1994;22:2071–2078. doi: 10.1093/nar/22.11.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal H, Elisha B G. Identification and characterization of an aadB gene cassette at a secondary site in a plasmid from Acinetobacter. FEMS Microbiol Lett. 1997;153:321–326. doi: 10.1111/j.1574-6968.1997.tb12591.x. [DOI] [PubMed] [Google Scholar]
- 21.Segal H, Elisha B G. Characterization of the Acinetobacter plasmid, pRAY, and the identification of regulatory sequences upstream of an aadB gene cassette on this plasmid. Plasmid. 1999;42:60–66. doi: 10.1006/plas.1999.1403. [DOI] [PubMed] [Google Scholar]
- 22.Seward R J, Lambert T, Towner K J. Molecular epidemiology of aminoglycoside resistance in Acinetobacter spp. J Med Microbiol. 1998;47:455–462. doi: 10.1099/00222615-47-5-455. [DOI] [PubMed] [Google Scholar]
- 23.Stokes H W, O’Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]